Abstract
We performed the first studies of analytic sensitivity, analytic specificity, and dynamic range for the new Xpert MTB/RIF assay, a nucleic acid amplification-based diagnostic system that detects Mycobacterium tuberculosis and rifampin (RIF) resistance in under 2 h. The sensitivity of the assay was tested with 79 phylogenetically and geographically diverse M. tuberculosis isolates, including 42 drug-susceptible isolates and 37 RIF-resistant isolates containing 13 different rpoB mutations or mutation combinations. The specificity of the assay was tested with 89 nontuberculosis bacteria, fungi, and viruses. The Xpert MTB/RIF assay correctly identified all 79 M. tuberculosis isolates and correctly excluded all 89 nontuberculosis isolates. RIF resistance was correctly identified in all 37 resistant isolates and in none of the 42 susceptible isolates. Dynamic range was assessed by adding 102 to 107 CFU of M. tuberculosis into M. tuberculosis-negative sputum samples. The assay showed a log-linear relationship between cycle threshold and input CFU over the entire concentration range. Resistance detection in the presence of different mixtures of RIF-resistant and RIF-susceptible DNA was assessed. Resistance detection was dependent on the particular mutation and required between 65% and 100% mutant DNA to be present in the sample for 95% certainty of resistance detection. Finally, we studied whether assay specificity could be affected by cross-contaminating amplicons generated by the GenoType MTBDRplus assay. M. tuberculosis was not detected until at least 108 copies of an MTBDRplus amplicon were spiked into 1 ml of sputum, suggesting that false-positive results would be unlikely to occur.
Conventional diagnostic methods for Mycobacterium tuberculosis are slow and/or lack sensitivity. A number of new diagnostic approaches have brought incremental improvements to detection and drug susceptibility testing; however, the technical complexity of these assays and their dependence on dedicated laboratory infrastructure have limited their adoption, especially in low-resource, high-burden settings (1, 11, 12, 21). The recently introduced Xpert MTB/RIF (manufactured and marketed by Cepheid, Sunnyvale, CA) assay simultaneously detects the presence of M. tuberculosis and its susceptibility to the important first-line drug rifampin (RIF) (7). A sample processing system and an automated heminested real-time PCR assay are integrated into a single disposable cartridge. The assay can be performed directly from a clinical sputum sample or from a decontaminated sputum pellet and can generally be completed in less than 2 h (7).
The Xpert MTB/RIF assay detects M. tuberculosis and RIF resistance by PCR amplification of the rifampin resistance-determining region (RRDR) of the M. tuberculosis rpoB gene and subsequent probing of this region for mutations that are associated with RIF resistance. Approximately 95% of RIF-resistant tuberculosis cases contain mutations in this 81-bp region (16). Our previous work has established that the Xpert MTB/RIF assay has a limit of detection (LOD), defined as the minimum number of bacilli that can be detected with 95% confidence) of 131 CFU per ml of clinical sputum (7). The assay was also able to identify RIF resistance in samples containing 23 common clinically occurring rpoB mutations. None of the 20 nontuberculosis mycobacteria (NTM) species tested, including the NTM species commonly described as causing human disease were falsely identified as M. tuberculosis (7), suggesting high specificity. Several small studies using clinical samples demonstrated 98% to 100% sensitivity overall, 72% sensitivity in smear-negative patients, and a specificity of 100% (7).
In the present study, we expand upon our previous work and report on several critical analytical assay performance characteristics, including dynamic range, sensitivity, specificity, RIF resistance detection in heterogeneous samples, and resiliency against cross-contamination by other nucleic acid amplification techniques (NAATs).
MATERIALS AND METHODS
Diagnostic system.
The Xpert MTB/RIF assay and the GeneXpert instrument have recently been described in detail (7). In brief, the assay consists of a single-use multichambered plastic cartridge preloaded with the liquid buffers and lyophilized reagent beads necessary for sample processing, DNA extraction, and heminested real-time PCR. Clinical sputum samples or decontaminated sputum pellets are treated with an NaOH and isopropanol-containing sample reagent (SR). The SR is added at a 2:1 ratio to the sputum sample or sputum pellet and incubated for 15 min at room temperature. The treated sample is transferred into the cartridge, the cartridge is loaded into the GeneXpert instrument, and an automatic process completes the remaining assay steps. The assay cartridge also contains lyophilized Bacillus globigii spores which serve as an internal sample processing and PCR control. The spores are automatically resuspended and processed during the sample processing step, and the resulting B. globigii DNA is amplified during the PCR step. The standard user interface indicates the presence or absence of M. tuberculosis, the presence or absence of RIF resistance, and a semiquantitative estimate of M. tuberculosis concentration (high, medium, low, and very low). Assays that are negative for M. tuberculosis and also negative for the B. globigii internal control are reported as invalid.
The PCR assay amplifies a 192-bp segment of the M. tuberculosis rpoB gene in a heminested real-time PCR. The internal control heminested B. globigii assay is multiplexed with the M. tuberculosis assay. M. tuberculosis is detected using five overlapping molecular beacon probes (probes A to E) that are complementary to the entire 81-bp RIF resistance-determining “core” region of the wild-type rpoB gene (5, 7, 14). Mutations in the rpoB gene target inhibit hybridization of one or more of the rpoB-specific molecular beacons, reducing or eliminating the signal from the corresponding probes. M. tuberculosis is identified when at least two of the five rpoB-specific molecular beacons give a positive signal with cycle threshold (CT) values that are ≤38 and that differ by no more than two cycles. B. globigii DNA is detected when the single B. globigii molecular beacon produces a CT of <38 cycles.
The difference in CT between the first (early CT) and last (later CT) M. tuberculosis-specific molecular beacon (ΔCT Max) is the basis of rpoB mutation and RIF resistance detection. RIF resistance is identified if the ΔCT Max is >3.5 cycles. RIF susceptibility is identified if the ΔCT Max is ≤3.5 cycles. A sample is considered RIF indeterminate when the last probe returns a CT of >38 and the first probe has a CT value of >34.5 cycles because the assay terminates at cycle 38, and a ΔCT Max of >3.5 cannot be measured. For the purpose of the current study, rpoB mutations that completely inhibit probe hybridization (and thus caused one or more molecular beacons to show CT values of >38) were defined as causing probe “dropouts,” whereas rpoB mutations that permit partial probe hybridization and produce a measurable ΔCT Max of >3.5 cycles were considered “delays.”
Dynamic range testing.
The laboratory M. tuberculosis strain H37Rv obtained from the American Type Culture Collection (ATCC) (Manassas, VA) was cultured and quantified as previously described (7). Excess discarded sputum samples were collected under institutional review board (IRB)-approved protocols from patients not suspected of tuberculosis as previously described (7). M. tuberculosis cells were spiked into 1-ml aliquots of M. tuberculosis-negative sputum samples to final concentrations ranging from 102 to 107 CFU/ml and were tested with the Xpert MTB/RIF assay according to the manufacturer's instructions.
Analytic sensitivity panel.
The 79 clinical M. tuberculosis isolates analyzed in the sensitivity study included 70 isolates from a collection of 211 highly characterized M. tuberculosis strains established by the United Nations Children's Fund/UNDP/World Bank/WHO Special Program for Research and Training in Tropical Diseases (3). Rifampin MIC was previously established by the agar proportion method on Lowenstein-Jensen medium, using a breakpoint of 40 μg/ml to establish resistance (13). M. tuberculosis DNA was isolated as described previously (17). The remaining nine clinical M. tuberculosis strains were selected from a panel of pansusceptible M. tuberculosis isolates obtained in a previous study (6). The phylogenetic lineage of each M. tuberculosis isolate within the collection was identified by testing genomic DNA for nine single-nucleotide polymorphisms (SNPs), which were used to classify each isolate into one of 10 SNP cluster groups and SNP cluster subgroups (6). The isolates were selected to include at least five examples of any unique SNP cluster group or subgroup. The details for each isolate, including geographic source, phylogenetic group, drug susceptibility profile, and drug resistance target genotype, are listed in Table S1 in the supplemental material.
Analytic specificity panel.
The 89 nontuberculosis bacteria, fungi, and viruses analyzed in the specificity study were obtained from the ATCC and BEI repositories (NIH Biodefense and Emerging Infections Research Resources Repository, NIAID, NIH), the Clinical Microbiology laboratory at the university hospital of the University of Medicine and Dentistry of New Jersey (UMDNJ), Zeptometrix Corporation (Buffalo, NY), or other collaborators (Table 1). Samples obtained from UMDNJ were verified to be of the appropriate species or genera as described in reference 2.
TABLE 1.
Analytic specificity panel
| Bacterial, fungal, or viral species | Strain or sourcea |
|---|---|
| Bacteria | |
| Acinetobacter baumanii | BEI NR-10146 |
| Acinetobacter calcoaceticus | Clinical strain |
| Actinomyces israelii | ATCC 12102 |
| Actinomyces meyeri | Clinical strain |
| Bacillus cereus | BEI NR-4198 |
| Bacillus subtilis | Clinical strain |
| Bordetella parapertussis | ATCC 151311D |
| Bordetella pertussis | Cepheid |
| Campylobacter jejuni | BEI NR-3057 |
| Chlamydia pneumoniae | Cepheid |
| Citrobacter freundii | Clinical strain |
| Corynebacterium diptheriae | PI 581 |
| Corynebacterium pseudodiptheriticum | ATCC 10700 |
| Corynebacterium xerosis | Clinical strain |
| Enterobacter aerogenes | Clinical strain |
| Enterobacter cloacae | Clinical strain |
| Enterococcus avium | Clinical strain |
| Enterococcus faecalis | Clinical strain |
| Enterococcus faecium | Clinical strain |
| Escherichia coli | Clinical strain |
| Escherichia coli O157H7 | ATCC 35150 |
| Fusobacterium nucleatum | ATCC 25586D |
| Haemophilus influenzae | ATCC 49247 |
| Haemophilus parahemolyticus | ATCC 10014 |
| Haemophilus parainfluenzae | Clinical strain |
| Klebsiella oxytoca | Clinical strain |
| Klebsiella pneumoniae | Clinical strain |
| Legionella pneumophila | ATCC 33152D |
| Leuconostoc mesenteroides | Clinical strain |
| Listeria grayi | ATCC 25401 |
| Listeria monocytogenes | BEI NR-4211 |
| Moraxella catarrhalis | ATCC 8176 |
| Morganella morganii | Clinical strain |
| Mycoplasma pneumoniae | ATCC 15531D |
| Neisseria gonorrhoeae | ATCC 49226 |
| Neisseria lactamica | ATCC 23971 |
| Neisseria meningitidis | Clinical strain |
| Neisseria mucosa | ATCC 69695 |
| Nocardia asteroides | ATCC 19247 |
| Nocardia cyriageorgica | ATCC BAA-1516 |
| Nocardia farcinica | ATCC 3318 |
| Pasteurella multocida | ATCC 3160 |
| Peptostreptococcus anaerobius | ATCC 49031D |
| Porphyromonas gingivalis | ATCC 33277D-5 |
| Prevotella melaninogenica | ATCC 25845D-5 |
| Propionibacterium acnes | Clinical strain |
| Proteus mirabilis | Clinical strain |
| Proteus vulgaris | BEI DD-460 |
| Providencia alcalifaciens | PI 368 |
| Pseudomonas aeruginosa | ATCC 27853 |
| Rhodococcus equi | ATCC 14187 |
| Salmonella enterica | BEI NR-515 |
| Salmonella typhi | Clinical strain |
| Serratia marcescens | Clinical strain |
| Shigella boydii | Clinical strain |
| Shigella flexneri | Clinical strain |
| Staphylococcus aureus | ATCC 25953 |
| Staphylococcus capitis | Clinical strain |
| Staphylococcus epidermidis | ATCC 12228 |
| Staphylococcus haemolyticus | Clinical strain |
| Staphylococcus hominis | Clinical strain |
| Staphylococcus lugdunensis | Clinical strain |
| Stenotrophomonas maltophilia | Clinical strain |
| Streptococcus equi | Clinical strain |
| Streptococcus pyogenes | ATCC 19615 |
| Streptococcus agalactiae | ATCC 12386 |
| Streptococcus constellatus | Clinical strain |
| Streptococcus mitis | Clinical strain |
| Streptococcus mutans | Clinical strain |
| Streptococcus pneumoniae | Clinical strain |
| Streptococcus uberis | Clinical strain |
| Veillonella parvula | ATCC 10790D-5 |
| Stenotrophomonas maltophilia | Clinical strain |
| Yersinia pestis | BEI NR-2644 |
| Fungi | |
| Candida albicans | ATCC 14053D |
| Cryptococcus neoformans | A. Casadevall lab |
| Histoplasma capsulatum | J. Nosanchuk lab |
| Kingella kingae | ATCC 23332D |
| Viruses | |
| Adenovirusb | ZM NATADV1-ST |
| Herpes simplex virus type 1 | Cepheid |
| Herpes simplex virus type 2 | Cepheid |
| Influenza A virusc | Cepheid |
| Influenza B virusc | Cepheid |
| Parainfluenza virus 2b | ZM NATPARA2-ST |
| Parainfluenza virus 3b | ZM NATPARA3-ST |
| Respiratory syncytial virus Ac | Cepheid |
| Respiratory syncytial virus Bc | Cepheid |
| Rhinovirus 6c | Cepheid |
| Rhinovirus 16c | Cepheid |
The 89 nontuberculosis bacteria, fungi, and viruses analyzed were obtained from the ATCC and BEI repositories (NIH Biodefense and Emerging Infections Research Resources Repository), the University of Medicine and Dentistry of New Jersey (UMDNJ), Zeptometrix Corporation (Buffalo, NY), Presque Isle Cultures (Erie, PA) (PI), or other collaborators. lab, laboratory. Clinical strains were from UMDNJ, and clinical strain DNA was from reference 2.
Whole-particle viral testing, sample from Zeptometrix Corporation.
The genomic source material was RNA.
Sensitivity and specificity panel testing.
The sensitivity and specificity studies were performed using production-lot cartridges preloaded with production-lot enzyme beads and reagent beads. However, unlike the commercially available assay, these studies used open cartridges that were not sealed with a sonically welded plastic top and were not preloaded with liquid reagents or the sample processing control bead containing B. globigii spores. The open cartridges preserved access to the cartridge chambers and permitted manual placement of test DNA into cartridge chamber 10, which would normally receive the DNA-containing eluent of lysed bacterial cells.
Five hundred microliters of TET buffer (50 mM Tris [pH 8.35], 0.1 mM EDTA, and 0.1% mM Tween 20) was added to cartridge chamber five. A 100-μl sample mixture containing either sensitivity or specificity panel DNA (or RNA for RNA viruses), 90 copies of genomic B. globigii DNA, and TET buffer was added directly into chamber 10. M. tuberculosis isolates were tested once at 45 genomes per reaction (10 times the Xpert MTB/RIF LOD for DNA) (7), and all other organisms were tested twice at 106 genome copies per reaction. Negative controls contained no DNA. Initial DNA concentrations were determined with a NanoDrop ND-1000 (Thermo Scientific) and diluted in TET buffer. When reliable genome size was not available, appropriate concentrations were determined by assuming genome sizes of 4.5 Mb/genome for bacteria and 22.5 Mb/genome for fungi. Testing of mixed samples containing DNA from a wild-type M. tuberculosis strain plus an M. tuberculosis strain containing either rpoB gene mutation 533ccg (ccg mutation at codon position 533) or 531ttg (ttg mutation at codon position 531) was performed at a final concentration of 90 copies per reaction. Strains used for mixture testing are indicated in Table S1 in the supplemental material. Preparations used for mixture testing were confirmed to be of appropriate concentration by quantitative real-time PCR of the M. tuberculosis lsr2 gene (4). Cartridges were loaded into the GeneXpert instrument and processed with a truncated version of the automated protocol that omitted the sample processing steps, proceeding immediately as though the lysis and elution of a normal sample had already been completed. This protocol preserved the remaining assay steps without change and is equivalent to version 1 of the MTB assay protocol, released on 8 April 2009.
Three organisms (Table 1) were obtained as whole viral particle NATTROL diagnostic standards (Zeptometrix), 100 μl of the viral particle stock was diluted 1:20 in TET buffer, and the mixture was tested with the Xpert MTB/RIF assay according to the manufacturer's instructions.
Amplicon contamination studies.
We created a modified MTBDRplus assay amplicon with defined mutations in the rpoB target, but unaltered primer binding sites. This distinguished between unintentional amplicon contamination and deliberate experimental contamination. Three smear-positive clinical sputum samples from individuals suspected to have tuberculosis were PCR amplified using the GenoType MTBDRplus kit (Hain Lifescience GmbH) as indicated in the product insert. The 168-bp DNA sequence of the full MTBDRplus amplicon was derived by outward sequencing of the product of the GenoType MTBDRplus assay from within the rpoB core. The MTBDRplus amplicon (minus the biotinylated ends) was then reconstructed via overlap extension of two 98-bp oligonucleotides (Hain-AT-do-F [5′-CACGCTCACGTGACAGACCGCCGGGCCCCAGCGCCCACAGTCGGCGCTTGTCGGTCAACCCCGACAGCGGGTTGTTCTGGACCATGAATTGGCTCAGC-3′] and Hain-AT-do-R [5′-CACGCTCACGTGACAGACCGCCGGGCCCCAGCGCCCACAGTCGGCGCTTGTCGGTCAACCCCGACAGCGGGTTGTTCTGGACCATGAATTGGCTCAGC-3′]). The construct included four mutations in the rpoB core region (511ccg, 516gtc, 526gac, and 531tgg) intended to block hybridization of all but one of the Xpert MTB/RIF assay's five molecular beacon probes.
Statistical analysis.
The minimum proportion of mutant DNA detectable in a wild-type mixture was determined by binary logistic regression. The data were converted to the percentages of RIF-resistant responses for each ratio of wild type to mutant DNA. Binary logistic regression was fitted through the tested concentrations, and lower and upper 95% confidence intervals (95% CIs) were generated for the curve. The 95% CI for the minimum proportion was determined by where the 95% probability level crossed the upper and lower 95% CIs. In all other cases, 95% CIs were calculated using a Student t test.
RESULTS
Dynamic range.
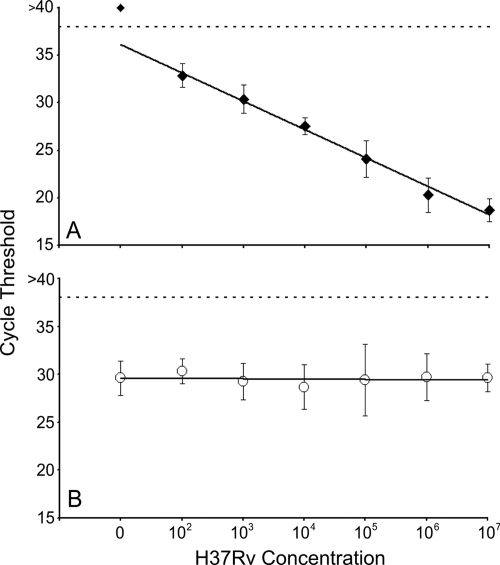
The concentration of M. tuberculosis bacteria in clinical sputum samples can vary from over 107 to less than 20 CFU/ml (10). We performed analytic studies to examine the CFU relationship between CT value and cell concentration over the potential range of M. tuberculosis cell concentrations. Clinical sputum samples were spiked with known numbers of M. tuberculosis bacteria ranging from 107 to 102 CFU/ml and tested in the assay. The assay correctly identified M. tuberculosis in all 28 sputum samples containing 107 to 103 CFU/ml and identified M. tuberculosis in 5 of the 6 samples containing 102 CFU/ml. The log-linear relationship between CT values and the number of M. tuberculosis cells present in each sample was maintained over the entire dynamic range, confirming the ability of the assay to provide semiquantitative estimates of input cell number (Fig. 1 A). The B. globigii internal control assay also functioned well over the entire range of added M. tuberculosis cells. The B. globigii assay was positive in all instances with an average cycle threshold of 29.7 (95% CI, 28.9 to 30.4) (Fig. 1B). Interestingly, the CT values of the B. globigii assay did not appear to be affected by the number of M. tuberculosis cells added to the sample. We suspect that this decoupling is due to the heminested PCR design of both assays.
FIG. 1.
Dynamic range studies. Log dilutions of M. tuberculosis H37Rv cells were added to 1 ml of M. tuberculosis-negative sputum to final concentrations ranging from 102 to 107 CFU/ml (n = 5 or 6 per dilution). (A) Average rpoB probe B cycle thresholds (CTs) were plotted for each M. tuberculosis concentration tested. Clinically relevant M. tuberculosis concentrations all fall within the linear range of the assay. (B) Average B. globigii internal control probe CTs were plotted for each concentration of M. tuberculosis tested. The B. globigii assay gave an average CT of 29.7 (95% CI, 28.9 to 30.4) for all dilutions and was not influenced by the concentration of M. tuberculosis in the sample. Dotted lines indicate maximum valid CT values for probe positivity.
Sensitivity.
Previous analytic studies examined assay performance using the laboratory M. tuberculosis strain H37Rv. We tested the ability of the assay to detect a more phylogenetically and geographically diverse collection of M. tuberculosis isolates (Table 2). The studies were performed at a relatively low target concentration (10 times the LOD with DNA) to test sensitivity under challenging conditions. The assay identified all 79 different M. tuberculosis strains as M. tuberculosis positive. We also tested the ability of the assay to identify different rpoB mutations and thereby RIF resistance in a variable clinical strain background. The assay detected RIF resistance in all 37 strains with known mutations in the rpoB core. The 42 RIF-susceptible isolates without known rpoB core mutations were all identified as RIF susceptible (Table 3). The RIF-susceptible isolates had an average ΔCT Max of 1.8 (95% CI, 1.74 to 1.92). The 37 RIF-resistant isolates contained 13 unique SNPs, three of which were present only in combination with another SNP (Table 2). Nine of 13 mutations caused at least one of the molecular beacon probes to drop out completely (CT of >38). Mutations 516tac, 526cgc, and 533ccg caused a delayed CT, resulting in ΔCT Max values of 9.7 (95% CI, 9.2 to 10.2; n = 2), 7.7 (95% CI, 7.4 to 8.0; n = 3), and 4.23 (95% CI, 3.7 to 4.8; n = 3), respectively. The 509agg mutation was present only in combination with the 526cgc mutation. The sample was correctly identified as RIF resistant on the basis of a delayed probe D CT corresponding to the 526cgc mutation. Interestingly, the 509agg mutation did not cause a dropout or delay of its corresponding probe.
TABLE 2.
Characteristics of M. tuberculosis test panel isolates
| Codon position(s)a | Mutation(s)b | No. of isolates in SNP cluster groupc |
Total no. of isolates with the mutation(s) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| I | II | IIIa | IIIb | IIIc | IV | V | VIa | VIb | |||
| 511 | CTG→ccg | 1 | 1 | ||||||||
| 516 | GAC→gtc | 1 | 1 | 2 | |||||||
| 516 | GAC→tac | 1 | 1 | 2 | |||||||
| 526 | CAC→agc | 1 | 1 | ||||||||
| 526 | CAC→cgc | 1 | 1 | 2 | |||||||
| 526 | CAC→gac | 1 | 1 | 1 | 3 | ||||||
| 526 | CAC→tac | 1 | 1 | 1 | 1 | 4 | |||||
| 531 | TCG→tgg | 1 | 1 | 1 | 3 | ||||||
| 531 | TCG→ttg | 2 | 2 | 1 | 1 | 1 | 4 | 1 | 1 | 13 | |
| 533 | CTG→ccg | 1 | 1 | 1 | 3 | ||||||
| 512, 531 | AGC→gcc, TCG→ttg | 1 | 1 | ||||||||
| 515, 516 | ATG→att, GAC→tac | 1 | 1 | ||||||||
| 509, 526 | AGC→agg, CAC→cgc | 1 | 1 | ||||||||
| Wild type | 5 | 4 | 4 | 4 | 4 | 4 | 6 | 7 | 4 | 42 | |
| Total no. | 9 | 15 | 6 | 7 | 5 | 5 | 17 | 10 | 5 | 79 | |
TABLE 3.
Xpert MTB/RIF result for direct testing of genomic DNA
| Organism | No. of strains with the following result with the Xpert MTB/RIF assay: |
||
|---|---|---|---|
|
M. tuberculosis positive |
M. tuberculosis negative | ||
| Resistance detected | Resistance not detected | ||
| M. tuberculosis strainsa | |||
| RIF-resistant | 37 | 0 | 0 |
| RIF-susceptible | 0 | 42 | 0 |
| Bacteriab | 0 | 0 | 74 |
| Fungib | 0 | 0 | 4 |
| Virusb | 0 | 0 | 11 |
| None (negative control) | 0 | 0 | 52 |
Specificity.
A previous study had shown that the Xpert MTB/RIF assay did not cross-react with 20 different NTM species tested at high copy numbers, suggesting that the assay would have high specificity (7). However, normal flora of the upper respiratory tract and respiratory tract pathogens can also be present in sputum samples of individuals suspected to have tuberculosis. We tested a panel of 89 organisms, including many that are found in the respiratory tract (Table 1) to further examine the specificity of the Xpert MTB/RIF assay. Each test was performed with 106 genomes per PCR to increase the likelihood of a false-positive test. Our results showed that the Xpert MTB/RIF assay did not mistakenly classify any of these organisms as M. tuberculosis (specificity of 100%) (Table 3). None of the organisms except Nocardia asteroides produced any CT of ≤38. Nocardia cyriageorgica was detected by a single probe with an average CT of 38.2. Partial homology between M. tuberculosis and N. asteroides rpoB primer binding sequences allowed for weak amplification of 106 N. asteroides genomes and hybridization of two probes with CT values of 34.9 and 37.2. Although two of the five molecular beacons detected this amplicon, the CT values of these molecular beacons were consistently >2 cycles apart from each other. This real-time PCR signal did not fulfill the criteria for M. tuberculosis detection by the Xpert MTB/RIF assay diagnostic algorithm and thus, were not reported as M. tuberculosis by the assay.
Detection of RIF resistance in mixed isolate samples.
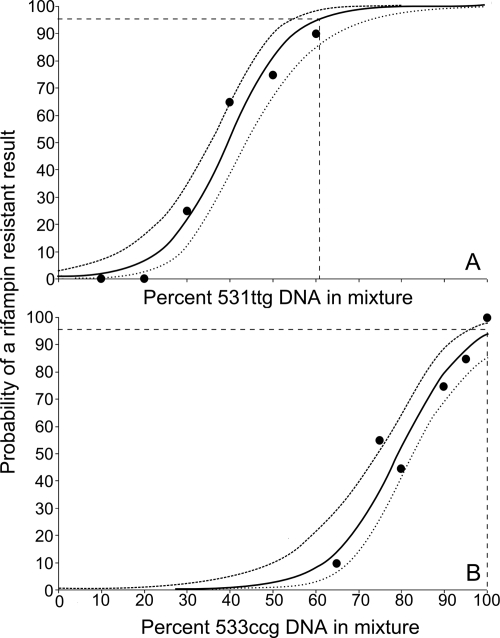
Patients infected with mixtures of both RIF-susceptible and RIF-resistant populations of M. tuberculosis could produce sputum containing a mixture of both strains, complicating resistance detection. We tested the ability of the assay to detect the RIF-resistant fraction of a mixed sample. M. tuberculosis DNA with a wild-type rpoB sequence was mixed in various ratios with M. tuberculosis DNA that contained rpoB mutations. Two different rpoB mutants were studied. One mutation (rpoB 531ttg) caused the molecular beacon complementary to the corresponding wild-type rpoB sequence to completely “drop out.” The second mutation (rpoB 533ccg) caused the molecular beacon complementary to the corresponding wild-type rpoB sequence to have a “delayed” CT, which was sufficient for the Xpert MTB/RIF diagnostic algorithm to identify the mutant as RIF resistant. Our mixing studies demonstrated that the proportion of mutant DNA required for the detection of RIF resistance was dependent on the type of mutation. For the “dropout” 531ttg mutation, the assay could detect the presence of resistance with 95% certainty when the mixture contained at least 65.6% 531ttg DNA (95% CI, 56.9 to 88.4) (Fig. 2 A). For the “delay” 533ccg mutation, the assay could not detect the presence of resistance with 95% certainty unless 100% of the DNA was mutant (95% CI, 96.8 to 100%) (Fig. 2B).
FIG. 2.
Minimum detectable fraction of mutant DNA. DNA extracted from RIF-resistant and RIF-susceptible M. tuberculosis isolates were mixed to six final concentrations of mutant DNA. Mutant DNA contained either an rpoB 531ttg mutation (A) or an rpoB 533ccg mutation (B). Sample mixtures equivalent to 20 times the assay limit of detection (LOD) of 4.5 genomes per reaction were added to the chamber receiving eluted DNA in the full cartridge protocol and were processed using a PCR-only protocol. The percentage of samples (n = 20) identified as RIF resistant was plotted at each concentration. As determined by logistic regression, there was a 95% probability of detecting RIF resistance when the DNA mixture contained 61.2% or 100% of the total DNA in the sample mixture for a 531ttg and 533ccg mutant, respectively. The curves from top to bottom indicate the upper, middle, and lower bounds of the 95% CI. Dashed lines indicate limit of detection.
Assay response to amplicon contamination.
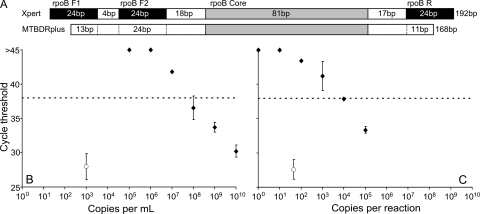
The GenoType MTBDRplus assay is a reverse-hybridization line probe assay that is increasingly used to test for mutations conferring isoniazid and/or rifampin resistance (8). The GenoType MTBDRplus protocol requires substantial manipulation of the assay's PCR product outside closed reaction tubes, increasing the risk for amplicon cross-contamination. The GenoType MTBDRplus and the Xpert MTB/RIF assays amplify a similar region of the rpoB gene. Thus, it was theoretically possible that MTBDRplus amplicon contamination of the Xpert MTB/RIF sample could adversely affect the specificity of the Xpert MTB/RIF assay. We sequenced the MTBDRplus amplicon and identified a 168-bp consensus sequence consistent with the approximately 166-bp amplicon length indicated by the GenoType MTBDRplus product insert. We determined that the MTBDRplus amplicon overlapped completely with the Xpert MTB/RIF assay's inner forward primer. However, the amplicon only overlapped the Xpert MTB/RIF assay's outer forward and reverse primers by 13 and 11 bp, respectively (Fig. 3 A). We spiked various numbers of a modified MTBDRplus amplicon into M. tuberculosis-negative sputum samples (treated with SR), simulating amplicon cross-contamination. Xpert MTB/RIF assays were then performed to test whether the spiked amplicon was falsely identified as tuberculosis. We found that concentrations of modified MTBDRplus less than approximately 108 copies per ml of sputum did not produce any false-positive results (Fig. 3B). We postulated that much of the assay specificity could be due to the efficient wash steps occurring within the Xpert MTB/RIF cartridge. Therefore, we also added modified MTBDRplus amplicon directly to the cartridge DNA chamber 10. At least 104 copies of MTBDRplus amplicon copies needed to be added to each reaction to cause a false-positive result (CT < 38), even with the filter and wash steps removed (Fig. 3C). These results suggest that MTBDRplus amplicon cross-contamination is unlikely to adversely affect Xpert MTB/RIF assay specificity.
FIG. 3.
Potential risk from amplicon contamination. (A) Alignment demonstrating the overlap between the GenoType MTBDRplus PCR amplicon (determined by sequencing) and the priming regions of the Xpert MTB/RIF assay. (B) MTBDRplus simulated amplicon (1010 to 105 copies per ml) was spiked into a mixture of sputum and SR prior to transfer to the cartridge sample chamber (n = 3 per dilution). At least 108 copies per ml were required to induce a false-positive result. Sputum spiked with 103 CFU of Mycobacterium bovis BCG per ml was processed according to the package insert as a control for sputum inhibition (open circle). (C) MTBDRplus amplicon was spiked into TET buffer that also contained B. globigii DNA added directly to the elution receiving chamber of an open cartridge and then processed with a PCR-only protocol (n = 2 per dilution). At least 104 copies per PCR were required to induce a false-positive result. The control (open circle) was 45 copies of M. tuberculosis DNA per reaction. Dotted lines indicate maximum valid CT values for probe positivity.
DISCUSSION
Near-patient molecular assays must be technically simple and robust compared to assays that are performed in specialized laboratories. These assays must also have a clinically relevant dynamic range and show high levels of sensitivity and specificity. Previous tests using clinical samples had suggested that the Xpert MTB/RIF assay would have an excellent dynamic range and that it would perform well in sensitivity and specificity studies (7); however, these properties had not been systematically examined. The analytical testing that we performed in the current study is a useful complement to clinical investigations because it allows for the simulation and testing of low-probability events in a controlled environment and at defined target concentrations. The Xpert MTB/RIF assay functioned over a wide dynamic range. A wide variety of M. tuberculosis strains and common rpoB mutations were reliably detected. The specificity against NTM has been established previously (7). Our study shows that specificity is also unlikely to be affected by the presence of nonmycobacterial species or cross-contamination with GenoType MTBDRplus amplicons. Our study also demonstrates that the identification of RIF resistance in mixed samples will depend on the nature of the rpoB mutation that is present in the RIF-resistant isolate.
Several prior studies of molecular beacon-based RIF resistance testing suggest that clinical isolates do not usually contain mixtures of RIF-susceptible and RIF-resistant organisms (5, 7, 19). However, mixed infections have been reported (18, 20), and studies of mixed samples are warranted. We found that the ability of the assay to detect mixed samples is dependent on the particular rpoB mutation that is present in the mixture. Mutations that effectively block the hybridization of the assay's molecular beacon to the mutant rpoB sequence appear to be more easily discernible than those that merely inhibit probe hybridization. For mixtures that contain mutant species with a “dropout” mutation, total fluorescence is almost completely dependent on the fraction of molecular beacon population binding to the available wild-type targets. This allows for a substantial amount of wild-type target to accumulate before the affected molecular beacon reports a CT within 3.5 cycles of the other unaffected molecular beacons. When the mutant species is a “delay” mutant, the molecular beacon-mutant target hybrid itself is a substantial contributor to total fluorescence. Under these circumstances, only a small amount of wild-type DNA appears to be required to overcome the delay in CT that occurs with only mutant DNA present in the sample. Fortunately, most of the common rpoB mutations that cause RIF resistance produce probe dropouts rather than delays in the Xpert MTB/RIF assay (7). However, our results suggest that the Xpert MTB/RIF assay should not be used to monitor patients for the emergence of RIF resistance during treatment. Approximately 5% of all RIF resistance is not encoded by mutations within the rpoB core region (16), and this last subset of RIF-resistant isolates will not be detectable either in pure culture or in mixed samples by the Xpert MTB/RIF assay.
The GenoType MTBDRplus assay was recently endorsed by the World Health Organization for rapid drug susceptibility testing of M. tuberculosis, and it will likely be present in tertiary clinical laboratories where the Xpert MTB/RIF assay might also be deployed (9). We found that the Xpert MTB/RIF assay is relatively resistant to contamination by MTBDRplus amplicons and could safely be used in the same laboratory environment. This is the result of inefficient amplification of the MTBDRplus amplicon, which has only limited overlap with the outer priming regions of the Xpert MTB/RIF assay. Additionally, the most likely route for amplicon entry will be by contaminating the clinical sample or sample-SR mixture. This mixture is filtered and washed within the GeneXpert instrument several times prior to PCR, significantly diluting a potential amplicon contaminant. Other commercial or in-house assays that amplify a larger region of the rpoB gene could provide a much more efficient amplification template and could result in increased probability of false-positive results if used without proper contamination controls. The Xpert itself contains amplicons within the sealed cartridge, reducing or eliminating the need for the precautions typically associated with other nucleic acid amplification tests.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health grants R41-AI52523 and R42-AI52523 and a grant from the Foundation for Innovative New Diagnostics.
We gratefully acknowledge the United Nation's Children's Fund/UNDP/World Bank/WHO Special Programme for Research and Training in Tropical Diseases for supplying the banked M. tuberculosis strains. Fungal DNA samples were the generous gifts of Arturo Casedevall and Joshua Nosanchuk at the Albert Einstein College of Medicine.
David Alland is one of a group of coinvestigators who invented molecular beacon technology and receive income from licensees. J.K., M.R.O., and M.J. are employed by Cepheid, which manufactures and markets the Xpert MTB/RIF assay.
Footnotes
Published ahead of print on 26 May 2010.
Supplemental material for this article may be found at http://jcm.asm.org/.
REFERENCES
- 1.Balasingham, S. V., T. Davidsen, I. Szpinda, S. A. Frye, and T. Tonjum. 2009. Molecular diagnostics in tuberculosis: basis and implications for therapy. Mol. Diagn. Ther. 13:137-151. [DOI] [PubMed] [Google Scholar]
- 2.Chakravorty, S., B. Aladegbami, M. Burday, M. Levi, S. A. Marras, D. Shah, H. H. El-Hajj, F. R. Kramer, and D. Alland. 2010. Rapid universal identification of bacterial pathogens from clinical cultures using a novel sloppy molecular beacon melting temperature signature technique. J. Clin. Microbiol. 48:258-267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chakravorty, S., B. Aladegbami, A. S. Motiwala, Y. Dai, H. Safi, M. Brimacombe, D. Helb, and D. Alland. 2008. Rifampin resistance, Beijing-W clade-single nucleotide polymorphism cluster group 2 phylogeny, and the Rv2629 191-C allele in Mycobacterium tuberculosis strains. J. Clin. Microbiol. 46:2555-2560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Colangeli, R., D. Helb, C. Vilcheze, M. H. Hazbon, C. G. Lee, H. Safi, B. Sayers, I. Sardone, M. B. Jones, R. D. Fleischmann, S. N. Peterson, W. R. Jacobs, Jr., and D. Alland. 2007. Transcriptional regulation of multi-drug tolerance and antibiotic-induced responses by the histone-like protein Lsr2 in M. tuberculosis. PLoS Pathog. 3:e87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.El-Hajj, H. H., S. A. Marras, S. Tyagi, F. R. Kramer, and D. Alland. 2001. Detection of rifampin resistance in Mycobacterium tuberculosis in a single tube with molecular beacons. J. Clin. Microbiol. 39:4131-4137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Filliol, I., A. S. Motiwala, M. Cavatore, W. Qi, M. H. Hazbon, M. Bobadilla del Valle, J. Fyfe, L. Garcia-Garcia, N. Rastogi, C. Sola, T. Zozio, M. I. Guerrero, C. I. Leon, J. Crabtree, S. Angiuoli, K. D. Eisenach, R. Durmaz, M. L. Joloba, A. Rendon, J. Sifuentes-Osornio, A. Ponce de Leon, M. D. Cave, R. Fleischmann, T. S. Whittam, and D. Alland. 2006. Global phylogeny of Mycobacterium tuberculosis based on single nucleotide polymorphism (SNP) analysis: insights into tuberculosis evolution, phylogenetic accuracy of other DNA fingerprinting systems, and recommendations for a minimal standard SNP set. J. Bacteriol. 188:759-772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Helb, D., M. Jones, E. Story, C. Boehme, E. Wallace, K. Ho, J. Kop, M. R. Owens, R. Rodgers, P. Banada, H. Safi, R. Blakemore, N. T. Lan, E. C. Jones-Lopez, M. Levi, M. Burday, I. Ayakaka, R. D. Mugerwa, B. McMillan, E. Winn-Deen, L. Christel, P. Dailey, M. D. Perkins, D. H. Persing, and D. Alland. 2010. Rapid detection of Mycobacterium tuberculosis and rifampin resistance by use of on-demand, near-patient technology. J. Clin. Microbiol. 48:229-237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hillemann, D., S. Rusch-Gerdes, and E. Richter. 2007. Evaluation of the GenoType MTBDRplus assay for rifampin and isoniazid susceptibility testing of Mycobacterium tuberculosis strains and clinical specimens. J. Clin. Microbiol. 45:2635-2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huang, W.-L., H.-Y. Chen, Y.-M. Kuo, and R. Jou. 2009. Performance assessment of the GenoType MTBDRplus test and DNA sequencing in detection of multidrug-resistant Mycobacterium tuberculosis. J. Clin. Microbiol. 47:2520-2524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Joloba, M. L., J. L. Johnson, A. Namale, A. Morrissey, A. E. Assegghai, R. D. Mugerwa, J. J. Ellner, and K. D. Eisenach. 2000. Quantitative sputum bacillary load during rifampin-containing short course chemotherapy in human immunodeficiency virus-infected and non-infected adults with pulmonary tuberculosis. Int. J. Tuber. Lung Dis. 4:528-536. [PubMed] [Google Scholar]
- 11.Migliori, G. B., A. Matteelli, D. Cirillo, and M. Pai. 2008. Diagnosis of multidrug-resistant tuberculosis and extensively drug-resistant tuberculosis: current standards and challenges. Can. J. Infect. Dis. Med. Microbiol. 19:169-172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nyendak, M. R., D. A. Lewinsohn, and D. M. Lewinsohn. 2009. New diagnostic methods for tuberculosis. Curr. Opin. Infect. Dis. 22:174-182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Parsons, L. M., A. Somoskovi, R. Urbanczik, and M. Salfinger. 2004. Laboratory diagnostic aspects of drug resistant tuberculosis. Front Biosci. 9:2086-2105. [DOI] [PubMed] [Google Scholar]
- 14.Piatek, A. S., S. Tyagi, A. C. Pol, A. Telenti, L. P. Miller, F. R. Kramer, and D. Alland. 1998. Molecular beacon sequence analysis for detecting drug resistance in Mycobacterium tuberculosis. Nat. Biotechnol. 16:359-363. [DOI] [PubMed] [Google Scholar]
- 15.Telenti, A., P. Imboden, F. Marchesi, D. Lowrie, S. Cole, M. J. Colston, L. Matter, K. Schopfer, and T. Bodmer. 1993. Detection of rifampicin-resistance mutations in Mycobacterium tuberculosis. Lancet 341:647-650. [DOI] [PubMed] [Google Scholar]
- 16.Van Der Zanden, A. G., E. M. Te Koppele-Vije, N. Vijaya Bhanu, D. Van Soolingen, and L. M. Schouls. 2003. Use of DNA extracts from Ziehl-Neelsen-stained slides for molecular detection of rifampin resistance and spoligotyping of Mycobacterium tuberculosis. J. Clin. Microbiol. 41:1101-1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.van Embden, J. D., M. D. Cave, J. T. Crawford, J. W. Dale, K. D. Eisenach, B. Gicquel, P. Hermans, C. Martin, R. McAdam, T. M. Shinnick, and P. M. Small. 1993. Strain identification of Mycobacterium tuberculosis by DNA fingerprinting: recommendations for a standardized methodology. J. Clin. Microbiol. 31:406-409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.van Rie, A., T. C. Victor, M. Richardson, R. Johnson, G. D. van der Spuy, E. J. Murray, N. Beyers, N. C. G. van Pittius, P. D. van Helden, and R. M. Warren. 2005. Reinfection and mixed infection cause changing Mycobacterium tuberculosis drug-resistance patterns. Am. J. Respir. Crit. Care Med. 172:636-642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Varma-Basil, M., H. El-Hajj, R. Colangeli, M. H. Hazbon, S. Kumar, M. Bose, M. Bobadilla-del-Valle, L. G. Garcia, A. Hernandez, F. R. Kramer, J. S. Osornio, A. Ponce-de-Leon, and D. Alland. 2004. Rapid detection of rifampin resistance in Mycobacterium tuberculosis isolates from India and Mexico by a molecular beacon assay. J. Clin. Microbiol. 42:5512-5516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Victor, T. C., H. Lee, S. N. Cho, A. M. Jordaan, G. van der Spuy, P. D. van Helden, and R. Warren. 2002. Molecular detection of early appearance of drug resistance during Mycobacterium tuberculosis infection. Clin. Chem. Lab Med. 40:876-881. [DOI] [PubMed] [Google Scholar]
- 21.Young, D. B., M. D. Perkins, K. Duncan, and C. E. Barry III. 2008. Confronting the scientific obstacles to global control of tuberculosis. J. Clin. Invest. 118:1255-1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.