Abstract
Fungal diseases in immunocompromised hosts pose significant threats to their prognoses. An accurate diagnosis and identification of the fungal pathogens causing the infection are critical to determine the proper therapeutic interventions, but these are often not achieved, due to difficulties with isolation and morphological identification. In an effort to ultimately carry out the simultaneous detection of all human pathogenic microbes, we developed a simple system to identify 26 clinically important fungi by using a combination of PCR amplification and DNA microarray assay (designated PCR-DM), in which PCR-amplified DNA from the internal transcribed spacer region of the rRNA gene was hybridized to a DNA microarray fabricated with species-specific probes sets using the Bubble Jet technology. PCR-DM reliably identified all 26 reference strains; hence, we applied it to cases of onychomycosis, taking advantage of the accessibility of tissue from skin. PCR-DM detected fungal DNA and identified pathogens in 92% of 106 microscopy-confirmed onychomycosis specimens. In contrast, culture was successful for only 36 specimens (34%), 3 of which had results inconsistent with the results of PCR-DM, but sequence analysis of the isolates proved that the PCR-DM result was correct. Thus, PCR-DM provides a powerful method to identify pathogenic fungi with high sensitivity and speed directly from tissue specimens, and this concept could be applied to other fungal or nonfungal infectious human diseases in less accessible anatomical sites.
The identification of a given pathogen is a fundamental step for appropriate treatment of infectious diseases. Isolating such pathogens by culture is generally performed to confirm the diagnoses of fungal diseases, but it is time-consuming and fungal outgrowth can take weeks. Among the common fungal diseases, candidiasis and aspergillosis can cause serious diseases in patients receiving immunosuppressive chemotherapy for cancer or suffering from immunodeficiency because of HIV infection. These invasive infections are often associated with poor prognoses, and accurate identification of fungal pathogens at the species level is important for the prompt initiation of antifungal treatments. It is thus desirable that a method which can systematically identify pathogens directly from patient specimens be developed. Taking advantage of the accessibility of tissue from skin, we selected fungal infection in the nail, onychomycosis, as a model for clinical application of the DNA microarray-based technology for the detection of a broad spectrum of species causing human fungal diseases. Most cutaneous fungal infections are superficial; are limited to the stratum corneum, hair, and nails; and are caused by Trichophyton spp. However, a spectrum of pathogens are also identified as causes of systemic infections (7, 14, 16, 23). Onychomycosis is a common disease of the nail caused by dermatophytes, yeasts, and other molds. Direct microscopic examination of nail specimens is generally performed to diagnose onychomycosis. However, attempts to isolate pathogens from nail specimens confirmed to contain fungal elements often yield negative results and fail to provide a species identification (19, 20). For these reasons, onychomycosis could serve as a relevant model to which we may apply our new DNA microarray system.
In recent years, numerous DNA-based methods have been developed to diagnose infectious diseases and to identify pathogens. PCR has proven to be useful for the rapid diagnosis of a variety of infectious diseases (3, 6, 22, 26). The rRNA gene cassette, particularly the large-subunit 28S rRNA gene, is well conserved among species and provides useful information for phylogenetic analysis (28). The internal transcribed spacer (ITS) regions of the rRNA gene have also been used for phylogenetic analyses (5, 25, 27) and to detect and identify human fungal pathogens (1, 2, 12, 30). The majority of the currently available methods rely on PCR with species-specific probes and therefore are limited in their multiplexing capabilities, resulting in high costs if all relevant species need to be considered or requiring a longer time if DNA sequences are to be determined. More recently, the DNA microarray assay was developed. It permits the simultaneous detection and analysis of thousands of genes in a short assay time and, recently, with greater cost-efficiency, and it has proven useful for the identification of pathogens in a variety of clinical settings (6, 8, 13).
Although the methods used to genetically identify pathogens have been vigorously studied, most of these were done using genomic DNA extracted from fungal isolates that were cultured from infected tissues (8, 29). This approach provides high specificity but does not exceed conventional methods in terms of time. In the present study, we generated a panel of oligonucleotide probes which correspond to unique sequences of the ribosomal ITS regions of fungal pathogens that are commonly encountered in clinical practice. Multiple sets of DNA microarrays were fabricated using the Bubble Jet technology, which ejects picoliter amounts of oligonucleotide probe solutions accurately as a unique matrix onto specially coated glass slides (21). The performance of this simple combination of PCR amplification and DNA microarray assay (PCR-DM) was evaluated for its sensitivity and specificity by using 87 reference fungal strains and 106 infected human nail tissues and comparing the results with those of the conventional culture technique.
MATERIALS AND METHODS
Microorganisms.
A total of 87 strains were obtained from the American Type Culture Collection (ATCC) and the Japan Collection of Microorganisms (JCM), used as precharacterized reference strains (see Table S1 in the supplemental material). Additionally, 40 clinical isolates from the Department of Dermatology, Keio University School of Medicine, were included in this study (Table 1).
TABLE 1.
Hybridization of 40 clinical isolates to PCR-DMa
| PCR-DM identification | No. of specimens identified |
|---|---|
| T. rubrum | 12 |
| T. mentagrophytes var. interdigitale | 3 |
| Arthroderma vanbreuseghemii | 5 |
| Trichophyton tonsurans | 4 |
| Arthroderma gypseum | 2 |
| Microsporum canis | 3 |
| E. floccosum | 1 |
| C. albicans | 5 |
| Candida parapsilosis | 1 |
| Candida dubliniensis | 1 |
| Total | 40 |
Three isolates were positive for universal probes but negative for species-specific hybridization and represented nontargeted species. Of those three, one was identified as a P. boydii isolate and two were identified as S. schenckii isolates.
DNA extraction.
The fungal strains were cultured on Sabouraud's dextrose agar slants at 27°C for 1 to 14 days. To extract DNA from the cultured fungi, a commercial kit, the EZ1 DNA blood kit (Qiagen, Hilden, Germany), with a BioRobot (Qiagen) was used. Before extraction, pretreatment was performed as follows: mycelium (approximately 0.5 by 0.5 cm) was scraped into a 2-ml screw-cap tube (Azygen Scientific, Union City, CA) containing 780 mg zilconia beads (a mixture of beads 1.2 mm, 2.3 mm, and 5.0 mm in diameter; Bio Medical Science Inc.). The mycelial suspension in 230 μl of buffer G2 was disrupted in a FastPrep FP100A instrument (MP Biomedicals) for 2 min at a speed of 5.5. The resulting fungal extracts were centrifuged at 1,500 × g for 2 min, and 200 μl of the supernatant was used for DNA extraction. To extract DNA from nail material, an EZ1 DNA tissue kit (Qiagen) was used. After disruption with zilconia beads, the solution was incubated at 56°C overnight after addition of 10 μl of proteinase K solution. Then, those samples were again disrupted in the FastPrep instrument. Centrifugation was done at 1,500 × g for 2 min before DNA extraction.
ITS amplification.
Fungus-specific universal primers ITS1 (5′-TCCGTAGGTGAACCTGCGG-3′) and ITS4 (5′-TCCTCCGCTTATTGATATGC-3′) were used to amplify the full ITS sequence, including the 5.8S rRNA gene (28). PCR was performed with 2 μl of template DNA in a total reaction volume of 10 μl under the following conditions: 94°C for 3 min; 45 cycles of denaturation (94°C, 45 s), annealing (55°C, 45 s), and extension (68°C, 1 min); and a final extension at 68°C for 5 min. A negative control was performed with each test run by replacing the template DNA with sterilized water in the PCR mixture. The PCR products were purified with a Wizard PCR purification kit (Promega, Madison, WI) for use in the labeling and sequencing reaction.
Capture probe design.
Species- or genus-specific oligonucleotide probes were designed from ITS sequences available in the GenBank database (EMBL and DDBJ databases) and our own sequencing data. By comparison of the sequences of the ITS1 and ITS2 regions of the target species, interspecies variations were carefully extracted to develop species-specific probes. Universal probes for fungi, filamentous fungi, and dermatophytes were designed on the basis of the conserved regions. Oligonucleotide probes of various lengths (20 to 29 bases) with unique sequences were designed, and a very narrow range of melting temperatures (Tms; ±3.5°C) was considered (see Table S1 in the supplemental material). To check for specificities against nonfungi, additional BLASTN searches of the GenBank database (EMBL and DDBJ databases) were performed.
DNA microarray fabrication.
The oligonucleotides used for the DNA microarray were synthesized at Canon, Inc. (Tokyo, Japan). The DNA microarray was made with 82 different oligonucleotide probes and control probes. Production of the array by use of the Bubble Jet technology (21) was significantly improved by developing a new printer head that can eject 8-picoliter droplets from multiple probe solutions automatically at extremely high speed.
Subjects and sample collection.
A total of 106 nail specimens treated with 15% KOH and 40% dimethyl sulfoxide were examined microscopically for the presence of fungal elements. These specimens were obtained from 83 symptomatic patients with onychomycosis (43 males and 40 females; ages, 28 to 90 years) who visited the outpatient clinic of the Department of Dermatology, Keio University Hospital, in 2006 and 2007. The protocol was approved by the Institutional Review Board, Keio University. The experiments were conducted according to the Declaration of Helsinki principles. The nail specimens were divided into three portions: the first portion was used for microscopic examination using 15% KOH with 40% dimethyl sulfoxide, the second portion was used for culture on Sabouraud's dextrose agar containing chloramphenicol (0.05%) with cycloheximide (0.5%) by incubation at 27°C for 4 to 6 weeks, and the third portion was used for PCR-DM. The clinical isolates were identified on the basis of the phenotypic characteristics of the colonies and microscopic examination of lactophenol cotton blue wet mounts.
Labeling of PCR products.
Thirty nanograms of purified PCR product was labeled with 0.5 μM Cy3-labeled ITS4 primer in 50 μl of an Ex Taq (Takara Bio, Otsu, Japan) reaction mixture. The reaction protocol was as follows: 10 s of initial denaturation at 98°C; 15 cycles of DNA denaturation at 98°C for 10 s, primer annealing at 55°C for 45 s, and elongation at 72°C for 1 min; and a final extension step at 72°C for 1 min. The labeled DNA was purified using a QIAquick PCR purification kit (Qiagen).
Hybridization of DNA microarray and image analysis.
The DNA microarray was placed in a hybridization apparatus (Hybridization Station; Genomic Solutions Inc.). The hybridization reaction was carried out in 6× SSPE (900 mM NaCl, 50 mM NaH2PO4, 6 mM EDTA, pH 7.4) containing 10% formamide and 0.05 mg/μl Cy3-labeled DNA at 65°C for 3 min and 55°C for 4 h. Washing was carried out with 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)-0.1% SDS at 50°C and 20°C, and then the microarray was manually rinsed with water. Each microarray included a position marker probe (5′-CGTACGATCGATGTAGCTAGCATGC-3′). These markers were detected with a Cy3-labeled complementary oligonucleotide (5′-Cy3-GCATGCTAGCTACATCGATCGTACG-3′ ) supplemented in the hybridization solution. After spin drying of the glass slides, the fluorescence signal was detected with a GenePix 4000B microarray scanner (Axon, Santa Clara, CA). Image processing and calculation of the signal intensities were performed with the ArrayPro program (version 4.5; Media Cybernetics Inc., Silver Spring, MD). Statistical analysis of the data was performed with Prism (version 5) software (Graphpad Software, San Diego, CA).
RESULTS
Probe design and DNA microarray fabrication.
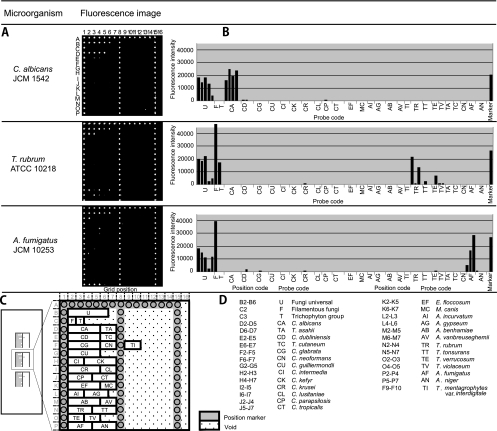
The fungal rRNA gene consists of the 18S, 28S, and 5.8S regions and flanking regions, such as ITS1, ITS2, ETS1, ETS2, and NTS. Of these regions, the 18S and 28S rRNA gene sequences have been used for phylogenetic studies. We chose ITS regions for our purpose because the sequence data in GenBank, as well as our own data, suggested that the 18S or 28S region is conserved at the genus level for some fungi and therefore is not optimal for our usage, which required identification of fungi at the species level (data not shown). We found that if multiple probes were used, the ITS region was useful for identification of all targeted fungi at the species level. The fungus-specific universal primer pair (primers ITS1 and ITS4) was used to amplify the full ITS sequence, including 5.8S rRNA (28). We designed 82 probes composed of 20 to 29 nucleotides to detect ITS1 or ITS2, or both, corresponding to 26 fungal species (see Table S1 in the supplemental material). Two to four probes were designed for each fungal species. These probes were spotted onto specially coated glass slides, where 82 specific probes, 3 sets of universal probes to detect all fungi, filamentous fungi, or Trichophyton spp., as well as position marker probes, were spotted in a unique matrix pattern (16 by 16) with 29 small blocks by the Bubble Jet technology (21) (Fig. 1 A, C, and D). The universal probes were intended to detect the presence of fungal DNA to which corresponding specific probes are not included in the microarray. Figure 1A shows the actual scanning image of three independent microarrays. Positive spots are readily seen to be differentiated from negative spots, and a distinct pattern was observed for each species that had been preidentified by morphology. The top panel of Fig. 1A, with strain JCM 1542, showed positive signals for B2 to B6 (fungal universal probe, probe U) and D2 to D5 (Candida albicans-specific probe, probe CA), with the former detecting fungal DNA and the pattern of the latter identifying the fungus as C. albicans. In the bottom panel, strain JCM 10253 showed positive signals for the B2 to B6 and C2 probes (filamentous fungal universal probe), indicating the presence of a filamentous fungus, and a positive pattern for the P2 to P4 probes revealed the identity as Aspergillus fumigatus. In the middle panel of Fig. 1A, strain ATCC 10218 is positive for the B2 to B6, C2, and C3 probes, again indicating a filamentous fungus, and a positive pattern for the N2 to N4 probes revealed the identity as Trichophyton rubrum.
FIG. 1.
Identification of representative fungal strains by PCR-DM. Data were obtained by PCR-DM after hybridization with Cy3-labeled target DNA from C. albicans (JCM 1542), T. rubrum (ATCC 10218), and A. fumigatus (JCM 10253). (A) Actual scanned fluorescence image; (B) graphs representing fluorescence intensities as arbitrary units. (C and D) Layout of the microarray, on which nine blocks are set per glass slide, but for simplicity, only three blocks are shown. Each block is composed of a unique matrix of oligonucleotide probes designed to target 26 clinically important fungi. The target fungi and the corresponding probe sequences are listed in Table S1 in the supplemental material.
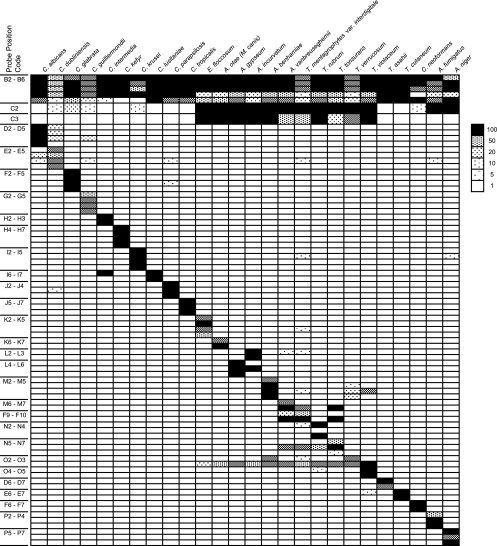
Note that some individual probes cross-react to other species in some cases, such as the O3 probe (Trichophyton verrucosum) to T. rubrum (Fig. 2). This cross-reactivity, however, constitutes a pattern for T. rubrum that is distinct from the patterns for other species. As for the T. verrucosum pattern, not only the O2 but also the O3 probe would be positive and the N2 to N4 T. rubrum-specific probes would never be positive. Thus, when each species is analyzed for its pattern of positivity, including cross-reactive probes, it displays a pattern that discriminates it from other targeted species.
FIG. 2.
PCR-DM specifically identifies 26 targeted fungal species. Normalized signal intensities of all hybridization experiments are listed by probe position and fungal species. Signals were normalized by dividing each absolute value by the background values. Averages were then taken from duplicates or triplicates within a chip and then the average of chip replicates from the same experiments. Normalized signal intensities of 1-, 5-, 10-, 20-, 50-, and 100-fold are indicated in the different pattern images. The abbreviations for the probe names are listed in Fig. 1.
These images were scanned digitally and analyzed by computer software, which calculates the fluorescence intensity of each spot (see Materials and Methods). Therefore, the fluorescence intensity of each spot can be quantified, and the amounts of amplified PCR products are shown as histograms (Fig. 1B).
Evaluation of PCR-DM using DNAs from 87 reference fungal strains.
DNA was extracted from 87 reference strains (see Table S1 in the supplemental material), the majority of which were preidentified by morphological analysis, and used to validate the performance of the PCR-DM method. The hybridization results for the PCR products from 26 reference strains are shown in Fig. 2. The use of multiple probes for each target allowed accurate identification of the reference strains, where the identities of 86 out of the 87 strains identified by PCR-DM matched the morphological identification. One strain (ATCC 32902) designated T. cutaneum by morphology was identified as Trichosporon asahii according to PCR-DM. Sequence analysis of the ITS regions of this DNA matched T. asahii but not Trichosporon cutaneum, indicating that the correct identification of this strain is most likely T. asahii. All other reference strains were processed for DNA sequence analyses, and the results were found to be concordant with the morphological and PCR-DM identifications.
Accurate identification of clinically isolated fungi by PCR-DM.
To examine the specificity of the designed probes and the applicability of the DNA microarray, 40 DNA specimens obtained from clinical isolates, including 3 nontargeted species, were analyzed by PCR-DM. These clinical isolates had been preidentified by their morphologies. Of 40 clinical specimens, the strain identifications were concordant for 37 (Table 1), and the results were confirmed by DNA sequence analyses (data not shown). Three nontarget clinical isolates could not be identified, but PCR-DM indicated the existence of fungal DNA that did not match any target probes on the microarray. Sequence analysis of the PCR products identified them as Pseudallescheria boydii (one case) and Sporothrix schenckii (two cases) (Table 1).
Clinical evaluation of PCR-DM using DNAs extracted from nail specimens.
One hundred six clinical nail specimens were collected from 86 patients, and they were confirmed to be positive for fungal elements by microscopy. Normal-appearing nail specimens were obtained from 50 healthy volunteers, who were confirmed to be KOH negative and served as healthy controls. These specimens were obtained by scraping and were divided into three portions: the first portion was used for KOH examination, the second one was subjected to conventional Sabouraud's dextrose culture, and the third one was used for DNA extraction. Bacteria and molds that outgrew the fungi at exceedingly early or late time points of culture were regarded as contaminants.
Culture of 106 clinical specimens yielded the outgrowth of several species of fungi, such as 27 isolates of T. rubrum, 4 of Trichophyton mentagrophytes var. interdigitale (an anthropophilic strain of Trichophyton interdigitale), 1 of Epidermophyton floccosum, and 4 of Candida spp., resulting in an overall success rate of 34% (36/106). Analyses of the remaining 70 specimens were unsuccessful due to no outgrowth, irrelevant bacterial growth, or contamination by nondermatophyte fungi.
Among the 106 specimens, 101 (95%) were found to be positive for fungal DNA, and the species in 98 (92%) specimens were identified by PCR-DM (Tables 2 and 3). Of the eight specimens whose isolates could not be determined, 5 were negative by PCR and 3 were positive by the use of fungal universal probes but were not target species and were thus unidentifiable by the DNA microarray (Tables 2 and 3).
TABLE 2.
Comparison of identification rate by conventional culture methods and PCR-DM method for 106 nail specimensa
| Conventional culture method finding | No. (%) identified |
||
|---|---|---|---|
| PCR-DM method |
Total | ||
| Determined | Undetermined | ||
| Determined | 36 | 0 | 36 (34) |
| Undetermined | 62 | 8b | 70c (66) |
| Total | 98 (92) | 8 (8) | 106 (100) |
Up to 10 pieces of nails were taken from each patient. All 106 specimens were microscopically positive for fungus elements.
The eight specimens include five that were PCR negative and three that had no species-specific hybridization.
The 70 specimens include 5 with suspected contaminations with a filamentous fungus, 5 with bacteria, and 60 culture negative.
TABLE 3.
Fungal species identified from nail specimens by PCR-DMa
| Species identified or parameter | Isolate for single species or isolate no. 1 for multiple species | Isolate no. 2 for multiple species | Isolate no. 3 for multiple species | No. (%) of specimens |
|---|---|---|---|---|
| Single species | T. rubrum | 55 | ||
| T. mentagrophytes var. interdigitale | 14 | |||
| E. floccosum | 1 | |||
| C. albicans | 4 | |||
| C. parapsilosis | 3 | |||
| Candida guilliermondii | 3 | |||
| Subtotal | 80 (75.5) | |||
| Multiple species | T. rubrum | T. mentagrophytes var. interdigitale | C. guilliermondii | 1 |
| T. rubrum | C. albicans | 1 | ||
| T. rubrum | C. parapsilosis | 3 | ||
| T. rubrum | C. guilliermondii | 7 | ||
| T. rubrum | T. cutaneum | 1 | ||
| T. mentagrophytes var. interdigitale | C. guilliermondii | 1 | ||
| C. albicans | C. parapsilosis | C. guilliermondii | 1 | |
| C. parapsilosis | C. guilliermondii | Filamentous fungus marker | 1 | |
| Candida intermedia | C. guilliermondii | Filamentous fungus marker | 1 | |
| C. guilliermondii | Filamentous fungus marker | 1 | ||
| Subtotal | 18 (17) | |||
| PCR negativeb | 5 | |||
| Undeterminedc | 3 | |||
| Subtotal | 8 (7.5) | |||
| Total | 106 (100) |
Up to 10 pieces of nails were taken from each patient. A total of 86 patients participated.
Hybridization experiments could not be performed.
Positive for universal probes but negative for species-specific hybridization.
Of 36 culture-positive specimens, 33 PCR-DM results matched the conventional phenotypic identification. Two specimens that were identified as T. mentagrophytes var. interdigitale by the phenotypic method were found to be T. rubrum by PCR-DM. Another specimen that was identified as T. rubrum by the phenotypic method was found to be T. mentagrophytes var. interdigitale by PCR-DM. When the PCR products of those specimens were sequenced, the identities for all of them were in agreement with those obtained by PCR-DM.
Eighty (75%) specimens were positive for single species, but 18 (17%) were positive with more than two species-specific probes (Table 3). Because of the low success rate of culture, it is not yet clear whether these results indicate multiple infections or the concomitant detection of commensal fungi that colonize the skin.
Given this finding, we analyzed whether quantification of PCR-amplified fungal DNA, as detected by fungal universal probes on PCR-DM, could discriminate active infection from colonization. Although the KOH-positive group displayed significantly larger amounts of fungus-derived PCR product, as measured by the fluorescence intensity, positive signals of similar intensity were also detected for a number of healthy controls (the KOH-negative group), showing that the sole detection of fungal DNA does not necessarily indicate fungal infection (see Fig. S1 in the supplemental material). This is an issue that has not yet been solved for the genetic identification of pathogens in general, where the sensitivity is so high that commensal microorganisms that are not necessarily pathogens are detected. Nevertheless, if the pathogenic (or nonpathogenic) microorganisms from tissues that are suspected to be infected could be identified, such information should be valuable for initiation of proper therapy targeting the specific pathogens.
DISCUSSION
DNA microarrays have been used to identify a variety of pathogens, including viruses (10, 11, 15), bacteria (4, 6), and fungi (8, 14, 24). They have been powerful tools to assess multiple parameters simultaneously in various clinical scenarios, such as identification of the pathogens causing urinary tract infections, acute upper respiratory tract infections, and sepsis (29). We have developed a method for fabricating a DNA microarray using the Bubble Jet device to eject picoliters of solution containing 5′-terminus-thiolated oligonucleotides onto a glass surface coated with maleimide groups (21). This technology was further improved to adapt the array for the high-throughput ejection of multiple probes and the mass production of microarrays at a low cost.
Our ultimate goal in developing this analysis system is to simultaneously detect pathogens causing all life-threatening infectious diseases as well as local but chronic diseases. We considered fungal disease to be an appropriate model because invasive infections have high rates of associated morbidity and mortality, often with a low rate of pathogen recovery, and local diseases are chronic and often hard to eradicate. Onychomycosis was chosen because of the tissue accessibility, the low rate of pathogen isolation, and the fact that systemic and local infections are caused by many of the same important pathogens, including Candida spp. and Aspergillus spp. (7, 23). In this initial study, ITS regions were chosen over 18 and 28S rRNA because of the variability that allowed identification of the targeted fungi to the species level. It is imaginable, however, that if many more different species of fungi need to be identified simultaneously, use of the ITS region alone may not be sufficient. Actually, we have confirmed by bioinformatic analyses that some zoophilic or rare species of Trichophyton might be difficult to distinguish by some of our probes. One potential of PCR-DM, however, is its expandability. In the near future, it may be possible to simultaneously amplify several regions of interest (e.g., ITS regions in combination with other rRNA regions) and then perform microarray analysis, which should allow us to increase the number of fungi that are to be analyzed in one test.
Utilization of the ITS regions of rRNA genes for DNA microarray analysis has been described previously (14). That system enabled identification of medically important Candida spp. and Aspergillus spp. DNA was obtained from clinical culture isolates but not directly from clinical tissue specimens. Our previous studies indicated that PCR is sensitive enough to directly amplify fungal DNA from infected tissue specimens (17, 18). After validating the performance of our PCR-DM system using reference strains, we actually applied it to identify fungal pathogens directly from infected human tissues. While a number of studies have established the efficacy of PCR-based identification directly from clinical specimens, most such studies were performed with species-specific primers with limited multiplexing capability or by sequencing each of the individual PCR products amplified by universal primers. Our PCR-DM system enabled us to simultaneously detect 26 clinically important fungal species and some nontargeted species from a single clinical specimen. Considering the existence of numerous fungi that are potentially pathogenic to humans, this PCR-DM system has great potential. The present study is the first to demonstrate the feasibility of application of a microarray-based approach to the detection and identification of fungi in the nail. DNA microarray analysis of nail specimens for onychomycosis was proven to be a more sensitive diagnostic method than conventional culture techniques. This system could be applied to systemic fungal infections, as well as local diseases, such as skin ulcers of unknown etiology or unusual pulmonary masses that require a differential diagnosis from lung cancer.
It is clear that a Trichophyton sp. is the pathogen when it is the only fungus detected; however, the result is less clear when several species are detected. It is reasonable to interpret that Trichophyton spp., when detected in the context of skin infection, might be the predominant pathogen, but it is difficult to determine the significance of other species detected simultaneously. It is likely that commensal fungi were detected in most cases, but dual infections cannot be completely ruled out. One specimen yielded both T. rubrum and T. mentagrophytes var. interdigitale, which is consistent with the findings presented in a previous report (9). Although data from PCR-DM can be quantified, a number of specimens from healthy controls yielded positive results by hybridization. This is again most likely due to commensal fungi, which is inevitable in the clinical setting. Hence, a positive indication of fungal DNA by PCR-DM does not necessarily indicate fungal disease. It is mandatory for an accurate diagnosis that clinical manifestations be carefully assessed in conjunction with the performance of PCR-DM. More strict PCR conditions might allow differentiation of commensal and pathogenic fungi, but this will need rigorous evaluation. Nevertheless, in the clinical setting, such as one involving immunocompromised patients, foreseeing a particular infection would be very difficult, and it would be helpful for the planning of therapeutic tactics if various anatomical sites be surveyed by PCR-DM for the profiles of microorganisms that may exist at those sites.
In conclusion, we developed a simple PCR-based DNA microarray method, PCR-DM, that was aimed at the detection of fungal pathogens directly from clinical specimens. Species identification was more sensitive and accurate by PCR-DM than by the conventional culture technique. We demonstrated that our culture-independent direct method is a powerful tool to identify fungal pathogens or to elucidate the profiles of microorganisms present in clinical specimens. Thus, PCR-DM could be applied for detection of other systemic and local human pathogens by fabricating carefully designed DNA microarrays.
Supplementary Material
Acknowledgments
This study was performed under the full financial sponsorship of Canon Inc., which had no influence over the design, results, or interpretation of the experiments.
Canon Inc. and Keio University have filed patents for the probes listed in Table S1 in the supplemental material.
We thank Mie Furuhashi and Minae Suzuki for their technical assistance.
Footnotes
Published ahead of print on 26 April 2010.
Supplemental material for this article may be found at http://jcm.asm.org/.
REFERENCES
- 1.Arabatzis, M., L. E. Bruijnesteijn van Coppenraet, E. J. Kuijper, G. S. de Hoog, A. P. Lavrijsen, K. Templeton, E. M. van der Raaij-Helmer, A. Velegraki, Y. Graser, and R. C. Summerbell. 2007. Diagnosis of common dermatophyte infections by a novel multiplex real-time polymerase chain reaction detection/identification scheme. Br. J. Dermatol. 157:681-689. [DOI] [PubMed] [Google Scholar]
- 2.Baek, S. C., H. J. Chae, D. Houh, D. G. Byun, and B. K. Cho. 1998. Detection and differentiation of causative fungi of onychomycosis using PCR amplification and restriction enzyme analysis. Int. J. Dermatol. 37:682-686. [DOI] [PubMed] [Google Scholar]
- 3.Brillowska-Dabrowska, A., D. M. Saunte, and M. C. Arendrup. 2007. Five-hour diagnosis of dermatophyte nail infections with specific detection of Trichophyton rubrum. J. Clin. Microbiol. 45:1200-1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bryant, P. A., D. Venter, R. Robins-Browne, and N. Curtis. 2004. Chips with everything: DNA microarrays in infectious diseases. Lancet Infect. Dis. 4:100-111. [DOI] [PubMed] [Google Scholar]
- 5.Caligiorne, R. B., P. Licinio, J. Dupont, and G. S. de Hoog. 2005. Internal transcribed spacer rRNA gene-based phylogenetic reconstruction using algorithms with local and global sequence alignment for black yeasts and their relatives. J. Clin. Microbiol. 43:2816-2823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fukushima, M., K. Kakinuma, H. Hayashi, H. Nagai, K. Ito, and R. Kawaguchi. 2003. Detection and identification of Mycobacterium species isolates by DNA microarray. J. Clin. Microbiol. 41:2605-2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hollenbach, E. 2008. Invasive candidiasis in the ICU: evidence based and on the edge of evidence. Mycoses 51(Suppl. 2):25-45. [DOI] [PubMed] [Google Scholar]
- 8.Hsiao, C. R., L. Huang, J. P. Bouchara, R. Barton, H. C. Li, and T. C. Chang. 2005. Identification of medically important molds by an oligonucleotide array. J. Clin. Microbiol. 43:3760-3768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kardjeva, V., R. Summerbell, T. Kantardjiev, D. Devliotou-Panagiotidou, E. Sotiriou, and Y. Gräser. 2006. Forty-eight-hour diagnosis of onychomycosis with subtyping of Trichophyton rubrum strains. J. Clin. Microbiol. 44:1419-1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim, C. J., J. K. Jeong, M. Park, T. S. Park, T. C. Park, S. E. Namkoong, and J. S. Park. 2003. HPV oligonucleotide microarray-based detection of HPV genotypes in cervical neoplastic lesions. Gynecol. Oncol. 89:210-217. [DOI] [PubMed] [Google Scholar]
- 11.Kozal, M. J., N. Shah, N. Shen, R. Yang, R. Fucini, T. C. Merigan, D. D. Richman, D. Morris, E. Hubbell, M. Chee, and T. R. Gingeras. 1996. Extensive polymorphisms observed in HIV-1 clade B protease gene using high-density oligonucleotide arrays. Nat. Med. 2:753-759. [DOI] [PubMed] [Google Scholar]
- 12.Lau, A., T. C. Sorrell, S. Chen, K. Stanley, J. Iredell, and C. Halliday. 2008. Multiplex tandem PCR: a novel platform for rapid detection and identification of fungal pathogens from blood culture specimens. J. Clin. Microbiol. 46:3021-3027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Leaw, S. N., H. C. Chang, R. Barton, J. P. Bouchara, and T. C. Chang. 2007. Identification of medically important Candida and non-Candida yeast species by an oligonucleotide array. J. Clin. Microbiol. 45:2220-2229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Leinberger, D. M., U. Schumacher, I. B. Autenrieth, and T. T. Bachmann. 2005. Development of a DNA microarray for detection and identification of fungal pathogens involved in invasive mycoses. J. Clin. Microbiol. 43:4943-4953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lipshutz, R. J., D. Morris, M. Chee, E. Hubbell, M. J. Kozal, N. Shah, N. Shen, R. Yang, and S. P. Fodor. 1995. Using oligonucleotide probe arrays to access genetic diversity. Biotechniques 19:442-447. [PubMed] [Google Scholar]
- 16.Lowther, A. L., A. K. Somani, M. Camouse, F. T. Florentino, and S. C. Somach. 2007. Invasive Trichophyton rubrum infection occurring with infliximab and long-term prednisone treatment. J. Cutan. Med. Surg. 11:84-88. [DOI] [PubMed] [Google Scholar]
- 17.Nagao, K., T. Ota, A. Tanikawa, Y. Takae, T. Mori, S. Udagawa, and T. Nishikawa. 2005. Genetic identification and detection of human pathogenic Rhizopus species, a major mucormycosis agent, by multiplex PCR based on internal transcribed spacer region of rRNA gene. J. Dermatol. Sci. 39:23-31. [DOI] [PubMed] [Google Scholar]
- 18.Nagao, K., T. Sugita, T. Ouchi, and T. Nishikawa. 2005. Identification of Trichophyton rubrum by nested PCR analysis from paraffin embedded specimen in trichophytia profunda acuta of the glabrous skin. Nippon Ishinkin Gakkai Zasshi 46:129-132. [DOI] [PubMed] [Google Scholar]
- 19.Naka, W., H. Hanyaku, S. Tajima, T. Harada, and T. Nishikawa. 1994. Application of neutral red staining for evaluation of the viability of dermatophytes and Candida in human skin scales. J. Med. Vet. Mycol. 32:31-35. [DOI] [PubMed] [Google Scholar]
- 20.Nishikawa, T., and W. Naka. 1994. Evaluation of antifungal effects of terbinafine and itraconazole using neutral red staining. Br. J. Dermatol. 130(Suppl. 43):4-6. [DOI] [PubMed] [Google Scholar]
- 21.Okamoto, T., T. Suzuki, and N. Yamamoto. 2000. Microarray fabrication with covalent attachment of DNA using Bubble Jet technology. Nat. Biotechnol. 18:438-441. [DOI] [PubMed] [Google Scholar]
- 22.Palacios, G., O. Jabado, D. Cisterna, F. de Ory, N. Renwick, J. E. Echevarria, A. Castellanos, M. Mosquera, M. C. Freire, R. H. Campos, and W. I. Lipkin. 2005. Molecular identification of mumps virus genotypes from clinical samples: standardized method of analysis. J. Clin. Microbiol. 43:1869-1878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Richardson, M., and C. Lass-Florl. 2008. Changing epidemiology of systemic fungal infections. Clin. Microbiol. Infect. 14(Suppl. 4):5-24. [DOI] [PubMed] [Google Scholar]
- 24.Spiess, B., W. Seifarth, M. Hummel, O. Frank, A. Fabarius, C. Zheng, H. Morz, R. Hehlmann, and D. Buchheidt. 2007. DNA microarray-based detection and identification of fungal pathogens in clinical samples from neutropenic patients. J. Clin. Microbiol. 45:3743-3753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sugita, T., R. Ikeda, and T. Shinoda. 2001. Diversity among strains of Cryptococcus neoformans var. gattii as revealed by a sequence analysis of multiple genes and a chemotype analysis of capsular polysaccharide. Microbiol. Immunol. 45:757-768. [DOI] [PubMed] [Google Scholar]
- 26.Turenne, C. Y., E. Witwicki, D. J. Hoban, J. A. Karlowsky, and A. M. Kabani. 2000. Rapid identification of bacteria from positive blood cultures by fluorescence-based PCR-single-strand conformation polymorphism analysis of the 16S rRNA gene. J. Clin. Microbiol. 38:513-520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Watanabe, S., M. Kawasaki, T. Mochizuki, and H. Ishizaki. 2004. RFLP analysis of the internal transcribed spacer regions of Sporothrix schenckii. Nippon Ishinkin Gakkai Zasshi 45:165-175. [DOI] [PubMed] [Google Scholar]
- 28.White, T. J., T. Bruns, S. Lee, and J. Taylor. 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics, p. 315-322. In M. A. Innis, D. H. Gelfand, J. J. Sninsky, and T. J. White (ed.), PCR protocols: a guide to methods and applications. Academic Press, Inc., San Diego, CA.
- 29.Wiesinger-Mayr, H., K. Vierlinger, R. Pichler, A. Kriegner, A. M. Hirschl, E. Presterl, L. Bodrossy, and C. Noehammer. 2007. Identification of human pathogens isolated from blood using microarray hybridisation and signal pattern recognition. BMC Microbiol. 7:78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yoshimura, R., Y. Ito, N. Morishita, J. Ninomiya, and I. Takiuchi. 2006. Comparative study between culture and PCR-RFLP analysis on identification of the causative agent of Tinea unguium. Nippon Ishinkin Gakkai Zasshi 47:11-14. (In Japanese.) [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.