Abstract
A morbidity-mortality event involving virulent Newcastle disease virus (NDV) in wild double-crested cormorants (Phalacrocorax auritus) occurred in North America in the summer of 2008. All 22 viruses isolated from cormorants were positively identified by the USDA-validated real-time reverse transcription-PCR assay targeting the matrix gene. However, the USDA-validated reverse transcription-PCR assay targeting the fusion gene that is specific for virulent isolates identified only 1 of these 22 isolates. Additionally, several of these isolates have been sequenced, and this information was used to identify genomic changes that caused the failure of the test and to revisit the evolution of NDV in cormorants. The forward primer and fusion probe were redesigned from the 2008 cormorant isolate sequence, and the revised fusion gene test successfully identified all 22 isolates. Phylogenetic analyses using both the full fusion sequence and the partial 374-nucleotide sequence identified these isolates as genotype V, with their nearest ancestor being an earlier isolate collected from Nevada in 2005. Histopathological analysis of this ancestral strain revealed morphological changes in the brain consistent with that of the traditional mesogenic pathotypes in cormorants. Intracerebral pathogenicity assays indicated that each of these isolates is virulent with values of >0.7 but not more virulent than earlier isolates reported from Canada.
The regulations of the World Organization for Animal Health (Biological Standards Commission Office, Office International des Epizooties [OIE], Paris, France, 2004) consider the previously classified mesogenic Newcastle disease virus (NDV) to be virulent on the basis of the sequence of the fusion protein cleavage site. This typically includes cormorant viruses (21). Cormorant viruses represent an interesting biological system because, together with pigeon viruses, they represent the two known natural reservoirs of virulent viruses in wild birds (6). NDV does not typically cause disease in adult cormorants, but as witnessed by the event described here and previous events, they can cause substantial mortality in juveniles. Additionally, this poses a major threat to the poultry industry, as wild birds, including cormorants, have been suspected of being the source of poultry outbreaks (3, 17).
To assess the risk that these viruses pose to poultry, it is important to continually evaluate the evolution and pathogenic characteristics of the viruses. The last analysis of the evolution of cormorant NDV was done in 2003 (21). There, on the basis of analysis of the fusion protein cleavage site sequence, 12 cormorant isolates recovered in Canada between 1995 and 2000 were found to form a separate clade. This may indicate significant evolution of an NDV population in wild birds that is generally considered stable. Here, we have evaluated the evolutionary and biological changes of the cormorant viruses over 16 years. The evolutionary changes measured were sufficient to cause changes in the most variable regions of the genome, including the region encoding the fusion protein amino terminal region. These changes in the fusion protein cleavage site obviously did not affect the outcome of the identification of the virus using the USDA-validated real-time reverse transcription-PCR (rRT-PCR) assay targeting the matrix gene (the M-gene assay), nor were the changes in this region sufficient to change the pathogenicity of the virus. Unfortunately, the USDA-validated rRT-PCR assay targeting the fusion gene (the F-gene assay) that is specific for virulent isolates identified only some of these cormorant isolates. If diagnostic laboratories do not perform additional tests, such as the test for the intracerebral pathogenicity index (ICPI), amino acid sequencing, or the mean death time (MDT) in embryonated chicken eggs (ECE), along with the F-gene assay, false-negative results could potentially prolong the identification of the source of an outbreak. Here, we report on the sequence characterization of the fusion proteins of 22 isolates from 2008 and 4 isolates from 2002 to 2006 and compare the findings to those for the fusion proteins of 19 viruses isolated from 1992 to 1998. In addition, we report on the development of a new set of primers and probes that recognize the fusion cleavage site of virulent cormorant NDVs, allowing their rapid characterization.
MATERIALS AND METHODS
Isolates and sequence data.
All newly characterized NDV isolates are part of the USDA Southeast Poultry Research Laboratory repository. This includes isolates obtained from the USDA Animal and Plant Health Inspection Services (APHIS) Wildlife Services, the USDA APHIS National Veterinary Services Laboratory (NVSL) repository, the U.S. Geological Survey National Wildlife Health Center (Madison, WI), and the Southeastern Cooperative for Wildlife Disease (Athens, GA). The samples obtained for this analysis were collected by state and federal wildlife officials working closely with laboratory diagnosticians; therefore, initial viral identification was possible and was based on a hemagglutination (HA) assay, followed by hemagglutination inhibition (HI) assays. However, since NDVs from cormorants typically do not hemagglutinate red blood cells well, if at all, egg death pattern analysis, nucleic acid sequencing, and the matrix gene rRT-PCR test were often done. NDV was isolated only from double-crested cormorants (Phalacrocorax auritus) (Table 1 ). All isolates with tree names that do not begin with “G” did not have previous GenBank submissions. These were sequenced and submitted to GenBank. The 374-nucleotide (nt) region of the fusion gene was sequenced (n = 30), including the full fusion gene for four isolates.
TABLE 1.
NDV sequences included in phylogenetic analysisa
| Tree and isolate | GenBank accession no. | Species |
|---|---|---|
| 374-nt partial fusion tree (Fig. 4) | ||
| G/1998/CA/254847453 | FJ705459 | Cr |
| G/1998/CA/111182194 | DQ837552 | Cr |
| G/1995/CA/254847455 | DQ837552 | Cr |
| G/1995/CA/111052705 | DQ833289 | Cr |
| G/1995/CA/24571080 | AF448486 | Cr |
| G/1995/CA/254847457 | FJ705461 | Cr |
| G/1995/CA/111227011 | DQ839244 | Cr |
| G/1995/CA/19068161 | AY063493 | Cr |
| G/1999/CA/111182200 | DQ837554 | Cr |
| G/1996/CA/111182206 | DQ837556 | Cr |
| G/1996/CA/19068165 | AY063492 | Cr |
| G/1997/US/284019479 | GU332654 | Cr |
| G/2000/CA/111227014 | DQ839245 | Cr |
| G/2000/CA/111052714 | DQ833292 | Cr |
| G/2001/CA/112363411 | DQ857316 | Cr |
| G/2001/CA/111182212 | DQ837558 | Cr |
| G/2001/CA/111052696 | DQ833286 | Cr |
| G/2006/US/284019497 | GU332663 | Cr |
| G/2003/CA/111052693 | DQ833285 | Cr |
| G/2002/US/284019469 | GU332649 | Cr |
| G/2003/CA/111052690 | DQ833284 | Cr |
| G/2003/US/254847459 | FJ705462 | Cr |
| G/1997/US/284019473 | GU332651 | Cr |
| G/1997/US/254847451 | FJ705458 | Cr |
| G/1997/US/254847449 | FJ705457 | Cr |
| G/1997/US/253317827 | GQ288388 | Cr |
| G/1997/US/284019475 | GU332652 | Cr |
| G/1997/US/284019477 | GU332653 | Cr |
| G/1997/US/284019471 | GU332650 | Cr |
| G/1997/US/253317778 | GQ288381 | Cr |
| G/2005/US/254847461 | FJ705463 | Cr |
| G/2008/US/284019505* | GU332667 | Cr |
| G/2008/US/284019503* | GU332666 | Cr |
| G/2008/US/284019529 | GU332676 | Cr |
| G/2008/US/284019483* | GU332656 | Cr |
| G/2008/US/284019489* | GU332659 | Cr |
| G/2008/US/284019481* | GU332655 | Cr |
| G/2008/US/284019526 | GU332675 | Cr |
| G/2008/US/284019532 | GU332677 | Cr |
| G/2008/US/284019499* | GU332664 | Cr |
| G/2008/US/284019523 | GU332674 | Cr |
| G/2008/US/284019507* | GU332668 | Cr |
| G/2008/US/284019487* | GU332658 | Cr |
| G/2008/US/284019485* | GU332657 | Cr |
| G/2008/US/284019495* | GU332662 | Cr |
| G/2008/US/284019491* | GU332660 | Cr |
| G/2008/US/284019511 | GU332670 | Cr |
| G/2008/US/284019514 | GU332671 | Cr |
| G/2008/US/284019501 | GU332665 | Cr |
| G/2008/US/284019517 | GU332672 | Cr |
| G/2008/US/284019520 | GU332673 | Cr |
| G/2008/US/284019493* | GU332661 | Cr |
| G/2008/US/284019509* | GU332669 | Cr |
| G/1990/CA/111227008 | DQ839243 | Cr |
| G/1990/CA/111227005 | DQ839242 | Cr |
| G/1992/CA/111052702 | DQ833288 | Cr |
| G/1992/CA/111052717 | DQ833293 | Cr |
| G/1992/US/254847447 | FJ705456 | Cr |
| G/1992/US/284019467 | GU332648 | Cr |
| G/1992/US/20799674 | AF503643 | Cr |
| G/1992/CA/111182182 | DQ837548 | Cr |
| G/1992/US/33772480 | AY289001 | Tk |
| G/1993/US/45511211 | AY562986 | An |
| G/1971/US/33772440 | AY288987 | Mx |
| G/2000/HN/33772456 | AY288993 | Ck |
| G/1972/US/45511218 | AY562987 | GF |
| G/2000/IT/33772459 | AY288994 | Ck |
| Full fusion tree (Fig. 5) | ||
| G/1999/AU/61393448 | AY935498 | |
| G/1999/AU/61393407 | AY935494 | |
| G/2002/AU/61393362 | AY935490 | |
| G/1998/AU/61393396 | AY935493 | |
| G/1998/AU/61393384 | AY935492 | |
| G/2000/IT/33772465 | AY288996 | Pg |
| G/2000/IT/33772462 | AY288995 | Dv |
| G/1997/AR/52426576 | AY734536 | Pg |
| G/1998/CN/15011276 | AF358785 | Pg |
| G/1990/KE/33772468 | AY288997 | Ck |
| G/1972/US/33772453 | AY288992 | Ck |
| G/2001/RU/58003480 | AY865652 | Tn |
| G/2000/CN/109631561 | DQ485258 | Ck |
| G/2000/CN/109631557 | DQ485256 | Ck |
| G/2000/CN/67550206 | DQ067447 | Ck |
| G/2000/TW/15011278 | AF358786 | Ck |
| G/1998/CN/18042244 | AF456437 | Go |
| G/1997/CN/5702360 | AF162714 | Go |
| G/2006/CN/146359011 | EF592503 | Mn |
| G/2005/CN/146358963 | EF592501 | Md |
| G/2006/CN/146359044 | EF592505 | Md |
| G/2006/CN/146359153 | EF592510 | Md |
| G/2006/CN/146359137 | EF592509 | Tl |
| G/2005/CN/146358939 | EF592500 | Md |
| G/2006/CN/146359067 | EF592506 | Mn |
| G/2005/CN/109255544 | DQ485271 | Ck |
| G/2005/CN/109255550 | DQ485274 | Dv |
| G/2006/CN/124295086 | EF211814 | Go |
| G/2001/CN/18042236 | AF456443 | Go |
| G/2003/CN/109290360 | DQ485230 | Ck |
| G/2003/CN/109631567 | DQ485261 | Ck |
| G/2003/CN/109290367 | DQ485231 | Ck |
| G/2002/CN/109290353 | DQ485229 | Ck |
| G/2001/CN/18042238 | AF456444 | Go |
| G/2005/KR/157674510 | EU140950 | Ck |
| G/2004/KR/157674506 | EU140948 | Ck |
| G/2005/KR/157674508 | EU140949 | Ck |
| G/2000/KR/157674504 | EU140947 | Ck |
| G/1999/CN/15011280 | AF358787 | Ck |
| G/1998/CN/15011295 | AF364835 | Ck |
| G/2000/CN/15011282 | AF358788 | Ck |
| G/2000/CN/28933797 | AF431744 | Go |
| G/2000/CN/18042226 | AF456438 | Go |
| G/1999/CN/68137216 | DQ080015 | Pg |
| G/2006/CN/146359113 | EF592508 | Md |
| G/2005/CN/146358987 | EF592502 | Go |
| G/2005/CN/146358620 | EF521889 | Go |
| G/2006/CN/146359087 | EF592507 | Go |
| G/2006/CN/146359028 | EF592504 | Go |
| G/2001/CN/18042234 | AF456442 | Go |
| G/2001/CN/18042232 | AF456441 | Go |
| G/2001/CN/30025969 | AY253912 | Pt |
| G/2002/CN/148534703 | EF579732 | Ck |
| G/2001/CN/77994255 | DQ227248 | |
| G/2003/CN/121282136 | EF175145 | Mv |
| G/2006/CN/119443716 | EF128055 | Dk |
| G/2006/CN/119443712 | EF128053 | Dk |
| G/2006/CN/119443714 | EF128054 | Dk |
| G/2005/CN/124295072 | EF211807 | Go |
| G/2004/CN/109255540 | DQ485269 | Ck |
| G/2005/CN/86371010 | DQ363536 | |
| G/2006/CN/148534707 | EF579734 | Go |
| G/2005/CN/124295076 | EF211809 | Go |
| G/2006/CN/124295082 | EF211812 | Go |
| G/1996/MX/33772474 | AY288999 | Ck |
| G/2005/CN/124295078 | EF211810 | Go |
| G/2006/CN/124295084 | EF211813 | Go |
| G/2006/CN/148534701 | EF579731 | Ck |
| G/2004/CN/148534705 | EF579733 | Ck |
| G/2002/US/154707827 | EF520718 | Ck |
| G/2002/US/45511218 | AY562987 | Ck |
| G/2000/MX/187711116 | EU518677 | Ck |
| G/1988/MX/187711123 | EU518680 | Ck |
| G/1996/MX/33772474 | AY288999 | Ck |
| G/2004/MX/187711129 | EU518682 | Dv |
| G/2005/MX/187711132 | EU518683 | Ck |
| G/2006/MX/187711135 | EU518684 | Ck |
| G/1971/US/33772440 | AY288987 | Mx |
| G/2006/CN/124295080 | EF211811 | Go |
| G/1992/US/253317820 | GQ288387 | Cr |
| G/1993/US/45511211 | AY562986 | An |
| G/2008/US/284019495 | GU332662 | Cr |
| G/2008/US/284019485 | GU332657 | Cr |
| G/2008/US/284019481 | GU332655 | Cr |
| G/2005/US/253317813 | GQ288386 | Cr |
| G/1997/US/253317778 | GQ288381 | Cr |
| G/1997/US/253317827 | GQ288388 | Cr |
| G/1995/CA/254847455 | DQ837552 | Cr |
| G/1995/CA/253317799 | GQ288384 | Cr |
| G/1998/CA/254847453 | FJ705459 | Cr |
| G/2006/US/284019497 | GU332663 | Cr |
| G/2003/US/254847459 | FJ705462 | Cr |
n = 91 sequences for Fig. 2 and n = 68 sequences for Fig. 3. The virus names used in the phylogenetic trees contain G (when they are available in GenBank)/year of collection/the two-digit ISO country code abbreviation/GI accession number (when it is available) or the in-house virus identification number. Isolates are listed in the order in which they appear on the tree. The two-letter species abbreviations are given, when they are known, as follows: Cr, cormorant; Tk, turkey; An, anhinga; Mx, mixed; Ck, chicken; GF, game fowl; Pg, pigeon; Dv, dove; Tn, tern; Go, goose; Mn, mandarin duck; Md, mallard duck; Tl, teal; Pt, parrot; Mv, muscovy duck; Dk, duck. *, isolates first characterized in this study.
Sequencing analysis was performed as described previously (11). Briefly, all sequencing reactions were performed with fluorescent dideoxynucleotide terminators in an automated sequencer (ABI 3730XL; Applied Biosystems Inc., Foster City, CA). Nucleotide sequence editing and analyses were conducted with a sequence analysis software package (LaserGene, version 5.07; DNAStar, Inc., Madison, WI). Using the full-length genome positions from the complete genome of the NDV LaSota vaccine strain (GenBank accession no. AF077761), the homologous regions sequenced were as follows: a 374-bp partial F gene (positions 4554 to 4917) and the complete coding region for the F gene (positions 4544 to 6205).
Pathogenicity assessment.
The pathogenic potential for selected isolates was evaluated using standard assay methods to determine the ICPI in 1-day-old chicks (2).
Animal experiment and tissue collection.
There were three groups of 10 4-week-old specific-pathogen-free (SPF) White Leghorn chickens. Each of the groups was inoculated bilaterally in the conjunctival sac with 0.1 ml of a viral inoculum of G/2005/US/254847461 (NV/2005) or G/2002/US/154707827 (CA/2002) or phosphate-buffered saline (PBS). The target dose of the inoculum was 105.0 50% embryo infectious doses (EID50s). The actual numbers of infectious doses, as determined by backtitration in embryonated chicken eggs, were 105.3 EID50s for NV/2005 and 105.3 EID50s for CA/2002.
The birds were monitored daily, and two birds from each group (PBS inoculated and infected) were sampled at days 3 and 6. Severely sick or terminally ill birds were sampled regardless of the scheduled sampling day. The birds were anesthetized and then euthanized with an intravenous injection of pentobarbital (Sigma-Aldrich, St. Louis, MO). Immediately postmortem, a necropsy was performed and tissues were harvested for histology.
Histopathology.
Sample tissues were fixed with 10% neutral buffered formalin for approximately 52 h. All sampled tissues were routinely processed into paraffin blocks, and 3- to 5-μm sections were cut for hematoxylin-eosin (H&E) staining. All tissues were examined histologically, and the severity of the lesions was recorded.
Real-time RT-PCR.
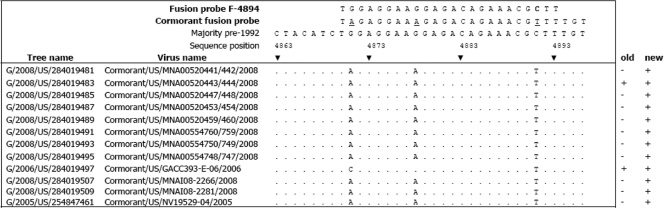
All viruses (n = 34) were tested using the USDA-validated M-gene and F-gene assay protocols (22). The F-gene assay was modified on the basis of the consensus sequence of an alignment (Fig. 1) of the 2008 cormorant NDV viruses reported here (n = 11). An alternate probe, 5′-6-carboxyfluorescein-ACAAAACGTTTCTGTCTCTTTCCTCTA-Black Hole Quencher-1, and an alternate forward primer, ACACTGACCACTTTACTCAC, were used.
FIG. 1.
Comparison of NDV sequences from recent cormorant virus isolates. The nucleotide alignment of the fusion protein cleavage site and adjacent sites for 11 new cormorant NDV isolates is shown. The sequences of both the original fusion probe and the cormorant virus-specific probes are shown above and are aligned with the majority sequence of isolates from before 1992, which the original fusion probe was designed to detect. The changes made in the new cormorant virus-specific fusion probe are underlined. Consensus matches (relative to the majority pre-1992) are shown as dots. The columns labeled “old” and “new” contain + or − to indicate the results of the respective fusion gene rRT-PCR test for each isolate.
Phylogenetic analysis.
The full coding region and 374-bp amino-terminal end-coding region of the F gene from the cormorant isolates were compared to reference sequences representing known genotypes. The 374-bp region of the F gene, which has commonly been used for phylogenetic analysis of NDV (1), was used for all (n = 68) isolates. The complete fusion-coding sequence was used to make phylogenetic trees of selected representative isolates and to localize the U.S. cormorant viruses among other class II genotype reference sequences (n = 91). Phylogenetic analyses were conducted in the MEGA, version 4 (MEGA4), software program (20). The evolutionary distances were computed using MEGA software program, based on neighbor-joining trees using the Kimura two-parameter method (13), with all codon positions being included. There were a total of 1,652 positions in the final data set of the complete fusion protein. The virus names used in the phylogenetic trees and included in Table 1 include G (indicating the GenBank accession number)/year of collection/the two-letter International Standards Organization (ISO) country code/GenInfo sequence identifier number (GI) accession number. GI is used because it is permanently linked to the sequence used in the tree and is not subject to the changes that may occur with GenBank accession numbers.
Nucleotide sequence accession numbers.
The NDV sequences characterized in the present study were submitted to GenBank and can be found under accession numbers GU332648 to GU332677.
RESULTS
Twenty-two isolates from the 2008 cormorant NDV morbidity-mortality event in Minnesota, as well as six other North American isolates collected since the most recent cormorant NDV phylogenetic characterization (21), were phylogenetically characterized. American white pelicans (Pelecanus erythrorhynchos) and ring-billed gulls (Larus delawarensis) were also involved in the morbidity-mortality event (Paul Wolf, personal communication); however, samples from these species were not included in this analysis. Numerous other isolates collected earlier (1992 to 1998) were similarly analyzed so that the most accurate phylogenetic relationship of the 2008 isolates could be determined. All NDV isolates used in the analyses are listed in Table 1.
Four of the isolates were pathotyped according to their ICPI, and each was virulent according to the OIE standard. The OIE standard states that virulent viruses have an ICPI of ≥0.7 or encode multiple basic amino acids at the C terminus of the F2 protein and have phenylalanine at residue 117. Each virus encoded a virulent fusion cleavage site motif (112R/KRQKRF117), and the ICPI for selected isolates (n = 4) ranged from 1.36 to 1.54 (Table 2).
TABLE 2.
CT values from three real-time RT-PCR assays and ICPI values
| Isolate | Isolate description |
CT by real-time PCRa |
ICPIb | ||
|---|---|---|---|---|---|
| Matrix | Fusion (old) | Fusion (new) | |||
| G/1997/US/253317778 | Cormorant/US/CAD9704285/1997 | 20.34 | 17.9 | 14.15 | 1.41 |
| G/2006/US/284019497 | Cormorant/US/GACC393-E-06/2006 | 15.14 | 21.87 | 15.73 | (1.49)e |
| G/2002/US/284019469 | Cormorant/US/FLCC415-A/2002 | 23.12 | 18.89 | 14.31 | |
| G/2008/US/284019509 | Cormorant/US/MNAI08-2281/2008 | 22.17 | 0 | 20.31 | |
| G/2008/US/284019507 | Cormorant/US/MNAI08-2266/2008 | 22.15 | 0 | 17.21 | |
| G/2008/US/284019491 | Cormorant/US/MNA00554760/759/2008 | 32.03 | 0 | 32.35 | |
| G/2008/US/284019493 | Cormorant/US/MNA00554750/749/2008 | 21.4 | 0 | 17.49 | 1.36 |
| G/2008/US/284019495 | Cormorant/US/MNA00554748/747/2008 | 21.93 | 0 | 14.48 | 1.54 |
| G/2008/US/284019489 | Cormorant/US/MNA00520459/460/2008 | 22.16 | 0 | 21.74 | |
| G/2008/US/284019487 | Cormorant/US/MNA00520453/454/2008 | 33.34 | 0 | 37.4 | |
| G/2008/US/284019485 | Cormorant/US/MNA00520447/448/2008 | 23.17 | 0 | 17.18 | 1.39 |
| G/2008/US/284019483 | Cormorant/US/MNA00520443/444/2008 | 33.17 | 33.7 | 27.65 | |
| G/2008/US/284019481 | Cormorant/US/MNA00520441/442/2008 | 22.08 | 0 | 15.89 | 1.45 |
| Cormorant/CN98CNN3-V1138/1998 | 20.99 | 20.24 | 13.97 | ||
| G/1998/CA/254847453 | Cormorant/CN/Sask/3-V1125/1998 | 19.36 | 19.38 | 16.71 | 1.53 (1.56)c |
| G/1997/US/254847449 | Cormorant/US/CA23071/1997 | 21.1 | 17.93 | 14.3 | |
| G/1992/CA/111182182 | Cormorant/CN/95DC2585/1995 | 20.43 | 21.58 | 17.94 | |
| G/1995/CA/254847457 | Cormorant/CN/95DC2345/1995 | 18.96 | 18.5 | 15.68 | 1.6 |
| G/1995/CA/111052705 | Cormorant/CN/Ont/2344/1995 | 21.64 | 21.82 | 16.91 | (1.71)c |
| G/1995/CA/254847455 | Cormorant/CN/Ont/2150/1995 | 19.31 | 19.17 | 14.65 | 1.59 (1.60)c |
| Cormorant/US92-45393/1992 | 16.48 | 22.66 | 15.44 | ||
| G/1992/US/284019467 | Cormorant/US92-43888/1992 | 15.52 | 24.94 | 15.71 | |
| G/1992/US/253317820 | Cormorant/US/MN92-40140/1992 | 13.47 | 17.19 | 12.76 | 1.39 (1.35)d |
| G/1992/US/20799674 | Cormorant/US/MI92-40068/1992 | 15.04 | 21.39 | 15.55 | (1.51)d |
| G/2005/US/254847461 | Cormorant/US/NV19529-04/2005 | 31.08 | 0 | 14.99 | 1.53 |
| G/2003/US/254847459 | Cormorant/US/WI18719-03/2003 | 21.61 | 19.61 | 15.95 | 1.41 |
| G/1997/US/284019479 | Cormorant/US/UT14854-02/1997 | 35.65 | 33.35 | 30.53 | |
| G/1997/US/284019477 | Cormorant/US/UT14854-01/1997 | 33.02 | 34.14 | 31.14 | |
| G/1997/US/284019475 | Cormorant/US/CA14803-02/1997 | 20.19 | 29.59 | 34.46 | |
| G/1997/US/284019473 | Cormorant/US/CA14798-03/1997 | 21.27 | 31.48 | 16.74 | |
| Cormorant/US/CA14798-01/1997 | 36.17 | 37.08 | 34.05 | ||
| G/1997/US/284019471 | Cormorant/US/CA14796-08/1997 | 21.7 | 32.55 | 25.95 | |
The matrix gene assay, the fusion gene assay and the revised fusion gene assay with a cormorant-specific probe were used.
ICPI values are given when they were available. Values in parentheses were reported by others, as specified in footnotes c to e.
Reported previously in reference 4.
Reported previously in reference 9.
Reported previously in reference 21.
Initial testing demonstrated that all isolates were positively identified by the USDA validated real-time RT-PCR M-gene assay (threshold cycle [CT] range, 13.47 to 36.17); however, only 1 of the 10 isolates recovered in 2008 and 4 of the 14 isolates recovered since 2002 tested were detected using the F-gene assay, after duplicate attempts (CT value, 0; Table 2). This is in contrast to the results for isolates that predate 2002, all of which were detected (n = 18). To determine whether mismatches at the probe site were responsible for the F-gene assay failure, representative viruses were sequenced and the sequences were compared to those of older isolates. A cormorant virus-specific probe was then designed by evaluating the alignment of 24 nt of cormorant NDV sequences (positions 4863 to 4896), which encode the 112R/KRQKRF117 motif, by comparison to the alignment obtained with the F-gene assay probe (Fig. 1). Most cormorant strains have 2 nucleotide mismatches with the fusion probe region of NDV poultry isolates, G at position 4871 to A and C at position 4891 to T. The nucleotide change at position 4871 results in an amino acid substitution, glycine to arginine, while the nucleotide change at position 4891 is silent. All of the 2008 cormorant isolates (but not isolate cormorant/US/GACC393-E-06/2006) have an additional change of G to A at position 4878 that appears to be responsible for the lack of detection by the F-4894 fusion probe. The F-4894 fusion probe was redesigned to be completely homologous with the 2008 isolates, most of which had three mismatches. These three probe mismatches were also present in cormorant/US/MNA00520443/444/2008, yet the original F-gene test was able to detect this isolate, although weakly, with a CT value of 33.7. Additionally, the forward primer for the existing F-gene test (F+4829) was replaced with a cormorant-specific primer to provide a better matched melting temperature (Tm) to the reverse primer (F-4939) and to avoid two of three common mismatches with cormorant NDV sequences (data not shown). The original F-gene test designed to specifically to detect virus during the 2002 California outbreak in poultry had primers with a Tm difference of 3.43°C (55.81 to 52.38°C). The newly designed cormorant-specific forward primer has a smaller Tm difference of 0.97°C (53.35 to 52.38°C) (data not shown). This new and improved cormorant-specific F-gene test detected all NDV isolates tested (n = 32), which, in addition to the 2008 isolates, included the older cormorant viruses (1992).
A total of 2,022 NDV sequences, from a total of over 3,000 in GenBank, have a virulent fusion protein cleavage site. These were analyzed for the presence of mismatches with the original fusion probe. Of these 2,022, a total of 1,836 sequences (90.8%) have three or fewer mismatches and 186 have four or more mismatches. The sequences with four or more mismatches were analyzed phylogenetically (Fig. 2). The majority of the sequences corresponded to the sequence of the previously reported genotype VIb virus of pigeons (12); however, viruses with changes likely to cause failure in the USDA-validated F-gene test were also found in genotype I viruses (those from the Australian outbreak from 1998 to 2000), genotype II viruses, and of course, the cormorant samples.
FIG. 2.
Phylogenetic analysis of NDV strains predicted to fail the USDA-validated F-gene test. The evolutionary history was inferred using the neighbor-joining method (19). The optimal tree with the sum of the branch length of 1.97627992 is shown. The tree is drawn to scale, with the branch lengths being in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the Kimura two-parameter method (13) and are in the units of the number of base substitutions per site. The codon positions included were the 1st, 2nd, 3rd, and noncoding postions. All positions containing gaps and missing data were eliminated from the data set (complete deletion option). There were a total of 373 positions in the final data set. Phylogenetic analyses were conducted in MEGA4 (20). The 2005 Nevada Cormorant NDV strain is marked with an asterisk. The GI and GenBank accession numbers of the isolates are listed in Table S1 in the supplemental material.
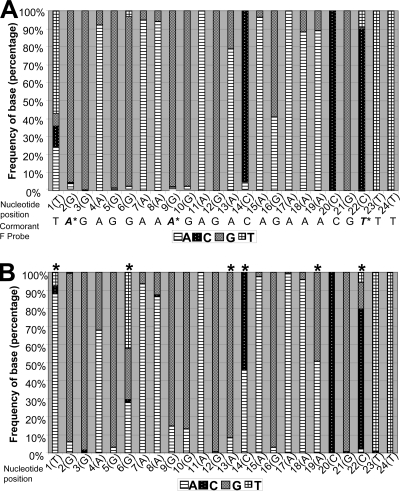
To determine the viability of the USDA-validated F-gene test, we analyzed the sequences of all available virulent isolates (as determined by reported virulence or the F-gene cleavage sequence). A total of 2,022 sequences from GenBank were compared, and the frequency of nucleotide distribution was analyzed using the BioEdit program (Fig. 3A). The sequences with less than four mismatches represented 90.8% of all the sequences available in GenBank, indicating that the test is still representative of the dominant nucleotides in NDVs worldwide and remains a valuable tool for the rapid assessment of virulence. In addition, all sequences containing four or more mismatches (n = 186) were analyzed separately, since, on the basis of our previous experience with the fusion test, those sequences are likely not to be detected by the USDA-validated F-gene test (Fig. 3B). Cormorant viruses represent a subpopulation of viruses that have significant changes at specific positions. For those viruses that had four or more mismatches, higher frequencies of differences were found at positions 1, 6, 13, 14, 19, and 22 (asterisks in Fig. 3), where the predominant nucleotide is either rare or variable. The cormorant-specific F-gene probe assay is not likely to be a substitute for the fusion test as a general identification assay, as the type of changes occurring in cormorant viruses are specific for the species.
FIG. 3.
Analysis of the probe region nucleotide frequencies. (A) Percent distribution of all nucleotides at positions 1 to 24 of the fusion probe in 2,022 virulent viruses in comparison the cormorant probe; (B) percent distribution of all nucleotides at positions 1 to 24 of the fusion probe in 186 virulent viruses that are expected to fail the test because they have four or more mismatches (asterisks indicate that the predominant nucleotide is different from that in the standard fusion probe).
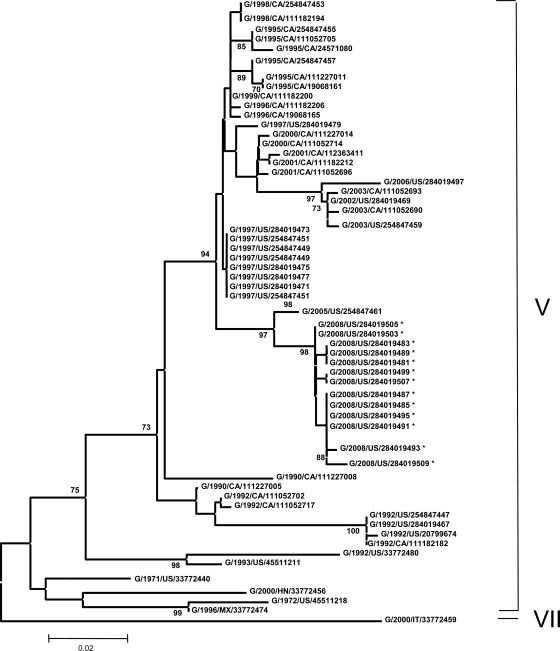
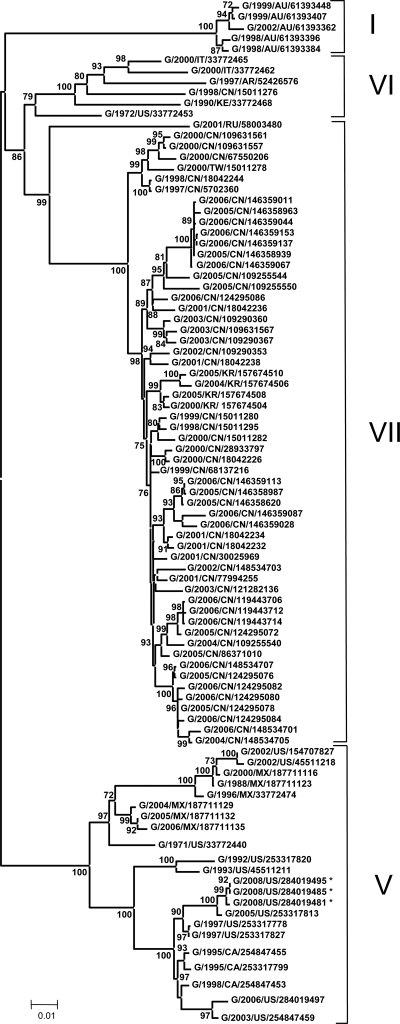
To determine the distribution of the 2008 viruses in comparison to known genotypes, sequencing and phylogenetic analysis were done. The phylogenetic analysis used only the 374-nucleotide partial fusion sequence to allow comparison with the largest possible number of cormorant isolates (Fig. 4). The 2008 cormorant viruses are tightly grouped and clearly fall within the genotype V viruses. Their nearest ancestor is a 2005 Nevada isolate (cormorant/US/NV19529-04/2005). The 2008 isolates are distinct but closely related to a branch of other genotype V viruses from the 1997 outbreak in the western United States. The most recent isolate before the 2008 event, cormorant/US/GACC393-E-06/2006 (2006 Georgia), is separated from the 2008 and 1997 viruses and is most related to the cormorant/US/WI18719-03/2003 and cormorant/US/FLCC415-A/2002 viruses.
FIG. 4.
Phylogenetic analysis of 374 nucleotides corresponding to the partial sequence of the F gene of 68 NDV isolates. The evolutionary history was inferred using the neighbor-joining method (19). The optimal tree with the sum of the branch length of 0.60932040 is shown. The percentages of replicate trees in which the associated taxa clustered together in the bootstrap test (500 replicates) are shown next to the branches (7). The tree is drawn to scale, with the branch lengths being in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the Kimura two-parameter method (13, 20) and are in the units of the number of base substitutions per site. The codon positions included were 1st, 2nd, 3rd, and noncoding positions. All positions containing gaps and missing data were eliminated from the data set (complete deletion option). There were a total of 354 positions in the final data set. Phylogenetic analyses were conducted in the MEGA4 (20). Not all isolates included in the analysis are shown on the tree because they group with other isolates on the same position on the branch. Isolates G/2008/US/284019511, -284019514, -284019501, -284019517, and -284019520 group with G/2008/US/284019485, -284019487, -284019491, and -284019495. Isolate G/2008/US/284019523 groups with G/2008/US/284019499 and -284019507. Isolates G/2008/US/284019526 and -284019532 group with G/2008/US/284019481, -284019483, and -284019489. Isolate G/2008/US/284019529 groups with G/2008/US/284019503 and -284019505. *, isolates first characterized in this study.
Phylogenetic analysis, including analysis of the full fusion gene sequences for three of the viruses recovered in 2008, gave results similar to those obtained with the 374-nucleotide partial fusion tree, with the isolates from 2008 being tightly grouped within the genotype V viruses (Fig. 5). This analysis included many strains isolated from other bird species that are of several different genotypes. As was seen with the 374-nucleotide partial fusion tree, a 2006 Georgia isolate and a 2003 Wisconsin isolate (isolate names in the tree, G/2006/US/284019497 and G/2003/US/254847459, respectively) were grouped and separated from the 2008 isolates, and a 2005 Nevada cormorant virus, NV19529 (isolate name in the tree, G/2005/US/254847461), was the nearest ancestor to the 2008 isolates.
FIG. 5.
Phylogenetic analysis of the fusion gene sequences of 91 NDV isolates. The evolutionary history was inferred using the neighbor-joining method (19). The optimal tree with the sum of the branch length of 1.00148249 is shown. The percentages of replicate trees in which the associated taxa clustered together in the bootstrap test (500 replicates) are shown next to the branches (7). The tree is drawn to scale, with the branch lengths being in the same units as those of the evolutionary distances used to infer the phylogenetic tree. *, isolates first characterized in this study.
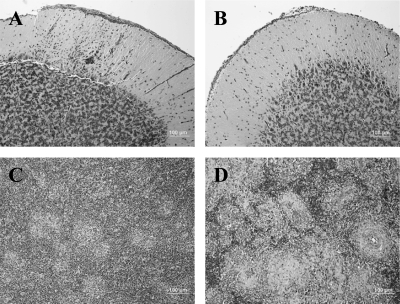
The pathogenesis of recent isolates was determined by determination of the ICPI (Table 2). Despite the sequence changes that rendered the F-gene assay undependable, no significant increase in pathogenicity in 1-day-old chicks was noted. This is not surprising, since the nucleotide change seen in the F-gene cleavage site of the 2008 isolates results in a conservative R-to-K amino acid substitution. Additionally, the isolate determined to be the most likely ancestor of the 2008 isolates, 2005 Nevada, was more thoroughly characterized by observation of in vivo pathogenesis in experimentally infected juvenile SPF chickens (Fig. 6). Mesogenic NDV is not thought to cause clinical disease in adult cormorants. During morbidity-mortality events, however, juveniles have a significant mortality rate and exhibit central nervous system signs, such as limb paralysis and head trembling (5, 14), that have been linked to cerebellar lesions (15, 16). In 4-week-old chickens inoculated with the 2005 Nevada cormorant virus, minimal morphological changes were noted in the eyelids, spleens, cecal tonsils, and hearts at 6 days postinfection (dpi) (Table 3). Furthermore, there was no evidence of any necrosis or lymphocytic depletion in the spleens determined by HE staining (Fig. 6C). A highly pathogenic NDV from the 2002 California outbreak in chickens was used as a positive control for disease. As an example, the spleens of chickens infected with the velogenic California 2002 isolate (18) showed multifocal to coalescing areas of necrosis and severe depletion of lymphocytes at 3 dpi (Fig. 6D).
FIG. 6.
Histopathological analysis of 4-week-old chickens infected with cormorant virus NV/2005. (A) Chicken inoculated with NV/2005, 6 dpi. Cerebellum, molecular layer. Multifocal areas of perivascular cuffing and gliosis. The vessels have hypertrophic endothelial cells. (B) Chicken inoculated with PBS. Cerebellum, molecular layer. There is no perivascular cuffing, and the vessels are minimally evident. (C) Chicken inoculated with NV/2005, 6 dpi. Spleen. The spleen is histologically normal, with no evidence of necrosis or lymphocytic depletion. (D) Chicken inoculated with CA/2002, 3 dpi. Spleen. The spleen has multifocal to coalescing areas of necrosis and severe depletion of lymphocytes.
TABLE 3.
Pathological scoring of disease at 6 dpi in chickens infected with 2005/Nevada
| Organa | Score at 6 dpia |
|---|---|
| Eyelid | + |
| Spleen | + |
| Thymus | − |
| Bursa | − |
| Harderian Gland | − |
| Proventriculus | − |
| Small intestine | − |
| Cecal tonsils | + |
| Large intestine | − |
| Pancreas | − |
| Air sacs | − |
| Trachea | − |
| Lung | − |
| Heart | + |
| Esophagus | − |
| Tongue | − |
| Pharynx | − |
| Crop | − |
| Brain | +++ |
| Liver | − |
| Kidney | − |
| Comb | − |
| Femur | − |
| Turbinates | − |
The results are based on hematoxylin-eosin staining, For the spleen, + is moderate hyperplasia; for the cecal tonsil, + is mild lymphocytic depletion; for the brain, +++ is vascular reactivity, perivascular cuffing, and gliosis.
With the 2005 Nevada virus, the only organ with morphological changes was the brain. At 6 dpi, there was encephalitis, especially in the cerebellum, characterized by multifocal areas of perivascular cuffing and gliosis (Fig. 6A). Control birds showed no perivascular cuffing in the cerebellum, and the vessels were minimally evident (Fig. 6B).
DISCUSSION
All of the 2008 cormorant isolates characterized here clearly belong to the class II genotype V group. This group likely originated from different bird species in the Americas and appears to have separated from genotype VI viruses in the 1970s (1). Numerous isolates from previously characterized NDVs in cormorants during the 1990s in Canada (21) as well as isolates from a California 1997 outbreak (unpublished) were included in the full fusion and 374-nucleotide fusion cleavage site phylogenetic analyses. This shows that the viruses that caused the 2008 morbidity-mortality event are significantly separated from viruses of both other recent outbreaks. However, interestingly, the 2008 isolates are more closely related to those from the 1997 California outbreak than the geographically closer Canadian isolates from 1990 to 2003 (21). The nearest relative of the 2008 isolates is the 2005 cormorant isolate from Nevada, which shares a common ancestor with both the 1997 California outbreak and the Canadian isolates from 1990 to 2003 but which is sufficiently removed from each to reliably determine which is the more related. Another recent isolate, 2006 Georgia, grouped with the 2003 Canadian and Wisconsin isolates but not the 2008 isolates and their 2005 Nevada ancestor. So, while phenotypic characterization using traditional pathotyping and histology shows no indication of evolution of the North American cormorant NDV strains since the last major outbreak in 1992, the phylogenetic analyses show a continued change in the genotype. Regardless, there is much concern over the ability of less virulent strains to evolve into more virulent strains, as has been surmised in outbreaks in Ireland (1990) (3) and Australia (1998 to 2000) (8) and in experiments in a laboratory setting (10).
The USDA NVSL pathotypes all NDV isolates by ICPI and/or amino acid sequencing. To date, a USDA-validated F-gene assay directed at the fusion cleavage site of NDV has been effective at differentiating virulent strains from those of low virulence (22). The F-gene assay is not an official pathotyping assay but is important for the rapid and efficient identification of virulent NDV, particularly during outbreaks. This differentiation test is key because the mere presence of NDV is not predictive of a disease outbreak because low-virulence strains circulate in wild birds and vaccine strains are administered to poultry (22). The F-gene assay failed to detect 9 of 10 cormorant isolates from 2008 and 1 isolate that our phylogenetic analyses showed was their nearest relative, the 2005 Nevada virus. This result is likely because this test was designed to specifically detect viruses from a 2002 outbreak in California (22). It appears that as NDV continues to evolve, no one PCR test will be effective for the detection of all viruses. The modified F-gene test with a new forward primer and probe successfully detected all older isolates tested and all 2008 isolates. However, the cormorant virus-specific probe is not likely to be a substitute for the F-gene test as a general identification test because the changes observed in cormorant viruses tend to be specific for the species. On the basis of our previous experience with the F-gene test, sequences containing four or more mismatches are not likely to be detected. In total, the new cormorant virus-specific F-gene test has been tested on a range of viruses isolated over a period of time from 1992 to 2008. Since the accurate identification of virulent isolates is important to continued monitoring efforts, modification of existing rRT-PCR tests should be expected as viruses continue to evolve.
Supplementary Material
Acknowledgments
We gratefully acknowledge Dawn Williams-Coplin and Tim Olivier for technical assistance and the South Atlantic Area Sequencing Facility for nucleotide sequencing.
This work was funded by USDA CRIS project number 6612-32000-049 and U.S. Poultry and Egg Association grant number 647.
The mention of trade names or commercial products in the manuscript is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the U.S. Department of Agriculture.
Footnotes
Published ahead of print on 27 January 2010.
Supplemental material for this article may be found at http://jcm.asm.org/.
REFERENCES
- 1.Aldous, E. W., J. K. Mynn, J. Banks, and D. J. Alexander. 2003. A molecular epidemiological study of avian paramyxovirus type 1 (Newcastle disease virus) isolates by phylogenetic analysis of a partial nucleotide sequence of the fusion protein gene. Avian Pathol. 32:239-256. [DOI] [PubMed] [Google Scholar]
- 2.Alexander, D. J. 1988. Newcastle disease diagnosis. Kluwer Academic Publisher, Boston, MA.
- 3.Alexander, D. J., G. Campbell, R. J. Manvell, M. S. Collins, G. Parsons, and M. S. McNulty. 1992. Characterisation of an antigenically unusual virus responsible for two outbreaks of Newcastle disease in the Republic of Ireland in 1990. Vet. Rec. 130:65-68. [DOI] [PubMed] [Google Scholar]
- 4.Allison, A. B., N. L. Gottdenker, and D. E. Stallknecht. 2005. Wintering of neurotropic velogenic Newcastle disease virus and West Nile virus in double-crested cormorants (Phalacrocorax auritus) from the Florida Keys. Avian Dis. 49:292-297. [DOI] [PubMed] [Google Scholar]
- 5.Banerjee, M., W. M. Reed, S. D. Fitzgerald, and B. Panigraphy. 1994. Neurotropic velogenic Newcastle disease in cormorants in Michigan: pathology and virus characterization. Avian Dis. 38:873-878. [PubMed] [Google Scholar]
- 6.Blaxland, J. D. 1951. Newcastle disease in shags and cormorants and its significance as a factor in the spread of this disease among domestic poultry. Vet. Rec. 68:731-733. [Google Scholar]
- 7.Felsenstein, J. 1985. Confidence limits on phylogenies: An approach using the bootstrap. Evolution 39:783-791. [DOI] [PubMed] [Google Scholar]
- 8.Gould, A. R., J. A. Kattenbelt, P. Selleck, E. Hansson, A. Della-Porta, and H. A. Westbury. 2001. Virulent Newcastle disease in Australia: molecular epidemiological analysis of viruses isolated prior to and during the outbreaks of 1998-2000. Virus Res. 77:51-60. [DOI] [PubMed] [Google Scholar]
- 9.Heckert, R. A., M. S. Collins, R. J. Manvell, I. Strong, J. E. Pearson, and D. J. Alexander. 1996. Comparison of Newcastle disease viruses isolated from cormorants in Canada and the USA in 1975, 1990 and 1992. Can. J. Vet. Res. 60:50-54. [PMC free article] [PubMed] [Google Scholar]
- 10.Islam, M. A., T. Ito, H. Takakuwa, A. Takada, C. Itakura, and H. Kida. 1994. Acquisition of pathogenicity of a Newcastle disease virus isolated from a Japanese quail by intracerebral passage in chickens. Jpn. J. Vet. Res. 42:147-156. [PubMed] [Google Scholar]
- 11.Kim, L. M., D. J. King, P. E. Curry, D. L. Suarez, D. E. Swayne, D. E. Stallknecht, R. D. Slemons, J. C. Pedersen, D. A. Senne, K. Winker, and C. L. Afonso. 2007. Phylogenetic diversity among low-virulence Newcastle disease viruses from waterfowl and shorebirds and comparison of genotype distributions to those of poultry-origin isolates. J. Virol. 81:12641-12653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim, L. M., D. J. King, H. Guzman, R. B. Tesh, A. P. Travassos da Rosa, R. Bueno, Jr., J. A. Dennett, and C. L. Afonso. 2008. Biological and phylogenetic characterization of pigeon paramyxovirus serotype 1 circulating in wild North American pigeons and doves. J. Clin. Microbiol. 46:3303-3310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kimura, M. 1980. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 16:111-120. [DOI] [PubMed] [Google Scholar]
- 14.Kuiken, T., F. A. Leighton, G. Wobeser, K. L. Danesik, J. Riva, and R. A. Heckert. 1998. An epidemic of Newcastle disease in double-crested cormorants from Saskatchewan. J. Wildl. Dis. 34:457-471. [DOI] [PubMed] [Google Scholar]
- 15.Kuiken, T., G. Wobeser, F. A. Leighton, D. M. Haines, B. Chelack, J. Bogdan, L. Hassard, R. A. Heckert, and J. Riva. 1999. Pathology of Newcastle disease in double-crested cormorants from Saskatchewan, with comparison of diagnostic methods. J. Wildl. Dis. 35:8-23. [DOI] [PubMed] [Google Scholar]
- 16.Meteyer, C. U., D. E. Docherty, L. C. Glaser, J. C. Franson, D. A. Senne, and R. Duncan. 1997. Diagnostic findings in the 1992 epornitic of neurotropic velogenic Newcastle disease in double-crested cormorants from the upper midwestern United States. Avian Dis. 41:171-180. [PubMed] [Google Scholar]
- 17.Mixson, M. A., and J. E. Pearson. 1992. Newcastle disease. Report of the Committee on Transmissible Diseases of Poultry and Other Avian Species, p. 357-360. Proc. 96th Annu. Meet. U.S. Anim. Health Assoc.
- 18.Pedersen, J. C., D. A. Senne, P. R. Woolcock, H. Kinde, D. J. King, M. G. Wise, B. Panigrahy, and B. S. Seal. 2004. Phylogenetic relationships among virulent Newcastle disease virus isolates from the 2002-2003 outbreak in California and other recent outbreaks in North America. J. Clin. Microbiol. 42:2329-2334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 20.Tamura, K., J. Dudley, M. Nei, and S. Kumar. 2007. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596-1599. [DOI] [PubMed] [Google Scholar]
- 21.Weingartl, H. M., J. Riva, and P. Kumthekar. 2003. Molecular characterization of avian paramyxovirus 1 isolates collected from cormorants in Canada from 1995 to 2000. J. Clin. Microbiol. 41:1280-1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wise, M. G., D. L. Suarez, B. S. Seal, J. C. Pedersen, D. A. Senne, D. J. King, D. R. Kapczynski, and E. Spackman. 2004. Development of a real-time reverse-transcription PCR for detection of Newcastle disease virus RNA in clinical samples. J. Clin. Microbiol. 42:329-338. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.