Abstract
An international multicenter study was conducted to assess the performance of a panel of simian immunodeficiency virus (SIV) RNA reference materials for plasma viral load determinations. Reliable quantification was demonstrated across an ∼6 log10 dynamic range. Availability of external reference materials will enable independent calibration of SIV plasma viral load assays.
Pandemic human immunodeficiency virus (HIV) has arisen by cross-species transfer from nonhuman primate species infected with simian immunodeficiency virus (SIV) (6, 28), adapted for transmission, and spread in human populations (29). Development of an effective vaccine to stem the spread of HIV/AIDS remains an urgent global health priority. The SIV/simian-human immunodeficiency virus (SHIV) macaque model supports (pre)clinical evaluations of candidate HIV vaccines to provide a scientific framework for rational vaccine design and evaluation (11, 15). Different candidate vaccine strategies may be investigated and viral pathogenesis elucidated (1, 2, 7, 18, 20, 24, 27). However, due to costs of undertaking biomedical research using macaque models and potentially conflicting issues of intellectual property, contemporaneous comparative vaccine efficacy studies at a single center are not feasible. If rational selection of the most promising vaccines is to be applied rapidly, it is important that outcomes of experimental studies are compared, a process facilitated by availability of external reference materials to calibrate key assays.
The concentration of HIV-1 virion RNA in plasma is an important correlate of disease progression, predictor of survival, and determinant of viral pathogenesis (9, 14, 19). Although first-generation assays to quantify HIV-1 RNA were variable (21), measurement of plasma viral RNA (vRNA) levels is now widely applied in clinical practice. Development, through international collaborative studies guided by SoGAT (Standardization of Gene Amplification Technologies), of an international standard (IS) for HIV-1 RNA has provided an ongoing quantified reference (16) and represents an important step in robust comparative standardization technology. Similar reference materials for HIV-2 are desirable (4), and an HIV-2 RNA IS is in development.
In the SIV/SHIV model, levels of plasma SIV RNA also predict survival and outcome (8, 13, 22, 30). Quantitative determinations of virion-associated SIV RNA are similarly employed to assess vaccine efficacy and modulation of virus infection in vaccine challenge studies and determine key aspects of HIV/SIV pathogenesis (1-3, 10, 12, 18, 23, 25, 27). However, no independent means of evaluating results reported by any one center and comparing them to those generated in another currently exists. As a first step in the development of reference materials to support SIV/SHIV macaque studies, an international collaborative study was undertaken to evaluate performance of a reference panel for quantitative assessments of SIV/SHIV vRNA load, focusing on EDTA-treated plasma.
Initially, bulk stocks of high-titer SIV-positive plasma were generated by inoculation of two purpose-bred cynomolgus macaques (Macaca fascicularis) with 10 MID50 (50% macaque infectious dose) SIVmac251/32H/L28 challenge stock, which replicates to >108 SIV RNA copies/ml at 10 to 14 days postinfection. The biological properties of this virus stock have been described previously (1). SIV RNA levels initially determined for each macaque (W341 and W342) by quantitative reverse transcriptase PCR (RT-PCR) (2) were subsequently analyzed using real-time PCR (1). From an initial 1/100 dilution of plasma from W342, five 10-fold dilution steps were performed in SIV-negative macaque plasma to provide an estimated (nominal) concentration range of 2 × 106 to 2 × 102 SIV RNA copies/ml. Multiple aliquots of each dilution (C1 to C5) were stored at −80°C.
This panel was distributed to five internationally recognized centers, four in Europe and one in the United States, employing six assays for quantitative SIV RNA determinations, four of which were real-time PCR (Table 1). Assay 5 (qRT-PCRv) provided additional validating information established using a virion-counting technique to determine input viral RNA copy number against a linear standard curve prepared by serial dilution of a SIVmac251 virus stock (5 × 105 to 1.25 ×102 particles/ml) in SIV-negative plasma. Preparations concentrated by ultracentrifugation prior to RNA extraction also included the HIV-1 CM240 virus as an internal control (20, 23, 24). The ExaVir/Cavidi assay (3), which measures RT activity calculated in femtograms/ml plasma, was expressed as RNA copies/ml. SIV RNA data generated from all six assays with the NIBSC panel, performed in a blinded manner at the respective centers, were returned to the coordinating partner (NIBSC).
TABLE 1.
Summary of techniques employed to quantify plasma SIV RNA
| Assay | Quantification systema | Target region | RNA extraction (plasma vol [μl]) | Calibration/standards | Upper and lower assay detection limits (RNA copies/ml) | Real-time PCR platform | Reference(s) |
|---|---|---|---|---|---|---|---|
| 1 | Real-time qRT-PCR | p27-gag | QIAamp, Qiagen (140) | External SIV251 RNA plasma series | 1 × 109 - 1.69 × 102 | Mx3000P (Stratagene) | 1 |
| 2 | Conventional qRT-PCRc | p27-gag | Trizol, propan-2-ol (200) | Coamplified synthetic RNA transcript | 1 × 107 - 1.6 × 102 | n/ab | 27 |
| 3 | Real-time qRT-PCR | p27-gag | QIAamp, Qiagen (200) | SIV251 RNA transcript | 1.6 × 106 - 1.69 × 102 | ABI Prism (Applied Biosystems) | 5, 18 |
| 4 | qRT assay | n/a | Viral binding gel (1,000) | BrdU/poly(A) system/HIV standard curve | 1.2 ×106 - 2.3 × 102 | n/a | 3 |
| 5 | Real-time qRT-PCRv | p15-gag | Nuclisens, bioMérieux (200) | SIV251 and internal HIV-1 control | 1 × 106 - 2.39 × 102 | ABI Prism (Applied Biosystems) | 20, 24, 25 |
| 6 | Real-time qRT-PCR | p27-gag | Trizol (200) | Coamplified synthetic RNA transcript | 1 × 1010 - 2.3 × 102 | iQ5 multicolor system (Bio-Rad) | 10 |
qRT-PCR, quantitative reverse transcriptase PCR; qRT-PCRc, conventional qRT-PCR system based on endpoint estimation of preamplified product; qRT-PCRv, quantitative RT-PCR validated by a virion estimation technique. qRT assay (assay 4), ExaVir Load v. 2 (Cavidi AB, Uppsala, Sweden). Assay 6 is a previously established real-time PCR method (10), with a modification of the SIV probe sequence to the Black Hole Quencher 2 dye in place of TAMRA.
n/a, not applicable.
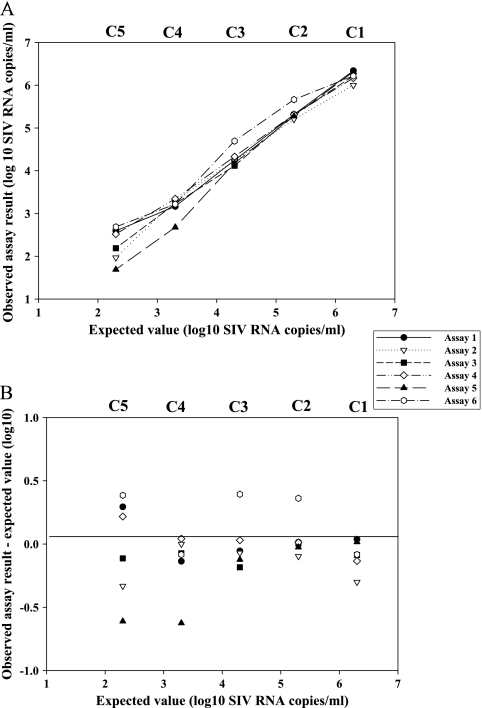
The purpose of this study was to evaluate a series of candidate reference materials for SIV RNA quantification in a range of assays. The returned data set from each center effectively represented a single assay estimate for each panel member. As a result, there were not sufficient data to perform a detailed analysis of the performance of the different participants or consistency between assay methods. However, the results (estimated log10 RNA copies/ml) from the different assay methods when plotted against the nominal expected values for the panel members (Fig. 1 A) indicated a good linear relationship between estimated (measured) and nominal expected values of the panel members across the 102 to 106 log10 SIV RNA copy number range. Fitting linear regression to the individual assay methods (log10 estimated copies/ml against nominal expected values in log10 copies/ml) gave a good fit, for both sequence-based amplification assays targeting conserved regions in p15/p27-gag and the RT assay, with r2 values ranging from 0.999 to 0.995.
FIG. 1.
(A) Comparison of SIV RNA levels evaluated using six different assays for the quantification of plasma viral load. Estimated copies/ml (log10) for each assay are plotted against the nominal or expected value of SIV RNA copies/ml (log10) for the panel members (C1, 6.3 log10; C2, 5.3 log10; C3, 4.3 log10; C4, 3.3 log10; C5, 2.3 log10) for each of the six assays: assay 1, qRT-PCR; assay 2, conventional qRT-PCR; assay 3, qRT-PCR; assay 4, qRT, assay 5; qRT-PCRv; assay 6, qRT-PCR (see Table 1 for details). (B) Difference between the observed estimated copies/ml (log10) and the expected nominal value copies/ml (log10) plotted against nominal expected copies/ml (log10) for individual assays (1 to 6) across panel members (C1 to C5).
Differences between the observed, estimated numbers of copies/ml (log10) and the expected numbers of copies/ml (log10) were also plotted against the expected copy/ml (log10) values (Fig. 1B). While variation between individual assays for different panel members was observed, this variation never exceeded 0.6 log10 for any combinations of assay and panel members, the majority of estimated copies/ml values falling within 0.5 log10 of the nominal expected value for all assays and panel members. For the panel series, the mean and standard deviation (SD) for all assays were calculated as follows: C1, log10 6.2, SD 0.11; C2, log10 5.34, SD 0.16; C3, log10 4.29, SD 0.20; C4, log10 3.15, SD 0.24; and C5, log10 2.26, SD 0.32. Hence, the standard deviation across the range of values increased as the copy number decreased, although this represents good agreement between the different assay methods over the wide dynamic range encountered among clinical samples. As expected, most variation was observed at the lowest copy number (panel member C5), which is in agreement with reports that greater variability in determinations of low viral copy number (<4 log10 copies/ml) is a recognized feature of viral quantification (17, 21, 26).
These reference materials may have broader utility in validation of a wider range of assays for quantitative vRNA determinations and to different target regions. Although based on SIVmac251/32H, for which the full genome sequence is available (www.hiv.lanl.gov; GenBank accession number D01065), the panel may be applicable to a broader range of viruses, including other SIVmac strains (SIV239), SIVsm-based viruses (SIVsmE660), HIV-2, and SHIVs, provided that issues of sequence variation are accounted for in the assay design and demonstrated not to impact adversely on individual assay performance.
Moreover, one of the main limitations to establishing reference materials of this kind is the availability of large amounts of high-titer material which can be titrated, characterized, and repeatedly sampled. Synthetic RNA transcripts are frequently used to establish, or internally control, quantitative viral load assays (5, 10, 12, 18, 25, 27). Ideally, external reference materials for independent validation should be as close in kind as possible to samples for which they are intended to provide a reference, such as viral RNA packaged into an intact virion particle prior to entering the extraction process. Reconstructed high-titer material using culture supernatant virus spiked into negative plasma can closely mimic the natural situation. In the current study, the availability of a bulk stock of high-titer plasma from a single macaque infected with a recognized stock of SIVmac251/32H/L28 made this plasma an ideal reference candidate representing closely the same composition as samples analyzed in SIV/macaque studies.
This is the first report to describe the development and evaluation of reference materials for quantitative SIV RNA determinations in plasma, which performed well in this international collaborative study. Provision of similar reagents based on this panel to the scientific community will facilitate standardization of vaccination and treatment clinical trials for HIV/AIDS.
Acknowledgments
We thank Alan Heath, NIBSC/HPA, for statistical advice and suggestions and Sal Butera and Shambavi Subbarao for their contributions.
The work was funded in part by grants from the United Kingdom Medical Research Council (G9025730 and G9419998) and EUFP6 grant Europrise (037611).
The findings and conclusions in this report are those of the authors and do not necessarily represent the views of the Centers for Disease Control and Prevention. Use of trade names is for identification only and does not constitute endorsement by the U.S. Department of Health and Human Services, the Public Health Service, or the Centers for Disease Control and Prevention.
Footnotes
Published ahead of print on 28 April 2010.
The authors have paid a fee to allow immediate free access to this article.
REFERENCES
- 1.Berry, N., R. Stebbings, D. Ferguson, C. Ham, J. Alden, S. Brown, A. Jenkins, A. Clayton, J. Lines, L. Davis, M. Page, R. Hull, J. Stott, and N. Almond. 2008. Resistance to superinfection of a vigorously replicating, uncloned stock of simian immunodeficiency virus (SIVmac251) stimulates replication of a live attenuated virus vaccine (SIVmacC8). J. Gen. Virol. 89:2240-2251. [DOI] [PubMed] [Google Scholar]
- 2.Clarke, S., N. Almond, and N. Berry. 2003. Simian immunodeficiency virus nef gene regulates the production of 2-LTR circles in vivo. Virology 306:100-108. [DOI] [PubMed] [Google Scholar]
- 3.Corrigan, G. E., E. Hansson, A. Mörner, N. Berry, C. Källander, and R. Thorstensson. 2006. Reverse transcriptase viral load correlates with RNA in SIV/SHIV infected macaques. AIDS Res. Hum. Retroviruses 22:917-923. [DOI] [PubMed] [Google Scholar]
- 4.Damond, F., A. Benard, J. Ruelle, A. Alabi, B. Kupfer, P. Gomes, B. Rodes, J. Albert, J. Boni, J. Garson, B. Ferns, S. Matheron, G. Chene, and F. Brun-Vezinet for the ACHIEV2E (a Collaboration on HIV-2 Infection) Study Group. 2008. Quality control assessment of human immunodeficiency virus type 2 (HIV-2) viral load quantification assays: results from an international collaboration on HIV-2 infection in 2006. J. Clin. Microbiol. 46:2088-2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Goletti, D., I. Macchia, P. Leone, M. Pace, L. Sernicola, M. R. Pavone-Cossut, M. T. Maggiorella, A. Cafaro, B. Ensoli, and F. Titti. 2006. Innate anti-viral immunity is associated with the protection elicited by the simian immunodeficiency virus (SIV) live attenuated virus vaccine in cynomolgus monkeys. Med. Sci. Monit. 12:330-340. [PubMed] [Google Scholar]
- 6.Hahn, B. H., G. M. Shaw, K. M. De Cock, and P. M. Sharp. 2000. AIDS as a zoonosis: scientific and public health implications. Science 287:607-614. [DOI] [PubMed] [Google Scholar]
- 7.Hessell, A. J., E. G. Rakasz, D. M. Tehrani, M. Huber, K. L. Weisgrau, G. Landucci, D. N. Forthal, W. C. Koff, P. Poignard, D. I. Watkins, and D. R. Burton. 2010. Broadly neutralizing monoclonal antibodies 2F5 and 4E10 directed against the human immunodeficiency virus type 1 gp41 membrane-proximal external region protect against mucosal challenge by simian-human immunodeficiency virus SHIVBa-L. J. Virol. 84:1302-1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hirsch, V. M., T. R. Fuerst, G. Sutter, M. W. Carroll, L. C. Yang, S. Goldstein, M. Piatek, W. R. Elkins, W. G. Alvord, D. C. Montefiori, B. Moss, and J. D. Lifson. 1996. Patterns of viral replication correlate with outcome in simian immunodeficiency virus (SIV)-infected macaques: effect of prior immunization with a trivalent SIV vaccine in modified vaccinia virus Ankara. J. Virol. 70:3741-3752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ho, D. D., A. U. Neumann, A. S. Perelson, W. Chen, J. M. Leonard, and M. Markovitz. 1995. Rapid turnover of plasma viraemia and CD4 lymphocytes in HIV-1 infection. Nature 373:123-126. [DOI] [PubMed] [Google Scholar]
- 10.Hoffmann-Lehman, R., R. K. Swenerton, V. Liska, C. M. Leutenegger, H. Lutz, H. M. McClure, and R. M. Ruprecht. 2000. Sensitive and robust one-tube real-time reverse transcriptase-polymerase chain reaction to quantify SIV RNA load: comparison of one-versus two-enzyme systems. AIDS Res. Hum. Retroviruses 16:1247-1257. [DOI] [PubMed] [Google Scholar]
- 11.Koff, W. C., P. R. Johnson, D. I. Watkins, D. R. Burton, J. D. Lifson, K. J. Hasenkrug, A. B. McDermott, A. Schultz, T. J. Zamb, R. Boyle, and R. C. Desrosiers. 2006. HIV vaccine design: insights from live attenuated SIV vaccines. Nat. Immunol. 7:19-23. [DOI] [PubMed] [Google Scholar]
- 12.Leutenegger, C. M., J. Higgins, T. B. Matthews, A. F. Tarantal, P. A. Luciw, N. C. Pedersen, and T. W. North. 2001. Real-time TaqMan PCR as a specific and more sensitive alternative to the branched-chain DNA assay for the quantification of simian immunodeficiency virus RNA. AIDS. Res. Hum. Retroviruses 17:243-251. [DOI] [PubMed] [Google Scholar]
- 13.Lifson, J. D., M. A. Nowak, S. Goldstein, J. L. Rossio, A. Kinter, G. Vasquez, T. A. Wiltrout, C. Brown, D. Schneider, L. Wahl, A. L. Lloyd, J. Williams, W. R. Elkins, A. S. Fauci, and V. M. Hirsch. 1997. The extent of early viral replication is a critical determinant of the natural history of simian immunodeficiency virus infection. J. Virol. 71:9508-9514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mellors, J. W., C. R. Rinaldo, P. Gupta, R. M. White, J. A. Todd, and L. A. Kingsley. 1996. Prognosis of HIV-1 infection predicted by the quantity of virus in plasma. Science 272:1167-1170. [DOI] [PubMed] [Google Scholar]
- 15.Morgan, C., M. Marthas, C. Miller, A. Duerr, C. Cheng-Mayer, R. Desrosiers, J. Flores, N. Haigwood, S. L. Hu, R. P. Johnson, J. Lifson, D. Montefiori, J. Moore, M. Robert-Guroff, H. Robinson, S. Self, and L. Corey. 2008. The use of nonhuman primate models in HIV vaccine development. PLoS Med. 5:e173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Morris, C., N. Berry, A. Heath, and H. Holmes. 2008. An international collaborative study to establish a replacement WHO international standard for HIV-1 RNA nucleic acid assays. Vox Sang. 95:218-225. [DOI] [PubMed] [Google Scholar]
- 17.Muyldermans, G., L. Debaisieux, K. Fransen, D. Marissens, K. Miller, D. Vaira, A. M. Vandamme, A. T. Vandenbroucke, C. Verhofstede, R. Schuurman, G. Zissis, and S. Lauwers. 2000. Blinded, multicenter quality control study for the quantification of human immunodeficiency virus type 1 RNA in plasma by the Belgian AIDS reference laboratories. Clin. Microbiol. Infect. 6:213-217. [DOI] [PubMed] [Google Scholar]
- 18.Negri, D. R., S. Baroncelli, S. Catone, A. Comini, Z. Michelini, M. T. Maggiorella, L. Sernicola, F. Crostarosa, R. Belli, M. G. Mancini, S. Farcomeni, Z. Fagrouch, M. Ciccozzi, S. Boros, P. Liljestrom, S. Norley, J. Heeney, and F. Titti. 2004. Protective efficacy of a multicomponent vector vaccine in cynomolgus monkeys after intrarectal simian immunodeficiency virus challenge. J. Gen. Virol. 85:1191-1201. [DOI] [PubMed] [Google Scholar]
- 19.Piatek, M., Jr., M. S. Saag, L. C. Yang, S. J. Clark, J. C. Kappes, K. C. Luk, B. Hahn, G. M. Shaw, and J. D. Lifson. 1993. High levels of HIV-1 in plasma during all stages of infection determined by competitive PCR. Science 259:1749-1754. [DOI] [PubMed] [Google Scholar]
- 20.Promadej-Lanier, N., P. Srinivasan, K. Curtis, D. R. Adams, C. Kim, W. Luo, H. Jia, S. Subbarao, R. A. Otten, and S. Butera. 2008. Systemic and mucosal immunological responses during repeated mucosal SHIV(162P3) challenges prior to and following infection in pigtailed macaques. Virology 375:492-503. [DOI] [PubMed] [Google Scholar]
- 21.Schuurman, R., D. Descamps, G. Jan Weverling, S. Kaye, J. Tinjnagel, I. Williams, R. van Leeuwen, R. Tedder, C. Boucher, F. Brun-Vezinet, and C. Loveday. 1996. Multicenter comparison of three commercial methods for quantification of human immunodeficiency virus type 1 RNA in plasma. J. Clin. Microbiol. 34:3016-3022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Staprans, S., P. J. Dailey, A. Rosenthal, C. Horton, R. M. Grant, N. Lerche, and M. B. Feinberg. 1999. Simian immunodeficiency virus disease course is predicted by the extent of virus replication during primary infection. J. Virol. 73:4829-4839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Subbarao, S., A. Ramos, C. Kim, D. Adams, M. Monsour, S. Butera, T. Folks, and R. A. Otten. 2007. Direct stringency comparison of two macaque models (single-high vs. repeat-low) for mucosal HIV transmission using an identical anti-HIV chemoprophylaxis intervention. J. Med. Primatol. 36:238-243. [DOI] [PubMed] [Google Scholar]
- 24.Subbarao, S., R. A. Otten, A. Ramos, C. Kim, E. Jackson, M. Monsour, D. R. Adams, S. Bashirian, J. Johnson, V. Soriano, A. Rendon, M. G. Hudgens, S. Butera, R. Janssen, L. Paxton, A. E. Greenberg, and T. M. Folks. 2006. Chemoprophylaxis with tenofovir disoproxil fumarate provided partial protection against infection with simian human immunodeficiency virus in macaques given multiple virus challenges. J. Infect. Dis. 194:904-911. [DOI] [PubMed] [Google Scholar]
- 25.Suryanarayana, K., T. A. Wiltrout, G. M. Vasquez, V. M. Hirsch, and J. D. Lifson. 1998. Plasma SIV RNA load determination by real-time quantification of product generation in reverse transcriptase polymerase chain reaction. AIDS Res. Hum. Retroviruses 14:183-189. [DOI] [PubMed] [Google Scholar]
- 26.Swanson, P., V. Soriano, S. G. Devare, and J. Hackett, Jr. 2001. Comparative performance of three viral load assays on human immunodeficiency virus type 1 (HIV-1) isolates representing group M (subtypes A to G) and group O: LCx HIV RNA quantitative, AMPLICOR HIV-1 MONITOR version 1.5, and Quantiplex HIV-1 RNA version 3.0. J. Clin. Microbiol. 39:862-870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.ten Haaft, P., B. Verstrepen, K. Uberla, B. Rosenwirth, and J. Heeney. 1998. A pathogenic threshold of virus load defined in simian immunodeficiency virus- or simian-human immunodeficiency virus-infected macaques. J. Virol. 72:10281-10285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Van Heuverswyn, F., and M. Peeters. 2007. The origins of HIV and implications for the global epidemic. Curr. Infect. Dis. Rep. 9:338-346. [DOI] [PubMed] [Google Scholar]
- 29.Wain, L. V., E. Bailes, F. Bibollet-Ruche, J. M. Decker, B. F. Keele, F. Van Heuverswyn, Y. Li, J. Takehisa, E. M. Ngole, G. M. Shaw, M. Peeters, B. H. Hahn, and P. M. Sharp. 2007. Adaptation of HIV-1 to its human host. Mol. Biol. Evol. 24:1853-1860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Watson, A., J. Ranchalis, B. Travis, J. McClure, W. Sutton, P. R. Johnson, S. L. Hu, and N. L. Haigwood. 1997. Plasma viraemia in macaques infected with simian immunodeficiency virus: plasma viral load early in infection predicts survival. J. Virol. 71:284-290. [DOI] [PMC free article] [PubMed] [Google Scholar]