Abstract
Salmonella enterica serovar Enteritidis (S. Enteritidis) is frequently associated with food-borne disease worldwide. Poultry-derived products are a major source. An epidemic of human infection with S. Enteritidis occurred in Uruguay, and to evaluate the extent of poultry contamination, we conducted a nationwide survey over 2 years that included the analysis of sera from 5,751 birds and 12,400 eggs. Serological evidence of infection with Salmonella group O:9 was found in 24.4% of the birds. All positive sera were retested with a gm flagellum-based enzyme-linked immunosorbent assay, and based on these results, the national prevalence of S. Enteritidis infection was estimated to be 6.3%. Salmonellae were recovered from 58 of 620 pools made up of 20 eggs each, demonstrating a prevalence of at least 1 in every 214 eggs. Surprisingly, the majority of the isolates were not S. Enteritidis. Thirty-nine isolates were typed as S. Derby, 9 as S. Gallinarum, 8 as S. Enteritidis, and 2 as S. Panama. Despite the highest prevalence in eggs, S. Derby was not isolated from humans in the period of analysis, suggesting a low capacity to infect humans. Microarray-based comparative genomic hybridization analysis of S. Derby and S. Enteritidis revealed more than 350 genetic differences. S. Derby lacked pathogenicity islands 13 and 14, the fimbrial lpf operon, and other regions encoding metabolic functions. Several of these regions are present not only in serovar Enteritidis but also in all sequenced strains of S. Typhimurium, suggesting that these regions might be related to the capacity of Salmonella to cause food-borne disease.
Salmonella enterica is a major cause of food-borne disease worldwide (14, 18, 46). Poultry-derived products, particularly chicken eggs, are considered a major source of human infection with Salmonella (2, 20, 38). Chickens can be infected with many different serovars of Salmonella. Of these, S. enterica serovars Pullorum and Gallinarum (S. Pullorum and S. Gallinarum, respectively) are host specific and represent a major concern for the poultry industry but have no impact on public health. Other S. enterica serovars frequently isolated from chickens, such as Typhimurium, Enteritidis, and Heidelberg, can infect a wider range of hosts and frequently reach the human food chain, causing food-borne disease.
A peculiar epidemiological feature of human salmonellosis is that epidemics are commonly associated with a particular prevalent serovar of S. enterica that shows temporal and geographical variation. Until the 1980s, S. enterica serovar Typhimurium (S. Typhimurium) was the serovar most commonly isolated from humans worldwide, but by the late 1980s, S. enterica serovar Enteritidis (S. Enteritidis) emerged as the most common cause of salmonellosis in Europe, and during the 1990s, it became the most prevalent serovar in many countries worldwide (9, 22, 33, 40, 43). The reasons for this worldwide serovar shift are still not understood, and several hypotheses have been proposed, including the existence of a rodent reservoir for S. Enteritidis or the epidemiological change induced by vaccination of poultry against the closely related bacterium S. Gallinarum (47).
In Uruguay, S. Typhimurium was the most frequently isolated serovar until 1994, and S. Enteritidis was only sporadically isolated (3, 24, 37). In 1995, a first outbreak of S. Enteritidis occurred, starting an epidemic that lasted almost 10 years. This outbreak was traced back to sandwiches prepared with contaminated mayonnaise that were distributed nationwide by a local catering service. According to data provided by the national public health authorities, the outbreak affected an estimated 600 individuals countrywide. From then on, several other outbreaks of various sizes occurred and S. Enteritidis was identified as the cause in 89% of Salmonella food poisoning episodes. In most of these cases (80%, according to official records), eggs or chicken meat was identified as the source of infection. From 1997 to 2004, S. Enteritidis was the most frequently identified serovar in Uruguay, accounting for more than 50% of the strains received each year at the National Salmonella Center and for more than 85% of the strains isolated from humans (3). After 2005, there was a dramatic reduction in the number of S. Enteritidis outbreaks, and this year was considered the end of the epidemic. Over the last 3 years, S. Typhimurium has become the serovar most frequently associated with isolated cases of food poisoning, and S. Derby and S. Panama have been sporadically isolated. Nevertheless, S. Enteritidis is still the serovar most frequently associated with outbreaks in the country.
S. Enteritidis frequently colonizes the alimentary tracts of chickens without causing disease. However, it can produce a systemic infection in young chicks that may further lead to the infection of egg contents (13, 51). With the aim of knowing the prevalence of S. Enteritidis infection in poultry, we designed and conducted a countrywide serological and microbiological survey of chicken flocks and commercially available eggs from 2000 to 2002, and the results are presented here. An unexpected result of the survey was a higher prevalence of S. Derby than S. Enteritidis in eggs, particularly because while the latter was identified as the etiological agent of the epidemic there were no reports of human infections with S. Derby in the same period of time. This suggested a low capacity of S. Derby isolates to infect humans; thus, we performed a genomic comparison of the two serovars to search for genetic differences that could be the basis of such marked differences in epidemiological behavior. We found that S. Derby lacks several genomic regions related to virulence, suggesting that these regions could be involved in the capacity of Salmonella to cause food-borne disease.
MATERIALS AND METHODS
Serological survey.
A serological survey for the detection of antibodies against Salmonella O:9 lipopolysaccharide (LPS) was conducted on a representative sample of Uruguayan breeding and commercial egg-laying flocks. The size of the sample was defined with statistical criteria, considering a whole population of approximately 2,500,000 laying hens and 250,000 breeders. Based on preliminary estimations, we assumed an expected prevalence of infection of 5% among laying birds and 2% among breeders. A sampling error of 0.2% and a confidence interval of 95% were taken into account in defining the extent of the sampling regimen. Based on these assumptions, a total of 34 flocks (20 layer and 14 breeder flocks) from the 664 flocks in the country were selected for sampling, and from these, 5,751 hens were randomly selected using a stratified random sampling methodology. The selected farms were located in 7 of the 19 Departments into which Uruguay is divided and were geographically representative of all of the major poultry production areas.
A questionnaire was administered along with the survey to build a database containing information from each farm surveyed, including the history of previous vaccination with a locally available product, i.e., 9R (Nobilis SG 9R; Hoechst Roussel-Intervet) or inactivated S. Enteritidis (SEI; Ciencia Lab, Merial) vaccine. Table 1 shows data from all of the sampled flocks and farms.
TABLE 1.
Description of serological and microbiological samplinga
| Type of hen and farm sampledb | No. of sera testedc | No. of eggs sampledd | Vaccinatione |
|---|---|---|---|
| Layers | |||
| A | 189 | 1,200 | No |
| B | 183 | 600 | 9R |
| C (5 flocks) | 949 | 800 | 9R |
| D | 189 | 600 | No |
| E | 186 | 700 | No |
| F | 0 | 600 | NA |
| G (2 flocks) | 339 | 600 | No |
| H | 181 | 700 | No |
| I | 0 | 500 | NA |
| J | 0 | 500 | NA |
| K | 183 | 500 | No |
| L | 189 | 700 | No |
| M | 0 | 400 | NA |
| N | 191 | 600 | SEI |
| Ñ | 198 | 400 | No |
| O | 0 | 400 | NA |
| P | 0 | 500 | NA |
| Q | 181 | 400 | No |
| R | 0 | 700 | NA |
| S | 182 | 600 | No |
| T | 189 | 400 | No |
| 7 | 185 | 0 | No |
| Layer-breeders | |||
| 14 | 153 | No | |
| 19 | 139 | No | |
| 20 | 151 | No | |
| 26 | 99 | 9R | |
| 27 | 99 | 9R | |
| 30 | 149 | No | |
| 31 | 151 | No | |
| 33 | 168 | No | |
| 34 | 157 | No | |
| Broiler-breeders | |||
| 2 | 150 | 9R | |
| 3 | 153 | No | |
| 4 | 158 | SEI | |
| 25 | 160 | 9R +SEI | |
| 32 | 150 | SEI |
Data from environmental and cloacal swabs are not included. Each flock was sampled for 60 cloacal swabs and 1 environmental swab.
A total of 41 flocks were tested.
A total of 5,751 sera from 34 flocks were tested.
A total of 12,400 eggs from 21 layer farms were tested.
NA, no available data. 9R, S. Gallinarum rough strain 9R vaccine. SEI, S. Enteritidis inactivated vaccine.
Blood was obtained from the wing vein, and serum was separated. Labeled coded samples were stored in freezer boxes at −20°C until assessed. Sera were evaluated double blind for serological evidence of infection using two different screening methods, an S. Enteritidis LPS enzyme-linked immunosorbent assay (ELISA) (a modification of the method described by Nicholas and Cullen in 1991 [35]) and agglutination by Rapid Slide Test (RST) using locally produced S. Pullorum-S. Gallinarum polyvalent somatic antigens as described by the Office International des Epizooties (48).
All sera yielding a positive result by any of the screening methods were also evaluated by a commercial ELISA kit that assesses antibodies against gm flagellin antigens (gm ELISA, Salmonella Enteritidis antibody test kit, FlockChek Se assay; IDEXX Laboratories, Inc., Westbrook, ME).
LPS ELISA.
Sera were analyzed using a modification of a previously described method (35). Briefly, microtiter plates were coated with 50 μl of LPS solution at 5 μg/ml (LPS from Salmonella Enteritidis; Sigma Chemical Co., St. Louis, MO) in coating buffer (0.05 M carbonate-bicarbonate buffer, pH 9.6) and incubated for 1 h at 37°C. The plates were washed five times in phosphate-buffered saline (PBS) containing 0.05% Tween 20. Test and control sera were diluted 1/1,000 in PBS-Tween buffer with 1% bovine serum albumin and assayed in duplicate using 50 μl per well. Plates were incubated for 1 h at 37°C and then washed as before. A solution of goat anti-chicken IgG conjugated to peroxidase (Bethyl Laboratories, Inc., Montgomery, TX) diluted in PBS-Tween-bovine serum albumin buffer was added (50 μl per well), and the mixture was incubated for 1 h at 37°C and washed as before. Finally, 50 μl of the substrate (0.4 mg/ml O-phenylenediamine dihydrochloride; ICN Biomedicals Inc., Irvine, CA) in pH 5 citrate-disodium phosphate buffer containing 0.02% hydrogen peroxide was added. The reaction was stopped after 15 min with 3 N sulfuric acid, and readings were taken at 490 nm in a laboratory microplate reader (MRX; Dinex Technologies, Inc., Chantilly, VA).
To define a cutoff value for the ELISA that distinguishes negative from positive samples, the sera from 50 specific-pathogen-free (SPF) chickens (obtained from the Central Veterinary Laboratories, Ministry of Agriculture, Uruguay) plus two positive controls (see below) were evaluated 10 times by two different operators. The average of all optical density (OD) values of the SPF chicken sera plus 2 standard deviations (SD) was calculated and defined as the threshold value.
For analysis of the sera taken during the survey, each ELISA plate included two positive-control and two negative-control sera. Positive controls were a positive-control serum (PC1) obtained from R. Davies (Central Veterinary Laboratories Weybridge, United Kingdom) and a pool of sera from chickens experimentally infected with S. Enteritidis (PC2). Negative controls were a pool of the sera from SPF birds (NC1) and a single SPF chicken serum (NC2). PC1/NC1 and PC2/NC2 ratios were calculated for the 10 plate determinations described above, and the average and SD values were registered. For plate validation, both ratios were calculated and the plate was validated when the ratios fell within the previously determined averages ± 2 SD.
RST.
The RST was performed by standard methods previously described (48). Briefly, equal volumes of crystal violet-stained polyvalent somatic antigen and serum were mixed in a glass slide; the results were read in 2 min maximum. Known positive and negative controls were examined together with each batch of samples.
gm ELISA.
Sera were processed using a commercial ELISA kit (FlockChek Se assay; IDEXX Laboratories, Inc., Westbrook, ME) specifically designed to detect antibodies to Salmonella Enteritidis in serum or egg yolk by following the manufacturer's instructions. The method is a gm flagellin-based assay that relies on anti-flagellar antibody in the sample to inhibit the subsequent binding of the enzyme conjugate, preventing color development in the positive sera. A flock was considered positive when it contained at least one positive serum result by this method.
Statistical analysis.
The hypothesis test for difference between proportions (two-proportion z test), Fisher's exact test, and the odds ratio (OR) were used to evaluate the differences between the proportion of positive flocks or positive sera corresponding to different groups of birds (e.g., breeders versus layers, vaccinated versus not vaccinated). An alpha value of 0.05 was used.
The prevalence of Salmonella in eggs was estimated by using a pooled prevalence estimate for a perfect test with exact confidence limits, using EpiTools software available from AusVet Animal Health Services, Australia.
Microbiological survey.
The prevalence of Salmonella in eggs was surveyed by evaluating a representative sample of eggs available commercially. Farms covering the whole country and different scales of production were randomly selected for evaluation, and based on data on their market share, 300 to 1,200 eggs were collected from each of them. A total of 12,400 eggs from 21 different poultry farms were evaluated; 15 of these farms had also been surveyed within the serological survey (Table 1).
The surface of the eggs was thoroughly cleaned with soap and water and disinfected twice with 70% ethanol. Egg contents were pooled in groups of 20 eggs per pool, placed in sterile plastic bags, and incubated for 24 h at 37°C. The day after, 10 ml per pool was added to 100 ml of peptone water and incubated for 18 h for pre-enrichment. One milliliter of each pre-enriched sample was added to 10 ml of tetrathionate broth and selenite-cystine broth, incubated for 24 h, and plated on both Brilliant Green agar and xylose lysine desoxycholate agar (Oxoid Ltd., Basingstoke, Hampshire, United Kingdom). Suspect colonies were further tested by standard metabolic and biochemical tests, and those that were identified as salmonellae were serotyped using somatic and flagellum-specific sera at the National Salmonella Center.
Sixty hens from each of the 34 poultry flocks were sampled by cloacal swab culture, and 38 environmental samples were taken, looking for Salmonella. At 16 farms, feed samples were also taken (100 g from each farm). Environmental contamination was determined by dragging swabs made moist with skim milk across the floor and other surfaces in contact with birds for 10 min (5). Cloacal samples were processed in pools of 20 swabs each (three pools per flock). Environmental and feed samples were cultivated by pre-enrichment in peptone water, enrichment in tetrathionate and selenite-cystine broth, and plating in MacConkey, Brilliant Green, and XLT4 (xylose lysine Tergitol-4) agar supplemented with novobiocin (Oxoid Ltd., Basingstoke, Hampshire, United Kingdom) (49).
Antibiotic susceptibility.
All Salmonella isolates were tested for their antibiotic susceptibility patterns by the standard disk diffusion method in Mueller-Hinton agar, and the results were interpreted in accordance with the criteria of the Clinical and Laboratory Standards Institute (8). The strains were screened for resistance to the following antibiotics (Oxoid, Basingstoke, United Kingdom): ampicillin, nalidixic acid, gentamicin, trimethoprim-sulfamethoxazole, tetracycline, and chloramphenicol. Strains showing ampicillin resistance were further tested against cephalothin, cefoxitin, cefuroxime, ceftriaxone, ceftazidime, ampicillin-sulbactam, and amoxicillin-clavulanic acid. The double-disk synergy test was used to screen for the production of extended-spectrum β-lactamases as described previously (27). Detection of inducible β-lactamases was performed by the disk approximation test, placing a 30-μg cefoxitin disk near ceftriaxone, ceftazidime, and cefuroxime disks as described previously (31). Strains showing nalidixic acid resistance were tested for resistance to ciprofloxacin. In all cases, Escherichia coli ATCC 25922 and ATCC 35218 were used as reference strains.
For quinolone-resistant strains, the presence of the qnr gene was investigated by PCR under previously described conditions (26).
Random amplified polymorphic DNA (RAPD) PCR.
All Salmonella isolates were genotyped by the RAPD PCR method using four arbitrary primers (P1254 [CCGCAGCCAA], 23L [CCGAAGCTGC], OPA-4 [AATCGGGCTG], and OPB-15 [CCAGGGTGTT]) as previously described for S. Enteritidis (3). The PCR products were electrophoresed in 2% agarose gels using 0.5× Tris-borate buffer. Only bands whose sizes fell between 200 and 2,000 bp were considered for defining amplification patterns. Patterns were considered different when they differed by more than two bands. A RAPD profile was assigned to each strain by using the combination of patterns obtained with each primer.
PCR for virulence genes.
The presence of the avrA, sopE, and spvC genes was evaluated by a multiplex PCR assay using invA-specific primers (GTGAAATTATCGCCACGTTCGGGCAA and TCATCGCACCGTCAAAGGAACC) as an internal control (32). The respective primers for spvC (CCCATAAATAGGCCTAATCT and TTACTCTGTCATCAAACGAT), sopE (CAGACCCGTGAAGCTATACT and AATTGCTGTGGAGTCGGCAT), and avrA (GTTATGGACGGAACGACATCGG and ATTCTGCTTCCCGCCGCC) were previously described (21, 36, 41). PCR was performed with 4 mM MgCl2, 0.3 μM avrA primers, 0.3 μM invA primers, 0.2 μM sopE primers, 0.6 μM spvC primers, and 5 U of Taq polymerase (Invitrogen) per 100 μl of PCR mixture. The reaction was performed using an annealing temperature of 51°C, annealing and denaturation times of 1 min each, and 35 cycles. Products were electrophoresed in 2.5% Tris-borate-EDTA agarose.
Comparative genomic hybridization (CGH) analysis.
One strain each of S. Derby and S. Enteritidis isolated from eggs was analyzed by CGH using the Salmonella generation III microarray (1, 11) and the S. Enteritidis PT4 P125109 sequenced strain (45) as references. The array is nonredundant and contains coding sequences from the following eight genomes: S. enterica serovar Typhi (S. Typhi) CT18, S. Typhi Ty2, S. Typhimurium LT2 (ATCC 700220), S. Typhimurium DT104 (NCTC 13348), S. Typhimurium SL1344 (NCTC 13347), S. Enteritidis PT4 P125109 (NCTC 13349), S. Gallinarum 287/91 (NCTC 13346), and S. bongori 12419 (ATCC 43975).
Total DNA (including plasmid DNA) was extracted from each strain using a Genome DNA extraction kit (Promega) and quantified by agarose gel electrophoresis. Each DNA sample was diluted to 0.1 μg/ml, sonicated for 10 s (level 2, Virsonic 300 sonicator), and then labeled with Cy5 (test) or Cy3 (control) by using the Bioprime kit (Gibco-BRL) according to the manufacturer's instructions. Labeled DNA from S. Enteritidis PT4 (control sample) and one of the query Salmonella strains (experimental sample) were mixed in equal volumes and concentrations. Dye swap labeling experiments were also performed with each test sample. Mixed labeled DNA was cleaned using an Amersham Autoseq G-50 column, denatured, and precipitated, and the resulting probes were hybridized to the microarray slide for 17 h at 49°C in a hybridization chamber (Genetix X2530). Washing procedures were stringent (two washes at 65°C in 2× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate]-0.1% sodium dodecyl sulfate for 30 min and two washes at 65°C in 0.1× SSC for 30 min).
Hybridization to microarray slides was detected by using a Genepix 4000B scanner (Axon Instruments, Inc.) and quantified by using Genepix Pro software (Axon Instruments, Inc.). Signal intensities were corrected by subtracting the local background values. Normalization was performed across all features on the microarray before any filtering took place. Data were median normalized, and the total list of 6,871 genes was filtered by removing those spots with a high background and genes without data in at least one of the replicates (three slides per strain, duplicate features per slide). After filtering, a list of 5,695 genes was obtained that corresponded to genes that presented a valid signal in at least one of the strains analyzed. Normalization and filtering were performed using GeneSpring microarray analysis software V7.2 (Silicon Genetics). Data analysis was performed on Excel files, following criteria previously described (4).
Calling of genes present in the PT4 genome (4,087 genes).
Spots showing low signal intensity when hybridized with PT4 DNA (i.e., the median contribution of the reference signal replicates to the total signal among the lowest 5% of the PT4 genes) were designated “uncertain.” For all of the other genes, the median of the ratios of the query strain to PT4 was registered and those with ratios higher than 0.67 were designated “present” in the query strain whereas those with ratios lower than 0.33 were designated “absent/divergent” in the query strain. Intermediate ratios were registered as “uncertain.”
Calling of genes absent from the PT4 genome (1,608 genes).
If the median contribution of all spots per gene was among the top 70% of the genes represented on the array and the query strain/PT4 signal ratio was higher than 2.5, the gene was defined as “present” in the query strain. If the median contribution was among the bottom 20% of the genes in the array, the gene was called “absent.” Spots that fell outside these categories were called “uncertain.”
For validation, we applied this method to predict present/absent genes in S. Typhi CT18 and S. Typhimurium DT104 sequenced strains and found an error rate of less than 1%.
Microarray data accession number.
Raw microarray data and grid files were submitted to ArrayExpress with accession number E-TABM-603 (http://www.ebi.ac.uk/microarray-as/ae/browse.html?keywords=E-TABM-603).
RESULTS
Serological survey.
The code names assigned to the farms, the characteristics of the birds, and the number of sera sampled per farm are indicated in Table 1. Two different screening methods, LPS ELISA and an RST, were used to assess a total of 5,751 sera as described in Materials and Methods. The results of the serological survey are presented in Tables 2 and 3 for layers and breeders, respectively, and taken together, we found that 979 (485 + 494) and 751 (524 + 227) sera tested positive as evaluated by LPS ELISA and RST, respectively. Similarly, we found that the sera of 858 layers (Table 2) and of 543 breeders (Table 3) tested positive by either of the two screening methods, which corresponds to 24.4% of the birds (1,401 out of 5,751) showing evidence of infection by at least one of these two methods (Table 4). Positive sera were found in all of the flocks sampled. All positive sera were retested by an S. Enteritidis-specific gm ELISA. As shown, 8 out of 20 flocks of layer hens (Table 2) and 4 out of 14 flocks of breeders (Table 3) had birds than tested positive. Thus, only 12 out of the 34 flocks analyzed presented at least one positive serum sample.
TABLE 2.
Serology results for layer hens
| Farm | No. of sera tested | No. positivea |
|||
|---|---|---|---|---|---|
| RST | LPS ELISA | Total | gm ELISA | ||
| A | 189 | 0 | 11 | 11 | 0 |
| B | 183 | 27 | 57 | 74 | 0 |
| C | 949 | 19 | 61 | 78 | 0 |
| D | 189 | 61 | 13 | 67 | 27 |
| E | 186 | 14 | 3 | 17 | 0 |
| F | 0 | NT | NT | NT | NT |
| G | 339 | 18 | 41 | 54 | 3 |
| H | 181 | 8 | 81 | 82 | 0 |
| I | 0 | NT | NT | NT | NT |
| J | 0 | NT | NT | NT | NT |
| K | 183 | 78 | 59 | 100 | 28 |
| L | 189 | 7 | 9 | 15 | 0 |
| M | 0 | NT | NT | NT | NT |
| N | 191 | 125 | 49 | 129 | 129 |
| Ñ | 198 | 14 | 4 | 18 | 0 |
| O | 0 | NT | NT | NT | NT |
| P | 0 | NT | NT | NT | NT |
| Q | 181 | 37 | 36 | 63 | 6 |
| R | 0 | NT | NT | NT | NT |
| S | 182 | 68 | 27 | 74 | 14 |
| T | 189 | 14 | 11 | 24 | 1 |
| 7 | 185 | 34 | 23 | 52 | 3 |
| Totalb | 3,714 | 524 | 485 | 858 | 211 |
Values indicate positive results for each test. NT, not tested (no flocks sampled for serology). One flock was tested for each farm, except for farms G and C, where two and five flocks, respectively, were sampled.
Total numbers correspond to 27 sampled flocks, including the 20 flocks sampled for serology.
TABLE 3.
Serology results for breeder hensa
| Farm | No. of sera tested | No. positive |
|||
|---|---|---|---|---|---|
| RST | LPS ELISA | Total | gm ELISA | ||
| Layer-breeders | |||||
| 14 | 153 | 0 | 54 | 54 | 0 |
| 19 | 139 | 82 | 106 | 113 | 1 |
| 20 | 151 | 0 | 4 | 4 | 0 |
| 26 | 99 | 0 | 5 | 5 | 0 |
| 27 | 99 | 6 | 30 | 33 | 0 |
| 30 | 149 | 3 | 8 | 11 | 0 |
| 31 | 151 | 0 | 63 | 63 | 0 |
| 33 | 168 | 0 | 37 | 37 | 0 |
| 34 | 157 | 0 | 7 | 7 | 0 |
| Broiler-breeders | |||||
| 2 | 150 | 10 | 0 | 10 | 0 |
| 3 | 153 | 5 | 7 | 11 | 2 |
| 4 | 158 | 6 | 0 | 6 | 0 |
| 25 | 160 | 15 | 40 | 46 | 5 |
| 32 | 150 | 100 | 133 | 143 | 141 |
| Totalb | 2,037 | 227 | 494 | 543 | 149 |
One flock was tested for each farm. Values indicate positive results for each test.
A total of 14 flocks were tested.
TABLE 4.
Estimated prevalence of infection based on different serological tests for different groups of birds
| Type of birds | No. of sera tested | No. of positive resultsa |
|
|---|---|---|---|
| Screening | gm ELISA | ||
| Laying hens | 3,714 | 858 (23.10) | 211 (5.68) |
| Layer-breeders | 1,266 | 327 (25.83) | 1 (0.08) |
| Broiler-breeders | 771 | 216 (28.02) | 148 (19.20) |
| Total | 5,751 | 1,401 (24.36) | 360 (6.26) |
Values indicate sera positive in screening tests (LPS ELISA and/or RST) or gm ELISA. Each value in parentheses is the estimated percent prevalence of positive infection.
Further analysis revealed that although the percentage of positive results obtained by either of the two screening methods did not differ between the different groups of birds (layers, layer-breeders, and broiler-breeders), the gm ELISA results were markedly different between groups (Table 4). The differences between any two groups were significant (P < 0.0001 by Fisher's exact test and P < 0.0002 by a difference-between-proportions test). The highest percentage of birds positive for anti-gm antibodies was found among broiler-breeders (19.2%), whereas layer-breeders presented the lowest estimated frequency (less than 0.1%). Laying hens had odds of being positive in the gm-ELISA that were more than 100 times those of layer-breeder hens (OR = 106.3; 95% confidence interval [CI], 14.83 to 762.0). Broiler-breeders had odds of being positive that were six times those of laying hens (OR = 6.67; 95% CI, 4.81 to 9.25).
Since previous vaccination of the flocks with any of the available vaccines (see Materials and Methods) might result in serum antibodies that could potentially interfere with the serological survey, farm owners were asked to report whether vaccination had been used on their own farms. Of 29 farms surveyed by serology, 9 had previously vaccinated their flocks. These farms comprised 13 flocks of which 9 had been vaccinated with the S. Gallinarum 9R vaccine, 3 with inactivated S. Enteritidis SEI vaccine, and 1 with both (Table 1). All birds from the nine flocks vaccinated with 9R alone tested negative by gm ELISA. Only two flocks vaccinated with SEI vaccine and one vaccinated with both vaccines included birds that tested positive by the gm ELISA. However, Salmonella was also isolated from eggs or from the environment at two of these farms. Overall, these results strongly suggest that previous vaccination did not bias the prevalence results obtained. As for the 20 farms were no vaccine had been used, they comprised 21 flocks, of which 9 had birds that tested positive by the gm ELISA. Considering all of the flocks together, nonvaccinated flocks had odds of testing positive by gm-ELISA (OR = 3.03; 95% CI, 0.6439 to 14.26) that were three times those of vaccinated flocks, although these differences lacked statistical significance. However, if the analysis was conducted considering the results of individual sera, it emerged that those animals from nonvaccinated flocks or from flocks vaccinated with SEI had odds of testing positive that were more than 30 times those of animals vaccinated with 9R vaccine (OR = 30.91; 95% CI, 12.76 to 74.87; P < 0.0001).
Bacterial cultures.
Salmonella could not be recovered from environmental samples, with the exception of a single S. Enteritidis isolate that was recovered from a broiler-breeder flock (farm 32) (Table 3) in which most of the birds (141 out of 150) tested positive by the gm ELISA. Similarly, all cloacal swab samples were negative for Salmonella.
Twelve thousand four hundred eggs from 21 different farms were assessed in pools of 20 for the presence of Salmonella within the eggs. Salmonella was recovered from 58 out of 620 pools, indicating that during the period of surveillance at least 1 of every 214 eggs (58 of 12,400) in the country was contaminated with S. enterica. This value represents an estimated prevalence of 0.0049 (95% CI, 0.0037 to 0.0063). The isolates were recovered from within eggs from 13 different poultry farms and were serotyped as S. Derby (O:1,4,[5],12:fg:[1,2]; 39 isolates), S. Gallinarum (O:1,9,12:−:−; 9 isolates), S. Enteritidis (O:1,9,12:g,m:−; 8 isolates), and S. Panama (O:1,9,12:l,v:1,5; 2 isolates) (Table 5). At 5 of the 13 farms, we isolated more than one Salmonella serovar. Thus, the prevalence of S. Enteritidis in eggs was estimated to be 0.0006 (95% CI, 0.0003 to 0.0013). Seven S. Enteritidis isolates were from eggs from farms that were also sampled for serology in this study.
TABLE 5.
Salmonella isolates from eggs and correlation with gm ELISA results
| Farm | Positive gm ELISAc | No. of egg pools analyzedb | No. positive fora: |
|||
|---|---|---|---|---|---|---|
| S. Enteritidis | S. Derby | S. Gallinarum | S. Panama | |||
| A | No | 60 | 3 | 2 | ||
| B | No | 30 | ||||
| C | No | 40 | 6 | 1 | ||
| D | Yes | 30 | 1 | |||
| E | No | 35 | 12 | |||
| F | NT | 30 | 1 | |||
| G | Yes | 30 | 1 | 5 | ||
| H | No | 35 | ||||
| I | NT | 25 | 1 | |||
| J | NT | 25 | ||||
| K | Yes | 25 | ||||
| L | No | 35 | 3 | |||
| M | NT | 20 | ||||
| N | Yes | 30 | 2 | 2 | ||
| Ñ | No | 20 | ||||
| O | No | 20 | ||||
| P | NT | 25 | 1 | |||
| Q | Yes | 20 | ||||
| R | NT | 35 | 8 | |||
| S | Yes | 30 | 4 | |||
| T | Yes | 20 | 4 | 1 | ||
| Total | 620 | 8 | 39 | 9 | 2 | |
Data indicate the number of positive pools (Salmonella isolation).
Each pool included 20 eggs.
No, all tested sera were negative. Yes, one or more tested sera were positive. NT, not tested (not sampled for serological studies).
Subtyping of strains isolated from eggs.
All S. Enteritidis isolates had the same RAPD amplification pattern for all of the primers tested (Fig. 1 A) and were susceptible to all of the antibiotics tested. All of the isolates were also positive for three virulence genes evaluated (sopE, avrA, and spvC), with the exception of a single isolate (32/02) that gave a negative PCR result for spvC (Table 6). All of the S. Derby isolates had a single RAPD profile and were susceptible to all of the antibiotics tested. PCR typing with the same three virulence genes showed that all of the S. Derby isolates were positive for avrA but negative for sopE and spvC (Table 6). Similarly, both S. Panama isolates had the same RAPD profile and were positive for avrA and negative for sopE and spvC. These two isolates were resistant to ampicillin, cephalothin, cefuroxime, and ceftazidime and susceptible to all of the other antibiotics tested.
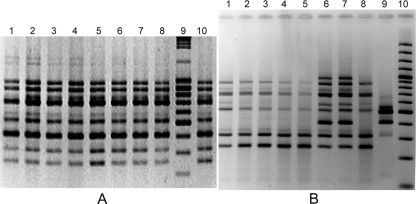
FIG. 1.
(A) S. Enteritidis RAPD patterns obtained using the OPA-4 primer. Lanes 1 to 8, S. Enteritidis egg isolates. Lane 9, 100-bp ladder. Lane10, environmental isolate. All strains correspond to RAPD profile I. (B) S. Gallinarum RAPD patterns obtained using the OPA-4 primer. Lanes 1 to 5 and 8 correspond to RAPD profile VI. Lanes 6 and 7 correspond to RAPD profile IV. Lane 9 corresponds to RAPD profile V.
TABLE 6.
Characterization of Salmonella strains isolated from eggs
| Serovar (no. of strains) and farm | No. of strains | RAPD profilea | Virulence typing (avrA sopE spvC) | ATB profileb |
|---|---|---|---|---|
| Enteritidis (8) | I | + + + | S | |
| F | 1 | I | + + + | S |
| G | 1 | I | + + + | S |
| N | 2 | I | + + + | S |
| S | 3 | I | + + + | S |
| S | 1 | I | + + − | S |
| Derby (39) | II | + − − | S | |
| A | 3 | II | + − − | S |
| C | 6 | II | + − − | S |
| E | 11 | II | + − − | S |
| L | 3 | II | + − − | S |
| N | 2 | II | + − − | S |
| P | 1 | II | + − − | S |
| R | 7 | II | + − − | S |
| T | 4 | II | + − − | S |
| N | 1 | II | + − − | S |
| R | 1 | II | + − − | S |
| Panama (2) | ||||
| T | 1 | III | + − − | R1 |
| D | 1 | III | + − − | R1 |
| Gallinarum (9) | ||||
| G | 2 | IV | + + + | S |
| G | 1 | V | + + + | R3 |
| G | 2 | VI | + + − | S |
| A | 2 | VI | + + − | R2 |
| C | 1 | VI | + + − | R2 |
| I | 1 | VI | + + − | R2 |
RAPD profiles correspond to a combination of amplification patterns obtained with four primers as described in Materials and Methods.
ATB profile, antibiotic resistance profile. S, susceptible to all drugs tested. R1, resistant to ampicillin, cephalothin, cefuroxime, and ceftazidime and susceptible to all other antibiotics tested. R2, resistant to nalidixic acid and susceptible to all other antibiotics tested. R3, intermediate susceptibility to ampicillin and susceptible to all other antibiotics tested.
Among the nine S. Gallinarum isolates, we found three antibiotic susceptibility patterns. One group (S) included four isolates that proved susceptible to all of the antibiotics tested. A second group (R2) included four isolates that were resistant to nalidixic acid and susceptible to all of the other antibiotics tested. PCR results of amplification of the qnr gene were negative in nalidixic acid-resistant isolates (results not shown). The third profile (R3) corresponded to a single strain that had intermediate susceptibility to ampicillin but showed susceptibility to all of the other antibiotics tested. RAPD analysis also allowed the discrimination of three different genetic profiles among these isolates (Fig. 1B). Profile IV comprised two susceptible isolates, profile V comprised a single isolate with intermediate susceptibility to ampicillin, and profile VI comprised six isolates, four of which were resistant and two susceptible to nalidixic acid. Three S. Gallinarum isolates were positive for avrA, sopE, and spvC, and the other six were negative for spvC (Fig. 2). All of these results are summarized in Table 6.
FIG. 2.
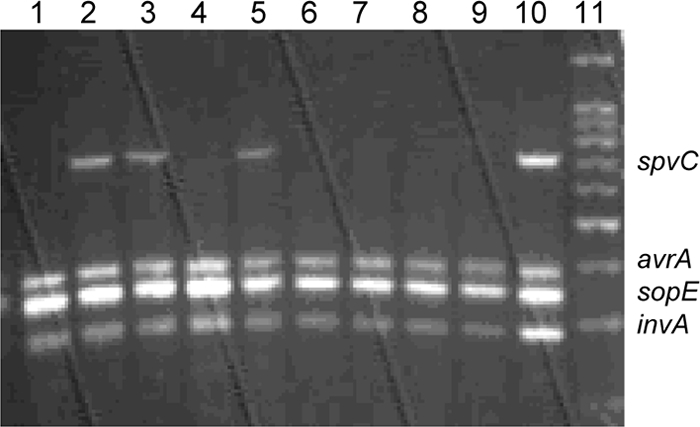
Multiplex PCR results for S. Gallinarum isolates. Lanes 1 to 5, isolates from farm G; lanes 6 and 7, isolates from farm A; lane 8, isolates from farm I; lane 9, isolates from farm C; lane 10, an S. Enteritidis isolate used as a positive control.
CGH analysis.
A particularly surprising finding was that S. Derby isolates markedly outnumbered the S. Enteritidis isolates from eggs, since during the S. Enteritidis epidemic there were no reported cases of human infection with S. Derby in Uruguay. These results could suggest that these S. Derby isolates have an impaired capacity to infect humans. To gain more knowledge about the differences between S. Enteritidis and S. Derby isolates and search for genetic differences that could be the basis of such marked differences in epidemiological behavior, we performed a microarray-based genomic comparison between one S. Enteritidis (SEN-251/01) egg isolate and one S. Derby (SDER-N11) egg isolate. The S. Enteritidis isolate selected is representative of the major genetic profile circulating in Uruguay, and it appears identical to the PT4 reference strain with regard to chromosomally located genes (4).
Of the 4,078 genes from the PT4 chromosome analyzed by CGH, 276 are absent from S. Derby (Table 7). Around 30% of these genes correspond to phage-encoded features, including S. Enteritidis phages SE10, SE12 (S. Enteritidis sopE phage), SE14, and SE20 (45). Another 20% of the absent genes correspond to surface structures, including the fimbrial sef and lpf operons, as well as various inner and outer membrane proteins, periplasmic and secreted proteins, and membrane transporters. Various genetic regions identified as related to virulence and pathogenicity were also predicted to be absent from S. Derby. These include the complete Salmonella pathogenicity island 13 (SPI-13) and SPI-14 regions, as well as part of SPI-6 (including various genes encoding Rhs family proteins and SafA lipoprotein) and part of SPI-10 that comprises the complete sef operon.
TABLE 7.
CGH results considering genes present in the PT4 chromosome that are predicted to be absent from the S. Derby strain analyzed
| SEN systematic | Presenta in: | Predicted function/annotation |
|---|---|---|
| 0030-0038c | CT18, LT2, DT104 | Sulfatases, secreted proteins |
| 0110A | NO | |
| 0139 | CT18 | Putative aldo/keto reductase |
| 0141 | CT18 | Putative LysR family transcriptional regulator |
| 0216 | NO | Putative viral enhancing factor |
| 0273-0274 | NO | Part of SPI-6, Rsh-asociated protein |
| 0281 | NO | Fimbrial subunit |
| 0308 | LT2, DT104 | Pseudogene |
| 0356 | NO | Putative autotransporter/virulence factor |
| 0392-0393 | CT18, LT2, DT104 | DeoR transcriptional regulator |
| 0641 | LT2, DT104, SL1344 | Putative inner membrane protein |
| 0800-0806c | CT18, LT2, DT104 | SPI-14 |
| 0910 | NO | Putative hypothetical protein |
| 0912 | NO | Putative Gifsy2 prophage tail fiber |
| 0916A-0921 | LT2, DT104 | Phage SE10 |
| 0988-0994c | CT18, LT2, DT104 | Secreted proteins, Na/Glu cotransporters, membrane transporters |
| 0998 | NO | Putative exported protein |
| 1000-1009 | NO | ROD-9b |
| 1013 | NO | Conserved hypothetical protein |
| 1107 | CT18 | Putative membrane protein |
| 1131-1155 | NO | Phage SE12 (SopE) |
| 1157 | NO | Hypothetical protein |
| 1158 | LT2, DT104 | Putative integrase (pseudogene) |
| 1172 | CT18, LT2, DT104 | Putative membrane protein |
| 1176 | CT18, LT2, DT104 | Transposase (pseudogene) |
| 1379-1395 | NO | Phage SE14 |
| 1423A | LT2, DT104 | Hypothetical protein |
| 1432-1436 | NO | Part of ROD-13 |
| 1485 | LT2, DT104 | FdnG |
| 1545-1546 | CT18, LT2, DT104 | RspA, YnfA |
| 1555 | CT18, LT2, DT104 | Conserved hypothetical protein |
| 1558 | CT18, LT2, DT104 | Putative ABC transporter membrane protein |
| 1751-1759 | NO | ROD-17 |
| 1922-1966 | NO | Phage SE20 |
| 1972-1987 | NO | Membrane, pilin-like, exported, and hypothetical proteins |
| 2085A-2085D | CT18 | Rfb |
| 2225-2226 | CT18, LT2 | Phage genes (pseudogenes) |
| 2378 | CT18 | Putative LPS modification acyltransferase |
| 2420 | NO | Putative exported protein |
| 2494 | CT18, DT104 | RatB, putative lipoprotein (pseudogene) |
| 2613-2614 | NO | Unknown |
| 2719 | CT18, LT2, DT104 | IagB, cell invasion protein |
| 2746A | LT2, DT104 | |
| 2781 | NO | Putative transposase |
| 2800 | LT2, DT104 | Hypothetical protein |
| 2870 | CT18, LT2, DT104 | Hypothetical protein |
| 2878-2879 | CT18, LT2, DT104 | Hipot virulence protein, endonuclease |
| 2960-2966c | LT2, DT104 | SPI-13 |
| 3112-3113 | LT2, DT104 | Inner membrane, cytoplasmic protein |
| 3167 | CT18, LT2, DT104 | Cytosine deaminase |
| 3305 | LT2, DT104 | Putative surface-exposed virulence protein (BigA) |
| 3381 | LT2, DT104 | Homology to death protein of phage P1 |
| 3459-3463c | LT2, DT104 | lpf operon |
| 3474-3476c | CT18, LT2, DT104 | Acetyltransferases |
| 3512 | CT18, LT2, DT104 | Putative lipoprotein |
| 3572-3573 | LT2, DT104 | Part of SPI-3 |
| 3577 | CT18, LT2, DT104 | Part of SPI-3 |
| 3643-3647c | LT2, DT104 | Galactonate operon |
| 3650-3651 | CT18, LT2, DT104 | LysR family transcriptional regulator |
| 3820 | CT18, LT2, DT104 | Putative lipase |
| 3862-3870c | CT18, LT2, DT104 | ABC transporters |
| 3887 | CT18, LT2, DT104 | Hypothetical protein |
| 3896-3898 | CT18 | ROD-34 |
| 3904-3910c | LT2, DT104 | PTSd system |
| 3924 | CT18, LT2, DT104 | Vitamin B12 receptor protein |
| 3978-3981 | CT18 | ROD-35 |
| 4165-4166 | CT18 | ROD-37 |
| 4199-4200 | CT18, LT2, DT104 | RelB, RelE |
| 4246-4251 | CT18 | SPI-10 sef operon |
| 4284-4294 | NO | Type I restriction modification system |
Indicates when the complete region or gene is present in the sequenced CT18, LT2, and/or DT104 strains.
ROD indicates the regions of difference between S. Enteritidis and S. Typhimurium previously described by Thomson et al. (45).
Indicates the genetic regions (including at least three contiguous genes) that are present in the sequenced strains of prevalent serovars Enteritidis and Typhimurium (LT2, DT104, DT2, and D23580).
PTS, phosphotransferase system.
The use of the pan-Salmonella array also allowed us to find a number of genes predicted to be present in S. Derby but which are absent from the S. Enteritidis PT4 P125109 chromosome. These include 152 features in the array that correspond to S. Typhi CT18, S. Typhimurium LT2, S. Typhimurium DT104, or S. bongori sequenced strains (Table 8). Among these genes, we found phage-borne genes (27%) plus genes encoding surface structures (26%). An important number of features present in S. Derby (19%) correspond to genes of unknown function.
TABLE 8.
CGH results considering genes absent from the PT4 chromosome that are predicted to be present in the S. Derby strain analyzeda
| Systematic | Presentb in: | Predicted function/annotation |
|---|---|---|
| AYGAL3309 | NO | Unknown |
| AYSL2646 | NO | Unknown |
| SBG0036-0038 | NO | Sulfatase, sulfatase regulatory, hypothetical proteins |
| SBG1292 | NO | MalR, maltose regulatory protein |
| SBG3911 | NO | Hypothetical protein |
| SDT2028-2032 | DT104 | Phage proteins |
| SDT2055 | DT104 | Unknown |
| SDT2675 | DT104 | Phage protein |
| STM0275-0278 | LT2, DT104 | Putative cytoplasmic, periplasmic proteins |
| STM0571-0577 | LT2, DT104 | PTSc system, inner membrane protein |
| STM1551 | LT2, DT104 | Putative cytoplasmic protein |
| STM1896 | LT2, DT104 | Putative cytoplasmic protein |
| STM2087-2089 | LT2, DT104 | Rfb (abequose transferase, synthetase) |
| STM2694 | LT2 | Fels-2 phage |
| STM2793 | LT2 | Fels-2 phage |
| STM3291 | LT2, DT104 | Putative cytoplasmic protein |
| STM4200-4208 | LT2, DT104 | Phage proteins |
| STM4210-4217 | LT2, DT104 | Putative cytoplasmic, inner membrane proteins |
| STM4417-4436 | LT2, DT104 | Inner membrane, DNA binding, enzymes |
| STM4488 | LT2, DT104 | Phage protein |
| STM4503 | LT2, DT104 | Putative inner membrane protein |
| STY0333 | CT18 | SPI-6, SafE |
| STY0290-0294 | CT18, LT2, DT104 | SPI-6 |
| STY0297-0298 | CT18, LT2, DT104 | SPI-6 |
| STY0302-0310 | CT18, LT2, DT104 | SPI-6 |
| STY0313-0314 | CT18 | SPI-6 |
| STY0756-0768 | CT18, LT2, DT104 | DNA recombinase, ABC transporter proteins, sugar transporter |
| STY0894 | CT18 | YliI |
| STY1413 | CT18 | Hypothetical protein |
| STY1444 | CT18, LT2, DT104 | Putative glycolate oxidase |
| STY1911 | CT18, LT2, DT104 | Hypothetical protein |
| STY2043-2044 | CT18 | Putative bacteriophage protein, putative endolysin |
| STY2349-2350 | CT18 | Putative exported proteins |
| STY2361 | CT18 | Putative exported protein |
| STY2364 | CT18 | Putative exported protein |
| STY2690 | CT18, LT2, DT104 | Hypothetical protein |
| STY3092 | CT18 | Hypothetical protein |
| STY3093 | CT18, LT2, DT104 | Hypothetical protein |
| STY3277 | CT18 | SPI-8, unknown |
| STY3291-3292 | CT18 | SPI-8, unknown |
| STY3605-3606 | CT18, LT2, DT104 | YigG, YigF |
| STY3617-3619 | CT18, LT2, DT104 | Conserved hypothetical, membrane proteins |
| STY3643-3645 | CT18 | Membrane proteins |
| STY3675 | CT18, LT2 | Phage terminase, ATPase subunit |
| STY3819 | CT18, LT2, DT104 | Possible membrane transport protein |
| STY3922 | CT18 | Probable fimbrial protein |
| STY3948-3950 | CT18 | Hypothetical proteins |
| STY4075 | CT18 | Hypothetical protein |
| STY4208 | CT18 | Putative lipoprotein |
| STY4601-4602 | CT18, LT2 | SopE phage (putative regulator of late gene expression) |
| STY4605-4607 | CT18, LT2 | SopE phage (structural genes) |
| STY4613-4617 | CT18, LT2 | SopE phage (structural genes) |
| STY4620 | CT18, LT2 | SopE phage (NucD2 putative lysozyme) |
| STY4623-4626 | CT18 | SopE phage (structural genes) |
| STY4641-4643 | CT18, LT2 | SopE phage (phage regulatory proteins) |
| STY4706 | CT18 | Conserved hypothetical protein |
| STY4825 | CT18 | Phage polarity suppression protein |
| STY4880 | CT18, LT2, DT104 | YjiW, conserved hypothetical protein |
| STY4922 | CT18 | YafM, conserved hypothetical protein |
| TY2.14 | NO | Hypothetical protein |
STY, S. Typhi CT18; STM, S. Typhimurium LT2; SDT, S. Typhimurium DT104; SBG, the strain of S. bongori that has been sequenced; TY, S. Typhi Ty2. AYSL and AYGAL, unannotated features present in the microarray from S. Typhimurium SL1344 and S. Gallinarum, respectively.
Indicates when the complete region or gene is present in the sequenced CT18, LT2, and/or DT104 strains.
PTS, phosphotransferase system.
DISCUSSION
S. Enteritidis has been reported for the last 10 to 15 years to be widely distributed in Argentina, Chile, and Brazil (6, 16, 17). Similarly, in Uruguay, S. Enteritidis started to be frequently isolated since 1995, and we report here the results of an epidemiological survey aimed at evaluating the extent of S. Enteritidis infection of poultry in the country. The survey was conducted over 2 years, and while it was taking place, an epidemic of food-borne disease was traced to poultry products containing S. Enteritidis. In the survey, serological analysis was emphasized because cloacal swabs were expected to yield false-negative results due to intermittent bacterial excretion or antibiotic use in feed or water or as a prophylactic treatment. The results presented, showing a lack of isolation of Salmonella from cloacal cultures together with a high percentage of isolation from eggs, support this assumption.
We used two different serological tests (RST and LPS ELISA) for screening that showed complementarity in the field and allowed us to efficiently detect potentially positive samples. The gm ELISA was used as a specific and confirmatory method to detect S. Enteritidis infection, inasmuch as this method would discriminate between sera of birds effectively infected with S. Enteritidis and those that were positive in the screening tests due to vaccination with the 9R strain (the most frequently used vaccine in the country) or due to infection with other serovars of Salmonella O:9. Nevertheless, given the antigenic formula of S. Derby (1,4,12:f,g) and the observed prevalence of this serovar in eggs, it could be that some of the positive results of the gm ELISA were, in fact, due to S. Derby infection. However, our results suggest that this is very unlikely, since S. Derby was isolated from six different farms that were also evaluated by serology and only two of these included flocks that yielded positive gm ELISA results. Furthermore, from one of these two farms (farm N), we also isolated S. Enteritidis. S. Enteritidis infection in layer birds was evidenced by the presence of anti-gm antibodies in sera, as well as by egg culture. S. Enteritidis was isolated from eggs from three layer farms that had also been evaluated by serology (farms G, N, and S), and flocks from these farms also gave positive results when tested with the gm ELISA. However, sera from birds at four additional laying farms also showed positive results by the gm ELISA, but eggs from these farms did not contain Salmonella (farms K and Q) or contained S. Panama (farms D and T).
Broiler-breeders revealed the highest levels of infection, as assessed by serological analysis. Furthermore, the single positive environmental sample was isolated on a broiler farm. Conversely, layer-breeders were free from serological evidence of infection, with the exception of a single positive serum in the gm ELISA. This low incidence might suggest that this population is not the origin of the infection in layers, which probably occurred through horizontal transfer and might thus eventually be controlled through cleaning and disinfection, water and feed safety assessment, and vaccination.
Vaccination with any of the available vaccines was declared for fewer than half of the flocks studied. Our results show that birds from flocks that were nonvaccinated or that had been vaccinated with SEI vaccine were more frequently infected with S. Enteritidis than birds immunized with the 9R vaccine (OR = 30.91; 95% CI, 12.76 to 74.87; P < 0.0001). Birds vaccinated only with 9R did not show evidence of S. Enteritidis infection. These results may reflect cross-protection afforded by core LPS or proteins of vaccine strain 9R, as has previously been suggested (15). An unexpected and particularly surprising finding was the coexistence of four different serovars within commercially available eggs. It is usually accepted that the high prevalence of a particular serovar occurs when it occupies the niche left by another serovar that was previously dominant (i.e., substitution of S. Enteritidis for S. Typhimurium as the main serovar in poultry) (2, 42). We have found Salmonella of different serovars even within eggs obtained from the same farm. The S. Gallinarum strains recovered were different from the 9R vaccine strain, since 9R has an electrophoretic pattern of a rough strain, whereas all of the strains recovered from eggs showed smooth electrophoretic profiles (results not shown). In addition, five out of nine S. Gallinarum isolates showed resistance traits which are not present in the vaccine strain.
We found a higher prevalence of S. Derby than S. Enteritidis in eggs. This finding is particularly intriguing because while the latter was identified as the etiological agent of the epidemic of food-borne disease, there were no reports of human infections with S. Derby in Uruguay in the same period of time. One reason for such a high prevalence could be that this serovar does not cause disease in chickens and thus it can be maintained without selection pressure in the population. On the other hand, we do not have a clear explanation for the lack of extensive human infection with S. Derby in Uruguay, but a reasonable hypothesis could be a low capacity of the S. Derby isolates to infect humans. PCR typing of virulence genes and CGH data revealed that S. Derby lacked an important number of genes previously related to virulence. Furthermore, CGH analysis showed that among the genes absent from S. Derby, there are genetic regions that are not only present in S. Enteritidis but also harbored by all of the S. Typhimurium isolates sequenced so far (strains LT2, DT104, SL1344, DT2, and DT23580). Considering that the majority of the worldwide human cases of salmonellosis are associated with either S. Enteritidis or S. Typhimurium, a detailed analysis of the genetic regions of difference between these prevalent serovars and serovar Derby might give clues about the basis of the ability of a particular serovar to cause human disease. Our analysis shows that nine of these regions of difference encompass at least three contiguous genes (Table 7). Some of those regions have a clear association with pathogenicity traits, including SPI-13, SPI-14, and the fimbrial lpf operon. BLAST analysis shows that these three regions are also present in serovars Newport, Dublin, Heidelberg, Paratyphi, Choleraesuis, Hadar, and Infantis. SPI-13 and SPI-14 were recently described as important in the pathogenesis of S. Gallinarum (44).
Other genetic regions absent from S. Derby but present in all of the sequenced S. Enteritidis and S. Typhimurium strains include a region associated with sulfatases (SEN0030-0038); the galactonate operon (SEN3643-3647); a region of several genes encoding ATP binding cassette (ABC) transporters, sugar kinases, and regulatory proteins (SEN3862-3870); and a region encoding various enzymes of the phosphotransferase system (SEN3904-3910). Sulfur is an essential element for bacterial growth and survival, and sulfatase genes are expressed under conditions of sulfur starvation, where the enzymes function in sulfate scavenging (28). Several authors have hypothesized that sulfatases may be involved in the host-pathogen interaction (10, 23, 30, 34). The role of sulfatases in the pathogenesis of Salmonella infections remains to be studied, but the presence of this genetic region may facilitate the survival of the pathogen in the human host, providing sulfur and carbon sources in tissues where free sulfur is limited.
Recently, it was reported that the monophasic serovar 4,12:d,−, which is highly adapted to poultry and is very rarely associated with human disease, lacks the galactonate and lpf operons (25). These genetic regions are present in S. Typhimurium and S. Enteritidis, which are serovars frequently associated with food-borne disease. Our results show that S. Derby lacks these operons, suggesting that the function of these genes could be associated with the ability to cause human disease.
ABC transporters are integral membrane proteins universally distributed among living organisms with functions in many different aspects of bacterial physiology. They are especially important in the import of essential nutrients and the export of toxic molecules across cellular membranes (12). The SEN3862-3870 region has homologues in all of the S. enterica subsp. enterica strains sequenced to date but is not present in the strains of S. bongori and S. enterica subsp. arizonae that have been sequenced. These observations suggest that ABC transporters might be involved in the ability to infect mammalian hosts.
The lack of the region encoding phosphotransferase system enzymes in serovar Derby is consistent with the hypothesis that these strains are attenuated for virulence in humans. It has been reported previously that some of these enzymes play an important role in the virulence of Salmonella and other pathogens (29) and that this system participates in the regulation of the expression of different virulence factors in Salmonella (39).
Our results strongly suggest that the strains of S. Derby circulating in Uruguay are impaired in the ability to cause human infection. If these strains acquire virulence determinants by lateral gene transfer, S. Derby may emerge as a zoonotic problem. This serovar has recently been recognized as a cause of human food-borne infection in Brazil (19), China (50), and Taiwan (7), being associated with multidrug resistance by the acquisition of Salmonella genomic island 1. S. Derby was isolated from human cases of gastroenteritis and invasive disease in Uruguay before 1991, and during 2008, a number of isolates were obtained from human disease. Further characterization of these isolates is currently ongoing.
Overall, our results show extensive flock contamination with S. enterica, to a higher degree than expected, with different serovars coexisting within the poultry population. In contrast, data derived from sampling of isolates from the national epidemic of human gastroenteritis infection show that most cases of human infection were due to a single S. Enteritidis serovar, suggesting major differences in the capacities of different serovars to infect human populations.
Acknowledgments
This work was jointly supported by INIA (National Institute for Agricultural Research) of Uruguay and by the Central Research Committee (CSIC) of the Universidad de la República Uruguay. Parts of this work were also supported by a project grant from the Wellcome Trust (078168/Z/05/Z).
Thanks to Elba Hernández and Alicia Rigoli for technical assistance and to Raquel Demarco for helpful suggestions for data analysis. Thanks to Robert Davies, Central Veterinary Laboratories, Weybridge, United Kingdom, for providing control sera.
Footnotes
Published ahead of print on 19 May 2010.
REFERENCES
- 1.Anjum, M. F., C. Marooney, M. Fookes, S. Baker, G. Dougan, A. Ivens, and M. J. Woodward. 2005. Identification of core and variable components of the Salmonella enterica subspecies I genome by microarray. Infect. Immun. 73:7894-7905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bäumler, A. J., B. M. Hargis, and R. M. Tsolis. 2000. Tracing the origins of Salmonella outbreaks. Science 287:50-52. [DOI] [PubMed] [Google Scholar]
- 3.Betancor, L., F. Schelotto, A. Martinez, M. Pereira, G. Algorta, M. A. Rodriguez, R. Vignoli, and J. A. Chabalgoity. 2004. Random amplified polymorphic DNA and phenotyping analysis of Salmonella enterica serovar Enteritidis isolates collected from humans and poultry in Uruguay from 1995 to 2002. J. Clin. Microbiol. 42:1155-1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Betancor, L., L. Yim, M. Fookes, A. Martinez, N. R. Thomson, A. Ivens, S. Peters, C. Bryant, G. Algorta, S. Kariuki, F. Schelotto, D. Maskell, G. Dougan, and J. A. Chabalgoity. 2009. Genomic and phenotypic variation in epidemic-spanning Salmonella enterica serovar Enteritidis isolates. BMC Microbiol. 9:237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Byrd, J. A., D. E. Corrier, J. R. DeLoach, and D. J. Nisbet. 1997. Comparison of drag-swab environmental protocols for the isolation of Salmonella in poultry houses. Avian Dis. 41:709-713. [PubMed] [Google Scholar]
- 6.Caffer, M. I., and T. Eiguer. 1994. Salmonella Enteritidis in Argentina. Int. J. Food Microbiol. 21:15-19. [DOI] [PubMed] [Google Scholar]
- 7.Chiu, C. H., H. L. Chen, L. S. Kao, C. Y. Yang, C. Chu, B. Doublet, K. Praud, and A. Cloeckaert. 2007. Variant Salmonella genomic island 1 antibiotic resistance gene clusters in Salmonella enterica serovar Derby isolates from humans in Taiwan. J. Antimicrob. Chemother. 59:325-326. [DOI] [PubMed] [Google Scholar]
- 8.Clinical and Laboratory Standards Institute. 2006. Performance standards for antimicrobial susceptibility testing; sixteenth informational supplement. M100-S16. Clinical and Laboratory Standards Institute, Wayne, PA.
- 9.Cogan, T. A., and T. J. Humphrey. 2003. The rise and fall of Salmonella Enteritidis in the UK. J. Appl. Microbiol. 94(Suppl.):114S-119S. [DOI] [PubMed] [Google Scholar]
- 10.Cohen, A. L., J. D. Oliver, A. DePaola, E. J. Feil, and E. F. Boyd. 2007. Emergence of a virulent clade of Vibrio vulnificus and correlation with the presence of a 33-kilobase genomic island. Appl. Environ. Microbiol. 73:5553-5565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cooke, F. J., J. Wain, M. Fookes, A. Ivens, N. Thomson, D. J. Brown, E. J. Threlfall, G. Gunn, G. Foster, and G. Dougan. 2007. Prophage sequences defining hot spots of genome variation in Salmonella enterica serovar Typhimurium can be used to discriminate between field isolates. J. Clin. Microbiol. 45:2590-2598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Davidson, A. L., E. Dassa, C. Orelle, and J. Chen. 2008. Structure, function, and evolution of bacterial ATP-binding cassette systems. Microbiol. Mol. Biol. Rev. 72:317-364, table of contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.De Buck, J., F. Van Immerseel, F. Haesebrouck, and R. Ducatelle. 2004. Colonization of the chicken reproductive tract and egg contamination by Salmonella. J. Appl. Microbiol. 97:233-245. [DOI] [PubMed] [Google Scholar]
- 14.de Jong, B., and K. Ekdahl. 2006. The comparative burden of salmonellosis in the European Union member states, associated and candidate countries. BMC Public Health 6:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Feberwee, A., T. S. de Vries, E. G. Hartman, J. J. de Wit, A. R. Elbers, and W. A. de Jong. 2001. Vaccination against Salmonella Enteritidis in Dutch commercial layer flocks with a vaccine based on a live Salmonella Gallinarum 9R strain: evaluation of efficacy, safety, and performance of serologic Salmonella tests. Avian Dis. 45:83-91. [PubMed] [Google Scholar]
- 16.Fernandes, S. A., A. C. Ghilardi, A. T. Tavechio, A. M. Machado, and A. C. Pignatari. 2003. Phenotypic and molecular characterization of Salmonella Enteritidis strains isolated in Sao Paulo, Brazil. Rev. Inst. Med. Trop. Sao Paulo. 45:59-63. [DOI] [PubMed] [Google Scholar]
- 17.Fernandez, J., A. Fica, G. Ebensperger, H. Calfullan, S. Prat, A. Fernandez, M. Alexandre, and I. Heitmann. 2003. Analysis of molecular epidemiology of Chilean Salmonella enterica serotype Enteritidis isolates by pulsed-field gel electrophoresis and bacteriophage typing. J. Clin. Microbiol. 41:1617-1622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Galanis, E., D. M. Lo Fo Wong, M. E. Patrick, N. Binsztein, A. Cieslik, T. Chalermchikit, A. Aidara-Kane, A. Ellis, F. J. Angulo, and H. C. Wegener. 2006. Web-based surveillance and global Salmonella distribution, 2000-2002. Emerg. Infect. Dis. 12:381-388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Geimba, M. P., E. C. Tondo, F. A. de Oliveira, C. W. Canal, and A. Brandelli. 2004. Serological characterization and prevalence of spvR genes in Salmonella isolated from foods involved in outbreaks in Brazil. J. Food Prot. 67:1229-1233. [DOI] [PubMed] [Google Scholar]
- 20.Hald, T., D. Vose, H. C. Wegener, and T. Koupeev. 2004. A Bayesian approach to quantify the contribution of animal-food sources to human salmonellosis. Risk Anal. 24:255-269. [DOI] [PubMed] [Google Scholar]
- 21.Haneda, T., N. Okada, N. Nakazawa, T. Kawakami, and H. Danbara. 2001. Complete DNA sequence and comparative analysis of the 50-kilobase virulence plasmid of Salmonella enterica serovar Choleraesuis. Infect. Immun. 69:2612-2620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Herikstad, H., Y. Motarjemi, and R. V. Tauxe. 2002. Salmonella surveillance: a global survey of public health serotyping. Epidemiol. Infect. 129:1-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hoffman, J. A., J. L. Badger, Y. Zhang, S. H. Huang, and K. S. Kim. 2000. Escherichia coli K1 aslA contributes to invasion of brain microvascular endothelial cells in vitro and in vivo. Infect. Immun. 68:5062-5067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hormaeche, C., R. De Marco, F. Schelotto, C. Alia de Montero, C. Rivas, D. Mutti, and N. Bello. 1977. Frecuencia de serotipos identificados en el Centro de Salmonelas de Montevideo. Rev. Urug. Patol. Clin. Microbiol. 15:43-47. [Google Scholar]
- 25.Huehn, S., C. Bunge, E. Junker, R. Helmuth, and B. Malorny. 2009. Poultry-associated Salmonella enterica subsp. enterica serovar 4,12:d:− reveals high clonality and a distinct pathogenicity gene repertoire. Appl. Environ. Microbiol. 75:1011-1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jacoby, G. A., N. Chow, and K. B. Waites. 2003. Prevalence of plasmid-mediated quinolone resistance. Antimicrob. Agents Chemother. 47:559-562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jarlier, V., M. H. Nicolas, G. Fournier, and A. Philippon. 1988. Extended broad-spectrum beta-lactamases conferring transferable resistance to newer beta-lactam agents in Enterobacteriaceae: hospital prevalence and susceptibility patterns. Rev. Infect. Dis. 10:867-878. [DOI] [PubMed] [Google Scholar]
- 28.Kertesz, M. A. 2000. Riding the sulfur cycle—metabolism of sulfonates and sulfate esters in gram-negative bacteria. FEMS Microbiol. Rev. 24:135-175. [DOI] [PubMed] [Google Scholar]
- 29.Kok, M., G. Bron, B. Erni, and S. Mukhija. 2003. Effect of enzyme I of the bacterial phosphoenolpyruvate: sugar phosphotransferase system (PTS) on virulence in a murine model. Microbiology 149:2645-2652. [DOI] [PubMed] [Google Scholar]
- 30.Lestrate, P., R. M. Delrue, I. Danese, C. Didembourg, B. Taminiau, P. Mertens, X. De Bolle, A. Tibor, C. M. Tang, and J. J. Letesson. 2000. Identification and characterization of in vivo attenuated mutants of Brucella melitensis. Mol. Microbiol. 38:543-551. [DOI] [PubMed] [Google Scholar]
- 31.Livermore, D. M. 1995. Beta-lactamases in laboratory and clinical resistance. Clin. Microbiol. Rev. 8:557-584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Malorny, B., J. Hoorfar, C. Bunge, and R. Helmuth. 2003. Multicenter validation of the analytical accuracy of Salmonella PCR: towards an international standard. Appl. Environ. Microbiol. 69:290-296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mishu, B., J. Koehler, L. A. Lee, D. Rodrigue, F. H. Brenner, P. Blake, and R. V. Tauxe. 1994. Outbreaks of Salmonella Enteritidis infections in the United States, 1985-1991. J. Infect. Dis. 169:547-552. [DOI] [PubMed] [Google Scholar]
- 34.Mougous, J. D., R. E. Green, S. J. Williams, S. E. Brenner, and C. R. Bertozzi. 2002. Sulfotransferases and sulfatases in mycobacteria. Chem. Biol. 9:767-776. [DOI] [PubMed] [Google Scholar]
- 35.Nicholas, R. A., and G. A. Cullen. 1991. Development and application of an ELISA for detecting antibodies to Salmonella Enteritidis in chicken flocks. Vet. Rec. 128:74-76. [DOI] [PubMed] [Google Scholar]
- 36.Pasmans, F., F. Van Immerseel, M. Heyndrickx, A. Martel, C. Godard, C. Wildemauwe, R. Ducatelle, and F. Haesebrouck. 2003. Host adaptation of pigeon isolates of Salmonella enterica subsp. enterica serovar Typhimurium variant Copenhagen phage type 99 is associated with enhanced macrophage cytotoxicity. Infect. Immun. 71:6068-6074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Peluffo, C., C. Hormaeche, R. De Marco, F. Schelotto, C. Alia de Montero, C. Rivas, D. Mutti, N. Bello, and M. Coubria. 1971. Frecuencia de tipos serológicos clasificados en el Centro Nacional de Salmonelas. Revista Uruguaya de Patología Clínica y Microbiología. 9:143-150. [Google Scholar]
- 38.Perales, I., and A. Audicana. 1988. Salmonella Enteritidis and eggs. Lancet 2(8620):1133. [DOI] [PubMed] [Google Scholar]
- 39.Poncet, S., E. Milohanic, A. Maze, J. N. Abdallah, F. Ake, M. Larribe, A. E. Deghmane, M. K. Taha, M. Dozot, X. De Bolle, J. J. Letesson, and J. Deutscher. 2009. Correlations between carbon metabolism and virulence in bacteria. Contrib. Microbiol. 16:88-102. [DOI] [PubMed] [Google Scholar]
- 40.Poppe, C. 1999. Epidemiology of Salmonella enterica serovar Enteritidis, p. 3-18. In A. M. Saed, R. K. Gast, M. E. Potter, and P. G. Wall (ed.), Salmonella enterica serovar Enteritidis in human and animals. Iowa State University Press, Ames.
- 41.Prager, R., W. Rabsch, W. Streckel, W. Voigt, E. Tietze, and H. Tschape. 2003. Molecular properties of Salmonella enterica serotype Paratyphi B distinguish between its systemic and its enteric pathovars. J. Clin. Microbiol. 41:4270-4278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rabsch, W., B. M. Hargis, R. M. Tsolis, R. A. Kingsley, K. H. Hinz, H. Tschape, and A. J. Bäumler. 2000. Competitive exclusion of Salmonella Enteritidis by Salmonella gallinarum in poultry. Emerg. Infect. Dis. 6:443-448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schroeder, C. M., A. L. Naugle, W. D. Schlosser, A. T. Hogue, F. J. Angulo, J. S. Rose, E. D. Ebel, W. T. Disney, K. G. Holt, and D. P. Goldman. 2005. Estimate of illnesses from Salmonella Enteritidis in eggs, United States, 2000. Emerg. Infect. Dis. 11:113-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shah, D. H., M. J. Lee, J. H. Park, J. H. Lee, S. K. Eo, J. T. Kwon, and J. S. Chae. 2005. Identification of Salmonella Gallinarum virulence genes in a chicken infection model using PCR-based signature-tagged mutagenesis. Microbiology 151:3957-3968. [DOI] [PubMed] [Google Scholar]
- 45.Thomson, N. R., D. J. Clayton, D. Windhorst, G. Vernikos, S. Davidson, C. Churcher, M. A. Quail, M. Stevens, M. A. Jones, M. Watson, A. Barron, A. Layton, D. Pickard, R. A. Kingsley, A. Bignell, L. Clark, B. Harris, D. Ormond, Z. Abdellah, K. Brooks, I. Cherevach, T. Chillingworth, J. Woodward, H. Norberczak, A. Lord, C. Arrowsmith, K. Jagels, S. Moule, K. Mungall, M. Sanders, S. Whitehead, J. A. Chabalgoity, D. Maskell, T. Humphrey, M. Roberts, P. A. Barrow, G. Dougan, and J. Parkhill. 2008. Comparative genome analysis of Salmonella Enteritidis PT4 and Salmonella Gallinarum 287/91 provides insights into evolutionary and host adaptation pathways. Genome Res. 18:1624-1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Voetsch, A. C., T. J. Van Gilder, F. J. Angulo, M. M. Farley, S. Shallow, R. Marcus, P. R. Cieslak, V. C. Deneen, and R. V. Tauxe. 2004. FoodNet estimate of the burden of illness caused by nontyphoidal Salmonella infections in the United States. Clin. Infect. Dis. 38(Suppl. 3):S127-S134. [DOI] [PubMed] [Google Scholar]
- 47.Ward, L. R., J. Threlfall, H. R. Smith, and S. J. O'Brien. 2000. Salmonella Enteritidis epidemic. Science 287:1753-1756. [DOI] [PubMed] [Google Scholar]
- 48.Wray, C. 1996. OIE manual of standards for diagnostic tests and vaccines, p. 642-650. Office International des Epizooties, Paris, France.
- 49.Wray, C., and R. Davies. 1994. Guidelines on detection and monitoring of Salmonella infected poultry flocks with particular reference to Salmonella Enteritidis. Veterinary Public Health Unit, World Health Organization, Graz, Austria.
- 50.Xia, S., R. S. Hendriksen, Z. Xie, L. Huang, J. Zhang, W. Guo, B. Xu, L. Ran, and F. M. Aarestrup. 2009. Molecular characterization and antimicrobial susceptibility of Salmonella isolates from infections in humans in Henan Province, China. J. Clin. Microbiol. 47:401-409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang-Barber, L., A. K. Turner, and P. A. Barrow. 1999. Vaccination for control of Salmonella in poultry. Vaccine 17:2538-2545. [DOI] [PubMed] [Google Scholar]