Abstract
We evaluated the ability of a commercial microarray system (Check KPC/ESBL; Check-Points Health BV) to detect clinically important class A β-lactamase genes. A total of 106 Gram-negative strains were tested. The following sensitivity and specificity results were recorded, respectively: for blaSHV, 98.8% and 100%; for blaTEM, 100% and 96.4%; and for blaCTX-M and blaKPC, 100% and 100%.
The spread of class A or group 2be extended-spectrum β-lactamases (ESBLs) represents an emerging public-health concern (1, 17). Among the organisms of the Enterobacteriaceae family (e.g., Klebsiella pneumoniae and Escherichia coli), the most frequently detected and clinically important ESBLs belong to the TEM, SHV, and CTX-M families (17). While TEM- and SHV-type ESBLs arise via substitutions in strategically positioned amino acids (e.g., Gly238 and Arg164) from the natural narrow-spectrum TEM-1, TEM-2, or SHV-1 β-lactamase genes, all currently identified CTX-M enzymes demonstrate an ESBL phenotype (7, 14).
The ability to rapidly identify narrow-spectrum β-lactamases (e.g., SHV-11 and TEM-1) and ESBLs (e.g., SHV-5 and SHV-12 or TEM-10) has important clinical implications. Usually, Enterobacteriaceae species producing narrow-spectrum enzymes are resistant to penicillins and narrow-spectrum cephalosporins, whereas those producing ESBLs manifest resistance to extended-spectrum oxyimino-cephalosporins and aztreonam (14). Since resistance to quinolones and aminoglycosides is frequently observed among ESBL producers, carbapenems represent one of the therapeutic options of last resort for life-threatening infections due to these organisms (6, 14).
In some geographic areas, the spread of carbapenemases belonging to class A (e.g., KPCs), class B (e.g., VIMs and IMPs), and class D (e.g., OXA-48) has significantly compromised the clinical use of carbapenems, consigning clinicians to the use of “last-line” antimicrobials such as colistin (2, 19). In particular, the KPC β-lactamases (primarily KPC-2 and KPC-3) are the serine carbapenemases that are most widespread in the United States, and strains producing these enzymes are responsible for numerous outbreaks with high mortality rates (3, 9, 13). Although nine KPC-type β-lactamases have been described, their susceptibility profiles are similar, rendering the differentiation of these variants less clinically relevant (13, 22).
Prompt and appropriate antibiotic treatment for infections due to ESBLs- and/or KPC-producing Enterobacteriaceae may positively affect the final outcome for infected patients (6, 13). Unfortunately, standard and confirmatory phenotypic tests may fail to identify ESBL- and, more frequently, KPC-producing organisms. For the latter group, the use of the modified Hodge test delays the final report by an additional 24 h (11, 12, 21). Therefore, a rapid and reliable method is needed to perform a quick and accurate analysis of the most important bla genes possessed by clinical isolates.
Microarray technologies are promising genotyping systems that possess a high multiplexing capacity and can be used for detecting different β-lactamase genes that are present in a single strain (8, 10, 23). This ability can assist clinicians in directing antimicrobial therapy. In the present work, we evaluated the ability of Check KPC/ESBL (Check-Points Health BV, Wageningen, Netherlands), the first rapid, commercially available, microarray-based diagnostic test system for detection and identification of bla genes belonging to the TEM, SHV, CTX-M, and KPC types. This system can detect single nucleotide polymorphisms found in the most important TEM- and SHV-type ESBLs (www.lahey.org/studies), including single mutations corresponding to amino acid positions Val84Ile, Glu104Lys, Arg164Ser/His/Cys, and Gly238Ser in TEMs and Gly238Ser/Ala and/or Glu240Lys in SHVs (7).
A total of 102 Enterobacteriaceae and four Acinetobacter baumannii isolates possessing different bla genes were tested (Table 1; see also Table S1 in the supplemental material). The majority of strains (n = 61) had previously been characterized (3-5, 15), whereas the bla genes of the remaining isolates were characterized by PCR amplification, standard DNA sequencing, and analytical isoelectric focusing (aIEF) as previously described (4). In this collection, isolates possessed an average of three different bla genes (range, one to five; see Table S1 in the supplemental material). The collection also included K. pneumoniae ATCC 700603, which produces the SHV-18 ESBL (20), and six E. coli DH10B control strains in which single bla genes are carried in different plasmid vectors (see Table S1 in the supplemental material).
TABLE 1.
Performance of the Check KPC/ESBL microarray assay in identification of β-lactamase genesa
| Species |
blaKPC |
blaSHV |
blaTEM |
blaCTX-M |
Isolates with all bla genes correctly reported |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| No. of isolates positive by PCR/Seq | Agreementi (%) | ESBLd |
Non-ESBL |
Total |
ESBLd |
Non-ESBL |
Total |
No. of isolates positive by PCR/Seq | Agreementi (%) | Agreementi (%) | |||||||
| No. of isolates positive by PCR/Seq | Agreementi (%) | No. of isolates positive by PCR/Seqc | Agreementi (%) | No. of isolates positive by PCR/Seq | Agreementi (%) | No. of isolates positive by PCR/Seq | Agreementi (%) | No. of isolates positive by PCR/Seqe | Agreementi (%) | No. of isolates positive by PCR/Seq | Agreementi (%) | ||||||
| K. pneumoniae (n = 79)b | 52 | 79/79 (100) | 41 | 41/41 (100) | 38 | 31/38 (81.6) | 79 | 72/79 (91.1) | 3 | 3/3 (100) | 64 | 64/64 (100) | 67 | 79/79 (100) | 8 | 79/79 (100) | 72/79 (91.1) |
| E. coli (n = 22)b | 4 | 22/22 (100) | 3 | 2/3 (66.7) | 2 | 2/2 (100) | 5 | 21/22 (95.5) | 2 | 2/2 (100) | 7 | 7/7 (100) | 9 | 21/22 (95.5) | 9 | 22/22 (100) | 20/22 (90.9) |
| A. baumannii (n = 4) | 0 | 4/4 (100) | 0 | 0 | 0 | 4/4 (100) | 2 | 2/2 (100) | 2 | 4/4 (100) | 2 | 4/4 (100) | 4/4 (100) | ||||
| Enterobacter cloacae (n = 1) | 1 | 1/1 (100) | 0 | 0 | 0 | 1/1 (100) | 0 | 1 | 1/1 (100) | 1 | 1/1 (100) | 0 | 1/1 (100) | 1/1 (100) | |||
| Total (n = 106) | 57 | 106/106 (100)f | 44 | 43/44 (97.7) | 40 | 33/40 (82.5) | 84 | 98/106 (92.5)g | 7 | 7/7 (100) | 72 | 72/72 (100) | 79 | 105/106 (99.1)h | 19 | 106/106 (100)f | 97/106 (91.5) |
Results represent comparisons of the Check KPC/ESBL microarray assay to PCR amplification followed by DNA sequencing (PCR/Seq).
Includes control isolates.
One E. coli isolate containing blaSHV-38 was reported as a non-ESBL SHV type; six K. pneumoniae isolates containing a blaSHV-non-ESBL gene (i.e., a blaSHV-1 or blaSHV-11 gene) were reported as representing SHV-ESBL strains; one K. pneumoniae isolate containing blaSHV-11 was reported as SHV negative (see Table S1 in the supplemental material).
Includes isolates containing blaESBL (e.g., blaSHV-12) or blaESBL plus blanon-ESBL genes (e.g., blaSHV-12 and blaSHV-11).
One blaTEM-negative E. coli isolate was reported as a non-ESBL TEM strain.
Sensitivity, 100%; specificity, 100%; positive predictive value, 100%; negative predictive value, 100%.
Sensitivity, 98.8%; specificity, 100%; positive predictive value, 100%; negative predictive value, 95.7%.
Sensitivity, 100%; specificity, 96.4%; positive predictive value, 98.8%; negative predictive value, 100%.
The value preceding the slash is the number of isolates whose Check KPC/ESBL microarray results agreed with their PCR/Seq results, and the value following the slash is the total number of isolates tested by both assays.
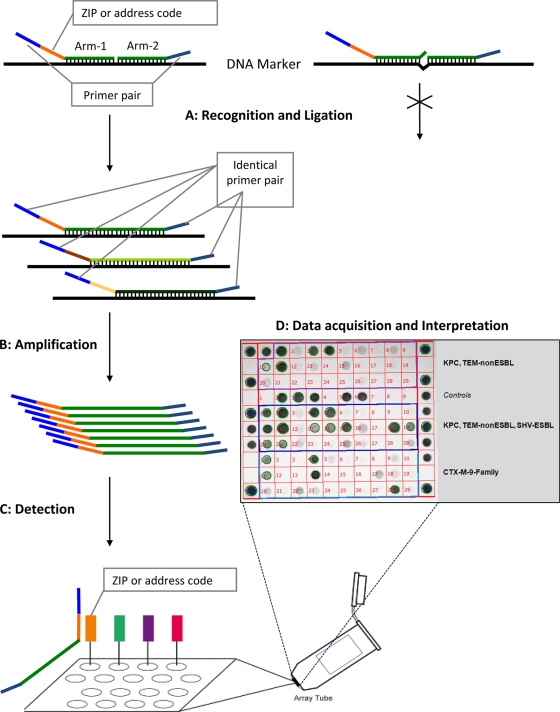
Genomic DNA of strains was extracted from overnight colonies grown on blood agar (BBL, Sparks, MD) by the use of a DNeasy blood and tissue kit (Qiagen Sciences, Germantown, MD). Microarray assays were performed according to the instructions of the manufacturer (Check-Points Health BV). Briefly, templates of the target bla DNA sequences are generated during the ligation step. These templates are then amplified, and the products are hybridized in specific array tubes. Tubes are then inserted in the array tube reader upon completion of the detection reaction, and images are acquired and interpreted with software supplied by the manufacturer (Fig. 1). For 50 isolates, the complete procedure (i.e., from genomic DNA extraction to results) can be performed in approximately 8 h.
FIG. 1.
Schematic flowchart representing the different steps used by the Check-Points KPC/ESBL platform to recognize specific bla genes. (Step A) Target DNA recognition and ligation (thermocycling conditions, 95°C for 3 min, 24 cycles of 95°C for 30 s and 65°C for 5 min, and 98°C for 2 min [total, 2.75 h]). Each target-specific probe consists of two oligonucleotide probe arms that are used to detect single nucleotide polymorphisms (SNPs). These two probe arms are connected by the ligase, generating a single probe molecule only when they perfectly match the target sequence. Only connected probe arms produce the labeled amplification products detected in step C. Every target-specific probe is equipped with the same consensus primer pair necessary for step B and a unique “ZIP code” necessary for step C. (Step B) PCR amplification of the target DNA sequences (thermocycling conditions, 95°C for 10 min, 35 cycles of 95°C for 5 s, 55°C for 30 s, and 72°C for 30 s, and 98°C for 2 min [total, 1.5 h]). Using a common primer pair, target ligated sequence templates labeled with specific “ZIP codes” are multiplied. (Step C) Detection (requiring approximately 1 to 2 h of processing, depending on the number of samples). Amplification products are targeted to specific addresses on the microarray. This targeting is dependent on the specific “ZIP code.” (Step D) Immediate acquisition of images by scanning of the microarray using the array tube reader and immediate interpretation of the acquired pictures by the use of dedicated software.
Overall, the Check KPC/ESBL system correctly identified representatives of the four bla gene families tested, including differentiation between non-ESBL and ESBL genes, in 97 of 106 isolates (91.5%). Specificities of 100% were recorded for the blaKPC, blaSHV, and blaCTX-M genes, whereas one false positive was reported for blaTEM genes (specificity of 96.4%). The system detected all blaKPC-, blaTEM-, and blaCTX-M-possessing isolates, including differentiation of ESBL from non-ESBL blaTEM-containing strains (Table 1). Notably, all blaCTX-M genes detected were classified into the appropriate family group (i.e., group I, CTX-M-1-like; group II, CTX-M-2-like; group III, CTX-M-8-like; group IV, CTX-M-9-like; group V, CTX-M-25/CTX-M-26) according to the classification method of Pitout et al. (16) (see Table S1 in the supplemental material).
Detection and recognition of the blaSHV genes showed 92.5% agreement, with sensitivity and specificity of 98.8% and 100%, respectively (Table 1). Only 1 in 44 blaESBL-positive strains (i.e., blaSHV-38-positive strains) was not identified (97.7% agreement). SHV-38 is a very rare chromosomal ESBL enzyme (group 2be) that was found in a single clinical isolate. It possesses a unique amino acid substitution (i.e., Ala146Val) and is capable of conferring resistance to ceftazidime and imipenem (18). The amino acid at position 146 is not included in those analyzed by the Check KPC/ESBL system.
Six strains with non-ESBL blaSHV genes were misclassified as ESBLs (Table 2). Notably, three of these were blaSHV-11-positive K. pneumoniae isolates (non-ESBL), which showed β-lactamase bands at pIs of 7.6 and 8.2 by aIEF and double spikes at positions 238 and/or 240 in the DNA sequencing traces of the blaSHV gene. This pattern is consistent with the possible production of an SHV-ESBL (along with the non-ESBL SHV-11) that was not detected with a cloning and DNA sequencing method that we previously employed (4). Therefore, blaSHV-positive total agreement and the overall agreement (i.e., all bla genes correctly reported) would improve by 2.8% if these three strains were classified as ESBL producers (Table 1).
TABLE 2.
Details of discrepancies between conventional PCR plus DNA sequencing results and Check KPC/ESBL genotyping results
| Strain | Source or reference | Gene identified by PCR and DNA sequencing | Gene identified by Check KPC/ESBL genotyping | Comment(s) |
|---|---|---|---|---|
| E. coli DH10B | This study | blaSHV-38 | SHV-non-ESBL | Rare chromosomal genotype found in a single K. pneumoniae isolate (18); mutation conferring ESBL phenotype not assayed with current microarray primers |
| K. pneumoniae VA-361a | 4 | blaKPC-2, blaTEM-1, blaSHV-11 | KPC, TEM-non-ESBL, SHV-ESBL | Possible production of an SHV-ESBL (e.g., SHV-5/SHV-12) |
| K. pneumoniae VA-388 | 4 | blaKPC-3, blaTEM-1, blaSHV-11 | KPC, TEM-non-ESBL | SHV genes were not detected |
| K. pneumoniae VA-392a | 4 | blaKPC-3, blaTEM-1, blaSHV-11 | KPC, TEM-non-ESBL, SHV-ESBL | Possible production of an SHV-ESBL (e.g., SHV-5/SHV-12) |
| K. pneumoniae VA-412a | 4 | blaKPC-2, blaTEM-1, blaSHV-11 | KPC, TEM-non-ESBL, SHV-ESBL | Possible production of an SHV-ESBL (e.g., SHV-5/SHV-12) |
| K. pneumoniae VA-414 | 4 | blaKPC-3, blaTEM-1, blaSHV-11 | KPC, TEM-non-ESBL, SHV-ESBL | SHV-11 is not an ESBL |
| K. pneumoniae 111b | 15 | blaTEM-10-like, blaSHV-1-like | TEM-ESBL, SHV-ESBL | SHV-1 is not an ESBL |
| K. pneumoniae 438b | 15 | blaTEM-2-like, blaSHV-1-like | TEM-non-ESBL, SHV-ESBL | SHV-1 is not an ESBL |
| E. coli 25 | This study | blaCTX-M-9-like | CTX-M-(IV),c TEM-non-ESBL | blaTEM genes were not detected by PCR analysis (including using internal primers); aIEF showed only one β-lactamase band at a pI of 6.7, possibly related to the CTX-M enzyme expression |
Analytical isoelectric focusing (aIEF) revealed β-lactamase bands with pIs of 7.6 and 8.2 and initial standard DNA sequencing of blaSHV showed double spikes in amino acid position 238 and/or 240 (4).
Partial DNA sequencing (i.e., from amino acid 35 to 274 for TEMs and from 8 to 249 for SHVs). The blaSHV and blaTEM sequences did not show substitutions conferring an ESBL phenotype.
Number in parentheses following CTX-M designation indicates the family group for CTX-M-type ESBLs (16).
The data presented above also support the previous observation that standard DNA sequencing of PCR amplification products fails to accurately detect more than one bla gene of a given family (4). In particular, many K. pneumoniae isolates possessing both blaSHV-11 (non-ESBL) and blaSHV-12 (ESBL) genes were initially identified incorrectly as blaSHV-11-positive isolates only with standard DNA sequence analysis (4). In contrast, the microarray can accurately identify the blaESBL gene (e.g., blaSHV-12) regardless of the coexistence of additional blanon-ESBL genes (e.g., blaSHV-1 and/or blaSHV-11) (see Table S1 in the supplemental material).
In conclusion, the results of the present work show that the microarray Check KPC/ESBL system is a highly accurate tool for detection of the clinically important β-lactamase genes found among contemporary Gram-negative organisms. Due to its rapid performance, this platform could be used in epidemiological or infection control studies in which large collections of isolates need to be characterized. Furthermore, the use of Check KPC/ESBL in clinical practice may lead to more appropriate use of antimicrobial agents, reduction of costs, and improved patient outcomes. More-extensive evaluations (e.g., using clinical isolates possessing bla genes not tested in this study) are needed to establish the full potential of this methodology for detecting different resistance genes.
Supplementary Material
Acknowledgments
This work was supported in part by the Veterans Affairs Merit Review Program (R.A.B.), the National Institutes of Health (grant RO3-AI081036 to R.A.B.), and the Geriatric Research Education and Clinical Center (grant VISN 10 to R.A.B.).
We thank Sarah Drawz for the critical revision of the manuscript and Francesco Luzzaro, Antonio Q. Toniolo, John Quale, David L. Paterson, Gerri S. Hall, and Stephen G. Jenkins for providing clinical isolates. We also thank Check-Points for the technical support and for providing the material necessary for the study.
Footnotes
Published ahead of print on 26 May 2010.
Supplemental material for this article may be found at http://jcm.asm.org/.
REFERENCES
- 1.Bush, K., and G. A. Jacoby. 2010. An updated functional classification of β-lactamases. Antimicrob. Agents Chemother. 54:969-976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cornaglia, G., and G. M. Rossolini. 2010. The emerging threat of acquired carbapenemases in Gram-negative bacteria. Clin. Microbiol. Infect. 16:99-101. [DOI] [PubMed] [Google Scholar]
- 3.Endimiani, A., J. M. Depasquale, S. Forero, F. Perez, A. M. Hujer, D. Roberts-Pollack, P. D. Fiorella, N. Pickens, B. Kitchel, A. E. Casiano-Colon, F. C. Tenover, and R. A. Bonomo. 2009. Emergence of blaKPC-containing Klebsiella pneumoniae in a long-term acute care hospital: a new challenge to our healthcare system. J. Antimicrob. Chemother. 64:1102-1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Endimiani, A., A. M. Hujer, F. Perez, C. R. Bethel, K. M. Hujer, J. Kroeger, M. Oethinger, D. L. Paterson, M. D. Adams, M. R. Jacobs, D. J. Diekema, G. S. Hall, S. G. Jenkins, L. B. Rice, F. C. Tenover, and R. A. Bonomo. 2009. Characterization of blaKPC-containing Klebsiella pneumoniae isolates detected in different institutions in the eastern USA. J. Antimicrob. Chemother. 63:427-437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Endimiani, A., F. Luzzaro, R. Migliavacca, E. Mantengoli, A. M. Hujer, K. M. Hujer, L. Pagani, R. A. Bonomo, G. M. Rossolini, and A. Toniolo. 2007. Spread in an Italian hospital of a clonal Acinetobacter baumannii strain producing the TEM-92 extended-spectrum β-lactamase. Antimicrob. Agents Chemother. 51:2211-2214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Endimiani, A., and D. L. Paterson. 2007. Optimizing therapy for infections caused by enterobacteriaceae producing extended-spectrum β-lactamases. Semin. Respir. Crit. Care Med. 28:646-655. [DOI] [PubMed] [Google Scholar]
- 7.Gniadkowski, M. 2008. Evolution of extended-spectrum β-lactamases by mutation. Clin. Microbiol. Infect. 14(Suppl.)1:11-32. [DOI] [PubMed] [Google Scholar]
- 8.Grimm, V., S. Ezaki, M. Susa, C. Knabbe, R. D. Schmid, and T. T. Bachmann. 2004. Use of DNA microarrays for rapid genotyping of TEM β-lactamases that confer resistance. J. Clin. Microbiol. 42:3766-3774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kitchel, B., J. K. Rasheed, J. B. Patel, A. Srinivasan, S. Navon-Venezia, Y. Carmeli, A. Brolund, and C. G. Giske. 2009. Molecular epidemiology of KPC-producing Klebsiella pneumoniae isolates in the United States: clonal expansion of multilocus sequence type 258. Antimicrob. Agents Chemother. 53:3365-3370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Leinberger, D. M., V. Grimm, M. Rubtsova, J. Weile, K. Schroppel, T. A. Wichelhaus, C. Knabbe, R. D. Schmid, and T. T. Bachmann. 2010. Integrated detection of extended-spectrum-β-lactam resistance by DNA microarray-based genotyping of TEM, SHV, and CTX-M genes. J. Clin. Microbiol. 48:460-471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Luzzaro, F., G. Gesu, A. Endimiani, G. Ortisi, S. Malandrin, L. Pagani, and G. M. Rossolini. 2006. Performance in detection and reporting β-lactam resistance phenotypes in Enterobacteriaceae: a nationwide proficiency study in Italian laboratories. Diagn. Microbiol. Infect. Dis. 55:311-318. [DOI] [PubMed] [Google Scholar]
- 12.Miriagou, V., G. Cornaglia, M. Edelstein, I. Galani, C. G. Giske, M. Gniadkowski, E. Malamou-Lada, L. Martinez-Martinez, F. Navarro, P. Nordmann, L. Peixe, S. Pournaras, G. M. Rossolini, A. Tsakris, A. Vatopoulos, and R. Canton. 2010. Acquired carbapenemases in Gram-negative bacterial pathogens: detection and surveillance issues. Clin. Microbiol. Infect. 16:112-122. [DOI] [PubMed] [Google Scholar]
- 13.Nordmann, P., G. Cuzon, and T. Naas. 2009. The real threat of Klebsiella pneumoniae carbapenemase-producing bacteria. Lancet Infect. Dis. 9:228-236. [DOI] [PubMed] [Google Scholar]
- 14.Paterson, D. L., and R. A. Bonomo. 2005. Extended-spectrum β-lactamases: a clinical update. Clin. Microbiol. Rev. 18:657-686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Paterson, D. L., K. M. Hujer, A. M. Hujer, B. Yeiser, M. D. Bonomo, L. B. Rice, and R. A. Bonomo. 2003. Extended-spectrum β-lactamases in Klebsiella pneumoniae bloodstream isolates from seven countries: dominance and widespread prevalence of SHV- and CTX-M-type β-lactamases. Antimicrob. Agents Chemother. 47:3554-3560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pitout, J. D., A. Hossain, and N. D. Hanson. 2004. Phenotypic and molecular detection of CTX-M-β-lactamases produced by Escherichia coli and Klebsiella spp. J. Clin. Microbiol. 42:5715-5721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pitout, J. D., and K. B. Laupland. 2008. Extended-spectrum β-lactamase-producing Enterobacteriaceae: an emerging public-health concern. Lancet Infect. Dis. 8:159-166. [DOI] [PubMed] [Google Scholar]
- 18.Poirel, L., C. Heritier, I. Podglajen, W. Sougakoff, L. Gutmann, and P. Nordmann. 2003. Emergence in Klebsiella pneumoniae of a chromosome-encoded SHV β-lactamase that compromises the efficacy of imipenem. Antimicrob. Agents Chemother. 47:755-758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Queenan, A. M., and K. Bush. 2007. Carbapenemases: the versatile β-lactamases. Clin. Microbiol. Rev. 20:440-458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rasheed, J. K., G. J. Anderson, H. Yigit, A. M. Queenan, A. Domenech-Sanchez, J. M. Swenson, J. W. Biddle, M. J. Ferraro, G. A. Jacoby, and F. C. Tenover. 2000. Characterization of the extended-spectrum β-lactamase reference strain, Klebsiella pneumoniae K6 (ATCC 700603), which produces the novel enzyme SHV-18. Antimicrob. Agents Chemother. 44:2382-2388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tenover, F. C., M. J. Mohammed, T. S. Gorton, and Z. F. Dembek. 1999. Detection and reporting of organisms producing extended-spectrum β-lactamases: survey of laboratories in Connecticut. J. Clin. Microbiol. 37:4065-4070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wolter, D. J., P. M. Kurpiel, N. Woodford, M. F. Palepou, R. V. Goering, and N. D. Hanson. 2009. Phenotypic and enzymatic comparative analysis of the novel KPC variant KPC-5 and its evolutionary variants, KPC-2 and KPC-4. Antimicrob. Agents Chemother. 53:557-562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhu, L. X., Z. W. Zhang, D. Liang, D. Jiang, C. Wang, N. Du, Q. Zhang, K. Mitchelson, and J. Cheng. 2007. Multiplex asymmetric PCR-based oligonucleotide microarray for detection of drug resistance genes containing single mutations in Enterobacteriaceae. Antimicrob. Agents Chemother. 51:3707-3713. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.