Abstract
An outbreak of norovirus (NoV) infection was identified in a kennel. Sequence analysis of a short fragment in the polymerase complex indicated the clonal origin of the strains, which were similar to the prototype canine NoV strain GIV.2/Bari/170/07-4/ITA (94.7% nucleotide identity). The findings demonstrate that canine NoV circulates in dogs in Greece and that it can spread easily across a group of animals.
The Caliciviridae are small nonenveloped viruses possessing a linear, single-stranded, positive-polarity RNA genome of 7.3 to 8.5 kb (5). The family includes four genera, Norovirus, Sapovirus, Lagovirus, and Vesivirus, along with as yet unclassified simian, bovine, and porcine caliciviruses (4, 8, 21).
Noroviruses (NoVs) seem to have a worldwide distribution, playing an important role as a cause of epidemic nonbacterial gastroenteritis in humans (5). NoVs are transmitted mainly by person-to-person contact or by contaminated food and water (3). NoVs are classified into at least five genogroups (genogroup I [GI] to genogroup V [GV]) and 34 genotypes (11, 12, 25, 28). Human NoVs are in GI, GII, and GIV. In addition, NoVs have been detected in pigs (GII), cows (GIII), and mice (GV) (7, 16, 25, 28). Recently, caliciviruses resembling GIV (Alphatron-like) human NoVs have been identified in small and large carnivores and classified as GIV.2 (10, 11). Also, a recombinant canine NoV strain, Bari/91/07/ITA, was found to have GIV.2 ORF1 (polymerase complex) but novel ORF2 (capsid protein) unrelated to any known NoV genogroups and genotypes (12).
A pilot study was initiated in Greece to investigate the presence of similar NoVs in dogs with gastroenteric symptoms from kennels and pet shops and in animals hospitalized in veterinary clinics. A total of 72 fecal samples, collected from 2- to 8-month-old dogs in different areas of Greece in 2008, were screened for common canine pathogens and for caliciviruses. Stool samples were tested by using a broadly reactive primer pair, p289-p290, amplifying a band of 315 bp for NoV and a band of 330 bp for vesivirus and sapovirus (6). These primers target highly conserved motifs, DYSKWDST and YGDD, of the RNA-dependent RNA polymerase (RdRp) region of the polymerase complex. Norovirus-specific primers JV12Y and JV13I, which target the same RdRp region, were used in order to confirm the presence of NoV in the samples, yielding amplicons of the expected size (24). By reverse transcription-PCR with primer pair p289-p290, 7 of 72 (9.7%) samples from Larissa and Thessaloniki (central and north Greece, respectively) were found to be positive for calicivirus. NoV infection was confirmed in 6 of 72 (8.3%) samples. All NoV-positive samples were identified from diarrheal animals housed in a kennel in Thessaloniki, with five dogs housed together and the other dog in a neighboring cage, thus suggesting a cluster of infection. The five dogs were 3 months old and members of the same litter. The other dog was 2.5 months old. All six dogs presented diarrhea and were also infected by canine coronavirus (CCoV).
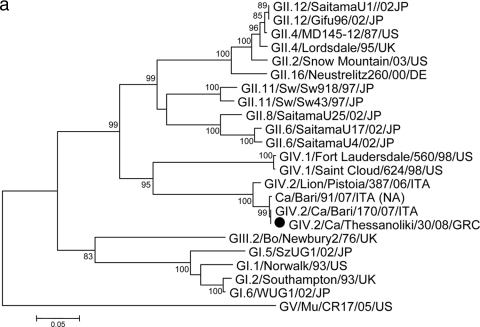
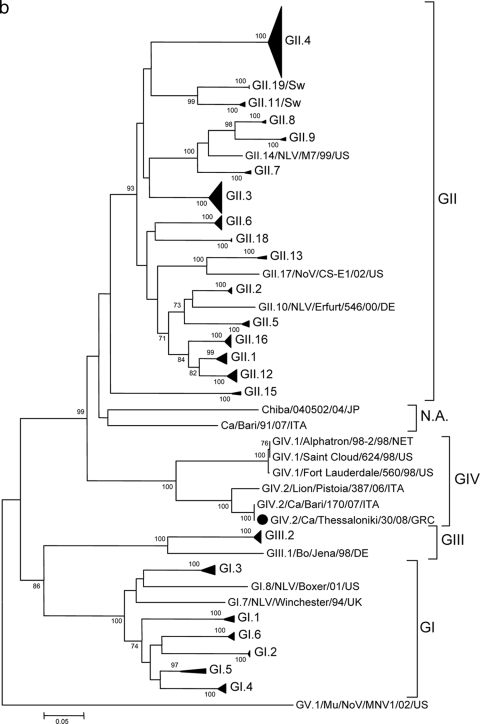
Sequence analysis of a short RNA fragment confirmed that the strains were identical in the region sequenced and highly similar to the prototype canine NoV strain GIV.2/Bari/170/07-4/ITA (94.7% nucleotide [nt] identity). For one such strain, Thessaloniki/30/2008/GRC, it was possible to determine the sequence of a 1.8-kb genome fragment encompassing the 3′ end of ORF1 (polymerase complex) and the 5′ end of ORF2 (capsid gene) http://www.ncbi.nlm.nih.gov/GenBank/index.html). The strain was found to resemble the canine virus strain GIV.2/Bari/170/07/ITA (Fig. 1 ) both in ORF1 (97.9% nt and 100% amino acid [aa] identities) and in ORF2 (97.9% nt and 99.4% aa identities).
FIG. 1.
(a) Phylogenetic tree constructed on the 268-aa sequence of the COOH terminus of the polymerase complex. The tree was constructed using a selection of NoV strains representative of GI to GV with MEGA software version 4.0 (22). The tree was generated using the neighbor-joining method without distance correction, with bootstrap analysis of more than 1,000 replicates. Strain designations follow the outlines of Wang et al. (25) and Zheng et al. (28). The GenBank accession numbers of the strains are as follows: SaitamaU1/02/JP, AB039775; Gifu96/02/JP, AB045603; MD145-12/87/US, AY032605; Lordsdale/95/UK, X86557; Snow Mountain/03/US, AY134748; Neustrelitz260/00/DE, AY772730; Sw/Sw918/97/JP, AB074893; Sw/Sw43/97/JP, AB074892; SaitamaU25/02/JP, AB039780; SaitamaU17/02/JP, AB039779; SaitamaU4/02/JP, AB039777; Fort Laudersdale/560/98/US, AF414426; Saint Cloud/624/98/US, AF414427; Lion/Pistoia/387/06/ITA, EF450827; Ca/Bari/91/07/ITA, FJ875027; Ca/Bari/170/07/ITA, EU224456; Bo/Newbury2/76/UK, AF097917; SzUG1/02/JP, AB039774; Norwalk/93/US, M87661; Southampton/93/UK, L07418; WUG1/02/JP, AB081723; and Mu/CR17/05/US, EU004682. (b) Phylogenetic tree constructed on the partial amino acid sequence of the capsid protein. The tree was constructed using a selection of NoV strains representative of GI to GV with MEGA software version 4.0 (22). The tree was generated using the neighbor-joining method without distance correction, with bootstrap analysis of more than 1,000 replicates. Strain designations follow the outlines of Wang et al. (25) and Zheng et al. (28). The GenBank accession numbers of the strains are as follows: NLV/M7/99/US, AY130761; NoV/CS-E1/02/US, AY502009; NLV/Erfurt/546/00/DE, AF427118; Chiba/040502/04/JP, AJ865586; Ca/Bari/91/07/ITA, FJ875027; Alphatron/98-2/98/NET, AF195847; Saint Cloud/624/98/US, AF414427; Fort Lauderdale/560/98/US, AF414426; Lion/Pistoia/387/06/ITA, EF450827; Ca/Bari/170/07/ITA, EU224456; Bo/Jena/98/DE, AJ011099; NLV/Boxer/01/US, AF538679; NLV/Winchester/94/UK, AJ277609; and Mu/NoV/MNV1/02/US, AY228235. Abbreviations: Bo, bovine; Mu, murine; Sw, swine; Ca, canine; NA, not assigned; NLV, Norwalk-like virus; MNV, murine norovirus; JP, Japan, US, United States; UK, United Kingdom; DE, Germany; ITA, Italy; GRC, Greece; NET, Netherlands.
There are a few studies documenting the detection and characterization of caliciviruses from dogs with diarrhea, and most viruses were characterized as either canine or feline vesiviruses (2, 9, 13, 14, 20). More recently, a canine NoV, strain Bari/170/07/ITA, was detected in a dog with diarrhea and vomiting which was concomitantly infected by canine parvovirus (10). In the polymerase complex and in the capsid protein, the NoV strain was found to resemble a NoV strain, Pistoia/387/06/ITA, detected in a captive lion cub (11) and classified as GIV.2. During a 1-year surveillance of dogs with enteritis in Italy, the prevalence of NoV infection was found to be 2.2% and the age of the infected dogs ranged between 60 and 70 days (4 of 187 sporadic cases). A recombinant NoV strain, Bari/91/07/ITA, was also found that resembled GIV.2 NoVs in its polymerase gene but that was genetically unrelated in its full-length capsid gene to NoVs from any established genogroup (<54.7% aa identity) except to an unclassified human NoV strain, Chiba/040502/04/Jp (58.1% aa identity) (12), thus suggesting that genetically diverse NoV strains may circulate in dogs.
The outbreak of NoV infection identified in the Greek kennel was caused by a strain highly similar to the prototype canine NoV strain GIV.2/Bari/170/07-4/ITA in partial sequences derived from both ORF1 and ORF2 genes. Accordingly, GIV.2 appears to be the typical classification for NoVs from carnivores, since GIV.2 viruses have been detected thus far only in those animals. Similar to previous observations (12), in the present study, NoV was detected in young puppies (75 to 90 days of age) but not in older animals. Pups at this age are highly susceptible to infectious pathogens since the lingering immunity of maternal origin is no longer able to protect them (18). Age-related patterns of distribution have been observed for porcine NoVs in pigs, with NoVs being detected almost exclusively and with high prevalence rates in asymptomatic finisher pigs (26). Also, sporadic cases and small to large outbreaks of NoV infection and disease have been reported in humans of all age groups (5), although the infection is uncommon in infants of less than 2 months of age (17). This has been related to the decrease of maternally derived immunity, virus blockage by histoblood group antigens (HBGA) secreted in the maternal milk, or the developmental nature of NoV receptors that are not expressed at high levels in the early stages of life (15, 27). Interestingly, the detection of the same strain in all puppies strongly suggests that canine NoV can easily spread within a community of dogs, i.e., in its homologous host, thus suggesting high infectivity, a finding that is consistent with contamination from a common source and with the high infectivity displayed by human NoVs (23).
All cases were mixed infection with CCoV. Experimental infections are necessary to elucidate the pathogenic role of canine NoV and to understand whether it can act by itself as a primary causative agent of gastrointestinal disease or whether it can trigger mechanisms of synergism in coinfections, as observed between coronaviruses and parvoviruses (1, 19).
In conclusion, this study provides evidence that canine NoV circulates in dogs and that it has the ability to spread among a group of animals. Serological assays seem necessary in order to estimate the prevalence of this novel virus in dogs, but at the same time, experimental infections may help in elucidating its pathogenic role.
Nucleotide sequence accession number.
The newly determined sequence reported here is available in the GenBank database under accession number GU354246.
Footnotes
Published ahead of print on 19 May 2010.
REFERENCES
- 1.Appel, M. J. 1988. Does canine coronavirus augment the effects of subsequent parvovirus infection? Vet. Med. 83:360-366. [Google Scholar]
- 2.Evermann, J. F., A. J. McKeirnan, A. W. Smith, D. E. Skilling, and R. L. Ott. 1985. Isolation and identification of caliciviruses from dogs with enteric infections. Am. J. Vet. Res. 46:218-220. [PubMed] [Google Scholar]
- 3.Fankhauser, R. L., S. S. Monroe, J. S. Noel, C. D. Humphrey, J. S. Bresee, U. D. Parashar, T. Ando, and R. I. Glass. 2002. Epidemiologic and molecular trends of “Norwalk-like viruses” associated with outbreaks of gastroenteritis in the United States. J. Infect. Dis. 186:1-7. [DOI] [PubMed] [Google Scholar]
- 4.Farkas, T., K. Sestak, C. Wei, and X. Jiang. 2008. Characterization of a rhesus monkey calicivirus representing a new genus of Caliciviridae. J. Virol. 82:5408-5416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Green, K. Y. 2007. Caliciviridae: the noroviruses. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 5th ed., vol. 1. Lippincott Williams & Wilkins, Philadelphia, PA.
- 6.Jiang, X., C. Espul, W. M. Zhong, H. Cuello, and D. O. Matson. 1999. Characterization of a novel human calicivirus that may be a naturally occurring recombinant. Arch. Virol. 144:2377-2387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Karst, S. M., C. E. Wobus, M. Lay, J. Davidson, and H. W. Virgin IV. 2003. STAT1-dependent innate immunity to a Norwalk-like virus. Science 299:1575-1578. [DOI] [PubMed] [Google Scholar]
- 8.L'Homme, Y., R. Sansregret, E. Plante-Fortier, A. M. Lamontagne, M. Ouardani, G. Lacroix, and C. Simard. 2009. Genomic characterization of swine caliciviruses representing a new genus of Caliciviridae. Virus Genes 39:66-75. [DOI] [PubMed] [Google Scholar]
- 9.Martella, V., A. Pratelli, M. Gentile, D. Buonavoglia, N. Decaro, P. Fiorente, and C. Buonavoglia. 2002. Analysis of the capsid protein gene of a feline-like calicivirus isolated from a dog. Vet. Microbiol. 85:315-322. [DOI] [PubMed] [Google Scholar]
- 10.Martella, V., E. Lorusso, N. Decaro, G. Elia, A. Radogna, M. D'Abramo, C. Desario, A. Cavalli, M. Corrente, M. Camero, C. A. Germinario, K. Banyai, B. Di Martino, F. Marsilio, L. E. Carmichael, and C. Buonavoglia. 2008. Detection and molecular characterization of a canine norovirus. Emerg. Infect. Dis. 14:1306-1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Martella, V., M. Campolo, E. Lorusso, P. Cavicchio, M. Camero, A. L. Bellacicco, N. Decaro, G. Elia, G. Greco, M. Corrente, C. Desario, S. Arista, K. Banyai, M. Koopmans, and C. Buonavoglia. 2007. Norovirus in captive lion cub (Panthera leo). Emerg. Infect. Dis. 13:1071-1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Martella, V., N. Decaro, E. Lorusso, A. Radogna, P. Moschidou, F. Amorisco, M. S. Lucente, C. Desario, V. Mari, G. Elia, K. Banyai, L. E. Carmichael, and C. Buonavoglia. 2009. Genetic heterogeneity and recombination in canine noroviruses. J. Virol. 83:11391-11396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Matsuura, Y., Y. Tohya, K. Nakamura, M. Shimojima, F. Roerink, M. Mochizuki, K. Takase, H. Akashi, and T. Sukimura. 2002. Complete nucleotide sequence, genome organization and phylogenetic analysis of the canine calicivirus. Virus Genes 25:67-73. [DOI] [PubMed] [Google Scholar]
- 14.Mochizuki, M., A. Kawanishi, H. Sakamoto, S. Tashiro, R. Fujimoto, and M. Ohwaki. 1993. A calicivirus isolated from a dog with fatal diarrhoea. Vet. Rec. 132:221-222. [DOI] [PubMed] [Google Scholar]
- 15.Morrow, A. L., G. M. Ruiz-Palacios, X. Jiang, and D. S. Newburg. 2005. Human-milk glycans that inhibit pathogen binding protect breast-feeding infants against infectious diarrhea. J. Nutr. 135:1304-1307. [DOI] [PubMed] [Google Scholar]
- 16.Oliver, S. L., A. M. Dastjerdi, S. Wong, L. El-Attar, C. Gallimore, D. W. G. Brown, J. Green, and J. C. Bridger. 2003. Molecular characterization of bovine enteric caliciviruses: a distinct third genogroup of norovirus (Norwalk-like viruses) unlikely to be of risk to humans. J. Virol. 77:2789-2798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pang, X. L., S. Honma, S. Nakata, and T. Vesikari. 2000. Human caliciviruses in acute gastroenteritis of young children in the community. J. Infect. Dis. 181(Suppl. 2):S288-S294. [DOI] [PubMed] [Google Scholar]
- 18.Pollock, R. V. H., and L. E. Carmichael. 1982. Maternally derived immunity to canine parvovirus infection: transfer, decline, and interference with vaccination. J. Am. Vet. Med. Assoc. 180:37-42. [PubMed] [Google Scholar]
- 19.Pratelli, A., M. Tempesta, F. P. Roperto, P. Sagazio, L. Carmichael, and C. Buonavoglia. 1999. Fatal coronavirus infection in puppies following canine parvovirus 2b infection. J. Vet. Diagn. Invest. 11:550-553. [DOI] [PubMed] [Google Scholar]
- 20.Schaffer, F. L., M. E. Soergel, J. W. Black, D. E. Skilling, A. W. Smith, and W. D. Cubitt. 1985. Characterization of a new calicivirus isolated from feces of a dog. Arch. Virol. 84:181-195. [DOI] [PubMed] [Google Scholar]
- 21.Smiley, J. R., K. O. Chang, J. Hayes, J. Vinje, and L. J. Saif. 2002. Characterization of an enteropathogenic bovine calicivirus representing a potentially new calicivirus genus. J. Virol. 76:10089-10098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tamura, K., J. Dudley, M. Nei, and S. Kumar. 2007. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596-1599. [DOI] [PubMed] [Google Scholar]
- 23.Teunis, P. F., C. L. Moe, P. Liu, S. E. Miller, L. Lindesmith, R. S. Baric, J. Le Pendu, and R. L. Calderon. 2008. Norwalk virus: how infectious is it? J. Med. Virol. 80:1468-1476. [DOI] [PubMed] [Google Scholar]
- 24.Vennema, H., E. de Bruin, and M. Koopmans. 2002. Rational optimization of generic primers used for Norwalk-like virus detection by reverse transcriptase polymerase chain reaction. J. Clin. Virol. 25:233-235. [DOI] [PubMed] [Google Scholar]
- 25.Wang, Q. H., M. G. Han, J. A. Funk, G. Bowman, D. A. Janies, and L. J. Saif. 2005. Genetic diversity and recombination of porcine sapoviruses. J. Clin. Microbiol. 43:5963-5972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang, Q. H., M. Souza, J. A. Funk, W. Zhang, and L. J. Saif. 2006. Prevalence of noroviruses and sapoviruses in swine of various ages determined by reverse transcription-PCR and microwell hybridization assays. J. Clin. Microbiol. 44:2057-2062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yamamoto, F. 2000. Molecular genetics of ABO. Vox Sang. 78(Suppl. 2):91-103. [PubMed] [Google Scholar]
- 28.Zheng, D. P., T. Ando, R. L. Fankhauser, R. S. Beard, R. I. Glass, and S. S. Monroe. 2006. Norovirus classification and proposed strain nomenclature. Virology 346:312-323. [DOI] [PubMed] [Google Scholar]