Abstract
Multilocus sequence analysis (MLSA) was used to clarify the taxonomic status of a virulent Borrelia organism previously isolated from patients with relapsing fever and from ticks in Spain that is designated the Spanish relapsing fever (SRF) Borrelia. This species has been used extensively in experimental infection models because of its continued virulence. Seven genes were amplified to analyze the phylogenetic relationships among several Spanish isolates of SRF Borrelia and other relapsing fever Borrelia species. The genes targeted in this study included rrs and flaB, which have commonly been used in phylogenetic studies; the rrf-rrl intergenic spacer (IGS), which is highly discriminatory; and four additional genes, p66, groEL, glpQ, and recC, which are located on the chromosome and which have therefore evolved in a clonal way. The species included in this study were Borrelia duttonii, B. recurrentis, B. crocidurae, and B. hispanica as Old World Borrelia species and B. turicatae and B. hermsii as New World Borrelia species. The results obtained by MLSA of the SRF Borrelia on the basis of 1% of the genomic sequence data analyzed confirmed that the SRF Borrelia isolates are B. hispanica. However, the prototype isolates of B. hispanica used in this study have an uncertain history and display unique phenotypic characteristics that are not shared with the SRF Borrelia. Therefore, we propose to use strain SP1, isolated from a relapsing fever patient in 1994 in southern Spain, as the type strain for B. hispanica.
The members of the Borrelia genus comprise two major groups: those causing Lyme disease and those causing relapsing fever (RF). Relapsing fever is distributed all over the world; and its agents are traditionally classified according to their geographic origins, vector, and infectivity in various animal species. In Spain, relapsing fever has been reported sporadically throughout the last century and has been associated with Borrelia hispanica (2, 11, 17, 19, 27, 28). From 1994 to 1996, spirochetes were isolated from the blood of two patients with RF symptoms and from Ornithodoros erraticus, and we have designated these spirochetes Spanish relapsing fever (SRF) Borrelia. This SRF organism is refractory to in vitro cultivation, and this represents a unique phenotypic characteristic not shared by B. hispanica (1). SRF Borrelia infects C3H/HeN, BALB/c, C57/B51, and Swiss outbred mice (1), and it is highly infectious. A murine model of SRF was developed (13) and has been used extensively in experimental studies on pathogenesis and the host response (5-7, 14, 16, 20). A preliminary genetic analysis comparing the rrs and flaB loci from B. hispanica to those from the Spanish isolates suggested that SRF could represent a new species (1).
To date, most of the ecological and epidemiological studies on relapsing fever Borrelia have used a single locus. Therefore, characterization of relapsing fever spirochetes has relied upon the amplification and sequencing of certain genes, such as rrs (24), flaB (12), and more recently, the noncoding intergenic spacer (IGS) rrs-rrl (4). However, the value of these approaches is limited. The rrs gene is highly conserved among relapsing fever Borrelia species, and it can discriminate among species but not among strains. The results with flaB have shown only minor differences among relapsing fever Borrelia species. On the other hand, although the intergenic spacer rrs-rrl is not able to discriminate between B. recurrentis and B. duttonii (29), it has been shown to discriminate between strains of other RF Borrelia species.
Multilocus sequence analysis (MLSA) is a powerful tool for the delineation of species and assignment of strains to defined species (23, 25, 26). The utilization of this method is increasing, especially with uncultured and slow-growing organisms that cannot be analyzed by DNA-DNA hybridization. MLSA has been used with the Borrelia genus in order to delineate or confirm species, such as B. spielmanii (25), B. californiensis (23), B. carolinensis (26), and B. bavariensis (21).
The purpose of the study described here was to clarify the taxonomic status of a group of SRF isolates from patients and ticks isolated in Spain. The selection of genes (rrs, rrf-rrl, flaB, glpQ, groEL, recC, p66) used for multilocus sequence analysis of this group of spirochetes was based on their suitability for phylogenetic purposes.
MATERIALS AND METHODS
Culture.
The relapsing fever Borrelia strains used in this study are listed in Table 1. B. duttonii, B. hermsii, B. crocidurae, and B. hispanica were grown in Barbour-Stoenner-Kelly H (BSK-H) medium (Sigma-Aldrich, St. Louis, MO) supplemented with 6% rabbit serum (Pel-Freez, Rogers, AR) at 33°C. Cultures were harvested in the mid-log phase by centrifugation and washed with sterile phosphate-buffered saline (Gibco, Grand Island, NY).
TABLE 1.
RF Borrelia strains used in this study
| Species | Strain | Geographic origin | Source |
|---|---|---|---|
| B. duttonii | 1120K3 | Congo | Ornithodoros moubata |
| B. duttonii | Ly | Tanzania | Human, CSFa |
| B. recurrentis | A1 | Ethiopia | Human blood |
| B. crocidurae | Achema | Mauritania | Ornithodoros sonrai |
| B. crocidurae | CR2Ab | Western Africa (?c) | Ornithodoros erraticus |
| B. hermsii | HS1 | USA | Ornithodoros hermsi |
| B. hermsii | DAH | USA | Human, CSF |
| B. turicatae | 91E135 | USA | Ornithodoros turicatae |
| B. hispanica | CR1 | ? | ? |
| B. hispanica | ORIX | ? | ? |
| B. hispanica | JH | ? | ? |
| B. hispanica | Sp1 | Spain | Human blood |
| B. hispanica | Sp2 | Spain | Human blood |
| B. hispanica | Sp3 | Spain | Ornithodoros erraticus |
CSF, cerebrospinal fluid.
Proposed in this study to be reclassified as B. duttonii CR2A.
?, unknown or uncertain.
Animal passages.
Six- to 8-week-old C3H/HeN mice (Charles River Laboratories, Wilmington, MA) were inoculated with stocks of three SRF isolates frozen at −80°C and killed during the first peak of spirochetemia. Blood was obtained by cardiac puncture, and spirochetes were harvested from the plasma by centrifugation and washed with fresh BSK-H medium.
DNA extraction, primers, and PCR conditions.
Genomic DNA was extracted using a DNeasy blood and tissue extraction kit (Qiagen, Valencia, CA), following the instructions of the manufacturer. Water was included as a negative control in every extraction to test for possible contamination. Primers were designed to amplify any relapsing fever Borrelia and are listed in Table 2. The conditions for the amplification of the rrs, flaB, rrf-rrl, p66, groEL, glpQ, and recC genes were as follows: an initial denaturalization step at 98°C for 30 s, followed by 35 cycles at 98°C for 10 s, 68°C for 15 s, and extension at 72°C for 15 s and with a final extension step at 72°C for 10 min. In the case of rrs fragment 2, rrs fragment 4, and flaB fragment 1, the annealing temperature was 57°C. The prevention of cross-contamination and false-positive results was achieved by the use of plugged tips, the performance of PCRs in a room separate from the room used for DNA extraction, and the use of specific separated areas dedicated for the production of reagents and analyses of the amplicons. A negative PCR control (water) was included in each run as well. The PCR products (20 μl) were separated in a 1% agarose gel and purified (30 μl) with a QIAquick PCR purification kit (Qiagen), following the manufacturer's recommendations. All the samples were submitted for direct sequencing to the DNA Sequencing Facility at Stony Brook University. Sequences were determined in both directions, using the same specific primers that were used in each PCR. All sequences were analyzed with the EditSeq module of DNAStar software (DNAStar, United Kingdom).
TABLE 2.
Primers used in this study
| Gene | Forward primer (5′-3′) | Reverse primer (5′-3′) | Amplicon size (bp) | Reference or source |
|---|---|---|---|---|
| flaB fragment 1 | TCATAAATCATAATACGTCAG | AATGTCCATGAAGCTTGTGA | 514 | This study |
| flaB fragment 2 | CTGAAGAGCTTGGAATGCAAC | AGGTACTTGATTTGCTTGTGC | 538 | This study |
| rrs fragment 1 | GTTTGATCCTGGCTTAGAAC | TTACAATCTTTCGACCTTCTT | 417 | This study |
| rrs fragment 2 | CACACTGGAACTGAGATACGG | TTCGCCTCTGGTATTCTTCCT | 419 | This study |
| rrs fragment 3 | TGCGTAAAATACCACAGCTCA | TGAGTCCCCATCTTTACATGC | 547 | This study |
| rrs fragment 4 | TACCAGGGCTTGACATATACA | GAGGTGATCCAGCCACACTTT | 561 | This study |
| rrf-rrl outer | GTATGTTTAGTGAGGGGGGTG | GGATCATAGCTCAGGTGGTTAG | 807a | 3 |
| rrf-rrl inner | AGGGGGGTGAAGTCGTAACAAG | GTCTGATAAACCTGAGGTCGGA | 765a | 3 |
| recC fragment 1 | AAGATATATAAAACAAACAAA | GTCAATTTCTCTAGTCTCTCC | 623 | This study |
| recC fragment 2 | ATTGAAACAAAGAGAATAATA | ATTTATTCATTACTTTTGTGT | 634 | This study |
| recC fragment 3 | GGAACGATTAGTAATTTTAA | TTTAAGGTATTGTATTTTTGT | 474 | This study |
| recC fragment 4 | ATATGAATGGGCAGAAATAAT | TTATTGATATCTGGATTTAGA | 495 | This study |
| recC fragment 5 | GCAAGCTCTGATAAAATTGAA | TTGATGTTTATGGGAATTATT | 737 | This study |
| groEL fragment 1 | TGGCTAAGGACATATATTTTA | ATCTTTGCCAACTCTGTCCAT | 512 | This study |
| groEL fragment 2 | TTCTGCAAATAATGATACTTC | AACATTCTCAAGAGTAAGTCC | 487 | This study |
| groEL fragment 3 | ATTGCTATACTTACTGGAGG | TAAATAGAACTCTCAAATCCA | 524 | This study |
| groEL fragment 4 | AAGGTTTTGAGATTGTGAAGA | TTACATCATTCCCATTCCAG | 323 | This study |
| p66 fragment 1 | TTTAGATTTGATATGGATGA | GATATGTGTCCAAGTATAGA | 899 | This study |
| p66 fragment 2 | TTCCTCAATAACATATGGTCT | ACACTTCCATTTTGATCTTT | 945 | This study |
| glpQ fragment 1 | CATTAATTATAGCTCACAGAG | AACAAGCATTATCAATTTTCC | 599 | This study |
| glpQ fragment 2 | TATGGCATAAACAACAAGGTA | AATCTGTAAATAGACCATCTA | 453 | This study |
The amplicon size may vary among species.
Phylogenetic analysis.
Available sequences from different loci (rrs, rrf-rrl, flaB, p66, recC, groEL, glpQ) of B. duttonii strain Ly, B. hermsii strain DAH, B. turicatae strain 91E135, and B. recurrentis strain A1 were used in the phylogenetic analysis, together with the sequences obtained in this study. Subsequent analysis of the genes was performed using the software package MEGA (version 3.1) (15), after multiple-sequence alignment by the CLUSTAL_X program (30). Distance options were according to the maximum composite likelihood model, and clustering with the neighbor-joining method was performed by using bootstrap values based on 2,000 replications.
Nucleotide sequence accession numbers.
The sequences generated in this study were deposited in GenBank under accession numbers GU350705 to GU350714 for the rrs gene sequences, GU350715 to GU350722 for the rrf-rrl gene sequences, GU357611 to GU357620 for the flaB gene sequences, GU357591 to GU357600 for the p66 gene sequences, GU357601 to GU357610 for the recC gene sequences, GU357581 to GU357590 for the groEL gene sequences, and GU357571 to GU357580 for the glpQ gene sequences.
RESULTS
Analysis of partial 16S rRNA gene.
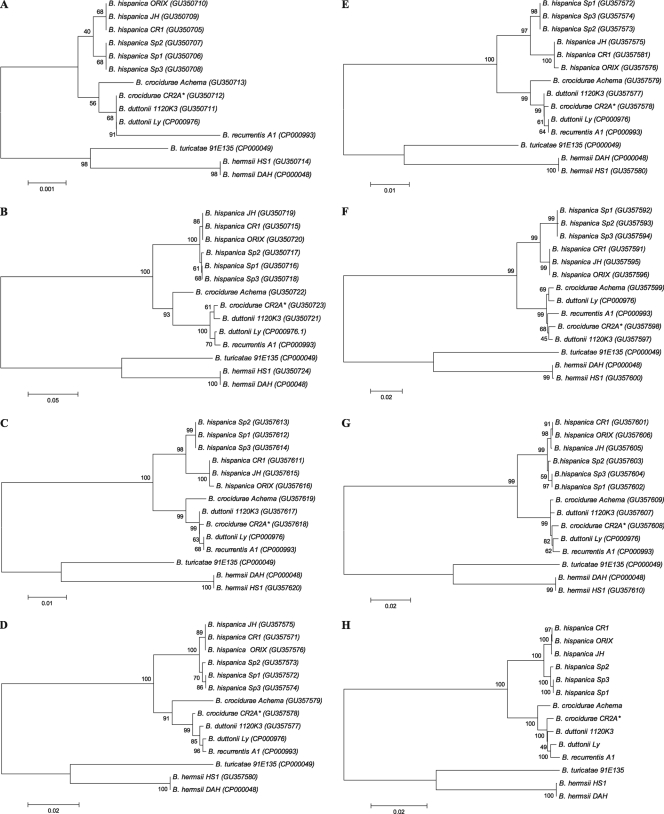
Four sets of primers were used to obtain a partial sequence of the rrs gene. All the species showed the same amplicon length, without either deletions or insertions. Comparison of the 1,513-bp portion of the rrs gene sequences revealed that all the strains from the same species analyzed produced a single cluster, with the exception of B. hispanica and SRF Borrelia, which were different by a single nucleotide and which clustered separately from each other (Fig. 1A). Although the differences among species are minor and therefore the value of the discrimination of species for genotypic purposes is limited, it is still useful to discriminate between species, especially between B. duttonii and B. recurrentis.
FIG. 1.
Phylogenetic analyses of 16S rRNA (A), IGS (B), flaB (C), glpQ (D), groEL (E), p66 (F), and recC (G). (H) Concatenated sequences. The GenBank accession numbers are indicated in parentheses in each tree. *, proposed in this study to be reclassified as B. duttonii CR2A. Scale bars, divergence, where 0.01 = 1% divergence.
Analysis of rrf-rrl IGS.
The sizes of the rrf-rrl spacer regions of the Borrelia species tested were determined by direct sequencing of the purified amplicons amplified by the primers shown in Table 2. The New World relapsing fever Borrelia species have the largest intergenic sequences (over 700 bp), whereas the Old World Borrelia species are shorter (459 to 521 bp). The length varies among species, with the exception of B. duttonii Ly and B. recurrentis A1, which display the same IGS length. The B. hispanica amplicon was 459 bp for all the strains tested and also for the SRF Borrelia. This is the shortest IGS among the Old World relapsing fever Borrelia species analyzed. The IGS sequence of SRF Borrelia strains Sp1 and Sp3 exhibited 100% sequence identity and 99.8% identity with strain Sp2. All the B. hispanica strains exhibited 100% sequence identity at this locus, 99.6% identity with SRF Borrelia strains Sp1 and Sp3, and 99.3% identity with SRF Borrelia strain Sp2. Interestingly, among the other Borrelia species analyzed, B. duttonii Ly is closer to B. recurrentis A1 than to B. duttonii 1120K3, and this highlights further the limitations of this locus to discriminate between these two species (Fig. 1B).
Analysis of partial flaB gene.
The size of the flaB amplicon was conserved among the Old World relapsing fever Borrelia species (937 bp), with the exception of B. hispanica, which showed a 3-bp deletion. All New World relapsing fever Borrelia species analyzed showed a 3-bp deletion, between positions 590 and 593 of the amplified fragment, compared with the sequences of the Old World RF species. The amplicons from SRF isolates showed 100% identity among themselves and 99% identity with B. hispanica strains. Interestingly, although this is a conserved locus, it showed more genetic differences between B. hispanica and SRF Borrelia than IGS. As happened with IGS, the partial amplification of the flaB gene failed to discriminate between B. duttonii strain Ly and B. recurrentis strain A1 (Fig. 1C).
Analysis of partial glpQ gene.
An 829-bp fragment was amplified and sequenced. No insertions or deletions were found in any of the sequences analyzed. All B. hispanica strains analyzed showed 100% identity among themselves and 99.5% identity with the SRF Borrelia. Again, the differences between B. duttonii and B. recurrentis were minor, and it is reflected by the fact that B. duttonii Ly is closer to B. recurrentis A1 than to B. duttonii 1120K3 (Fig. 1D).
Analysis of partial groEL gene.
Four set of primers were used to amplify and sequence a 1,596-bp fragment of the groEL gene. B. hermsii had a 3-bp insertion, for a total length of 1,599 bp. The B. hispanica strains showed 100% identity among themselves, whereas the identity with the different SRF isolates ranged from 99.81 to 99.74%. This locus cannot be used to discriminate between B. duttonii strains and B. recurrentis (Fig. 1E).
Analysis of partial p66 gene.
A fragment that ranged from 1,588 bp to 1,594 bp, depending on the species analyzed, was amplified using two set of primers. All the B. hispanica strains showed 100% identity among themselves and were clearly separate from the cluster formed by SRF isolates (Fig. 1F). Actually, the identity between these two was just 98.2%, which is a remarkable difference when it is taken into account that other African relapsing fever Borrelia species, such as B. duttonii, B. recurrentis, and B. crocidurae, showed p66 gene sequence identity of greater than 98.8% among themselves. Also, all the Old World relapsing fever Borrelia, with the exception of B. hispanica, showed a 6-bp deletion (AACCAA) between positions 1333 and 1336 of the amplified fragment.
Analysis of partial recC gene.
A 2,468-bp partial sequence of the recC gene was sequenced. All the B. hispanica strains clustered together, and strains CR1 and ORIX had identical sequences (Fig. 1G). The identity between them and the SRF strains ranged from 99.42% to 99.59%. The recC gene did not discriminate well among the B. duttonii and B. recurrentis species; also, the genetic differences were minor, with the identity between them being greater than 99.4%.
Analysis of concatenated sequences.
The distance matrix for phylogenetic analysis was generated by alignment of the partial rrs gene, the partial p66 gene, the partial flaB gene, the partial groEL gene, the partial recC gene, the IGS, and the partial glpQ gene. The concatenated sequences had ≈9,300 nucleotides, which represents about 1% of the chromosome. The phylogenetic tree of the concatenated sequences is shown in Fig. 1H. Analysis of the concatenated sequences of the SRF Borrelia with the six control species of relapsing fever Borrelia clearly exhibited the separation between the Old World and the New World relapsing fever Borrelia. Among the Old World relapsing fever species, it is clear that B. hispanica is well separated from the cluster formed by B. crocidurae, B. duttonii, and B. recurrentis. The nucleotide sequence difference for B. crocidurae versus B. duttonii and B. recurrentis is approximately 1%. Interestingly, this approach cannot distinguish between B. duttonii and B. recurrentis. The different isolates from the SRF Borrelia cluster together and form a cluster different from B. hispanica. Two of these isolates, Sp1 and Sp3, have the same concatenated sequence. The differences among the B. hispanica strains were minor. The similarities of the concatenated sequences of relapsing fever Borrelia species are shown in Table 3.
TABLE 3.
Similarities of concatenated sequences of relapsing fever spirochetesa
| Strain | % similaritya |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| B. recurrentis A1 |
B. duttonii |
B. crocidurae |
B. hispanica |
B. turicatae 91E135 | B. hermsii HS1 | ||||
| Ly | 1120K3 | CR2Ab | Achema | CR1 | Sp1 | ||||
| A1 | 100 | ||||||||
| Ly | 99.49 | 100 | |||||||
| 1120K3 | 99.39 | 99.46 | 100 | ||||||
| CR2A | 99.36 | 99.43 | 99.54 | 100 | |||||
| Achema | 98.76 | 98.92 | 99.01 | 98.97 | 100 | ||||
| CR1 | 96.5 | 96.64 | 96.76 | 96.73 | 96.87 | 100 | |||
| Sp1 | 96.55 | 96.63 | 96.75 | 96.72 | 96.88 | 99.39 | 100 | ||
| 91E135 | 84.69 | 84.78 | 84.87 | 84.88 | 84.92 | 84.90 | 84.77 | 100 | |
| HS1 | 84.74 | 84.90 | 84.95 | 84.98 | 84.95 | 84.97 | 84.94 | 91.28 | 100 |
Similarities were calculated from the distance matrix (P-distance values) in a pairwise deletion procedure.
Proposed in this study to be reclassified as B. duttonii CR2A.
DISCUSSION
The species designation of relapsing fever Borrelia has historically been based upon the species names of their tick vectors (10), their geographical distribution, and their host range. In the last decade of the 20th century, the development of molecular tools enabled the common use of PCRs for phylogenetic purposes. Lately, MLSA approaches have been used for the species designation of isolates in the Borrelia genus, particularly with Lyme disease Borrelia (25, 26). In the present study, we have used an MLSA approach based on the sequences of seven different genes, rrs, flaB, groEL, recC, rrs-rrl, glpQ, and p66, to reveal the phylogenetic relationship between B. hispanica and SRF Borrelia. In these analyses, B. hermsii and B. turicatae were included as representative species of the American relapsing fever group and also served as outgroups for the Old World relapsing fever Borrelia.
Multilocus sequence analysis of the relapsing fever Borrelia showed that both groups, the Old World and the New World Borrelia, are well differentiated. The New World species included in the MLSA were clearly separated in different clusters for all the genes analyzed, and, as expected, for the concatenated sequences, too.
The MLSA analysis revealed that the SRF Borrelia cluster showed only minor genetic differences with the cluster of B. hispanica. The identity between the two clusters determined using the seven concatenated genes is 99.4%. The SRF Borrelia consistently clustered together for all the loci analyzed, which it is not surprising, since these SRF isolates shared the same geographical origin. On the other hand, all three B. hispanica strains tested were very close to each other and showed a lack of variation. The nearly exact genetic identity among the strains of B. hispanica suggests that these strains have a common origin. Although the three isolates of B. hispanica (strains JH, CR1, and ORIX) have been stored for a long time in at least two laboratories in Europe, their history is not known; therefore, given their striking identity, it could be possible that these three strains of B. hispanica are actually the same isolate. Furthermore, the identity among the B. hispanica strains was higher than the identity among the SRF isolates, which were isolated at the same location during a short period of time.
The aim of this study was to clarify the taxonomic status of the SRF isolates that were thought to represent a new species (1). Interestingly, although the two studies reached a different conclusion, the results of both studies using the rrs and flaB genes were similar (1). These two genes are known to discriminate between species but not between strains because these two genes are well conserved among relapsing fever spirochetes. In both studies, flaB and rrs display differences, and in the case of flaB, this suggests that the SRF Borrelia could be a new species. The p66 gene, a membrane protein that has a surface location and that would therefore be subjected to environmental pressures, also displayed noticeable sequence differences between the B. hispanica and the SRF Borrelia clusters. On the contrary, the noncoding intergenic spacer between the 16S and 23S genes that has been shown to be particularly valuable for characterizing relapsing fever spirochetes and the glpQ gene does not discriminate between strains, suggesting that these are the same species. The concatenated sequence, which takes into account all the loci analyzed, showed that while B. hispanica and the SRF Borrelia display some genetic differences, with an overall identity of 99.4%, the differences are not sufficient to justify a new species designation for the SRF Borrelia.
Apart from the minor genetic differences, the SRF Borrelia isolates have unique phenotypic characteristics that are not shared by B. hispanica strains. The SRF Borrelia has so far remained refractory to in vitro cultivation, and it is highly infectious for C3H/HeN, BALB/c, C57BL/6, and outbred mice, whereas the available strains of B. hispanica (strains ORIX, CR1, and JH) are cultivable and grow only in SCID mice. Nonetheless, it is possible that these phenotypic differences between B. hispanica and the SRF Borrelia are the result of an adaptation process in the laboratory through years of passage in guinea pigs and culture media. In summary, the SRF isolates have a better-documented history than the B. hispanica isolates. Therefore, we propose that an SRF isolate be used as the type strain for B. hispanica.
In addition to establishing the taxonomic status of the SRF Borrelia, this study has reached other conclusions as well. The Old World Borrelia species analyzed in this study were closely related. There are two main clusters; one cluster included B. duttonii, B. recurrentis, and B. crocidurae and the other cluster included B. hispanica and the SRF Borrelia. This is in agreement with the results of previous phylogenetic studies (8, 29). The MLSA scheme could not discriminate between B. recurrentis and B. duttonii with an identity greater than 99.39%. The close identity between the two species suggests that both species are indeed the same. This is not surprising, given that previous phylogenetic and genomic sequencing data suggest that either B. recurrentis has evolved from B. duttonii or they have a common ancestral origin (9, 18). Moreover, the B. recurrentis genome is a degraded subset of the genome of B. duttonii (18).
Our results also support the hypothesis that B. duttonii/B. recurrentis and B. crocidurae are clonal variants. However, all the genes analyzed, especially IGS and glpQ, showed greater differences between the B. duttonii/B. recurrentis complex and B. crocidurae. These results are in agreement with those described in previous reports (24, 29). Also, the fact that B. duttonii Ly and B. recurrentis A1 cluster more closely suggests that the Ly ancestor is closer to the B. recurrentis ancestor than to the B. duttonii 1120K3 ancestor. In our study, we included a second strain of B. crocidurae named CR2A, and the rrs gene of this organism was identical to the rrs sequences of other B. duttonii sequences. Actually, for all the loci analyzed, this strain clusters together with the B. duttonii/B. recurrentis complex, including when the IGS region, which has been reported to be valuable for the typing of relapsing fever spirochetes (4), is used for analysis. Consequently, it has been demonstrated in this study that B. crocidurae CR2A is actually a B. duttonii strain and that it should be relocated to the correct taxon. Interestingly, B. crocidurae CR2A was isolated from an O. erraticus and was therefore named B. crocidurae, according to traditional phylogenetic criteria. Nowadays, this traditional phylogeny has largely been rewritten, with recent peculiarities including B. duttonii in Togo, East Africa (22), a B. crocidurae-like sequence in a patient and in an Ornithodoros moubata organism in the same geographical area (29), and the result obtained with B. crocidurae CR2A in this work, suggesting that these spirochetes may coexist in the same area and may have extended vector compatibility. We propose that it be renamed B. duttonii CR2A.
Acknowledgments
This work was supported by grants from the National Institutes of Health to J.L.B. (grants R37-AI-027044, RO1-AR-040445, and U54-AI-071558 [Lipkin]).
Footnotes
Published ahead of print on 12 May 2010.
REFERENCES
- 1.Anda, P., W. Sanchez-Yebra, M. del Mar Vitutia, E. Perez Pastrana, I. Rodriguez, N. S. Miller, P. B. Backenson, and J. L. Benach. 1996. A new Borrelia species isolated from patients with relapsing fever in Spain. Lancet 348:162-165. [DOI] [PubMed] [Google Scholar]
- 2.Aznar, P. 1926. Algunas investigaciones clinicas y experimentales sobre la fiebre recurrente española. Arch. Inst. Nac. Hig. Alfonso XIII 4:121-127. [Google Scholar]
- 3.Bunikis, J., U. Garpmo, J. Tsao, J. Berglund, D. Fish, and A. G. Barbour. 2004. Sequence typing reveals extensive strain diversity of the Lyme borreliosis agents Borrelia burgdorferi in North America and Borrelia afzelii in Europe. Microbiology 150:1741-1755. [DOI] [PubMed] [Google Scholar]
- 4.Bunikis, J., J. Tsao, U. Garpmo, J. Berglund, D. Fish, and A. G. Barbour. 2004. Typing of Borrelia relapsing fever group strains. Emerg. Infect. Dis. 10:1661-1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Connolly, S. E., and J. L. Benach. 2001. Cutting edge: the spirochetemia of murine relapsing fever is cleared by complement-independent bactericidal antibodies. J. Immunol. 167:3029-3032. [DOI] [PubMed] [Google Scholar]
- 6.Connolly, S. E., and J. L. Benach. 2005. The versatile roles of antibodies in Borrelia infections. Nat. Rev. Microbiol. 3:411-420. [DOI] [PubMed] [Google Scholar]
- 7.Connolly, S. E., D. G. Thanassi, and J. L. Benach. 2004. Generation of a complement-independent bactericidal IgM against a relapsing fever Borrelia. J. Immunol. 172:1191-1197. [DOI] [PubMed] [Google Scholar]
- 8.Cutler, S. J., J. Moss, M. Fukunaga, D. J. Wright, D. Fekade, and D. Warrell. 1997. Borrelia recurrentis characterization and comparison with relapsing-fever, Lyme-associated, and other Borrelia spp. Int. J. Syst. Bacteriol. 47:958-968. [DOI] [PubMed] [Google Scholar]
- 9.Cutler, S. J., J. C. Scott, and D. J. M. Wright. 2008. Phylogenetic origins of Borrelia recurrentis. Int. J. Med. Microbiol. 298:193-202.17765656 [Google Scholar]
- 10.Davis, G. E. 1952. The relapsing fevers: tick-spirochete specificity studies. Exp. Parasitol. 1:406-410. [Google Scholar]
- 11.De Buen, E., and P. De la Cámara. 1931. Notas sobre 59 casos de fiebre recurrente española. Bol. Tecn. Dir. San. 6:193-207. [Google Scholar]
- 12.Fukunaga, M., Y. Ushijima, L. Y. Aoki, and A. Talbert. 2001. Detection of Borrelia duttonii, a tick-borne relapsing fever agent in central Tanzania, within ticks by flagellin gene-based nested polymerase chain reaction. Vector Borne Zoonotic Dis. 1:331-338. [DOI] [PubMed] [Google Scholar]
- 13.Garcia-Monco, J. C., N. S. Miller, P. B. Backenson, P. Anda, and J. L. Benach. 1997. A mouse model of Borrelia meningitis after intradermal injection. J. Infect. Dis. 175:1243-1245. [DOI] [PubMed] [Google Scholar]
- 14.Gebbia, J. A., J. C. Monco, J. L. Degen, T. H. Bugge, and J. L. Benach. 1999. The plasminogen activation system enhances brain and heart invasion in murine relapsing fever borreliosis. J. Clin. Invest. 103:81-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kumar, S., K. Tamura, and M. Nei. 2004. MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief. Bioinform. 5:150-163. [DOI] [PubMed] [Google Scholar]
- 16.LaRocca, T. J., L. I. Katona, D. G. Thanassi, and J. L. Benach. 2008. Bactericidal action of a complement-independent antibody against relapsing fever Borrelia resides in its variable region. J. Immunol. 180:6222-6228. [DOI] [PubMed] [Google Scholar]
- 17.León de Lope, M., A. A. Leal Luna, J. Gálvez Aceval, J. Royo Aguado, E. Romero Velasco, and F. Martínez Luengas. 1985. Borreliosis: descripcion de 4 casos. Med. Clin. (Barc.) 85:111-113. [PubMed] [Google Scholar]
- 18.Lescot, M., S. Audic, C. Robert, T. T. Nguyen, G. Blanc, S. J. Cutler, P. Wincker, A. Couloux, J. M. Claverie, D. Raoult, and M. Drancourt. 2008. The genome of Borrelia recurrentis, the agent of deadly louse-borne relapsing fever, is a degraded subset of tick-borne Borrelia duttonii. PLoS Genet. 4:e1000185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.López-Cortés, L., F. Lozano de Léon, J. M. Gómez-Mateos, A. Sánchez-Porto, and C. Obrador. 1989. Tick-borne relapsing fever in intravenous drug abusers. J. Infect. Dis. 159:804. [PubMed] [Google Scholar]
- 20.Malkiel, S., C. J. Kuhlow, P. Mena, and J. L. Benach. 2009. The loss and gain of marginal zone and peritoneal B cells is different in response to relapsing fever and Lyme disease Borrelia. J. Immunol. 182:498-506. [DOI] [PubMed] [Google Scholar]
- 21.Margos, G., S. A. Vollmer, M. Cornet, M. Garnier, V. Fingerle, B. Wilske, A. Bormane, L. Vitorino, M. Collares-Pereira, M. Drancourt, and K. Kurtenbach. 2009. A new Borrelia species defined by multilocus sequence analysis of housekeeping genes. Appl. Environ. Microbiol. 75:5410-5416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nordstrand, A., I. Bunikis, C. Larsson, K. Tsogbe, T. G. Schwan, M. Nilsson, and S. Bergstrom. 2007. Tickborne relapsing fever diagnosis obscured by malaria, Togo. Emerg. Infect. Dis. 13:117-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Postic, D., M. Garnier, and G. Baranton. 2007. Multilocus sequence analysis of atypical Borrelia burgdorferi sensu lato isolates—description of Borrelia californiensis sp. nov., and genomospecies 1 and 2. Int. J. Med. Microbiol. 297:263-271. [DOI] [PubMed] [Google Scholar]
- 24.Ras, N. M., B. Lascola, D. Postic, S. J. Cutler, F. Rodhain, G. Baranton, and D. Raoult. 1996. Phylogenesis of relapsing fever Borrelia spp. Int. J. Syst. Bacteriol. 46:859-865. [DOI] [PubMed] [Google Scholar]
- 25.Richter, D., D. Postic, N. Sertour, I. Livey, F. R. Matuschka, and G. Baranton. 2006. Delineation of Borrelia burgdorferi sensu lato species by multilocus sequence analysis and confirmation of the delineation of Borrelia spielmanii sp. nov. Int. J. Syst. Evol. Microbiol. 56:873-881. [DOI] [PubMed] [Google Scholar]
- 26.Rudenko, N., M. Golovchenko, L. Grubhoffer, and J. H. Oliver, Jr. 2009. Borrelia carolinensis sp. nov., a new (14th) member of the Borrelia burgdorferi sensu lato complex from the southeastern region of the United States. J. Clin. Microbiol. 47:134-141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sánchez Porto, A., M. Crespo Anguita, C. Iglesias, and M. Lucio Villegas. 1991. Brote de borreliosis: a proposito de 7 casos. Enf. Infecc. Microbiol. Clin. 9:125. [PubMed] [Google Scholar]
- 28.Sánchez-Yebra, W., Y. Díaz, P. Molina, J. Sedeno, P. Giner, M. Vitutia, and P. Anda. 1997. Fiebre recurrente transmitida por garrapatas. Descripcion de 5 casos. Enf. Infecc. Microbiol. Clin. 15:77-81. [PubMed] [Google Scholar]
- 29.Scott, J. C. 2005. Typing African relapsing fever spirochetes. Emerg. Infect. Dis. 11:1722-1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The CLUSTAL_X Windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]