Abstract
We report on a leukemic patient who suffered from a persistent, generalized, and eventually fatal Staphylococcus epidermidis infection during prolonged aplasia. Over a 6-week period, we isolated a genetically and phenotypically unstable S. epidermidis strain related to an epidemic clone associated with hospital infections worldwide. Strikingly, the strain showed a remarkable degree of variability, with evidence of selection and increasing predominance of biofilm-producing and oxacillin-resistant variants over time. Thus, in the early stages of the infection, the strain was found to generate subpopulations which had spontaneously lost the biofilm-mediating ica locus along with the oxacillin resistance-conferring mecA gene. These deletion mutants were obviously outcompeted by the ica- and mecA-positive wild-type genotype, with the selection and predominance of strongly biofilm-forming and oxacillin-resistant variants in the later stages of the infection. Also, a switch from protein- to polysaccharide intercellular adhesin/poly-N-acetylglucosamine (PIA/PNAG)-mediated-biofilm production was detected among ica-positive variants in the course of the infection. The data highlight the impact of distinct S. epidermidis clonal lineages as serious nosocomial pathogens that, through the generation and selection of highly pathogenic variants, may critically determine disease progression and outcome.
Coagulase-negative staphylococci (CoNS) contribute significantly to catheter-related bloodstream infections after hematopoietic stem cell transplantation (HSCT) (15). Recent studies identified defined clonal lineages for CoNS that persist in health care units and might spread even within or between hospitals (3, 8, 19). Various multilocus sequence typing (MLST) studies have identified an epidemic Staphylococcus epidermidis clone, named sequence type ST27 (or, alternatively, ST2, according to a revised MLST scheme) (12), as the cause of a majority of nosocomial S. epidermidis infections in numerous hospital settings (8, 10, 12, 20). In S. epidermidis, biofilm formation has been recognized as a crucial factor for the colonization of medical devices (reviewed in references 13 and 14). Biofilms can be generated by various mechanisms, and polysaccharide intercellular adhesin/poly-N-acetylglucosamine (PIA/PNAG)-mediated-biofilm formation is a very common one (13). The enzymes that facilitate PIA/PNAG synthesis are encoded by the four-gene operon icaADBC, which is present in many disease-associated S. epidermidis strains, including ST27/ST2 strains (9, 10). Here, we report on a leukemic patient who received an allogeneic HSCT and experienced a generalized S. epidermidis infection. We describe the affiliation of the strain to the epidemic ST27/ST2 clone and investigate genomic instability and variations in biofilm expression and oxacillin resistance that occurred in the course of the infection.
Case Report
A 23-year-old male patient was treated with an allogeneic cord-blood HSCT for a second relapse of a high-risk common-B-cell acute lymphoblastic leukemia. On day 5 posttransplant, the patient became febrile. Piperacillin-tazobactam treatment was started empirically and, when the fever persisted, was supplemented with caspofungin treatment on day 13. S. epidermidis grew in three of the four blood culture bottles (Table 1) (isolates 1 and 2). The central venous line was removed.
TABLE 1.
Microbiological and genetic properties of S. epidermidis isolates
| Isolate sample | Day after transplantation | Sample source | Biofilm formationa | Result for icaA and icaC PCR | Oxacillin susceptibilityb | Result for mecA PCR | SCCmec type | MLST | No. of IS256-specific fragments |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 13 | Blood culture | Negative | Negative | S | Negative | VIc | ST27/ST2d | 4 |
| 2 | Blood culture | Positive (protein) | Positive | R | Positive | VI | ST27/ST2 | 5 | |
| 3 | 29 | Blood culture | Negative | Negative | S | Negative | VIc | ST27/ST2d | 4 |
| 4 | Blood culture | Positive (protein) | Positive | R | Positive | VI | ST27/ST2 | 5 | |
| 5 | 34 | Blood culture | Positive (protein) | Positive | R | Positive | VI | ST27/ST2 | 5 |
| 6 | Blood culture | Negative | Negative | S | Negative | VIc | ST27/ST2d | 4 | |
| 7 | 40 | Blood culture | Positive (protein) | Positive | R | Positive | VI | ST27/ST2 | 5 |
| 8 | Blood culture | Negative | Negative | S | Negative | VIc | ST27/ST2d | 4 | |
| 9 | 52 | Blood culture | Strongly positive (PIA) | Positive | R | Positive | VI | ST27/ST2 | 5 |
| 10 | Blood culture | Strongly positive (PIA) | Positive | R | Positive | VI | ST27/ST2 | 5 | |
| 11 | 55 | CSF | Strongly positive (PIA) | Positive | R | Positive | VI | ST27/ST2 | 5 |
| 12 | CSF | Strongly positive (PIA) | Positive | R | Positive | VI | ST27/ST2 | 5 |
“PIA” and “protein” indicate PIA-mediated biofilms and protein-mediated biofilms, respectively.
S, sensitive; R, resistant.
Isolates that displayed an SCCmec type-VI pattern, but for which the mecA gene was not detected.
Isolates that lacked the arcC gene, which is part of both MLST schemes.
The patient was treated with vancomycin (2 × 1 g intravenously [i.v.]) for 10 days. Blood cultures were negative during treatment. Five days (day 29) after vancomycin treatment was stopped, he developed bacteremia again; four of four bottles were positive for S. epidermidis (Table 1) (isolates 3 and 4). The central venous line was removed, and i.v. treatment with vancomycin and catheter-lock therapy was begun. Blood cultures remained positive for S. epidermidis (Table 1) (isolates 5 to 8); because of low trough levels of vancomycin (i.e., 4.4 μg/ml), the dosage was increased to 2 × 1.5 g. The central line was replaced again. The patient then developed a painful bilateral swelling above the tibial muscles with redness and tenderness. The level of creatinine kinase increased from 21 U/liter to 400 U/liter. Magnetic resonance imaging (MRI) revealed TH2 hyperintensity along the musculus peroneus longus and musculus extensor hallucis, with collection of fluid in the muscle, but no abnormalities of the fascias, favoring the diagnosis of myositis. A repeat MRI 5 days later showed involvement of the anterior and posterior tibial muscles (data not shown).
On day 42, still in aplasia, histological results from biopsies at different sites in both tibial muscles showed necrosis and clusters of coccoid bacteria. The culture yielded S. epidermidis. Amikacine was added to vancomycin. Daily blood cultures then were sterile until day 52, when another blood culture yielded S. epidermidis (Table 1) (isolates 9 and 10). On day 55, the patient complained of a headache and presented nuchal rigidity. An MRI of the brain showed two 4- to 6-mm size-enhancing lesions in the prefrontal cortex, which is consistent with the presence of septic emboli. A spinal tap produced a clear cerebrospinal fluid (CSF) that had a slightly elevated cell count (5.3 × 106 cells/liter), a protein load of 576 mg/liter, and the presence of S. epidermidis (Table 1) (isolates 11 and 12). Because of its good penetration into the central nervous system, rifampin (2 × 600 mg i.v.) was added to vancomycin, and treatment with amikacine was stopped. After 70 days of nonengraftment, a haplo-identical rescue transplantation from the mother was performed. The patient remained febrile, but daily blood cultures were negative. On day 72, the patient developed acute respiratory failure with rapidly progressive acute respiratory distress syndrome (ARDS). Despite mechanical ventilation and extracorporal oxygenation, the patient died 2 days later of multiorgan failure. The autopsy did not reveal any sign of leukemia or any sign of endocarditis.
MATERIALS AND METHODS
Strain cultivation and characterization.
Samples were cultured on blood agar plates, and representative colonies from various morphotypes were selected and analyzed by standard microbiological techniques and 16S rRNA gene sequencing, respectively. PCR detection of the icaA and icaC genes was performed as described previously (21).
Biofilm testings.
Biofilm formation was tested on 96-well, polystyrene tissue culture plates as described previously (21), using tryptone soya broth (TSB; Oxoid) as a growth medium (17.0 g/liter pancreatic digest of casein, 3.0 g/liter papaic digest of soybean meal, 5.0 g/liter sodium chloride, 2.5 g/liter dipotassium hydrogen phosphate, 2.5 g/liter glucose). S. epidermidis RP62A and Staphylococcus carnosus TM300 were used as positive and negative controls, respectively. To determine whether the biofilm was mediated by PIA production or by proteins, biofilms were treated with sodium periodate and proteinase K, respectively (7).
In vitro detection of biofilm-negative variants.
Generation of biofilm-negative variants in vitro was established using Congo red agar (CRA) as a biofilm indicator medium (4), and frequencies were calculated as described previously (21). Briefly, a single bacterial colony was picked and diluted in phosphate-buffered saline. Appropriate aliquots of this dilution were used to inoculate 50 ml tryptone soya broth, and CFU in the inoculum were determined by plating and incubating the corresponding aliquots on tryptone soy agar. Following incubation overnight at 37°C with agitation, the optical density of the liquid culture was determined and serial dilutions were plated onto CRA. Upon incubation for 18 h at 37°C and at least another 12 h at room temperature, biofilm-forming colonies appear on CRA as black colonies, while biofilm-negative variants are red. Total CFU on the plates as well as black and red colonies were counted, and the frequency of the occurrence of biofilm-negative variants per cell and generation was calculated using the equation 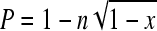 , in which x is defined as the number of red colonies divided by the number of total CFU. n represents the number of generations which are determined by the equation n = (log N − log N0)/(log 2), where N0 is the number of bacterial cells in the inoculum and N is the total number of bacterial cells in the liquid culture after incubation.
, in which x is defined as the number of red colonies divided by the number of total CFU. n represents the number of generations which are determined by the equation n = (log N − log N0)/(log 2), where N0 is the number of bacterial cells in the inoculum and N is the total number of bacterial cells in the liquid culture after incubation.
DNA fingerprinting by PFGE.
Genomic DNA for pulsed-field gel electrophoresis (PFGE) was prepared and resolved in a CHEF-DR III apparatus as described previously (5).
MLST.
MLST was performed as described previously (20) and by using the revised scheme according to Miragaia et al. (12).
SCCmec typing and IS256 detection.
Staphylococcal cassette chromosome mec (SCCmec) typing and mecA PCR were performed as previously described (11). To detect IS256 copies, chromosomal DNA was isolated and then EcoRI digested, blotted, and hybridized with an IS256-specific gene probe (21).
RESULTS
Primary cultivation of blood and CSF samples resulted regularly in the appearance of colonies differing in size and color. They were picked and identified as S. epidermidis by 16S rRNA sequencing. Antibiotic resistance analyses and PCR detection of the methicillin/oxacillin resistance-mediating mecA gene revealed the simultaneous presence of both oxacillin-resistant/mecA-positive and oxacillin-susceptible/mecA-negative colonies in blood culture samples for days 13 to 40, while at a later stage of infection (days 52 and 55), isolates were uniformly oxacillin resistant (Table 1).
The mecA gene is usually located on SCCmec cassettes that represent large, structurally diverse, integrated genomic islands of the staphylococcal chromosome, which can be detected by a multiplex PCR approach (11). SCCmec typing identified a type VI cassette in all isolates, but with no mecA gene in the oxacillin-sensitive variants (Table 1). Notably, SCCmec type VI-specific PCR fragments were still detectable in the mecA-negative/oxacillin-sensitive variants, suggesting that the SCCmec cassette was lost in part and that the deleted fragment included the mecA gene.
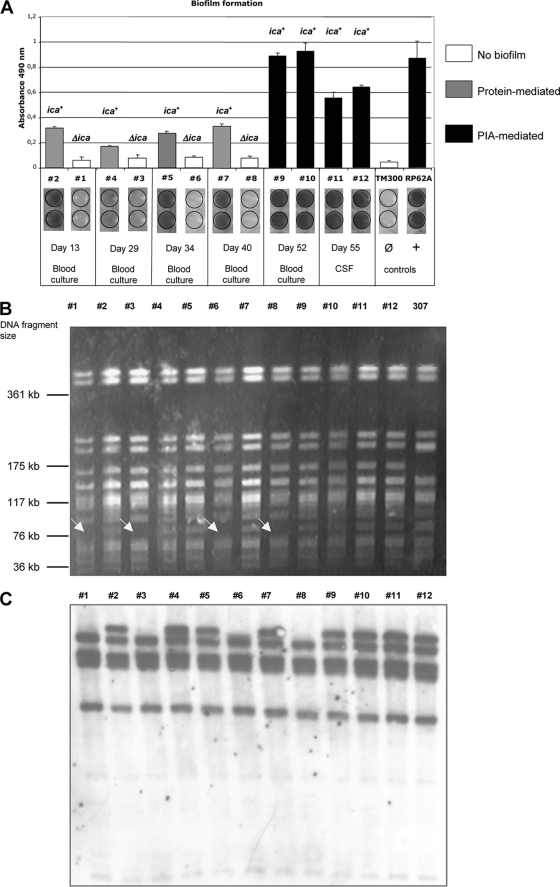
Interestingly, differences in oxacillin resistance were clearly associated with various biofilm-forming capacities. Thus, in biofilm-forming tests in vitro, mecA-negative isolates were found to be biofilm negative, while mecA-positive isolates were biofilm positive, which was in good agreement with the detection of the ica genes in biofilm-positive isolates (Fig. 1A and Table 1). ica- and mecA-positive colonies coexisted with ica- and mecA-negative colonies in samples obtained at the beginning of the infection, whereas later (days 52 and 55), only biofilm-forming, ica- and mecA-positive colonies prevailed (Fig. 1A). Even more striking was the difference in the amounts of biofilm produced by ica-positive isolates. Thus, isolates 2, 4, 5, and 7 (days 13 to 40) generated less biofilm than isolates 9 to 12, which were grown from blood and cerebrospinal fluid samples from a later stage of infection (days 52 and 55). Proteinase K and metaperiodate treatment of the biofilms demonstrated that weaker biofilms were mediated by protein but that the strong biofilms recorded at the end were due to PIA production (Fig. 1A).
FIG. 1.
(A) Biofilm-forming capacity of the S. epidermidis isolates (#1 to #12) on polystyrene tissue culture plates. S. epidermidis RP62A and Staphylococcus carnosus TM300 represent positive and negative controls, respectively. (B) PFGE of SmaI-restricted genomic DNA. #1 to #12, S. epidermidis patient isolates; 307, S. epidermidis strain 307 (see Discussion for details). Arrows indicate variants in which a DNA fragment of ∼100 kb in size is lacking. (C) IS256-specific Southern hybridization of EcoRI-restricted chromosomal DNA. #1 to #12, S. epidermidis patient isolates.
As the mobile genetic element IS256 has been reported to be involved in variations of biofilm formation in S. epidermidis (1, 21), the isolates were also tested for this element. Southern hybridization of chromosomal DNA revealed the presence of multiple IS256 copies in all isolates, with uniform hybridization patterns within mecA- and ica-positive and mecA- and ica-negative variants, but with aberrations between those groups. Thus, the ica-positive variants exhibited five IS256-specific bands, whereas ica-negative isolates displayed only four such bands (Fig. 1C).
The findings so far strongly suggested a mixed infection by at least two different S. epidermidis strains. Molecular typing by PFGE, however, revealed nearly identical restriction patterns pointing to a clonal origin of the isolates (Fig. 1B). The only difference recorded was the regular absence of a 100-kb DNA fragment in mecA- and ica-negative isolates (identified by arrows in Fig. 1B). When two different MLST schemes were applied, all isolates were identified as ST27 (20) or, alternatively, as ST2 (12), confirming again the clonality of the isolates. It is noteworthy that all mecA- and ica-negative variants also lacked the arcC gene, which is part of both MLST schemes (Table 1). To further establish the clonality of the isolates, we analyzed whether or not mecA- and ica-negative variants may arise from a mecA- and ica-positive genotype in vitro. For this purpose, we grew the biofilm-forming, ica- and mecA-positive isolate 2 on Congo red agar, an indicator medium on which biofilm-formers appear as black colonies, while biofilm-negative variants are red. A single black colony was picked, and appropriate dilutions were spread on CRA plates. Following incubation, both black and red colonies arose on the indicator medium, with red colonies occurring at a frequency of 10−2 per cell and generation. Biofilm tests as well as ica- and mecA-specific PCRs demonstrated a biofilm-negative phenotype in the red colonies, which was again accompanied by lack of the ica and mecA genes, respectively. Finally, the PFGE restriction patterns of the variants were found to be identical to those of the ica- and mecA-negative deletion mutants detected in the patient (data not shown). In summary, these experiments demonstrate the genomic instability of the ica and mecA genes as a molecular background for the observed heterogeneity rather than a mixed infection by different strains.
DISCUSSION
S. epidermidis is primarily a commensal bacterium with a low level of pathogenic potential, and often, it is difficult to decide whether a specific isolate is the cause of an infection or represents an unspecific contamination. The appearance of various colony morphotypes in the primary cultures obtained from our patient mimicked a mixed culture, suggesting contamination. However, molecular typing revealed a common clonal origin for all the isolates. The strain was identified as ST27/ST2, an S. epidermidis sequence type that is currently emerging as an epidemic nosocomial pathogen in medical facilities worldwide (8, 10, 12, 20). It is tempting to speculate that the “success” and wide distribution of ST27/ST2 strains are linked to the specific genetic makeup of these strains, which may be illustrated by the fact that the vast majority of the strains carry the ica operon and are capable of producing biofilms (10). Moreover, these strains seem to acquire and readily exchange mobile genetic elements, such as various SCCmec cassettes and IS elements, the latter being important genetic factors associated with genome flexibility (10, 12).
The hypervariable ST27/ST2 S. epidermidis strain detected in our patient generated a multitude of phenotypic and genetic variants. The prolonged persistence in the patient's blood cultures and the ensuing medical complications might, at least in part, be explained by bacterial biofilm production, which is often associated with reduced in vivo antibiotic susceptibility. Biofilm formation and genetic adaptability are crucial factors employed by S. epidermidis to cope with changing environmental conditions, such as antibacterial host response and antibiotic treatment. In our case, biofilm formation and oxacillin susceptibility proved to be variable features. They were associated with the conservation or deletion of the ica and mecA genes. Interestingly, biofilm and resistance variants coexisted only at the beginning of the infection. Later in infection, ica- and mecA-negative variants appeared to be outcompeted by strongly biofilm-producing and oxacillin-resistant variants (Fig. 1A). Interestingly, ica-positive variants isolated early in infection (between days 13 and 40) produced protein-mediated rather than PIA-mediated biofilms, although the icaADBC operon was present in those isolates (Fig. 1A). Preliminary transcriptome studies revealed no significant differences in icaADBC operon transcription between the ica-positive variants obtained early and late in infection (S. M. K. Schoenfelder and W. Ziebuhr, unpublished data). The molecular basis for the lack of PIA production remains to be elucidated but may be linked to differing metabolic regulation. Also, the factors involved in protein-mediated-biofilm formation at the beginning of the infection are not established yet and require further analysis. Possible candidates for future studies are the accumulation associated protein Aap, the biofilm-associated protein Bhp, and other cell wall-associated proteins known to be involved in biofilm production (13).
Although the molecular details are unknown, recent in vitro studies clearly demonstrate that S. epidermidis is capable of switching between different modes of biofilm formation (i.e., PIA versus protein) (7). Our data suggest that this mechanism may also have biological relevance in the dynamic process of an infection. It is generally thought that PIA-mediated-biofilm production provides a selective advantage in established, device-associated infection, while representing a loss of fitness when the isolates colonize the skin (2, 16). In line with this idea, competition experiments in a device-related animal model using ica mutants and PIA-producing strains revealed growth defects in the ica mutants (2). Over time, the mutants were outcompeted by their ica wild-type counterparts, giving rise to the selection of strong biofilm formers in the course of the infection (2), which is in perfect agreement with the observed predominance of PIA-producing variants in our patient later in the infection. In contrast, in a human skin colonization model, a PIA-producing variant was shown to be outcompeted by its isogenic ica mutant, suggesting that PIA is not required or even detrimental when S. epidermidis colonizes the skin (16). In light of these findings, it is tempting to speculate that PIA production might be controlled by the environment that the bacteria encounter and that in our patient, the PIA-negative variants early in infection might have switched off PIA production, as their gene expression status still resembles that of skin colonizers. In an established device-related infection in the bloodstream, PIA might be required and beneficial for escaping the innate immune system and for withstanding the action of antibiotics (13, 18), which would explain the strong PIA production later in infection. However, more experimental work is needed to substantiate this hypothesis and to elucidate the regulatory mechanisms involved in this process.
The genetic mechanisms causing the spontaneous generation of ica- and mecA-negative variants is an interesting part of the study. The variants were found to lack a large DNA fragment that also includes the ica genes and part of the SCCmec type VI cassette (including mecA) (Fig. 1B and Table 1). Interestingly, these deletions could also be triggered in vitro, where they occurred at a surprisingly high frequency of 10−2 per cell and generation. Typically, S. epidermidis biofilm variations are known to occur in the order of magnitude of 10−5 to 10−6 per cell and generation and were found to be associated with classical phase variation events, mostly by inactivation of biofilm genes or their corresponding regulators (1, 6, 21). Deletion of large genomic DNA fragments at such a high frequency is a novel phenomenon, and at the present stage of experimental work, we have no compelling evidence for an underlying mechanism. However, the deletion was accompanied by a diminished number of IS256 copies (Fig. 1C and Table 1). We made similar observations in a recent in vitro study using another ST27/ST2-related strain, called S. epidermidis 307 (10). The strain was isolated from a leukemia patient in Germany in 1994 and had PFGE restriction patterns that were very similar to those seen in isolates obtained from our current patient, demonstrating a close genetic relationship (Fig. 1B) (isolate 307). Interestingly, in S. epidermidis 307, the deletions of the ica genes were correlated with IS256 activity (M. Eckart and W. Ziebuhr, unpublished data). These data and the differences observed in the IS256-specific hybridization patterns among the patient's isolates lead us to speculate that similar IS256-mediated mechanisms may also occur in vivo.
This report, together with previously published data, demonstrates that specific S. epidermidis clonal lineages may pose a risk to vulnerable patients. The identification of strains belonging to these clonal lineages requires specific attention because, due to their genetic and phenotypic variability, these strains can easily be misidentified as contaminants. Positive blood cultures with CoNS are generally challenging for clinicians, and usually, antibiotic treatment is initiated only when more than one bottle is positive and the patient has a central venous catheter or indwelling foreign material, e.g., an artificial heart valve. ST27/ST2 isolates further complicate this difficult decision-making process, as phenotypically mixed cultures of CoNS are often not regarded as true pathogens, resulting in inappropriate or even no treatment. Given the virulence of these clones, this misinterpretation may put patients at a significant risk. Notably, the biofilm-forming capacity of ST27/ST2 isolates is a serious problem that requires specific treatment efforts. Thus, our patient had a central venous line which was changed repeatedly. However, in spite of an adequate management, blood cultures remained positive. Only upon addition of rifampin, an antibiotic known to act against biofilm-forming staphylococci (17, 22), did blood cultures eventually turn negative, indicating a critical role for biofilm formation in the persistence of the infection. The worldwide occurrence of ST27/ST2 strains in hospital settings suggests that the pathogenic potential of specific S. epidermidis clonal lineages should be reconsidered. The timely and correct identification of these clones may prompt adequate interventions and improve patient management. Systematic surveillance programs may help to closely monitor and prevent the further spread of these strains, thereby lowering the risk and costs of health care-associated infections in the future.
Acknowledgments
We thank M. Battegay for critical review of and valuable comments on the manuscript.
W. Ziebuhr is supported by grants from the Deutsche Forschungsgemeinschaft (Transregional Collaborative Research Centre 34), by the Federal Ministry of Education and Research (Network Pathogenomics-plus; grant PTJ-BIO//03U213B), and by the Department for Employment and Learning (Northern Ireland) through its “Strengthening the All-Island Research Base” program. S. M. K. Schoenfelder holds a European Social Fund Scholarship awarded through Queen's University Belfast, and M. Weisser and U. Flückiger thank the Stiftung Forschung Infektionskrankheiten for financial support.
Footnotes
Published ahead of print on 26 May 2010.
REFERENCES
- 1.Conlon, K. H., H. Humphreys, and J. P. O'Gara. 2004. Inactivations of rsbU and sarA by IS256 represent novel mechanisms of biofilm phenotypic variation in Staphylococcus epidermidis. J. Bacteriol. 186:6208-6219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Flückiger, U., M. Ulrich, A. Steinhuber, G. Döring, D. Mack, R. Landmann, C. Goerke, and C. Wolz. 2005. Biofilm formation, icaADBC transcription, and polysaccharide intercellular adhesin synthesis by staphylococci in a device-related infection model. Infect. Immun. 73:1811-1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Foka, A., V. Chini, E. Petinaki, F. Kolonitsiou, E. D. Anastassiou, G. Dimitracopoulos, and I. Spiliopoulou. 2006. Clonality of slime-producing methicillin-resistant coagulase-negative staphylococci disseminated in the neonatal intensive care unit of a university hospital. Clin. Microbiol. Infect. 12:1230-1233. [DOI] [PubMed] [Google Scholar]
- 4.Freeman, D. J., F. R. Falkiner, and C. T. Keane. 1989. New method for detecting slime production by coagulase negative staphylococci. J. Clin. Pathol. 42:872-874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Goering, R. V., and M. A. Winters. 1992. Rapid method for epidemiological evaluation of gram-positive cocci by field inversion gel electrophoresis. J. Clin. Microbiol. 30:577-580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Handke, L. D., K. M. Conlon, S. R. Slater, S. Elbaruni, F. Fitzpatrick, H. Humphreys, W. P. Giles, M. E. Rupp, P. D. Fey, and J. P. O'Gara. 2004. Genetic and phenotypic analysis of biofilm phenotypic variation in multiple Staphylococcus epidermidis isolates. J. Med. Microbiol. 53:367-374. [DOI] [PubMed] [Google Scholar]
- 7.Hennig, S., S. Nyunt Wai, and W. Ziebuhr. 2007. Spontaneous switch to PIA-independent biofilm formation in an ica-positive Staphylococcus epidermidis isolate. Int. J. Med. Microbiol. 297:117-122. [DOI] [PubMed] [Google Scholar]
- 8.Klingenberg, C., A. Ronnestad, A. S. Anderson, T. G. Abrahamsen, J. Zorman, A. Villaruz, T. Flaegstad, M. Otto, and J. E. Sollid. 2007. Persistent strains of coagulase-negative staphylococci in a neonatal intensive care unit: virulence factors and invasiveness. Clin. Microbiol. Infect. 13:1100-1111. [DOI] [PubMed] [Google Scholar]
- 9.Kozitskaya, S., S. H. Cho, K. Dietrich, R. Marre, K. Naber, and W. Ziebuhr. 2004. The bacterial insertion sequence element IS256 occurs preferentially in nosocomial Staphylococcus epidermidis isolates: association with biofilm formation and resistance to aminoglycosides. Infect. Immun. 72:1210-1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kozitskaya, S., M. E. Olson, P. D. Fey, W. Witte, K. Ohlsen, and W. Ziebuhr. 2005. Clonal analysis of Staphylococcus epidermidis isolates carrying or lacking biofilm-mediating genes by multilocus sequence typing. J. Clin. Microbiol. 43:4751-4757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Milheiriço, C., D. C. Oliveira, and H. de Lencastre. 2007. Update to the multiplex PCR strategy for assignment of mec element types in Staphylococcus aureus. Antimicrob. Agents Chemother. 51:3374-3377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Miragaia, M., J. C. Thomas, I. Couto, M. C. Enright, and H. de Lencastre. 2007. Inferring a population structure for Staphylococcus epidermidis from multilocus sequence typing data. J. Bacteriol. 189:2540-2552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.O'Gara, J. P. 2007. ica and beyond: biofilm mechanisms and regulation in Staphylococcus epidermidis and Staphylococcus aureus. FEMS Microbiol. Lett. 270:179-188. [DOI] [PubMed] [Google Scholar]
- 14.Otto, M. 2009. Staphylococcus epidermidis—the ‘accidental’ pathogen. Nat. Rev. Microbiol. 7:555-567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Poutsiaka, D. D., L. L. Price, A. Ucuzian, G. W. Chan, K. B. Miller, and D. R. Snydman. 2007. Blood stream infection after hematopoietic stem cell transplantation is associated with increased mortality. Bone Marrow Transplant. 40:63-70. [DOI] [PubMed] [Google Scholar]
- 16.Rogers, K. L., M. E. Rupp, and P. D. Fey. 2008. The presence of icaADBC is detrimental to the colonization of human skin by Staphylococcus epidermidis. Appl. Environ. Microbiol. 74:6155-6157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Villain-Guillot, P., M. Gualtieri, L. Bastide, and J. P. Leonetti. 2007. In vitro activities of different inhibitors of bacterial transcription against Staphylococcus epidermidis biofilm. Antimicrob. Agents Chemother. 51:3117-3121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vuong, C., J. M. Voyich, E. R. Fischer, K. R. Braughton, A. R. Whitney, F. R. DeLeo, and M. Otto. 2004. Polysaccharide intercellular adhesin (PIA) protects Staphylococcus epidermidis against major components of the human innate immune system. Cell. Microbiol. 6:269-275. [DOI] [PubMed] [Google Scholar]
- 19.Widerstrom, M., T. Monsen, C. Karlsson, and J. Wistrom. 2006. Molecular epidemiology of meticillin-resistant coagulase-negative staphylococci in a Swedish county hospital: evidence of intra- and interhospital clonal spread. J. Hosp. Infect. 64:177-183. [DOI] [PubMed] [Google Scholar]
- 20.Wisplinghoff, H., A. E. Rosato, M. C. Enright, M. Noto, W. Craig, and G. L. Archer. 2003. Related clones containing SCCmec type IV predominate among clinically significant Staphylococcus epidermidis isolates. Antimicrob. Agents Chemother. 47:3574-3579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ziebuhr, W., V. Krimmer, S. Rachid, I. Loessner, F. Götz, and J. Hacker. 1999. A novel mechanism of phase variation of virulence in Staphylococcus epidermidis: evidence for control of the polysaccharide intercellular adhesin synthesis by alternating insertion and excision of the insertion sequence element IS256. Mol. Microbiol. 32:345-356. [DOI] [PubMed] [Google Scholar]
- 22.Zimmerli, W., A. F. Widmer, M. Blatter, R. Frei, and P. E. Ochsner. 1998. Role of rifampin for treatment of orthopedic implant-related staphylococcal infections: a randomized controlled trial. Foreign-Body Infection (FBI) Study Group. JAMA 279:1537-1541. [DOI] [PubMed] [Google Scholar]