Abstract
Williamsia serinedens has been isolated from soil but has not yet been implicated in human disease. We report the first case of perinatal sepsis caused by a dual-morphotype form of Williamsia serinedens in a 31-year-old pregnant woman hospitalized with preterm labor.
CASE REPORT
A 31-year-old woman who was 23 weeks pregnant was admitted to our hospital for management of preterm labor in the setting of cervical insufficiency and twin gestation. She had originally presented to her obstetrician 1 week earlier for routine prenatal care, at which time she was found to have cervical dilation at 5 cm. At the time of admission, the patient was asymptomatic with normal vital signs and unremarkable physical examination. Pelvic ultrasonography revealed a shortened cervix and twin fetuses with normal heart tones, motion, and size. The patient was started on tocolytic therapy utilizing indomethacin and magnesium sulfate, as well as ceftazidime for empirical management of chorioamnionitis.
Eight weeks into her hospitalization, the patient reported generalized myalgias and malaise, although no fever was documented initially. After routine hematology revealed a leukocyte (WBC) count of 9,900 WBCs/mm3 with 34% band forms, an amniocentesis was performed, revealing only 22 WBCs/mm3 (reference range, <30 WBCs/mm3) with culture yielding no growth. Empirical meropenem was initiated, and two sets of blood cultures collected 25 h apart, one from her percutaneously inserted central catheter (PICC) and the other from venipuncture, yielded aerobic, Gram-positive bacilli which could not be identified by our laboratory. The patient's antibiotic therapy was then changed to ampicillin and vancomycin, her PICC line was removed, and the tip was cultured, yielding no growth.
Despite empirical therapy, the patient's clinical course worsened with the development of headache, fever, hypotension, and persistence of left shift. After a lumbar puncture revealed only 1 WBC/mm3 (reference range, <4 WBC/mm3), cesarean delivery was performed at 32 weeks gestation with successful delivery of twins. The twins had identical Apgar scores of 7 and 9. Placental pathology revealed mild villous edema but no structural anomaly or inflammation. The patient improved thereafter with resolution of fever and sterilization of blood cultures. She completed a 2-week course of intravenous vancomycin and oral ampicillin. Blood cultures collected from each of her neonates on the day of delivery yielded no growth. As of this submission, both she and her infants have recovered and are healthy.
The isolate was transferred from our laboratory to the Tennessee Department of Health laboratory, where it was identified as an aerobic actinomycete. Morphologically, it was observed as a pleomorphic Gram-positive, non-acid-fast coccobacillus, whose macrocolonies on brain heart infusion (BHI) agar at 4 to 7 days growth were orange, of soft texture, and with mycelial edges. It was submitted to the Centers for Disease Control and Prevention actinomycete reference laboratory, where it was reported as Williamsia serinedens by 16S rRNA gene sequence analysis. The isolate was forward from the CDC laboratory to the unit of bacterial taxonomy at the Institute of Medical Microbiology and Immunology of the University of Bonn, Germany, for further characterization.
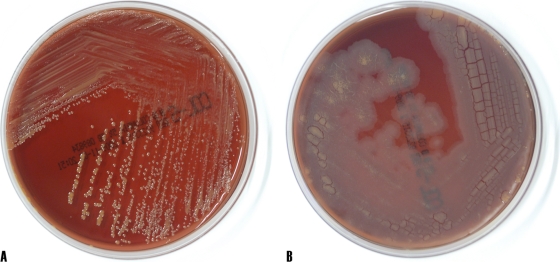
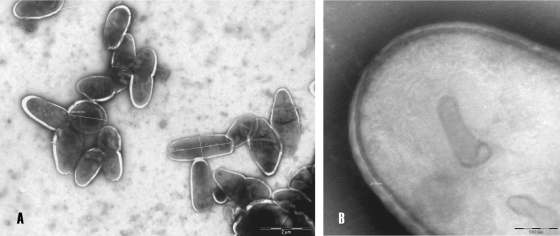
Morphological examination of the isolate IMMIB W-9660 revealed the following findings. The organism was aerobic, catalase positive, and oxidase negative. Its colonies on Columbia agar plates containing 5% sheep blood (BD Stacker Plates), were smooth, round, slightly convex, orange, and 1 to 3 mm in diameter (Fig. 1AA). Gram stain revealed Gram-positive coccobacilli. Detailed morphology of the cell was observed using a transmission electron microscope (model Philips CM-10) for cultures grown on LB broth medium at 34°C for 48 h. Negative staining of cells was performed with 1% phosphotungstic acid. The cells ranged from coccoid with a diameter of 1.4 μm to irregular rods with a length of 2.5 μm and a width of 0.86 μm (Fig. 2A) and were surrounded with 0.021- to 0.028-μm-thick regular walls (Fig. 2B).
FIG. 1.
Morphological appearance of Williamsia serinedens isolate IMMIB W-9660 on Columbia blood agar demonstrating S morphotype (A) and R morphotype (B).
FIG. 2.
Transmission electron micrographs of negatively stained cells of isolate IMMIB W-9660T (S morphotype) grown on LB broth at 34°C for 48 h (A) and cell wall detail (B). Bars, 2 μm (A) and 100 nm (B).
We then studied the effect of different temperatures (5, 10, 20, 27, 30, 34, and 40°C) on the growth of isolate IMMIB W-9660. Abnormalities in colony morphology were monitored daily for up to 30 days. At 10 to 34°C, the isolate exhibited growth; however, the organism failed to grow at 5°C and at 40°C. Prolonged incubation (10 to 30 days) at room temperature led to dissociation of the smooth colonies into a flat, rough colony morphology (Fig. 1B). Repeated subculture of the flat rough colonies confirmed stability of the rough morphotype in that they did not revert to smooth colonies.
Phenotypic characterization of isolate IMMIB W-9660 was performed using tests to determine carbon source utilization and hydrolysis of various substrates as described previously (12). There were no differences in the biochemical characteristics of the two variants, and the results were in accordance with those reported for W. serinedens DSM 45037T, with the exception that, for both morphotypes, nonutilization of p-hydroxybenzoic acid, sodium lactate, and 1,2-propandiol as carbon sources was confirmed.
Further identification was performed by 16S rRNA gene sequencing. Genomic DNA extraction and PCR-mediated amplification of the 16S rRNA gene were carried out using established procedures (8). The purified PCR products were sequenced using a Taq DyeDeoxy Terminator cycle sequencing kit (Applied Biosystems) as described by the manufacturer. A Genetic Analyzer (310; Applied Biosystems) was used for electrophoresis of the sequence reaction products. Phylogenetic and molecular evolutionary analyses were conducted using the ARB database (5). The phylogenetic tree was generated by the neighbor-joining method (9). Topologies of the neighbor-joining tree were evaluated using bootstrap analyses, based on 1,000 resamplings (2). It can be seen in Fig. 3 that both variants (S and R) of isolate IMMIB W-9660 were recovered within the Williamsia 16S rRNA gene clade and clustered together with W. serinedens, an association supported by a 100% bootstrap value in the neighbor-joining analysis. The 16S rRNA gene sequences of the S variant (1,480 bp) and R variant (1,477 bp) of isolate IMMIB W-9660 showed the highest sequence similarity (99.9% similarity) with the type strain of W. serinedens and with the type strain of W. deligens (97.9% similarity).
FIG. 3.
Phylogenetic analysis using the neighbor-joining method based on 16S rRNA gene sequences of representative Williamsia species showing the position of the S and R morphotypes of isolate IMMIB W-9660. The numbers at the nodes indicate the level of bootstrap support of >70% based on analyses of 1,000 resampled data sets. Bar, 1.0% sequence divergence. Arcanobacterium haemolyticum was used as an outgroup.
Antimicrobial drug susceptibility pattern and MIC determinations were performed using the Etest strips (AB Biodisk). Mueller-Hinton (MH) agar was inoculated with a 3-McFarland standard suspension of the isolate, and Etest strips containing amikacin (256 μg ml−1), ampicillin (256 μg ml−1), doxycycline (256 μg ml−1), imipenem (32 μg ml−1), linezolid (256 μg ml−1), meropenem (32 μg ml−1), oxacillin (256 μg ml−1), penicillin G (256 μg ml−1), tobramycin (1,024 μg ml−1), trimethoprim-sulfamethoxazole (32 μg ml−1), and vancomycin (256 μg ml−1) were placed on the plate. After 48 h of incubation at 30°C in ambient air, MIC results showed that both S and R morphotypes were susceptible to amikacin (0.38 μg ml−1), ampicillin (0.094 μg ml−1), doxycycline (0.50 μg ml−1), imipenem (0.008 μg ml−1), linezolid (0.125 μg ml−1), meropenem (0.023 μg ml−1), penicillin G (0.023 μg ml−1), tobramycin (0.50 μg ml−1), and vancomycin (0.38 μg ml−1). Both morphotypes were resistant to oxacillin (>256 μg ml−1) and trimethoprim-sulfamethoxazole (>32 μg ml−1).
The genus Williamsia was proposed by Kämpfer et al. to accommodate actinomycetes with atypical cell morphology that are unable to grow at 5 or at 45°C and possess mycolic acids with carbon chain lengths of 50 to 56 (4). Based on mycolic acid structure, Williamsia takes an intermediate position between Rhodococcus (mycolic acid chain lengths of 34 to 45) and Gordonia (mycolic acid chain lengths of 54 to 66) (4). The genus Williamsia currently comprises six recognized species: Williamsia muralis, isolated from indoor building material of a children's day care center in Finland (4); Williamsia maris, isolated from deep sediments of the Sea of Japan (10); Williamsia deligens, isolated from human blood (11); Williamsia serinedens, isolated from an oil-contaminated soil (13); Williamsia marianensis, isolated from deep-sea sediment (7); and Williamsia faeni, isolated from a hay meadow (3).
With the exception of W. deligens, most isolates have come only from environmental samples, and there is a paucity of evidence suggesting human pathogenicity. Two cases have been reported in the literature in which Williamsia species have been implicated as true pathogens: one case of respiratory tract infection and one case of endophthalmitis, both of which were caused by W. muralis (1, 6). Although W. deligens has been isolated from human blood, no infections have been reported (11). To our knowledge, this is the first report of bacteremia and clinical sepsis caused by W. serinedens.
As the ecology and epidemiology of Williamsia species are not well understood, it is difficult to know what factors led to our patient's sepsis with Williamsia serinedens, although her percutaneously inserted central catheter represents the most probable source of infection. Nevertheless, it is pertinent to note that our isolate's ability to undergo an S/R phase variation may be linked to its pathogenicity. We speculate that the S morphotype, which probably represents the principal environmental form, spontaneously dissociates into the R morphotype, its pathogenic form, after prolonged incubation in ambient air. In this regard, the R variant may be selected in vivo. It is important to note that the phenomenon of the S/R variation has been observed in W. deligens, which was first isolated from human blood. The phenomenon of dual colony morphology of Williamsia strains isolated from clinical specimens raises the question of a possible link between colony morphology and virulence of Williamsia species. However, as the molecular basis for the S or R appearance of Williamsia species is presently unknown, further study is needed to identify the factors associated with the S-to-R switch of Williamsia and to determine how pathogenicity is conveyed.
This is the first report of sepsis caused by Williamsia species and, to our knowledge, the first case of a Williamsia serinedens infection. As the phylogenetic tree of the Corynebacterineae continues to evolve and expand through the development of molecular identification techniques, it is likely that as more species emerge, identification of human infection will be more frequently recognized.
Nucleotide sequence accession numbers.
The 16S rRNA gene sequences from the W. serinedens IMMIB W-9660 (S variant) and IMMIB W-9660 (R variant) strains were submitted to GenBank and assigned the accession numbers FN673549 and FN673550, respectively.
Footnotes
Published ahead of print on 12 May 2010.
REFERENCES
- 1.del Mar Tomas, M., R. Moure, J. A. Saez Nieto, S. Fojon, A. Fernandez, M. Diaz, R. Villanueva, and G. Bou. 2005. Williamsia muralis pulmonary infection. Emerg. Infect. Dis. 11:1324-1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Felsenstein, J. 1985. Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783-791. [DOI] [PubMed] [Google Scholar]
- 3.Jones, A. L., G. D. Payne, and M. Goodfellow. 2009. Williamsia faeni sp. nov., a novel actinomycete isolated from a hay meadow. Int. J. Syst. Evol. Microbiol. [Epub ahead of print.] doi: 10.1099/ijs.0.015826-0. [DOI] [PubMed]
- 4.Kampfer, P., M. A. Andersson, F. A. Rainey, R. M. Kroppenstedt, and M. Salkinoja-Salonen. 1999. Williamsia muralis gen. nov., sp. nov., isolated from the indoor environment of a children's day care centre. Int. J. Syst. Bacteriol. 49(Pt. 2):681-687. [DOI] [PubMed] [Google Scholar]
- 5.Ludwig, W., O. Strunk, R. Westram, L. Richter, H. Meier, Yadhukumar, A. Buchner, T. Lai, S. Steppi, G. Jobb, W. Forster, I. Brettske, S. Gerber, A. W. Ginhart, O. Gross, S. Grumann, S. Hermann, R. Jost, A. Konig, T. Liss, R. Lussmann, M. May, B. Nonhoff, B. Reichel, R. Strehlow, A. Stamatakis, N. Stuckmann, A. Vilbig, M. Lenke, T. Ludwig, A. Bode, and K. H. Schleifer. 2004. ARB: a software environment for sequence data. Nucleic Acids Res. 32:1363-1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Murray, R. J., M. Aravena-Roman, and P. Kampfer. 2007. Endophthalmitis due to Williamsia muralis. J. Med. Microbiol. 56:1410-1412. [DOI] [PubMed] [Google Scholar]
- 7.Pathom-Aree, W., Y. Nogi, I. C. Sutcliffe, A. C. Ward, K. Horikoshi, A. T. Bull, and M. Goodfellow. 2006. Williamsia marianensis sp. nov., a novel actinomycete isolated from the Mariana Trench. Int. J. Syst. Evol. Microbiol. 56:1123-1126. [DOI] [PubMed] [Google Scholar]
- 8.Rainey, F. A., N. Ward-Rainey, R. M. Kroppenstedt, and E. Stackebrandt. 1996. The genus Nocardiopsis represents a phylogenetically coherent taxon and a distinct actinomycete lineage: proposal of Nocardiopsaceae fam. nov. Int. J. Syst. Bacteriol. 46:1088-1092. [DOI] [PubMed] [Google Scholar]
- 9.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 10.Stach, J. E., L. A. Maldonado, A. C. Ward, A. T. Bull, and M. Goodfellow. 2004. Williamsia maris sp. nov., a novel actinomycete isolated from the Sea of Japan. Int. J. Syst. Evol. Microbiol. 54:191-194. [DOI] [PubMed] [Google Scholar]
- 11.Yassin, A. F., and H. Hupfer. 2006. Williamsia deligens sp. nov., isolated from human blood. Int. J. Syst. Evol. Microbiol. 56:193-197. [DOI] [PubMed] [Google Scholar]
- 12.Yassin, A. F., F. A. Rainey, H. Brzezinka, J. Burghardt, H. J. Lee, and K. P. Schaal. 1995. Tsukamurella inchonensis sp. nov. Int. J. Syst. Bacteriol. 45:522-527. [DOI] [PubMed] [Google Scholar]
- 13.Yassin, A. F., C. C. Young, W. A. Lai, H. Hupfer, A. B. Arun, F. T. Shen, P. D. Rekha, and M. J. Ho. 2007. Williamsia serinedens sp. nov., isolated from an oil-contaminated soil. Int. J. Syst. Evol. Microbiol. 57:558-561. [DOI] [PubMed] [Google Scholar]