Abstract
Innate immunity operates as a first line of defense in multicellular organisms against infections caused by different classes of microorganisms. Antimicrobial peptides (AMPs) are synthesized constitutively in barrier epithelia to protect against microbial attack and are also upregulated in response to infection. Here, we implicate Drifter/Ventral veinless (Dfr/Vvl), a class III POU domain transcription factor, in tissue-specific regulation of the innate immune defense of Drosophila. We show that Dfr/Vvl is highly expressed in a range of immunocompetent tissues, including the male ejaculatory duct, where its presence overlaps with and drives the expression of cecropin, a potent broad-spectrum AMP. Dfr/Vvl overexpression activates transcription of several AMP genes in uninfected flies in a Toll pathway- and Imd pathway-independent manner. Dfr/Vvl activates a CecA1 reporter gene both in vitro and in vivo by binding to an upstream enhancer specific for the male ejaculatory duct. Further, Dfr/Vvl and the homeodomain protein Caudal (Cad) activate transcription synergistically via this enhancer. We propose that the POU protein Dfr/Vvl acts together with other regulators in a combinatorial manner to control constitutive AMP gene expression in a gene-, tissue-, and sex-specific manner, thus promoting a first-line defense against infection in tissues that are readily exposed to pathogens.
Innate immunity is an evolutionarily conserved host defense system used by multicellular organisms to protect against harmful effects of microorganisms. The innate immune system is multilayered with protective physical and chemical barriers, as well as constitutive and induced responses to infection. Most multicellular organisms produce antimicrobial peptides (AMPs), which are capable of killing microbes directly, including bacteria, fungi, viruses, and protozoa (1, 2, 10). AMPs are produced constitutively in local barrier epithelial structures that are in direct contact with microbes, creating a microbicidal surface that inhibits infection and defends the body from invading microbes (80). Additionally, by virtue of pathways that have been studied extensively with Drosophila in recent years, AMPs are produced in response to infection, both systemically and locally (36, 74). Signaling via the Toll and IMD pathways leads to nuclear translocation of the Drosophila NF-κB/Rel factors Dorsal, Dif, and Relish (Rel), and subsequent activation of different and sometimes overlapping sets of AMP genes (reviewed in references 20, 27, 36, and 74).
Mammals harbor an innate immune system that resembles that of insects (reviewed in reference 34). In fact, the mammalian innate immune system utilizes Toll-like receptor (TLR) signaling (3, 34) in pathways homologous to the Toll pathway, which was first discovered in Drosophila. Recognition of microbes by the innate immune system is based on binding of pathogen-encoded molecules to soluble or membrane-bound sensors in the host, such as the TLRs, nucleotide-binding oligomerization domains (NODs), and peptidoglycan recognition proteins (PGRPs) (54, 56, 69). In mammals, the innate immune response and the TLRs also signal to and communicate with the adaptive immune system (72). Similar to the role of the Toll pathway in insects, mammalian TLR signaling functions in the production of AMPs in response to an infection (7, 35, 37, 47, 76).
Mammalian AMPs, such as cathelicidins and α- and β-defensins (65), are capable of killing microbes directly, as are insect AMPs (10), by virtue of mechanisms that have been studied extensively in vitro (66, 80). A comprehensive picture of the in vivo mode of action of AMPs is missing, but in a knockout mouse model, cathelicidin/LL-37/CRAMP has been shown to protect against infections in the skin and the urinary tract and against tuberculosis (16, 37, 49). Similarly, mice overexpressing a human intestinal β-defensin were protected against enteric salmonellosis (60), while matrilysin-deficient mice, which lack mature α-defensins, showed impaired antibacterial capacity (78). In addition, it has been suggested that mammalian AMPs may act as secondary signals to other layers of the immune system (35, 47, 80). Despite our knowledge of the importance of AMPs in defending against microbial infection, surprisingly little is known about how tissue-specific constitutive AMP gene expression is regulated.
In insects, like in mammals, AMP genes are constitutively expressed in a tissue-specific manner, in addition to their coordinated expression in response to infection. In a study of the tissue specificity of seven different Drosophila AMP genes, Tzou et al. (73) reported that individual AMP genes are both expressed constitutively in restricted sets of tissues and induced more broadly in barrier epithelia after direct contact with bacteria. Although the transcription factors regulating the systemically induced response to bacteria are well known (i.e., NF-κB/Rel factors, see above and references 20 and 36), much less is known about the control of tissue-specific expression of AMPs and the transcription factors that interact to coordinate the constitutive and inducible responses (74).
Presumably, protection of epithelial surfaces against infection was part of an ancient defense system for multicellular organisms, and many aspects of this defense are likely to have been conserved through evolution. Based on this hypothesis, we used Drosophila as a model to identify transcription factors that activate constitutive expression of AMP genes. One candidate regulatory protein identified previously is the Drosophila homeodomain protein Caudal (Cad). Cad was shown to be involved in both positive and negative regulation of AMP genes in different epithelial tissues (58, 59). We recently isolated another transcription factor, the POU protein Drifter/Ventral veinless (Dfr/Vvl), in a yeast screen for transcriptional regulators of Drosophila AMP gene expression (32). Here, we have investigated the role(s) of Dfr/Vvl in AMP gene expression in vivo.
The POU domain proteins belong to a subclass of the family of homeodomain proteins that are characterized by a bipartite DNA-binding domain, referred to as the POU domain (26, 71), which was first described for the mammalian transcription factors Pit1 and Oct1/Oct2 and the Caenorhabditis elegans Unc-86 protein (26). In the Drosophila genome, five genes encoding POU proteins have been identified (11). The Drosophila class III POU factor Dfr/Vvl is an essential factor during development. Dfr/Vvl mutants die during embryogenesis due to defects in tracheal development and in commissural development in the central nervous system (CNS), the brain, and motoneurons, which is consistent with expression of Dfr/Vvl in the developing embryo, the brain, oenocytes, and wing discs of developing larvae (5, 6, 8, 12, 15, 18, 28, 38, 43, 81). Dfr/Vvl mutations affect wing development by virtue of regulation of the vestigial gene (13), and Dfr/Vvl was shown to regulate the neuronal expression of Dopa decarboxylase (Ddc) (13, 30, 31), a gene recently shown to also play a role in epithelial immunity (17).
In the present study, we show that Dfr/Vvl is localized to the nuclei of tissues known to express AMPs constitutively and that there is a strong correlation between nuclear Dfr/Vvl and constitutive expression of the broad-spectrum AMP gene CecA1 in restricted parts of the male reproductive organs. We show that overexpression of Dfr/Vvl in flies was sufficient to upregulate several AMP genes in an infection-independent manner. AMP gene upregulation was not dependent on functional Toll or Imd pathways and did not lead to upregulation of Rel expression, indicating that Dfr/Vvl is involved in constitutive, rather than infection-induced, expression of AMP genes. Dfr/Vvl binding and activation of CecA1 expression were mapped to an upstream enhancer required for constitutive AMP expression in the male ejaculatory duct. We further show that Dfr/Vvl interacted synergistically with the homeodomain protein Cad to activate CecA1 and that this synergism required specific sequences in the CecA1 ejaculatory duct enhancer. These results indicate that Dfr/Vvl is a crucial regulator of AMP gene expression and suggest that homeodomain proteins may interact in multicellular organisms to establish a constitutive, protective antimicrobial barrier in tissues and organs that are frequently invaded by potentially harmful microorganisms.
MATERIALS AND METHODS
Fly stocks, culture, and infections.
Fly stocks were from the Bloomington stock center, unless specified. Flies for overexpression of Dfr/Vvl (w1118; UAS-Dfrwt/MKRS and w; UAS-dsdfr/CyO wg) and for downregulation of Dfr/Vvl by RNA interference (RNAi) (w; UAS-dsdfr/TM3 Sb and w; dfr B1.29 UAS dsdfr/TM6B Tb Ubi-GFP) were a gift from S. Certel and are described in references 14 and 15). The CecA1-lac Z constructs are described in references 53 and 55. The w; CecA1-GFP-drs stocks were a gift from J. L. Imler (73). Imd1 and MyD88KG03447 flies carry hypomorphic alleles with strong immune defects in the IMD and Toll pathways, respectively. Flies with a recombination of hs-Gal4 and CecA1-GFP transgenes were created by recombining w; P(w[+mC] = GAL4Hsp70.PB)2 and w; CecA1-GFP-drs on the second chromosome. The following GAL4 drivers were used: daughterless-GAL4 (w1118; P{da-GAL4.w−}3), Collagen IV-GAL4 (w1118; P{Cg-GAL4.A}2), Hemolectin-GAL4 (w1118; P{Hml-GAL4.G}5-6), Larval serum protein 2-GAL4 (w1118; P{Lsp2-GAL4.H}3), hs-GAL4 (w; P{w[+mC] = GAL4-Hsp70.PB}89-2-1), 69B-GAL4 (w1118; P{GawB} 69B), Caudal-GAL4 (P{UAS-cad.R}), c564-GAL4 (w1118; P{GawB}c564), and c729-GAL4 (w1118; P{GawB}c729). Flies were maintained on standard cornmeal agar medium in vials and kept in humid culture rooms at 25°C, with a 12-h-light/12-h-dark cycle. Flies were kept in mixed populations for at least 5 days after eclosion to reduce differences in AMP gene expression levels due to differences in mating experiences. Recordings of AMP gene expression were done with males and females separately. Infections were done with a 1:1 mixture of overnight cultures of Micrococcus luteus and Enterobacter cloacae β-12, which were washed once in phosphate-buffered saline (PBS; pH 7). Larvae or flies were injected with the bacterial suspension using a fine glass capillary needle with a microinjector (TriTech Research, Los Angeles, CA). Injected larvae and flies were kept under humid conditions at 25°C for the desired time.
Overexpression of Dfr/Vvl in flies.
Overexpression of Dfr/Vvl using the UAS-GAL4 system (9) via the strong, ubiquitous da-GAL4 driver resulted in lethality, probably reflecting sensitivity to excess Dfr/Vvl during development. Tissue-specific GAL4 drivers expressed in hemocytes and/or lymph glands (Cg-GAL4 and Hml-GAL4) led to melanotic tumors, and the progeny died as first or second instar larvae. When Dfr/Vvl was overexpressed in the fat body of third instar larvae (Lsp 2-GAL4), most of the larvae died prior to pupariation. The use of an hs-GAL4 driver promoted GAL4 expression at levels that were tolerable in combination with UAS-Dfrwt, when crosses were maintained at 25°C, and then flies were either transferred to 29°C for at least 5 days to promote continuous leaky expression or heat shocked twice for 30 min at 37°C with a 120-min recovery at 29°C in between, to give a peak of high-level expression.
To determine whether ectopic Dfr/Vvl expression was sufficient to activate AMP gene expression, different GAL4 lines were used to drive UAS-Dfr/Vvl expression in combination in transgenic flies harboring a CecA1-GFP reporter construct. Strong GAL4 drivers, such as Da-GAL4 and 69B-GAL4, were lethal, and no escapers could be retrieved. Using c564-GAL4 (fat body) and Cad-GAL4 (gut and abdominal tissues), flies eclosed but died after 1 or 2 days. Dissection of newly eclosed flies revealed severe malformations and even complete absence of the tissues/organs in which Dfr/Vvl had been overexpressed, indicating that growth and differentiation processes were greatly disturbed. Instead hs-GAL4 was recombined with CecA1-GFP on the second chromosome, crossed with UAS-Dfr/MKRS Sb and reared at 18°C to minimize Dfr/Vvl expression, and this allowed female escapers to hatch. Heat shock at 37°C for 60 min promoted CecA1-GFP in the fat body and intestine of these flies, but not in control flies.
Downregulation of Dfr/Vvl in flies.
Downregulation of Dfr/Vvl was done using flies carrying a UAS-dsDfr/Vvl construct recombined onto a chromosome carrying a null allele of Dfr/Vvl, (dfrB129 UAS-dsDfr/Vvl) (15). The 791-bp fragment from the 5′ gene of Dfr/Vvl used to create the inverted repeat construct is specific for the Dfr/Vvl gene (12), without any predicted off-targets in the Drosophila genome. Several different GAL4 drivers were tested, with Omb-GAL4 as a positive control for downregulation of Dfr/Vvl, which gave the expected Dfr/Vvl mutant wing phenotype. The da-GAL4 driver resulted in embryonic lethality at 25°C and 29°C when combined with dfrB129 UAS-dsDfr/Vvl. When da-GAL4/dfrB129; UAS-dsDfr/Vvl flies were raised at 18°C, the third instar larvae displayed several phenotypic defects, such as melanized spots and regions in the cuticle, malpighian tubules, and gastric ceca, and abnormal morphology of the fat body, and all animals died prior to eclosion. The use of a hemocyte- and fat body-specific GAL4 driver, c729-GAL4, resulted in melanized tissues and death prior to eclosion. Expression of dsDfr/Vvl from an hs-GAL4 driver (w; P(w[+mC] = GAL4Hsp70.PB)89-2-1) was tolerable when crosses were maintained at 25°C. Eclosed offspring were aged for 5 days at 29°C to allow leaky GAL4 expression, optimal GAL4/UAS interaction, and subsequent depletion of Dfr/Vvl mRNA. The efficiency of the downregulation varied, and it was not possible to consistently reproduce it, probably due to differences in the expression levels of dsDfr/Vvl, but most likely also due to autoregulation of the Dfr/Vvl gene, which previously has been demonstrated (12).
Gateway cloning.
Expression plasmids of Dfr/Vvl and Cad were made by PCR-based isolation of coding sequences from respective cDNA, followed by directional TOPO cloning into Gateway pENTRY vectors, according to the manufacturer's instructions (Invitrogen). Destination vectors (pAW) designed for use in Drosophila were obtained from Terence D. Murphy, and recombination was made using Gateway LR Clonase enzyme mix according to the manufacturer's instructions (Invitrogen).
Mutation of putative Dfr/Vvl binding sites in the CecA1-luc plasmid.
Mutations in the putative Dfr/Vvl binding sites were created by inverse PCR with phosphorylated primers. To induce the mutation (mutated bases shown in italics) in CecA1mutE1-luc, the primers E1F (5′-CCCCAATTATTTTTTATTGTCATTTAATGC-3′) and E1R (5′-ACTGAGCACGTACGAATCTGA-3′) were used; in CecA1mutE2A-luc, E2AF (GGGATTGTCATTTAATGCCTATTGAA) and E2AR (5′-AAATAATTGTTTACTGAGCACG-3′) were used; in CecA1mutE2-luc, E2BF (5′-GGGAATGCCTATTGAATTTTTCAAAC-3′) and E2BR (5′-TGACAATAAAAAATAATTGTTTAC-3′) were used; in CecA1mutE3-luc, E3F (5′-GGGAGTGCCTTTAGTAAAATATTGTAG-3′) and E3R (5′-TTAAGTTTGAAAAATTCAATAGG-3′) were used; and in CecA1Δ-luc, CecΔF (5′-CCTTTAGTAAAATATTGTAGTG-3′) and CecΔR (5′-GCACGTACGAATCTGAAAACTT-3′) were used. The PCR consisted of the following: 98°C for 2 min, followed by 5 cycles of 50°C for 30 s, 72°C for 4 min, and 98°C for 30 s, and then 30 cycles of 55°C for 30 s, 72°C for 4 min, and 98°C for 30 s, and a final extension of 7 min at 72°C, using Phusion high-fidelity DNA polymerase (Finnzymes). PCR products were gel purified, ligated overnight at 4°C, and transformed into chemically competent subcloning efficiency DH5α (Invitrogen), and recovered plasmids were sequenced to confirm recovery of the desired mutations.
Cell cultures and transfections.
Drosophila mbn-2 cells (23) were grown at 25°C and transfected with CecA1-luciferase constructs, as described in reference 59. Transfections were done with 3 μg of pPacPL-Drifter (39) and 3 μg pAW-Caudal plasmids, or empty pAct5CPL vector, mixed with 100 ng of CecA1-luciferase reporter constructs (59) and 100 ng of pPacPL-lacZ as an internal standard.
RT-PCR.
Reverse transcriptase quantitative PCR (RT-qPCR) and semiquantitative RT-PCR were carried out essentially as previously described (32). When possible, primers and/or probes covered intron/exon boundaries to ensure specific amplification of cDNA. Sequences of probes/primers for Dfr/Vvl (CG10037), Cad (CG1759), Rel (CG11992), DptA (CG12763), CecA1 (CG1365), Drs (CG10810), AttA (CG10146), AttB (CG18372), Def (CG1385), Mtk (CG8175) Anp (CG1361), and Rp49 (CG7939) are available upon request. RNA extraction, DNase treatment, reverse transcription, and PCR were carried out as previously described (32). All samples were analyzed in triplicate, and the measured mRNA concentration was normalized relative to the control Rp49 values. The normalized data were used to quantify the relative levels of a given mRNA according to cycle threshold (2−ΔΔCT) analysis (64).
Protein extraction and immunoblotting.
Protein extraction and immunoblotting were performed essentially as described previously (70), using 50 μg of fly protein extract per lane. After transfer to a Hybond-P membrane (Amersham Biosciences), the membrane was blocked in Tris-buffered saline (TBS) plus 10% milk for 3 h, incubated with rat anti-Dfr/Vvl antibody (1:1,000) overnight at 4°C, and incubated with enhanced chemiluminescence (ECL) rat IgG, horseradish peroxidase (HRP)-linked antibody (from goat) (1:10,000) (GE Healthcare). Washing and development using an ECL Western blot detection kit was performed according to the manufacturer's instructions (Amersham Biosciences). Signals were recorded with a charge-coupled-device (CCD) camera and results analyzed with IR LAS-1000 Pro software.
Bacterial expression and DNA-binding experiments.
Full-length Dfr/Vvl protein fused to the glutathione-S-transferase epitope (GST-Dfr/Vvl) (13) was produced in protease-deficient Escherichia coli BL21 cells (Invitrogen). Untagged GST protein was expressed from the pGEX-5X1 vector (GE Healthcare). Overexpression was induced with 0.5 mM isopropyl-β-d-thiogalactopyranoside (IPTG) at an optical density at 600 nm (OD600) of 0.5 and carried out at 30°C for 4 h. Extraction and purification using glutathione-Sepharose beads were according to the manufacturer's instructions (GE Healthcare). Binding of GST-Dfr/Vvl fusion or GST alone (control protein) to the CecA1 promoter was examined using an electrophoretic mobility shift assay (EMSA). Single-stranded oligonucleotides were annealed and end labeled with [γ-32P]dATP (GE Healthcare) using T4 kinase (Invitrogen). Oligonucleotide sequences were −663 to −644 bp, 5′-d(CCGAAGTAATTCTGCACTGC) (control); −493 to −454 bp, 5′-d(TAAATAAGTTTTCAGATTCGTACGTGCTCAGTAAACAATT) (E1); −458 to −419 bp, 5′-d(CAATTATTTTTTATTGTCATTTAATGCCTATTGAATTTTT) (E2); and −423 to −380 bp, 5′-d(TTTTTCAAACTTAATTTAGTGCCTTTAGTAAAATATTGTAGTGA) (E3). The EMSA was carried out as previously described (75), using 400 ng of the purified GST-Dfr/Vvl fusion or GST alone, 60 μg of bovine serum albumin, 3 μg of poly(dI-dC), and 1 ng of labeled probe in 20 μl EMSA buffer (75).
Immunostaining and β-Gal staining.
Antibody staining of dissected larval and adult tissues was performed as described previously (50). Cryostat sections of adults were prepared as previously described (55), and immunostaining of the sectioned tissues as described for dissected tissues. Primary rabbit antibody against the Dfr/Vvl protein (kindly provided by Sarah Certel) was used at a 1:2,000 dilution; secondary antibodies were Cy2-conjugated goat anti-rat IgG (1:200) and Cy3-conjugated donkey anti-rat IgG (1:500; both from Jackson Immuno Research Laboratories, Inc.). Simultaneous visualization of Dfr/Vvl immunostaining and CecA1-GFP reporter gene expression was performed with a Zeiss Axioscope equipped for epi-illuminations and recorded with a Nikon Coolpix 4500. For observation of CecA1-lacZ reporter gene expression in male reproductive organs, flies were dissected, fixed, and stained for β-galactosidase (β-Gal) activity using X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) as the substrate, as described in reference 55.
RESULTS
Drifter/Ventral veinless is expressed and localized to the nuclei of immunocompetent tissues.
We previously isolated the POU protein Dfr/Vvl in a yeast screen for regulators of Drosophila AMP genes (32). To reveal sites of possible in vivo regulation of AMPs by Dfr/Vvl, we investigated the distribution of Dfr/Vvl protein in uninfected and infected wild-type (wt) larvae and flies. Interestingly, we found that in uninfected flies, Dfr/Vvl was highly expressed in immune-responsive tissues, such as fat body and barrier epithelia of the digestive, respiratory, and reproductive tracts (Fig. 1 and 2). In larvae, Dfr/Vvl immunostaining was predominantly nuclear in fat body tissue (Fig. 1A), salivary glands (Fig. 1B), trachea (Fig. 1C), gastric ceca (Fig. 1F), epidermis (Fig. 1J), oenocytes, and the ring gland (data not shown). In some tissues, such as the proventriculus (Fig. 1G) and hindgut (Fig. 1H), Dfr/Vvl immunostaining was very intense and more uniformly distributed between the cytoplasm and nucleus. In others, such as the malpighian tubules, staining was consistently uniform (Fig. 1D) or exclusively nuclear (Fig. 1E) in different regions. In the midgut, Dfr/Vvl staining was weak (Fig. 1F and H), and we did not observe any Dfr/Vvl staining in lymph glands or hemocytes (data not shown). In addition, strong nuclear Dfr/Vvl immunoreactivity was found in the brain and several imaginal discs (data not shown), as previously reported by others (6, 18).
FIG. 1.
Dfr/Vvl is localized to the nuclei of fat body, digestive system, and barrier epithelia in uninfected larvae. Shown are immunostaining of Dfr/Vvl (red) in dissected tissues from third instar larvae and staining of nuclei with DAPI (4′,6-diamidino-2-phenylindole) (blue). Strong nuclear Dfr/Vvl staining was evident in the fat body (fb) (A and A′), salivary glands (sg) (B and B′), and trachea (tr) (C and C′). In the malpighian tubules (malp), different regions reproducibly conferred either equal distributions between the nuclei and cytoplasm (D and D′) or predominantly nuclear distribution (E and E′). In the digestive system, Dfr/Vvl staining was intense in the proventriculus (pv) (F, G, and G′), in gastric ceca (gc) (F), and in the hindgut (hg) (H and H′). In the midgut (mg), Dfr/Vvl staining was weak and diffuse (F and H). Also shown are differential interference contrast micrograph of the larval cuticle (cut) and epidermis (epi), with two ventral denticle belts (db) visible (J) and fluorescence micrographs of the same specimen showing nuclear localization of Dfr/Vvl (J′) and DAPI (J") in epidermal cells. Tissues/organs were named according to M. Demerec (45).
FIG. 2.
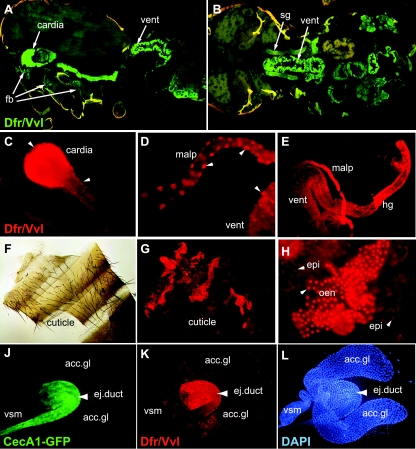
Dfr/Vvl is expressed in immunocompetent barrier epithelia of uninfected flies. Frozen cryostat sections of the thorax and abdomen of a fly in sagittal (A) and horizontal (B) sections revealed strong Dfr/Vvl immunostaining (green) in the digestive tract, especially in the cardia, ventriculus (vent), and salivary glands (sg). In addition, Dfr/Vvl was apparent in the fat body (fb). Anterior is to the left. (C to E) Dissected organs from adults confirmed strong Dfr/Vvl immunoreactivity (red) in the digestive system, in the cardia, malpighian tubules (malp), ventriculus (vent), and hindgut (hg). Dissected cuticle of a male fly in bright field (F) and in epifluorescence (G and H) revealed intense nuclear Dfr/Vvl immunostaining in oenocytes lying just beneath the cuticle (G) and in higher magnification (H). (H) Weak staining in nuclei of epidermal cells (epi) was also evident. Arrowheads point to nuclear Dfr/Vvl staining. Dissected male reproductive organs from an uninfected fly show the distribution of CecA1-GFP (J), the Dfr/Vvl protein (K), and nuclear DAPI (L) staining. Expression was strong in the ejaculatory duct (ej.duct) but absent from the accessory glands (acc.gl.), seminal vesicle (vsm), and testis (not shown).
In adults, Dfr/Vvl immunostaining was apparent in the digestive tract in sections of whole flies, represented by immunofluorescence in the cardia (foregut), ventriculus (midgut), and salivary glands (Fig. 2A and B). Strong Dfr/Vvl immunostaining was also observed with dissected tissues of the digestive systems, including the cardia (Fig. 2C), malpighian tubules (Fig. 2D and E), midgut/ventriculus (Fig. 2D and E), and hindgut (Fig. 2E) but not observed with the esophagus or rectum (data not shown). As in larvae, some regions revealed an exclusively nuclear Dfr/Vvl staining, such as in the upper region of the malpighian tubules (Fig. 2D), while in other regions of the digestive system the staining was present in both the nucleus and cytoplasm. We did not observe any tissues with predominant cytoplasmic staining. In male flies, intense nuclear Dfr/Vvl staining was observed in the ejaculatory duct (Fig. 2K) but not in the accessory glands, seminal vesicle, or testis of the male reproductive organs (Fig. 2K and L). Dfr/Vvl was not detected in any part of the female reproductive tract (data not shown), but otherwise the distributions of Dfr/Vvl immunostaining were similar in male and female flies. In addition, nuclear Dfr/Vvl staining was evident in the fat body (Fig. 2A), epidermis, and oenocytes located just beneath the cuticle (Fig. 2F to H), as well as in the adult trachea (data not shown). Significantly, most of the tissues and organs that are known sites of constitutive AMP expression in both larvae and adults (22, 73)—proventriculus/cardia, midgut, malphigian tubules, salivary glands, trachea, and male reproductive organs—revealed nuclear localization of Dfr/Vvl in uninfected animals. Immunostaining in bacterium-infected larvae and adult flies revealed the same distribution of Dfr/Vvl as that found in uninfected animals. In addition, we did not observe any differences in the nucleocytoplasmic localization of Dfr/Vvl before and after infection (data not shown), indicating that Dfr/Vvl is not prone to signal-dependent redistribution. These results suggested that Dfr/Vvl could regulate AMP gene expression constitutively in specific tissues to set up a protective antimicrobial barrier.
Constitutive CecA1 expression in male reproductive organs overlaps with Dfr/Vvl.
To further characterize the role of Dfr/Vvl in constitutive, tissue-specific expression of AMP genes, we focused on the male reproductive organs. Recent findings have shown that the reproductive tract of males is readily accessible to pathogen attack (24) and that sexual transmission of Serratia marcescens from contaminated males during mating is sufficient to cause infection and death in females (44). In agreement with the receptiveness to infection, the male reproductive tract is a site of robust expression of a number of AMP genes in Drosophila, including CecA1 (73) and the male-specific andropin gene (62). Dfr/Vvl is constitutively expressed and localized in the nuclei of the male ejaculatory duct (Fig. 2K and 3 A′). In the same flies, expression of a CecA1 promoter-driven green fluorescent protein gene (CecA1-GFP) (Fig. 2J and 3A), reporter construct, or CecA1-lacZ gene (see Fig. 5A) overlapped with nuclear Dfr/Vvl staining in the ejaculatory duct and was absent from other parts of the male reproductive organs.
FIG. 3.
Constitutive CecA1 expression in the ejaculatory duct is Toll pathway and Imd pathway independent. Constitutive expression of CecA1-GFP in the ejaculatory duct of uninfected wt (A), Myd88 mutant (B), and imd mutant (C) male flies corresponds to nuclear Dfr/Vvl staining (A′, B′, and C′) and nuclear DAPI staining (A", B", and C").
FIG. 5.
Identification of a Dfr/Vvl-dependent CecA1 regulatory region. (A to D) Constitutive CecA1 expression in male reproductive organs in transgenic flies carrying different CecA1-lacZ constructs. Reporter β-galactosidase staining was specifically localized to the ejaculatory duct (ej.duct) but was absent from the accessory glands (acc.glands), seminal vesicle (vsm), and testis. Expression was evident from constructs carrying 760 bp upstream of the CecA1 transcription start site (21) (A, B, and C), even if the regulatory R1 and κB sequences had been deleted (55) (B) or the GATA site mutated (53) (C). A construct containing 111 bp of the CecA1 upstream sequence did not confer any β-Gal staining in the ejaculatory duct (D), indicating that necessary elements must be located in the bp −760 to −111 region. (E) Quantitative measurements of luciferase activity in extracts of mbn-2 cells after expression of Dfr/Vvl, Cad, or Dfr/Vvl and Cad proteins, with a panel of CecA1-luc reporter constructs, carrying progressive 5′ deletions (top graph) or specific mutations in putative Dfr/Vvl binding sites (bottom graph), as indicated. The upstream region and 5′ untranslated region (UTR) of the CecA1 gene (bp −751 to +71) is represented by a horizontal line. The EMSA control probe (C) and the location of the proximal regulatory region, containing the R1, κB, and GATA motifs, are indicated (not to scale). The extent of the deletion in CecA1Δ-luc (bp −466 to −403) is indicated by Δ and by parentheses in the sequences below. The sequences of probes E1 to E3 used in the EMSA are shown (note that each overlaps with the previous probe by 5 bp). The putative Dfr/Vvl binding sites are shown in red, and mutated base pairs in the CecA1-luc constructs are underlined. The results are presented as the mean values of at least three independent experiments (± standard error of the mean [SEM]). (F) EMSA with purified, GST-tagged Dfr/Vvl, and GST protein alone, using four different 32P-labeled CecA1 probes, E1 (bp −493 to −454), E2 (bp −458 to −419), E3 (bp −423 to −380) covering partly overlapping parts of the bp −484 to −377 region 5′ of CecA1, and, as a control, an upstream fragment (C; bp −663 to −644). GST-tagged Dfr/Vvl protein, but not GST alone, bound with high affinity to the E1 and E3 probes, while only weak binding was observed with the E2 probe. Dfr/Vvl protein did not bind to the control (C) fragment (bp −663 to −644), and no binding was observed for lanes without any protein added (−).
The fact that CecA1-GFP (Fig. 2J and 3A) and CecA1-lacZ (see Fig. 5A) were highly expressed in the ejaculatory duct of all uninfected males indicates that the expression is not dependent on signaling in response to infection but rather reflects constitutive, tissue-specific regulation of the CecA1 promoter. To test this hypothesis directly, CecA1-GFP expression was analyzed in mutants of the Toll and Imd pathways. Both nuclear Dfr/Vvl immunostaining and CecA1-GFP were prominent in male flies mutant for Myd88 (Fig. 3B), a component of the Toll pathway, and in flies with a mutation in imd (Fig. 3C), a component of the Imd pathway. In fact, the CecA1-GFP staining was somewhat stronger in ejaculatory ducts from the Myd88 and imd mutant flies than in those from wt flies. Thus, constitutive CecA1 expression in the ejaculatory duct is not dependent on signals propagated through the Toll or Imd pathways, suggesting that this expression is involved in establishing AMP synthesis in the ejaculatory duct prior to, and irrespective of, infection in this or other parts of the organism.
The POU protein Dfr/Vvl can activate AMP gene expression in vivo.
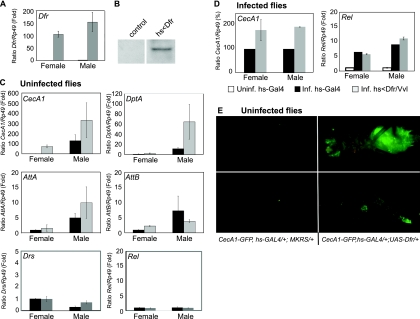
To determine if Dfr/Vvl plays a direct role in AMP expression, we overexpressed Dfr/Vvl in whole animals and examined AMP gene expression. Expression of Dfr/Vvl, using a UAS-Dfr/Vvl transgene (15) and several different GAL4 drivers led to abnormal development, melanization, and lethality prior to pupation, as described in Materials and Methods. However, Dfr/Vvl expression in hs-GAL4/UAS-Dfr/Vvl did not kill the flies, although it promoted more than 100-fold overexpression of Dfr/Vvl mRNA when flies were kept at 29°C for 5 days, as measured by RT-qPCR (Fig. 4A). Consistent with this increase in RNA expression, a substantial increase in Dfr/Vvl protein levels was observed (Fig. 4B). Overexpression of Dfr/Vvl by means of leaky expression from the heat-shock promoter at 29°C promoted relatively modest and various upregulation of AMP genes (data not shown). A more robust AMP gene response was gained by keeping the flies at a lower temperature and giving them heat shock at 37°C. The latter procedure led to a 70-fold upregulation of CecA1 and an about 2-fold upregulation of the DptA, AttA, and AttB genes in uninfected female flies (Fig. 4C). In males, which express higher basal levels of these AMP genes, overexpression of Dfr/Vvl led to upregulation of CecA1, DptA, AttA, and Drs, ranging between 2- and 5-fold (Fig. 4C). In addition to the genes shown in Fig. 4, the expression of Defensin (Def), Metchnikowin (Mtk), and the male-specific Andropin (Anp) mRNA was analyzed by semiquantitative RT-PCR in extracts from hs-GAL4/UAS-Dfr/Vvl flies. The levels of Def and Anp mRNA increased in response to Dfr/Vvl in both females and males, while the expression of Mtk mRNA was unaffected, compared to the control (data not shown). Thus, at least seven different AMP genes, representing different families of AMPs, were upregulated in response to Dfr/Vvl overexpression, indicating that Dfr/Vvl is a potent activator of AMP gene expression independently of infection. In addition, moderate overexpression of Dfr/Vvl, by means of leaky expression of hs-GAL4/UAS-Dfr/Vvl, primed flies for enhanced CecA1 expression during infection (Fig. 4D). This indicates that Dfr/Vvl plays a role as an activator also during infection. This corroborates the finding of strong nuclear Dfr/Vvl immunostaining in the fat body (Fig. 1A and 2A), the tissue known to respond most strongly to infection-induced expression of the CecA1 gene (55, 61). This raised the question of whether some of the Dfr/Vvl-driven AMP gene regulation could be the result of transcriptional upregulation of the NF-κB/Rel factor Relish. However, Rel mRNA expression was not affected by Dfr/Vvl overexpression in either uninfected or infected flies (Fig. 4C and D), but as expected, Rel mRNA was upregulated more than 5-fold in response to infection (Fig. 4). Activation of Rel transcription is known to depend on the Imd pathway, leading to Relish cleavage, nuclear translocation, and autoregulation of the Rel gene. Therefore, AMP gene activation in response to Dfr/Vvl is most likely not via transcriptional activation of Rel but rather reflects a direct effect of Dfr/Vvl.
FIG. 4.
Overexpression of Dfr/Vvl-induced expression of AMP genes in an infection-independent manner. (A) Quantification of Dfr/Vvl mRNA by RT-qPCR in Dfr/Vvl-overexpressing flies (hs-GAL4/UAS-Dfrwt) (gray bars) compared to control flies (hs-GAL4/+) (black bars). The level of Dfr/Vvl mRNA in uninfected female control flies was set to 1, using Rp49 levels as an internal control. (B) Western blot analysis of nuclear extracts from control and Dfr/Vvl-overexpressing flies, with genotypes as described in the legend for panel A. (C) RT-qPCR analysis of AMP gene expression in uninfected Dfr/Vvl-overexpressing flies (hs-GAL4/UAS-Dfrwt) (gray bars) compared to control flies (hs-GAL4+>) (black bars). The relative mRNA levels of each gene (CecA1, Dpt, AttA, AttB, Drs, and Rel) in extracts of female or male flies of each genotype were calculated relative to the expression of the same gene in female control flies, which was set to 1, using Rp49 mRNA as an internal control. (D) RT-qPCR analysis of CecA1 and Rel levels in infected females and males overexpressing Dfr/Vvl (hs-GAL4/UAS-Dfrwt) (gray bars) compared to control (hs-GAL4/+) flies (black bars). The level of CecA1 mRNA in infected control flies was set to 100%. The level of Rel mRNA in infected flies was calculated relative to the expression of the same gene in uninfected female control (hs-GAL4/+) flies, set to 1. (E) Ubiquitous overexpression of Dfr/Vvl (hs-GAL4, CecA1-GFP; UAS-Dfrwt/+) by heat shock treatment promoted CecA1-GFP expression in the fat body of the head, thorax, and abdomen (top right panel) and in several regions of the midgut (bottom right panel) of uninfected flies. In control flies (hs-GAL4, CecA1-GFP; +/+) (left panels), no CecA1-GFP expression was induced by the heat shock.
To investigate in which tissue(s) the strong upregulation of CecA1 gene expression takes place, ubiquitous overexpression of Dfr/Vvl was combined with a CecA1-GFP reporter construct. This genotype (hs-GAL4 CecA1-GFP/+; UAS-Dfr/Vvl/+) was lethal during development, especially to male flies, but female flies could be recovered by raising the flies at 18°C. Upon heat shock, overexpression of Dfr/Vvl promoted strong CecA1-GFP expression in the fat body and midgut of the uninfected flies (Fig. 4E). No GFP expression was observed without the heat shock induction, even if flies were kept at 29°C after eclosion, or in heat-shocked control flies (hs-GAL4 CecA1-GFP/+) (Fig. 4E). The fat body and midgut are not sites of constitutive AMP gene expression but prone to strong infection-induced expression. The fact that Dfr/Vvl immunostaining was observed with the fat body and midgut of uninfected flies (Fig. 2A and B), in which CecA1 normally is not expressed, suggests either that these levels of Dfr/Vvl are suboptimal or that the CecA1 gene is prone to negative regulation by other factor(s) in these tissues in uninfected flies, which is overcome by Dfr/Vvl overexpression.
AMP mRNA levels differ considerably between males and females (Fig. 4C). It is known that expression of many immune defense genes is induced in females in response to mating (42, 48, 52). However, our expression analysis revealed that the relative expression of CecA1 was 100-fold higher and that those of the DptA, AttA, AttB, and Drs genes were 5- to 10-fold higher in uninfected males than in females, even for flies that had been kept in mixed populations for more than 5 days (Fig. 4C). This suggests either a generally higher rate of AMP gene expression in males or that these genes are highly expressed in male-specific organs, consistent with the intense expression of CecA1-GFP seen in the male ejaculatory duct of uninfected males (Fig. 2J) and of AttA-GFP and Drs-GFP (data not shown) (73).
In summary, transiently elevated expression of the Dfr/Vvl protein was sufficient for activation of several AMP genes in an infection-independent manner and also promoted enhanced levels of CecA1 expression during infection. The response of individual genes to Dfr/Vvl overexpression varied in a manner that was gene specific and sex specific, suggesting that the regulatory regions of each AMP gene differ in organization and response to Dfr/Vvl binding.
In addition to these experiments examining the effects of overexpression of Dfr/Vvl, we have attempted exhaustively to downregulate Dfr/Vvl expression using a combination of mutant alleles as well as different driver lines to express double-stranded (ds) RNA constructs. It was not possible to consistently reproduce substantial knockdown of Dfr/Vvl gene function, and we have therefore been unable to analyze the expression of AMPs following the loss of Dfr/Vvl expression. For a detailed description, see Material and Methods. We believe that it is impossible to significantly reduce Dfr/Vvl levels because this gene is essential both during development and for adult survival and is therefore under tight regulatory control that prevents loss of expression. Hypomorphic alleles of Dfr/Vvl do cause a modest reduction of Dfr/Vvl mRNA levels, but this did not have any reproducible effects on AMP gene expression, most likely due to feedback regulation and compensatory effects.
Dfr/Vvl directly activates CecA1 gene expression.
Together, the results presented above suggest that Dfr/Vvl regulates constitutive AMP gene expression, in particular, the expression of CecA1 in the male ejaculatory duct. To identify the cis-acting regulatory elements controlling CecA1 expression in male reproductive organs, we analyzed CecA1-lacZ reporter gene expression in transgenic animals. We and others have previously reported that the upstream region of the CecA1 gene contains regulatory elements required for tissue-specific expression (55, 59). Here we specifically analyzed the expression of different CecA1-lacZ constructs in uninfected male flies. Constructs carrying −760 bp of upstream sequence conferred strong, tissue-specific β-galactosidase (β-Gal) expression in the ejaculatory duct (Fig. 5 A). This expression was not affected by deletions or mutations in any of the previously characterized proximal regulatory elements, called R1, κB, and GATA (Fig. 5B and C). These cis-regulatory elements are all involved in the activation of CecA1 in the fat body and hemocytes in response to infection (21, 33, 55, 75). Consistent with this, a 5′ deletion construct, carrying 111 bp of upstream sequence, including the R1, κB, and GATA sites, did not promote detectable β-Gal expression in male reproductive organs (Fig. 5D), but flies carrying this construct revealed strong β-Gal expression in the fat body upon infection (data not shown) (55). Thus, constitutive CecA1-lacZ expression in the male ejaculatory duct requires a regulatory element(s), located in the bp −760 to −111 region that is independent of those required for induction following infection.
Next, we asked whether Dfr/Vvl directly activates CecA1 transcription via the regulatory element(s) located between bp −760 and −111. Expression of a series of CecA1-luciferase constructs, carrying progressive 5′ deletions, was analyzed in the absence and presence of Dfr/Vvl in cell transfection experiments. Reporter gene constructs carrying 751 bp, 519 bp, and 484 bp of the CecA1 5′ upstream region, relative to the transcription start site, were strongly activated by Dfr/Vvl. The constructs were activated to similar extents, suggesting that the Dfr/Vvl-responsive region(s) was not located between bp −751 and −484 (Fig. 5E). The three shortest constructs, carrying 377 bp or less of the CecA1 5′ region, conferred very weak or no activation of the reporter gene in the presence of Dfr/Vvl (Fig. 5E). These results suggest that Dfr/Vvl interacts with regulatory element(s) in the region from bp −484 to −377 to promote activation of CecA1 expression. Thus, both expression of CecA1 in the ejaculatory duct and trans activation by Dfr/Vvl require sequences in the distal promoter region, indicating that this region contains an ejaculatory duct enhancer.
Further analysis of the ejaculatory duct enhancer (bp −484 and −377) revealed four putative Dfr/Vvl binding sites, sharing the consensus motif CANTAAA (highlighted in red in Fig. 5E; Table 1). To determine if these sites were sufficient for Dfr/Vvl induction of CecA1 expression, we used site-directed mutagenesis to change each site individually in the 760-bp CecA1-luc construct. Individual mutation of each of these sites (called E1, E2A, E2B, and E3) caused a significant decrease (25 to 40%) in observed luciferase activity, suggesting that each site is important but that more than one of the sites are required for full activation of CecA1 expression. Consistent with this prediction, a deletion within the ejaculatory duct enhancer containing all four sites (bp −466 to −403; CecA1Δ-luc) eliminated Dfr/Vvl-driven expression. Together, these data suggest that the region from bp −484 to −377 constitutes a regulatory enhancer that contains several Dfr/Vvl-responsive elements, which all contribute to the expression of CecA1 in the male ejaculatory duct. A similar transfection analysis using Drs-luc constructs revealed that 0.92 kb 5′ of the Drs gene did not respond to Dfr/Vvl activation, while a construct with 2.2 kb of the 5′ upstream sequence, which includes a Dfr/Vvl consensus sequence (Table 1), was upregulated more than 20-fold by Dfr/Vvl (data not shown).
TABLE 1.
Sequences with a perfect match to the Dfr/Vvl binding consensus motif CANTAAA in the regulatory regions of Drosophila immune-related genes
| Gene | Distance (bp) or UTRa | Sequenceb |
|---|---|---|
| CecA1 | −503 | CAATAAA |
| −464 | CAGTAAA | |
| −447 | CAATAAA | |
| −438 | CATTAAA | |
| −407 | CACTAAA | |
| CecB | −861 | CAATAAA |
| −692 | CAATAAA | |
| −302 | CACTAAA | |
| CecC | −1,612 | CAATAAA |
| −1,564 | CAATAAA | |
| −820 | CAGTAAA | |
| Anp | −800 | CATTAAA |
| −419 | CATTAAA | |
| AttA | 3′ UTR | CAATAAA |
| AttB | −285 | CATTAAA |
| AttC | 3′ UTR | CAATAAA |
| AttD | −625 | CATTAAA |
| 3′ UTR | CAATAAA | |
| Def | −648 | CAATAAA |
| −317 | CAGTAAA | |
| DiptA | −933 | CAGTAAA |
| 3′ UTR | CAATAAA | |
| DiptB | −920 | CAATAAA |
| Dro | −994 | CATTAAA |
| −218 | CATTAAA | |
| 3′ UTR | CATTAAA | |
| Drs | −1,211 | CACTAAA |
| Ddc | −1,283 | CATTAAA |
| −863 | CAATAAA |
Numbers refer to distance in base pairs upstream of the transcription initiation site/cap site.
Sequences shown represent motifs identified on either DNA strand in either orientation. The motifs present in the 3′ UTR of the AttA, DiptA, and Dro genes are within 1 kb 5′ of another AMP gene and might therefore be involved in transcriptional regulation of the downstream gene. It cannot be ruled out, however, that the sites in the 3′ UTR are involved in posttranscriptional regulation of the upstream gene, since the motif resembles a poly(A) site.
Dfr/Vvl binds directly to the male ejaculatory duct enhancer.
To test if the −484 and −377 region is a target for direct binding of Dfr/Vvl, we expressed and purified GST-tagged Dfr/Vvl protein for in vitro DNA-binding experiments. As shown in Fig. 5F, Dfr/Vvl bound to the Dfr/Vvl-dependent upstream enhancer in EMSAs. Specific binding occurred with DNA probes E1, E2, and E3 (covering the regions from bp −493 to −454, −458 to −419, and −423 to −380, respectively). No binding was observed using a negative-control fragment upstream of the Dfr/Vvl-responsive region (C; bp −663 to −644). Thus, Dfr/Vvl protein can bind directly to three independent sequence elements within the male ejaculatory duct enhancer that contains four independent Dfr/Vvl consensus sequences. In summary, Dfr/Vvl is likely required for constitutive CecA1 expression in male reproductive organs, and this tissue-specific expression requires the distal region of the CecA1 gene. Dfr/Vvl interacts with several independent sequence elements within the region from bp −484 to −377 and promotes CecA1 expression via this ejaculatory duct enhancer.
The POU protein Dfr/Vvl and the homeodomain protein Cad activate CecA1 in a synergistic manner.
Dfr/Vvl protein bound the CecA1 regulatory element from bp −484 to −377 (Fig. 5F). The homeodomain protein Caudal (Cad) was previously shown to interact directly with a Cad protein DNA recognition element (CDRE) present in this region (59). Cad bound most strongly to sequences within the E2 and E3 fragments (S2 and S5 in reference 59), and mutations that directly overlap with Dfr/Vvl binding sites in E2A and E3 (S1 and S3) abolished Cad binding, while mutations overlapping with Dfr/Vvl binding sites in E1 and E2B did not interfere with Cad binding (59). Thus, Dfr/Vvl and Cad have the potential to activate CecA1 expression by binding to the same enhancer region (bp −484 to −377), possibly independently or in a complex. The mbn-2 cell line does not express Dfr/Vvl but does contain endogenous Cad (data not shown). To eliminate the possibility that endogenous Cad was responsible for the high expression of CecA1-luc following Dfr/Vvl transfection, we pretreated mbn-2 cells with double-stranded Cad RNA. We found that knockdown of Cad expression by RNA interference had no effect on Dfr/Vvl-induced CecA1-luc expression (data not shown), demonstrating that Dfr/Vvl can activate CecA1 expression independently of Cad. Consistent with this observation, Dfr/Vvl (Fig. 5F) and Cad (59) can independently bind DNA in this region.
To investigate if Dfr/Vvl and Cad support or interfere with each other's trans activation, cotransfection experiments were carried out. There was a 45-fold activation of CecA1-luc expression (Fig. 5E) when Dfr/Vvl and Cad were coexpressed, while each promoted 23-fold and 6-fold activation alone, respectively. This indicates a strong synergism between these two transcription factors in activation of CecA1 gene expression. This synergistic activation of CecA1 required sequences upstream of −377 bp (Fig. 5E and data not shown). Taken together, these findings demonstrate that Dfr/Vvl and Cad can bind and act together to activate the transcription of the CecA1 gene. Further, CecA1 and other AMP genes are strongly activated in tissues in which both Cad and Dfr/Vvl are present, such as the male ejaculatory duct (Fig. 2E) (59), suggesting that they may coordinately regulate expression of other AMP genes as well. In fact, Dfr/Vvl and Cad binding sites in CecA1 overlap. To test which sites were required for Dfr/Vvl/Cad synergism, we transfected mbn-2 cells expressing our mutated CecA1-luc constructs with Cad or both Cad and Dfr/Vvl. We found that mutation of E2B alone was sufficient to eliminate the synergistic effect of Cad and Dfr/Vvl (Fig. 2E), although this mutation alone did not eliminate the ability of Cad or Dfr/Vvl to individually induce CecA1-luc expression. Interestingly, Ryu et al. (59) showed that Cad binds to E2A (S2) and E3 (S5), but not to E1 (S1) and E2B (S3), indicating that the synergistic activation relies on binding of Dfr/Vvl and Cad to alternating target sites within this region and that Dfr/Vvl binding to the E2B sequence is crucial. Single mutations of the E1, E2A, and E3 had no effect on the ability of Cad and Dfr/Vvl to synergistically induce CecA1-luc expression (Fig. 5E).
To summarize the results presented here and by Ryu et al. (59), Dfr/Vvl and Cad can bind independently to the same enhancer region in CecA1. This region contains several binding sites for both factors, and it is required for CecA1 expression in the ejaculatory gut. Coexpression of Dfr/Vvl and Cad promotes synergistic activation of CecA1 expression. It will be interesting in the future to investigate if a similar mode of Dfr/Vvl-Cad synergism is acting in several of the tissues where their expression overlaps.
DISCUSSION
In the present study, we show that Dfr/Vvl, a POU domain transcription factor, is an important regulator of tissue-specific expression of AMP genes in Drosophila. In general, POU domain transcription factors interact with promoter sequences based on the octamer consensus element ATGCAAAT (4, 67). Dfr/Vvl has, however, been shown to bind three distinct Dfr/Vvl recognition elements that are only weakly related to the octamer consensus motif and to each other (5, 12, 13, 30). We analyzed the 5′ regulatory region of CecA1 in silico to search for putative Dfr/Vvl-binding sites. There were several sites matching reported Dfr/Vvl consensus binding elements, including the POU-binding octamer sequence, but none were located within the Dfr/Vvl-responsive region between bp −484 and −377. Instead, we identified 4 sequence elements within this region that shared the consensus sequence CANTAAA. In addition, this sequence motif is present in 1 to 3 copies in the regulatory regions of most Drosophila AMP genes (Table 1). Importantly, this motif resembles a recently predicted Dfr/Vvl target sequence, identified using comparative genomics of 12 Drosophila species (68; M. Kellis, personal communication). Individual mutation of each of these four sites significantly reduced CecA1-luc expression; however, a deletion encompassing these four sites was required to completely eliminate Dfr/Vvl-induced expression (Fig. 5E). Together, these results suggest that several of these sites are required for full Dfr/Vvl induction of CecA1 gene expression.
In accordance with the expression analysis, the CecA1 gene, which is highly responsive to Dfr/Vvl trans-activation in vivo and in cell transfection assays, contains in total 5 consensus CANTAAA motifs within the 500-bp 5′ region (Table 1). The DiptA, AttA, AttB, and Drs genes, which were moderately upregulated by Dfr/Vvl overexpression in vivo, each contain one consensus motif in its regulatory region. Thus, there is a close correlation between the presence of the Dfr/Vvl target sequences and the responsiveness to Dfr/Vvl in vivo and in cell transfection experiments.
Most tissues that have been reported to express one or several AMP genes (22, 73) express substantial levels of Dfr/Vvl (Fig. 1 and 2), as revealed by immunostaining. In all tissues with positive Dfr/Vvl staining, it was either exclusively nuclear or uniformly distributed between the cytoplasm and nucleus, suggesting that Dfr/Vvl is constitutively present in the nucleus. This is supported by the presence of a putative nuclear localization signal (NLS) in Dfr/Vvl (data not shown). The predominantly nuclear localization of Dfr/Vvl contrasts with the behavior of the NF-κB/Rel proteins, which respond to signaling by nuclear translocation (25, 29, 70). This supports the notion that Dfr/Vvl is a regulator of constitutive tissue-specific expression of AMP genes. The finding that CecA1-GFP expression in response to overexpression of Dfr/Vvl was observed predominantly in the fat body and midgut indicates that Dfr/Vvl has the potential to activate CecA1 in these immunoresponsive tissues and suggests that Dfr/Vvl may play a role as a competence factor during infection.
We propose that Dfr/Vvl, together with the homeodomain protein Cad, binds to multiple target sequences within the male ejaculatory duct enhancer in the CecA1 gene to constitutively activate AMP expression (Fig. 6). The following observations support this model and indicate that Dfr/Vvl regulates constitutive expression of AMP genes in male reproductive organs: (i) CecA1 is constitutively expressed in the male ejaculatory duct (Fig. 3A to C and Fig. 5A to C); (ii) Dfr/Vvl is expressed and localized in the nuclei of the ejaculatory duct cells (Fig. 2K and Fig. 3A to C); (iii) the CecA1 upstream enhancer (bp −484 to −377) to which Dfr/Vvl binds is crucial for constitutive expression of CecA1 in the ejaculatory duct (Fig. 5A to F) (59); and (iv) overexpression of Dfr/Vvl caused a massive upregulation of CecA1 expression in vivo. The in vitro DNA binding results and transfection studies (Fig. 5E and F) indicate that the regulation of CecA1 expression by Dfr/Vvl is direct. Further, Dfr/Vvl and Cad (59) bind independently to sites in the upstream enhancer, and this appears to be the basis for the synergistic effect on the transcriptional activation by these two proteins. Consistent with this observation, we have shown that one site, here called E2B, is required for the synergism observed in Cad and Dfr/Vvl cotransfection experiments (Fig. 5E) and that mutations in the enhancer region lead to dramatically reduced CecA1 expression (Fig. 5E) (59).
FIG. 6.
An upstream enhancer regulates constitutive, tissue-specific expression of CecA1. Shown are a schematic representation of the genomic region upstream of the CecA1 coding sequence and a model for the organization of cis-regulatory elements present in this region. A tissue-specific enhancer that directs expression in the ejaculatory duct, named the male ejaculatory duct enhancer, is proposed to be targeted by Dfr/Vvl (stars) and Cad (hexagons), each binding to different sequence elements (E1 to E3) within the enhancer. Binding of Dfr/Vvl to the CANTAAA sequence motif in the enhancer activates expression of CecA1. The proximal regulatory region (gray box), which is a target of infection-induced signaling and binding of Rel and GATA factors (gray dimer), does not appear to be required for the constitutive expression in the ejaculatory duct, since CecA1-lacZ constructs with deletions/mutations in this region conferred ejaculatory duct-specific reporter gene expression. The model is based on results from this and previous studies (21, 29, 53, 55, 59, 70, 75).
Our results, together with data reported by Ryu et al. (59), indicate that the high constitutive expression of CecA1 in the ejaculatory duct is the result of Dfr/Vvl and Cad acting together in this tissue. This is, to our knowledge, the first report of a functional interaction between a homeodomain protein and a POU domain protein in regulation of genes involved in the Drosophila immune defense. Dfr/Vvl and Cad show overlapping tissue distribution in tissues other than the male reproductive organs, such as the intestine and other parts of the digestive tract (Fig. 1 and 2) (46), suggesting that they may interact in AMP gene regulation in other tissues as well. Interestingly, Cad has been shown to be a regulator of innate immune homeostasis in the intestine, by acting as a repressor of AMP genes in the presence of the commensal microbial flora (58). This Cad-dependent repression is abolished upon infection with pathogens. Ubiquitous overexpression of Dfr/Vvl activated a CecA1-GFP reporter gene in the midgut of uninfected flies (Fig. 4E). This would imply that Dfr/Vvl overexpression abolished Cad inhibition of CecA1, most likely by direct interaction between Dfr/Vvl and Cad at the CecA1 regulatory region. It was recently shown that the human class III POU protein Brn-4 and human Caudal homolog Cdx-2 physically interact and confer both synergistic and redundant effects on the regulation of proglucagon gene expression in intestinal endocrine cells (77). It will be of great interest in the future to investigate the mechanism of Dfr/Vvl and Cad coactivation and to determine whether other POU and homeodomain proteins interact in regulation of immune defense genes as well.
To date, more than 900 gene-encoded AMPs have been identified in eukaryotic organisms and more than 30 β-defensin genes have been identified in the human genome. Human AMPs are expressed predominantly in leukocytes and on mucosal surfaces, skin, and other epithelia. The expression of AMPs in analogous barrier epithelia in humans and insects suggests that this branch of the innate immune defense has been functionally important and possibly sustained during evolution. Whether the regulation of AMP genes in epithelial tissues shows some evolutionary conservation awaits investigation. The Drosophila POU protein Dfr/Vvl is strongly expressed in several barrier epithelia, such as the epidermis, trachea, and the intestinal epithelium, and in the ejaculatory duct, which also is covered with an epithelial lining. Interestingly, a large number of the human, rodent, and porcine β-defensin gene family members have been shown to be preferentially expressed in the male reproductive tract, particularly in the epididymis and testis, with suggested dual functions in immunity and sperm maturation (41, 51, 63, 79). In addition, the AMP cathelicidin/LL37/hCAP18/CRAMP is expressed in male reproductive organs in many species (reviewed in reference 19). Intriguingly, the human form of cathelicidin, hCAP18, is expressed in the epithelium of the epididymis and present in seminal plasma and on the surface of spermatozoa (40). Thus, nature has provided both mammals and insects with natural antibiotics at the site of sperm production and storage, suggesting a key role in immunity and/or conception. The human genome contains several families of POU protein transcription factors that exhibit differentiated expression patterns ranging from brain to epithelial specific (57). This opens up the possibility of POU proteins having important functions as AMP gene modulators in vertebrate innate immunity.
Acknowledgments
We thank Sarah Certel, Won-Jae Lee, Jean-Luc Imler, Tony Ip, Stephen T. Crews, the Drosophila Genomics Resource Center, and the Bloomington Stock Center for providing valuable reagents and fly stocks and Gunnel Björklund for technical assistance.
This work was supported by the Swedish Cancer Society, the Swedish Research Council, and the Foundation of Strategic Research to Y.E. and the National Institutes of Health to L.P.
Footnotes
Published ahead of print on 10 May 2010.
REFERENCES
- 1.Aerts, A. M., I. E. Francois, B. P. Cammue, and K. Thevissen. 2008. The mode of antifungal action of plant, insect and human defensins. Cell. Mol. Life Sci. 65:2069-2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Agerberth, B., and G. H. Gudmundsson. 2006. Host antimicrobial defence peptides in human disease. Curr. Top. Microbiol. Immunol. 306:67-90. [DOI] [PubMed] [Google Scholar]
- 3.Akira, S., S. Uematsu, and O. Takeuchi. 2006. Pathogen recognition and innate immunity. Cell 124:783-801. [DOI] [PubMed] [Google Scholar]
- 4.Andersen, B., and M. G. Rosenfeld. 2001. POU domain factors in the neuroendocrine system: lessons from developmental biology provide insights into human disease. Endocr. Rev. 22:2-35. [DOI] [PubMed] [Google Scholar]
- 5.Anderson, M. G., S. J. Certel, K. Certel, T. Lee, D. J. Montell, and W. A. Johnson. 1996. Function of the Drosophila POU domain transcription factor drifter as an upstream regulator of breathless receptor tyrosine kinase expression in developing trachea. Development 122:4169-4178. [DOI] [PubMed] [Google Scholar]
- 6.Anderson, M. G., G. L. Perkins, P. Chittick, R. J. Shrigley, and W. A. Johnson. 1995. drifter, a Drosophila POU-domain transcription factor, is required for correct differentiation and migration of tracheal cells and midline glia. Genes Dev. 9:123-137. [DOI] [PubMed] [Google Scholar]
- 7.Biragyn, A., P. A. Ruffini, C. A. Leifer, E. Klyushnenkova, A. Shakhov, O. Chertov, A. K. Shirakawa, J. M. Farber, D. M. Segal, J. J. Oppenheim, and L. W. Kwak. 2002. Toll-like receptor 4-dependent activation of dendritic cells by beta-defensin 2. Science 298:1025-1029. [DOI] [PubMed] [Google Scholar]
- 8.Boube, M., M. Llimargas, and J. Casanova. 2000. Cross-regulatory interactions among tracheal genes support a co-operative model for the induction of tracheal fates in the Drosophila embryo. Mech. Dev. 91:271-278. [DOI] [PubMed] [Google Scholar]
- 9.Brand, A. H., and N. Perrimon. 1993. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118:401-415. [DOI] [PubMed] [Google Scholar]
- 10.Bulet, P., R. Stocklin, and L. Menin. 2004. Anti-microbial peptides: from invertebrates to vertebrates. Immunol. Rev. 198:169-184. [DOI] [PubMed] [Google Scholar]
- 11.Bürglin, T. R., and G. Ruvkun. 2001. Regulation of ectodermal and excretory function by the C. elegans POU homeobox gene ceh-6. Development 128:779-790. [DOI] [PubMed] [Google Scholar]
- 12.Certel, K., M. G. Anderson, R. J. Shrigley, and W. A. Johnson. 1996. Distinct variant DNA-binding sites determine cell-specific autoregulated expression of the Drosophila POU domain transcription factor drifter in midline glia or trachea. Mol. Cell. Biol. 16:1813-1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Certel, K., A. Hudson, S. B. Carroll, and W. A. Johnson. 2000. Restricted patterning of vestigial expression in Drosophila wing imaginal discs requires synergistic activation by both Mad and the drifter POU domain transcription factor. Development 127:3173-3183. [DOI] [PubMed] [Google Scholar]
- 14.Certel, S. J., and W. A. Johnson. 1996. Disruption of mesectodermal lineages by temporal misexpression of the Drosophila POU-domain transcription factor, drifter. Dev. Genet. 18:279-288. [DOI] [PubMed] [Google Scholar]
- 15.Certel, S. J., and S. Thor. 2004. Specification of Drosophila motoneuron identity by the combinatorial action of POU and LIM-HD factors. Development 131:5429-5439. [DOI] [PubMed] [Google Scholar]
- 16.Chromek, M., Z. Slamova, P. Bergman, L. Kovacs, L. Podracka, I. Ehren, T. Hökfelt, G. H. Gudmundsson, R. L. Gallo, B. Agerberth, and A. Brauner. 2006. The antimicrobial peptide cathelicidin protects the urinary tract against invasive bacterial infection. Nat. Med. 12:636-641. [DOI] [PubMed] [Google Scholar]
- 17.Davis, M. M., D. A. Primrose, and R. B. Hodgetts. 2008. A member of the p38 mitogen-activated protein kinase family is responsible for transcriptional induction of Dopa decarboxylase in the epidermis of Drosophila melanogaster during the innate immune response. Mol. Cell. Biol. 28:4883-4895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.de Celis, J. F., M. Llimargas, and J. Casanova. 1995. Ventral veinless, the gene encoding the Cf1a transcription factor, links positional information and cell differentiation during embryonic and imaginal development in Drosophila melanogaster. Development 121:3405-3416. [DOI] [PubMed] [Google Scholar]
- 19.Durr, U. H., U. S. Sudheendra, and A. Ramamoorthy. 2006. LL-37, the only human member of the cathelicidin family of antimicrobial peptides. Biochim. Biophys. Acta 1758:1408-1425. [DOI] [PubMed] [Google Scholar]
- 20.Engström, Y. 1999. Induction and regulation of antimicrobial peptides in Drosophila. Dev. Comp. Immunol. 23:345-358. [DOI] [PubMed] [Google Scholar]
- 21.Engström, Y., L. Kadalayil, S. C. Sun, C. Samakovlis, D. Hultmark, and I. Faye. 1993. Kappa B-like motifs regulate the induction of immune genes in Drosophila. J. Mol. Biol. 232:327-333. [DOI] [PubMed] [Google Scholar]
- 22.Ferrandon, D., A. C. Jung, M. Criqui, B. Lemaitre, S. Uttenweiler-Joseph, L. Michaut, J. Reichhart, and J. A. Hoffmann. 1998. A drosomycin-GFP reporter transgene reveals a local immune response in Drosophila that is not dependent on the Toll pathway. EMBO J. 17:1217-1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gateff, E., L. Gissmann, R. Shrestha, N. Plus, H. Pfister, J. Schröder, and H. Zur Hausen. 1980. Characterization of two tumorous blood cell lines of Drosophila melanogaster and the viruses they contain, p. 517-533. In E. Kurstak, K. Maramorosch, and A. Dübendorfer (ed.), Invertebrate systems in vitro. Elsevier/North Holland Biomedical Press, Amsterdam, Netherlands.
- 24.Gendrin, M., D. P. Welchman, M. Poidevin, M. Herve, and B. Lemaitre. 2009. Long-range activation of systemic immunity through peptidoglycan diffusion in Drosophila. PLoS Pathog. 5:e1000694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Govind, S., and R. Steward. 1991. Dorsoventral pattern formation in Drosophila: signal transduction and nuclear targeting. Trends Genet. 7:119-125. [DOI] [PubMed] [Google Scholar]
- 26.Herr, W., R. A. Sturm, R. G. Clerc, L. M. Corcoran, D. Baltimore, P. A. Sharp, H. A. Ingraham, M. G. Rosenfeld, M. Finney, G. Ruvkun, et al. 1988. The POU domain: a large conserved region in the mammalian pit-1, oct-1, oct-2, and Caenorhabditis elegans unc-86 gene products. Genes Dev. 2:1513-1516. [DOI] [PubMed] [Google Scholar]
- 27.Hultmark, D. 2003. Drosophila immunity: paths and patterns. Curr. Opin. Immunol. 15:12-19. [DOI] [PubMed] [Google Scholar]
- 28.Inbal, A., D. Levanon, and A. Salzberg. 2003. Multiple roles for u-turn/ventral veinless in the development of Drosophila PNS. Development 130:2467-2478. [DOI] [PubMed] [Google Scholar]
- 29.Ip, Y. T., M. Reach, Y. Engström, L. Kadalayil, H. Cai, S. Gonzalez-Crespo, K. Tatei, and M. Levine. 1993. Dif, a dorsal-related gene that mediates an immune response in Drosophila. Cell 75:753-763. [DOI] [PubMed] [Google Scholar]
- 30.Johnson, W. A., and J. Hirsh. 1990. Binding of a Drosophila POU-domain protein to a sequence element regulating gene expression in specific dopaminergic neurons. Nature 343:467-470. [DOI] [PubMed] [Google Scholar]
- 31.Johnson, W. A., C. A. McCormick, S. J. Bray, and J. Hirsh. 1989. A neuron-specific enhancer of the Drosophila dopa decarboxylase gene. Genes Dev. 3:676-686. [DOI] [PubMed] [Google Scholar]
- 32.Junell, A., H. Uvell, L. Pick, and Y. Engström. 2007. Isolation of regulators of Drosophila immune defense genes by a double interaction screen in yeast. Insect Biochem. Mol. Biol. 37:202-212. [DOI] [PubMed]
- 33.Kadalayil, L., U. M. Petersen, and Y. Engström. 1997. Adjacent GATA and kappa B-like motifs regulate the expression of a Drosophila immune gene. Nucleic Acids Res. 25:1233-1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kimbrell, D. A., and B. Beutler. 2001. The evolution and genetics of innate immunity. Nat. Rev. Genet. 2:256-267. [DOI] [PubMed] [Google Scholar]
- 35.Lai, Y., and R. L. Gallo. 2008. Toll-like receptors in skin infections and inflammatory diseases. Infect. Disord. Drug Targets 8:144-155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lemaitre, B., and J. Hoffmann. 2007. The host defense of Drosophila melanogaster. Annu. Rev. Immunol. 25:697-743. [DOI] [PubMed] [Google Scholar]
- 37.Liu, P. T., S. Stenger, H. Li, L. Wenzel, B. H. Tan, S. R. Krutzik, M. T. Ochoa, J. Schauber, K. Wu, C. Meinken, D. L. Kamen, M. Wagner, R. Bals, A. Steinmeyer, U. Zugel, R. L. Gallo, D. Eisenberg, M. Hewison, B. W. Hollis, J. S. Adams, B. R. Bloom, and R. L. Modlin. 2006. Toll-like receptor triggering of a vitamin D-mediated human antimicrobial response. Science 311:1770-1773. [DOI] [PubMed] [Google Scholar]
- 38.Llimargas, M., and J. Casanova. 1997. ventral veinless, a POU domain transcription factor, regulates different transduction pathways required for tracheal branching in Drosophila. Development 124:3273-3281. [DOI] [PubMed] [Google Scholar]
- 39.Ma, Y., K. Certel, Y. Gao, E. Niemitz, J. Mosher, A. Mukherjee, M. Mutsuddi, N. Huseinovic, S. T. Crews, W. A. Johnson, and J. R. Nambu. 2000. Functional interactions between Drosophila bHLH/PAS, Sox, and POU transcription factors regulate CNS midline expression of the slit gene. J. Neurosci. 20:4596-4605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Malm, J., O. Sorensen, T. Persson, M. Frohm-Nilsson, B. Johansson, A. Bjartell, H. Lilja, M. Stahle-Backdahl, N. Borregaard, and A. Egesten. 2000. The human cationic antimicrobial protein (hCAP-18) is expressed in the epithelium of human epididymis, is present in seminal plasma at high concentrations, and is attached to spermatozoa. Infect. Immun. 68:4297-4302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Maxwell, A. I., G. M. Morrison, and J. R. Dorin. 2003. Rapid sequence divergence in mammalian beta-defensins by adaptive evolution. Mol. Immunol. 40:413-421. [DOI] [PubMed] [Google Scholar]
- 42.McGraw, L. A., G. Gibson, A. G. Clark, and M. F. Wolfner. 2004. Genes regulated by mating, sperm, or seminal proteins in mated female Drosophila melanogaster. Curr. Biol. 14:1509-1514. [DOI] [PubMed] [Google Scholar]
- 43.Meier, S., S. G. Sprecher, H. Reichert, and F. Hirth. 2006. ventral veins lacking is required for specification of the tritocerebrum in embryonic brain development of Drosophila. Mech. Dev. 123:76-83. [DOI] [PubMed] [Google Scholar]
- 44.Miest, T. S., and M. C. Bloch-Qazi. 2008. Sick of mating: sexual transmission of a pathogenic bacterium in Drosophila melanogaster. Fly (Austin) 2:215-219. [DOI] [PubMed] [Google Scholar]
- 45.Miller, A. 1994. The internal anatomy and histology of the imago of Drosophila melanogaster, p. 420-534. In M. Demerec (ed.), Biology of Drosophila. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 46.Mlodzik, M., and W. J. Gehring. 1987. Expression of the caudal gene in the germ line of Drosophila: formation of an RNA and protein gradient during early embryogenesis. Cell 48:465-478. [DOI] [PubMed] [Google Scholar]
- 47.Mookherjee, N., K. L. Brown, D. M. Bowdish, S. Doria, R. Falsafi, K. Hokamp, F. M. Roche, R. Mu, G. H. Doho, J. Pistolic, J. P. Powers, J. Bryan, F. S. Brinkman, and R. E. Hancock. 2006. Modulation of the TLR-mediated inflammatory response by the endogenous human host defense peptide LL-37. J. Immunol. 176:2455-2464. [DOI] [PubMed] [Google Scholar]
- 48.Mueller, J. L., J. L. Page, and M. F. Wolfner. 2006. An ectopic expression screen reveals Drosophila seminal fluid proteins' protective and toxic effects. Genetics 175:777-783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nizet, V., T. Ohtake, X. Lauth, J. Trowbridge, J. Rudisill, R. A. Dorschner, V. Pestonjamasp, J. Piraino, K. Huttner, and R. L. Gallo. 2001. Innate antimicrobial peptide protects the skin from invasive bacterial infection. Nature 414:454-457. [DOI] [PubMed] [Google Scholar]
- 50.Patel, N. H. 1994. Imaging neuronal subsets and other cell types in whole-mount Drosophila embryos and larvae using antibody probes. Methods Cell Biol. 44:445-487. [DOI] [PubMed] [Google Scholar]
- 51.Patil, A. A., Y. Cai, Y. Sang, F. Blecha, and G. Zhang. 2005. Cross-species analysis of the mammalian beta-defensin gene family: presence of syntenic gene clusters and preferential expression in the male reproductive tract. Physiol. Genomics 23:5-17. [DOI] [PubMed] [Google Scholar]
- 52.Peng, J., P. Zipperlen, and E. Kubli. 2005. Drosophila sex-peptide stimulates female innate immune system after mating via the Toll and Imd pathways. Curr. Biol. 15:1690-1694. [DOI] [PubMed] [Google Scholar]
- 53.Petersen, U. M., L. Kadalayil, K. P. Rehorn, D. K. Hoshizaki, R. Reuter, and Y. Engström. 1999. Serpent regulates Drosophila immunity genes in the larval fat body through an essential GATA motif. EMBO J. 18:4013-4022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Philpott, D. J., and S. E. Girardin. 2004. The role of Toll-like receptors and Nod proteins in bacterial infection. Mol. Immunol. 41:1099-1108. [DOI] [PubMed] [Google Scholar]
- 55.Roos, E., G. Björklund, and Y. Engström. 1998. In vivo regulation of tissue-specific and LPS-inducible expression of the Drosophila Cecropin genes. Insect Mol. Biol. 7:51-62. [DOI] [PubMed] [Google Scholar]
- 56.Royet, J., and R. Dziarski. 2007. Peptidoglycan recognition proteins: pleiotropic sensors and effectors of antimicrobial defences. Nat. Rev. Microbiol. 5:264-277. [DOI] [PubMed] [Google Scholar]
- 57.Ryan, A. K., and M. G. Rosenfeld. 1997. POU domain family values: flexibility, partnerships, and developmental codes. Genes Dev. 11:1207-1225. [DOI] [PubMed] [Google Scholar]
- 58.Ryu, J. H., S. H. Kim, H. Y. Lee, J. Y. Bai, Y. D. Nam, J. W. Bae, D. G. Lee, S. C. Shin, E. M. Ha, and W. J. Lee. 2008. Innate immune homeostasis by the homeobox gene caudal and commensal-gut mutualism in Drosophila. Science 319:777-782. [DOI] [PubMed] [Google Scholar]
- 59.Ryu, J. H., K. B. Nam, C. T. Oh, H. J. Nam, S. H. Kim, J. H. Yoon, J. K. Seong, M. A. Yoo, I. H. Jang, P. T. Brey, and W. J. Lee. 2004. The homeobox gene Caudal regulates constitutive local expression of antimicrobial peptide genes in Drosophila epithelia. Mol. Cell. Biol. 24:172-185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Salzman, N. H., D. Ghosh, K. M. Huttner, Y. Paterson, and C. L. Bevins. 2003. Protection against enteric salmonellosis in transgenic mice expressing a human intestinal defensin. Nature 422:522. [DOI] [PubMed] [Google Scholar]
- 61.Samakovlis, C., D. A. Kimbrell, P. Kylsten, A. Engström, and D. Hultmark. 1990. The immune response in Drosophila: pattern of cecropin expression and biological activity. EMBO J. 9:2969-2976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Samakovlis, C., P. Kylsten, D. A. Kimbrell, A. Engström, and D. Hultmark. 1991. The andropin gene and its product, a male-specific antibacterial peptide in Drosophila melanogaster. EMBO J. 10:163-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sang, Y., A. A. Patil, G. Zhang, C. R. Ross, and F. Blecha. 2006. Bioinformatic and expression analysis of novel porcine beta-defensins. Mamm. Genome 17:332-339. [DOI] [PubMed] [Google Scholar]
- 64.Schmittgen, T. D. 2001. Real-time quantitative PCR. Methods 25:383-385. [DOI] [PubMed] [Google Scholar]
- 65.Selsted, M. E., and A. J. Ouellette. 2005. Mammalian defensins in the antimicrobial immune response. Nat. Immunol. 6:551-557. [DOI] [PubMed] [Google Scholar]
- 66.Shai, Y. 2002. Mode of action of membrane active antimicrobial peptides. Biopolymers 66:236-248. [DOI] [PubMed] [Google Scholar]
- 67.Singh, H., R. Sen, D. Baltimore, and P. A. Sharp. 1986. A nuclear factor that binds to a conserved sequence motif in transcriptional control elements of immunoglobulin genes. Nature 319:154-158. [DOI] [PubMed] [Google Scholar]
- 68.Stark, A., M. F. Lin, P. Kheradpour, J. S. Pedersen, L. Parts, J. W. Carlson, M. A. Crosby, M. D. Rasmussen, S. Roy, A. N. Deoras, J. G. Ruby, J. Brennecke, E. Hodges, A. S. Hinrichs, A. Caspi, B. Paten, S. W. Park, M. V. Han, M. L. Maeder, B. J. Polansky, B. E. Robson, S. Aerts, J. van Helden, B. Hassan, D. G. Gilbert, D. A. Eastman, M. Rice, M. Weir, M. W. Hahn, Y. Park, C. N. Dewey, L. Pachter, W. J. Kent, D. Haussler, E. C. Lai, D. P. Bartel, G. J. Hannon, T. C. Kaufman, M. B. Eisen, A. G. Clark, D. Smith, S. E. Celniker, W. M. Gelbart, and M. Kellis. 2007. Discovery of functional elements in 12 Drosophila genomes using evolutionary signatures. Nature 450:219-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Steiner, H. 2004. Peptidoglycan recognition proteins: on and off switches for innate immunity. Immunol. Rev. 198:83-96. [DOI] [PubMed] [Google Scholar]
- 70.Stöven, S., I. Ando, L. Kadalayil, Y. Engström, and D. Hultmark. 2000. Activation of the Drosophila NF-kappaB factor Relish by rapid endoproteolytic cleavage. EMBO Rep. 1:347-352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sturm, R. A., and W. Herr. 1988. The POU domain is a bipartite DNA-binding structure. Nature 336:601-604. [DOI] [PubMed] [Google Scholar]
- 72.Trinchieri, G., and A. Sher. 2007. Cooperation of Toll-like receptor signals in innate immune defence. Nat. Rev. Immunol. 7:179-190. [DOI] [PubMed] [Google Scholar]
- 73.Tzou, P., S. Ohresser, D. Ferrandon, M. Capovilla, J. M. Reichhart, B. Lemaitre, J. A. Hoffmann, and J. L. Imler. 2000. Tissue-specific inducible expression of antimicrobial peptide genes in Drosophila surface epithelia. Immunity 13:737-748. [DOI] [PubMed] [Google Scholar]
- 74.Uvell, H., and Y. Engström. 2007. A multilayered defense against infection: combinatorial control of insect immune genes. Trends Genet. 23:342-349. [DOI] [PubMed] [Google Scholar]
- 75.Uvell, H., and Y. Engström. 2003. Functional characterization of a novel promoter element required for an innate immune response in Drosophila. Mol. Cell. Biol. 23:8272-8281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Vora, P., A. Youdim, L. S. Thomas, M. Fukata, S. Y. Tesfay, K. Lukasek, K. S. Michelsen, A. Wada, T. Hirayama, M. Arditi, and M. T. Abreu. 2004. Beta-defensin-2 expression is regulated by TLR signaling in intestinal epithelial cells. J. Immunol. 173:5398-5405. [DOI] [PubMed] [Google Scholar]
- 77.Wang, P., T. Liu, Z. Li, X. Ma, and T. Jin. 2006. Redundant and synergistic effect of Cdx-2 and Brn-4 on regulating proglucagon gene expression. Endocrinology 147:1950-1958. [DOI] [PubMed] [Google Scholar]
- 78.Wilson, C. L., A. J. Ouellette, D. P. Satchell, T. Ayabe, Y. S. Lopez-Boado, J. L. Stratman, S. J. Hultgren, L. M. Matrisian, and W. C. Parks. 1999. Regulation of intestinal alpha-defensin activation by the Metalloproteinase matrilysin in innate host defense. Science 286:113-117. [DOI] [PubMed] [Google Scholar]
- 79.Yenugu, S., V. Chintalgattu, C. J. Wingard, Y. Radhakrishnan, F. S. French, and S. H. Hall. 2006. Identification, cloning and functional characterization of novel beta-defensins in the rat (Rattus norvegicus). Reprod. Biol. Endocrinol 4:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zasloff, M. 2002. Antimicrobial peptides of multicellular organisms. Nature 415:389-395. [DOI] [PubMed] [Google Scholar]
- 81.Zelzer, E., and B. Z. Shilo. 2000. Interaction between the bHLH-PAS protein Trachealess and the POU-domain protein Drifter, specifies tracheal cell fates. Mech. Dev. 91:163-173. [DOI] [PubMed] [Google Scholar]