Abstract
Abdominal-class homeodomain-containing (Hox) factors form multimeric complexes with TALE-class homeodomain proteins (Pbx, Meis) to regulate tissue morphogenesis and skeletal development. Here we have established that Pbx1 negatively regulates Hoxa10-mediated gene transcription in mesenchymal cells and identified components of a Pbx1 complex associated with genes in osteoblasts. Expression of Pbx1 impaired osteogenic commitment of C3H10T1/2 multipotent cells and differentiation of MC3T3-E1 preosteoblasts. Conversely, targeted depletion of Pbx1 by short hairpin RNA (shRNA) increased expression of osteoblast-related genes. Studies using wild-type and mutated osteocalcin and Bsp promoters revealed that Pbx1 acts through a Pbx-binding site that is required to attenuate gene activation by Hoxa10. Chromatin-associated Pbx1 and Hoxa10 were present at osteoblast-related gene promoters preceding gene expression, but only Hoxa10 was associated with these promoters during transcription. Our results show that Pbx1 is associated with histone deacetylases normally linked with chromatin inactivation. Loss of Pbx1 from osteoblast promoters in differentiated osteoblasts was associated with increased histone acetylation and CBP/p300 recruitment, as well as decreased H3K9 methylation. We propose that Pbx1 plays a central role in attenuating the ability of Hoxa10 to activate osteoblast-related genes in order to establish temporal regulation of gene expression during osteogenesis.
Embryonic patterning of the skeleton is a complex process responsible for regulating the positional identity, shape, and size of skeletal primordial elements by an intricate orchestration of signaling molecules and transcriptional regulators. The most widely studied transcriptional regulators governing embryonic patterning are the Hox homeodomain DNA-binding proteins. Several emerging lines of evidence have demonstrated that these Hox proteins play a wider role, extending their functions from patterning the developing embryo to regulating formation and maintenance of mature tissues such as bone (13, 18, 19, 21). In adults, bone turnover is a continual process of resorption and renewal that depends on the commitment of mesenchymal progenitors to the osteoblast lineages. This process is mediated by a variety of extracellular signals that are also necessary for embryonic skeletal development, including canonical Wnt signals and bone morphogenetic proteins (BMPs) (5, 12, 27). Homeodomain proteins, including the abdominal-class Hox family, have been identified as downstream targets and regulators of osteogenic BMP signaling (23, 25, 26). One of the more compelling questions regarding Hox function is the mechanism(s) by which these proteins regulate transcription in a cell-specific manner to promote mature tissue regeneration.
Several abdominal-class Hox proteins are upregulated in bone repair, including Hoxa2, Hoxd9, Hoxa11, and Hoxa13 (15, 24), suggesting that the embryonic program is reactivated during new bone formation. For example, Hoxa10 is strongly induced in osteoblasts immediately following BMP2 treatment, coincident with the robust expression of Runx2, the master regulator of osteoblast commitment (2). Hoxa10 has been shown to bind to several phenotypic bone promoters at Hox consensus sequences and to enhance osteocalcin (bglap2; Ocn) transcriptional activity in reporter assays in vitro (19). Although the exogenous expression of Hoxa10 can induce the transcription of several bone genes in normal osteoblasts, Hoxa10 binds to chromatin of endogenous bone genes preceding the binding of Runx2 (19). Hoxa10 has been postulated to mark chromatin for activation by allowing Runx2 binding and subsequent gene expression. It is important to note that during osteoblast differentiation phenotypic genes are temporally expressed, and this sequential pattern of expression suggests that several chromatin remodeling factors are required for osteoblastic gene activation. Perhaps due to the critical but varied roles of Hox proteins, a large complement of coregulatory molecules serve as coactivators or corepressors of Hox-mediated transcriptional activity, functioning by direct protein-protein interactions or by modulating Hox DNA-binding activity (31).
One of the best characterized groups of Hox-interacting proteins is the three-amino-acid loop extension (TALE) family of homeodomain-containing transcription factors, which includes the pre-B-cell leukemia homeobox protein 1 (Pbx1). Like Hox proteins, Pbx1 plays a critical role in patterning of the axial skeleton, with deletion of the gene resulting in severe skeletal malformations due to defects in chondrocyte maturation (40). Pbx1 regulation of skeletal development has been shown to be both dependent and independent of Hox protein involvement (6, 31). Several TALE proteins are closely related, but loss of Pbx2 or Pbx3 does not confer a skeletal phenotype (36, 41); thus, Pbx1 may have a distinct role during skeletal development. Furthermore, mapping of polymorphisms related to bone mineral density in Chinese populations revealed a strong correlation between nucleotide variation in the PBX1 gene and decreased bone mass (10). It has been suggested that during mesenchymal cell commitment to the myogenic lineage, Pbx proteins may act as “pioneer” transcription factors that penetrate repressive chromatin and mark specific genes for activation by MyoD (38). In this study we addressed the functional role of Pbx1 in osteogenesis and the Hoxa10-dependent activation of the osteocalcin and bone sialoprotein (Bsp) genes as classical models for osteoblast-specific gene expression.
MATERIALS AND METHODS
Cell culture.
Human embryonic kidney cells (HEK-293T), mouse preosteoblasts (MC3T3-E1), and mesenchymal cells (C3H10T1/2) were obtained from ATCC (Manassas, VA). HEK-293T and MC3T3-E1 cells were maintained in minimum essential medium alpha medium (αMEM) (Invitrogen/Gibco Carlsbad, CA) supplemented with 10% fetal bovine serum (FBS) (HyClone, Logan, UT), 2 mM l-glutamine, and 100 U/ml of penicillin and 100 μg/ml streptomycin (Pen/Strep). C3H10T1/2 cells were maintained in Dulbecco's modified Eagle's medium (DMEM) (Invitrogen/Gibco) containing 10% FBS, 2 mM l-glutamine, and Pen/Strep. Primary rat osteoprogenitors were isolated from embryonic (day 21) rat calvaria as previously described (16). Osteoprogenitor cells were maintained in Dulbecco's minimal essential medium (MEM) (Invitrogen/Gibco) containing 10% FBS, 2 mM l-glutamine, and Pen/Strep. To induce osteogenic differentiation, media were further supplemented with BMP2 (100 ng/ml) (generously provided by John Wozney [Wyeth Research, Women's Health and Musculoskeletal Biology, Cambridge, MA]) and/or 280 μM ascorbic acid and 10 mM β-glycerophosphate (βGP) (Sigma Aldrich, St. Louis, MO). Media were replaced every 2 days for the duration of all experiments. All cells were maintained at 37°C in a humidified 5% CO2 environment.
Expression constructs and short hairpin RNA (shRNA) virus generation.
The Xpress-Pbx1 plasmid used in this study was previously described (19). The Flag-Pbx1 construct was generated by excising the Pbx1 fragment from pCMV-Pbx1 (generously provided by Corey Largman, University of California, San Francisco) by BamHI digestion followed by ligation into linearized pCDNA3 (Invitrogen). To create the Pbx1-pSuper construct used for lentiviral packaging, a PCR fragment of Pbx1-pCDNA3.1, containing the cytomegalovirus (CMV) promoter, Pbx1 sequence, and poly(A) signal with additional EcoRI and ClaI overhangs, was generated and ligated into linearized pSuper vector. Pbx1-shRNA pGIPZ plasmid was obtained from Open Biosystems (Thermo Fisher Scientific, Huntsville, AL). All plasmids were either entirely or partially sequenced to ensure fidelity at the University of Massachusetts Medical School sequencing facility (Worcester, MA).
For viral packing, lentiviral vectors containing Pbx1 or Pbx1-shRNA sequences were cotransfected along with pMD2.G and pCMVΔR-8.91 (generously provided by Didier Trono; Addgene plasmids 12259 and 12263) viral plasmids into HEK293T cells. Cells were propagated for approximately 48 h posttransfection, and the supernatant was collected, analyzed for titer in HEK293T cells, and subsequently used for infections.
Antibodies.
The following antibodies were used for Western blotting: anti-Pbx1 (P-20, sc-889; Santa Cruz Biotechnology Inc., Santa Cruz, CA), anti-Hoxa10 (N-20, sc-17158; Santa Cruz), anti-Runx2 (monoclonal, clone 8G5; MBL International, Woburn, MA), anti-histone deacetylase 1 (anti-HDAC1) (05-641; Upstate/Millipore, Billerica, MA), and anti-HDAC3 (B-12, 39500; Active Motif, Carlsbad, CA). The following antibodies were used for immunoprecipitations and/or chromatin immunoprecipitations and immunohistochemistry: anti-RNA polymerase II (Pol II) (clone 8WG16; Covance), anti-Runx2 (M-70, sc-10758; Santa Cruz), anti-Hoxa10 (N-20X, sc-17158X; Santa Cruz), anti-Pbx1(4342; Cell Signaling Technology, Danvers, MA), CBP (C-20, sc-10758; Santa Cruz), anti-p300 (N-20, sc-584; Santa Cruz), anti-penta-acetylated histone H4 (06-946; Upstate), anti-histone H3 acetylated lysine 9 (07-352; Upstate), anti-histone H4 acetylated lysine 16 (ab1762; Abcam, Cambridge, MA), anti-HDAC2 (C-19, sc-6296; Santa Cruz), anti-HDAC6 (L-18, sc-5258; Santa Cruz), and anti-histone H3 dimethyl lysine 9 (ab9050; Abcam).
Immunohistochemistry.
Long bones of 2-day-old mice were fixed with PLP fixative (2% paraformaldehyde, 0.075 M lysine, 0.037 M sodium phosphate, 0.01 M periodate, pH 7.4) for 48 h. The fixed long bones were then demineralized by successive treatment with 18% EDTA (disodium salt dihydrate, pH 7.4) for 14 days, dehydrated, and embedded in paraffin using standard procedures. Paraffin-embedded tissues were sectioned (5 μm), adhered to glass slides, and rehydrated, and antigens were recovered by treatment with retrieval buffer (1 mM Tris, 0.5 mM EGTA, pH 9.0). Sections were blocked with phosphate-buffered saline supplemented with 1% bovine serum albumin, 0.05% saponin, and 0.2% gelatin. Serial sections were incubated overnight at 4°C with anti-Pbx1 (Cell Signaling), anti-Hoxa10 (N-20), or anti-Runx2 (M-70) diluted (1:100) in phosphate-buffered saline (PBS) supplemented with 0.1% bovine serum albumin and 0.3% Triton X-100. Labeling was visualized with species-specific horseradish peroxidase-conjugated secondary antibody (P448, 1:200; Dako). Sections were counterstained with methyl green (0.5% methyl green, 0.1 M sodium acetate trihydrate, pH 4.2) or toluidine blue (0.1% toluidine blue, 7% ethanol, 150 mM sodium chloride, pH 2.3).
Transfection and reporter assays.
A 1.1-kbp DNA fragment encompassing the proximal, tissue-specific promoter of the rat osteocalcin gene was inserted into pGL3 basic vector as previously described (16). A pGL3 basic vector containing a 938-bp region of the rat bone sialoprotein proximal promoter region was previously described (34) (generously provided by the late Jaro Sodek, University of Toronto). Alteration of the Pbx or Hox DNA consensus sequence was performed by using a QuikChange site-directed mutagenesis kit (Stratagene, Cedar Creek, TX). Cells were transfected with 500 ng of reporter plasmid and the indicated concentrations of Hoxa10, Pbx1, or pCDNA 3.1 expression vector using FuGENE6 transfection reagent according to the manufacturer's instructions (Roche). At 16 to 24 h posttransfection, cells were scraped into PBS, collected by centrifugation, and lysed with 5× passive lysis buffer (Promega, Madison, WI). In each experiment, cells were also cotransfected with phRL-null Renilla luciferase plasmids for normalization. Firefly and Renilla luciferase activities were assayed using the dual-luciferase reporter assay system according to manufacturer's instructions (Promega). Experiments were performed in triplicate and results displayed as mean values ± standard errors of the means (SEM).
RNA isolation and RT-qPCR.
Total RNA was isolated using Trizol reagent (Invitrogen) according to the manufacturer's specifications. DNase I-treated total cellular RNA was primed with oligo(dT) and reverse transcribed into cDNA using the Superscript first-strand cDNA synthesis kit (Invitrogen) according to the manufacturer's instructions. Gene expression was determined by quantitative real-time reverse transcription-PCR (RT-qPCR) using Power SYBR green PCR master mix (Applied Biosystems Inc., Foster City, CA) and gene-specific primers in an ABI Prism 7000 thermocycler. For each gene, expression levels were normalized to GAPDH (glyceraldehyde-3-phosphate dehydrogenase). Experiments were performed in triplicate and results displayed as mean values ± SEM.
Immunoblotting.
Cells were lysed in direct lysis buffer (2% SDS, 10 mM dithiothreitol [DTT], 10% glycerol, 12% urea, 10 mM Tris-HCl [pH 7.5], 1 mM phenylmethylsulfonyl fluoride [PMSF], 1× protease inhibitor cocktail [Roche], 25 μM MG132 [proteasome inhibitor]) and boiled for 5 min. Samples were quantified, and equal amounts of protein were resolved by SDS-PAGE. Proteins were transferred to a polyvinylidene difluoride membrane and were subjected to immunoblotting with the appropriate antibodies. Immunoreactive proteins were detected using Western Lightning chemiluminescence reagent (Perkin-Elmer, Boston, MA).
ChIP assays.
Chromatin immunoprecipitation (ChIP) assays were performed as previously described (17). Briefly, formaldehyde cross-linking was performed for 15 min, and cells were collected in 1× PBS and then lysed in lysis buffer (25 mM Tris-HCl [pH 8.0], 5 mM MgCl2, 10 mM EDTA, 1% SDS, 1% Triton X-100, 162.5 mM NaCl, 25 μM MG-132, and 1× Complete protease inhibitor). Lysates were sonicated to obtain DNA fragments with an average size of 0.2 kbp to 0.6 kbp. In some experiments, chromatin was treated with PstI restriction endonuclease (NEB, Danvers, MA) for 2 h at 37°C. Immunoprecipitations were performed with the appropriate antibodies or immunoglobulin G (IgG) as a control. Immune complexes were collected, followed by recovery of DNA.
Sequential ChIP studies were performed using the primary pulldown from one antibody, which was divided into equal aliquots for the second pulldown with antibodies specific for coregulatory molecules. Instead of the elution step (1% SDS and 100 mM Na2HCO3) after washing, immunocomplexes were eluted in 10 mM DTT. The eluate was further diluted 1:40 in ChIP dilution buffer (0.01% SDS, 1.1% Triton X-100, 1.2 mM EDTA, 167 mM Tris-HCl [pH 8.1], 167 mM NaCl) and used for the secondary immunoprecipitations.
Aliquots of each recovered DNA sample were assayed by quantitative PCR to detect the proximal Ocn promoter region spanning bp −198 to −28 upstream of the transcription start site. As a control for nonspecific binding/proper DNA fragmentation, a region of the Ocn distal promoter from bp −1377 to −1184 was also analyzed. The oligonucleotides used were as follows: proximal promoter, 5′-GGC AGC CTC TGA TTG TGT CC-3′ (−198 to −179) and 5′-TAT ATC CAC TGC CTG AGC GG-3′ (−47 to −28); Pbx site, 5′-GTC TCT AGG GCC AGC CAG TGC-3′(−305 to −285) and 5′-AGG CTG CCG GGT CCT GAC AT-3′(−211 to −190); Hox site, 5′-CCT ATT GCG CAC ATG ACC CCC A-3′ (−112 to −91) and 5′-GCA CCG AGT TGC TGT GTG GGA-3′(−5 to + 15); and distal promoter, 5′-TGT CTT CAG GCA CAC CAG AAG-3′ (−1377 to −1356) and 5′-AAA TCT GCA GCC GTT CCC CCA GT −3′ (−1207 to −1184). Samples were normalized to the initial input and expressed as percent chromatin pulldown (compared to input).
Immunoprecipitations.
Initially, protein A/G plus agarose beads (Santa Cruz) (100 μl; 1:1 beads/PBS) were noncovalently complexed with 20 to 100 μg of specific antibody. Unbound antibodies were washed away with phosphate-buffered saline (PBS), and antibody-bound protein A/G agarose was resuspended in 100 μl PBS. Disuccinimidyl suberate (DSS) cross-linker (Pierce) was dissolved in dimethyl sulfoxide (DMSO) and added to the antibody-protein A/G solution at a final concentration of 1 mM. The cross-linking reaction was performed at room temperature for 1 h. Excess DSS was removed by washing the resin four times with 400 μl of Tris-buffered saline (TBS) (50 mM Tris, 150 mM NaCl, pH 7.2), four times with 0.1 M glycine (pH 2.8) to remove free antibody, and finally three times with TBS. The cross-linking efficiency was evaluated by A280. Approximately 107 cells per immunoprecipitation were lysed in 500 μl of Nonidet P-40 (NP-40) lysis buffer (150 mM NaCl, 50 mM Tris [pH 8.0], 1% NP-40, 1× Complete protease inhibitor [Roche], 25 μM MG132 [Sigma Aldrich]) for 15 min at 4°C. Cell lysates were sonicated, followed by centrifugation at 16,000 × g for 15 min at 4°C. The supernatant was transferred to a clean microcentrifuge tube and precleared with 40 μl of protein A/G plus agarose beads at 4°C for 30 min. To precipitate immunocomplexes, 50 μl of antibody-protein A/G plus agarose complex was added and incubated at 4°C with agitation for 1 to 4 h. Beads were washed three times with 1× PBS containing 1× protease inhibitors and eluted in 50 μl 0.1 M glycine (pH 2.8). Samples were neutralized by addition of 50 μl Tris-HCl (pH 9.2), denatured by addition of 6× sample buffer followed by boiling for 5 min, and then analyzed by Western blotting.
Statistical analysis.
Statistical analysis was performed using Prism software (Graphpad Software, La Jolla, CA). Gene expression data were repeated at least three times and analyzed by one or two-way analysis of variance (ANOVA) followed by the Bonferroni posttest. Chromatin immunoprecipitation data were repeated at least two times and analyzed by Student's t test.
RESULTS
Pbx1 is highly expressed in osteoprogenitors and decreases during osteoblast differentiation.
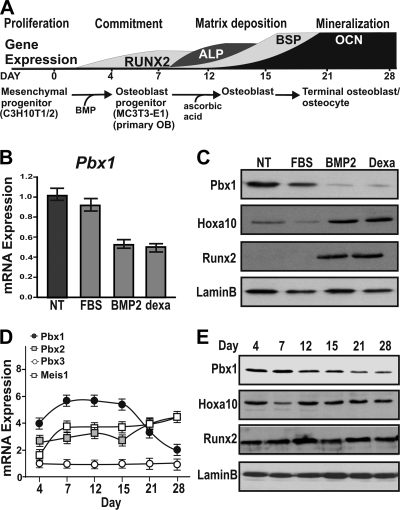
Pbx1 is strongly associated with skeletal patterning during embryonic development (40); however, the expression profile of Pbx1 from mesenchymal progenitors through stages of osteoblast differentiation is not clear. The commitment to the osteoblast phenotype is normally characterized by expression of specific genes (e.g., Runx2, the alkaline phosphatase gene [Alp], Bsp, and Ocn) at defined time points (Fig. 1A), and we sought to determine the expression of Pbx1 through these defined stages. We used two different progenitor cell lines representing distinct stages of commitment to the osteoblast lineage. When treated with BMP2 or dexamethasone to induce osteoblast differentiation, C3H10T1/2 cells, a model of multipotential mesenchymal progenitors, exhibited decreased Pbx1 mRNA expression (Fig. 1B) and protein levels (Fig. 1C). Analysis of lineage-committed MC3T3-E1 preosteoblastic cells revealed that Pbx1 expression was relatively high during the proliferative stage (day 4) but decreased consistently after day 15 with maturation of the osteoblast precursors (Fig. 1D). Correspondingly, Pbx1 protein levels were reduced at later stages of the differentiation time course (Fig. 1E). In contrast, mRNAs of other TALE family proteins (Pbx2, Pbx3, and Meis1) were detectable at low levels and did not decline during osteoblast differentiation (Fig. 1D). These findings suggest that unlike other TALE family members, Pbx1 is specifically downregulated during differentiation to a mature osteoblast.
FIG. 1.
Pbx1 expression decreases during osteoblast differentiation. (A) Schematic illustrating temporal gene expression during osteoblast differentiation. Osteoblast-like differentiation was induced in C3H10T1/2 cells by treatment with either 2% FBS (control), 100 ng/ml BMP2, or 10 nM dexamethasone for 7 days. (B) Relative mRNA expression of Pbx1 in C3H10T1/2 cells was determined by RT-qPCR. (C) Protein expression was determined by Western blotting using an anti-Pbx1 antibody. MC3T-3E1 cells were treated with 280 μM ascorbic acid-5 mM β-glycerol phosphate to induce osteogenic differentiation for a total period of 28 days. (D) Total RNA was isolated, and relative expression of TALE protein family genes was determined by real-time quantitative PCR using gene-specific primers. (E) Relative protein expression of Pbx1 over the 28-day time course was determined by Western blotting using an anti-Pbx1 antibody. Error bars indicate SEM.
Pbx1 is expressed in the bone environment.
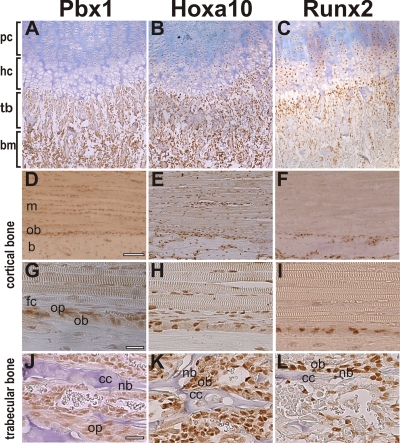
We sought to determine if the pattern of Pbx1 expression observed in maturing osteoblasts was indicative of protein expression in native bone. Immunohistochemical staining of mouse long bones revealed that Pbx1 was expressed in bone marrow cells and proliferating chondrocytes but could not be observed in hypertrophic chondrocytes (Fig. 2A ). Pbx1 was also expressed in preosteoblasts and active osteoblasts on newly forming trabecular bone (Fig. 2A, G, and J). On the periosteal bone surface, robust Pbx1 staining could be observed in muscle cells and osteoprogenitors but not in mature osteoblasts (cells directly adjacent to new bone) (Fig. 2D and J). In contrast, Hoxa10 could be observed in most cells within cartilage, bone, and muscle (Fig. 2B, E, H, and K), while Runx2 was restricted primarily to hypertrophic chondrocytes and osteoblasts and was not expressed in bone marrow or muscle (Fig. 2C, F, I, and L). These findings suggest that in bone Pbx1 is present in osteoprogenitor cells but is reduced in mature osteoblasts, supporting the notion that Pbx1 is downregulated during osteoblast maturation.
FIG. 2.
Pbx1 is expressed in bone but not in mature osteoblasts. Two-day-old mouse long bones were demineralized and stained for Pbx1, Hoxa10, or Runx2 as indicated. (A, B, and C) Mouse femur region displaying proliferating chondrocytes (pc), trabecular bone (tb), bone marrow (bm), and hypertrophic chondrocytes (hc). (D, E, and F) Cortical bone demonstrating muscle (m), periosteal region (p), and mature bone (b). Scale bar, 200 μm. (G to L) High-magnification images (scale bars, 50 μm) for cortical bone with adjacent periosteum and muscle, showing fibrous capsule (fc), osteoprogenitors (op) with robust Pbx1 staining, and mature osteoblasts (ob) (G, H, and I), and for trabecular bone, demonstrating newly formed bone (nb) with many osteoprogenitors (op) surrounding calcified cartilage (cc) (J, K, and L).
Pbx1 functionally impairs osteoblast differentiation.
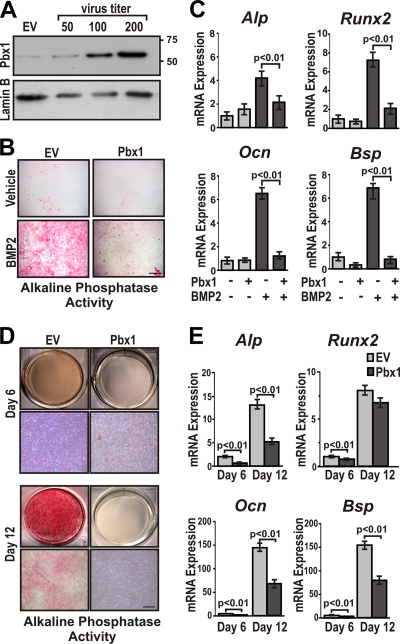
To understand the contribution of Pbx1 in commitment and maintenance of the osteoblast lineage, Pbx1 was exogenously expressed in C3H10T1/2 cells under the control of a CMV promoter using recombinant lentivirus. Increased virus titer demonstrated a dose-dependent correlation with Pbx1 protein levels (Fig. 3A), and high expression of Pbx1 was maintained over the 12-day experimental period (data not shown). C3H10T1/2 cells were infected with Pbx1 or control (green fluorescent protein [GFP]) lentivirus and treated with BMP2 to induce osteoblast differentiation. Pbx1 overexpression decreased alkaline phosphatase activity by 50% compared to controls (Fig. 3B). Examination of a panel of osteoblast markers (Alp, Runx2, Ocn, and Bsp) by RT-qPCR also showed significant (P < 0.01) reductions (10 to 25% of control values) in expression of these endogenous genes in response to Pbx1 (Fig. 3C). Similarly, C2C12 cells (a mesenchymal/myogenic progenitor with osteoblastic potential) examined under the same conditions demonstrated a significant (P < 0.01) decrease in BMP2-induced osteoblastogenesis in response to Pbx1 (see Fig. S1 at http://labs.umassmed.edu/steinlab/). Our studies with the committed preosteoblast MC3T3-E1 cells also echoed a reduction in osteoblast-specific differentiation by Pbx1 expression from the onset of alkaline phosphatase activity (day 6) through the matrix maturation stage (day 12) (Fig. 3D). Due to the committed osteogenic phenotype of MC3T3-E1 cells, Pbx1 had no effect on expression of the early osteogenic marker Runx2 (day 12), while mature osteoblast genes Bsp and Ocn were significantly (P < 0.01) decreased, by 50% (Fig. 3E). Taken together, these findings indicate that Pbx1 is a negative regulator of mesenchymal progenitor commitment to the osteoblast phenotype as well as the maturation of osteoprogenitors.
FIG. 3.
Overexpression of Pbx1 results in decreased expression of osteoblast-related genes in mesenchymal progenitor cells. (A) C3H10T1/2 cells were infected with several concentrations (∼50, 100, and 200 PFU/cell) of recombinant lentivirus encoding Pbx1. Relative expression of Pbx1 was determined by Western blotting using an anti-Pbx1 antibody (N-20; Santa Cruz) followed by detection with anti-rabbit-horseradish peroxidase (HRP)/ECL reagent. (B) and C) C3H10T1/2 cells were infected with ∼100 PFU/cell of recombinant lentivirus (EV or Pbx1). Cells were then treated with BMP2 or vehicle (PBS) for 7 days. (B) C3H10T1/2 cells overexpressing Pbx1 demonstrated less alkaline phosphatase activity. Scale bar, 500 μm. (C) Total RNA was isolated from C3H10T1/2 cells, and relative expression of osteoblast-related genes was determined by RT-qPCR using gene-specific primers. (D and E) MC3T3-E1 cells were infected with ∼100 PFU/cell of recombinant lentivirus (EV or Pbx1) and treated with 280 μM ascorbic acid-5 mM β-glycerol phosphate to induce osteogenic differentiation for a period of 6 and 12 days. (D) MC3T3-E1 cells were fixed and stained for alkaline phosphatase activity. (E) Total RNA was isolated from MC3T3-E1 cells, and relative expression of osteoblast-related genes was determined by RT-qPCR using gene-specific primers. Statistical significance was determined by one-way ANOVA followed by a Bonferroni posttest. Data are presented as the means from three experiments ± SEM.
Pbx1 negatively regulates osteogenic genes: depletion of Pbx1 by shRNA increases osteoblast differentiation.
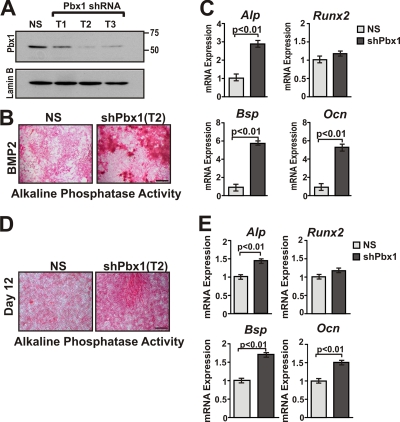
The reduction of osteoblast markers by Pbx1 overexpression in differentiated cells suggested that Pbx1 may have a biological role in attenuating osteoblast-specific gene expression. To address this question, several Pbx1 shRNAs were generated and exogenously expressed in C3H10T1/2 cells using recombinant lentivirus. All three Pbx1 shRNA sequences (T1, T2, and T3) successfully decreased Pbx1 protein levels (Fig. 4A). This decrease in Pbx1 levels was maintained for the period of the experiments (>20 days) (data not shown), and the shRNA construct generating the most efficient knockdown (T2) was used for subsequent studies. C3H10T1/2 cells with reduced Pbx1 expression exhibited increased (3-fold) alkaline phosphatase activity and expression compared to cells infected with the nonsilencing (NS) control (Fig. 4B). Additionally in Pbx1-shRNA-infected cells, Ocn and Bsp mRNA levels, markers of the mature osteoblast, were upregulated 5- to 6-fold compared to NS control cells (Fig. 4C), while Runx2 mRNA levels did not change significantly. Pbx1-shRNA resulted in a similar enhancement of osteoblast differentiation in MC3T3-E1 cells, with significant (P < 0.01) increases in Alp activity and Alp, Ocn, and Bsp mRNA levels compared to those in NS-infected controls but no change in Runx2 mRNA (Fig. 4D and E). These findings are consistent with the Pbx1 expression profile in osteoblasts (Fig. 1 and 2) and forced-expression studies (Fig. 3) and establish that Pbx1 is a potent negative regulator of osteoblastic genes expressed during early and late stages of osteoblast differentiation.
FIG. 4.
Depletion of Pbx1 by shRNA results in increased expression of osteoblast-related genes in mesenchymal cells. (A) to C) C3H10T1/2 cells were infected with ∼100 PFU/cell of shRNA-encoding recombinant lentivirus (nonsilencing [NS] or three different Pbx1-specific targets [T1, T2, and T3]). C3H10T1/2 cells were then treated with 100 ng/ml BMP2 or vehicle (PBS) for a period of 7 days. (A) Relative protein levels of Pbx1 in treated C3H10T1/2 cells were determined by Western blotting using an anti-Pbx1 antibody. (B) C3H10T1/2 cells treated with Pbx1-shRNA, demonstrating increased alkaline phosphatase activity. Scale bar, 500 μm. (C) Relative expression of osteoblast-related genes, monitored by RT-qPCR, was significantly increased in Pbx1-shRNA-infected C3H10T1/2 cells. (D and E) MC3T3-E1 cells were infected with shRNA-containing lentivirus (as described above) and treated with 280 μM ascorbic acid-5 mM β-glycerol phosphate for a period of 12 days to induce osteogenesis. (D) MC3T3-E1 cells treated with Pbx1-shRNA demonstrated increased alkaline phosphatase activity. (E) Relative expression of osteoblast-related genes in MC3T3-E1 cells was determined by RT-qPCR. Statistical significance was determined by one-way ANOVA followed by a Bonferroni posttest. Data are presented as the means from three experiments ± SEM.
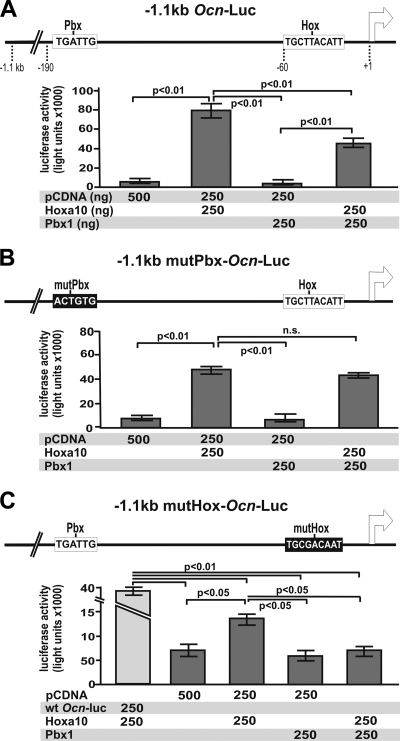
A Pbx1 regulatory element is required to inhibit Hoxa10-mediated activation of osteocalcin and bone sialoprotein promoters.
The potent effects on osteoblast gene expression mediated by alteration of Pbx1 expression levels suggested that Pbx1 may directly regulate bone-related gene promoter activity. Pbx1 has been demonstrated to bind directly to DNA and modulate the binding of Hox proteins for several promoters (22). We sought to determine if the negative regulation of the osteoblast phenotype by Pbx1 was dependent on the putative Pbx regulatory elements (33) located in the proximal promoters of several bone-related genes, including Bsp and Ocn. To address this question, Pbx1 and Hoxa10 expression plasmids were cotransfected with luciferase reporter constructs containing approximately 1 kbp of the Ocn (−1.1 kbp Ocn-Luc) (Fig. 5A) or Bsp (−938 bp Bsp-Luc) promoter region (see Fig. S2 at http://labs.umassmed.edu/steinlab/). MC3T3-E1 preosteoblasts transfected with Hoxa10 in addition to the Ocn or Bsp luciferase reporter exhibited significantly (P < 0.01) increased luciferase activity (5- to 6-fold) (Fig. 5A and data not shown). However, cotransfection of cells with Hoxa10 and Pbx1 along with either reporter construct (Ocn or Bsp) resulted in a 75% decrease in luciferase activity compared to that in cells transfected with Hoxa10 alone (Fig. 5A; also see Fig. S2 at http://labs.umassmed.edu/steinlab/). There was no change in luciferase activity in cells transfected with Pbx1 alone. These results indicate that Pbx1 acts to reduce Hoxa10-mediated transcriptional activation at bone-specific promoters.
FIG. 5.
Alteration of Pbx1 consensus sequence in the osteocalcin promoter results in decreased attenuation of Hoxa10 activity. MC3T3E1 preosteoblasts were transfected with 500 ng of the indicated luciferase reporter plasmids as well as the indicated amounts (ng) of pCDNA3, pCDNA-Hoxa10, and/or pCDNA-Pbx1, and relative luciferase activity was assessed with a luminometer. (A) Luciferase reporter construct (pGL3-Basic) containing the −1.1-kb region of rat Ocn (−1.1 kb Ocn-Luc). Hoxa10-mediated luciferase reporter activity was significantly decreased upon cotransfection of Pbx1. (B and C) The −1.1-kb Ocn-luc plasmid was modified by site-directed mutagenesis to alter the putative Pbx1 DNA consensus sequence (−1.1 kb mutPbx-Ocn-Luc) or the Hox DNA consensus sequence (−1.1 kb mutHox-Ocn-Luc). (B) Alteration of the Pbx1 DNA consensus sequence resulted in loss of Pbx1 repression of Hoxa10-mediated Ocn-luciferase activity. (C) Alteration of Hox sequence resulted in a significant decrease in Ocn-driven luciferase activity and no significant activation in the presence of Pbx1. Statistical significance was determined by two-way ANOVA followed by a Bonferroni posttest. Data are presented as the means from three experiments ± SEM.
We next identified that the Pbx1-binding site is functionally active by determining that in vitro-translated (IVTT) recombinant Pbx1 and MC3T3-E1 nuclear protein extracts could bind to the radioactively labeled probe (Ocn promoter sequence GGCAGCCTCTGATTGTGTCC) by electrophoretic mobility shift assay (see Fig. S3 at http://labs.umassmed.edu/steinlab/). The presence of the Pbx1 consensus sequence or Pbx1 antibody reduced binding of Pbx1 to the labeled oligonucleotide in both recombinant protein and MC3T3-E1 nuclear extracts.
We next addressed whether the putative Pbx site is directly involved in binding Pbx1 and plays a role in Hoxa10 repression. An Ocn reporter plasmid with the Pbx site mutated was cotransfected with Hoxa10, which resulted in similar levels of Hoxa10-mediated activation as with the wild-type (WT) Ocn plasmid (Fig. 5B). However, cotransfection of the mutant Pbx-Ocn promoter with both Hoxa10 and Pbx1 did not show the expected decrease in Hoxa10 stimulated activity (Fig. 5B). Similar results were observed for the equivalently mutated Bsp promoter (see Fig. S2 at http://labs.umassmed.edu/steinlab/). Thus, the Pbx-binding sequence is required for Pbx1-mediated repression of Hoxa10 transcriptional activity. This conclusion is further supported by mutation of the Hox-binding sequence in the Ocn promoter, which resulted in significantly (P < 0.01) reduced luciferase activity when cotransfected with Hoxa10 expression plasmid (Fig. 5C, bars 1 and 3). The weak level of activity of the mutHox-Ocn reporter (Fig. 5C) could be due to the contribution from increased Runx2-mediated transcriptional activity, as Hoxa10 can directly upregulate Runx2 expression (19). However, all Hox-mediated activation is significantly (P < 0.05) decreased by cotransfection of Pbx1 (Fig. 5C). Taken together, these results provide direct evidence that Pbx1-DNA interactions are required to block Hoxa10-mediated transcriptional activation of osteoblast-related genes.
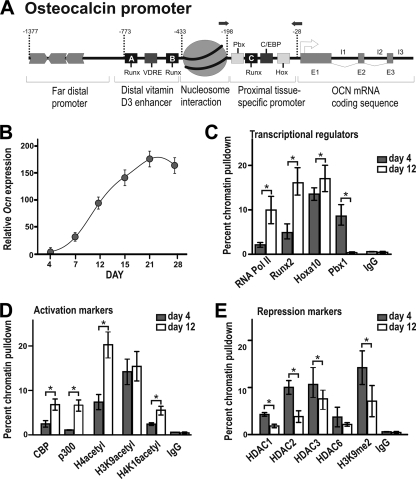
Pbx1 is associated with repression of chromatin remodeling on the osteocalcin promoter.
Our previous findings identified Hoxa10 as an early activator of bone gene expression and demonstrated that Hoxa10 functions by facilitating the remodeling of chromatin (18). Given the role of Pbx1 as a mediator of Hox function, we sought to establish if Pbx1 is associated with chromatin remodeling of the temporally expressed osteocalcin gene during osteoblast differentiation. The Ocn gene promoter (Fig. 6A) and the Bsp promoter (see Fig. S4A and B at http://labs.umassmed.edu/steinlab/) are strongly regulated by chromatin structure and are tightly packaged as heterochromatin in proliferating cells (day 4) (32). During transcriptional activation, the proximal promoter must adopt an open conformation to allow binding of transcriptional regulators, resulting in an exponential increase in gene expression (day 12) (Fig. 6B). By chromatin immunoprecipitation analysis, Pbx1 is strongly associated with the Ocn and Bsp promoters prior to expression (day 4) but not during exponential gene expression (day 12) (Fig. 6C; also see Fig. S4C at http://labs.umassmed.edu/steinlab/). This result was in contrast to the pattern observed with RNA polymerase II and known transcriptional activators (e.g., Runx2 and Hoxa10), all of which demonstrated increased Ocn promoter occupancy with increased Ocn gene expression (Fig. 6C). Several mediators and covalent modifications of histone proteins associated with chromatin remodeling, including CBP/p300, acetylated histone H4, and acetylated H4 lysine 16 (H4K16), were found to increase with osteoblast differentiation (Fig. 6D; also see Fig. S4D at http://labs.umassmed.edu/steinlab/), although acetylation of H3 lysine 9 (H3K9) was unchanged. In contrast repressive marks on chromatin, including histone deacetylases (HDAC) 1, 2, and 3, as well as methylation of histone H3K9, demonstrated a decrease upon osteoblast differentiation (Fig. 6E; also see Fig. S4E at http://labs.umassmed.edu/steinlab/). These results strongly suggest that Pbx1 is associated with negative regulation of chromatin remodeling at the osteocalcin and bone sialoprotein promoters.
FIG. 6.
Pbx1 displays a pattern of gene-repressive functionality on the osteocalcin promoter. (A) Diagram of rat osteocalcin promoter displaying relative binding sites and primer sites used for chromatin immunoprecipitation analysis. Rat calvarial osteoblasts were isolated from embryonic day 18.5 rat pups and collected during the proliferative stage (day 4) or cultured in differentiating conditions and collected during exponential increase in osteocalcin expression (day 12). (B) Calvarial osteoblasts were analyzed at the indicated differentiation stages by RT-qPCR to determine levels of osteocalcin gene expression. (C to E) The ChIP analysis was performed on cleared lysates from primary osteoblasts using ∼5 μg of the indicated antibody. Recovered DNA was then quantified by qPCR using primers specific for the proximal promoter region of the osteocalcin gene to determine relative occupancy of transcriptional activators (C), activation markers (D), or repressive markers (E). Statistical significance was determined by Student's t test (*, P < 0.05 versus matched control). ChIP experiments were repeated at least two times with similar results, and one representative experiment is presented (± standard deviation [SD]).
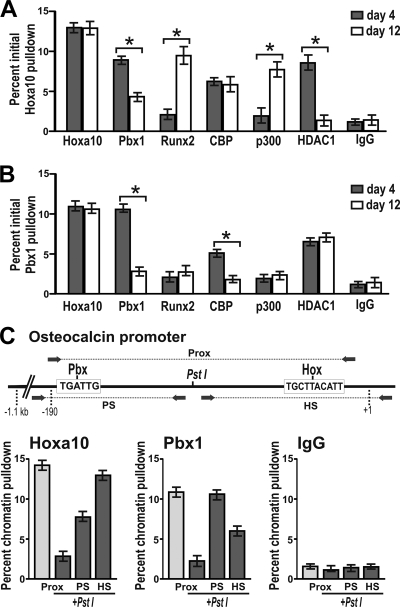
To address whether there is a direct association of Hoxa10 and Pbx1 with regulators of chromatin remodeling at the osteocalcin promoter in differentiated osteoblasts, two-step chromatin immunoprecipitation (ChIP-reChIP) experiments were performed using a primary pulldown with an anti-Hoxa10 or anti-Pbx1 antibody. The chromatin fraction associated with Hoxa10 at the Ocn promoter demonstrated a high level of association with Pbx1, as the secondary pulldown with anti-Pbx1 antibody resulted in a signal that was approximately 60% of the input (Hoxa10) (Fig. 7A). Consistent with the primary ChIP analysis (Fig. 6C), the association of Pbx1 with the Hoxa10 chromatin fraction was greatly reduced at day 12. In comparison, Runx2 was weakly associated with the Hoxa10 chromatin fraction at day 4 and subsequently increased at day 12. This increased binding of Runx2 in association with Hoxa10 on the Ocn promoter at day 12 reflects upregulation of the Ocn gene, which is also demonstrated by the 4- to 5-fold-increased association of the transcriptional coactivator p300.
FIG. 7.
Pbx1 cooccupancy with repressive histone-modifying enzymes and Hoxa10 on the osteocalcin promoter in proliferating cells. Cross-linked cell lysates from primary calvarial osteoblasts were immunoprecipitated using either anti-Hoxa10 or anti-Pbx1 antibody, and the resulting fraction was further immunoprecipitated using the specified antibodies (ChIP-reChIP). (A) The Hoxa10-immunoprecipitated fraction was further immunoprecipitated using Hoxa10, Runx2, Pbx1, CBP, p300, HDAC1, or control (IgG) antibodies and quantified by qPCR. (B) The Pbx1-immunoprecipitated fraction was further immunoprecipitated using Hoxa10, Runx2, Pbx1, CBP, p300, HDAC1, or control (IgG) antibodies and quantified by qPCR. (C) Cross-linked chromatin from proliferating calvarial osteoblasts was treated with PstI to cleave DNA between the Hoxa10- and Pbx1-binding sites, immunoprecipitated with anti-Hoxa10 or anti-Pbx1 antibody, and evaluated with primer sets specific for digested (Pbx site [PS] or Hox site [HS]) or undigested (proximal promoter [Prox]) fragments. Statistical significance was determined by Student's t test (*, P < 0.05 versus matched control). the ChIP experiment was repeated three times with similar results, and the data presented are from one representative experiment (± SD).
In contrast to Hoxa10, the Pbx1-Ocn chromatin fraction was only weakly associated with Runx2, CBP, or p300 (by secondary pulldown) at either day 4 or day 12 (Fig. 7B), reflecting the role of Pbx1 as a repressor of Ocn gene expression. However, the Pbx1 chromatin fraction was strongly associated with HDAC1 at the osteocalcin promoter when the gene was silent during proliferation (day 4) and during bone-specific gene expression (day 12), and neither HDAC1 nor Pbx1 was strongly associated with Hoxa10 at day 12. Due to the cooccupancy of Pbx1 with negative regulators of chromatin remodeling at the osteocalcin promoter during proliferation and differentiation (Fig. 7B), we sought to determine if association of Pbx1 with these factors (including Hoxa10) was by direct protein-protein interactions. Chromatin digested with PstI restriction endonuclease to separate the Pbx and Hox binding sites was immunoprecipitated with Hoxa10, Pbx1, or control (IgG) antibodies (Fig. 7C). DNA containing the Hoxa10-binding site could be detected in both Hoxa10 and Pbx1 pulldowns by qPCR. Similar results were observed for the Pbx1-binding site fragment, suggesting that Hoxa10 and Pbx1 may interact through direct protein-protein interactions on the Ocn promoter.
To detect other protein-protein interactions, whole-cell lysates from confluent MC3T3-E1 cells were immunoprecipitated with Hoxa10, Runx2, Pbx1, HDAC1, or control (IgG) antibodies (Fig. 8A ). Western blotting with anti-Hoxa10 and anti-Pbx1 antibodies demonstrated that Hoxa10 and Pbx1 form protein complexes with Runx2 and HDAC1 in osteoblasts. Furthermore, immunoprecipitation using anti-HDAC1 followed by Western blotting with anti-Pbx1 indicated that Pbx1 was present in complexes containing HDAC1 (Fig. 8B). Taken together, these findings identify Pbx1 functions on bone gene promoters (i) in osteoprogenitors prior to their expression by interaction with HDACs and (ii) in postproliferative mature osteoblasts, where Pbx1 dissociating from bone gene promoters results in Hoxa10-mediated changes in chromatin remodeling to promote gene transcription.
FIG. 8.
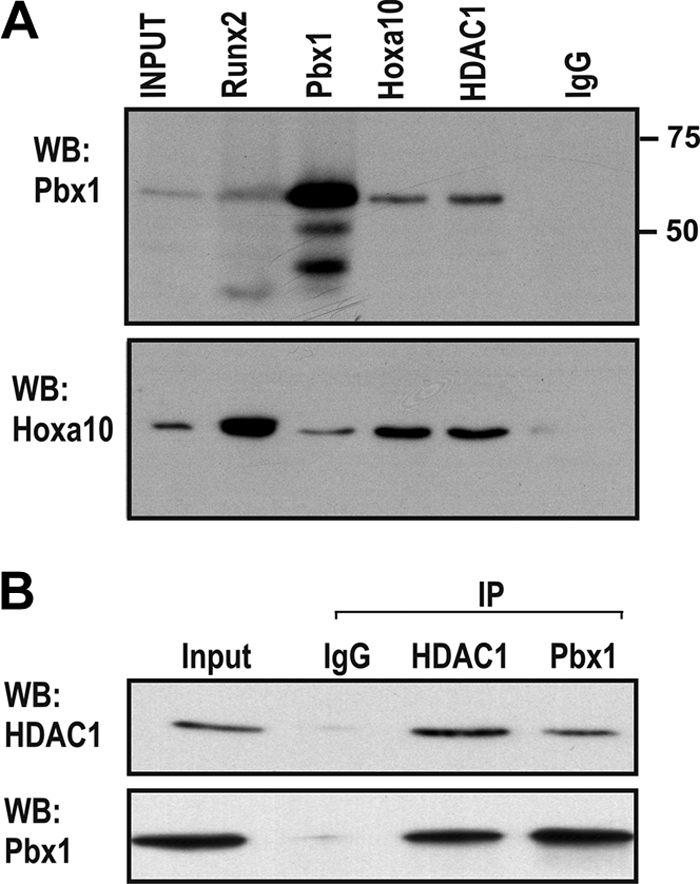
Pbx1 protein-protein interactions in osteoblasts. (A) Whole-cell lysates from confluent MC3T3-E1 cells were immunoprecipitated with Hoxa10, Runx2, Pbx1, HDAC1, or control (IgG) antibodies. Proteins were resolved by SDS-PAGE, and Hoxa10 and Pbx1 were detected by Western blotting with specific antibodies. (B) Lysates from MC3T3-E1 were used for immunoprecipitation using anti-HDAC1 followed by Western blotting with anti-Pbx1 or specific HDAC antibody.
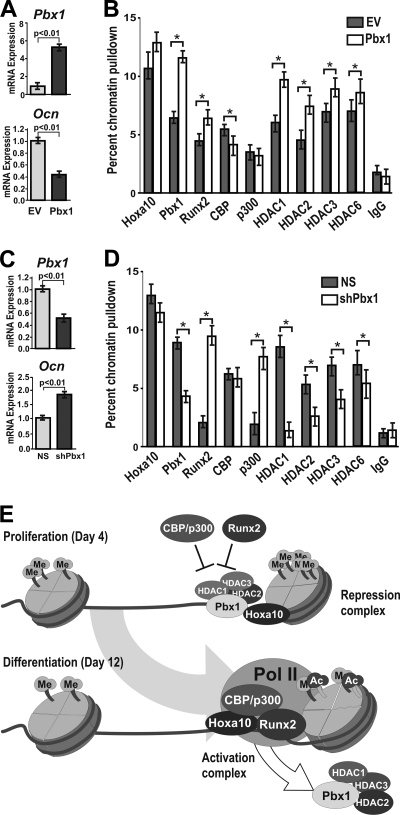
Pbx1 gain of function and loss of function alter the chromatin remodeling profile at bone-related gene promoters.
Having established that Pbx1 was directly associated with regulators of chromatin remodeling, at both the protein-protein and promoter-regulation levels, we hypothesized that modification of Pbx1 levels in osteoblasts should alter the chromatin profile at the osteocalcin promoter. Primary calvarial osteoblasts were treated with recombinant lentivirus to exogenously overexpress Pbx1 (Fig. 9A) and analyzed by ChIP (Fig. 9B). With increased Pbx1 expression, a corresponding increase in Pbx1 together with significant increases in occupancy of histone deacetylases HDAC1 and HDAC2 at the Ocn promoter was evident. In primary osteoblasts with depleted Pbx1 expression (Pbx1-shRNA) (Fig. 9C), Pbx1 binding to Ocn chromatin was decreased compared to that in NS-infected cells, as would be predicted based on reduced cellular levels of Pbx1 (Fig. 9D). While there was no effect on Hoxa10 binding, a large increase in both Runx2 and p300 occupancy at the osteocalcin promoter in cells deficient in Pbx1 as well as a reduction in occupancy of HDAC1 and HDAC2 was found, consistent with findings from ChIP-reChIP studies (Fig. 7). These results indicate that Pbx1 may be important for recruiting these HDAC factors to the osteocalcin promoter to mark the gene for expression while maintaining a repressive state during proliferation. The data suggest that Pbx1 contributes to the switching of chromatin remodeling factors from repression to open chromatin for activation, as illustrated in Fig. 9E.
FIG. 9.
Modification of Pbx1 levels in osteoblasts results in alteration of histone-modifying enzymes at the osteocalcin promoter. (A and B) Rat calvarial osteoblasts were isolated from embryonic day 18.5 rat pups and infected with Pbx1 or empty (EV) lentiviral constructs, and cells were collected at 48 h after infection after just reaching confluence (day 6). (A) Relative expression of the Pbx1 and Ocn genes was monitored by RT-qPCR. (B) ChIP analysis was performed on cleared lysates from primary osteoblasts using ∼5 μg of the indicated antibody. Recovered DNA was then quantified by qPCR using primers specific for the proximal promoter region of the osteocalcin gene to determine the relative occupancy of the indicated proteins. (C) Isolated osteoblasts (as described above) were infected with Pbx1-shRNA or nonsilencing shRNA (NS) lentiviral constructs, and relative expression of Pbx1 and Ocn was determined by RT-qPCR. (D) ChIP analysis was performed on cleared lysates from treated primary osteoblasts (as described above) to determine the relative occupancy of the indicated proteins on the osteocalcin promoter. Statistical significance was de- termined by Student's t test (*, P < 0.05 versus matched control). ChIP experiments were repeated two times with similar results, and the data presented are representative of one experiment (± SD). (E) Schematic model of Pbx-mediated repression of the osteocalcin gene.
DISCUSSION
Here we have identified an integral role for the Hox-interacting protein Pbx1 as an important regulator of the osteoblast phenotype. Pbx1 acts to coordinate the temporal expression of the Ocn and Bsp genes by Hoxa10 during osteogenesis. It is apparent that Hox and TALE genes have defined roles in embryonic development; however, our results indicate that these proteins can have analogous or concordant roles in the maintenance of adult tissues. It has been suggested that Pbx1, as well as other TALE family proteins, can act in concert with (1, 35) or independent of (4, 8) Hox proteins in its role of regulating gene expression through direct binding to cognate regulatory elements. We have provided several lines of evidence that Pbx1 is a critical negative regulator of osteoblast differentiation by demonstrating that (i) Pbx1 is highly expressed in osteoprogenitors and its expression at the transcript and protein levels decreases upon commitment to the osteoblast phenotype and again during late stages of differentiation; (ii) exogenous expression of Pbx1 or depletion by targeted-shRNA expression inhibits or increases, respectively, osteoblastogenesis and the expression of osteoblast-related genes; (iii) Pbx1 acts directly on the Ocn and Bsp promoters to repress Hoxa10-mediated expression of the gene by a mechanism in which Pbx1 recruits negative regulators of chromatin remodeling; and (iv) Pbx1 has the ability to act as a molecular switch functioning with Hoxa10 in proliferating cells to maintain osteoblast-specific genes in a repressed state and facilitate Hoxa10 activation of gene promoters by dissociating from the chromatin in differentiated cells. These key findings highlight a novel regulatory role for Pbx1 to repress and mark osteoblast genes for expression and limit Hoxa10-mediated gene transcription for regulating the temporal activation of postproliferative gene expression.
Our data suggest that Pbx1 has a role in attenuating osteogenesis by repressing osteoblast gene expression at several stages. Increased Pbx1 expression in mesenchymal precursors decreased expression of early osteoblast genes (Runx2 and Alp) and late-stage genes (Bsp and Ocn), whereas decreased Pbx1 levels resulted in increased osteoblast gene expression. The observation of enhanced commitment to the osteoblast phenotype in the absence of Pbx1 is concordant with reports of premature mineralization of skeletal elements, particularly at the hypertrophic chondrocyte zone, in growth plates in Pbx1-null mice (40). In contrast to Pbx1-null mice, which die at embryonic day 15, mice with knockouts of other TALE family genes (Pbx2 and Pbx3) survive after birth and show only slight skeletal defects, with no apparent effects on osteoblast differentiation (7, 41). Meis1-null mice die due to defects in fetal hematopoiesis and do not develop the skeletal abnormalities observed in Pbx1−/− mice (20), together suggesting that Pbx1 may have a specific role in the commitment of precursors to the osteoblast lineage. It is interesting to note that conditional Pbx1 deletion in hematopoietic stem cells results in loss of self-renewal and the premature initiation of transcriptional mechanisms, resulting in maturation of progenitor cells to a differentiated state (14). Likewise, Pbx1 depletion from the osteoprogenitor stage to the osteoblast appears to have a temporal role in promoting differentiation. Thus, Pbx function is to limit differentiation by maintaining a mesenchymal phenotype, similar to the role of the protein in maintaining the embryonic mesoderm (7).
Our previous studies demonstrated that Hoxa10 functions as an immediate-early response to the osteogenic BMP2 signal and is capable of inducing osteogenic gene expression. Therefore, Hoxa10 is an important cofactor to establish the osteoblast phenotype (19). In this study, our findings show that Pbx1 occupies bone gene promoters coincident with Hoxa10, resulting in tempered Hoxa10-mediated gene expression. In committed osteoprogenitors where Pbx1 expression is robust, it is clear that Pbx1 and Hoxa10 both bind to the osteocalcin promoter preceding gene expression. These findings are consistent with our earlier studies that showed a significant increase in Hoxa10 occupancy of bone promoters that are expressed in mature osteoblasts (19). Here we show that Hoxa10 is associated with chromatin of the actively transcribed genes (day 12).
The regulation of osteoblast genes by Pbx1 and Hoxa10 appears to be facilitated by the conserved organization of regulatory elements in the promoters of osteoblast-related genes. The proximal promoter regions of several genes generally associated with bone formation feature multiple homeodomain-binding DNA motifs, including some that preferentially bind Pbx1 (tTGAC/T) (33). Strikingly in the Bsp and Ocn promoters, the Pbx1 consensus sequence and the Hoxa10-binding site flank functional Runx2-binding elements (16, 37) (Fig. 6A; also see Fig. S4A at http://labs.umassmed.edu/steinlab/). The regulation of the Ocn promoter by Hoxa10 involves the accumulation of Hoxa10 on the Ocn promoter, peaking with active gene transcription. This finding establishes a key event for Hoxa10-mediated gene activation by dissociation of Pbx1 from the Ocn and Bsp genes when they are actively transcribed. Coincident with the dissociation of Pbx1, there is an increase in Runx2 and RNA Pol II, which marks the onset of active gene transcription. Thus, we conclude from our studies of primary osteoblasts progressing through stages of differentiation that dissociation of Pbx1 may be an important step (or switching mechanism) in allowing RNA Pol II and/or Runx2 recruitment to these promoters, resulting in gene transcription.
Chromatin remodeling through the recruitment of histone-modifying enzymes that regulate the methylation or acetylation of lysine and arginine residues is a fundamental underlying mechanism regulating transcription (29, 30). Hoxa10 as well as Pbx1 has a defined role in the patterning of the axial skeleton (6, 43, 44) by regulating chromatin remodeling that leads to phenotypic gene expression in several tissues (4, 28). Pbx1 has been demonstrated to be involved in chromatin remodeling events leading to the activation of MyoD-dependent promoters (such as myogenin), and the protein is constitutively bound to silent chromatin prior to initiation of muscle differentiation (4, 11). Supporting the concept that Pbx1 exerts functional activity through epigenetic control, the pattern of histone modifications and histone-modifying enzymes at the osteocalcin promoter indicates that Pbx1 is associated with a repressed chromatin state and that the chromatin adopts an “active” conformation marked by higher levels of H3K4 methylation and acetylation upon Pbx1 dissociation. Several studies have reported that HOX proteins (including Hoxa10) can mediate a modified chromatin acetylation status leading to transcriptional activation or repression by interacting with p300/CBP (3, 9, 42). The ChIP-reChIP experiments presented in this study demonstrate that Hoxa10 is strongly associated with CBP at the osteocalcin promoter during both proliferation and differentiation and with p300 during differentiation, while Pbx1 is localized on the osteocalcin promoter coincident with histone deacetylases and Hoxa10 in proliferating cells. This result would further imply that Pbx1 is a repressor of chromatin remodeling and gene transcription in osteoblasts. Supporting this model, it has been previously demonstrated that Hox-Pbx complexes can promote histone deacetylation by recruiting class I histone deacetylases to repress gene transcription (39). It is therefore intriguing that when colocated at the osteocalcin promoter, Hoxa10 and Pbx1 have the characteristics of both repressional and activational regulators, containing both HDACs and histone acetyltransferases (CBP/p300). Our findings suggest that even though Pbx1 is a strong inhibitor of osteoblastogenesis, it may have a temporal role in assisting in the activation of chromatin remodeling and support a switching mechanism from repression to activation.
In conclusion, in this study we have demonstrated functional interactions between Pbx1 and Hoxa10 in progenitor cells and osteoblasts for control of the temporal expression of bone-specific genes at multiple levels, including transcriptional regulation at the promoter level and initiation of transcriptional activity through chromatin remodeling. This characterization of the Pbx1 and Hoxa10 interactions on the osteocalcin promoter has provided a clearer understanding of the commitment to an osteogenic lineage preceding Runx2 recruitment in mesenchymal progenitors. This concept has implications for bone repair and molecular intervention to correct bone disorders.
Acknowledgments
We are grateful to A. Imbalzano for advice during these studies. We also thank Judy Rask for editorial assistance, members of the laboratory for helpful discussions, and Sadiq Hussain for technical support.
The studies reported here were supported in part by grants from the National Institutes of Health (DE12528, AR39588, and AR48818). Core resources supported by Diabetes Endocrinology Research Center (DERC) grant DK32520 from the National Institute of Diabetes and Digestive and Kidney Diseases were used; Jane Lian and Gary Stein are members of the University of Massachusetts DERC (DK32520).
The contents of this work are solely our responsibility and do not necessarily represent the official views of the National Institutes of Health.
Footnotes
Published ahead of print on 3 May 2010.
REFERENCES
- 1.Allen, T. D., Y. X. Zhu, T. S. Hawley, and R. G. Hawley. 2000. TALE homeoproteins as HOX11-interacting partners in T-cell leukemia. Leuk. Lymphoma 39:241-256. [DOI] [PubMed] [Google Scholar]
- 2.Balint, E., D. Lapointe, H. Drissi, C. van der Meijden, D. W. Young, A. J. van Wijnen, J. L. Stein, G. S. Stein, and J. B. Lian. 2003. Phenotype discovery by gene expression profiling: mapping of biological processes linked to BMP-2-mediated osteoblast differentiation. J. Cell. Biochem. 89:401-426. [DOI] [PubMed] [Google Scholar]
- 3.Bei, L., Y. Lu, S. L. Bellis, W. Zhou, E. Horvath, and E. A. Eklund. 2007. Identification of a HoxA10 activation domain necessary for transcription of the gene encoding beta3 integrin during myeloid differentiation. J. Biol. Chem. 282:16846-16859. [DOI] [PubMed] [Google Scholar]
- 4.Berkes, C. A., D. A. Bergstrom, B. H. Penn, K. J. Seaver, P. S. Knoepfler, and S. J. Tapscott. 2004. Pbx marks genes for activation by MyoD indicating a role for a homeodomain protein in establishing myogenic potential. Mol. Cell 14:465-477. [DOI] [PubMed] [Google Scholar]
- 5.Canalis, E., V. Deregowski, R. C. Pereira, and E. Gazzerro. 2005. Signals that determine the fate of osteoblastic cells. J. Endocrinol. Invest. 28:3-7. [PubMed] [Google Scholar]
- 6.Capellini, T. D., G. Di Giacomo, V. Salsi, A. Brendolan, E. Ferretti, D. Srivastava, V. Zappavigna, and L. Selleri. 2006. Pbx1/Pbx2 requirement for distal limb patterning is mediated by the hierarchical control of Hox gene spatial distribution and Shh expression. Development 133:2263-2273. [DOI] [PubMed] [Google Scholar]
- 7.Capellini, T. D., R. Zewdu, G. Di Giacomo, S. Asciutti, J. E. Kugler, A. Di Gregorio, and L. Selleri. 2008. Pbx1/Pbx2 govern axial skeletal development by controlling Polycomb and Hox in mesoderm and Pax1/Pax9 in sclerotome. Dev. Biol. 321:500-514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chang, C. P., K. Stankunas, C. Shang, S. C. Kao, K. Y. Twu, and M. L. Cleary. 2008. Pbx1 functions in distinct regulatory networks to pattern the great arteries and cardiac outflow tract. Development 135:3577-3586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chariot, A., L. C. van, M. Chapelier, J. Gielen, M. P. Merville, and V. Bours. 1999. CBP and histone deacetylase inhibition enhance the transactivation potential of the HOXB7 homeodomain-containing protein. Oncogene 18:4007-4014. [DOI] [PubMed] [Google Scholar]
- 10.Cheung, C. L., B. Y. Chan, V. Chan, S. Ikegawa, I. Kou, H. Ngai, D. Smith, K. D. Luk, Q. Y. Huang, S. Mori, P. C. Sham, and A. W. Kung. 2009. Pre-B-cell leukemia homeobox 1 (PBX1) shows functional and possible genetic association with bone mineral density variation. Hum. Mol. Genet. 18:679-687. [DOI] [PubMed] [Google Scholar]
- 11.de la Serna, I., Y. Ohkawa, C. A. Berkes, D. A. Bergstrom, C. S. Dacwag, S. J. Tapscott, and A. N. Imbalzano. 2005. MyoD targets chromatin remodeling complexes to the myogenin locus prior to forming a stable DNA-bound complex. Mol. Cell. Biol. 25:3997-4009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Deng, Z. L., K. A. Sharff, N. Tang, W. X. Song, J. Luo, X. Luo, J. Chen, E. Bennett, R. Reid, D. Manning, A. Xue, A. G. Montag, H. H. Luu, R. C. Haydon, and T. C. He. 2008. Regulation of osteogenic differentiation during skeletal development. Front. Biosci. 13:2001-2021. [DOI] [PubMed] [Google Scholar]
- 13.Duverger, O., and M. I. Morasso. 2008. Role of homeobox genes in the patterning, specification, and differentiation of ectodermal appendages in mammals. J. Cell. Physiol. 216:337-346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ficara, F., M. J. Murphy, M. Lin, and M. L. Cleary. 2008. Pbx1 regulates self-renewal of long-term hematopoietic stem cells by maintaining their quiescence. Cell Stem Cell 2:484-496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gersch, R. P., F. Lombardo, S. C. McGovern, and M. Hadjiargyrou. 2005. Reactivation of Hox gene expression during bone regeneration. J. Orthop. Res. 23:882-890. [DOI] [PubMed] [Google Scholar]
- 16.Gutierrez, S., A. Javed, D. Tennant, M. van Rees, M. Montecino, G. S. Stein, J. L. Stein, and J. B. Lian. 2002. CCAAT/enhancer-binding proteins (C/EBP) β and δ activate osteocalcin gene transcription and synergize with Runx2 at the C/EBP element to regulate bone-specific expression. J. Biol. Chem. 277:1316-1323. [DOI] [PubMed] [Google Scholar]
- 17.Hassan, M. Q., A. Javed, M. I. Morasso, J. Karlin, M. Montecino, A. J. van Wijnen, G. S. Stein, J. L. Stein, and J. B. Lian. 2004. Dlx3 transcriptional regulation of osteoblast differentiation: temporal recruitment of Msx2, Dlx3, and Dlx5 homeodomain proteins to chromatin of the osteocalcin gene. Mol. Cell. Biol. 24:9248-9261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hassan, M. Q., S. Saini, J. A. Gordon, A. J. van Wijnen, M. Montecino, J. L. Stein, G. S. Stein, and J. B. Lian. 2009. Molecular switches involving homeodomain proteins, HOXA10 and RUNX2 regulate osteoblastogenesis. Cells Tissues Organs 189:122-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hassan, M. Q., R. Tare, S. H. Lee, M. Mandeville, B. Weiner, M. Montecino, A. J. van Wijnen, J. L. Stein, G. S. Stein, and J. B. Lian. 2007. HOXA10 controls osteoblastogenesis by directly activating bone regulatory and phenotypic genes. Mol. Cell. Biol. 27:3337-3352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hisa, T., S. E. Spence, R. A. Rachel, M. Fujita, T. Nakamura, J. M. Ward, D. E. Devor-Henneman, Y. Saiki, H. Kutsuna, L. Tessarollo, N. A. Jenkins, and N. G. Copeland. 2004. Hematopoietic, angiogenic and eye defects in Meis1 mutant animals. EMBO J. 23:450-459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Iimura, T., and O. Pourquie. 2007. Hox genes in time and space during vertebrate body formation. Dev. Growth Differ. 49:265-275. [DOI] [PubMed] [Google Scholar]
- 22.LaRonde-LeBlanc, N. A., and C. Wolberger. 2003. Structure of HoxA9 and Pbx1 bound to DNA: Hox hexapeptide and DNA recognition anterior to posterior. Genes Dev. 17:2060-2072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lengerke, C., S. Schmitt, T. V. Bowman, I. H. Jang, L. Maouche-Chretien, S. McKinney-Freeman, A. J. Davidson, M. Hammerschmidt, F. Rentzsch, J. B. Green, L. I. Zon, and G. Q. Daley. 2008. BMP and Wnt specify hematopoietic fate by activation of the Cdx-Hox pathway. Cell Stem Cell 2:72-82. [DOI] [PubMed] [Google Scholar]
- 24.Leucht, P., J. B. Kim, R. Amasha, A. W. James, S. Girod, and J. A. Helms. 2008. Embryonic origin and Hox status determine progenitor cell fate during adult bone regeneration. Development 135:2845-2854. [DOI] [PubMed] [Google Scholar]
- 25.Li, X., and X. Cao. 2006. BMP signaling and skeletogenesis. Ann. N. Y. Acad. Sci. 1068:26-40. [DOI] [PubMed] [Google Scholar]
- 26.Li, X., S. Nie, C. Chang, T. Qiu, and X. Cao. 2006. Smads oppose Hox transcriptional activities. Exp. Cell Res. 312:854-864. [DOI] [PubMed] [Google Scholar]
- 27.Lian, J. B., G. S. Stein, A. Javed, A. J. van Wijnen, J. L. Stein, M. Montecino, M. Q. Hassan, T. Gaur, C. J. Lengner, and D. W. Young. 2006. Networks and hubs for the transcriptional control of osteoblastogenesis. Rev. Endocr. Metab. Disord. 7:1-16. [DOI] [PubMed] [Google Scholar]
- 28.Lu, Y., I. Goldenberg, L. Bei, J. Andrejic, and E. A. Eklund. 2003. HoxA10 represses gene transcription in undifferentiated myeloid cells by interaction with histone deacetylase 2. J. Biol. Chem. 278:47792-47802. [DOI] [PubMed] [Google Scholar]
- 29.Margueron, R., P. Trojer, and D. Reinberg. 2005. The key to development: interpreting the histone code? Curr. Opin. Genet. Dev. 15:163-176. [DOI] [PubMed] [Google Scholar]
- 30.Martin, C., and Y. Zhang. 2005. The diverse functions of histone lysine methylation. Nat. Rev. Mol. Cell Biol. 6:838-849. [DOI] [PubMed] [Google Scholar]
- 31.Moens, C. B., and L. Selleri. 2006. Hox cofactors in vertebrate development. Dev. Biol. 291:193-206. [DOI] [PubMed] [Google Scholar]
- 32.Montecino, M., J. Lian, G. Stein, and J. Stein. 1996. Changes in chromatin structure support constitutive and developmentally regulated transcription of the bone-specific osteocalcin gene in osteoblastic cells. Biochemistry 35:5093-5102. [DOI] [PubMed] [Google Scholar]
- 33.Noyes, M. B., R. G. Christensen, A. Wakabayashi, G. D. Stormo, M. H. Brodsky, and S. A. Wolfe. 2008. Analysis of homeodomain specificities allows the family-wide prediction of preferred recognition sites. Cell 133:1277-1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ogata, Y., S. Nakao, R. H. Kim, J. J. Li, S. Furuyama, H. Sugiya, and J. Sodek. 2000. Parathyroid hormone regulation of bone sialoprotein (BSP) gene transcription is mediated through a pituitary-specific transcription factor-1 (Pit-1) motif in the rat BSP gene promoter. Matrix Biol. 19:395-407. [DOI] [PubMed] [Google Scholar]
- 35.Ota, T., H. Asahina, S. H. Park, Q. Huang, T. Minegishi, N. Auersperg, and P. C. Leung. 2008. HOX cofactors expression and regulation in the human ovary. Reprod. Biol. Endocrinol. 6:49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rhee, J. W., A. Arata, L. Selleri, Y. Jacobs, S. Arata, H. Onimaru, and M. L. Cleary. 2004. Pbx3 deficiency results in central hypoventilation. Am. J. Pathol. 165:1343-1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Roca, H., M. Phimphilai, R. Gopalakrishnan, G. Xiao, and R. T. Franceschi. 2005. Cooperative interactions between RUNX2 and homeodomain protein-binding sites are critical for the osteoblast-specific expression of the bone sialoprotein gene. J. Biol. Chem. 280:30845-30855. [DOI] [PubMed] [Google Scholar]
- 38.Sagerstrom, C. G. 2004. PbX marks the spot. Dev. Cell 6:737-738. [DOI] [PubMed] [Google Scholar]
- 39.Saleh, M., I. Rambaldi, X. J. Yang, and M. S. Featherstone. 2000. Cell signaling switches HOX-PBX complexes from repressors to activators of transcription mediated by histone deacetylases and histone acetyltransferases. Mol. Cell. Biol. 20:8623-8633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Selleri, L., M. J. Depew, Y. Jacobs, S. K. Chanda, K. Y. Tsang, K. S. Cheah, J. L. Rubenstein, S. O'Gorman, and M. L. Cleary. 2001. Requirement for Pbx1 in skeletal patterning and programming chondrocyte proliferation and differentiation. Development 128:3543-3557. [DOI] [PubMed] [Google Scholar]
- 41.Selleri, L., J. DiMartino, D. J. van, A. Brendolan, M. Sanyal, E. Boon, T. Capellini, K. S. Smith, J. Rhee, H. Popperl, G. Grosveld, and M. L. Cleary. 2004. The TALE homeodomain protein Pbx2 is not essential for development and long-term survival. Mol. Cell. Biol. 24:5324-5331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shen, W. F., K. Krishnan, H. J. Lawrence, and C. Largman. 2001. The HOX homeodomain proteins block CBP histone acetyltransferase activity. Mol. Cell. Biol. 21:7509-7522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wahba, G. M., S. L. Hostikka, and E. M. Carpenter. 2001. The paralogous Hox genes Hoxa10 and Hoxd10 interact to pattern the mouse hindlimb peripheral nervous system and skeleton. Dev. Biol. 231:87-102. [DOI] [PubMed] [Google Scholar]
- 44.Wellik, D. M. 2007. Hox patterning of the vertebrate axial skeleton. Dev. Dyn. 236:2454-2463. [DOI] [PubMed] [Google Scholar]