Abstract
Maintenance of sex-specific germ cells requires balanced activation and repression of genetic hierarchies to ensure gender-appropriate development in mammals. Figla (factor in the germ line, alpha) encodes a germ cell-specific basic helix-loop-helix transcription factor first identified as an activator of oocyte genes. In comparing the ovarian proteome of normal and Figla null newborn mice, 18 testis-specific or -enhanced proteins were identified that were more abundant in Figla null ovaries than in normal ovaries. Transgenic mice, ectopically expressing Figla in male germ cells, downregulated a subset of these genes and demonstrated age-related sterility associated with impaired meiosis and germ cell apoptosis. Testis-associated genes, including Tdrd1, Tdrd6, and Tdrd7, were suppressed in the transgenic males with a corresponding disruption of the sperm chromatoid body and mislocalization of MVH and MILI proteins, previously implicated in posttranscriptional processing of RNA. These data demonstrate that physiological expression of Figla plays a critical dual role in activation of oocyte-associated genes and repression of sperm-associated genes during normal postnatal oogenesis.
Mouse gestation takes ∼20 days, and at embryonic day 10.5 (E10.5) the bipotential mammalian gonad commits to becoming a testis or an ovary, depending on the presence of the male-specifying Y chromosome. Subsequent gender-specific genetic hierarchies modulate the transition of the undifferentiated gonad to an ovary or testis, which produces haploid gametes through the reductive divisions of meiosis (5). However, sexual fate is plastic among metazoans (e.g., flies, worms, and mice), and this reversible commitment indicates an ongoing need for genetic controls to maintain one sex with concomitant nonexpression of genes that support the opposite identity (21, 33). Recently, Foxl2, a forkhead transcription factor expressed in somatic cells, has been implicated in maintaining female gonad sexual identify in adult mice; its ablation in postnatal ovaries leads to sex reversal via upregulation of Sox9 in somatic tissue independent of oocytes (51). It seems likely that genetic hierarchies expressed within oocytes complement these somatic signals to maintain appropriate germ cell identity by activating oocyte-associated genes and repressing sperm-associated genes during postnatal oogenesis.
Members of the basic helix-loop-helix (bHLH) family of transcription factors are key regulators of cell growth and differentiation that can either activate or repress transcription of target genes (19). Figla (factor in the germ line, alpha; official name, folliculogenesis-specific basic helix-loop-helix) encodes a germ cell-specific bHLH transcription factor that was initially identified by its ability to coordinate the expression of the oocyte-specific zona pellucida genes (28). Although Figla transcripts are first detected in oocytes at E14.5, few differences are observed between normal and null transcriptomes prior to birth (20), at which time null oocytes do not form primordial follicles, a defining structure in the female gonad, and become sterile. Low levels of FIGLA are detected in adult testes (28), but Figla null males have normal testicular histology and are fertile (46).
In comparing the proteome of normal and Figla null newborn ovaries, we unexpectedly identified 20 testis-associated proteins that were more abundant in the null ovaries. These gene products were detected at relatively lower levels in normal ovaries, suggesting that FIGLA physiologically represses genes that are expressed normally in the testis. This hypothesis was validated by ectopic expression of Figla in male germ cells that resulted in downregulation of a subset of these genes and multiple defects in postnatal spermatogenesis.
MATERIALS AND METHODS
Two-dimensional difference gel electrophoresis.
Proteins were extracted from newborn normal and Figla null ovaries and analyzed by two-dimensional difference gel electrophoresis (2D-DIGE; Etta 2D DIGE system) (2) and mass spectrometry (Proteomics Lab, Washington University, St. Louis, MO). 2D gel experiments used instrumentation and protocols from Amersham Biosciences/GE Healthcare Biosciences (Piscataway, NJ).
Establishment of transgenic mice.
The 2.4-kb human TSPY1 promoter (22, 44) and a 4.3-kb genomic fragment containing the coding region (3.64 kb) and 3′-flanking region (0.66 kb) of mouse Figla were cloned into pGL2 (Promega, Madison, WI) at SmaI and HindIII restriction enzyme sites. After confirmation of the DNA sequence across the junctions, the transgene was gel purified and injected into the male pronucleus of one-cell embryos (38). Six founders were identified, and germ line transmission of the transgene to their progeny was followed by PCR and Southern analyses of tail DNA (primer sequences are available upon request). The abundance of Figla transcripts was determined in postpubertal male gonads by quantitative reverse transcription-PCR (qRT-PCR). The two transgenic lines with the highest levels of expression were further analyzed and had similar phenotypes. Normal and TSPY1-Figla transgenic males were mated with normal and transgenic females (five pairs), and litters from each fertile pair were analyzed for length of gestation and size. Differences were compared statistically using Student's t test.
RT-PCR.
Mouse tissues or mixed germs cells were isolated and homogenized in individual microcentrifuge tubes, and total RNA was extracted using an RNeasy microkit (Qiagen, Valencia, CA). After DNase I digestion, cDNA was generated from normalized RNA using the SuperScript III first-strand synthesis system (Invitrogen, Eugene, OR). Samples were assayed by qRT-PCR using an ABI Prism 7900HT thermal cycler and prevalidated TaqMan primers and probes to identify Figla and genes expressed in the testes. Gapdh transcripts were used as an endogenous template control (Applied Biosystems, Foster City, CA). The primers and probes used for these procedures are available upon request.
In situ hybridization.
The expression of testis-associated mRNA was detected by in situ hybridization of biotin-Green Star hyperlabeled oligonucleotide probes (Gene Detect, Auckland, New Zealand) using the company's online protocol, which included detection with an antidigoxigenin antibody conjugated with a tyramide signal amplification kit (DakoCytomation, Glostrup, Denmark).
Computer-assisted sperm analysis (CASA).
Sperm were evaluated on an HTM-IVOS motility analyzer (version 10.8; Hamilton Thorne Biosciences, Beverly, MA) with the following settings: phase contrast; frame rate, 60 Hz; minimum contrast, 30; low and high static size gates, 0.8 and 6.25; low and high intensity gates, 0.25 and 1.50; low and high static elongation gates, 20 and 70; default cell size, 5 pixels; default cell intensity, 55; magnification, ×0.78. Sperm (60 μl; 6 × 107 ml−1) were loaded into a prewarmed, 20 μm microcell counting chamber (Conception Technologies, San Diego, CA) and observed under 4× magnification. For each genotype, sperm from two cauda epididymides were examined, and data were averaged from 20 fields with a total of >500 sperm. After incubation (30 min, 37°C) in M16 medium (Specialty Media, Chemicon International, Phillipsburg, NJ), sperm motility, viability, velocity (straight line velocity [VSL] and curvilinear velocity [VCL]) and other movement characteristics (linearity [LIN]and straightness [STR]) were determined, and total motility or progressive motility was defined as the percentage of sperm with an average path velocity of >7.5 or >25 μm/s, respectively.
Sperm binding assay and in vitro fertilization.
Caudal epididymal sperm isolated from normal and TSPY1-Figla transgenic mice were placed in M16 medium (Chemicon, Millipore, Temecula, CA) and incubated under capacitating conditions (1 h, 37°C, 5% CO2). Sperm binding was performed using ovulated eggs from FVB female mice (17). At the end of the 30-min assay, samples were fixed in 2% paraformaldehyde and stained with Hoechst 33242 (5 mg/ml; Sigma-Aldrich, St. Louis, MO) prior to imaging by confocal microscopy (37). For in vitro fertilization, 2 × 105 ml−1 normal or TSPY1-Figla sperm were incubated with ovulated eggs in cumulus obtained from gonadotropin-stimulated FVB mice (38). After 24 and 48 h, embryos were isolated, fixed, stained with Hoechst 33342, and imaged by confocal microscopy. Fertilization was confirmed by the presence of pronuclei within the egg cytoplasm and by progression to two-cell embryos.
Histology.
Testes were isolated, placed in Bouin's fixative at room temperature for 2 to 3 days, and dehydrated through graded ethanol solutions. After embedding in paraffin, 5-μm sections were stained with hematoxylin or periodic acid-Schiff's (PAS) reagent, counterstained with eosin, and digitally imaged on a Zeiss Axioplan 2 microscope. Histological sections from at least three animals of each age and genotype were examined.
TUNEL assay.
To detect apoptosis, sections were deparaffinized by heating at 60°C followed by washing in xylene and rehydration through a graded series of ethanol. Slides were then microwaved (1 min; 100 mM citrate buffer [pH 6.0]) and blocked (30 min; 3% bovine serum albumin-phosphate-buffered saline), and an in situ cell death detection kit (Roche Applied Biosciences, Indianapolis, IN) was used for terminal deoxynucleotidyltransferase-mediated dUTP-biotin nick end labeling (TUNEL) assays.
Chromosome spreads of mouse spermatocytes.
Mixed germ cell preparations were obtained by enzymatic digestion of testes from 5-month-old animals (25), and chromosome spreads of spermatocytes were stained with (i) rabbit polyclonal anti-γH2AX (10 μg/ml; Upstate Biotechnology, Lake Placid, NY) using Alexa-555-labeled donkey anti-rabbit IgG (1:200; Molecular Probes, Invitrogen, Eugene, OR) as second antibody or (ii) biotin-labeled rabbit anti-SYCP3 (1:100; Abcam, Cambridge, MA) using fluorescein isothiocyanate (FITC)-labeled ExtrAvidin (1:200; eBiosciences) to visualize staining. Nuclei were detected with Hoechst 33242.
Analysis of TSPY1-Figla transgenic testes.
Proteins were extracted from mixed testicular germ cells for use in immunoblot assays (27) using as probes rabbit polyclonal antibodies (1:500) to TDRD1, TDRD6, or TDRD7 (10). Proteins were localized by immunofluorescence of lyophilized, frozen testicular sections using the following primary antibodies: rabbit polyclonal antibodies to TDRD1 (1:500), TDRD6 (1:500), TDRD7 (1:500), MVH (10 μg/ml; Abcam), MILI (10 μg/ml; Abcam), γH2AX (10 μg/ml; Upstate), and biotin-labeled rabbit anti-SYCP3 (8 μg/ml; Novus Biologicals, Littleton, CO). Alexa-555-labeled donkey anti-rabbit IgG (1:200; Molecular Probes) or FITC-labeled ExtrAvidin (1:200; eBiosciences) was used to detect primary antibody staining. Nuclei of testes were detected with Hoechst 33242 (10 μg/ml).
Electron microscopy.
Cells from mouse testes were fixed in 1.5% glutaraldehyde buffered in sodium cacodylate, pH 7.4, for 2 h at 4°C. After extensive washing in cacodylate buffer, cells were pelleted by centrifugation and embedded in melted (37°C) agarose. Samples were dehydrated through a graded series of ethanol and processed for embedding in LR-White resin (32). Ultrathin sections were counterstained with uranyl acetate followed by lead citrate and imaged in a Phillips CM120 transmission electron microscope (FEI Company).
RESULTS
Overexpression of testis-associated genes in Figla null ovaries.
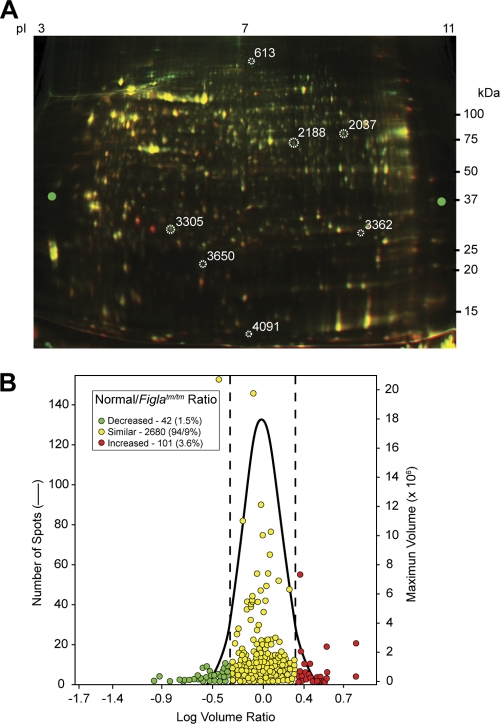
To identify potential downstream targets of FIGLA, proteins were extracted from newborn normal and Figla null ovaries and systematically analyzed by 2D-DIGE and mass spectrometry (Fig. 1A). Of the 2,823 protein spots identified, 101 were significantly (>2-fold; P < 0.05) more abundant in Figla null ovaries and 42 were more abundant in normal ovaries (Fig. 1B). Sixty-six proteins from the former and 48 (some spots contained two proteins) from the latter were identified by microscale liquid chromatography and tandem mass spectrometry (MS/MS). Twenty of the proteins found in greater abundance in Figla null newborn ovaries were expressed in the testes of normal mice (Table 1). To confirm the results from the 2D gels, the abundance of the transcripts encoding testis proteins was assayed by quantitative RT-PCR in normal tissues and in Figla null ovaries. The abundance of 18 of the 20 transcripts was greater in normal testis than in normal ovary, and their abundance in Figla null ovaries ranged from 5 to 342% of that observed in normal testis (Table 1). These results raised the possibility that during oogenesis FIGLA not only activates genes in oocytes but also participates in repression, either directly or indirectly, of genes that are normally expressed during spermatogenesis.
FIG. 1.
2D-DIGE of proteins in normal and Figla null newborn mouse ovaries. (A) Ovarian protein extracts of normal (green) and Figla null (red) mice were labeled with Cy5 and Cy3, respectively, and separated by 2D-DIGE. Molecular masses (in kDa) are shown on the right, and the pI gradient was estimated from the label on the isoelectric focusing gel. (B) Comparison of log volume ratios of proteins expressed in normal and Figla null ovaries. While most (2,680) proteins were similar (94.9% [yellow]), 101 protein spots were increased in normal ovaries (3.6% [red]) and 42 (1.5% [green]) were increased in Figla null ovaries.
TABLE 1.
Abundance of mRNA encoded by genes potentially downregulated by FIGLA
| Genea | Description | No. of peptides based on MS/MS | mRNA abundance inb: |
Null vs normal ovaries (fold change) | Null ovary vs normal testes (% change) | KO availablec | ||
|---|---|---|---|---|---|---|---|---|
| Normal ovary | Normal testis | Figla null ovary | ||||||
| 1700019M22Rik | RIKEN cDNA 1700019M22 | 3 | 0.0 | 19.8 ± 1.6 | 3.2 ± 1.2 | ∞ | 16 | |
| Ttll13 | Tubulin tyrosine ligase-like family member 13 | 2 | 1.8 ± 0.2 | 17.3 ± 8.3 | 59.1 ± 3.7 | 33.6 | 342 | |
| 1700009N14Rik | RIKEN cDNA 1700009N14 | 2 | 3.7 ± 0.7 | 27.1 ± 8.3 | 59.7 ± 6.7 | 16.2 | 220 | |
| Cyp17a1 | Cytochrome P450, family 17, subfamily a, polypeptide 1 | 10 | 2.3 ± 0.4 | 139.6 ± 7.0 | 29.1 ± 5.2 | 12.6 | 21 | 29 |
| Gsg1 | Germ cell-specific gene 1 | 10 | 4.7 ± 1.8 | 100.0 ± 0.4 | 52.2 ± 11.3 | 11.0 | 52 | |
| Tnp2 | Transition protein 2 | 4 | 8.5 ± 6.4 | 58.0 ± 6.0 | 92.8 ± 2.9 | 10.9 | 160 | 1 |
| Cypt4 | Cysteine-rich perinuclear theca 4 | 5 | 4.5 ± 0.1 | 28.3 ± 12.4 | 22.6 ± 3.7 | 5.0 | 80 | |
| Irgc1 | Immunity-related GTPase family, cinema 1 | 1 | 0.4 ± 0.1 | 24.2 ± 3.8 | 1.5 ± 0.9 | 4.3 | 6 | |
| Cypt3 | Cysteine-rich perinuclear theca 3 | 4 | 3.5 ± 1.8 | 61.5 ± 5.8 | 13.2 ± 8.8 | 3.7 | 21 | |
| Crisp2 | Cysteine-rich secretory protein 2 | 6 | 4.5 ± 2.0 | 308.7 ± 11.7 | 15.8 ± 7.2 | 3.5 | 5 | |
| Tdrd6 | Tudor domain-containing 6 | 3 | 0.5 ± 0.2 | 15.1 ± 4.1 | 1.9 ± 0.3 | 3.5 | 12 | 52 |
| Slc2a3 | Solute carrier family 2 (facilitated glucose transporter), member 3 | 2 | 9.9 ± 1.6 | 55.3 ± 2.0 | 30.8 ± 6.8 | 3.1 | 56 | 15 |
| 1700011E24Rik | RIKEN cDNA 1700011E24 | 2 | 1.3 ± 0.5 | 6.3 ± 0.6 | 4.0 ± 0.3 | 3.1 | 63 | |
| Adad1 | Adenosine deaminase domain-containing 1 (testis specific) | 3 | 24.9 ± 1.1 | 119.5 ± 16.9 | 66.1 ± 15.3 | 2.7 | 55 | 11 |
| Dkkl1 | Dickkopf-like 1 | 2 | 9.2 ± 5.8 | 30.4 ± 3.0 | 24.0 ± | 2.6 | 79 | |
| Spata19 | Spermatogenesis-associated 19 | 5 | 1.1 ± 0.0 | 32.4 ± 1.7 | 2.4 ± 0.3 | 2.2 | 7 | |
| Ly6k | Lymphocyte antigen 6 complex, locus K | 4 | 15.9 ± 0.3 | 11.4 ± 0.2 | 31.5 ± 0.3 | 2.0 | 276 | |
| Klhl10 | Kelch-like 10 (Drosophila) | 3 | 11.7 ± 0.1 | 33.2 ± 2.6 | 23.0 ± 1.1 | 2.0 | 69 | 54 |
| Lrrc27 | Leucine-rich repeat-containing 27 | 4 | 25.3 ± 8.3 | 125.6 ± 8.7 | 47.5 ± 0.5 | 1.9 | 38 | |
| 4930511I11Rik | RIKEN cDNA 4930511I11 | 3 | 2.2 ± 0.3 | 2.5 ± 0.7 | 3.3 ± 1.2 | 1.5 | 135 | |
Genes shown in boldface were downregulated in 7-week-old mouse TSPY1-Figla testes.
Compared to the Gapdh control, which was assigned a value of 1. Values are means ± SEM.
Reference in which a KO (knockout) homozygous null mouse line has been reported.
TSPY1-Figla transgenic mice.
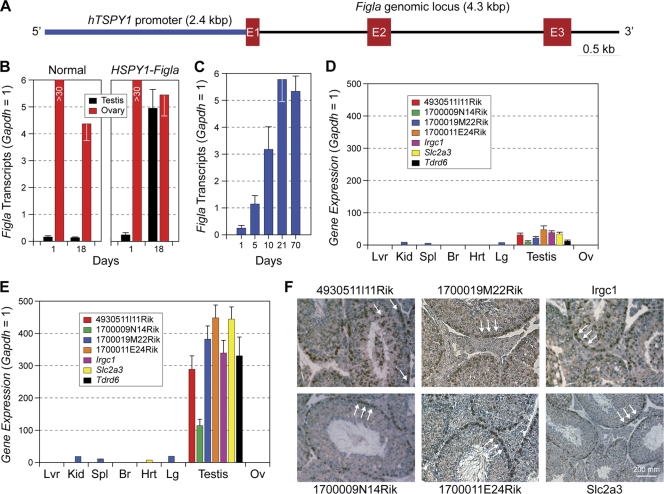
To determine its effect on the expression of the genes encoding these testis-associated proteins, FIGLA was ectopically expressed in transgenic mice by using a human sperm-specific promoter (TSPY1; testis-specific protein Y encoded) (Fig. 2A), which drives gene expression in spermatogonia and early primary spermatocytes (22, 44). Transgenic male and female mice, maintained on a pure FVB background, appeared normal at birth, grew to adulthood, and were initially fertile. Only low levels of FIGLA transcripts were detected by qRT-PCR in normal testes (Fig. 2B), but transcripts were abundant in TSPY1-Figla transgenic testis as early as day 1 after birth (D1), reaching a maximum at D21 and persisting in adults (Fig. 2C).
FIG. 2.
Ectopic expression of Figla in male germ cells. (A) Transgene containing the Figla genomic locus (4.3 kbp) downstream of the human TSPY1 promoter (2.4 kbp), which is expressed in spermatogonia and early spermatocytes. Exons are represented by Arabic numbered red boxes. (B) Figla transcripts in normal and TSPY1-Figla transgenic ovaries and testes at D1 and D18 were detected by qRT-PCR and are expressed as means ± SEM of total RNA isolated from three biological samples. Gapdh was used as an endogenous template control. (C) The same experiment as shown in panel B, but using testicular RNA from D1 to 10-week-old TSPY1-Figla transgenic mice. (D) Transcript abundance levels of seven testis genes (means ± SEM) were determined by qRT-PCR relative to Gapdh in total RNA isolated from different organs harvested from 2-month-old TSPY1-Figla transgenic mice. Lvr, liver; Kid, kidney; Br, brain; Hrt, heart; Lg, lung; Ov, ovary. (E) The same experiment as shown in panel D, but using total RNA isolated from different organs in normal mice. (F) In situ hybridization of normal testes using antisense probes specific for 4930511l11Rik, 1700009N14Rik, 1700019M22Rik, 1700011E24Rik, Irgc1, and Slc2a3. Arrows highlight localization of transcripts.
The abundance of transcripts encoding the 20 testis proteins (Fig. 1; Table 1) was then determined by qRT-PCR of transgenic testes. No differences were observed at D5, but transcripts from Irgc1 (immunity-related GTPase family, cinema 1) and Slc2a3 (solute carrier family 2, member 3, or Glut3) were 18% and 23% lower, respectively, than normal at D21. At 7 weeks, seven testis-specific (or testis-enriched) genes, including Slc2a3, Tdrd6 (Tudor domain-containing 6), Irgc1, and four genes of unknown function (1700011E24Rik, 1700009N14Rik, 4930511I11Rik, and 1700019M22Rik) were at least 5-fold lower in transgenic testes than in normal controls (Fig. 2D and E). This decrease in the TSPY1-Figla transgenic testes corresponded to the increase in cognate protein expression observed in Figla null ovaries in the initial 2D-DIGE screen (Fig. 1A, circles).
Each of the seven genes was testis specific or enhanced (Fig. 2F), and by using digoxigenin-labeled antisense probes to six of the genes, transcripts were detected exclusively in germ lineages, with no apparent signal in Sertoli or interstitial cells of the testes. Only background levels of signal were observed with control sense probes (data not shown). Slc2a3, 1700011E24Rik, and 1700009N14Rik were primarily located in spermatogonia and spermatocytes, whereas 4930511I11Rik, 1700019M22Rik, and lrgc1 were detected in all stages of male germ cells, including elongating spermatids (Fig. 2F). Expression of Tdrd6 has been previously reported to be germ cell specific (18) and has been shown to participate in the chromatoid body, a defining structure of the round spermatid. Taken together, these data suggest that at least seven sperm-specific or -enriched genes are downstream targets of FIGLA and were repressed by ectopic expression of FIGLA in male germ cells in 7-week-old TSPY1-Figla transgenic mice. The persistence of the remaining 13 transcripts indicates a specificity that cannot be accounted for by generalized loss of germ cells and suggests that corepressors and modifications of chromatin structures not present in sperm may play a role in repression of these genes in growing oocytes.
Infertility of TSPY1-Figla transgenic mice.
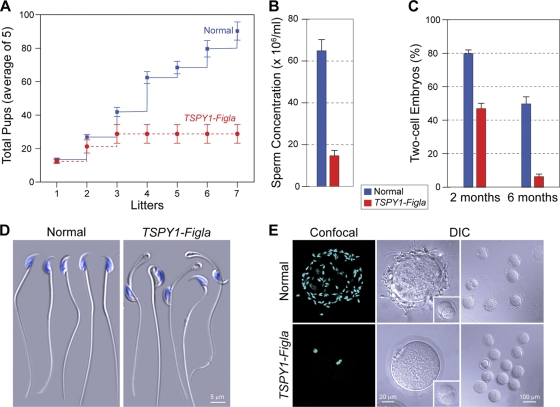
Although initially fecund, all TSPY1-Figla transgenic males became sterile by 5 months (some as early as 3 months) of age. Average litter sizes (± standard errors of the means [SEM]) for normal and transgenic males were comparable (13.3 ± 0.5 and 10.2 ± 1.2, respectively), but transgenic males sired no more than three litters (Fig. 3A) and passed the transgene to <20% of their offspring. In contrast, TSPY1-Figla females had normal litter sizes (10.0 ± 0.4) and, as expected, 50% of their litters were transgenic.
FIG. 3.
Age-related sterility of TSPY1-Figla transgenic males. (A) Normal and TSPY1-Figla transgenic males were mated for >8 months with normal females (five pairs) to assess fertility. Numbers of cumulative litters of normal and transgenic mice are expressed as the mean ± SEM. (B) Sperm concentrations from 5-month-old normal and transgenic TSPY1-Figla epididymides were determined by CASA and are expressed as the mean ± SEM. (C) Comparison of successful IVF (mean ± SEM) using normal and transgenic sperm isolated from 2- and 6-month-old males. (D) Five-month-old normal and TSPY1-Figla transgenic mouse epididymal sperm were imaged by difference interference contrast (DIC) microscopy. Nuclei were stained with Hoechst 33242. (E) A 30-min sperm binding assay was used to compare normal and TSPY1-Figla sperm binding to normal ovulated eggs. After washing with a wide-bore pipette, no more than two to five sperm remained bound to control two-cell embryos (insets). Nuclei were stained with Hoechst 33242. Confocal and DIC images were obtained.
Fewer sperm (∼20%) were recovered from the cauda epididymis of 5-month-old TSPY1-Figla transgenic mice compared to age-matched normal male mice ([64.5 ± 5.2] × 106 ml−1 versus [14.6 ± 2.5] × 106 ml−1) (Fig. 3B). After adjustment to the same concentration (2 × 105 ml−1), normal and transgenic epididymal sperm were assayed for in vitro fertilization (IVF) of ovulated eggs in cumulus. Successful fertilization was scored by the presence of two pronuclei and subsequent progression to two-cell embryos. Even at 2 months, there was a decrease in successful IVF (81.7% ± 2.4% versus 52.6% ± 7.4%) of normal compared to transgenic mice (Fig. 3C) that was dramatically accentuated by 6 months (50.00% ± 4.6% versus 6.4% ± 1.4%).
Unexpectedly, ∼50% of the transgenic sperm had severe tail angulation at the cytoplasmic droplet (Fig. 3D), characteristic of defects in volume regulation (55). CASA documented decreases in motility (61.5% ± 4.6% versus 18.0% ± 1.7%), progressive motion (39.3% ± 4.9% versus 11.3% ± 2.2%), and hyperactivity (49.0% ± 5.9% versus 14.0% ± 2.1%) in transgenic compared to normal mice with little effect on viability (79.3% ± 1.5% versus 67.5% ± 5.5%). To assay gamete recognition, epididymal sperm from 5-month-old normal and TSPY1-Figla transgenic mice were capacitated and incubated with normal ovulated eggs. At the end of 30 min, eggs were washed with a wide-bore pipette until no more than two to five sperm remained bound to the control two-cell embryos. Compared to normal sperm, there was a dramatic decrease in the number of transgenic sperm bound to normal ovulated eggs (Fig. 3E). Thus, defects in spermatogenesis imposed by ectopic expression of Figla result in progressive, age-related male infertility.
Histopathology of TSPY1-Figla transgenic mice.
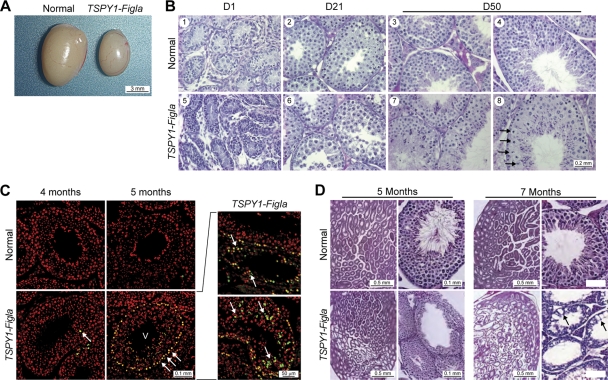
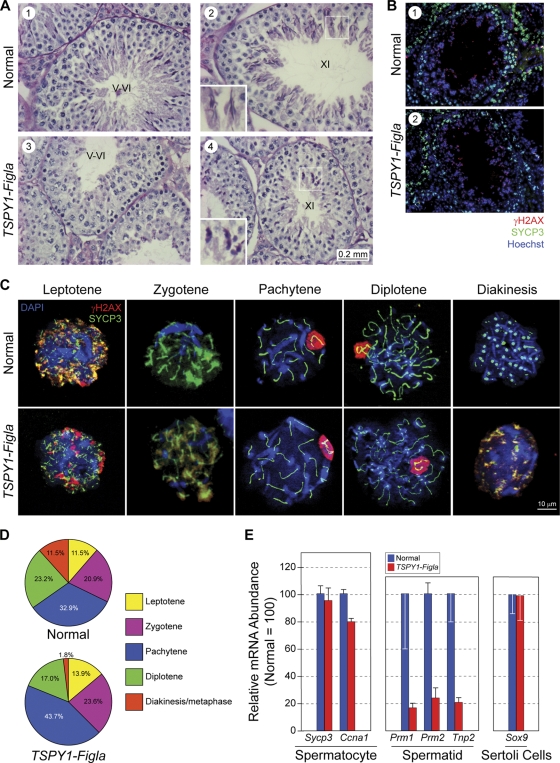
To investigate the cellular and molecular basis of the age-related defects in spermatogenesis, gonads of TSPY1-Figla testes were isolated and examined in greater detail. By 7 weeks (Fig. 4A), the testes of transgenic males were 73% of normal size (73.6 ± 3.1 versus 100 ± 2.1 mg). Prior to D50, transgenic epididymal sperm appeared morphologically unaffected, and TSPY1-Figla testes had normal histology with spermatogonia, spermatocytes, and maturing spermatids present within seminiferous tubules (Fig. 4B). However, by 3 weeks, TSPY1-Figla transgenic tubules contained fewer male germ cells at all stages of development, and by 7 to 8 weeks, a few tubules (<5%) contained abnormally large spermatocytes (Fig. 4B, panel 8, arrows).
FIG. 4.
Testicular dysmorphology of TSPY1-Figla mice. (A) At 7 weeks, the TSPY1-Figla testes were smaller than normal and the average weight (mean ± SEM) was 73.6 ± 3.1 mg, compared to 100 ± 2.1 mg for normal mice. (B) Cross-sections of seminiferous tubules of normal and transgenic mice at D1, D21, and D50 after fixing in Bouin's solution and staining with hematoxylin and counterstaining with eosin. Arrows (panel 8) indicate enlarged spermatocytes. (C) TUNEL assay at 4 and 5 months of normal and TSPY1-Figla transgenic testes. Apoptotic cells, Hoechst 33242-stained nuclei, and merged images are green, red, and yellow, respectively. Roman numerals refer to the stages of mouse spermatogenesis. Arrows indicate apoptotic germ cells. (D) Testicular histology (fixed in Bouin's solution and stained with periodic acid-Schiff's reagent and eosin) of normal and TSPY1-Figla transgenic animals at 5 and 7 months. Abnormal spermatogenesis and vacuolation (arrows) that were accentuated by age were observed in transgenic tubules.
Testes were assayed for germ cell apoptosis in a fluorescent TUNEL assay based on nuclear DNA fragmentation (16). Very few apoptotic germ cells were detected in normal testes, but they were present in transgenic testis beginning at 4 months and became remarkably more abundant after 5 months (Fig. 4C). A large number of apoptotic spermatogonia A and intermediate spermatogonia germ cells were found in the edge of stage IV to VI seminiferous tubules. In addition, later-stage male germ cells, defined by their location within the germinal epithelium, were apoptotic as well (Fig. 4C, panels on right). In contrast, TUNEL-positive cells were not observed in somatic Sertoli and Leydig cells in 5-month-old or older TSPY1-Figla transgenic mice.
Secondary to germ cell loss, the seminiferous tubules of aging TSPY1-Figla males became increasingly vacuolar. At 5 months, only patches of histopathology were observed, with 20 to 30% of tubules being affected, but by 7 months vacuolation was widespread and more than half of the seminiferous tubules exhibited histological signs of degeneration (Fig. 4D). Eventually the integrity of seminiferous tubules was disrupted and germ and somatic cells were observed in the interstitial regions of the testes (data not shown). Similar histopathology has been reported in other genetic mutations that deplete male germ cells (3, 6, 34).
Meiotic defects in spermatogenesis.
Beginning at 5 months, the defects in spermatogenesis became apparent and multiple germ cells within transgenic tubules did not complete meiosis. Consequently, stage IV to VII seminiferous tubules of 5-month-old TSPY1-Figla mice (Fig. 5A) contained few postmeiotic cell types (round spermatids, elongated spermatids, mature spermatozoa), and in a small subset of seminiferous tubules in which meiosis progressed further (stages XI and XII) there were noticeable morphological defects, including spermatids with less-elongated heads (Fig. 5A, panels 2 and 4, insets).
FIG. 5.
Meiosis in TSPY1-Figla male mice. (A) Testicular histology of normal and TSPY1-Figla transgenic mice at 5 months. Few postmeiotic cells were present in stage V and VI tubules from transgenic mice (panels 1 and 3), and those in stage XI had foreshortened heads (panels 2 and 4, insets). Stages in the seminiferous epithelial cycle are indicated by Roman numerals. (B) Testicular sections from 4-month-old normal and TSPY1-Figla transgenic mouse testicular tubules were stained with antibodies to SYCP3 (green), a synaptonemal protein, and γH2AX (red), a phosphorylated histone variant, indicative of double-strand breaks and regions of asynapsis. Both proteins were detected in peripherally located spermatocytes, and γH2AX was also detected in the more centrally located elongating spermatids. Nuclei were stained with Hoechst 33242. (C) Immunofluorescence of chromatin spreads from mixed germ cells isolated from 4-month-old normal or TSPY1-Figla transgenic mouse testes and probed with antibodies to SYCP3 (green) and γH2AX (red). Nuclei were detected with Hoechst 33242. (D) Pie graphs indicate the percentage of spermatocytes in each of the substages of the prophase of the first meiotic division of normal mice (680 cells) and TSPY1-Figla transgenic mice (798 cells). (E) qRT-PCR analysis (mean ± SEM) of expression levels of selected genes in spermatocytes, spermatids, and Sertoli cells from 5-month-old normal and TSPY1-Figla transgenic mouse testes.
Meiotic recombination occurs during the prophase of meiosis I when each pair of homologous chromosomes is joined by a synaptonemal complex that includes SYCP3 (encoded by Synaptonemal complex protein 3). γH2AX, the phosphorylated form of the variant histone H2AX, is recruited to the site of double-strand breaks and localizes repair enzymes for their subsequent resolution to ensure successful recombination. γH2AX persists in regions of asynapsis in the XY sex body until the diplotene stage, and later, during spermiogenesis, it localizes to double-strand breaks incurred by the repackaging of DNA onto protamines (7, 26). Using immunofluorescence and antibodies to SYCP3 and γH2AX, costaining was observed in peripherally located primary spermatocytes of normal and TSPY1-Figla transgenic mice, and γH2AX was detected in the more centrally located elongating spermatids. No significant differences were observed in the staining patterns of these two antibodies, which are used to characterize meiotic progression in prophase I (Fig. 5B).
Nuclear spreads of germ cells were then obtained from normal and TSPY1-Figla transgenic testes and stained with antibodies to SYCP3 and γH2AX (Fig. 5C). In normal mice, the substages of meiosis I were distributed among leptotene (11.5%), zygotene (20.9%), pachytene (32.9%), diplotene (23.2%), and diakinesis/metaphase I (11.5%). In TSPY1-Figla transgenic germ cells, there was an ∼6% decrease of diplotene-stage cells (17.0% versus 23.2%), and fewer than 2% germ cells were observed in the transition diakinesis/metaphase stages. As a result, the proportions of leptotene (13.9%) and zygotene (23.6%) stages increased and the pachytene stage was dominant in transgenic spermatocytes (43.7%). Thus, meiosis progression is significantly delayed or arrested in the prophase of meiosis I in older TSPY1-Figla transgenic mice (Fig. 5D).
To complement these histological observations, total RNA of mixed germ cells was isolated from 5-month-old TSPY1-Figla transgenic and normal mouse testes for analysis by qRT-PCR. Sycp3 and Ccna1 (cyclin A1) are expressed during the first meiotic division (35, 39). The abundance of their transcripts was comparable in TSPY1-Figla transgenic and normal mice, indicating a similar complement of spermatocytes (Fig. 5E). In contrast, the abundance of transcripts encoding three spermatid-specific proteins, Prm1 (protamine 1), Prm2 (protamine 2), and Tnp2 (transition nuclear protein 2), were dramatically decreased (>5 times) in TSPY1-Figla transgenic compared to normal testes. Somatic cell histology appeared normal, and expression of Sox9, a Sertoli cell marker, was unaffected in the transgenic testes (Fig. 5E). Taken together, these data suggest that postmeiotic defects in TSPY1-Figla transgenic male mice result in defective spermatogenesis.
Expression of Tdrd genes and formation of the sperm chromatoid body.
In the original proteomic screen (Fig. 1A), TDRD6 was more abundant in Figla null than normal newborn ovaries and was decreased in the male germ cells of TSPY1-Figla transgenic mice (Fig. 2D). Intriguingly, the phenotype observed in Tdrd6 null male mice (52) partially recapitulates that of TSPY1-Figla mice. Multiple members of the Tudor domain family of genes, including Tdrd1, Tdrd6, and Tdrd7, are expressed in male germ cells (18). The TDRD proteins participate along with MVH (official name, Ddk4, DEAD [Asp-Glu-Ala-Asp] box polypeptide 4), an RNA helicase, and MILI (official name, Piwi-like homologue 2), a Piwi-interacting RNA (piRNA) binding protein in forming the chromatoid body initially derived from the male nuage and characteristic of the postmeiotic, maturing spermatid (12, 56).
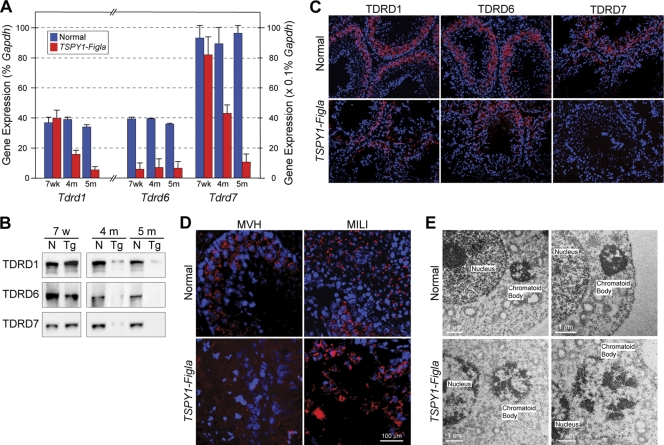
To explore the possibility that downregulation of one or more of the Tdrd genes contributes to the defects in spermatogenesis, we examined expression of Tdrd1, Tdrd6, and Tdrd7 in mixed populations of germ cells from TSPY1-Figla transgenic testes by qRT-PCR. Although the abundance of Tdrd6 transcripts was significantly decreased at 7 weeks in TSPY1-Figla transgenic testes, neither Tdrd1 nor Tdrd7 was affected. At 4 months, when only minimal germ cell apoptosis was observed (Fig. 4C), the abundance of Tdrd6 transcripts was reduced more than 3-fold in mixed germ cell populations, and those of Tdrd1 and Tdrd7 transcripts were reduced 68% and 70%, respectively, from normal controls. Although at 5 months the abundance levels of the three Tdrd transcripts were <20% that of normal controls (Fig. 6A), the onset of more widespread apoptosis (Fig. 4C) and localized loss of cellular integrity (Fig. 4D) suggest that germ cell loss plays an increasingly large role in decreased TDRD expression.
FIG. 6.
Tdrd expression and chromatoid body formation. (A) Expression levels of Tdrd1, Tdrd6, and Tdrd7 were determined as a percentage of Gapdh expression by qRT-PCR (mean ± SEM) in mixed germ cells isolated from normal and TSPY1-Figla transgenic mouse testes at 7 weeks, 4 months, and 5 months. (B) Immunoblot of extracts of mixed germ cells from normal and TSPY1-Figla transgenic mice at 7 weeks, 4 months, and 5 months that were probed with antibodies to TDRD1, TDRD6, and TDRD7. (C) Immunofluorescence of 4-month-old normal and TSPY1-Figla transgenic mouse testes probed with antibodies to TDRD1, TDRD6, and TDRD7 (red). Nuclei were stained with Hoechst 33242 (blue). (D) Immunofluorescence of normal and TSPY1-Figla testes with antibodies to MVH (red) or MILI (red) as markers of the chromatoid body. Nuclei were stained with Hoechst 33242 (blue). (E) Electron microscopy of normal and TSPY1-Figla testes (two biological samples), demonstrating that the normally compact electron-dense chromatoid body in close proximity to the nucleus becomes less dense and more diffuse in transgenic sperm.
A corresponding decrease of TDRD1, TDRD6, and TDRD7 proteins was observed on immunoblots of mixed germ cell populations isolated from TSPY1-Figla transgenic testes at 4 and 5 months (Fig. 6B). Using immunofluorescence, each of the TDRD proteins normally localize to germ cells, but the abundance of each protein was substantially decreased in transgenic males by 4 months (Fig. 6C). Using antibodies to MVH or MILI as markers, the chromatoid body was observed as a discrete dot in normal male germ cells. However, with the downregulation of the TDRD proteins in TSPY1-Figla mice, these two markers appeared diffusely present in the cytoplasm, consistent with the loss of chromatoid body integrity (Fig. 6D).
To confirm disruption of the chromatoid body, electron photomicrographs of testicular cells were obtained from normal and TSPY1-Figla transgenic mice at 5 months (Fig. 6E). The electron-dense chromatoid body, clearly visible in normal testicular cells, was greatly attenuated in cells from TSPY1-Figla transgenic testes. These data are consistent with ectopic expression of FIGLA in male germ cells with downregulated testis genes, some of which affect formation of the chromatoid body and lead to postmeiotic defects in spermatogenesis.
DISCUSSION
Following embryonic sex determination in mice (∼E10.5), male and female germ cells establish genetic hierarchies to maintain germ cell identity during mitotic proliferation, meiosis, and maturation. By comparing protein profiles of normal versus Figla null ovaries and by ectopically expressing FIGLA in male germ cells, we have provided evidence that Figla, initially implicated as an activator of oocyte-specific genetic hierarchies (28, 46), also inhibits sperm-associated developmental pathways during postnatal oogenesis.
Ectopic expression of Figla in male germ cells in transgenic mice downregulates a set of testis-associated genes that were initially identified by upregulation in Figla null newborn ovaries. The consequent testicular pathology and pre- and postmeiotic effects on spermatogenesis result in male sterility and suggest a role for FIGLA in the downregulation of male-associated genes during oogenesis, when Figla is normally expressed. Nanos2 also has a sexually dimorphic effect on gene expression, although its effect is earlier in gametogenesis and in the opposite sex. Nanos2 encodes an RNA binding protein first observed in germ cells within the colonized gonad and, in its absence, most male germ cells undergo apoptosis (50). Expression levels of both Figla and Nobox, an oocyte-specific homeobox transcription factor (36), were increased in Nanos null male gonads at E16.5. To confirm the effect, Nanos2 was ectopically expressed in female cells, where it suppressed Stra8 expression and inhibited meiosis but also activated several male genes, including Tdrd1 and Dnmt3l (DNA cytosine-5-methyltransferase 3-like) during embryogenesis (47). Nanos2 is required for maintenance of stem cells as premeiotic spermatogonia (42), whereas Figla regulates events in growing oocytes that are arrested in the prophase of the first meiotic division (46), well after the establishment of sexual identity. Nevertheless, both Figla and Nanos2 appear to support female and male germ cell developmental programs, respectively, at least in part by suppressing genetic hierarchies promoting the developmental programs of the opposite sex.
The observed absence of male germ cell gene expression within normal oocytes could result from the lack of activation or from repression of testis genes. While limiting access to DNA binding sites (e.g., nucleosome positioning, histone modifications, DNA methylation) can prevent gene activation, it would not account for the ability of ectopically expressed FIGLA to repress male genes during spermatogenesis. However, changes in dimerization partners or independent cofactors that interact with FIGLA could prevent expression of testis genes during oogenesis. These effects could be direct or indirect. Myogenic regulatory factors (MRFs), a family of canonical bHLH transcription factors, are instructive and can both activate and repress downstream target genes (49), depending on dimerization partners (30), epigenetic modifications (14, 31, 43, 45), and recruitment of additional proteins to the transcriptional complex (4, 40). FIGLA is likely to utilize a similar strategic repertoire for its physiological effects during oogenesis.
The developmental imperative of spermatogenesis is the production of haploid gametes capable of fertilization, which is necessary for propagation of the species. Following onset of sexual maturity, TSPY1-Figla transgenic male mice were initially fertile, but the intervals between litters became delayed, and none of the male mice sired litters after 5 months of mating. Figla was robustly expressed in male germ cells by 3 weeks of age in TSPY1-Figla transgenic mice, and decreased testicular size by 2 months was accompanied by repression of at least seven testis-associated genes. However, significant abnormalities in spermatogenesis were not observed until 5 months. The initial delay and progressive nature in the severity of the phenotype could have several etiologies. Figla encodes a transcription factor, and therefore its presence is manifest only secondarily through the repression of target genes. At least some of these target genes also encode transcription factors which, while affecting a broader network of genes, could further delay appearance of the full phenotype. In addition, Figla is not fully expressed until several weeks after birth, and depending on the stability of cognate mRNA and protein the phenotype might not become evident until gene products are degraded below a functional threshold.
Multiple molecular defects of spermatogenesis were observed in the transgenic mice (Table 1). Tnp2, which was upregulated in Figla null ovaries, was less abundant, as were Prm1 and Prm2, in TSPY1-Figla transgenic male mice. During normal spermatogenesis, DNA is repackaged from somatic histones via transition proteins (TNP1 and TNP2) onto protamines (PRM1 and PRM2) to form a highly condensed nucleus characteristic of the mature male gamete. Males lacking TNP2 have decreased fecundity (depending on genetic background) and defects in spermatogenesis, including incomplete chromatin condensation, sperm head abnormalities, and reduced sperm motility (1, 57). Mice that are haploid insufficient in either protamine 1 or 2 have normal sperm counts, but dysmorphology and altered chromatin integrity results in a sterile phenotype (8, 9). A similar sterile phenotype with haploid insufficiency is observed in chimeric mice established from embryonic stem cells with a null allele of Cyp17a1 (cytochrome P450, family 17, subfamily a, polypeptide 1) (29), which is also upregulated in Figla null ovaries. All four of these genes affect the later stages of spermatogenesis with defects in morphology and motility, and sperm with the Cyp17a1 null allele have the same severe tail angulation observed with TSPY1-Figla sperm.
The infertility observed in TSPY1-Figla transgenic male mice arises both from defective spermatogenesis and decreased sperm production in older mice. Spermatogenic cell apoptosis was detected by a TUNEL assay beginning in 4-month-old mouse testes and mostly affected spermatogonia germ cells. However, cell death of stem cells may not fully account for decreased sperm counts in older TSPY1-Figla mice, which also have widespread and progressive testicular degeneration beginning at 5 months. Few elongated spermatids were detected in stage XI and XII seminiferous tubules and there was a dramatic decrease in epididymal sperm, which had concomitant defects in sperm motility and apparent abnormalities in sperm volume regulation. The cumulative effect of these abnormalities in spermatogenesis affects normal gamete development and causes a striking reduction in rates of successful fertilization in vitro and in vivo.
Perhaps most striking, ectopic expression of Figla severely disrupts the chromatoid body, a cytoplasmic structure characteristic of postmeiotic spermatids. It is derived from the nuage, which are granulated, acellular, cytoplasmic structures interspersed among mitochondria in both growing oocytes and in the early stages of spermatogenesis. In male germ cells, the chromatoid body is defined as an electron-dense structure initially scattered in the cytoplasm of spermatocytes which becomes condensed as a single structure in round spermatids and persists in elongating haploid spermatids prior to sequestration in the residual body (12, 13). Specific RNA and proteins have been associated with this structure, including MVH, MILI, TDRD1, TDRD6, and TDRD7 (18, 23). Proteins encoded by Mvh and Mili interact with one another, and disruption of either gene blocks spermatogenesis in the zygotene or early pachytene stage of MI prophase (24, 48). TDRD1, TDRD6, and TDRD7 also interact with one another, and their localization to the chromatoid body requires the presence of MVH (18). More recent data have demonstrated that TDRD1 interacts with MILI, and the absence of TDRD1 in null mice activates germ line transposons and affects the profile of piRNAs (41, 53). TDRD6 also binds to MILI and MIWI (official name, Piwi1), and altered miRNA expression profiles are observed in Tdrd6 null testes (52). In addition, both TDRD1 and TDRD6 interact with MVH, and mice lacking either protein have small testes with disorganized spermatogenesis and male sterility. Tdrd1 null testes have meiotic defects in prophase I, whereas Tdrd6 null testes are devoid of elongated spermatids. Although the chromatoid body is present in normal round spermatids, it is significantly disrupted in both Tdrd1 and Tdrd6 null male mice (10, 52).
FIGLA, ectopically expressed in male germ cells, effectively downregulates Tdrd1, Tdrd6, and Tdrd7, leading to a phenotype that encompasses those described in Tdrd1 and Tdrd6 null mice, including male sterility, meiotic defects in diakinesis/metaphase stages, a postmeiotic defect in elongated round spermatids, loss of the chromatoid body structure, and displacement of MVH and MILI proteins. The absence of the chromatoid body integrity and mislocalization of the chromatoid body-associated proteins in TSPY1-Figla transgenic testes provide further evidence that TDRD proteins are required for spermatogenesis and chromatoid body architecture. Thus, our study provides evidence that FIGLA represses multiple male germ cell-associated genes, the expression of which could disrupt normal oogenesis.
These data are consistent with FIGLA, a bHLH transcription factor, sustaining the female phenotype by activating female and repressing male germ cell genetic hierarchies in growing oocytes during postnatal ovarian development. Combining the gain of function of TSPY1-Figla transgenic males with the loss of function of Figla null females provides complementary models for further analysis of the genetic hierarchies that maintain a female-specific phenotype during oogenesis.
Acknowledgments
We appreciate the critical comments by Mary Ann Handel and the gift of the antibodies to TDRD proteins from Shinichiro Chuma.
This research was supported by the Intramural Research Program of the National Institutes of Health, NIDDK.
Footnotes
Published ahead of print on 17 May 2010.
REFERENCES
- 1.Adham, I. M., K. Nayernia, E. Burkhardt-Gottges, O. Topaloglu, C. Dixkens, A. F. Holstein, and W. Engel. 2001. Teratozoospermia in mice lacking the transition protein 2 (Tnp2). Mol. Hum. Reprod. 7:513-520. [DOI] [PubMed] [Google Scholar]
- 2.Alban, A., S. O. David, L. Bjorkesten, C. Andersson, E. Sloge, S. Lewis, and I. Currie. 2003. A novel experimental design for comparative two-dimensional gel analysis: two-dimensional difference gel electrophoresis incorporating a pooled internal standard. Proteomics 3:36-44. [DOI] [PubMed] [Google Scholar]
- 3.Ballow, D., M. L. Meistrich, M. Matzuk, and A. Rajkovic. 2006. Sohlh1 is essential for spermatogonial differentiation. Dev. Biol. 294:161-167. [DOI] [PubMed] [Google Scholar]
- 4.Barrera, L. O. 2006. The transcriptional regulatory code of eukaryotic cells: insights from genome-wide analysis of chromatin organization and transcription factor binding. Curr. Opin. Cell Biol. 18:291-298. [DOI] [PubMed] [Google Scholar]
- 5.Brennan, J., and B. Capel. 2004. One tissue, two fates: molecular genetic events that underlie testis versus ovary development. Nat. Rev. Genet. 5:509-521. [DOI] [PubMed] [Google Scholar]
- 6.Buaas, F. W., A. L. Kirsh, M. Sharma, D. J. McLean, J. L. Morris, M. D. Griswold, D. G. de Rooij, and R. E. Braun. 2004. Plzf is required in adult male germ cells for stem cell self-renewal. Nat. Genet. 36:647-652. [DOI] [PubMed] [Google Scholar]
- 7.Burgoyne, P. S., S. K. Mahadevaiah, and J. M. Turner. 2009. The consequences of asynapsis for mammalian meiosis. Nat. Rev. Genet. 10:207-216. [DOI] [PubMed] [Google Scholar]
- 8.Cho, C., H. Jung-Ha, W. D. Willis, E. H. Goulding, P. Stein, Z. Xu, R. M. Schultz, N. B. Hecht, and E. M. Eddy. 2003. Protamine 2 deficiency leads to sperm DNA damage and embryo death in mice. Biol. Reprod. 69:211-217. [DOI] [PubMed] [Google Scholar]
- 9.Cho, C., W. D. Willis, E. H. Goulding, H. Jung-Ha, Y. C. Choi, N. B. Hecht, and E. M. Eddy. 2001. Haploinsufficiency of protamine-1 or -2 causes infertility in mice. Nat. Genet. 28:82-86. [DOI] [PubMed] [Google Scholar]
- 10.Chuma, S., M. Hosokawa, K. Kitamura, S. Kasai, M. Fujioka, M. Hiyoshi, K. Takamune, T. Noce, and N. Nakatsuji. 2006. Tdrd1/Mtr-1, a tudor-related gene, is essential for male germ-cell differentiation and nuage/germinal granule formation in mice. Proc. Natl. Acad. Sci. U. S. A. 103:15894-15899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Connolly, C. M., A. T. Dearth, and R. E. Braun. 2005. Disruption of murine Tenr results in teratospermia and male infertility. Dev. Biol. 278:13-21. [DOI] [PubMed] [Google Scholar]
- 12.Eddy, E. M. 1975. Germ plasm and the differentiation of the germ cell line. Int. Rev. Cytol. 43:229-280. [DOI] [PubMed] [Google Scholar]
- 13.Fawcett, D. W., E. M. Eddy, and D. M. Phillips. 1970. Observations on the fine structure and relationships of the chromatoid body in mammalian spermatogenesis. Biol. Reprod. 2:129-153. [DOI] [PubMed] [Google Scholar]
- 14.Fulco, M., R. L. Schiltz, S. Iezzi, M. T. King, P. Zhao, Y. Kashiwaya, E. Hoffman, R. L. Veech, and V. Sartorelli. 2003. Sir2 regulates skeletal muscle differentiation as a potential sensor of the redox state. Mol. Cell 12:51-62. [DOI] [PubMed] [Google Scholar]
- 15.Ganguly, A., R. A. McKnight, S. Raychaudhuri, B. C. Shin, Z. Ma, K. Moley, and S. U. Devaskar. 2007. Glucose transporter isoform-3 mutations cause early pregnancy loss and fetal growth restriction. Am. J. Physiol. Endocrinol. Metab. 292:E1241-E1255. [DOI] [PubMed] [Google Scholar]
- 16.Gavrieli, Y., Y. Sherman, and S. A. Ben-Sasson. 1992. Identification of programmed cell death in situ via specific labeling of nuclear DNA fragmentation. J. Cell Biol. 119:493-501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hoodbhoy, T., S. Joshi, E. S. Boja, S. A. Williams, P. Stanley, and J. Dean. 2005. Human sperm do not bind to rat zonae pellucidae despite the presence of four homologous glycoproteins. J. Biol. Chem. 280:12721-12731. [DOI] [PubMed] [Google Scholar]
- 18.Hosokawa, M., M. Shoji, K. Kitamura, T. Tanaka, T. Noce, S. Chuma, and N. Nakatsuji. 2007. Tudor-related proteins TDRD1/MTR-1, TDRD6 and TDRD7/TRAP: domain composition, intracellular localization, and function in male germ cells in mice. Dev. Biol. 301:38-52. [DOI] [PubMed] [Google Scholar]
- 19.Jones, S. 2004. An overview of the basic helix-loop-helix proteins. Genome Biol. 5:226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Joshi, S., H. Davies, L. P. Sims, S. E. Levy, and J. Dean. 2007. Ovarian gene expression in the absence of FIGLA, an oocyte-specific transcription factor. BMC Dev. Biol. 7:67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kai, T., and A. Spradling. 2004. Differentiating germ cells can revert into functional stem cells in Drosophila melanogaster ovaries. Nature 428:564-569. [DOI] [PubMed] [Google Scholar]
- 22.Kido, T., and Y. F. Lau. 2005. A Cre gene directed by a human TSPY promoter is specific for germ cells and neurons. Genesis 42:263-275. [DOI] [PubMed] [Google Scholar]
- 23.Kotaja, N., and P. Sassone-Corsi. 2007. The chromatoid body: a germ-cell-specific RNA-processing centre. Nat. Rev. Mol. Cell Biol. 8:85-90. [DOI] [PubMed] [Google Scholar]
- 24.Kuramochi-Miyagawa, S., T. Kimura, T. W. Ijiri, T. Isobe, N. Asada, Y. Fujita, M. Ikawa, N. Iwai, M. Okabe, W. Deng, H. Lin, Y. Matsuda, and T. Nakano. 2004. Mili, a mammalian member of Piwi family gene, is essential for spermatogenesis. Development 131:839-849. [DOI] [PubMed] [Google Scholar]
- 25.La Salle, S., F. Sun, X. D. Zhang, M. J. Matunis, and M. A. Handel. 2008. Developmental control of sumoylation pathway proteins in mouse male germ cells. Dev. Biol. 321:227-237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Leduc, F., V. Maquennehan, G. B. Nkoma, and G. Boissonneault. 2008. DNA damage response during chromatin remodeling in elongating spermatids of mice. Biol. Reprod. 78:324-332. [DOI] [PubMed] [Google Scholar]
- 27.Li, L., B. Baibakov, and J. Dean. 2008. A subcortical maternal complex essential for pre-implantation mouse embryogenesis. Dev. Cell 15:416-425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liang, L.-F., S. M. Soyal, and J. Dean. 1997. FIGα, a germ cell specific transcription factor involved in the coordinate expression of the zona pellucida genes. Development 124:4939-4949. [DOI] [PubMed] [Google Scholar]
- 29.Liu, Y., Z. X. Yao, C. Bendavid, C. Borgmeyer, Z. Han, L. R. Cavalli, W. Y. Chan, J. Folmer, B. R. Zirkin, B. R. Haddad, G. I. Gallicano, and V. Papadopoulos. 2005. Haploinsufficiency of cytochrome P450 17α-hydroxylase/17,20 lyase (CYP17) causes infertility in male mice. Mol. Endocrinol. 19:2380-2389. [DOI] [PubMed] [Google Scholar]
- 30.Lu, J., R. Webb, J. A. Richardson, and E. N. Olson. 1999. MyoR: a muscle-restricted basic helix-loop-helix transcription factor that antagonizes the actions of MyoD. Proc. Natl. Acad. Sci. U. S. A. 96:552-557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mal, A., M. Sturniolo, R. L. Schiltz, M. K. Ghosh, and M. L. Harter. 2001. A role for histone deacetylase HDAC1 in modulating the transcriptional activity of MyoD: inhibition of the myogenic program. EMBO J. 20:1739-1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Newman, G. R. 1999. LR White embedding for immunoelectron microscopy. Histochem. J. 31:79. [DOI] [PubMed] [Google Scholar]
- 33.Otori, M., T. Karashima, and M. Yamamoto. 2006. The Caenorhabditis elegans homologue of deleted in azoospermia is involved in the sperm/oocyte switch. Mol. Biol. Cell 17:3147-3155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pan, J., S. Eckardt, N. A. Leu, M. G. Buffone, J. Zhou, G. L. Gerton, K. J. McLaughlin, and P. J. Wang. 2009. Inactivation of Nxf2 causes defects in male meiosis and age-dependent depletion of spermatogonia. Dev. Biol. 330:167-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pfeifer, C., P. D. Thomsen, and H. Scherthan. 2001. Centromere and telomere redistribution precedes homologue pairing and terminal synapsis initiation during prophase I of cattle spermatogenesis. Cytogenet. Cell Genet. 93:304-314. [DOI] [PubMed] [Google Scholar]
- 36.Rajkovic, A., S. A. Pangas, D. Ballow, N. Suzumori, and M. M. Matzuk. 2004. NOBOX deficiency disrupts early folliculogenesis and oocyte-specific gene expression. Science 305:1157-1159. [DOI] [PubMed] [Google Scholar]
- 37.Rankin, T., P. Talbot, E. Lee, and J. Dean. 1999. Abnormal zonae pellucidae in mice lacking ZP1 result in early embryonic loss. Development 126:3847-3855. [DOI] [PubMed] [Google Scholar]
- 38.Rankin, T. L., Z.-B. Tong, P. E. Castle, E. Lee, R. Gore-Langton, L. M. Nelson, and J. Dean. 1998. Human ZP3 restores fertility in Zp3 null mice without affecting order-specific sperm binding. Development 125:2415-2424. [DOI] [PubMed] [Google Scholar]
- 39.Ravnik, S. E., and D. J. Wolgemuth. 1999. Regulation of meiosis during mammalian spermatogenesis: the A-type cyclins and their associated cyclin-dependent kinases are differentially expressed in the germ-cell lineage. Dev. Biol. 207:408-418. [DOI] [PubMed] [Google Scholar]
- 40.Remenyi, A., H. R. Scholer, and M. Wilmanns. 2004. Combinatorial control of gene expression. Nat. Struct. Mol. Biol. 11:812-815. [DOI] [PubMed] [Google Scholar]
- 41.Reuter, M., S. Chuma, T. Tanaka, T. Franz, A. Stark, and R. S. Pillai. 2009. Loss of the Mili-interacting Tudor domain-containing protein-1 activates transposons and alters the Mili-associated small RNA profile. Nat. Struct. Mol. Biol. 16:639-646. [DOI] [PubMed] [Google Scholar]
- 42.Sada, A., A. Suzuki, H. Suzuki, and Y. Saga. 2009. The RNA-binding protein NANOS2 is required to maintain murine spermatogonial stem cells. Science 325:1394-1398. [DOI] [PubMed] [Google Scholar]
- 43.Sartorelli, V., P. L. Puri, Y. Hamamori, V. Ogryzko, G. Chung, Y. Nakatani, J. Y. Wang, and L. Kedes. 1999. Acetylation of MyoD directed by PCAF is necessary for the execution of the muscle program. Mol. Cell 4:725-734. [DOI] [PubMed] [Google Scholar]
- 44.Schubert, S., B. Skawran, F. Dechend, K. Nayernia, A. Meinhardt, I. Nanda, M. Schmid, W. Engel, and J. Schmidtke. 2003. Generation and characterization of a transgenic mouse with a functional human TSPY. Biol. Reprod. 69:968-975. [DOI] [PubMed] [Google Scholar]
- 45.Simone, C., S. V. Forcales, D. A. Hill, A. N. Imbalzano, L. Latella, and P. L. Puri. 2004. p38 pathway targets SWI-SNF chromatin-remodeling complex to muscle-specific loci. Nat. Genet. 36:738-743. [DOI] [PubMed] [Google Scholar]
- 46.Soyal, S. M., A. Amleh, and J. Dean. 2000. FIGα, a germ-cell specific transcription factor required for ovarian follicle formation. Development 127:4645-4654. [DOI] [PubMed] [Google Scholar]
- 47.Suzuki, A., and Y. Saga. 2008. Nanos2 suppresses meiosis and promotes male germ cell differentiation. Genes Dev. 22:430-435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tanaka, S. S., Y. Toyooka, R. Akasu, Y. Katoh-Fukui, Y. Nakahara, R. Suzuki, M. Yokoyama, and T. Noce. 2000. The mouse homolog of Drosophila Vasa is required for the development of male germ cells. Genes Dev. 14:841-853. [PMC free article] [PubMed] [Google Scholar]
- 49.Tapscott, S. J. 2005. The circuitry of a master switch: Myod and the regulation of skeletal muscle gene transcription. Development 132:2685-2695. [DOI] [PubMed] [Google Scholar]
- 50.Tsuda, M., Y. Sasaoka, M. Kiso, K. Abe, S. Haraguchi, S. Kobayashi, and Y. Saga. 2003. Conserved role of nanos proteins in germ cell development. Science 301:1239-1241. [DOI] [PubMed] [Google Scholar]
- 51.Uhlenhaut, N. H., S. Jakob, K. Anlag, T. Eisenberger, R. Sekido, J. Kress, A. C. Treier, C. Klugmann, C. Klasen, N. I. Holter, D. Riethmacher, G. Schutz, A. J. Cooney, R. Lovell-Badge, and M. Treier. 2009. Somatic sex reprogramming of adult ovaries to testes by FOXL2 ablation. Cell 139:1130-1142. [DOI] [PubMed] [Google Scholar]
- 52.Vasileva, A., D. Tiedau, A. Firooznia, T. Muller-Reichert, and R. Jessberger. 2009. Tdrd6 is required for spermiogenesis, chromatoid body architecture, and regulation of miRNA expression. Curr. Biol. 19:630-639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang, J., J. P. Saxe, T. Tanaka, S. Chuma, and H. Lin. 2009. Mili interacts with tudor domain-containing protein 1 in regulating spermatogenesis. Curr. Biol. 19:640-644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yan, W., L. Ma, K. H. Burns, and M. M. Matzuk. 2004. Haploinsufficiency of kelch-like protein homolog 10 causes infertility in male mice. Proc. Natl. Acad. Sci. U. S. A. 101:7793-7798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yeung, C. H., A. Wagenfeld, E. Nieschlag, and T. G. Cooper. 2000. The cause of infertility of male c-ros tyrosine kinase receptor knockout mice. Biol. Reprod. 63:612-618. [DOI] [PubMed] [Google Scholar]
- 56.Yokota, S. 2008. Historical survey on chromatoid body research. Acta Histochem. Cytochem. 41:65-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhao, M., C. R. Shirley, Y. E. Yu, B. Mohapatra, Y. Zhang, E. Unni, J. M. Deng, N. A. Arango, N. H. Terry, M. M. Weil, L. D. Russell, R. R. Behringer, and M. L. Meistrich. 2001. Targeted disruption of the transition protein 2 gene affects sperm chromatin structure and reduces fertility in mice. Mol. Cell. Biol. 21:7243-7255. [DOI] [PMC free article] [PubMed] [Google Scholar]