Abstract
This study identifies KLF3 as a transcriptional regulator of muscle genes and reveals a novel synergistic interaction between KLF3 and serum response factor (SRF). Using quantitative proteomics, KLF3 was identified as one of several candidate factors that recognize the MPEX control element in the Muscle creatine kinase (MCK) promoter. Chromatin immunoprecipitation analysis indicated that KLF3 is enriched at many muscle gene promoters (MCK, Myosin heavy chain IIa, Six4, Calcium channel receptor α-1, and Skeletal α-actin), and two KLF3 isoforms are upregulated during muscle differentiation. KLF3 and SRF physically associate and synergize in transactivating the MCK promoter independently of SRF binding to CArG motifs. The zinc finger and repression domains of KLF3 plus the MADS box and transcription activation domain of SRF are implicated in this synergy. Our results provide the first evidence of a role for KLF3 in muscle gene regulation and reveal an alternate mechanism for transcriptional regulation by SRF via its recruitment to KLF binding sites. Since both factors are expressed in all muscle lineages, SRF may regulate many striated- and smooth-muscle genes that lack known SRF control elements, thus further expanding the breadth of the emerging CArGome.
Expression of Muscle creatine kinase (MCK) is restricted to terminally differentiated striated muscle, where it is one of the most abundantly expressed genes. MCK transcription is regulated by an upstream enhancer (−1256 to −1050), a proximal promoter (−358 to +7), and an intron 1 modulatory region (+740 to +1731). Despite several decades of analysis, the study of MCK is still revealing novel mechanisms of transcriptional control with direct relevance to many other striated-muscle genes.
The MCK proximal promoter can drive muscle-specific expression on its own, but it also synergizes with the upstream enhancer to drive much higher expression in both skeletal and cardiac muscle in cell culture (1, 27) and in transgenic mice (14, 54). The MCK promoter contains a conserved CArG site, an E box, and a GC-rich MPEX site, and the last two elements contribute to expression in both types of striated muscle (25, 41). Interestingly, in contrast to the MCK enhancer, where high sequence conservation occurs only within control elements, the promoter is highly conserved throughout (∼70% identical between human and mouse), suggesting complex regulatory functions and control elements that have yet to be identified.
In a previous study, we used quantitative proteomics to identify factors binding the MCK promoter MPEX site and demonstrated that one of these candidates, MAZ, regulates muscle-specific genes through both established and divergent binding motifs (25). Here, we investigate the role of Kruppel-like factor 3 (KLF3), which was also identified as a candidate MPEX-binding factor (MPEX-BF) by proteomic analysis. We demonstrate that KLF3 binds MPEX, as well as functional CACCC boxes in the MCK promoter, and that KLF3 is enriched at endogenous muscle gene promoters (MCK, Myosin heavy chain IIa [MyHCIIa], Six4, Calcium channel receptor α-1 [CaChR], and Skeletal α-actin [Skαact]).
KLF3 (BKLF/TEF2), originally identified as a prominent CACCC box binding factor in erythroid cells, has been shown to bind C(A/C)CACCC with particularly high affinity (12). It is a widely expressed member of the Kruppel-like factor subfamily of Sp/KLF factors that contacts DNA via 3 Cys2-His2-type zinc fingers. Although KLF3 can exhibit positive transcriptional activity (12), it has been characterized as a potent repressor of transcription. This occurs through an N-terminal repression domain (RD) that recruits both the C-terminal binding protein 2 (CtBP2) corepressor and the four-and-a-half LIM domain (FHL) protein FHL3 (60). In addition to contact with CtBP2, sumoylation of KLF3 residues K10 and K197 is also required for full repression (46). Sequence-tagged transcripts in the GenBank database include several spliceoforms of KLF3, but their functions are not known. KLF3 knockout mice display adipogenic defects, but no defective muscle phenotype has been detected, possibly due to functional redundancy within the Sp/KLF family. This family contains 26 known proteins, many of them with ubiquitous or widespread expression patterns (56). Although at least six KLF family members are thought to play roles in striated and smooth muscle (21), the role(s) of KLF3 in these tissues has not been characterized.
In the process of investigating the role of KLF3 in skeletal muscle, we discovered that its expression is initiated during terminal differentiation, a period when the expression of many new transcription factors and structural proteins is activated. Furthermore, chromatin immunoprecipitation (ChIP) analysis demonstrated that KLF3 is enriched at the promoters of many of these newly activated genes. Why would KLF3, a strong transcriptional repressor with no characterized transcription activation domain (TAD), be enriched at muscle genes at the onset of differentiation? One possibility is that it might cooperate with other factors to create a positive transcriptional complex that binds KLF3 control elements. Using transactivation studies to find potential KLF3 interaction partners, we found that serum response factor (SRF) exhibits strong synergy with KLF3 in transactivating the MCK promoter.
SRF is a MADS box transcription factor that is a critical mediator of cardiac-, skeletal-, and smooth-muscle gene expression (16, 36, 43). Conditional knockouts of SRF display severe defects in both cardiac and skeletal muscle (6, 32, 34, 44, 45). The MADS domain of SRF mediates both dimerization and binding to a DNA motif known as the CArG box (CC[A/T]TATA[A/T]GG), as well as interactions with cofactors (43, 63). SRF has been shown to associate with a number of proteins, including GATA-4/6, Nkx factors, myogenin/MyoD, TEF-1, Ets factors, CRP1/2, TFIIF, Barx-2b, HOP, and Smads (3, 5, 7, 19, 20, 22, 29, 37, 42, 48, 52, 55). Myocardin and two myocardin-related transcription factors (MRTF-A and MRTF-B) have also emerged as critical mediators of SRF activity (47). Because SRF controls gene expression in response to both growth and differentiation signals (36), the identification of its target genes has been of long-standing interest. Identification of the CArGome (the full spectrum of all known and novel CArG sites in the genome) brought to light a number of new targets of SRF activity, and 23% encode contractile/cytoskeletal proteins (57). More recently, SRF-binding sites throughout the genome were identified using ChIP combined with human promoter microarrays (10). Intriguingly, in the latter study, 33% of validated SRF gene targets did not contain a CArG box within 4 kb upstream and 1 kb downstream of the transcription start site (10).
Here, we identify a positive transcriptional interaction between KLF3 and SRF that is independent of SRF binding to CArG motifs. This discovery could provide a mechanism for how SRF target genes are regulated in the absence of known SRF-binding sites. It also implies that transcription of the CArG site-independent subset of SRF target genes can be partially controlled by signal transduction pathways that modify KLF3 function. Synergistic interactions between these pathways provide possibilities for fine-tuning the regulation of many striated- and smooth-muscle genes.
MATERIALS AND METHODS
Plasmids and antibodies.
The reporter plasmids −80MCKCAT, e−80MCKCAT, −358MCKCAT, e−358MCKCAT, (MPEX-mt)−80MCKCAT, and pUCSV2PAP have been described previously (1, 25, 54), as have constructs containing full-length mouse KLF3 cDNA in pMT2 (12), full-length and all truncated mouse KLF3 cDNAs in pMT3 (46), FLAG-tagged mouse KLF3 cDNA in pMT3 (46), and full-length KLF4 cDNA in pcDNA3 (a generous gift of Gary Owens [35]). Full-length human SRF cDNA in pCGN (11) was a generous gift of Robert Schwartz. The pcDNA constructs encoding the human SRF MADS domain (112-265) and mouse SRF lacking the C-terminal TAD (Δ266-508) were generous gifts of Robert Schwartz and Michael Parmacek (15). The MEF2C expression construct was a generous gift of Eric Olson (38). FLAG-KLF3(Δ8-89), FLAG-KLF3(Δ8-119), SRF(Δ8-177), and SRF(Δ8-133) were generated from FLAG-KLF3 or SRF-pCGN by deletion mutagenesis using a QuikChange Site-Directed Mutagenesis Kit (Stratagene) according to the manufacturer's directions, as were the other deletions/mutations (see Fig. 3B and 4A). For CAC2-mt constructs, the CACCC box was mutated (underlined) to GACGG. All constructs were sequenced to verify the integrity of the modified DNA.
FIG. 3.
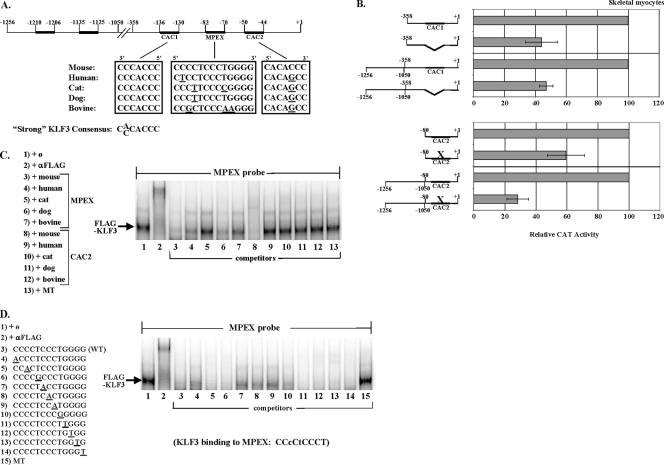
Multiple KLF binding motifs are important for the activity of the MCK promoter in skeletal myocytes. (A) Sequence conservation of KLF binding motifs in the MCK promoter. The position of KLF binding motifs in the MCK enhancer (−1256 to −1050) and proximal promoter (−358 to +1) are shown (dark bars), with sequence alignment of MPEX and C(A/C)CACCC boxes within the MCK promoters from multiple mammalian species. Forward and reverse sequences are indicated, and bases that differ from the mouse sequences are underlined. (B) CAC1 and CAC2 are important for MCK promoter activity in skeletal myocytes. Skeletal myocytes were transfected with constructs containing the CAT reporter under the control of either the 358-bp MCK proximal promoter, the 80-bp MCK minimal promoter, or the equivalent constructs containing the MCK enhancer, and the PAP reference plasmid. The activities of the wild-type constructs compared to constructs containing a deletion of CAC1 or a mutation in CAC2 are shown. The data are plotted as the mean value and standard deviation of the CAT/PAP ratio determined for each culture dish, and the activity of each corresponding wild-type construct is set at 100. (C and D) KLF3 recognizes divergent MPEX sequences, but not the divergent CAC2 motif present in nonmouse species. Labeled mouse MPEX probe was mixed with 2 μg of nuclear extracts from COS-7 cells overexpressing FLAG-KLF3 and analyzed via gel shift assays. The KLF3-specific band (supershifted by antibodies to FLAG) (lane 2) is indicated. In panel C, oligonucleotides containing the MPEX and CAC2 sequences from the mouse, human, cat, dog, and bovine MCK promoters were tested alongside a mutant MPEX sequence (MT) as competitors for KLF3 binding. In panel D, oligonucleotides containing the wild-type mouse MPEX sequence (WT) or with single-base-pair changes (underlined) at each position were tested alongside a mutant MPEX sequence (MT) as competitors for KLF3 binding. (The oligonucleotides containing changes in bases 2 [C→T] and 4 [C→T] are the human and dog versions of MPEX, respectively, tested in panel C, lanes 4 and 6.) A summary of the KLF3 recognition sequence within MPEX is shown, with less stringent base requirements indicated in lowercase letters.
FIG. 4.
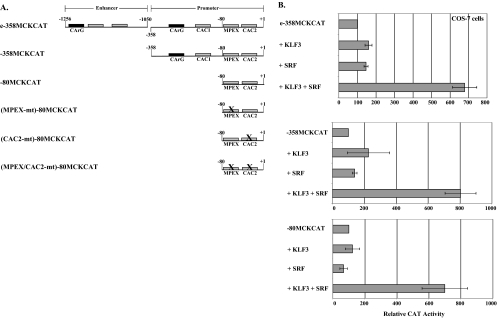
KLF3 and SRF synergize in transactivating the MCK promoter, and the synergy is independent of CArG sites. (A) Diagrams of the constructs used in panel B and Fig. 5 and 6. CAT reporter constructs contain the MCK enhancer linked to the proximal promoter (e−358MCKCAT), the MCK proximal promoter (−358MCKCAT), the MCK minimal promoter (−80MCKCAT), or the MCK minimal promoter containing mutations in MPEX [(MPEX-mt)−80MCKCAT], CAC2 [(CAC2-mt)−80MCKCAT], or both sites [(MPEX/CAC2-mt)−80MCKCAT]. (B) KLF3 and SRF synergize in a CArG-independent fashion. COS-7 cells were transfected with e−358MCKCAT, −358MCKCAT, or −80MCKCAT. Each reporter construct was transfected alone or with 0.4 μg of KLF3 expression vector, 1 μg of SRF expression vector, or both. The data are plotted as the mean value and standard deviation of relative CAT activity, with the activity of the reporter construct alone set at 100.
The antibodies used in this study were as follows: rabbit polyclonal antisera to KLF3 and rabbit preimmune sera (12), rabbit polyclonal antibody to SRF (G-20; sc-335; Santa Cruz Biotechnology, Inc.), anti-FLAG M2 monoclonal antibody (F3165; Sigma), nonimmune rabbit serum (R9133; Sigma), and mouse GATA-2 antibody (CG2-96; sc-267; Santa Cruz Biotechnology, Inc.).
Cell culture.
Mouse MM14 skeletal myoblasts were grown on 100-mm gelatin-coated tissue culture dishes in proliferation medium (Ham's F-10C supplemented with 15% horse serum and 2 ng/ml basic fibroblast growth factor) as previously described (9, 40). For transient transfections, log-phase cultures at ∼4 × 105 cells/dish were induced to differentiate by rinsing the cultures twice with saline G and then switching them to differentiation medium (Ham's F-10C supplemented with 1.5% horse serum and 6 μg/ml insulin). The cells were maintained for 48 h in differentiation medium prior to being harvested. For myocyte nuclear extracts used in Fig. 1 B and 2 B, 100-mm dishes were plated with ∼1 × 105 log-phase cells/dish, grown to near confluence (∼4 × 106 cells/dish), and then allowed to differentiate in proliferation medium without additional fibroblast growth factor for 4 days prior to being harvested. All cultures contained >90% terminally differentiated myocytes, as assessed by immunostaining a parallel culture with the myosin-specific antibody MF-20. For the early-differentiated myocyte nuclear extracts used in Fig. 2B, cells were grown in proliferation medium as described above and then switched to differentiation medium for 20 h prior to being harvested. For myoblast nuclear extracts, cells were grown in proliferation medium as described above and harvested at ∼5 × 105 cells/dish. COS-7 cells were grown on 100-mm gelatin-coated tissue culture dishes in Dulbecco's modified Eagle's medium (DMEM) with 10% fetal bovine serum (FBS) and antibiotics (100 U/ml penicillin and 0.1 mg/ml streptomycin).
FIG. 1.
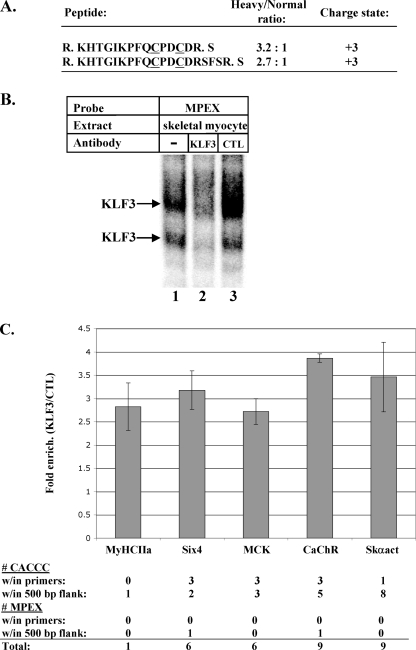
KLF3 binds the MCK promoter and other muscle gene promoters in skeletal myocytes. (A) Peptides corresponding to KLF3 are enriched in MPEX versus MPEX-mt DNA affinity-purified samples. Factors binding the MPEX site in the MCK promoter were selectively enriched from skeletal myocytes and identified by ICAT-based quantitative proteomics; the experimental details are described in our previous study (25). Peptides were analyzed by microcapillary liquid chromatography-electrospray ionization-tandem mass spectroscopy (μLC-ESI-MS/MS), and protein identifications were assigned using the SEQUEST algorithm to search a mouse protein sequence database. ICAT-labeled cysteine residues are underlined, and the dots represent sites of tryptic cleavage. The PeptideProphet probability scores for each peptide were >0.9. The relative abundance of each peptide in heavy (isolated from MPEX beads) versus normal (isolated from MPEX-mt beads) ICAT-labeled samples was calculated using XPRESS and is expressed as a ratio. (B) KLF3 in skeletal myocytes binds the MPEX sequence. Labeled mouse MPEX probe was mixed with 2 μg of skeletal myocyte nuclear extracts and analyzed via gel shift interference assay. KLF3-specific antisera reduced formation of the indicated complexes, whereas nonimmune sera (cytotoxic T lymphocytes [CTL]) had no effect. (C) KLF3 occupies muscle gene promoters in skeletal myocytes. ChIP assays were performed using MM14 skeletal myocytes and KLF3-specific antisera or preimmune sera. Immunoprecipitated chromatin was analyzed by qPCR using primers specific for the promoters of MyHCIIa, Six4, MCK, CaChR, and Skαact. The data are represented as fold enrichment (enrich.) of the indicated promoter region by KLF3-specific antisera relative to preimmune sera (CTL). Each bar represents the average and standard deviation (SD) from 3 independent ChIP experiments, with 3 replicate PCRs per experiment. The number of KLF3 consensus motifs (CACCC) and KLF3-binding MPEX motifs (based on gel shift analysis) (Fig. 3D) is shown for the region encompassed by the PCR primers and within 500 bp 5′ or 3′ of the primers.
FIG. 2.
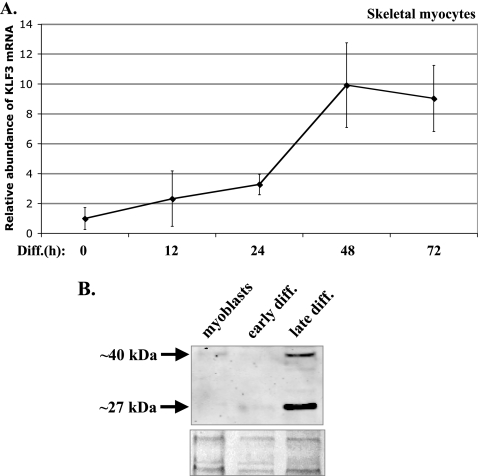
KLF3 expression is initiated during skeletal myocyte differentiation. (A) KLF3 transcripts increase during myocyte differentiation (Diff.). RNA was isolated from undifferentiated skeletal myoblasts (0 h) and myocytes differentiated for 12, 24, 48, and 72 h. qRT-PCR was performed using primers specific for KLF3 mRNA or 18S rRNA. The data are represented as the fold change in KLF3/18S RNA relative to myoblasts. Student's t test P values were 0.01 for 0 h versus 24 h and 0.05 for 24 h versus 48 h (n = 4). The error bars represent SD. (B) KLF3 protein increases during myocyte differentiation. Cytoplasmic extracts were made from undifferentiated skeletal myoblasts and myocytes differentiated for 1 day (early diff.) and 4 days (late diff.) and subjected to Western analysis using antisera to KLF3 (top panel). The predicted size of mouse KLF3 is 38 kDa. The bands in the lower gel represent Ponceau S staining for total protein as a loading control.
Transient transfections.
For reporter assays, proliferating MM14 or COS-7 cells were transfected, using a standard calcium phosphate technique (1), with 8 μg of reporter plasmids containing MCK test regions driving the chloramphenicol acetyltransferase (CAT) gene and either 2 μg of the pUCSV2PAP reference plasmid or various amounts of the expression plasmids described above. Four hours later, the cells were glycerol shocked; MM14 cells were switched to differentiation medium (as described above). Cells were harvested 48 h after glycerol shock and analyzed for CAT activity (1). The results of independent transfections are shown as the average of at least two transfections of four plates each.
Preparation of nuclear extracts.
Crude nuclear extracts from cultured cells were prepared as previously described (13) using a cocktail of several protease inhibitors (P8340; Sigma). The total protein in the extracts was quantitated by the Bradford method (2).
Gel mobility shift assays.
Gel shift assays were carried out as previously described (25). Incubations with antisera or unlabeled oligonucleotide competitors were carried out at room temperature for 20 min prior to the addition of probe. For MCK oligonucleotides used as probes or competitors, the forward sequences were as follows: mouse MPEX, 5′-GGGCCCCTCCCTGGGGACAGCCCCTCCTGGCT-3′; human MPEX, 5′-AGAACTCCTCCCTGGGGACAACCCCTCCCAGC-3′; cat MPEX, 5′-AGCTCCCTTCCCCGGGGGGCAGCCCCTCCCAG-3′; dog MPEX, 5′-AGCTCCCTTCCCTGGGGGCAGCCCCTCCCAGC-3′; bovine MPEX, 5′-AGCCCGCTCCCAAGGGGCAGCCCTTCCCAGCC-3′; MT, 5′-GGGCCACACACTGTGGCCCGACACGCATGGCT-3′; mouse CAC2, 5′-GCTAGTCACACCCTGTAGGCTCCTC-3′; human CAC2, 5′-AGCCAATAGCACAGCCTAGGTCCCC-3′; cat CAC2, 5′-CCTCCCAGCTGCACAGCCCGGCCCC-3′; dog CAC2, 5′-CCAGCCAATAGCACAGCCCGGCCCC-3′; bovine CAC2, 5′-CAGCCAATCACACAGCCCAGGCCCC-3′; and mouse CAC1, 5′-CCTGGGTCCGGGGTGGGCACGGTGC-3′.
For MPEX oligonucleotides, the MPEX sequence is underlined, and for CAC2 and CAC1 oligonucleotides, the CACCC box (altered in nonmouse species) is underlined. For MT, mutations from the mouse MPEX sequence are underlined. Single-base-pair mutations in the mouse MPEX sequence are shown in Fig. 3 D.
ChIP assays.
ChIP assays were performed with cultured MM14 skeletal myocytes using the Fast ChIP method (39) with some modifications. Cells were fixed in 1% formaldehyde in Ham's F10-C for 10 min and subjected to Dounce homogenization 10 times prior to sonication. The cells were sonicated for 10 rounds of 15-s pulses at 100% power output on a Model 100 Sonic Dismembrator (Fisher Scientific) to shear the DNA to fragments of ∼200 to 800 bp, as determined by agarose gel electrophoresis. Chromatin was immunoprecipitated using KLF3-specific antisera or preimmune sera at a final concentration of 6 μg/μl. Quantitative PCR (qPCR) was performed using forward and reverse primers (250 nM) and the 2× SensiMix DNA Kit (Quantace Ltd). The reaction conditions were 40 cycles of 94°C for 15 s, 51.8°C for 30 s, and 72°C for 30 s. The mouse primer sequences were as follows: MyHCIIa promoter, F (5′-TCTCTCCCATGTTCCTCTAGTGT-3′ [−783 to −761]) and R (5′-AGTTGCATGCCTTCAACAAT-3′ [−485 to −504]); Six4 promoter, F (5′-CAGAAGCTTGGCCAGAAAAG-3′ [−215 to −234]) and R (5′-ATAGCTGCTTTCTGCCGTTC-3′ [+16 to −4]); MCK promoter, F (5′-CGCCAGCTAGACTCAGCACT-3′ [−238 to −219]) and R (5′-GAGGAGCCTACAGGGTGTGA-3′ [−32 to −51]); CaChR promoter, F (5′-GCTTCCTTGACAGCGAGTG-3′ [−857 to −839]) and R (5′-TGCACCAGCTAGCAAACATT-3′ [−645 to −664]); and Skαact promoter, F (5′-GTGAGCCTTGGAGCCAGTT-3′ [−276 to −258]) and R (5′-GTCCCCTTGCACAGGTTTT-3′ [−10 to −28]).The PCR products were analyzed on a 1.5% agarose gel to verify the correct size of the product and the specificity of primer annealing.
Coimmunoprecipitations.
COS-7 cells were transfected with 1 μg SRF constructs, 2 μg FLAG-KLF3 constructs, or both using Lipofectamine LTX reagent (Invitrogen) according to the manufacturer's protocol. Forty-eight hours after transfection, the plates were washed twice with cold phosphate-buffered saline (PBS), and cells were harvested in PBS plus protease inhibitors. The cells were pelleted, washed with PBS plus protease inhibitors, and then incubated in lysis buffer (50 mM Tris, 150 mM NaCl, 1% NP-40, 1 mM Na3VO4, 1 mM dithiothreitol [DTT], and protease inhibitors) for 30 min on ice. The lysates were mixed with equal parts adjustment buffer (50 mM Tris, 150 mM NaCl), and then the cell debris was pelleted and the lysates were removed to new tubes. The lysates were incubated with or without antibodies for 2 h at 4°C with rotation and then incubated with protein A-Sepharose beads (17-5280-01; Amersham) for 2 h at 4°C with rotation. The immunoprecipitates were washed three times with wash buffer (1:1 lysis buffer-adjustment buffer) and then resuspended in 2× SDS-PAGE loading dye and boiled for 5 min before being stored at −20°C.
Western analysis.
Samples were heated at 70°C for 10 min prior to being loaded on NuPAGE 4 to 12% Bis-Tris gels (Invitrogen). The gels were run in 1× NuPAGE MES (morpholineethanesulfonic acid) SDS running buffer using the XCell II system (Invitrogen), according to the manufacturer's instructions. Proteins were transferred to nitrocellulose membranes in 1× NuPAGE transfer buffer using the XCell II system according to the manufacturer's instructions. The membranes were washed once in PBS, blocked in Sea Block blocking buffer (37527; Thermo Scientific) for 1 h at room temperature, and then incubated with primary antibodies in blocking buffer overnight at 4°C with rotation. The membranes were washed three times with PBS and then three times with wash buffer (0.05% Tween 20 in PBS) for 10 min each time. The membranes were incubated with secondary antibody [goat anti-rabbit IgG(H+L), DyLight 680 conjugated; 35568; Thermo Scientific] for 1 h at room temperature in the dark and then washed three times with PBS and six times with wash buffer for 5 min each time. Protein bands were visualized on a Li-Cor Odyssey Infrared Imaging System.
Quantitative reverse transcriptase (qRT) PCR.
MM14 skeletal myoblasts were allowed to differentiate for 0, 12, 24, 48, and 72 h. RNA was extracted using the Qiagen RNeasy kit, according to the manufacturer's instructions, and parallel plates for each time point were fixed and stained for MyHC expression. The RNA was DNase treated and reverse transcribed as previously described (25). Quantitative PCR was performed using 40 ng cDNA, forward and reverse primers (300 nM each), and SYBR green PCR Master Mix (Applied Biosystems). The reaction conditions were 40 cycles of 94°C for 15 s, 52.8 to 55.5°C (depending on the primer melting temperature [Tm]) for 30 s, and 72°C for 30 s. The primers used were specific to mouse KLF3 (F, 5′-TGCAAGAGAACCATCCTTCC-3′; R, 5′-GGTGCATTTGTACGGCTTTT-3′) and 18S rRNA (F, 5′-CGCCGCTAGAGGTGAAATTCT-3′; R, 5′-CGAACCTCCGACTTTCGTTCT-3′). Each PCR was performed in triplicate on RNAs from 4 separate plates per time point. The PCR products were analyzed on a 1.5% agarose gel to verify the correct size of the product and the specificity of primer annealing.
RESULTS
KLF3 binds the MPEX sequence and is enriched at the MCK promoter and other muscle gene promoters.
MPEX is a GC-rich sequence within the MCK promoter that is critical for high-level promoter activity in both skeletal and cardiac myocytes (25). The MPEX sequence contains putative binding sites for a number of transcription factors, including the prodigious Sp/KLF family, with 26 known members. After selectively enriching MPEX-BFs from MM14 skeletal myocytes, we used a quantitative proteomic strategy to identify MPEX-BF candidates (described in a previous study [25]). Briefly, we looked for proteins that were enriched in binding to beads linked to wild-type versus mutant MPEX oligonucleotides. KLF3 was one of nine transcription factor candidates identified, with two unique peptides enriched ∼3-fold in wild-type versus mutant MPEX samples (Fig. 1A). Details regarding experimental conditions and data filtering are described in our previous study (25).
KLF3 has been shown to bind with high affinity to CACCC boxes and, to a lesser extent, to other GC-rich sequences (12). To confirm the ability of KLF3 to bind MPEX, gel shift interference assays were performed using KLF3-specific antisera and nuclear extracts from skeletal myocytes. Antisera to KLF3 significantly reduced the formation of two complexes (Fig. 1B, lane 2), whereas nonspecific antisera had no effect (lane 3). The presence of two KLF3-containing complexes could indicate the existence of different isoforms, a cofactor interaction, or proteolytic cleavage of KLF3 in differentiated skeletal myocytes.
To determine whether KLF3 binds the MCK promoter in vivo, ChIP assays were performed using chromatin from skeletal myocytes. Immunoprecipitation with KLF3-specific antisera yielded ∼2.5-fold enrichment of the MCK promoter over that obtained with preimmune sera (Fig. 1C). When we assessed other muscle genes for KLF3 occupancy, we found that MyHCIIa, Six4, MCK, CaChR, and Skαact were all enriched to similar extents (∼2.5- to 4-fold) (Fig. 1C). Although these genes contain different numbers of KLF3-binding motifs within the primer-designated regions, we believe the similar enrichments are due to the fact that ChIP analysis also detects enrichment of sheared genomic regions flanking the 5′ and 3′ primer locations. Because ChIP detects binding to large regions, it is unclear which of these motifs are bound by KLF3, and with the exception of the KLF3-binding motifs in the MCK promoter (see below), the functional significance of these sites has not been characterized. Moreover, other members of the Sp/KLF family are also capable of recognizing these sequences and may compete with KLF3 for binding. These caveats may help to explain why KLF3 enrichment at the MyHCIIa promoter, which contains only a single CACCC box, is equivalent to enrichment at promoters that contain as many as 9 CACCC boxes within the ∼1,200-bp region including and flanking our PCR primers. Nonetheless, these results indicate a role for KLF3 in the regulation of many muscle genes during skeletal myocyte differentiation.
KLF3 expression increases during skeletal muscle differentiation.
To examine the temporal regulation of KLF3 during muscle differentiation, we performed qRT-PCR on mRNA isolated from skeletal myoblasts and myocytes at various stages of differentiation. KLF3 transcripts increased 2-fold by 12 h of differentiation and 10-fold by 48 h (Fig. 2A). To determine whether the KLF3 protein, in addition to transcripts, is upregulated during muscle differentiation, we compared KLF3 protein levels in nuclear extracts from skeletal myoblasts, early differentiated myocytes, and late differentiated myocytes by Western analysis. The predicted molecular mass of KLF3 is 38 kDa, and KLF3 from erythroid cells and fetal liver has been shown to run at ∼40 kDa (12). Whereas no KLF3 protein was detectable in myoblasts or at the early stages of differentiation, two bands of ∼40 and ∼27 kDa were detected in late differentiated myocytes (Fig. 2B), consistent with the expression of full-length KLF3 and a smaller product. Interestingly, an alternatively spliced KLF3 transcript has been found in a number of tissues, including lung, bone marrow, kidney, and uterus (available in the GenBank database under accession numbers BU679969.1, AV760739.2, CR742435.1, and AW796043.1, respectively), that would encode a protein of 25.4 kDa, close to the ∼27-kDa protein we found in skeletal myocytes. However, this reported KLF3 spliceoform lacks the exons encoding the zinc fingers; therefore, it would be unable to bind DNA. Thus, neither KLF3-specific complex in our gel shift study (Fig. 1B) would correspond to this splice product. Nonetheless, the initiation of expression of full-length KLF3 in terminally differentiated myocytes suggests that KLF3 may play an important role in muscle differentiation.
Multiple KLF binding sites regulate the MCK promoter in skeletal myocytes.
The CACCC box is recognized by many factors, including members of the Sp/KLF family. Since KLF3 binds these motifs preferentially over other GC-rich sequences, we searched the MCK enhancer and promoter for these strong KLF3-binding motifs. There are three CACCC boxes within the MCK enhancer and two flanking the MPEX sequence within the MCK promoter (Fig. 3A). Since the upstream CACCC box in the enhancer (−1210 to −1206) overlaps a functional MEF3 motif (24) and since deletion of a 63-bp region including two adjacent CACCC boxes (−1135 to −1125) had no effect on enhancer activity in skeletal myocytes (51), we confined our analysis to the KLF3-binding sites present in the MCK promoter. Interestingly, both of these sites conform to the high-affinity KLF3-binding sequence C(A/C)CACCC present in many erythroid genes (12). While the upstream motif (CAC1) appears to be fully conserved among mammalian species, the downstream motif (CAC2) is present only in the mouse (Fig. 3A). To determine whether either of these motifs is important for MCK promoter activity, we tested the effects of altering these sequences in transient transfections of skeletal myocytes. Deletion of CAC1 reduced the activity of a reporter construct containing the MCK promoter by ∼55%, while mutation of CAC2 reduced activity by ∼40% (Fig. 3B). Interestingly, while deletion of CAC1 had the same effect in the presence of the MCK enhancer, mutation of CAC2 had a much more deleterious effect in this context, resulting in a 70% decrease in activity (Fig. 3B). These data suggested that CAC2 might synergize with an element(s) in the MCK enhancer.
Since the MPEX sequence in the MCK promoter lacks a CACCC box and is not fully conserved among mammals, we examined whether mouse KLF3 could recognize human, dog, cat, and bovine MPEX sequences by comparing them as competitors in gel shift assays. For these experiments, we used nuclear extracts from COS cells (which do not express detectable KLF3 [12]) transfected with FLAG-tagged mouse KLF3. The human and dog sequences competed almost as well as the mouse sequence for KLF3 binding, whereas the bovine sequence competed less well, and the cat sequence did not compete at all (Fig. 3C). We also tested the CAC2 sequence in the nonmouse species (which contains a single-base-pair change from the CACCC box present in the mouse [Fig. 3A]), but this sequence did not compete for KLF3 binding (Fig. 3C).
To better determine the spectrum of sequences that KLF3 recognizes, we designed oligonucleotides containing single-base-pair changes at each position of the mouse MPEX sequence and tested them as competitors in gel shift assays (Fig. 3C, lanes 4 and 6 [the human and dog oligonucleotides have single-base-pair changes at positions 2 and 4, respectively]) (Fig. 3D). We found that for 6 out of 11 C/G base pairs, a single-base-pair change to A/T greatly reduced the ability of KLF3 to recognize the MPEX sequence. The KLF3 recognition sequence within MPEX (with less stringent base requirements indicated in lowercase) is shown in Fig. 3D. It is important to emphasize that this sequence was determined using only single-base-pair changes and does not represent a comprehensive analysis of KLF3 binding preferences. For example, while the mutant oligonucleotides in Fig. 3D, lanes 5 and 6, each exhibit strong competition, it is not known whether an oligonucleotide containing both mutations would compete. However, the GC-rich nature of MPEX appears to be important for recognition by KLF3. This is consistent with previous reports of KLF3 binding preferences in erythroid cells (12).
KLF3 and SRF synergize in transactivating the MCK promoter.
Since KLF3 binds positive elements in the MCK promoter (MPEX and CACCC boxes) and since its expression is initiated upon muscle differentiation, it seemed likely that KLF3 serves as a positive regulator of muscle gene transcription. However, since no TAD has been described for KLF3, we asked whether this factor mediates positive effects on transcription by associating with other transcriptional regulators of MCK. COS-7 cells were used as an informative system in which to test these interactions, since these cells are devoid of KLF3, which is expressed in most cells and tissues.
One of the factors we tested for synergy was SRF, which has been shown to bind CArG sites in the MCK enhancer and promoter (53, 62). Targeted deletion of SRF results in greatly decreased levels of many muscle-specific transcripts, including MCK (6, 44). Despite the presence of two CArG sites and six KLF binding motifs in the MCK enhancer-promoter (Fig. 4 A), overexpression of KLF3 or SRF alone had little effect on the activity of this construct (Fig. 4A and B, e−358MCKCAT). However, coexpression of KLF3 and SRF resulted in ∼7-fold-higher activity of the MCK enhancer-promoter (Fig. 4B). Although COS-7 cells contain low levels of SRF, as determined by gel supershift assays (data not shown), the endogenous levels are apparently not sufficient for detectable synergy with overexpressed KLF3. In the absence of the MCK enhancer, KLF3 and SRF still synergize in activating the MCK promoter, which contains a single CArG site and three KLF binding motifs (Fig. 4A and B, −358MCKCAT). Surprisingly, we also found that KLF3 and SRF synergize to the same extent in activating even the MCK minimal promoter, which contains two KLF binding motifs and no CArG site (Fig. 4A and B, −80MCKCAT). Since there are no CArG sites within the rest of the reporter plasmid, this result strongly suggests that KLF3 can function to recruit SRF to KLF binding sites within the MCK promoter.
To determine if related transcription factors can synergize with either KLF3 or SRF, we tested KLF4 with SRF and MEF2C with KLF3. Although KLF4, which contains a TAD (64) and also recognizes CACCC boxes, activates the MCK minimal promoter quite strongly, it displayed no synergy with SRF (Fig. 5 A). Likewise, we tested the transcriptional response of KLF3 in conjunction with MEF2C, a key muscle transcription factor belonging to the same MADS box family as SRF. As expected, MEF2C alone had no effect on the MCK minimal promoter, which contains no MEF2 (A/T-rich) binding motifs, but MEF2C also exhibited no synergy in the presence of KLF3 (Fig. 5B). Since MADS box factors are quite divergent outside the MADS domain, this suggested that other regions of SRF (possibly in addition to the MADS domain) may be important for synergizing with KLF3.
FIG. 5.
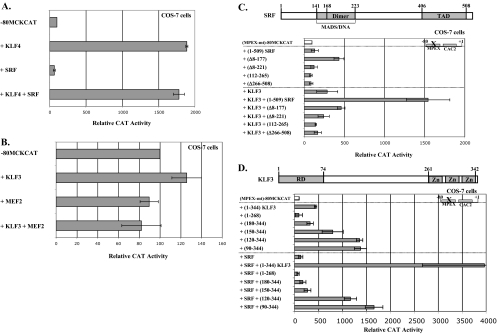
Synergy is specific to KLF3 and SRF, requiring the N terminus and zinc fingers of KLF3 and the MADS domain and TAD of SRF. (A) SRF does not synergize with KLF4. COS-7 cells were transfected with −80MCKCAT alone or with 5 μg of KLF4 expression vector, 1 μg of SRF expression vector, or both. (B) KLF3 does not synergize with MEF2. COS-7 cells were transfected with −80MCKCAT alone or with 0.4 μg of KLF3 expression vector, 0.5 μg of MEF2C expression vector, or both. (C) The MADS box and TAD of SRF are important for synergy with KLF3. COS-7 cells were transfected with (MPEX-mt)−80MCKCAT alone or with 0.4 μg of KLF3 expression vector, 1 μg of expression vectors containing various truncated versions of SRF, or both. The diagram of SRF shows the MADS box and C-terminal TAD. (D) Full-length KLF3 is required for synergy with SRF. COS-7 cells were transfected as for panel C, except that expression vectors containing various truncated versions of KLF3 were tested with and without full-length SRF. The diagram of KLF3 shows the N-terminal RD and C-terminal zinc fingers. For panels A to D, the data are plotted as the mean value and standard deviation of relative CAT activity, with the activity of the reporter construct alone set at 100.
To determine the regions of SRF and KLF3 that are required for synergy, we tested various truncated forms of the two factors. For these experiments, we used a reporter construct containing the MCK minimal promoter with a mutation in the MPEX site [Fig. 4A, (MPEX-mt)−80MCKCAT], since KLF3 and SRF were found to cooperate best in activating this construct, which contains a single strong KLF binding motif (Fig. 6 C).
FIG. 6.
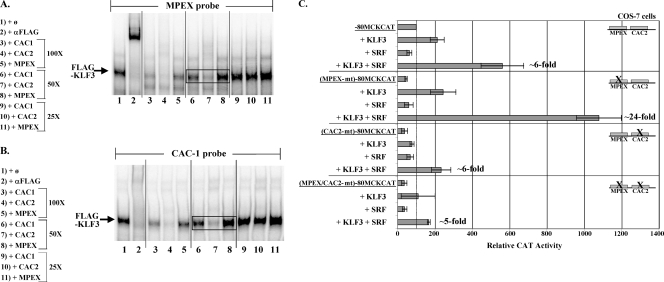
KLF3-SRF synergy is stronger with CAC2 than with MPEX in the MCK promoter. (A and B) KLF3 binds preferentially to CAC2 over MPEX and CAC1. Labeled mouse MPEX or CAC1 probe was mixed with 2 μg of nuclear extracts from COS-7 cells overexpressing FLAG-KLF3 and analyzed via gel shift assays. The KLF3-specific complex (supershifted by antibodies to FLAG [A, lane 2] or prevented by antibodies to FLAG [B, lane 2]) is indicated. Decreasing concentrations of oligonucleotides containing mouse CAC1, CAC2, or MPEX (100-, 50-, and 25-fold molar excess over probe) were tested as competitors. CAC2 competed more strongly than MPEX or CAC1 for KLF3 binding (boxed bands in lanes 6 to 8). (C) KLF3 transactivates CAC2 more strongly than MPEX, and KLF3-SRF synergy is stronger with CAC2 versus MPEX. COS-7 cells were transfected with various reporter constructs (diagrammed in Fig. 4A) with or without KLF3 and SRF expression vectors, as in Fig. 4. The levels of activity over reporter constructs alone are indicated for KLF3-plus-SRF-transfected cells. The data are plotted as the mean value and standard deviation of relative CAT activity, with the activity of the reporter construct alone set at 100.
We found that SRF lacking amino acids (aa) 8 to 177 (Δ8-177) was capable of transactivating the reporter by ∼4-fold (Fig. 5C), consistent with a previous report demonstrating that the first 171 aa of SRF contain an inhibitory domain (28) (see Discussion). We also found that truncated forms of SRF lacking this N-terminal region (Δ8-177), the N-terminal region including the full MADS box (Δ8-221), or the C-terminal region including the TAD (Δ266-508) were unable to synergize with KLF3 (Fig. 5C). Consistent with the requirement for the TAD, the MADS box of SRF alone (112 to 265) also displayed no synergy (Fig. 5C). These results suggest that both the MADS box and TAD of SRF are required for synergy with KLF3.
As expected, when the three zinc fingers of KLF3 are removed, the remaining protein (1 to 268) does not activate the MCK minimal promoter (Fig. 5D). However, when the N-terminal 179 aa of KLF3 are removed, the remaining protein (180 to 344) is almost as active as the full-length form (1 to 344), presumably due to the loss of the RD (Fig. 5D). Interestingly, when aa 120 to 179 are added back (120 to 344), the protein is ∼3-fold more active than the full-length form (Fig. 5D), suggesting that sequences in the middle of KLF3 are important for activity.
As shown above, full-length SRF alone does not activate the MCK minimal promoter, since the reporter construct contains no SRF-binding sites (Fig. 5D). Importantly, when the three zinc fingers of KLF3 are removed, the remaining protein (1 to 268) is unable to synergize with SRF (Fig. 5D), consistent with the proposition that KLF3-SRF synergy acts through association with KLF3-binding sites. Additionally, since none of the N-terminally truncated forms of KLF3 is able to synergize, sequences within aa 1 to 90 (which includes the RD of KLF3) are also required for synergy with SRF (Fig. 5D). It is worth pointing out that all the truncated forms of KLF3, with the exception of KLF3 lacking the zinc finger domain, were transcriptionally active, suggesting that the truncations did not affect the structure of the remaining protein.
KLF3-SRF synergy is strongest on CAC2 in the MCK promoter.
The mouse MCK minimal promoter contains two disparate KLF binding sites: the MPEX sequence and a CACCC box (Fig. 3A, CAC2). Since KLF3 and SRF synergize in transactivating the MCK minimal promoter, we wanted to determine if these two factors synergize more strongly on one site than the other.
To first determine which sequence is preferentially bound by KLF3, we compared CAC1, CAC2, and MPEX as competitors in gel shift assays (all sequences from the mouse MCK proximal promoter [Fig. 3A]). Consistent with previous reports that KLF3 binds better to CACCC boxes than to other GC-rich sequences (12), both CAC1 and CAC2 compete better than the MPEX site for KLF3 binding. However, CAC2 competes more strongly than CAC1 despite the fact that both sequences conform to the “strong” KLF3-binding consensus of C(A/C)CACCC (Fig. 6A and B, lanes 3 to 5 and 6 to 8). KLF3 exhibited the same binding preference for competitor (CAC2 > CAC1 > MPEX) regardless of whether the MPEX sequence or CAC1 was used as a probe. These data suggest that sequences flanking the C(A/C)CACCC box also play a role in determining KLF3-binding affinity.
To determine if KLF3-SRF synergy is stronger on CAC2 than MPEX, we performed transfections with constructs containing mutations in either site in the MCK minimal promoter. Consistent with the gel shift results, KLF3 activated a construct containing a single functional CACCC box better than one containing a single functional MPEX site (Fig. 6C). Whereas KLF3 activated the wild-type MCK minimal promoter (−80MCKCAT) and (CAC2-mt)−80MCKCAT by ∼2-fold, it activated (MPEX-mt)−80MCKCAT by ∼5-fold. It is not entirely clear why transactivation is stronger in the absence of an intact MPEX site; perhaps binding to MPEX sequesters KLF3 in a less productive conformation for interactions with coactivators. The fact that the FLAG antibody causes a supershift of KLF3 bound to the MPEX probe (Fig. 6A), whereas it prevents binding of KLF3 to the CAC-1 probe (Fig. 6B) or CAC-2 probe (data not shown), implies that KLF3 almost certainly adopts different conformations when bound to the MPEX sequence versus the CACCC box. Regardless, an intact CAC2 element appears to be necessary for maximal transactivation by KLF3. KLF3-SRF synergy was also stronger on (MPEX-mt)−80MCKCAT than on the other constructs (Fig. 6C).
The fact that KLF3 synergizes with SRF even when both KLF binding sites are mutated [Fig. 6C, (MPEX/CAC2-mt)−80MCKCAT] is surprising and suggests the presence of a cryptic KLF binding site(s) elsewhere in the MCK minimal promoter. This is a strong possibility, since the promoter is highly GC rich. Additionally, there are two CACCC boxes within the coding sequence of the CAT reporter gene. However, it is important to note that the amount of transactivation in this case is still much less than that achieved with an intact CAC2 site, indicating that KLF3 transactivates most strongly through CAC2.
KLF3 physically associates with SRF.
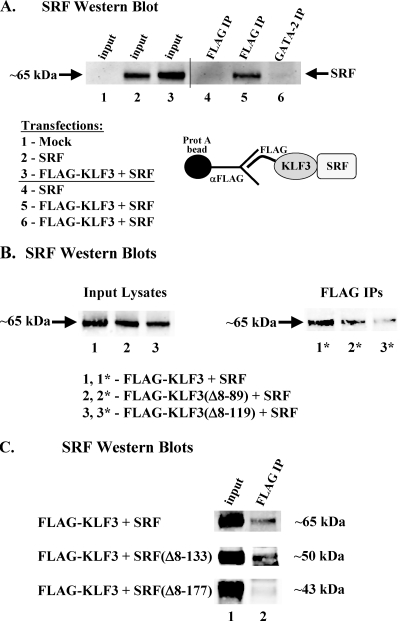
To determine whether KLF3 and SRF can interact in the absence of DNA, we made cytoplasmic extracts from COS-7 cells transfected with SRF alone or with FLAG-KLF3 plus SRF. Coimmunoprecipitations were performed using FLAG antibodies, and immunoprecipitated proteins were probed with antibodies to SRF.
SRF is not detectable in mock-transfected lysates (Fig. 7 A, lane 1) and is seen only in lysates from cells transfected with SRF (lanes 2 and 3). As expected, when cells were transfected with SRF alone, no SRF was immunoprecipitated using the FLAG antibody (lane 4). Only when cells were transfected with both FLAG-KLF3 and SRF was SRF immunoprecipitated, consistent with a physical interaction between these two factors (lane 5). As a further control, SRF was not immunoprecipitated by antibodies to an unrelated protein, GATA-2 (lane 6).
FIG. 7.
KLF3 physically associates with SRF. (A to C) COS-7 cells were transfected with various combinations of FLAG-KLF3, FLAG-KLF3(Δ8-89), FLAG-KLF3(Δ8-119), SRF, SRF(Δ8-133), or SRF(Δ8-177), and cytoplasmic extracts were immunoprecipitated with FLAG antibodies. Proteins coimmunoprecipitating with FLAG-KLF3 were subjected to Western analysis using antisera to SRF. The predicted size of full-length SRF is 65 kDa. All truncated proteins were the expected size and expressed at levels similar to that of the equivalent full-length protein, except for SRF(Δ8-177), which was expressed at ∼3-fold-higher levels than full-length SRF (data not shown). (A) KLF3 interacts with SRF. Lanes 1 to 3, 0.5% input lysates from cells that were mock transfected or transfected with SRF or FLAG-KLF3 plus SRF; lane 4, FLAG-immunoprecipitated proteins from SRF-transfected cells; lane 5, FLAG-immunoprecipitated proteins from cells transfected with FLAG-KLF3 plus SRF; lane 6, GATA-2 IgG-precipitated proteins from cells transfected with FLAG-KLF3 plus SRF (negative control). A diagram of molecular interactions is shown. Prot, protein. (B) Sequences C terminal to the RD of KLF3 are important for interaction with SRF. Lanes 1 to 3, 0.5% input lysates from cells that were transfected with FLAG-KLF3 plus SRF, FLAG-KLF3(Δ8-89) plus SRF, or FLAG-KLF3(Δ8-119) plus SRF; lanes 1* to 3*, FLAG-immunoprecipitated proteins from cells transfected with each of the above combinations. (C) The MADS domain of SRF is required for interaction with KLF3. Lane 1, 0.5% input lysates from cells that were transfected with FLAG-KLF3 plus SRF, FLAG-KLF3 plus SRF(Δ8-133), or FLAG-KLF3 plus SRF(Δ8-177); lane 2, FLAG-immunoprecipitated proteins from cells transfected with each of the above combinations. The predicted sizes of full-length and truncated forms of SRF are shown.
To determine whether the physical interaction between KLF3 and SRF requires N-terminal regions of KLF3, similar studies were carried out with extracts from COS cells transfected with SRF plus FLAG-KLF3 or SRF plus FLAG-KLF3 with aa 8 to 89 [FLAG-KLF3(Δ8-89)] or aa 8 to 119 [FLAG-KLF3(Δ8-119)] deleted.
We found that when cells were transfected with both SRF and FLAG-KLF3(Δ8-89), SRF was still immunoprecipitated, albeit at reduced levels, indicating that this N-terminal region of KLF3, while critical for transcriptional synergy, is not absolutely required for the physical interaction with SRF (Fig. 7B, lane 2*). However, when aa 8 to 119 were deleted from KLF3, interaction with SRF was almost completely eliminated (Fig. 7B, lane 3*). Thus, sequences immediately C terminal to the KLF3 RD are important for contacting or stabilizing contact with SRF.
To determine the interacting regions of SRF, we performed reciprocal experiments with extracts from COS cells transfected with FLAG-KLF3 plus SRF, or FLAG-KLF3 plus SRF with aa 8 to 133 [SRF(Δ8-133)] or aa 8 to 177 [SRF(Δ8-177)] deleted. When cells were transfected with both FLAG-KLF3 and SRF(Δ8-133), SRF was still immunoprecipitated similarly to the full-length form (Fig. 7C). However, when aa 8 to 177 (including part of the MADS domain) were deleted from SRF, the interaction with KLF3 was abolished (Fig. 7C), indicating that sequences in the MADS domain are required for this interaction.
DISCUSSION
Using isotope-coded affinity tag (ICAT)-based quantitative proteomics, multiple candidate factors were identified as binding to a single positive element (MPEX) in the MCK promoter (25). One of these factors, MAZ, was subsequently found to play an important role in muscle gene regulation (25). In the current study, we describe the identification of KLF3 as another candidate MPEX-binding factor. Despite its widespread expression, the analysis of KLF3 has thus far been confined mostly to erythroid and adipose cells, and to the best of our knowledge, our study presents the first reported evidence of a role for KLF3 in skeletal muscle.
While interpretive complexities have plagued many cell lines, MM14 skeletal myoblasts are a well-established system for understanding myogenesis (9). Using these cells, we show that KLF binding sites are important for activity of the MCK promoter, that KLF3 transcripts and protein increase during muscle differentiation, and that KLF3 is enriched at the promoters of endogenous muscle genes. Taken together, these data strongly suggest that KLF3 plays a role in muscle gene regulation.
Although KLF3 knockout mice have no apparent muscle defects (56), there is an enormous potential for compensation by the large family of Sp/KLF factors (many of which are expressed in muscle) and by other factors, such as MAZ, that transactivate through the same DNA-binding motifs (such as the CCCTCCC motif in MPEX) (25). Thus, the absence of a clearly defective muscle phenotype in KLF3 knockout mice may not be surprising. In this context, it is interesting that the initial studies of MyoD-null mice also showed no muscle defects, due to compensation by another family member (Myf5) (49). Nevertheless, extensive in vitro data, including ChIP analysis, have consistently demonstrated the seminal role of MyoD in muscle development—a role that could only be demonstrated in vivo by disrupting both MyoD and Myf5 (50) and that was not fully understood until mice containing a triple knockout of MyoD, Myf5, and MRF4 were analyzed (30).
In an effort to find a factor(s) that might cooperate with KLF3, we discovered that KLF3 and SRF synergize in transactivating the MCK promoter. Furthermore, we found that this synergy is independent of CArG sites. This result was surprising in light of the fact that SRF has traditionally been thought to mediate its effects only through binding to CArG boxes in target genes (36, 43). Indeed, in cases where SRF has been shown to synergize with other transcription factors, synergy requires CArG sites (20, 52). Thus, while SRF is able to recruit factors such as GATA-4 and Nkx2.5 to CArG sites (52), the recruitment of SRF to other control elements has not been previously demonstrated.
It has been reported that when the first 171 aa (including part of the MADS domain) are removed from SRF, the protein is more active than the full-length form, indicating that the N terminus contains an inhibitory domain (28). Consistent with this hypothesis, we found that SRF lacking the first 177 aa gave stronger activation of the MCK promoter than full-length SRF. However, it has not been clear how SRF could mediate transactivation in the absence of an intact DNA-binding domain. Our study suggests a novel mechanism for this observation by recruitment of SRF to another transcription factor's binding site. Indeed, while the MADS domain of SRF is required for both interaction and synergy with KLF3, in the previous study (28) SRF might be recruited by other transcription factors and mediate synergistic effects in the absence of this region (possibly via the TAD).
KLF3-SRF synergy is at least partly specific to these factors, as KLF4 cannot substitute for KLF3, and MEF2 (another MADS box factor) cannot substitute for SRF. There is precedent for such specific interactions between members of the Sp/KLF family and the MADS box family. In a previous study, Sp1 and MEF2 were shown to interact, and this association was much weaker when MEF2 was replaced with SRF (18). However, in this earlier study, synergy between Sp1 and MEF2 was tested only in the presence of binding sites for both factors; thus, it is unclear whether Sp1 recruits MEF2 to Sp1 sites (which include CACCC, as well as CT and CG, boxes) or whether MEF2 recruits Sp1 to A/T-rich motifs. Analyses of Sp1 interactions with the MCK promoter and possible synergy with SRF and MEF2 are under way.
Since the KLFs are quite divergent outside their C-terminal zinc fingers, and since MADS box factors are divergent outside the MADS domain, the inability of other factors to substitute in KLF3-SRF synergy suggested that other regions of KLF3 and SRF are necessary for this positive interaction. Consistent with this idea, the SRF MADS box alone is not sufficient for synergy with KLF3, and the TAD of SRF is also required. Conversely, we found that the zinc finger domain and, unexpectedly, the RD of KLF3 were required for synergy with SRF. It is worth pointing out that the mode of KLF3-SRF synergy does not consist of SRF simply obscuring the RD of KLF3. If this were the case, KLF3 lacking the RD should exhibit activity similar to that of full-length KLF3 plus SRF, but in fact, it is only ∼40% as active as the latter. We hypothesized that sequences within the KLF3 RD may be important for contacting SRF; however, our coimmunoprecipitation results demonstrated that physical association between KLF3 and SRF requires sequences C terminal to the RD. Collectively, these results led us to propose a model in which association with SRF results in a conformational change in the RD of KLF3, allowing it to recruit positive transcriptional cofactors rather than negative ones.
KLF3 has been shown to associate with CtBP2 and FHL3, which mediate its repressive effects on gene transcription (58, 60). Whereas CtBP1 and CtBP2 have been best characterized as transcriptional corepressors (8, 59), the FHL proteins have been shown to behave as strong repressors of transcription, as well as strong activators (17, 60). CtBP1 and CtBP2, as well as FHL1, FHL2, and FHL3, are highly expressed in skeletal and cardiac muscle (31), and CtBP1 and CtBP2 compound-null mice exhibit muscle defects (23). Whether FHL factors are recruited by KLF3-SRF complexes to promote muscle gene expression remains an open question.
As myoblasts undergo differentiation, SRF protein levels do not change, but the role of SRF changes dramatically from the activation of immediate-early genes to the activation of muscle-specific genes, such as Myogenin, Skeletal troponin T, and Skeletal α-actin (61). It has been demonstrated that protein kinase C α (PKC α) phosphorylation of SRF at Ser 162 allows SRF, in association with Ets factors, to activate immediate-early genes through CArG boxes with flanking Ets-binding sites. At the same time, the SRF-Ets complexes are prevented from activating muscle genes, which contain CArG boxes without flanking Ets-binding motifs (26). When myoblasts withdraw from the cell cycle, phosphorylation of SRF by PKC α declines, allowing it to bind CArG boxes in muscle genes and help drive the program of muscle differentiation (26). Interestingly, we found that the KLF3 protein is undetectable in proliferating myoblasts and is expressed only in fully differentiated myocytes. Thus, SRF would not be recruited by KLF3 to muscle gene promoters until differentiation is well under way. Presumably, when SRF assumes its myogenic role, its interaction with KLF3 changes the configuration of the KLF3 N-terminal domain (as stated above), facilitating the recruitment of coactivators instead of corepressors. In the course of this study, we found that KLF3-SRF synergy is stronger on a promoter containing only a single CACCC box than on the same promoter containing only a single MPEX sequence (Fig. 6C). Furthermore, KLF3 appears to adopt different conformations when bound to a CACCC probe versus an MPEX probe, since the gel shift complex is prevented versus supershifted by KLF3 antibodies (Fig. 6A and B). Taken together, these results suggest that the conformation of KLF3 bound to a CACCC box may be more productive for interactions with SRF and associated coactivators than KLF3 bound to other sequences.
The MRTFs, which include myocardin, MRTF-A, and MRTF-B, have emerged as important coregulators of SRF transcriptional activity through contacts within α-helix II of SRF (4). Like SRF, these coactivators contain strong C-terminal TADs; however, the mechanism by which they enhance SRF transactivation is unknown. In addition, the TAD of SRF has been shown to contact the RAP74 subunit of TFIIF, and this interaction is required for RAP74-mediated SRF activation of the c-fos promoter (29). Whether these interactions take place when SRF is tethered to KLF3 remains an interesting question for future studies.
In cardiac muscle, interactions with cofactors, such as GATA-4 and Nkx2.5, serve to stabilize SRF binding to CArG boxes to drive a program of sarcomeric gene transcription (52). Our proposed mechanism of SRF-KLF3 synergy might represent a novel way for SRF to transactivate genes, such as MCK, that are expressed in cardiac muscle but lack GATA and Nkx2.5 binding sites. Although our coimmunoprecipitation results indicate that KLF3 and SRF can interact in the absence of DNA, it is unclear whether a preassembled complex of KLF3 and SRF associates with KLF binding motifs or whether SRF is recruited by DNA-bound KLF3 to target promoters/enhancers. Regardless, recruitment by KLF3 may help to stabilize SRF at regions that lack cooperative binding sites for SRF cofactors. This mechanism may also allow the cooperative formation of multiple SRF-binding complexes, as shown for the Skeletal α-actin promoter (33).
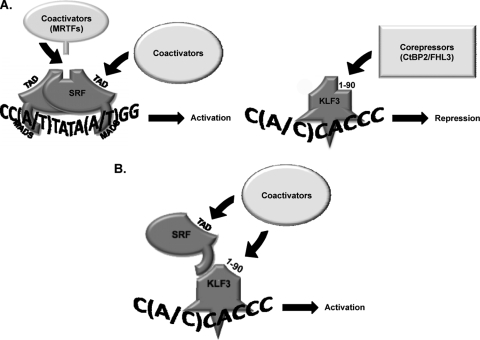
Collectively, the data presented in this study suggest a novel mode of KLF3 and SRF activity. In the established model of SRF activity, SRF contacts CArG sites in its target genes via α-helix I of its MADS domain (Fig. 8 A). α-helix II of SRF recruits the MRTF coactivators, and the TAD of SRF may recruit other families of coactivators to promote transactivation of target genes. In the established model of KLF3 activity, KLF3 contacts CACCC boxes (and, to a lesser extent, GC-rich sequences like MPEX) in its target genes via its 3 C-terminal zinc fingers, and the N-terminal RD (aa 1 to 90) recruits corepressors, such as CtBP2 and FHL3, to repress target genes (Fig. 8A).
FIG. 8.
Model of KLF3-SRF synergy on CACCC boxes. (A) Established models of KLF3 and SRF activities. SRF contacts CArG sites in its target genes via α-helix I of its MADS domain. α-Helix II of SRF recruits the MRTF coactivators, and the TAD of SRF may recruit other families of coactivators to promote target gene expression. KLF3 contacts CACCC boxes in its target genes via its three C-terminal zinc fingers, and the N-terminal RD (aa 1 to 90) recruits corepressors, such as CtBP2 and FHL3 to repress target gene expression. (B) Proposed model of KLF3-SRF synergy. KLF3 recruits SRF to CACCC boxes, possibly through α-helix I of the SRF MADS domain, which has been shown to mediate both DNA binding and interaction with a number of accessory factors. Association with SRF changes the conformation of the KLF3 N terminus (aa 1 to 90) to allow recruitment of coactivators instead of corepressors. The TAD of SRF may also aid in the recruitment of coactivators.
In our proposed model of KLF3-SRF synergy, KLF3 recruits SRF to CACCC boxes, most likely through α-helix I of the SRF MADS domain, which has been shown to mediate both DNA binding and interaction with a number of accessory factors (Fig. 8B). Association with SRF changes the conformation of the KLF3 N terminus (aa 1 to 90) to allow recruitment of coactivators instead of corepressors. Synergy also requires the TAD of SRF, which may aid in the recruitment of coactivators. While KLF3 and SRF associate in solution, we have not been able to detect a complex containing both factors bound to a KLF binding site probe. Unfortunately, protein-protein interactions are often difficult to detect in gel shift assays, which may not mimic in vivo conditions closely enough for any but the most stable interactions to be detected.
Intriguingly, in a genome-wide study identifying regions of SRF enrichment, 33% of validated SRF targets did not contain a CArG box within 4 kb upstream and 1 kb downstream of the transcription start site (10). Since many of these genes contain CACCC boxes and GC-rich sequences in their promoters, our results provide a novel explanation for this phenomenon, where SRF exerts its effects on target genes through recruitment to KLF binding sites. This mode of action may be broadly applicable to all tissues in which SRF and KLF3 are coexpressed—primarily skeletal, cardiac, and smooth muscle. If this mechanism is revealed to be widespread, SRF may have a larger role in regulating transcription than is indicated simply by the presence of CArG boxes in its target genes, and many genes within the CArGome (57) may be transcriptionally regulated via KLF control elements.
Our results do not preclude the possibility of KLF3 synergizing with SRF when both factors are bound to their cognate sites. This is a strong likelihood with genes such as MCK, which contain multiple CArG boxes and KLF binding motifs. The greater effect of mutating the MCK promoter CAC2 site in the presence of the enhancer suggests that a factor(s) binding CAC2, such as KLF3, might be cooperating with a factor(s) binding enhancer elements, such as SRF. Furthermore, it is interesting that one documented splice variant of KLF3 encodes amino acids 1 to 232, which include the RD and internal sequences but not the zinc fingers. Therefore, this truncated form of KLF3 would, in theory, be capable of interacting with other proteins but not of binding DNA. If this 1-to-232 splice variant is the smaller form of KLF3 we see in skeletal myocytes, it might be recruited by other factors to activate or suppress target gene expression.
Finally, we want to stress that in regard to MCK transcription, KLF3 is only one of the factors that can bind MPEX and CACCC boxes. In a previous study, we characterized MAZ as a regulator of MCK and other muscle genes through MPEX and related sequences (25), and KLF4—another MPEX-binding factor candidate identified in our proteomic screen—is also a strong transactivator of MCK through these elements. The ability of multiple factors to recognize the same or very similar sequences allows both robustness and fine-tuning of gene regulation. Whether an element is bound by MAZ, KLF4, KLF3-SRF, or any of a host of other factors in vivo is likely to depend on the cell type (skeletal versus cardiac muscle or fast versus slow fibers), as well as various physiological stimuli. Understanding the spatiotemporal dynamics of these interactions remains a significant challenge for future studies.
Acknowledgments
We thank J. Angello, J. Buskin, D. Helterline, Q. Nguyen, P. Tai, and R. Welikson for technical assistance and/or critical discussions and P. Tai for artwork in Fig. 8. We are grateful to J. Klimek and D. Martin at the Institute for Systems Biology proteomics facility for help with mass spectrometry.
This work was supported by NIH-RO1-AR18860 (to S.D.H.); NIH-1P01-NS046788 (to S.D.H.); NIH-T32-HL007312, Experimental Pathology of Cardiovascular Disease (to C.L.H.); contract no. NO1-HV-28179 from the National Heart, Lung, and Blood Institute (to S.D.H. and C.L.H.); and grant no. P50GMO76547 from the National Institute of General Medical Sciences (to J.A.R.).
Footnotes
Published ahead of print on 19 April 2010.
REFERENCES
- 1.Amacher, S. L., J. N. Buskin, and S. D. Hauschka. 1993. Multiple regulatory elements contribute differentially to muscle creatine kinase enhancer activity in skeletal and cardiac muscle. Mol. Cell. Biol. 13:2753-2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 3.Camoretti-Mercado, B., D. J. Fernandes, S. Dewundara, J. Churchill, L. Ma, P. C. Kogut, J. F. McConville, M. S. Parmacek, and J. Solway. 2006. Inhibition of transforming growth factor beta-enhanced serum response factor-dependent transcription by SMAD7. J. Biol. Chem. 281:20383-20392. [DOI] [PubMed] [Google Scholar]
- 4.Cen, B., A. Selvaraj, and R. Prywes. 2004. Myocardin/MKL family of SRF coactivators: key regulators of immediate early and muscle specific gene expression. J. Cell. Biochem. 93:74-82. [DOI] [PubMed] [Google Scholar]
- 5.Chang, D. F., N. S. Belaguli, D. Iyer, W. B. Roberts, S. P. Wu, X. R. Dong, J. G. Marx, M. S. Moore, M. C. Beckerle, M. W. Majesky, and R. J. Schwartz. 2003. Cysteine-rich LIM-only proteins CRP1 and CRP2 are potent smooth muscle differentiation cofactors. Dev. Cell 4:107-118. [DOI] [PubMed] [Google Scholar]
- 6.Charvet, C., C. Houbron, A. Parlakian, J. Giordani, C. Lahoute, A. Bertrand, A. Sotiropoulos, L. Renou, A. Schmitt, J. Melki, Z. Li, D. Daegelen, and D. Tuil. 2006. New role for serum response factor in postnatal skeletal muscle growth and regeneration via the interleukin 4 and insulin-like growth factor 1 pathways. Mol. Cell. Biol. 26:6664-6674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen, F., H. Kook, R. Milewski, A. D. Gitler, M. M. Lu, J. Li, R. Nazarian, R. Schnepp, K. Jen, C. Biben, G. Runke, J. P. Mackay, J. Novotny, R. J. Schwartz, R. P. Harvey, M. C. Mullins, and J. A. Epstein. 2002. Hop is an unusual homeobox gene that modulates cardiac development. Cell 110:713-723. [DOI] [PubMed] [Google Scholar]
- 8.Chinnadurai, G. 2003. CtBP family proteins: more than transcriptional corepressors. Bioessays 25:9-12. [DOI] [PubMed] [Google Scholar]
- 9.Clegg, C. H., T. A. Linkhart, B. B. Olwin, and S. D. Hauschka. 1987. Growth factor control of skeletal muscle differentiation: commitment to terminal differentiation occurs in G1 phase and is repressed by fibroblast growth factor. J. Cell Biol. 105:949-956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cooper, S. J., N. D. Trinklein, L. Nguyen, and R. M. Myers. 2007. Serum response factor binding sites differ in three human cell types. Genome Res. 17:136-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Croissant, J. D., J. H. Kim, G. Eichele, L. Goering, J. Lough, R. Prywes, and R. J. Schwartz. 1996. Avian serum response factor expression restricted primarily to muscle cell lineages is required for alpha-actin gene transcription. Dev. Biol. 177:250-264. [DOI] [PubMed] [Google Scholar]
- 12.Crossley, M., E. Whitelaw, A. Perkins, G. Williams, Y. Fujiwara, and S. H. Orkin. 1996. Isolation and characterization of the cDNA encoding BKLF/TEF-2, a major CACCC-box-binding protein in erythroid cells and selected other cells. Mol. Cell. Biol. 16:1695-1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dignam, J. D., R. M. Lebovitz, and R. G. Roeder. 1983. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 11:1475-1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Donoviel, D. B., M. A. Shield, J. N. Buskin, H. S. Haugen, C. H. Clegg, and S. D. Hauschka. 1996. Analysis of muscle creatine kinase gene regulatory elements in skeletal and cardiac muscles of transgenic mice. Mol. Cell. Biol. 16:1649-1658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Du, K. L., H. S. Ip, J. Li, M. Chen, F. Dandre, W. Yu, M. M. Lu, G. K. Owens, and M. S. Parmacek. 2003. Myocardin is a critical serum response factor cofactor in the transcriptional program regulating smooth muscle cell differentiation. Mol. Cell. Biol. 23:2425-2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Duprey, P., and C. Lesens. 1994. Control of skeletal muscle-specific transcription: involvement of paired homeodomain and MADS domain transcription factors. Int. J. Dev. Biol. 38:591-604. [PubMed] [Google Scholar]
- 17.Fimia, G. M., D. De Cesare, and P. Sassone-Corsi. 2000. A family of LIM-only transcriptional coactivators: tissue-specific expression and selective activation of CREB and CREM. Mol. Cell. Biol. 20:8613-8622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grayson, J., R. Bassel Duby, and R. S. Williams. 1998. Collaborative interactions between MEF-2 and Sp1 in muscle specific gene regulation. J. Cell. Biochem. 70:366-375. [PubMed] [Google Scholar]
- 19.Groisman, R., H. Masutani, M. P. Leibovitch, P. Robin, I. Soudant, D. Trouche, and A. Harel-Bellan. 1996. Physical interaction between the mitogen-responsive serum response factor and myogenic basic-helix-loop-helix proteins. J. Biol. Chem. 271:5258-5264. [DOI] [PubMed] [Google Scholar]
- 20.Gupta, M., P. Kogut, F. J. Davis, N. S. Belaguli, R. J. Schwartz, and M. P. Gupta. 2001. Physical interaction between the MADS box of serum response factor and the TEA/ATTS DNA-binding domain of transcription enhancer factor-1. J. Biol. Chem. 276:10413-10422. [DOI] [PubMed] [Google Scholar]
- 21.Haldar, S. M., O. A. Ibrahim, and M. K. Jain. 2007. Kruppel-like Factors (KLFs) in muscle biology. J. Mol. Cell Cardiol. 43:1-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Herring, B. P., A. M. Kriegel, and A. M. Hoggatt. 2001. Identification of Barx2b, a serum response factor-associated homeodomain protein. J. Biol. Chem. 276:14482-14489. [DOI] [PubMed] [Google Scholar]
- 23.Hildebrand, J. D., and P. Soriano. 2002. Overlapping and unique roles for C-terminal binding protein 1 (CtBP1) and CtBP2 during mouse development. Mol. Cell. Biol. 22:5296-5307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Himeda, C. L., J. A. Ranish, J. C. Angello, P. Maire, R. Aebersold, and S. D. Hauschka. 2004. Quantitative proteomic identification of Six4 as the Trex binding factor in the muscle creatine kinase enhancer. Mol. Cell. Biol. 24:2132-2143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Himeda, C. L., J. A. Ranish, and S. D. Hauschka. 2008. Quantitative proteomic identification of MAZ as a transcriptional regulator of muscle-specific genes in skeletal and cardiac myocytes. Mol. Cell. Biol. 28:6521-6535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Iyer, D., D. Chang, J. Marx, L. Wei, E. N. Olson, M. S. Parmacek, A. Balasubramanyam, and R. J. Schwartz. 2006. Serum response factor MADS box serine-162 phosphorylation switches proliferation and myogenic gene programs. Proc. Natl. Acad. Sci. U. S. A. 103:4516-4521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jaynes, J. B., J. E. Johnson, J. N. Buskin, C. L. Gartside, and S. D. Hauschka. 1988. The muscle creatine kinase gene is regulated by multiple upstream elements, including a muscle specific enhancer. Mol. Cell. Biol. 8:62-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Johansen, F. E., and R. Prywes. 1993. Identification of transcriptional activation and inhibitory domains in serum response factor (SRF) by using GAL4-SRF constructs. Mol. Cell. Biol. 13:4640-4647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Joliot, V., M. Demma, and R. Prywes. 1995. Interaction with RAP74 subunit of TFIIF is required for transcriptional activation by serum response factor. Nature 373:632-635. [DOI] [PubMed] [Google Scholar]
- 30.Kassar-Duchossoy, L., B. Gayraud-Morel, D. Gomès, D. Rocancourt, M. Buckingham, V. Shinin, and S. Tajbakhsh. 2004. Mrf4 determines skeletal muscle identity in Myf5:Myod double-mutant mice. Nature 431:466-471. [DOI] [PubMed] [Google Scholar]
- 31.Katsanis, N., and E. M. Fisher. 1998. A novel C-terminal binding protein (CTBP2) is closely related to CTBP1, an adenovirus E1A-binding protein, and maps to human chromosome 21q21.3. Genomics 47:294-299. [DOI] [PubMed] [Google Scholar]
- 32.Lahoute, C., A. Sotiropoulos, M. Favier, I. Guillet-Deniau, C. Charvet, A. Ferry, G. Butler-Browne, D. Metzger, D. Tuil, and D. Daegelen. 2008. Premature aging in skeletal muscle lacking serum response factor. PLoS One 3:e3910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee, T. C., K. L. Chow, P. Fang, and R. J. Schwartz. 1991. Activation of skeletal alpha-actin gene transcription: the cooperative formation of serum response factor-binding complexes over positive cis-acting promoter serum response elements displaces a negative-acting nuclear factor enriched in replicating myoblasts and nonmyogenic cells. Mol. Cell. Biol. 11:5090-5100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li, S., M. P. Czubryt, J. McAnally, R. Bassel-Duby, J. A. Richardson, F. F. Wiebel, A. Nordheim, and E. N. Olson. 2005. Requirement for serum response factor for skeletal muscle growth and maturation revealed by tissue-specific gene deletion in mice. Proc. Natl. Acad. Sci. U. S. A. 102:1082-1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu, Y., S. Sinha, and G. Owens. 2003. A transforming growth factor-beta control element required for SM alpha-actin expression in vivo also partially mediates GKLF-dependent transcriptional repression. J. Biol. Chem. 278:48004-48011. [DOI] [PubMed] [Google Scholar]
- 36.Miano, J. M. 2003. Serum response factor: toggling between disparate programs of gene expression. J. Mol. Cell Cardiol. 35:577-593. [DOI] [PubMed] [Google Scholar]
- 37.Mo, Y., W. Ho, K. Johnston, and R. Marmorstein. 2001. Crystal structure of a ternary SAP-1/SRF/c-fos SRE DNA complex. J. Mol. Biol. 314:495-506. [DOI] [PubMed] [Google Scholar]
- 38.Molkentin, J. D., B. L. Black, J. F. Martin, and E. N. Olson. 1995. Cooperative activation of muscle gene expression by MEF2 and myogenic bHLH proteins. Cell 83:1125-1136. [DOI] [PubMed] [Google Scholar]
- 39.Nelson, J. D., O. Denisenko, and K. Bomsztyk. 2006. Protocol for the fast chromatin immunoprecipitation (ChIP) method. Nat. Protoc. 1:179-185. [DOI] [PubMed] [Google Scholar]
- 40.Neville, C., N. Rosenthal, M. McGrew, N. Bogdanova, and S. Hauschka. 1997. Skeletal muscle cultures. Methods Cell Biol. 52:85-116. [PubMed] [Google Scholar]
- 41.Nguyen, Q. G., J. N. Buskin, C. L. Himeda, M. A. Shield, and S. D. Hauschka. 2003. Differences in the function of three conserved E boxes of the muscle creatine kinase gene in cultured myocytes and in transgenic mouse skeletal and cardiac muscle. J. Biol. Chem. 278:46494-46505. [DOI] [PubMed] [Google Scholar]
- 42.Nishida, W., M. Nakamura, S. Mori, M. Takahashi, Y. Ohkawa, S. Tadokoro, K. Yoshida, K. Hiwada, K. Hayashi, and K. Sobue. 2002. A triad of serum response factor and the GATA and NK families governs the transcription of smooth and cardiac muscle genes. J. Biol. Chem. 277:7308-7317. [DOI] [PubMed] [Google Scholar]
- 43.Niu, Z., A. Li, S. X. Zhang, and R. J. Schwartz. 2007. Serum response factor micromanaging cardiogenesis. Curr. Opin. Cell Biol. 19:618-627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Parlakian, A., C. Charvet, B. Escoubet, M. Mericskay, J. D. Molkentin, G. Gary-Bobo, L. J. De Windt, M. A. Ludosky, D. Paulin, D. Daegelen, D. Tuil, and Z. Li. 2005. Temporally controlled onset of dilated cardiomyopathy through disruption of the SRF gene in adult heart. Circulation 112:2930-2939. [DOI] [PubMed] [Google Scholar]
- 45.Parlakian, A., D. Tuil, G. Hamard, G. Tavernier, D. Hentzen, J. P. Concordet, D. Paulin, Z. Li, and D. Daegelen. 2004. Targeted inactivation of serum response factor in the developing heart results in myocardial defects and embryonic lethality. Mol. Cell. Biol. 24:5281-5289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Perdomo, J., A. Verger, J. Turner, and M. Crossley. 2005. Role for SUMO modification in facilitating transcriptional repression by BKLF. Mol. Cell. Biol. 25:1549-1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pipes, G. C., E. E. Creemers, and E. N. Olson. 2006. The myocardin family of transcriptional coactivators: versatile regulators of cell growth, migration, and myogenesis. Genes Dev. 20:1545-1556. [DOI] [PubMed] [Google Scholar]
- 48.Qiu, P., X. H. Feng, and L. Li. 2003. Interaction of Smad3 and SRF-associated complex mediates TGF-beta1 signals to regulate SM22 transcription during myofibroblast differentiation. J. Mol. Cell Cardiol. 35:1407-1420. [DOI] [PubMed] [Google Scholar]
- 49.Rudnicki, M. A., T. Braun, S. Hinuma, and R. Jaenisch. 1992. Inactivation of MyoD in mice leads to up-regulation of the myogenic HLH gene Myf-5 and results in apparently normal muscle development. Cell 71:383-390. [DOI] [PubMed] [Google Scholar]
- 50.Rudnicki, M. A., P. N. Schnegelsberg, R. H. Stead, T. Braun, H. H. Arnold, and R. Jaenisch. 1993. MyoD or Myf-5 is required for the formation of skeletal muscle. Cell 75:1351-1359. [DOI] [PubMed] [Google Scholar]
- 51.Salva, M. Z., C. L. Himeda, P. W. Tai, E. Nishiuchi, P. Gregorevic, J. M. Allen, E. E. Finn, Q. G. Nguyen, M. J. Blankinship, L. Meuse, J. S. Chamberlain, and S. D. Hauschka. 2007. Design of tissue-specific regulatory cassettes for high-level rAAV-mediated expression in skeletal and cardiac muscle. Mol. Ther. 15:320-329. [DOI] [PubMed] [Google Scholar]
- 52.Sepulveda, J. L., S. Vlahopoulos, D. Iyer, N. Belaguli, and R. J. Schwartz. 2002. Combinatorial expression of GATA4, Nkx2-5, and serum response factor directs early cardiac gene activity. J. Biol. Chem. 277:25775-25782. [DOI] [PubMed] [Google Scholar]
- 53.Shield, M. A. 1995. Ph.D. thesis. University of Washington, Seattle, WA.
- 54.Shield, M. A., H. S. Haugen, C. H. Clegg, and S. D. Hauschka. 1996. E box sites and a proximal regulatory region of the muscle creatine kinase gene differentially regulate expression in diverse skeletal muscles and cardiac muscle of transgenic mice. Mol. Cell. Biol. 16:5058-5068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shin, C. H., Z. P. Liu, R. Passier, C. L. Zhang, D. Z. Wang, T. M. Harris, H. Yamagishi, J. A. Richardson, G. Childs, and E. N. Olson. 2002. Modulation of cardiac growth and development by HOP, an unusual homeodomain protein. Cell 110:725-735. [DOI] [PubMed] [Google Scholar]
- 56.Sue, N., B. H. Jack, S. A. Eaton, R. C. Pearson, A. P. Funnell, J. Turner, R. Czolij, G. Denyer, S. Bao, J. C. Molero-Navajas, A. Perkins, Y. Fujiwara, S. H. Orkin, K. Bell-Anderson, and M. Crossley. 2008. Targeted disruption of the basic Krüppel-like factor gene (Klf3) reveals a role in adipogenesis. Mol. Cell. Biol. 28:3967-3978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sun, Q., G. Chen, J. W. Streb, X. Long, Y. Yang, C. J. Stoeckert, Jr., and J. M. Miano. 2006. Defining the mammalian CArGome. Genome Res. 16:197-207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Turner, J., and M. Crossley. 1998. Cloning and characterization of mCtBP2, a corepressor that associates with basic Krüppel-like factor and other mammalian transcriptional regulators. EMBO J. 17:5129-5140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Turner, J., and M. Crossley. 2001. The CtBP family: enigmatic and enzymatic transcriptional corepressors. Bioessays 23:683-690. [DOI] [PubMed] [Google Scholar]
- 60.Turner, J., H. Nicholas, D. Bishop, J. M. Matthews, and M. Crossley. 2003. The LIM protein FHL3 binds basic Krüppel-like factor/Krüppel-like factor 3 and its corepressor C-terminal-binding protein 2. J. Biol. Chem. 278:12786-12795. [DOI] [PubMed] [Google Scholar]
- 61.Vandromme, M., C. Gauthier-Rouvière, G. Carnac, N. Lamb, and A. Fernandez. 1992. Serum response factor p67SRF is expressed and required during myogenic differentiation of both mouse C2 and rat L6 muscle cell lines. J. Cell Biol. 118:1489-1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Vincent, C. K., A. Gualberto, C. V. Patel, and K. Walsh. 1993. Different regulatory sequences control creatine kinase M gene expression in directly injected skeletal and cardiac muscle. Mol. Cell. Biol. 13:1264-1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Vlieghe, D., A. Sandelin, P. J. De Bleser, K. Vleminckx, W. W. Wasserman, F. van Roy, and B. Lenhard. 2006. A new generation of JASPAR, the open-access repository for transcription factor binding site profiles. Nucleic Acids Res. 34(Database issue):D95-D97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yet, S. F., M. M. McA'Nulty, S. C. Folta, H. W. Yen, M. Yoshizumi, C. M. Hsieh, M. D. Layne, M. T. Chin, H. Wang, M. A. Perrella, M. K. Jain, and M. E. Lee. 1998. Human EZF, a Krüppel-like zinc finger protein, is expressed in vascular endothelial cells and contains transcriptional activation and repression domains. J. Biol. Chem. 273:1026-1031. [DOI] [PubMed] [Google Scholar]