Abstract
Adipogenesis is governed by a well-documented cascade of transcription factors. However, less is known about non-transcription factors that govern early stages of adipogenesis. Here we show that cellular retinol-binding protein type I (CRBP-I), a small cytosolic binding protein for retinol and retinaldehyde, is specifically restricted to preadipocytes in white adipose tissue. The absence of CRBP-I in mice (CRBP-I-KO mice) leads to increased adiposity. Despite increased adiposity, CRBP-I-KO mice remain more glucose tolerant and insulin sensitive during high-fat-diet feeding. 3T3-L1 cells deficient in CRBP-I or mouse embryonic fibroblasts derived from CRBP-I-KO mice had increased adipocyte differentiation and triglyceride (TG) accumulation. This was due to increased expression and activity of PPARγ, while other transcription factor pathways in early and late differentiation remained unchanged. Conversely, the overexpression of CRBP-I in 3T3-L1 cells results in decreased TG accumulation. In conclusion, CRBP-I is a cytosolic protein specifically expressed in preadipocytes that regulates adipocyte differentiation in part by affecting PPARγ activity.
Adipose tissue plays an essential role in the maintenance of whole-body energy homeostasis. It is the major tissue for storage of excess energy in the form of triglycerides (TG) and functions as an endocrine organ secreting metabolically active adipokines affecting energy metabolism (2, 12, 31, 38). Excess accumulation of adipose tissue leads to obesity and is considered a major risk factor for insulin resistance, type 2 diabetes, and cardiovascular diseases (8, 30).
Adipogenesis is a complex process governed by a multifaceted transcriptional regulatory cascade. Members of the CCAAT/enhancer binding protein family (C/EBP) and peroxisome proliferator-activated receptor gamma (PPARγ) act together to regulate adipocyte differentiation (3, 31). Several transcription factors present in preadipocytes act to repress or promote the conversion of preadipocytes into adipocytes (3). Apart from the well-described preadipocyte marker Pref-1, little is known about proteins that regulate adipogenesis in the preadipogenic stage without acting as transcription factors (3, 42).
In recent years, data have emerged indicating a role of retinoids in adipose tissue function. Adipose tissue has an active role in retinoid metabolism and is the second largest active storage site for retinoids, with the liver being the largest (25, 46). Retinol binding protein 4 (RBP4), secreted by adipocytes, has been associated with insulin resistance (52). Furthermore, retinoic acid (RA) can inhibit adipocyte differentiation by activating retinoic acid receptors (RAR) and subsequently repressing C/EBPβ transcriptional activity (37). Similarly, the administration of RA to obese mice has been associated with weight loss (1, 4, 19). A different retinoid, retinaldehyde, has been shown to inhibit adipocyte differentiation by repressing PPARγ and retinoid X receptor (RXRα) activities (53).
Intracellularly, retinol is the precursor for both retinaldehyde and RA and is bound to cellular retinol-binding proteins (CRBP) (24, 32). We have previously described the presence of CRBP-III in adipose tissue (29, 48, 54) and demonstrated that CRBP-III is involved in energy metabolism (54). However, less is known about the role of CRBP-I in adipose tissue. Here, we show that CRBP-I is specifically expressed in preadipocytes but not in differentiated adipocytes. In mice, CRBP-I deficiency leads to increased adiposity but a favorable metabolic phenotype compared to the adiposity and metabolic phenotype of wild-type (WT) mice when fed a high-fat diet (HFD). Similarly, suppression of CRBP-I expression in vitro enhanced adipocyte differentiation, while overexpression led to reduced TG accumulation. While PPARγ activity and expression levels were significantly increased, retinaldehyde and retinoic acid levels were not different in cells with suppressed expression of CRBP-I compared to their levels in control cells. Our findings indicate an important role for CRBP-I as a non-transcriptionally active preadipocyte factor that is involved in adipocyte differentiation.
MATERIALS AND METHODS
Mouse studies.
CRBP-I knockout (CRBP-I-KO) mice have been described previously (9, 29). CRBP-I-KO mice were backcrossed to the C57BL/6J genetic background for seven generations prior to experiments. WT and CRBP-I-KO mice were placed on a high-fat diet (HFD) at 5 weeks of age for 20 weeks (D12492, 60% calories from fat; Research Diets, New Brunswick, NJ). Blood samples for insulin and adipokine measurements were obtained after 18 weeks on the HFD. At the end of the dietary regimen, all mice were sacrificed, white adipose tissue weight recorded, and tissue stored at −70°C until analysis. Adipose tissue was fixed in 10% formalin overnight and embedded in paraffin, and sections (5 μm) were stained with hematoxylin and eosin. Adipocyte size was determined using ImageJ Pro (National Institutes of Health) and measuring at least 350 cells per sample. All experiments involving mice were approved by the Institutional Animal Care and Use Committee at Columbia University.
Commercially available enzyme-linked immunosorbent assays (ELISAs) (Millipore) were used to measure serum insulin and adipokine levels. The ELISA for adiponectin measures high- and low-molecular-mass forms of adiponectin. Retinol binding protein 4 (RBP4) levels were determined using Western blot analysis. Commercially available kits were used to measure serum free fatty acid (Wako, United States) and triglyceride (Thermo Scientific) levels.
Glucose and insulin tolerance tests.
Mice were fasted for 6 h, and d-glucose (1 g/kg of body weight [BW]) for glucose tolerance tests (GTT) and insulin (0.75 IU/kg BW) for insulin tolerance tests (ITT) were administered intraperitoneally (i.p.). Blood glucose levels in blood obtained from tail bleeds were monitored using a Glucometer (Lifescan). For assessment of insulin levels in response to glucose injection, serum insulin levels were measured at 15, 30, and 45 min following i.p. injection of glucose (1g/kg BW).
Cell culture.
3T3-L1 preadipocytes were maintained and differentiated as previously established (5). Mouse embryonic fibroblasts (MEFs) were obtained from mice at embryonic day 12.5. Two independent cell lines from WT and CRBP-I-KO mice were established according to established methods (22, 43). Differentiation of MEFs was induced by adding 0.5 μM dexamethasone, 10 μg/ml insulin, 0.5 mM 3-isobutyl-1-methylxanthine, and 10 μM troglitazone to the medium 2 days after the cells reached confluence.
Control cells, cells in which short hairpin RNAs (shRNAs) were used to suppress CRBP-I expression (CRBP-I-shRNA cells), and CRBP-I-shRNA cells in which CRBP-I was reexpressed (rescue cells) were treated with vehicle or retinoic acid (2 μM) between days 0 and 2 of differentiation, and TG levels measured at day 10 of differentiation. In a separate experiment, control cells, shRNA-treated cells, and rescue cells were treated with vehicle or retinol (2 μM) between days 0 and 2 of differentiation, and TG levels measured at day 10 of differentiation.
Cellular TG levels.
Triglycerides (TG) were extracted from cultured cells using hexane/isopropanol (3:2 [vol/vol]). The lipid extracts were dried under a gentle stream of nitrogen and dissolved in cold isopropanol, and TG levels measured using a commercially available kit (Thermo Scientific). Bodipy dye staining of neutral lipids in differentiated 3T3-L1 cells was performed as previously described (11).
Lentivirus shRNA.
The following Mission shRNAs (Sigma) were used to suppress CRBP-I expression in 3T3-L1 cells: CRBP-I-shRNA1, CCGGCGAGGAGATAACCTTGGTCTTCTCGAGAAGACCAAGGTTATCTCCTCGTTTTTG, and CRBP-I-shRNA2, CGGTGTGATCTGCAAGCAAGTGTTCTCGAGAACACTTGCTTGCAGATCACATTTTTG. The shRNAs were cloned into the BLOCK-iT lentivirus interfering RNA (RNAi) system driven by a U6 promoter (Invitrogen), and lentivirus for the different shRNAi's were generated. 3T3-L1 preadipocytes were infected with the lentivirus (multiplicity of infection of 10 to 50) and subsequently selected with blasticidin. For ectopic expression, whole-length CRBP-I cDNA was integrated into the pLenti4 vector (Invitrogen) using homologous recombination, and lentivirus produced as described above. 3T3-L1 wild-type and CRBP-I knockdown cells (CRBP-I-shRNA1) were infected with the lentivirus and selected for 10 days with zeocin. As CRBP-I-shRNA1 targets the 3′ untranslated region (UTR), CRBP-I cDNA expression will not be affected by the shRNA.
Cell proliferation assay.
The cell proliferation assay was performed as previously reported (10). Briefly, cells were synchronized in Dulbecco's modified Eagle's medium supplemented with 1% fetal bovine serum for 24 h prior to the experiment. The medium was changed to regular growth medium, and [3H]thymidine added to the cells. After 4 h, the medium was removed, and the cells washed with phosphate-buffered saline (PBS) and treated with 5% ice-cold trichloroacetic acid (TCA) to remove unincorporated [3H]thymidine. The cells were solubilized in 1 M NaOH, and samples analyzed in a liquid scintillation counter.
Cell sorting.
White adipose tissue was digested with collagenase to obtain adipocytes and the stromal-vascular fraction (SVF) as previously described (51). For separation of different cell types in the SVF, cells were labeled with specific antibodies, including CD11b for macrophages and Pecam (CD31) for endothelial cells (51). Cells were sorted on a FACSCalibur (Columbia University Irving Cancer Center Core Facility), RNA extracted, and real-time PCR performed for CRBP-I expression.
ChIP assay.
WT and CRBP-I-shRNA 3T3-L1 cells were induced to differentiate 2 days after reaching confluence (see above). Chromatin immunoprecipitation (ChIP) was performed essentially as previously described on cells at days 1 and 2 of differentiation (17). Briefly, cells were cross-linked with formaldehyde for 10 min at room temperature. After chromatin disruption using a Bioruptor, DNA concentrations were assessed for each time point and cell line. Immunoprecipitation of protein was performed using anti-Krüppel-like factor 5 (KFL5; Millipore), anti-C/EBPβ (Santa Cruz), or nonspecific anti-rabbit IgG. Quantitative real-time PCR of the DNA was performed using SYBR green (Fermentas). The primer sequences used for the PPARγ promoter region that binds C/EBPβ and KLF5 (26) are forward, 5′ TATCTGGTGTTTCATAACTTAGAGATTAAG 3′, and reverse, 5′ ATTACAAGGAAAACGTTGCTACATTGTCTC 3′. The results are expressed as changes in the protein levels in CRBP-I-shRNA cells compared to the levels in control cells.
Quantitative real-time PCR.
Total RNA was extracted from tissues and cells at indicated times of differentiation using STAT-60 (Tel-Test), and cDNA was generated using Superscript reverse transcriptase (Invitrogen). The real-time PCR mixture contained gene-specific primers, 1× SYBR green PCR master mix (Fermentas), and 10 μl of diluted cDNA. Primer sequences are available upon request.
Determination of retinoid levels.
Control, CRBP-I-shRNA, and rescue cells were harvested, pelleted, and immediately flash-frozen in liquid nitrogen. Retinoid levels were quantified as previously described using liquid chromatography tandem mass spectrometry with atmospheric pressure chemical ionization in positive-ion mode on an API-4000 (Applied Biosystems, Foster City, CA) (13, 14). The results were expressed as pmoles per g total protein.
Lipolysis assay.
Lipolysis assays were performed in differentiated control and CRBP-I-shRNA 3T3-L1 cells. Cells were treated with isoproterenol (10 μM), and glycerol concentrations in the medium measured at 2 and 4 h following the treatment.
Luciferase assays.
For determination of RARα and PPARγ activities, all cells were differentiated for 24 h and subsequently transfected with 2 μg retinoic acid response element (RARE)-Luc (kind gift of N. Noy, Case Western Reserve University) or 3× peroxisome proliferator response element (PPRE)-Luc (kindly provided by M. A. Lazar, University of Pennsylvania) constructs using Lipofectamine (Invitrogen) for 18 h. PPRE-luciferase activity assays were conducted at day 4 of differentiation, and RARE-luciferase reporter activity was measured at day 2 of differentiation. Luciferase activities were measured using a Dual-Luciferase reporter assay (Promega) following the manufacturer's instructions. All transfections were normalized by cotransfection with a Renilla luciferase construct.
Statistical analysis.
Student's t test was used for comparison between two groups, and significance was considered to be a P value of <0.05. Two-way analysis of variance was used to detect differences in gene expression across different time points and cell lines. For significant outcomes, post hoc analyses were performed.
RESULTS
CRBP-I-KO mice have increased adiposity but improved glucose tolerance and insulin sensitivity during diet-induced obesity.
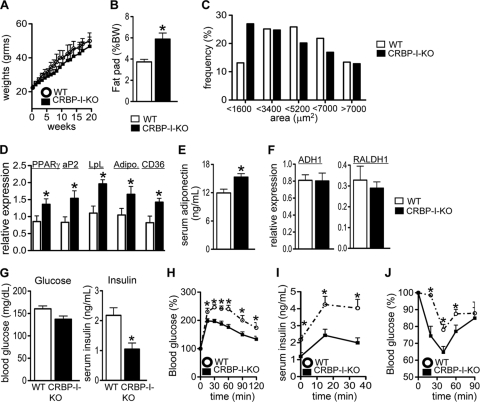
To gain insight into the role of CRBP-I in adipose tissue biology, wild-type (WT) and CRBP-I-KO mice were placed on a high-fat diet (HFD) for 20 weeks. Body weights were not significantly different between the two groups throughout the dietary regimen (Fig. 1 A). However, CRBP-I-KO mice developed increased adiposity compared to that of WT mice (Fig. 1B). The ratio of the epididymal fat pad to body weight was significantly increased in CRBP-I-KO mice compared to this ratio in WT mice at the end of the dietary regimen (Fig. 1B). Histological examination revealed that epididymal adipose tissue from CRBP-I-KO mice contained a higher frequency of smaller adipocytes than epididymal adipose tissue from WT mice (Fig. 1C). We next determined whether the increased adiposity was associated with changes in adipocyte differentiation. The expression of the adipogenic transcription factor PPARγ was significantly increased in white adipose tissue of CRBP-I-KO mice compared to its expression in white adipose tissue of WT mice (Fig. 1D). Furthermore, the expression levels of PPARγ target genes and markers of late differentiation (adipocyte fatty-acid binding protein [aP2], CD36, lipoprotein lipase [LpL], and adiponectin) were significantly increased in white adipose tissue of CRBP-I-KO mice compared to their expression levels in white adipose tissue of WT mice (Fig. 1D). In agreement with increased adiponectin gene expression, serum adiponectin levels were significantly higher in CRBP-I-KO mice than in WT mice (Fig. 1E). The levels of other adipokines, including leptin and resistin, and the RBP4 and serum lipid levels were not different (Table 1 and data not shown). The expression levels for retinoic acid (RA) synthesis genes were not different between the two mouse models (Fig. 1F). Taken together, these data indicate that the increased adiposity in CRBP-I-KO mice correlates with increased adipogenic gene expression and may reflect enhanced adipocyte differentiation.
FIG. 1.
CRBP-I-KO mice had increased adiposity but remained glucose and insulin tolerant compared to WT mice after being fed a high-fat diet (HFD). (A) Body weight gain for WT and CRBP-I-KO mice fed an HFD. n = 10 WT mice, and n = 9 CRBP-I-KO mice. (B) CRBP-I-KO mice displayed increased adiposity after 20 weeks on an HFD. n = 10 WT mice, and n = 9 CRBP-I-KO mice. (C) White adipose tissue from CRBP-I-KO mice had a higher percentage of small adipocytes than white adipose tissue from WT mice. n = 8 WT mice, and n = 8 CRBP-I-KO mice. (D) Expression of PPARγ and downstream target genes in white adipose tissue of CRBP-I-KO and WT mice. n = 7 WT mice, and n = 7 CRBP-I-KO mice. Adipo., adiponectin. (E) Serum adiponectin levels in CRBP-I-KO mice compared to the levels in WT mice after 18 weeks of an HFD. n = 15 WT mice, and n = 14 CRBP-I-KO mice. (F) Expression of genes involved in intracellular retinaldehyde and RA synthesis in white adipose tissue in WT and CRBP-I-KO mice. ADH1, alcohol dehydrogenase type 1; RALDH1, retinaldehyde dehydrogenase type 1. n = 7 WT mice, and n = 7 CRBP-I-KO mice. (G) Fasting serum glucose and insulin levels in mice on an HFD. While blood glucose levels were not different, serum insulin levels were significantly reduced in CRBP-I-KO mice. n = 10 WT mice, and n = 9 CRBP-I-KO mice. (H) Results of glucose tolerance test (GTT) in WT and CRBP-I-KO mice fed an HFD. (I) Glucose-stimulated insulin secretion in WT and CRBP-I-KO mice. (J) Results of insulin tolerance test (ITT) in WT and CRBP-I-KO mice fed an HFD. n = 10 WT mice, and n = 9 CRBP-I-KO mice. *, significantly different at P < 0.05. Data are represented as the means ± standard errors of the means.
TABLE 1.
Concentrations of several serum analytes of wild-type and CRBPI-KO mice fed an HFDa
| Serum analyte | Concn (mean ± SEM) in: |
|
|---|---|---|
| WT mice | CRBP-I-KO mice | |
| Resistin (ng/μl) | 3.0 ± 0.4 | 3.2 ± 0.4 |
| Leptin (ng/μl) | 16.1 ± 2.6 | 12.2 ± 2.9 |
| Triglyceride (mg/dl) | 91.3 ± 6.9 | 86.4 ± 6.7 |
| Free fatty acids (nmol/liter) | 0.80 ± 0.6 | 0.75 ± 0.04 |
Serum leptin, resistin, free fatty acid, and TG levels were measured after mice were fed an HFD for 18 weeks.
Changes in adiposity are often associated with alterations in glucose and insulin homeostasis (31). While the blood glucose levels in WT and CRBP-I-KO mice after a 6-h fast were similar, the serum insulin levels were significantly lower in CRBP-I-KO mice than in WT mice (Fig. 1G). Glucose tolerance tests (GTT) revealed significantly lower blood glucose levels in CRBP-I-KO mice than in WT mice in response to glucose challenge (Fig. 1H). Furthermore, serum insulin levels in response to a glucose challenge remained significantly lower in CRBP-I-KO mice than in WT mice (Fig. 1I), indicating that CRBP-I-KO mice remained more glucose tolerant during diet-induced obesity. Similar to the GTT results, CRBP-I-KO mice were more sensitive to insulin, with a significantly greater hypoglycemic response to the injection of insulin than WT mice (Fig. 1J). Together, these results indicate that CRBP-I-KO mice are more glucose tolerant and insulin sensitive than WT mice during HFD feeding despite having increased fat.
CRBP-I is a preadipocyte gene that represses adipocyte differentiation in vitro.
We and others have previously shown that CRBP-I is expressed in white adipose tissue, specifically, in the stromal-vascular fraction (46, 48). Further analysis revealed that within the stromal-vascular fraction, CRBP-I expression is restricted to preadipocytes, and no expression was detected in endothelial cells or macrophages (data not shown). To assess CRBP-I expression during adipocyte differentiation, we used the well-established 3T3-L1 preadipocyte cell line. CRBP-I expression is highest during the first 2 days of differentiation and decreases thereafter, and it is no longer detected at 5 days of differentiation (Fig. 2 A), indicating that CRBP-I may play an important role during early differentiation.
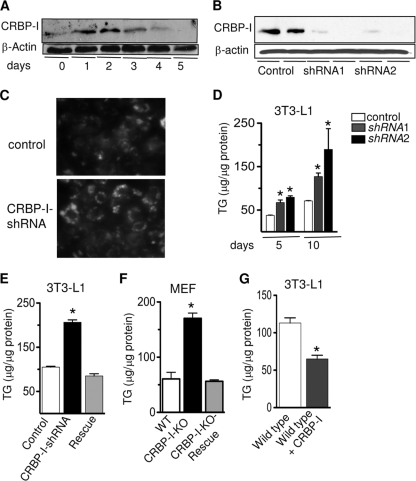
FIG. 2.
Reducing CRBP-I expression in 3T3-L1 preadipocytes results in increased adipocyte differentiation and TG accumulation. (A) Western blot analysis of CRBP-I protein during differentiation of 3T3-L1 cells. (B) CRBP-I protein levels in control 3T3-L1 preadipocytes and 3T3-L1 cells expressing specific shRNAs targeting CRBP-I. Two cell lines were established, CRBP-I-shRNA1 and CRBP-I-shRNA2. (C) Staining of lipid droplets showed increased lipids in cells with repressed CRBP-I expression. Lipid droplets were stained with Bodipy 493/503 dye and visualized by confocal microscopy. (D) Intracellular TG levels in control and CRBP-I-shRNA cells at 5 and 10 days after start of differentiation. *, significantly different compared to results for control cells at the indicated time point (P < 0.05). (E) TG levels in control cells, CRBP-I-shRNA cells, and CRBP-I-shRNA cells expressing ectopic CRBP-I (designated “rescue”). *, significantly different compared to results for control and rescue cells (P < 0.05). (F) TG levels were significantly increased in CRBP-I-KO MEFs compared to TG levels in WT or CRBP-I-KO rescue MEFs 10 days after the start of differentiation (*, P < 0.05). (G) Intracellular TG levels in wild-type 3T3-L1 cells compared to TG levels in 3T3-L1 cells with overexpression of CRBP-I. *, significantly different at P < 0.05. The experiments were performed in triplicate. Data are presented as the means ± standard errors of the means. Results in panels D to F are representative of three independent experiments.
We next sought to provide a mechanistic understanding of the enhanced adiposity of CRBP-I-KO mice by determining the role of CRBP-I in adipocyte differentiation. We hypothesized that if CRBP-I is involved in repressing adipocyte differentiation, then suppression of CRBP-I expression will lead to enhanced adipocyte differentiation. 3T3-L1 preadipocytes were infected with lentivirus that harbored either shRNAs against CRBP-I (shRNA1 and shRNA2) or a control shRNA. CRBP-I protein levels were successfully reduced in two independent cell lines (Fig. 2B). Remarkably, cells with decreased expression of CRBP-I showed enhanced lipid accumulation as determined by staining of intracellular lipids (Fig. 2C). Strikingly, the decreased expression of CRBP-I led to significantly increased intracellular TG levels at 5 and 10 days after the start of differentiation (Fig. 2D), indicative of enhanced adipocyte differentiation. To confirm that the increased intracellular TG accumulation was due to the diminished expression of CRBP-I, we reexpressed CRBP-I in the CRBP-I knockdown cell line, generating a rescue cell line (data not shown). Intracellular TG levels in CRBP-I-shRNA cells with ectopic CRBP-I expression were not different from the TG levels in control cells (Fig. 2E), indicating that the increased TG accumulation was due specifically to reduced CRBP-I levels.
To further confirm these findings using an independent genetic model, we generated mouse embryonic fibroblast (MEF) cell lines from WT and CRBP-I-KO mice. Consistent with our in vivo and 3T3-L1 shRNA data, adipocyte differentiation was significantly increased in CRBP-I-KO MEFs compared to the adipocyte differentiation in wild-type MEFs. Intracellular TG levels were increased nearly 3-fold in CRBP-I-KO mice compared to the TG levels in wild-type MEFs (Fig. 2F). Similar to the findings for 3T3-L1 CRBP-I knockdown cells, reexpression of CRBP-I in the CRBP-I-KO MEFs resulted in normalization of intracellular TG levels comparable to the levels of TG in wild-type MEFs (Fig. 2F). Moreover, overexpression of CRBP-I in wild-type 3T3-L1 cells resulted in significantly reduced TG levels compared to the levels in control cells (Fig. 2G). Together, these findings indicate that CRBP-I negatively regulates adipocyte differentiation.
CRBP-I participates in adipocyte differentiation.
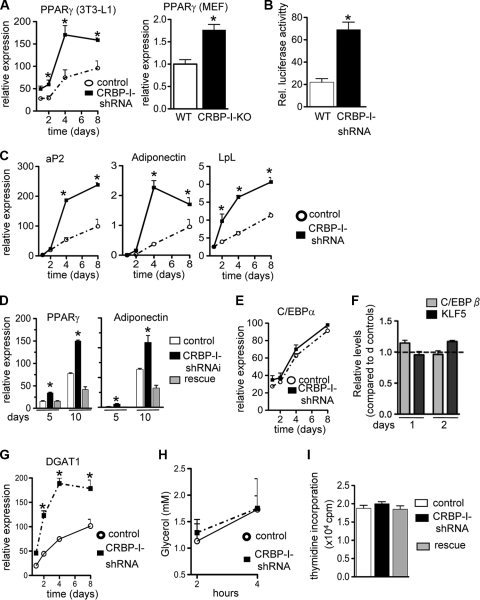
Next, we wanted to determine the stage in the transcriptional cascade that is affected by a CRBP-I deficiency during adipocyte differentiation. Consistent with our results in vivo (Fig. 1D), PPARγ expression was significantly increased during the differentiation process in CRBP-I-shRNA cells and CRBP-I-KO MEFs compared to its expression in the respective control cells (Fig. 3 A). Furthermore, we determined that PPARγ activity was significantly increased when CRBP-I levels were diminished (Fig. 3B). There was a 2-fold increase in PPARγ activity, assessed using a PPARγ transactivation reporter assay, in CRBP-I-shRNA cells compared to the PPARγ activity in control cells (Fig. 3B). In agreement with these data, the expression levels of PPARγ target genes (encoding aP2, adiponectin, and LpL) were significantly increased in cells with repressed CRBP-I expression compared to their expression levels in control cells (Fig. 3C). Consistently, PPARγ expression levels in the rescue cells were reduced to levels similar to those in control cells (Fig. 3D). Similarly, the levels of adiponectin expression in the rescue cells were reduced to the levels measured in control cells (Fig. 3D). In contrast to the increased expression of PPARγ, the expression levels for C/EBPα, another essential transcription factor during adipocyte differentiation, were not altered by decreased CRBP-I expression (Fig. 3E). To understand the lack of C/EBPα induction, we analyzed the PPARγ promoter using ChIP assays. It has previously been shown that KLF5 and C/EBPβ regulate the PPARγ promoter in concert (26). However, the occupancy of the PPARγ promoter by KLF5 or C/EBPβ did not appear to be different between control and CRBP-I-shRNA cells (Fig. 3F).
FIG. 3.
Repressed expression of CRBP-I in 3T3-L1 cells leads to increased expression and activity of PPARγ. (A) PPARγ mRNA levels during differentiation (indicated in days) in CRBP-I-shRNA cells (left panel) and CRBP-I-KO MEFs (right panel) compared to PPARγ mRNA levels in their respective control cells. MEFs were analyzed at 10 days after the start of differentiation. *, significantly different at P < 0.05. (B) Results of luciferase assays of PPARγ transcriptional activity using 3× PPRE-Luc in control and CRBP-I-shRNA cells at 4 days after start of differentiation. Rel., relative. *, significantly different at P < 0.05. (C) Expression of PPARγ downstream target genes at different times of differentiation in control and CRBP-I-shRNA cells. (D) PPARγ and adiponectin expression levels in control, shRNA, and rescue (reexpression of CRBP-I) cells. *, significantly different from results for control and rescue cells (P < 0.05). (E) Expression levels of C/EBPα during adipocyte differentiation in control and CRBP-I-KO shRNA cells. (F) Results of ChIP of C/EBPβ and KLF5 binding to the PPARγ promoter in control versus CRBP-I-shRNA cells at days 1 and 2 of differentiation. Real-time PCR of the PPARγ promoter region known to bind C/EBPβ and KLF5 was performed. Results are shown as the ratio of the level in CRBP-I-shRNA cells and the level in control cells for days 1 and day 2. (G) Expression of diacylglycerol acyltransferase type 1 (DGAT1) was significantly increased during adipocyte differentiation in CRBP-I-shRNA cells compared to its expression in control cells. (H) Results of lipolysis assay in WT and CRBP-I-shRNA cells. Glycerol release into the medium was measured at 2 and 4 hours after stimulation of lipolysis with isoproterenol. (I) Repressed expression of CRBP-I did not affect cell proliferation of 3T3-L1 cells. Results of proliferation assays for control and CRBP-I-shRNA cells are shown. Experiments were conducted in duplicate for each time point and cell line. Results in panels A to D are representative of three independent experiments. *, significantly different at P < 0.05. Data are represented as the means ± standard errors of the means.
The increased TG accumulation in CRBP-I-shRNA cells was accompanied by increased expression of DGAT1 (Fig. 3G), another marker of differentiated adipocytes, indicating increased TG synthesis in these cells. The rates of lipolysis were not different between control and CRBP-I-shRNA cells (Fig. 3H).
Based on the expression pattern during adipocyte differentiation (Fig. 2A), CRBP-I may function in the early stages of adipocyte differentiation. We hypothesized that the suppression of CRBP-I may alter the expression of genes present early in adipocyte differentiation and/or may affect genes that in turn lead to downstream changes in PPARγ expression and activity. We evaluated Pref-1, a prominent preadipocyte protein that plays an inhibitory role in adipocyte differentiation and whose absence leads to enhanced adipocyte differentiation (16, 40, 41, 47). However, in our study, the expression levels of Pref-1 and its downstream target gene product Sox9 (50) were not different between control and CRBP-I-shRNA cells (data not shown). Similarly, the expression levels of preadipocyte transcription factors that affect PPARγ expression and transcriptional activity, including C/EBPβ, GATA2, and ID2 (28, 44, 45), were not altered in CRBP-I-shRNA cells (data not shown). In addition, the loss of CRBP-I expression did not affect preadipocyte proliferation (Fig. 3I). Taken together, our data indicate that a CRBP-I deficiency augments PPARγ activity, an effect that cannot be readily explained by changes in known proteins that repress or promote adipocyte differentiation during the early stages of differentiation.
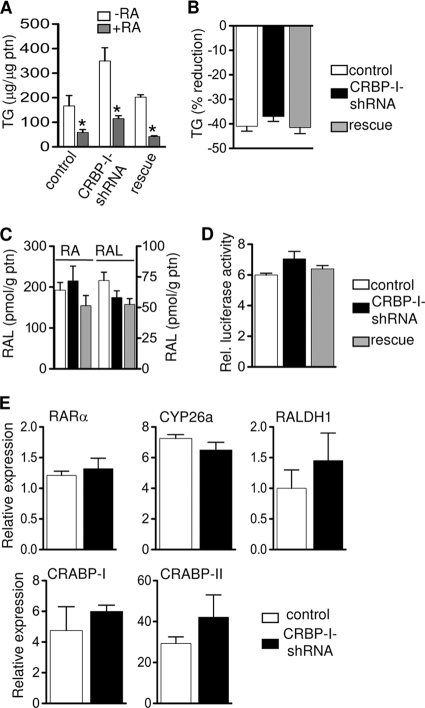
CRBP-I binds retinol and its immediate oxidation product retinaldehyde (27). The proposed role of CRBP-I is to participate in the metabolism of retinol to retinyl ester or to the metabolically active compound RA (21). RA in particular, when added early in differentiation, has an inhibitory effect on adipocyte differentiation (37, 39). Thus, we tested the hypothesis that CRBP-I regulates adipocyte differentiation through sensitizing cells to the inhibitory effects of RA on adipocyte differentiation. We treated control, CRBP-I-shRNA, and rescue cells with vehicle or RA during early differentiation. TG accumulation was inhibited by RA independently of the expression levels of CRBP-I, indicating that in the absence of CRBP-I, cells remain sensitive to the inhibitory effects of RA (Fig. 4 A). An alternative hypothesis is that CRBP-I is important in maintaining the levels of retinaldehyde and RA necessary to repress adipocyte differentiation. We treated cells with retinol and determined the levels of TG accumulation as a measure of adipocyte differentiation. Retinol treatment decreased adipocyte differentiation to the same extent in the presence or absence of CRBP-I (Fig. 4B). Furthermore, intracellular retinaldehyde and RA levels were not different among control, CRBP-I-shRNA, and rescue cells (Fig. 4C). This finding is also corroborated by the results of RARE-luciferase assays showing that the transcriptional activity of RA was not altered (Fig. 4D) and by the finding that the expression levels of RA-regulated genes remained unchanged (Fig. 4E). Taken together, the retinaldehyde and retinoic acid levels and the ability of retinol or retinoic acid to suppress adipocyte differentiation were not different in the presence or absence of CRBP-I.
FIG. 4.
Control, shRNA, and rescue cells were treated with vehicle or RA (2 μM) (A) or vehicle or retinol (2 μm) (B) during early adipocyte differentiation (days 0 to 2), and TG levels measured at day 10 of differentiation. *, significantly different from results for untreated cells at P < 0.05. RA, retinoic acid. Data are representative of three independent experiments. (C) RA and retinaldehyde (RAL) concentrations in control (white bars), CRBP-I-shRNA (black bars), and rescue (grey bars) cells. Experiments were performed in triplicate for each cell line. (D) RARα transcriptional activity (RARE-luciferase activity) in control, CRBP-I-shRNA, and rescue cells was assessed at day 2 of differentiation. (E) Comparison of expression levels of genes involved in retinoid metabolism at day 2 of differentiation. The results in panels D and E are representative of two independent experiments. Data are represented as the means ± standard errors of the means. CYP26a, cytochrome P45026a; RALDH1, retinaldehyde dehydrogenase type 1.
DISCUSSION
We have demonstrated that CRBP-I deficiency leads to a proadipogenic phenotype with significantly enhanced adipocyte differentiation and increased intracellular TG accumulation due to augmented PPARγ activity. CRBP-I has been assigned the role of a chaperone in retinoid metabolism, primarily protecting retinol from being metabolized by unspecific enzymes (9, 18, 20, 21). In this role, CRBP-I maintains normal levels of retinol for retinyl ester synthesis in the liver and the eye and controls RA synthesis (9, 18, 20, 21, 33). We examined an exhaustive list of possible explanations for our observations, including intracellular retinoid levels, transcriptional activity of RA, expression of genes involved in intracellular retinoid metabolism, the sensitivity of our cell models to retinol and RA treatment, and changes in cell proliferation.
Because it has previously been demonstrated that the two downstream products of retinol, retinaldehyde and RA, are potent inhibitors of adipocyte differentiation (37, 39, 53), we expected that the observed effects of CRBP-I deficiency would correlate with reduced levels of retinaldehyde or RA. In contrast to our expectations, the intracellular levels of retinaldehyde and RA remained the same in the absence of CRBP-I. Decreased delivery of RA to the nucleus could offer an explanation for our phenotype (1). However, the levels of cellular retinoic acid-binding protein II (CRABP-II) and fatty acid binding protein 5 (FABP5) (data not shown), two proteins that are thought to be important for RA delivery to the nucleus (1, 34), were not different in the presence or absence of CRBP-I. Additionally, the transcriptional activity of RA and expression levels of RA target genes were unaffected by the absence of CRBP-I. Furthermore, the cells remained sensitive to the inhibitory action of retinol and RA on differentiation. Finally, we ruled out the possibility that the absence of CRBP-I increases cell proliferation. Based on these results, the actions of CRBP-I in adipocyte differentiation cannot be readily explained by changes in retinaldehyde and RA levels. Recently, in an elegant study, Lazar and coworkers demonstrated that the enzyme retinol saturase (RetSat) promotes adipocyte differentiation (36). In a finding similar to that in our study, the function of RetSat was not readily explained by its enzymatic role of generating 13,14-dihydroretinol from retinol.
Importantly, CRBP-I deficiency is tightly associated with enhanced PPARγ activity. CRBP-I is expressed in preadipocytes prior to the expression of PPARγ and C/EBPα, the two master regulators of adipocyte differentiation. The expression levels and transcriptional activity of PPARγ were significantly increased in the absence of CRBP-I, while C/EBPα expression was not affected. Consistently, in our CRBP-I-KO model, the levels of PPARγ and its downstream targets were increased, but not the levels of C/EBPα or other adipogenic transcription factors. We evaluated several known transcription factors upstream of PPARγ that are known to affect PPARγ activity but did not detect significant changes. Taken together, the absence of CRBP-I specifically leads to an increase in PPARγ activity. While CRBP-I is a cytosolic protein (27), it remains to be determined if CRBP-I regulates the activities or levels of PPARγ cofactors.
The proadipogenic phenotype observed in our shRNA knockdown cell lines and CRBP-I-KO MEFs also translated into increased adiposity in vivo. CRBP-I-KO mice challenged with an HFD displayed increased adiposity compared to that of WT mice on the same diet. Importantly, despite the expansion of the fat depot, CRBP-I-KO mice exhibited a better glucose homeostasis than WT mice on a high-fat diet. Increased adipose PPARγ expression, an increased frequency of small adipocytes, and increased plasma adiponectin levels in CRBP-I-KO mice compared to the findings for WT mice offer an explanation for the observed positive impact on metabolism. Our observations are in line with several examples where increased adiposity was associated with increased insulin sensitivity (15, 49). For example, the overexpression of adiponectin in adipose tissue of leptin-deficient mice results in increased adiposity but also in an improved metabolic profile compared to that of controls (15). The treatment of animals and humans with PPARγ agonists leads to insulin sensitivity and greater adiposity with smaller adipocytes (23).
To our knowledge, CRBP-I and Pref-1 are the only non-transcription factors present in preadipocytes that modulate adipocyte differentiation. It is important to note that while overexpression of CRBP-I resulted in a significant reduction in TG levels, it did not lead to complete inhibition of adipocyte differentiation. This is in partial contrast to findings for Pref-1, which acts as a dominant factor completely inhibiting adipocyte differentiation when overexpressed (41). A possible explanation for this observation is that CRBP-I function requires other factors that must be present at a specific stochiometry with CRBP-I during adipocyte differentiation. Interestingly, it has recently been shown that the knockdown of PPARγ in adipocytes leads to significant increases in Pref-1 and CRBP-I, reflecting dedifferentiation of the cells (35). As CRBP-I expression is restricted to preadipocytes in the adipose tissue, its gene may prove to be an additional useful marker gene for preadipocytes and, potentially, a therapeutic target to treat insulin resistance and type 2 diabetes, similar to the consideration given to another member of the FABP family, FABP4/aP2 (6, 7).
In conclusion, our data indicate an important role of CRBP-I in adipocyte differentiation. CRBP-I is a protein specific to preadipocytes, and its gene could thus become an additional useful marker gene for preadipocytes. Furthermore, in the absence of CRBP-I, PPARγ expression and activity are increased, leading to enhanced adipocyte differentiation. Overall, this represents a novel function of CRBP-I in adipose tissue and provides new insights into the regulation of early adipocyte differentiation.
Acknowledgments
We thank H. N. Ginsberg at Columbia University for helpful discussions and critical review of the manuscript, J. L. Napoli at the University of California—Berkeley for performing the retinoid analyses in our cell models, and G. Reyes at Columbia University for technical assistance.
The work was supported by grants RO1 DK-067512 (S. Vogel) and DK-063608-06 (S. Vogel) from the NIDDK and the German Academic Exchange Service (S.K.F.).
Footnotes
Published ahead of print on 24 May 2010.
REFERENCES
- 1.Berry, D. C., and N. Noy. 2009. All-trans-retinoic acid represses obesity and insulin resistance by activating both peroxisome proliferation-activated receptor beta/delta and retinoic acid receptor. Mol. Cell. Biol. 29:3286-3296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Duncan, R. E., M. Ahmadian, K. Jaworski, E. Sarkadi-Nagy, and H. S. Sul. 2007. Regulation of lipolysis in adipocytes. Annu. Rev. Nutr. 27:79-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Farmer, S. R. 2006. Transcriptional control of adipocyte formation. Cell Metab. 4:263-273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Felipe, F., M. L. Bonet, J. Ribot, and A. Palou. 2004. Modulation of resistin expression by retinoic acid and vitamin A status. Diabetes 53:882-889. [DOI] [PubMed] [Google Scholar]
- 5.Frost, S. C., and M. D. Lane. 1985. Evidence for the involvement of vicinal sulfhydryl groups in insulin-activated hexose transport by 3T3-L1 adipocytes. J. Biol. Chem. 260:2646-2652. [PubMed] [Google Scholar]
- 6.Furuhashi, M., and G. S. Hotamisligil. 2008. Fatty acid-binding proteins: role in metabolic diseases and potential as drug targets. Nat. Rev. Drug Discov. 7:489-503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Furuhashi, M., G. Tuncman, C. Z. Gorgun, L. Makowski, G. Atsumi, E. Vaillancourt, K. Kono, V. R. Babaev, S. Fazio, M. F. Linton, R. Sulsky, J. A. Robl, R. A. Parker, and G. S. Hotamisligil. 2007. Treatment of diabetes and atherosclerosis by inhibiting fatty-acid-binding protein aP2. Nature 447:959-965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gesta, S., M. Bluher, Y. Yamamoto, A. W. Norris, J. Berndt, S. Kralisch, J. Boucher, C. Lewis, and C. R. Kahn. 2006. Evidence for a role of developmental genes in the origin of obesity and body fat distribution. Proc. Natl. Acad. Sci. U. S. A. 103:6676-6681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ghyselinck, N. B., C. Bavik, V. Sapin, M. Mark, D. Bonnies, C. Hindelang, A. Dierich, C. B. Nilsson, H. Hakansson, P. Sauvant, V. Azais-Braesco, M. Frasson, S. Picaud, and P. Chambon. 1999. Cellular retinol-binding protein I is essential for vitamin A homeostasis. EMBO J. 18:4903-4914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gupta, P., S. W. Park, M. Farooqui, and L. N. Wei. 2007. Orphan nuclear receptor TR2, a mediator of preadipocyte proliferation, is differentially regulated by RA through exchange of coactivator PCAF with corepressor RIP140 on a platform molecule GRIP1. Nucleic Acids Res. 35:2269-2282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kadereit, B., P. Kumar, W. J. Wang, D. Miranda, E. L. Snapp, N. Severina, I. Torregroza, T. Evans, and D. L. Silver. 2008. Evolutionarily conserved gene family important for fat storage. Proc. Natl. Acad. Sci. U. S. A. 105:94-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kahn, B. B., and J. S. Flier. 2000. Obesity and insulin resistance. J. Clin. Invest. 106:473-481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kane, M. A., N. Chen, S. Sparks, and J. L. Napoli. 2005. Quantification of endogenous retinoic acid in limited biological samples by LC/MS/MS. Biochem. J. 388:363-369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kane, M. A., A. E. Folias, C. Wang, and J. L. Napoli. 2008. Quantitative profiling of endogenous retinoic acid in vivo and in vitro by tandem mass spectrometry. Anal. Chem. 80:1702-1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim, J. Y., E. van de Wall, M. Laplante, A. Azzara, M. E. Trujillo, S. M. Hofmann, T. Schraw, J. L. Durand, H. Li, G. Li, L. A. Jelicks, M. F. Mehler, D. Y. Hui, Y. Deshaies, G. I. Shulman, G. J. Schwartz, and P. E. Scherer. 2007. Obesity-associated improvements in metabolic profile through expansion of adipose tissue. J. Clin. Invest. 117:2621-2637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee, K., J. A. Villena, Y. S. Moon, K. H. Kim, S. Lee, C. Kang, and H. S. Sul. 2003. Inhibition of adipogenesis and development of glucose intolerance by soluble preadipocyte factor-1 (Pref-1). J. Clin. Invest. 111:453-461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lefterova, M. I., Y. Zhang, D. J. Steger, M. Schupp, J. Schug, A. Cristancho, D. Feng, D. Zhuo, C. J. Stoeckert, Jr., X. S. Liu, and M. A. Lazar. 2008. PPARgamma and C/EBP factors orchestrate adipocyte biology via adjacent binding on a genome-wide scale. Genes Dev. 22:2941-2952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Matt, N., C. K. Schmidt, V. Dupe, C. Dennefeld, H. Nau, P. Chambon, M. Mark, and N. B. Ghyselinck. 2005. Contribution of cellular retinol-binding protein type 1 to retinol metabolism during mouse development. Dev. Dyn. 233:167-176. [DOI] [PubMed] [Google Scholar]
- 19.Mercader, J., J. Ribot, I. Murano, F. Felipe, S. Cinti, M. L. Bonet, and A. Palou. 2006. Remodeling of white adipose tissue after retinoic acid administration in mice. Endocrinology 147:5325-5332. [DOI] [PubMed] [Google Scholar]
- 20.Molotkov, A., N. B. Ghyselinck, P. Chambon, and G. Duester. 2004. Opposing actions of cellular retinol-binding protein and alcohol dehydrogenase control the balance between retinol storage and degradation. Biochem. J. 383:295-302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Napoli, J. L. 2000. A gene knockout corroborates the integral function of cellular retinol-binding protein in retinoid metabolism. Nutr. Rev. 58:230-236. [DOI] [PubMed] [Google Scholar]
- 22.Nofsinger, R. R., P. Li, S. H. Hong, J. W. Jonker, G. D. Barish, H. Ying, S. Y. Cheng, M. Leblanc, W. Xu, L. Pei, Y. J. Kang, M. Nelson, M. Downes, R. T. Yu, J. M. Olefsky, C. H. Lee, and R. M. Evans. 2008. SMRT repression of nuclear receptors controls the adipogenic set point and metabolic homeostasis. Proc. Natl. Acad. Sci. U. S. A. 105:20021-20026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nolan, J. J., B. Ludvik, P. Beerdsen, M. Joyce, and J. Olefsky. 1994. Improvement in glucose tolerance and insulin resistance in obese subjects treated with troglitazone. N. Engl. J. Med. 331:1188-1193. [DOI] [PubMed] [Google Scholar]
- 24.Noy, N. 2000. Retinoid-binding proteins: mediators of retinoid action. Biochem. J. 348:481-495. [PMC free article] [PubMed] [Google Scholar]
- 25.O'Byrne, S. M., N. Wongsiriroj, J. Libien, S. Vogel, I. J. Goldberg, W. Baehr, K. Palczewski, and W. S. Blaner. 2005. Retinoid absorption and storage is impaired in mice lacking lecithin:retinol acyltransferase (LRAT). J. Biol. Chem. 280:35647-35657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Oishi, Y., I. Manabe, K. Tobe, K. Tsushima, T. Shindo, K. Fujiu, G. Nishimura, K. Maemura, T. Yamauchi, N. Kubota, R. Suzuki, T. Kitamura, S. Akira, T. Kadowaki, and R. Nagai. 2005. Kruppel-like transcription factor KLF5 is a key regulator of adipocyte differentiation. Cell Metab. 1:27-39. [DOI] [PubMed] [Google Scholar]
- 27.Ong, D. E., M. E. Newcomer, and F. Chytil. 1994. Cellular retinoid-binding proteins, p. 283-317. In M. B. Sporn, A. B. Roberts, and D. S. Goodman (ed.), The retinoids, biology, chemistry, and medicine, 2nd ed. Raven, New York, NY.
- 28.Park, K. W., H. Waki, C. J. Villanueva, L. A. Monticelli, C. Hong, S. Kang, O. A. MacDougald, A. W. Goldrath, and P. Tontonoz. 2008. Inhibitor of DNA binding 2 is a small molecule-inducible modulator of peroxisome proliferator-activated receptor-gamma expression and adipocyte differentiation. Mol. Endocrinol. 22:2038-2048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Piantedosi, R., N. Ghyselinck, W. S. Blaner, and S. Vogel. 2005. Cellular retinol-binding protein type III is needed for retinoid incorporation into milk. J. Biol. Chem. 280:24286-24292. [DOI] [PubMed] [Google Scholar]
- 30.Qatanani, M., and M. A. Lazar. 2007. Mechanisms of obesity-associated insulin resistance: many choices on the menu. Genes Dev. 21:1443-1455. [DOI] [PubMed] [Google Scholar]
- 31.Rosen, E. D., and B. M. Spiegelman. 2006. Adipocytes as regulators of energy balance and glucose homeostasis. Nature 444:847-853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ross, A. C. 1993. Cellular metabolism and activation of retinoids: roles of cellular retinoid-binding proteins. FASEB J. 7:317-327. [DOI] [PubMed] [Google Scholar]
- 33.Saari, J. C., M. Nawrot, G. G. Garwin, M. J. Kennedy, J. B. Hurley, N. B. Ghyselinck, and P. Chambon. 2002. Analysis of the visual cycle in cellular retinol-binding protein type I (CRBPI) knockout mice. Invest. Ophthalmol. Vis. Sci. 43:1730-1735. [PubMed] [Google Scholar]
- 34.Schug, T. T., D. C. Berry, N. S. Shaw, S. N. Travis, and N. Noy. 2007. Opposing effects of retinoic acid on cell growth result from alternate activation of two different nuclear receptors. Cell 129:723-733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schupp, M., A. G. Cristancho, M. I. Lefterova, E. A. Hanniman, E. R. Briggs, D. J. Steger, M. Qatanani, J. C. Curtin, J. Schug, S. A. Ochsner, N. J. McKenna, and M. A. Lazar. 2009. Re-expression of GATA2 cooperates with peroxisome proliferator-activated receptor-gamma depletion to revert the adipocyte phenotype. J. Biol. Chem. 284:9458-9464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schupp, M., M. I. Lefterova, J. Janke, K. Leitner, A. G. Cristancho, S. E. Mullican, M. Qatanani, N. Szwergold, D. J. Steger, J. C. Curtin, R. J. Kim, M. Suh, M. R. Albert, S. Engeli, L. J. Gudas, and M. A. Lazar. 2009. Retinol saturase promotes adipogenesis and is downregulated in obesity. Proc. Natl. Acad. Sci. U. S. A. 106:1105-1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schwarz, E. J., M. J. Reginato, D. Shao, S. L. Krakow, and M. A. Lazar. 1997. Retinoic acid blocks adipogenesis by inhibiting C/EBPbeta-mediated transcription. Mol. Cell. Biol. 17:1552-1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sethi, J. K., and A. J. Vidal-Puig. 2007. Thematic review series: adipocyte biology. Adipose tissue function and plasticity orchestrate nutritional adaptation. J. Lipid Res. 48:1253-1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shao, D., and M. A. Lazar. 1997. Peroxisome proliferator activated receptor gamma, CCAAT/enhancer-binding protein alpha, and cell cycle status regulate the commitment to adipocyte differentiation. J. Biol. Chem. 272:21473-21478. [DOI] [PubMed] [Google Scholar]
- 40.Smas, C. M., D. Kachinskas, C. M. Liu, X. Xie, L. K. Dircks, and H. S. Sul. 1998. Transcriptional control of the pref-1 gene in 3T3-L1 adipocyte differentiation. Sequence requirement for differentiation-dependent suppression. J. Biol. Chem. 273:31751-31758. [DOI] [PubMed] [Google Scholar]
- 41.Smas, C. M., and H. S. Sul. 1993. Pref-1, a protein containing EGF-like repeats, inhibits adipocyte differentiation. Cell 73:725-734. [DOI] [PubMed] [Google Scholar]
- 42.Sul, H. S. 2009. Pref-1: role in adipogenesis and mesenchymal cell fate. Mol. Endocrinol. 23:1717-1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Todaro, G. J., and H. Green. 1963. Quantitative studies of the growth of mouse embryo cells in culture and their development into established lines. J. Cell Biol. 17:299-313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tong, Q., G. Dalgin, H. Xu, C. N. Ting, J. M. Leiden, and G. S. Hotamisligil. 2000. Function of GATA transcription factors in preadipocyte-adipocyte transition. Science 290:134-138. [DOI] [PubMed] [Google Scholar]
- 45.Tong, Q., J. Tsai, G. Tan, G. Dalgin, and G. S. Hotamisligil. 2005. Interaction between GATA and the C/EBP family of transcription factors is critical in GATA-mediated suppression of adipocyte differentiation. Mol. Cell. Biol. 25:706-715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tsutsumi, C., M. Okuno, L. Tannous, R. Piantedosi, M. Allan, D. S. Goodman, and W. S. Blaner. 1992. Retinoids and retinoid-binding protein expression in rat adipocytes. J. Biol. Chem. 267:1805-1810. [PubMed] [Google Scholar]
- 47.Villena, J. A., C. S. Choi, Y. Wang, S. Kim, Y. J. Hwang, Y. B. Kim, G. Cline, G. I. Shulman, and H. S. Sul. 2008. Resistance to high-fat diet-induced obesity but exacerbated insulin resistance in mice overexpressing preadipocyte factor-1 (Pref-1): a new model of partial lipodystrophy. Diabetes 57:3258-3266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vogel, S., C. L. Mendelsohn, J. Mertz, R. Piantedosi, C. Waldburger, M. E. Gottesman, and W. S. Blaner. 2001. Characterization of a new member of the fatty acid-binding protein family that binds all-trans-retinol. J. Biol. Chem. 276:1353-1360. [DOI] [PubMed] [Google Scholar]
- 49.Wang, M. Y., P. Grayburn, S. Chen, M. Ravazzola, L. Orci, and R. H. Unger. 2008. Adipogenic capacity and the susceptibility to type 2 diabetes and metabolic syndrome. Proc. Natl. Acad. Sci. U. S. A. 105:6139-6144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang, Y., and H. S. Sul. 2009. Pref-1 regulates mesenchymal cell commitment and differentiation through Sox9. Cell Metab. 9:287-302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Weisberg, S. P., D. McCann, M. Desai, M. Rosenbaum, R. L. Leibel, and A. W. Ferrante, Jr. 2003. Obesity is associated with macrophage accumulation in adipose tissue. J. Clin. Invest. 112:1796-1808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yang, Q., T. E. Graham, N. Mody, F. Preitner, O. D. Peroni, J. M. Zabolotny, K. Kotani, L. Quadro, and B. B. Kahn. 2005. Serum retinol binding protein 4 contributes to insulin resistance in obesity and type 2 diabetes. Nature 436:356-362. [DOI] [PubMed] [Google Scholar]
- 53.Ziouzenkova, O., G. Orasanu, M. Sharlach, T. E. Akiyama, J. P. Berger, J. Viereck, J. A. Hamilton, G. Tang, G. G. Dolnikowski, S. Vogel, G. Duester, and J. Plutzky. 2007. Retinaldehyde represses adipogenesis and diet-induced obesity. Nat. Med. 13:695-702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zizola, C. F., G. J. Schwartz, and S. Vogel. 2008. Cellular retinol-binding protein type III is a PPARgamma target gene and plays a role in lipid metabolism. Am. J. Physiol. Endocrinol. Metab. 295:E1358-E1368. [DOI] [PMC free article] [PubMed] [Google Scholar]