Abstract
To prevent aneuploidy, cells require a mitotic surveillance mechanism, the spindle assembly checkpoint (SAC). The SAC prevents metaphase/anaphase transition by blocking the ubiquitylation and destruction of cyclin B and securin via the Cdc20-activated anaphase-promoting complex or cyclosome (APC/C)-mediated proteolysis pathway. This checkpoint involves the kinetochore proteins Mad2, BubR1, and Cdc20. Mad2 and BubR1 are inhibitors of the APC/C, but Cdc20 is an activator. Exactly how the SAC regulates Cdc20 via unattached kinetochores remains unclear; in vertebrates, most current models suggest that kinetochore-bound Mad2 is required for initial binding to Cdc20 to form a stable complex that includes BubR1. Here, we show that the Mad2 kinetochore dimerization recruitment mechanism is conserved and that the recruitment of Cdc20 to kinetochores in Drosophila requires BubR1 but not Mad2. BubR1 and Mad2 can bind to Cdc20 independently, and the interactions are enhanced after cells are arrested at mitosis by the depletion of Cdc27 using RNA interference (RNAi) in S2 cells or by MG132 treatment in syncytial embryos. These findings offer an explanation of why BubR1 is more important than Mad2 for SAC function in flies. These findings could lead to a better understanding of vertebrate SAC mechanisms.
The spindle assembly checkpoint (SAC) is a mitotic surveillance mechanism that negatively regulates the activation of the anaphase-promoting complex or cyclosome (APC/C)-mediated proteolysis pathway to prevent the destruction of two key substrates, cyclin B and securin, thereby inhibiting the metaphase-to-anaphase transition until bipolar attachment of all chromosomes has been achieved (35). A number of conserved kinetochore proteins have been identified as SAC components, such as Mad1, Mad2, Bub1, BubR1, Bub3, Mps1, Zw10, and Rod and Aurora B kinase (reviewed by Musacchio and Salmon [35]). In vertebrates, it is believed that a diffusible inhibitory “wait anaphase” signal is generated from unattached kinetochores or lack of spindle tension (27, 45, 47) and that its primary target is Cdc20/Fzy (Fzy is the Drosophila Cdc20 homolog that we refer to as Cdc20 here), which is an essential APC/C activator (35). Mad2, BubR1 (Mad3 in Saccharomyces cerevisiae), Bub3, and Cdc20 have been found in the mitotic checkpoint complex (MCC) or its subcomplexes Bub3-BubR1-Cdc20 and Mad2-Cdc20 (42, 50, 56). Kinetochore-dependent recruitment and activation of Mad2 have been illustrated in a “template” model (12) and later a modified “two-state” model (28, 32, 35, 36, 40, 57). This model suggests that a kinetochore-bound and conformationally rearranged Mad2 is required for Cdc20 binding and that it leads to the formation of the Mad2-Cdc20 complex (8, 9, 12, 16, 48, 49). This is further supported by a more recent report that unattached kinetochores from purified HeLa cell chromosomes can catalytically generate a diffusible Cdc20 inhibitor when presented with kinetochore-bound Mad2 and that these purified chromosomes can also promote BubR1 binding to APC/C-Cdc20 by acting directly on Mad2 but not BubR1 (27). In vitro assays also suggest that Mad2 is required for Cdc20 binding to BubR1 (7, 10, 19). Fluorescence recovery after photobleaching analysis has suggested that the ∼50% of green fluorescent protein (GFP)-Cdc20 that associates with slow-phase kinetics on PtK2 cell kinetochores is Mad2 dependent (22). However, contradictory reports also exist to suggest that Mad2 might not be required for Cdc20 kinetochore localization in Xenopus and PtK2 cells (22) and that BubR1 might play a crucial role for this in human cell lines (33). In contrast to the above-mentioned slow-phase GFP-Cdc20, the remaining ∼50% of GFP-Cdc20 that associates with fast kinetics on prometaphase or metaphase kinetochores is Mad2 independent, and its kinetics parallel those of GFP-BubR1 in PtK2 cells. GFP-Cdc20 is still detectable on kinetochores through anaphase, where both Mad2 and BubR1 are greatly reduced (22, 25). Moreover, the direct requirement for the kinetochore in the formation of the SAC-inhibitory complexes has been challenged by a non-kinetochore-based formation hypothesis, with MCC found to be present in HeLa cells during S phase (50) and complex formation in yeast previously shown to be independent of intact kinetochores (17, 43). Therefore, despite the importance of Cdc20 in understanding SAC mechanisms, exactly how the SAC regulates Cdc20 via unattached kinetochores remains unclear in vertebrates.
Drosophila melanogaster is a well-established model used to study the spindle assembly checkpoint (2, 3, 6, 39). More recently, phenotypes of two mad2-null Drosophila mutant alleles, mad2Δ and mad2P, have been characterized, showing that Mad2 protein is not essential for normal mitotic progression but remains essential for SAC when microtubule attachment, chromosome alignment, and congression are abnormal (5). This contrasts with its counterpart in mouse and human (14, 34, 54) and is also different from the lethality phenotypes reported for bubR1 and cdc20 mutations in Drosophila (3, 11). It has also been reported that Mad2 is less important for SAC than BubR1 and that it is regulated differently in Drosophila S2 culture cells (39). These observations led to the tentative conclusion that Drosophila Mad2 may possess different kinetochore molecular mechanisms and function differently from its homologs in mouse and human (14, 34, 54, 58). We therefore tested Mad2 kinetochore function and further investigated the mechanisms required for Cdc20 kinetochore recruitment and localization using Drosophila transgenic and mutant lines, as well as culture cells. We have characterized a new mad2-null mutant allele, mad2EY, and demonstrated that Drosophila possesses a highly conserved Mad2 kinetochore dimerization mechanism required for SAC function. However, Mad2 is not required for Cdc20 kinetochore recruitment and localization. Instead, there is an essential role for BubR1 in this mechanism during normal mitosis and SAC activation.
MATERIALS AND METHODS
Mutant fly stocks.
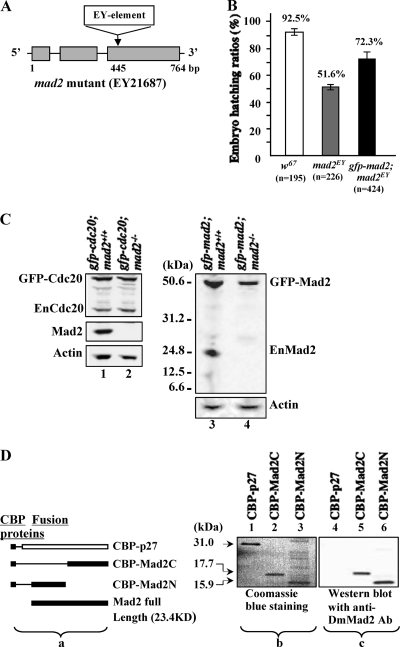
A mad2EY mutant (EY21687 or CG17498; stock no. 22495) stock was purchased from the Baylor Gene Disruption Project (BDGP). This mutant contains an EY element insertion 445 bp in front of the third exon region of the gene on the third chromosome (Fig. 1A). Western blot analysis confirmed this as a null mutation of the gene, as no endogenous or truncated form of Mad2 was detected (Fig. 1C, lanes 2 and 4). The flies could be maintained as a stable laboratory stock under normal cultivation conditions (18 to 25°C). The female mutant flies lay only 60% as many eggs as the w67 control line (data not shown). Only around 51.6% of the eggs laid hatch, compared to a 92.5% hatching ratio in the control line (Fig. 1B). The bubR1K03113 (y[1] w[67c23]; P{w[+mC] = lacW} Bub1[k03113]/CyO) (stock number 10526) mutant line was obtained from the Bloomington stock center. The mutation is caused by a P{lacW} insertion in the 5′ untranslated region of the bubR1 gene 48 bp upstream of the initiator ATG codon (see Fig. 5A). The fly carries a recessive lethal phenotype. The lethality occurs during third-instar larval or pupal stages. The Curly of Oster (CyO) balancer in this line has been replaced with an SM5-TM6 Tb compound balancer to allow identification and isolation of homozygous first- or second-instar larvae for dissection of brains for neuroblast studies at their third-instar larval stage of development. The lack of detectable BubR1 protein from neuroblast samples, as shown by Western blotting (see Fig. 5B, lane 3, top), confirmed this as a null line.
FIG. 1.
Characterization of the mad2EY mutant. (A) Schematic drawing showing the mad2EY mutant caused by an EY element insertion into the third exon at bp 445 of the mad2 gene region (4). (B) The mad2EY mutant can be maintained as laboratory homozygous stock with approximately 50%-reduced viability. This reduced-viability phenotype can be partially rescued by introducing an ectopically expressed transgene of gfp-mad2 (TM2-20) or mad2-Ypet (TM2-27; data not shown) on the second chromosome. The error bars indicate standard deviations. (C) Western blot results. (Lanes 1 and 2) GFP-Cdc20 protein in w67 and mad2EY-null mutant syncytial embryos, respectively. GFP-Cdc20 expression was around 2-fold that of endogenous Cdc20. (Lane 3) GFP-Mad2 and endogenous Mad2 expression in w67 embryos. Lanes 2 and 4 show no detectable endogenous Mad2, or any of its truncated forms, from mad2EY mutant embryo samples probed with an affinity-purified anti-full-length-DmMad2 antibody. (Lane 4) GFP-Mad2 was expressed at a level equivalent to that of the endogenous Mad2 in control samples (lane 3). Actin bands acted as loading controls. (D) (a) Schematic drawing showing the CBP-tagged fusion proteins used to test the anti-DmMad2 antibody. (b) Coomassie blue-stained 10% SDS-PAGE gel showing three purified fusion proteins, with molecule sizes indicated. The sample loading was as follows: lane 1, CBP-p27 (∼0.014 mg); lane 2, CBP-Mad2C (∼0.012 mg); lane 3, CBP-Mad2N (∼0.016 mg). (c) Western blot results showing that anti-DmMad2 antibody (Ab) can detect both the N- and C-terminal halves of truncated Mad2 fusions but not CBP-p27. Lane 4, CBP-p27; lane 5, CBP-Mad2C; lane 6, CBP-Mad2N. The samples loaded were 10 times less than those loaded for Coomassie blue staining. CBP-p27 (CBP plus full-length p27 protein) was used as the control to rule out nonspecific CBP signals.
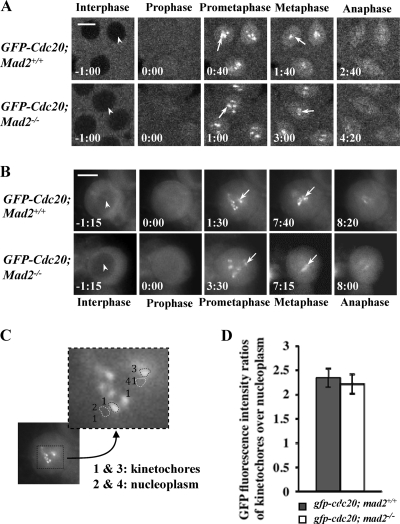
FIG. 5.
Cdc20 kinetochore recruitment and localization is Mad2 independent. (A and B) A gfp-cdc20 transgene on the second chromosome was introduced into the mad2 wild-type (w67) and the mad2EY-null mutant lines, and time-lapse images were acquired for comparison: confocal images show GFP-Cdc20 in syncytial embryos (A), and DeltaVision fluorescence microscope images show GFP-Cdc20 in third-instar larval neuroblasts (B). (Top rows) Wild-type embryos. (Bottom rows) mad2EY-null (mad2−/−) mutant embryos. The arrowheads indicate the exclusion of GFP-Cdc20 signal from the interphase nucleus. The arrows indicate kinetochore-associated GFP-Cdc20. Bar = 5 mm. (C) Sample image showing a selection of the region of interest on individual kinetochores or nearby nucleoplasm regions for fluorescence intensity quantification and ratio analysis of the GFP-Cdc20 kinetochore signal at its peak time. (D) Quantitative comparison of GFP-Cdc20 kinetochore/nucleoplasm ratio signals between the wild type and mad2EY-null mutant; 16 kinetochores from 4 different embryos or neuroblasts were analyzed. The error bars indicate standard deviations.
P-element-mediated transgenic constructs.
Full-length Drosophila cdc20 (CG4274) (44), mad2 (CG17498), and egfp (see Fig. 8A) cDNAs were modified by PCR to incorporate necessary restriction enzyme sites into the DNA fragments for sequential cloning. For egfp, a PCR fragment was generated using primers 5′-GAGCTCGGTACCATGAGTAAAGGAGAAGAACTT-3′ (forward primer with additional SacI-KpnI sites [underlined] just before the egfp ATG) and 5′-GCGGAATTCCTTGTACAGCTCGTCCA-3′ (reverse primer with an additional EcoRI site [underlined] added to the 3′ end of egfp, replacing the stop codon). For mad2, the primers used were 5′-GCGGAATTCATGTCAACTGCCCAGGCGACC-3′ (forward primer with an additional EcoRI site [underlined] upstream of the mad2 ATG) and 5′-GCGGGTACCTTAAGTGCTCATCTTGTAGTT-3′ (reverse primer with an additional KpnI site [underlined] after the stop codon). For Cdc20 (Fzy), the primers used were 5′-GCGGAATTCTCGCAGTTCAATTTTGTGAGC-3′ (forward primer with an additional EcoRI site [underlined] upstream of the Fzy ATG) and 5′-GCGGGTACCTCAACGGATGCTCTGTCGGAA-3′ (reverse primer with an additional KpnI site [underlined] just after the stop codon). Appropriate PCR products were first cloned into the pBluescript vector at SacI and KpnI sites to assemble the full-length SacI-KpnI-gfp-mad2 (or gfp-fzy)-KpnI cDNA construct and for sequencing verification (using DNA sequencing and service, University of Dundee, United Kingdom). Fusion DNA fragments of gfp-mad2 (or gfp-fzy) were then isolated and subcloned into the pWR-pubq P-element-mediated Drosophila transformation vector (44), which placed these cDNAs under the control of a polyubiquitin promoter. To make gfp-mad2ΔC, plasmid DNA of gfp-mad2 in the pBluescript vector was used as a DNA template; the PCR primers used were 5′-GAGCTCGGTACCATGAGTAAAGGAGAAGAACTT-3′ (forward primer with additional SacI-KpnI sites [underlined] just before the egfp ATG) and 5′-GCGACTAGTCACGGTGTCCACCTTGTGCA-3′ (reverse primer with an SpeI site [underlined] added to the 3′ end of mad2 after a 24-bp deletion). The sequence of the KpnI-gfp-mad2C-SpeI DNA fragment was verified before it was directly subcloned into the pWR-pubq vector as mentioned above.
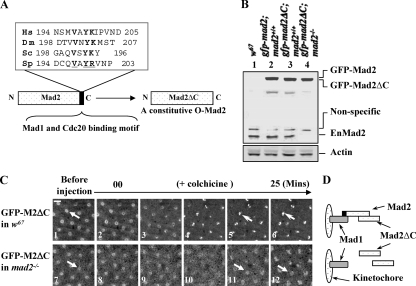
FIG. 8.
C-Mad2 and O-Mad2 can form heterodimers on kinetochores in living syncytial embryos, and O-Mad2 kinetochore localization is essential for Mad2-dependent SAC function. (A) Schematic drawing and sequence alignments showing the last 10 C-terminal amino acids of Mad2 from different species; the conserved residues are underlined and shown in boldface. The DmMad2ΔC construct with the deletion of the last 10 residues is illustrated. (B) Western blot results showing GFP-Mad2 and GFP-Mad2ΔC expression levels in syncytial embryos compared with endogenous Mad2 levels. Lane 1, w67 sample; lane 2, GFP-Mad2 and endogenous Mad2 levels in w67 embryo; lane 3, GFP-Mad2ΔC and endogenous Mad2 levels in w67 embryo; lane 4, GFP-Mad2ΔC level in mad2EY-null mutant embryo. Actin bands acted as loading controls. (C) GFP-Mad2ΔC can localize to kinetochores (top row, open arrows) in w67 embryos when endogenous Mad2 was present (top row, closed arrow indicates a nucleus before colchicine treatment) but not in a mad2EY-null mutant embryo (bottom row, closed arrows); both living embryos were treated with colchicine as described in Materials and Methods. Bar = 10 mm. (D) Schematic drawing showing the likely kinetochore behavior of GFP-Mad2ΔC in both scenarios.
pCal-n+mad2N and pCal-n+mad2C plasmid DNA construction.
A full-length mad2 cDNA PCR product was generated using the primers 5′-GCGGAATTCAAATGTCAACTGCCCAGGC-3′ (forward primer with the addition of an EcoRI site [underlined] upstream of the mad2 ATG) and 5′-GCGAAGCTTCGTGCTCATCTTGTAGTTG-3′ (reverse primer with the addition of a HindIII site [underlined] to the end of the mad2 sequence after the stop codon). The EcoRI-mad2-HindIII PCR fragment was first subcloned into the pGEM-T (Promega) cloning vector. The unique intrinsic XhoI site in the middle of mad2 was used to release two fragments: EcoRI-mad2N-XhoI (303 bp) and XhoI-mad2C-HindIII (298 bp). These two DNA fragments were then subcloned into a pCal-n (Stratagene) expression vector at the EcoRI-XhoI and XhoI-HindIII sites, respectively, resulting in two truncated mad2 fragments in frame with the upstream calmodulin binding peptide (CBP) sequence from the pCal-n vector. The two generated plasmid DNAs were checked by sequencing and transformed into BL21 competent cells for fusion protein expression under IPTG (isopropyl-β-d-thiogalactopyranoside) induction. Samples were later used for Western blotting. Fusion proteins containing CBP and the Mad2 N- or C-terminal half protein (16.0 and 17.7 kDa, respectively) were detectable by Mad2 antibody (Fig. 1D). CBP-p27 affinity-purified protein was used as a negative control.
Transgenic flies.
The constructs described above were used to generate a number of transgenic Drosophila lines that expressed GFP-Mad2, GFP-Cdc20, and GFP-Mad2ΔC fusion proteins in a w67 genetic background, as described previously (44). His2BmRFP transgenic fly lines driven by a ubiquitin promoter were a kind gift from Yohanns Bellaïche at UMR 144 CNRS/Institut Curie, Paris, France.
Time-lapse confocal microscopy.
Transgenic embryos of the appropriate genotype were observed using time-lapse confocal microscopy as described previously (44). A Leica TCS SP2 confocal microscope was used for imaging. The images were transported into Adobe Photoshop or MetaMorph offline for quantification of the fluorescence intensities. Movies of the embryos were then compiled in MetaMorph.
Antibodies.
The affinity-purified Cdc20 (44) and Cdc27 rabbit polyclonal antibodies used were described in previous reports (23). Anti-Cdc20 antibody is not suitable for immunofluorescence staining of the syncytial embryos under various fixation conditions (reference 44 and data not shown). Mad2 antibody was generated in rabbit using an affinity-purified, bacterially expressed MBP-full-length-Mad2 fusion protein as an antigen. Antigen injection and bleeding were commercially performed by Eurogentec. The antibodies were affinity purified and stored as described previously (44). BubR1 antibody (a kind gift from C. Sunkel) was preincubated with Drosophila BubR1 homozygous null mutant 3rd instar larval acetone powder for 4 h at room temperature prior to use in order to further reduce nonspecific binding. Mouse monoclonal antibody (AC-15) to beta-actin (Abcam) was used as a loading control.
Coimmunoprecipitation.
GFP-agarose beads (GFP [B-2] AC; sc-9996; mouse monoclonal IgG; Santa Cruz Biotechnology) or Dynal protein A beads (Dynal Biotech) covalently cross-linked to Cdc20 antibody were used for coimmunoprecipitation. Cdc20 antibody (rabbit polyclonal; 0.4 mg/ml) in 0.1 M NaPO4 (pH 8.1) was cross-linked with the beads according to the manufacturer's protocol. The beads were then washed 5 times in 2.5 volumes of 0.1 M Na citrate (pH 3.1) to further remove excess/non-cross-linked antibody, followed by five 2.5-volume washes with 0.1 M NaPO4. Prior to use, the beads were blocked at room temperature for 15 min in 100% Odyssey blocking buffer (Infrared Imaging Systems).
(i) Cdc20 coimmunoprecipitation from Schneider S2 cell extracts.
S2 cells were RNA interference (RNAi) treated for 72 h (24). The cells were then lysed in ice-cold Tris lysis buffer (50 mM Tris, pH 8.0, 25 mM NaCl, 1 mM EDTA, pH 8.0, 0.25% NP-40) containing protease inhibitors (1 mM phenylmethylsulfonyl fluoride [PMSF]; 1 mM benzamidine; 0.2 mg/ml [each] aprotinin, leupeptin, and pepstatin) at a concentration of 4 × 107 cells/ml. The cells were placed on ice for 15 min prior to centrifugation at maximum speed for 15 min at 4°C in a benchtop centrifuge. Four milliliters of Cdc20-Dynal beads was added to 100 ml of S2 supernatant and mixed by rotation at 4°C overnight. The beads were then pulled from solution with a magnet and washed 5 times in 2.5 volumes of ice-cold Tris lysis buffer before being resuspended in 1× SDS-PAGE sample buffer and heated at 95°C for 10 min.
(ii) Cdc20 coimmunoprecipitation from syncytial-embryo extracts.
Thirty-minute- to 1.5-hour-old embryos were collected, dechorionated using 60% domestic bleach for 1 min, and thoroughly washed with 1× phosphate-buffered saline (PBS). Some of the embryos were treated with 200 mM MG132 in 1× PBS for 20 min at room temperature before they were used to make embryo extracts. This treatment causes about 80% of embryos to arrest the nuclear division cycle in mitosis (data not shown). Preparation of embryo extracts and coimmunoprecipitation were carried out as described above.
SDS-PAGE was performed as described previously (44). Western blots were transferred to nitrocellulose (Schleicher and Schuell), with blocking and quantification of Western blots performed according to standard Odyssey protocol.
Colchicine treatment.
Colchicine was used at a final intracellular concentration of around 0.25 mM, introduced by microinjection (stock solution, 250 mM in injection buffer; about 0.1% embryo volume per injection) as described previously (44). Images were taken for 20 to 30 min after injection to encompass at least one full nuclear-division cycle.
RESULTS
mad2EY (EY21687) is a null mutant.
mad2EY homozygotes can be maintained as a stable laboratory stock under normal cultivation conditions (18 to 25°C) with reduced viability. Female mutant flies lay 60% of the number of eggs laid by w67 control flies (data not shown). Only around 51.6% of the eggs laid hatched, in contrast to a 92.5% for w67 control flies (Fig. 1B). Ectopically expressed GFP-Mad2 can partially restore the viability phenotype (Fig. 1B). Western blot analysis confirmed that there were no detectable full-length or truncated forms of the endogenous Mad2 in homozygous syncytial embryo samples (Fig. 1C, lanes 2 and 4). The affinity-purified Mad2 antibody was raised against a full-length Drosophila Mad2 protein. The antibody is capable of detecting both the N-terminal and C-terminal halves of the Mad2 proteins (Fig. 1D).
The mad2EY mutant cannot induce a SAC.
Wild-type embryos immediately arrest cell cycle progression at metaphase in response to SAC activation by colchicine treatment, and this can be illustrated by using GFP-Cdc20 as a cell cycle progression marker. When the embryo responds to the arrest, GFP-Cdc20 immediately accumulates and persists on arrested kinetochores for more than 30 min over the time course of the observations (equivalent to three nuclear division cycles) upon treatment with colchicine (Fig. 2, top, shows only 16 min for comparison). However, under the same treatment conditions, the checkpoint arrest was bypassed in mad2EY mutant syncytial embryos, as indicated by the continuing cycles of GFP-Cdc20 on and off kinetochores and of chromatin condensation/decondensation (Fig. 2, middle), although cytokinesis appears to be defective, as would be expected in embryos lacking microtubules. This impaired SAC can be fully restored by introducing the above-mentioned ectopically expressed GFP-Mad2 (Fig. 2, bottom). The data suggest that the mad2EY mutant is a new null mutant.
FIG. 2.
Mad2 is essential for colchicine-invoked SAC function. gfp-cdc20 mad2+/+, gfp-cdc20 mad2EY (mad2−/− null mutant), and gfp-mad2 mad2EY embryos were treated with colchicine by microinjection. Time-lapse confocal images were taken before (01, 07, and 13) or after (02 to 06, 08 to 12, and 14 to 18) injection. GFP-Cdc20 or GFP-Mad2 kinetochore signals were used as cell cycle progression markers. (Top row) The arrows indicate the arrested kinetochores in images 02 to 06. The area marked by an arrow in image 01 indicates that before colchicine treatment, GFP-Cdc20 was excluded from the late-interphase nucleus. (Middle row) In the absence of endogenous Mad2, GFP-Cdc20 signals continued to oscillate in and out of the nucleus and on and off the kinetochores in the presence of colchicine (indicated by arrows in images 07 to 12). Separation of daughter nuclei failed in image 10. (Bottom row) Arrowheads in images 14 to 18 indicate arrested kinetochores with accumulated GFP-Mad2 fusion proteins. The arrowhead in image 13 indicates GFP-Mad2 accumulation in a late-interphase nucleus. Bar = 5 mm.
GFP-Cdc20 and GFP-Mad2 can mimic dynamic distribution and localization of their endogenous proteins and are functional.
To determine whether Mad2 is required for Cdc20 kinetochore recruitment and localization, we first examined and compared the subcellular localizations of Cdc20 and Mad2 using GFP-Cdc20 or GFP-Mad2 in living transgenic syncytial embryos. The GFP-Cdc20 (24) or GFP-Mad2 fusion protein was expressed in transgenic-fly lines under the control of a polyubiquitin promoter. GFP-Mad2 can fully rescue the checkpoint defect in null mutant embryos, as discussed above (Fig. 2, bottom), though it only partially restores the viability of mad2EY-null mutant phenotypes (Fig. 1B). The reason for partial rescue is likely that, as GFP-Mad2 is under regulation by a polyubiquitin promoter, its spatial and temporal expression may not be identical with that of the endogenous Mad2 during development. It is also possible that targeting Cdc20 may require different concentrations of Mad2 to rescue the viability and SAC defect in mad2−/− flies. As GFP-Mad2 is fully functional in SAC, it is not a problem to use it to study the kinetochore recruitment and interaction mechanisms of Mad2, BubR1, and Cdc20. GFP-Mad2 in living syncytial embryos shows the same distribution as the endogenous Mad2 in S2 cells and is consistent with previous reports (6), in that Mad2 is mainly found in the nucleolus in interphase and starts to associate with kinetochores in prophase (compare Fig. 3A and D). Similarly, GFP-Mad2 can be detected on centrosomes during mitosis (compare Fig. 3C and D). GFP-Cdc20 has been successfully used as a surrogate for localization of the endogenous Cdc20 protein (22, 44) and is functional, as it can restore the viability of a weak allele of a Cdc20 mutant (P{EP}FzyEP1028, from Szeged, Hungary) (data not shown). Consistent with previous reports (44), GFP-Cdc20 enters the nucleus after nuclear envelope breakdown (NEB), and its entry was clearly visible in all of our experiments, whether in syncytial embryos or neuroblasts. Entry of GFP-Cdc20 into the nucleus after NEB occurs just as chromatin assumes a prophase configuration, as illustrated using coexpressed his2BmRFP as a chromatin marker (Fig. 3B2 and B3 for GFP-Cdc20, B9 and B10 for DNA, and B16 and B17 for merged images). Thus, the entry of GFP-Cdc20 into the nucleus was used as a prophase marker to allow comparison of the timing of cell cycle progression in subsequent experiments. GFP-cdc20 was chosen as a cell cycle marker in our experiments rather than GFP-cyclin B from transgenic flies, which we had previously characterized (23), for a number of reasons. The GFP-cyclin B transgenic flies express this fusion protein at a relatively low level, and it requires four copies of the transgene to detect the protein, and even then, it is not detectable/visible until around syncytial nuclear-division cycles 10 and 11. A further advantage of using these flies was that it allowed us to maintain genetic consistency throughout the experiments.
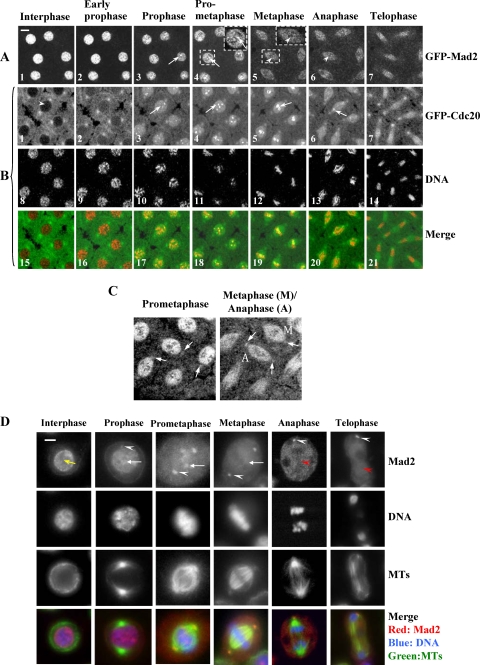
FIG. 3.
Comparison of the dynamic kinetochore localization of GFP-Mad2 and GFP-Cdc20 in living syncytial embryos and endogenous Mad2 localization in fixed S2 cells. (A) GFP-Mad2 kinetochore signals could be detected only at prophase and prometaphase (arrowheads in A3, A4, and insets), disappearing from metaphase and anaphase kinetochores (arrowheads in A5, inset, and A6). (B) GFP-Cdc20 could be readily observed on prophase and prometaphase kinetochores (B3 and B4, arrows) and persisted on metaphase and anaphase kinetochores (B5 and B6, arrows). GFP-Cdc20 was excluded from the interphase nucleus (arrowhead), entering the nucleus by early prophase. Chromatin morphologies were determined using coexpressed His2BmRFP as a marker (B8 to B14). The developmental stage of the syncytial embryos was nuclear division cycles 7 and 8. Time-lapse images were recorded using a Leica TCS SP2 confocal system at 22°C. The data are representative of at least 10 experiments. (C) Confocal images showing GFP-Mad2 association with prometaphase, metaphase (M), and anaphase (A) centrosomes (arrows) taken from another GFP-Mad2 transgenic syncytial time-lapse movie in which centrosomes were better focused during the confocal laser scanning. (D) Cold-methanol-fixed Drosophila S2 cells stained with anti-tubulin antibody (Dm 1a) (green), anti-full-length-DmMad2 antibody (affinity purified) (red), and DNA stained with propidium iodide (blue). White arrowheads, Mad2 on centrosomes; arrows, Mad2 in interphase nucleus, at prometaphase kinetochores, or no longer visible on metaphase kinetochores; red arrowheads, Mad2 associated with midbody microtubules. Bars = 5 mm.
Confocal images indicated that GFP-Mad2 and GFP-Cdc20 both localize to prophase and prometaphase kinetochores (Fig. 3A3, A4, and inset; B3, B4, B17, and B18). GFP-Mad2 disappears from metaphase kinetochores (Fig. 3A5 and inset), whereas some of the the GFP-Cdc20 signal persists on kinetochores through anaphase (Fig. 3B5, B6, B19, and B20; see Movie S1 in the supplemental material for GFP-Cdc20 kinetochore dynamic localization throughout the cell cycle) (44), as has been observed, though not remarked upon, in HeLa cells (22, 26). These data suggest that Cdc20 kinetochore localization might not require colocalization of Mad2, at least after metaphase.
The sensitivities of kinetochore accumulation of GFP-Mad2 and GFP-Cdc20 in response to SAC activation are different.
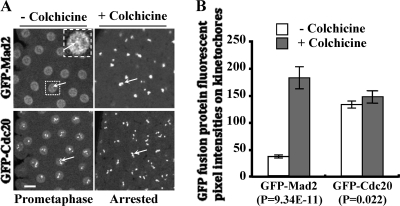
Mad2 is prominently localized to unattached prometaphase kinetochores, and this localization is enhanced upon SAC activation (6, 21, 53). We would expect the response of GFP-Cdc20 to SAC activation to be similar to that of GFP-Mad2 if Cdc20 kinetochore localization were Mad2 dependent. However, when we used gfp-cdc20 mad2+/+ or gfp-mad2 mad2+/+ transgenic embryos to measure the kinetochore fluorescence intensities of GFP-Cdc20 or GFP-Mad2 in the presence of endogenous Cdc20 and Mad2 proteins, respectively, the results showed that the GFP-Cdc20 fusion protein did not increase significantly on the kinetochores after colchicine treatment, despite arrest of the cell cycle (Fig. 4A, top, and B). In contrast, a 4-fold increase of kinetochore-associated GFP-Mad2 was detected under the same conditions (Fig. 4A, bottom, and B). Quantification of the Western blot results suggested that GFP-Cdc20 is expressed at a level about 1.4-fold that of the endogenous Cdc20 and GFP-Mad2 is expressed at a level 1.2-fold that of the endogenous Mad2 (data not shown). The ratio of endogenous Cdc20 to Mad2 levels in the syncytial embryo (30- to 90-min-old embryos) was about 2.0, and this increased to 4.9-fold in gfp-cdc20 mad2+/+ embryos and decreased to 1.1 in gfp-mad2 mad2+/+ embryos when GFP-Cdc20 or GFP-Mad2 levels were calculated (data not shown). Therefore, the different increments of GFP-Cdc20 and GFP-Mad2 on kinetochores in response to colchicine treatments are independent of the relative expression levels of these proteins. The presence of endogenous Mad2 in gfp-cdc20 mad2+/+ embryos suggests that Mad2 is less likely to play a catalytic role for GFP-Cdc20 kinetochore localization in response to SAC activation, as GFP-Cdc20 did not increase significantly at the kinetochores before (at normal prometaphase) or after colchicine treatments in these flies (Fig. 4A, top, and B) or compared to gfp-cdc20 mad2−/− (Mad2-null) flies (Fig. 2, top and middle). A similar increment of human Mad2 at the kinetochore has also been observed when the SAC is active in HeLa cells (20). This suggests that the sensitivities of kinetochore accumulation of GFP-Mad2 and GFP-Cdc20 in response to SAC activation are different.
FIG. 4.
Quantitative comparison of the kinetochore increment signals of GFP-Mad2 and GFP-Cdc20 in response to SAC activation. (A) (Left) Images of normal prometaphase embryos taken when GFP-Mad2 or GFP-Cdc20 kinetochore signals had reached their peaks. GFP-Mad2 kinetochore signals in the inset image (top right) could be viewed only after both brightness and contrast were increased. (Right) Images of colchicine-treated embryos taken when kinetochore GFP-Mad2 or GFP-Cdc20 (arrows) had accumulated to its highest level. The images were acquired using a Leica TCS P2 confocal microscope at consistent minimum laser power and settings to avoid signal saturation of colchicine-treated samples. (B) Comparison of the quantitative results of the kinetochore fluorescence intensities (FL) (n = 16 kinetochores from 4 different embryos) of GFP-Mad2 and GFP-Cdc20 from the confocal images in panel A. Bar = 10 mm. The error bars indicate standard deviations.
Recruitment and localization of Cdc20 on kinetochores is Mad2 independent.
To test directly whether Mad2 plays a part in the localization of Cdc20 to the kinetochore at the SAC, we introduced a copy of the gfp-cdc20 transgene into the mad2EY mutant background, in which the fusion protein expression level is around 1.5- to 2-fold greater than that of the endogenous Cdc20 (Fig. 1C, lanes 1 and 2). We used this to examine GFP-Cdc20 localization in syncytial embryos (Fig. 5A) and neuroblasts (Fig. 5B) during normal mitosis with or without the presence of endogenous Mad2. Interestingly, given the importance of Mad2 to the SAC mechanism, the dynamic localization patterns of GFP-Cdc20 in living cell compartments, especially on and off the kinetochores, were indistinguishable in the mad2EY-null mutant and the wild-type backgrounds in both syncytial embryos (Fig. 5A) and neuroblasts (Fig. 5B). Comparison of quantification results of the peak time fluorescence intensity ratios of GFP-Cdc20 at kinetochores to the nearby nucleoplasm in wild-type or Mad2-null mutant neuroblasts showed no differences in the levels of recruitment of GFP-Cdc20 onto kinetochores in mitosis (Fig. 5A and B). Thus, it is clear that Cdc20 kinetochore recruitment and localization are Mad2 independent during normal mitosis.
BubR1 is required for proper localization of GFP-Cdc20 onto kinetochores in the Drosophila neuroblast.
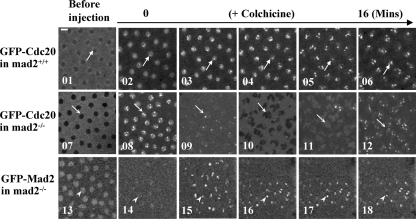
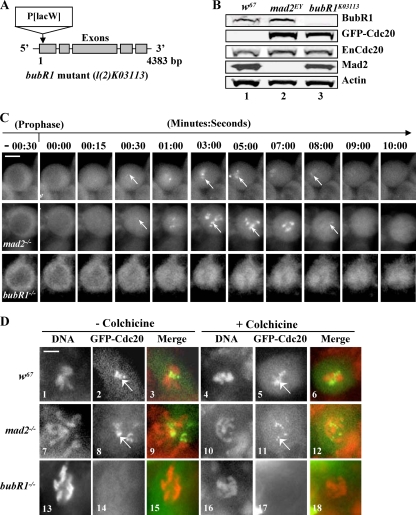
We next tested whether BubR1 was required for Cdc20 kinetochore localization. A bubR1-null mutant (Bub1K03113; CG7838) (Fig. 6A) with a lethality phenotype at the larval/pupal transition stage was obtained from the Bloomington stock center (3, 29). Western blotting confirmed the lack of detectable BubR1 in the neuroblast samples from the homozygous mutant (Fig. 6B, lane 3, top). A copy of the gfp-cdc20 transgene was also introduced into the mutant on the third chromosome, and the fusion protein expression was confirmed by Western blotting (Fig. 6B, lane 3). The behavior of GFP-Cdc20 in homozygous neuroblasts was then examined, and the entry of GFP-Cdc20 into the nucleus was used as a prophase marker to time the cell cycle progressions. GFP-Cdc20 was visibly associated with wild-type or mad2EY mutant kinetochores approximately 30 s after it entered the nucleus in neuroblasts; it remained clearly detectable on kinetochores for 8.5 minutes and then disappeared (22°C) (Fig. 6C, top and middle) (44). It was also detectable on fixed wild-type or mad2EY mutant neuroblast kinetochores (Fig. 6D, 1 to 12). In mutant bubR1 neuroblasts, GFP-Cdc20 also entered the nucleus, but in contrast, no detectable kinetochore signal was observed (Fig. 6C, bottom). This was further confirmed with formaldehyde-fixed mitotic bubR1 mutant neuroblast kinetochores (Fig. 6D, 13 to 15) even after colchicine treatment (Fig. 6D, 16 to 18). Thus, it is clear that BubR1 plays an essential role in Cdc20 kinetochore recruitment and localization.
FIG. 6.
BubR1 is required for proper Cdc20 kinetochore recruitment and localization. (A) The bubR1K03113 mutation was generated by a P-element insertion at the beginning of the first exon of the gene. (B) Western blot results. Lane 1, Control wild-type (w67) samples showing endogenous BubR1, Cdc20, and Mad2 protein expression levels; lane 2, GFP-Cdc20 expression levels in mad2EY-null mutant background; lane 3, no detectable BubR1 signals from homozygous BubR1K03113 third-instar larval brain samples (top). The GFP-Cdc20 fusion proteins were expressed at levels around 1.5- to 2-fold greater than their endogenous counterparts in both mad2EY- and bubR1-null mutant brain samples (lanes 2 and 3). Actin bands acted as loading controls. (C) The top and middle rows show clear kinetochore GFP-Cdc20 signals in w67 and mad2EY-null mutant neuroblasts. The arrows indicate kinetochore-associated GFP-Cdc20 signals. GFP-Cdc20 kinetochore signals began to be visible around 15 s after NEB. The bottom row shows no detectable GFP-Cdc20 kinetochore signals after NEB in a bubR1 mutant living neuroblast. (D) GFP-Cdc20 signals (arrows) on kinetochores can be easily identified in mitotic neuroblasts from formaldehyde-fixed w67 or mad2EY-null mutant third-instar larval brains in the presence or absence of colchicine. (Top row) Samples from w67: 1 to 3, normal metaphase chromosome and kinetochores; 4 to 6, with colchicine treatment. (Middle row) Samples from mad2EY-null mutant: 7 to 9, normal prometaphase-like chromosome and kinetochores; 10 to 12, with colchicine treatment. (Bottom row) Samples from bubR1K03113: 14 and 17, no detectable GFP-Cdc20 kinetochore signals in homozygous bubR1 mutant third-instar larval brains; 13 to 15, prometaphase-like chromosome with no colchicine treatment; 16 to 18, with colchicine treatment. Mitotic chromosomes were identified using anti-phosphohistone 3 antibody (data not shown). Green, GFP-Cdc20; red, DNA (propidium iodide). Bar = 5 mm.
BubR1 and Mad2 can form a complex with Cdc20 independently of each other.
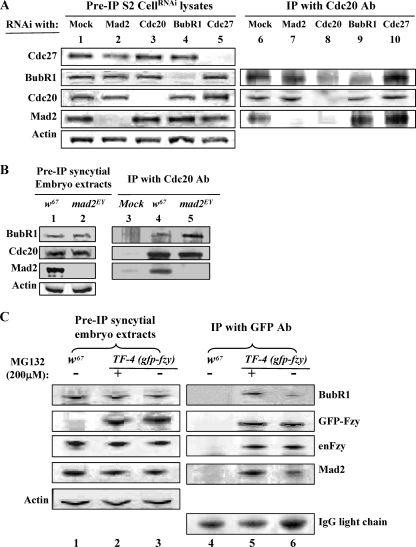
In order to further understand the mechanism of the interaction between BubR1, Cdc20, and Mad2, we performed coimmunoprecipitation experiments using anti-Drosophila Cdc20 antibody with samples collected from S2 culture cells, which were treated with either Mad2 or BubR1 double-stranded RNAi (dsRNAi); from a sample subjected to depletion of Cdc27, a core component of the APC/C; or from syncytial embryos treated with 200 mM MG132 (39) to increase the metaphase-arrested cell population (18, 44) (Fig. 7A, lanes 1 to 5, and C). The results indicated that BubR1 and Mad2 could bind to Cdc20 independently of each other (Fig. 7A, lanes 7 and 9). These interactions were mutually enhanced upon metaphase arrest in extracts made from S2 cells and syncytial embryos (Fig. 7A, lane 10, and C). Formation of a complex between BubR1 and Cdc20 independently of Mad2 was also confirmed by its ability to be coimmunoprecipitated with Cdc20 in extracts made from mad2EY-null syncytial embryos (Fig. 7B, lane 5).
FIG. 7.
Mad2 and BubR1 can bind independently to Cdc20, and their binding is mutually enhanced in metaphase extracts. (A) Lanes 1 to 5 show levels of Mad2, Cdc20, BubR1, and Cdc27 in lysates of cells with mock or RNAi treatment; the levels of the cognate protein were reduced by more than 90% after 72 h of RNAi treatment in each case. Lane 1, mock depletion control samples; lanes 6 to 10, protein levels coimmunoprecipitated (co-IP) with anti-Cdc20 antibody from these lysates; lane 6, control samples with mock depletion showing both BubR1 and Mad2 coprecipitated with Cdc20; lane 7, in the absence of Mad2, BubR1 can still bind to Cdc20; lane 8, no detectable BubR1 and Mad2 coprecipitated when Cdc20 had been depleted; lane 9, in the absence of BubR1, Mad2 can also coprecipitate with Cdc20 to levels similar to that of the control sample (lane 6); lane 10, more BubR1 and Mad2 coprecipitated with Cdc20 from metaphase extracts subjected to Cdc27RNAi depletion. (B) Lanes 1 and 2 show BubR1, Cdc20, and Mad2 protein levels in control (w67) and mad2EY mutant embryo extracts. Extracts were made from overnight embryos. Lane 3, nonspecific coprecipitation of BubR1, Cdc20, and Mad2 with anti-GFP antibody beads; lane 4, both BubR1 and Mad2 proteins coprecipitated with Cdc20 from w67 control samples; lane 5, BubR1 can still coprecipitate with Cdc20 in the absence of Mad2 (sample made from mad2EY-null mutant embryos). Actin bands acted as a loading control. (C) Lanes 1 to 3 show protein levels of Mad2, BubR1, Cdc20, and GFP-Cdc20 in lysates from syncytial embryos with a relevant genotype; actin bands acted as the loading control. Lane 4, nonspecific coprecipitation background with anti-GFP antibody beads; lanes 5 and 6, both BubR1 and Mad2 interactions with Cdc20 were enhanced after embryos (30 min to 1.5 hours old) were treated with 200 mM MG132 for 20 min (lane 5) compared to embryos that had not been treated with MG132 (lane 6). IgG light-chain bands were used as loading controls. The data are representative of three separate experiments.
A conserved mechanism of Drosophila Mad2 recruitment to the kinetochore at the SAC.
To understand the significance of the above findings, it is important to determine whether Drosophila Mad2 plays a conserved SAC role. We therefore tested whether Drosophila shares one of the crucial Mad2 kinetochore recruitment mechanisms in which Mad2 molecules dimerize in different conformations, as demonstrated in other systems (12, 13, 37). A GFP-tagged DmMad2ΔC construct was created by deletion of the 10 C-terminal amino acids of Mad2 (Fig. 8A), which are required for binding to Mad1 and Cdc20 (12). This mutant was used to generate transgenic lines. The deletion of these 10 residues affects the structural stability of the C-Mad2 conformation while leaving the stability of O-Mad2 unaffected, thus creating a constitutively open form of Mad2 (12). This transgene (TM2ΔC-16) on the second chromosome was then introduced into the mad2+/+ wild-type (w67) or the mad2EY-null mutant fly line. The protein expression level of GFP-Mad2ΔC in syncytial embryos was around 1.5- to 2-fold greater than that of the endogenous Mad2 (Fig. 8B, lanes 3 and 4). GFP-Mad2ΔC kinetochore recruitment with or without the presence of the endogenous Mad2 protein under SAC activation by colchicine treatment was then determined in syncytial embryos. The images in Fig. 8C (top) show that DmMad2ΔC (closed arrow; DMMad2ΔC in the nucleus before the SAC) was indeed still recruited to the kinetochore by the SAC in wild-type embryos (open arrows), since endogenous wild-type Mad2 can be recruited onto kinetochores by Mad1 and acts as the closed form of Mad2, dimerizing with Mad2ΔC (shown as a schematic in Fig. 8D). If the stable, closed form is essential for Mad2 recruitment to the kinetochore, we predicted that, in the absence of wild-type Mad2, DmMad2ΔC would not be recruited, nor would it be able to rescue SAC function, unlike wild-type GFP-Mad2 (Fig. 2, bottom). This is what we observed in the mad2EY mutants (Fig. 8C, bottom. closed arrows, and D). Thus, the mechanism of recruitment of Mad2 to the kinetochore under the SAC in Drosophila syncytial embryos has the properties predicted from earlier biochemical works in yeast and human (12, 13, 37). This also further proved that Mad2 kinetochore localization and modification are essential to establish proper spindle checkpoint function.
DISCUSSION
mad2EY (EY21687) is a null mutant.
Any significant interpretation of the GFP-Cdc20 kinetochore localization signals in the mad2EY mutant line relies on the absence of the endogenous Mad2 protein. The mad2EY mutant contains an EY element insertion 445 bp in front of the third exon region of the mad2 gene (Fig. 1A). Therefore, some of the N-terminally truncated forms of Mad2 may still be translated but not detected, as the antibody used could potentially lack the ability to detect the N-terminal region of the Mad2 protein, despite the fact that it was affinity purified and raised against the full-length Drosophila Mad2 protein. This has been tested and can be ruled out, as the antibody can detect both the N- and C-terminal halves of Mad2 fusions (Fig. 1D). Thus, mad2EY is a null mutant. Functional-analysis results also proved that mad2EY lacks the ability to respond to SAC activation (Fig. 2). Moreover, the essential motif for binding of Mad2 to Cdc20 or Mad1 is its last 10 C-terminal amino acids (12, 31); even if truncated N-terminal forms of Mad2 were to remain present, they should not interfere with GFP-Cdc20 kinetochore localization, as they would not be kinetochore localized. This was supported by the fact that the Mad2ΔC construct (which lacks these 10 amino acids) could no longer be recruited onto kinetochores when it was expressed in the mad2EY mutant background (Fig. 8C and D).
Cdc20 kinetochore recruitment is dependent on BubR1 but not Mad2.
We have demonstrated in this study with in vivo results that Cdc20 kinetochore recruitment and localization are Mad2 independent during normal mitosis and after SAC activation (Fig. 5A and B), although we cannot completely rule out the possibility that undetectable residual Mad2 may localize to the kinetochores, where it could act in a catalytic fashion to localize Cdc20. Previous reports have also revealed that Cdc20 is still detectable on SAC-activated kinetochores after depletion of Mad1 in Xenopus (52) and that a Cdc20 fusion protein, Cdc20Δ1-167-GFP, lacking the N-terminal Mad2 interaction domain was still able to localize to kinetochores in PtK2 cells (22, 26). These data suggest that our findings might not be relevant only to Drosophila.
The finding that BubR1 is required for proper Cdc20 kinetochore recruitment and localization (Fig. 5A and B) is supported by previous observations that cell lines carrying BubR1 mutations from cancer patients display reduced Cdc20 kinetochore signals (33). Interestingly, the dynamic localization patterns of GFP-BubR1 in Drosophila neuroblasts (5) or BubR1-RFP (our unpublished data) are very similar to GFP-Cdc20 localizations in both embryos and neuroblasts observed in this study (Fig. 3B, 5, and 6C). This is different, however, from findings in PtK2 cells, where it has been reported that Cdc20 localizes to kinetochores in prophase, when BubR1 has apparently not yet entered the nucleus, and persists on kinetochores through anaphase, while BubR1 was greatly reduced (22). These data suggest that proteins other than BubR1 might also be involved in Cdc20 kinetochore localization, especially after metaphase in vertebrates.
Our observations that Mad2 and BubR1 can form a complex with Cdc20 independently of one other in S2 culture cells and syncytial-embryo extracts (Fig. 7A7, A9, and A10) is consistent with previous findings in human HeLa cell and Xenopus extracts (1, 15, 51). These interactions were mutually enhanced in the extracts made from metaphase-arrested culture cells or embryos, consistent with more recent findings in HeLa cells that increased levels of BubR1, Bub3, Mad2, and Cdc20 were associated with APC/C when the checkpoint is active. This suggests that Mad2 is a stoichiometric component of APC/CMCC (20). However, our above-mentioned findings are inconsistent with some other reports showing that the binding of Cdc20 to an N-terminal region of mouse BubR1 (amino acids 1 to 477) is also Mad2 dependent using HeLa cell extracts or yeast two-hybrid analysis (10, 38, 46). It has also been reported that Cdc20p cannot coimmunoprecipitate with Mad3 (BubR1 in yeast) in extracts made from a nocodazole-treated yeast Mad2-null strain (19) and that immunodepletion of Mad2 prevents Cdc20-BubR1 interaction in human HeLa cell and Xenopus extracts (7, 38), which suggested a catalytic role of Mad2 in promoting an interaction between Cdc20 and BubR1 (27). At present, we do not fully understand the reason for these inconsistencies. However, it is possible that the eukaryotic BubR1 contains multiple binding regions and that the interaction requires a conformational change of Cdc20 in vivo (51), and therefore, the full-length BubR1 may behave differently from the truncated form of the mouse BubR11-477 or Mad3 in yeast. Sequence analyses of mouse, Xenopus, chicken, and human have revealed a conserved amino acid region (FDE) between residues 519 and 529, before the kinase domain of the mouse BubR1 (10). This region can strongly interact with Cdc20 in a Mad2-independent manner, although it possesses no apparent checkpoint function (10, 51). A similar region, but not conserved, from yeast Bub1 also binds to Cdc20 (10). The much shorter BubR1 functional homolog of yeast Mad3 lacks this motif region, as well as the consequential entire kinase domain (10, 46). It would be interesting to determine whether the similar region of Drosophila BubR1 could also bind to Cdc20 to facilitate their interaction on kinetochores. The kinetochore interaction of Cdc20 and BubR1 is likely transient, as both BubR1 and Cdc20 are rapidly turned over on PtK2 cell kinetochores (26). Our finding that Mad2 is not required for Cdc20 kinetochore recruitment and localization does not rule out the idea that Mad2 may still interact with Cdc20 on kinetochores. This transient kinetochore interaction between BubR1 and Cdc20 may provide a conformational change of Cdc20 required for interaction with Mad2, and perhaps this then stabilizes the interaction between Cdc20 and BubR1 to form the MCC, either on or off kinetochores. In support of this, we have observed that GFP-Cdc20 kinetochore signals continued to cycle upon colchicine treatment in a Mad2-null syncytial embryo in which the checkpoint arrest had been bypassed (Fig. 2, middle, 07 to 12). In contrast, GFP-Cdc20 signals persisted on metaphase-arrested kinetochores in Mad2 wild-type syncytial embryos under the same conditions (Fig. 2, top, 01 to 06). FRAP analyses in living PtK2 cells have revealed that both GFP-Cdc20 and GFP-BubR1 have biphasic exponential recruitment kinetics on the kinetochore. In contrast, GFP-Mad2 and GFP-Bub3 display only a single exponential kinetics similar to the slower phase of Cdc20 and BubR1 (22). This slower phase of GFP-Cdc20 recruitment has been shown to depend on Mad2 and has been suggested to represent complex formation with Mad2. However, the significance of the fast-phase kinetochore dynamics associated with the other 50% of Cdc20 remained unexplored (22). Our results suggest that the slower-phase GFP-Cdc20 might be converted from the fast phase once Cdc20 is recruited onto the kinetochore and bound to Mad2 if the two phases are not completely independent of each other in PtK2 cells.
A conserved Mad2 kinetochore recruitment mechanism.
Crystallographic and biochemical studies in Xenopus egg extracts, HeLa cells, and S. cerevisiae have revealed that Mad2 exists in multiple conformations and that they are important for Mad2 interaction on kinetochores (12, 30, 32, 37, 55). These studies have allowed the formulation of a “Mad2 template model” in which Mad1-bound C-Mad2 interacting with O-Mad2 on the kinetochore functions as a platform for assembly of the Cdc20-Mad2 complex. This is a crucial mechanism for SAC function (12, 40, 41, 57). We have demonstrated here that Drosophila Mad2 preserves this same kinetochore-Mad2 interaction mechanism for SAC function (Fig. 8C and D).
In summary, we have found that BubR1 appears to be central and causal in recruiting GFP-Cdc20 to the kinetochore during normal mitosis and following activation of the SAC. The levels of recruitment of GFP-Cdc20 to the kinetochore are similar during normal mitosis and after SAC activation. Mad2 is not required for recruitment of GFP-Cdc20 to the kinetochore either during normal mitosis or following SAC activation. It is not essential for normal mitotic progression, but it has a crucial role in the SAC, with GFP-Mad2 levels increasing markedly on the kinetochore when spindle microtubule damage occurs, perhaps to facilitate the formation and stabilization of the MCC. Thus, SAC activation markedly alters the relative amounts of GFP-Cdc20 and GFP-Mad2 associated with the kinetochore. We propose that the main function of Mad2 is not to recruit Cdc20 to the kinetochore at prophase but to retain Cdc20 on the kinetochore once the SAC is activated.
Supplementary Material
Acknowledgments
This work was supported by a Wellcome Trust project grant to J.-Y.H.
We thank Maureen Sinclair for technical support; Mark Levasseur, Sally Hanton, and Ashleigh Herriott for critical reading; Bernardo Orr, from Claudio E. Sunkel's laboratory (IBMC, Porto, Portugal), for anti-Dm BubR1 antibody; Roger Karess (CNRS, Gif-sur-Yvette, France) for supplying a GFP-Mad2 transgenic line driven by the intrinsic Mad2 promoter for comparison; and Yohanns Bellaïche (UMR 144 CNRS/Institut Curie, Paris, France) for his2BmRFP transgenic lines. We thank Jordan Raff (Gurdon Institute, Cambridge, United Kingdom) for his support and for the anti-Dm Cdc27, -Mad2, and -Fizzy antibodies, as well as GFP-Cdc20 transgenic lines.
Footnotes
Published ahead of print on 26 April 2010.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Abrieu, A., J. A. Kahana, K. W. Wood, and D. W. Cleveland. 2000. CENP-E as an essential component of the mitotic checkpoint in vitro. Cell 102:817-826. [DOI] [PubMed] [Google Scholar]
- 2.Basto, R., F. Scaerou, S. Mische, E. Wojcik, C. Lefebvre, R. Gomes, T. Hays, and R. Karess. 2004. In vivo dynamics of the rough deal checkpoint protein during Drosophila mitosis. Curr. Biol. 14:56-61. [DOI] [PubMed] [Google Scholar]
- 3.Basu, J., H. Bousbaa, E. Logarinho, Z. Li, B. C. Williams, C. Lopes, C. E. Sunkel, and M. L. Goldberg. 1999. Mutations in the essential spindle checkpoint gene bub1 cause chromosome missegregation and fail to block apoptosis in Drosophila. J. Cell Biol. 146:13-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bellen, H. J., R. W. Levis, G. Liao, Y. He, J. W. Carlson, G. Tsang, M. Evans-Holm, P. R. Hiesinger, K. L. Schulze, G. M. Rubin, R. A. Hoskins, and A. C. Spradling. 2004. The BDGP gene disruption project: single transposon insertions associated with 40% of Drosophila genes. Genetics 167:761-781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Buffin, E., D. Emre, and R. E. Karess. 2007. Flies without a spindle checkpoint. Nat. Cell Biol. 9:565-572. [DOI] [PubMed] [Google Scholar]
- 6.Buffin, E., C. Lefebvre, J. Huang, M. E. Gagou, and R. E. Karess. 2005. Recruitment of Mad2 to the kinetochore requires the Rod/Zw10 complex. Curr. Biol. 15:856-861. [DOI] [PubMed] [Google Scholar]
- 7.Chen, R. H. 2002. BubR1 is essential for kinetochore localization of other spindle checkpoint proteins and its phosphorylation requires Mad1. J. Cell Biol. 158:487-496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen, R. H., A. Shevchenko, M. Mann, and A. W. Murray. 1999. The spindle checkpoint of budding yeast depends on a tight complex between the Mad1 and Mad2 proteins. Mol. Biol. Cell 10:2607-2618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chung, E., and R. H. Chen. 2002. Spindle checkpoint requires Mad1-bound and Mad1-free Mad2. Mol. Biol. Cell 13:1501-1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Davenport, J., L. D. Harris, and R. Goorha. 2006. Spindle checkpoint function requires Mad2-dependent Cdc20 binding to the Mad3 homology domain of BubR1. Exp. Cell Res. 312:1831-1842. [DOI] [PubMed] [Google Scholar]
- 11.Dawson, I. A., S. Roth, M. Akam, and S. Artavanis-Tsakonas. 1993. Mutations of the fizzy locus cause metaphase arrest in Drosophila melanogaster embryos. Development 117:359-376. [DOI] [PubMed] [Google Scholar]
- 12.De Antoni, A., C. G. Pearson, D. Cimini, J. C. Canman, V. Sala, L. Nezi, M. Mapelli, L. Sironi, M. Faretta, E. D. Salmon, and A. Musacchio. 2005. The Mad1/Mad2 complex as a template for Mad2 activation in the spindle assembly checkpoint. Curr. Biol. 15:214-225. [DOI] [PubMed] [Google Scholar]
- 13.DeAntoni, A., V. Sala, and A. Musacchio. 2005. Explaining the oligomerization properties of the spindle assembly checkpoint protein Mad2. Philos. Trans. R Soc. Lond. B Biol. Sci. 360:637-648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dobles, M., V. Liberal, M. L. Scott, R. Benezra, and P. K. Sorger. 2000. Chromosome missegregation and apoptosis in mice lacking the mitotic checkpoint protein Mad2. Cell 101:635-645. [DOI] [PubMed] [Google Scholar]
- 15.Fang, G. 2002. Checkpoint protein BubR1 acts synergistically with Mad2 to inhibit anaphase-promoting complex. Mol. Biol. Cell 13:755-766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fang, G., H. Yu, and M. W. Kirschner. 1998. The checkpoint protein MAD2 and the mitotic regulator CDC20 form a ternary complex with the anaphase-promoting complex to control anaphase initiation. Genes Dev. 12:1871-1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fraschini, R., A. Beretta, L. Sironi, A. Musacchio, G. Lucchini, and S. Piatti. 2001. Bub3 interaction with Mad2, Mad3 and Cdc20 is mediated by WD40 repeats and does not require intact kinetochores. EMBO J. 20:6648-6659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goshima, G., R. Wollman, S. S. Goodwin, N. Zhang, J. M. Scholey, R. D. Vale, and N. Stuurman. 2007. Genes required for mitotic spindle assembly in Drosophila S2 cells. Science 316:417-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hardwick, K. G., R. C. Johnston, D. L. Smith, and A. W. Murray. 2000. MAD3 encodes a novel component of the spindle checkpoint which interacts with Bub3p, Cdc20p, and Mad2p. J. Cell Biol. 148:871-882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Herzog, F., I. Primorac, P. Dube, P. Lenart, B. Sander, K. Mechtler, H. Stark, and J. M. Peters. 2009. Structure of the anaphase-promoting complex/cyclosome interacting with a mitotic checkpoint complex. Science 323:1477-1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Howell, B. J., D. B. Hoffman, G. Fang, A. W. Murray, and E. D. Salmon. 2000. Visualization of Mad2 dynamics at kinetochores, along spindle fibers, and at spindle poles in living cells. J. Cell Biol. 150:1233-1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Howell, B. J., B. Moree, E. M. Farrar, S. Stewart, G. Fang, and E. D. Salmon. 2004. Spindle checkpoint protein dynamics at kinetochores in living cells. Curr. Biol. 14:953-964. [DOI] [PubMed] [Google Scholar]
- 23.Huang, J., and J. W. Raff. 1999. The disappearance of cyclin B at the end of mitosis is regulated spatially in Drosophila cells. EMBO J. 18:2184-2195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang, J. Y., and J. W. Raff. 2002. The dynamic localisation of the Drosophila APC/C: evidence for the existence of multiple complexes that perform distinct functions and are differentially localised. J. Cell Sci. 115:2847-2856. [DOI] [PubMed] [Google Scholar]
- 25.Kallio, M., J. Weinstein, J. R. Daum, D. J. Burke, and G. J. Gorbsky. 1998. Mammalian p55CDC mediates association of the spindle checkpoint protein Mad2 with the cyclosome/anaphase-promoting complex, and is involved in regulating anaphase onset and late mitotic events. J. Cell Biol. 141:1393-1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kallio, M. J., V. A. Beardmore, J. Weinstein, and G. J. Gorbsky. 2002. Rapid microtubule-independent dynamics of Cdc20 at kinetochores and centrosomes in mammalian cells. J. Cell Biol. 158:841-847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kulukian, A., J. S. Han, and D. W. Cleveland. 2009. Unattached kinetochores catalyze production of an anaphase inhibitor that requires a Mad2 template to prime Cdc20 for BubR1 binding. Dev. Cell 16:105-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lénart, P., and J. M. Peters. 2006. Checkpoint activation: don't get mad too much. Curr. Biol. 16:R412-R414. [DOI] [PubMed] [Google Scholar]
- 29.Logarinho, E., H. Bousbaa, J. M. Dias, C. Lopes, I. Amorim, A. Antunes-Martins, and C. E. Sunkel. 2004. Different spindle checkpoint proteins monitor microtubule attachment and tension at kinetochores in Drosophila cells. J. Cell Sci. 117:1757-1771. [DOI] [PubMed] [Google Scholar]
- 30.Luo, X., G. Fang, M. Coldiron, Y. Lin, H. Yu, M. W. Kirschner, and G. Wagner. 2000. Structure of the Mad2 spindle assembly checkpoint protein and its interaction with Cdc20. Nat. Struct. Biol. 7:224-229. [DOI] [PubMed] [Google Scholar]
- 31.Luo, X., Z. Tang, J. Rizo, and H. Yu. 2002. The Mad2 spindle checkpoint protein undergoes similar major conformational changes upon binding to either Mad1 or Cdc20. Mol. Cell 9:59-71. [DOI] [PubMed] [Google Scholar]
- 32.Luo, X., Z. Tang, G. Xia, K. Wassmann, T. Matsumoto, J. Rizo, and H. Yu. 2004. The Mad2 spindle checkpoint protein has two distinct natively folded states. Nat. Struct. Mol. Biol. 11:338-345. [DOI] [PubMed] [Google Scholar]
- 33.Matsuura, S., Y. Matsumoto, K. Morishima, H. Izumi, H. Matsumoto, E. Ito, K. Tsutsui, J. Kobayashi, H. Tauchi, Y. Kajiwara, S. Hama, K. Kurisu, H. Tahara, M. Oshimura, K. Komatsu, T. Ikeuchi, and T. Kajii. 2006. Monoallelic BUB1B mutations and defective mitotic-spindle checkpoint in seven families with premature chromatid separation (PCS) syndrome. Am. J. Med. Genet. A 140:358-367. [DOI] [PubMed] [Google Scholar]
- 34.Michel, L. S., V. Liberal, A. Chatterjee, R. Kirchwegger, B. Pasche, W. Gerald, M. Dobles, P. K. Sorger, V. V. Murty, and R. Benezra. 2001. MAD2 haplo-insufficiency causes premature anaphase and chromosome instability in mammalian cells. Nature 409:355-359. [DOI] [PubMed] [Google Scholar]
- 35.Musacchio, A., and E. D. Salmon. 2007. The spindle-assembly checkpoint in space and time. Nat. Rev. Mol. Cell Biol. 8:379-393. [DOI] [PubMed] [Google Scholar]
- 36.Nasmyth, K. 2005. How do so few control so many? Cell 120:739-746. [DOI] [PubMed] [Google Scholar]
- 37.Nezi, L., G. Rancati, A. De Antoni, S. Pasqualato, S. Piatti, and A. Musacchio. 2006. Accumulation of Mad2-Cdc20 complex during spindle checkpoint activation requires binding of open and closed conformers of Mad2 in Saccharomyces cerevisiae. J. Cell Biol. 174:39-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nilsson, J., M. Yekezare, J. Minshull, and J. Pines. 2008. The APC/C maintains the spindle assembly checkpoint by targeting Cdc20 for destruction. Nat. Cell Biol. 10:1411-1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Orr, B., H. Bousbaa, and C. E. Sunkel. 2007. Mad2-independent spindle assembly checkpoint activation and controlled metaphase-anaphase transition in Drosophila S2 cells. Mol. Biol. Cell 18:850-863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Peters, J. M. 2008. Checkpoint control: the journey continues. Curr. Biol. 18:R170-R172. [DOI] [PubMed] [Google Scholar]
- 41.Peters, J. M. 2006. The anaphase promoting complex/cyclosome: a machine designed to destroy. Nat. Rev. Mol. Cell Biol. 7:644-656. [DOI] [PubMed] [Google Scholar]
- 42.Poddar, A., J. A. Daniel, J. R. Daum, and D. J. Burke. 2004. Differential kinetochore requirements for establishment and maintenance of the spindle checkpoint are dependent on the mechanism of checkpoint activation in Saccharomyces cerevisiae. Cell Cycle 3:197-204. [PubMed] [Google Scholar]
- 43.Poddar, A., P. T. Stukenberg, and D. J. Burke. 2005. Two complexes of spindle checkpoint proteins containing Cdc20 and Mad2 assemble during mitosis independently of the kinetochore in Saccharomyces cerevisiae. Eukaryot. Cell 4:867-878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Raff, J. W., K. Jeffers, and J. Y. Huang. 2002. The roles of Fzy/Cdc20 and Fzr/Cdh1 in regulating the destruction of cyclin B in space and time. J. Cell Biol. 157:1139-1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rieder, C. L., R. W. Cole, A. Khodjakov, and G. Sluder. 1995. The checkpoint delaying anaphase in response to chromosome monoorientation is mediated by an inhibitory signal produced by unattached kinetochores. J. Cell Biol. 130:941-948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sczaniecka, M., A. Feoktistova, K. M. May, J. S. Chen, J. Blyth, K. L. Gould, and K. G. Hardwick. 2008. The spindle checkpoint functions of Mad3 and Mad2 depend on a Mad3 KEN box-mediated interaction with Cdc20-APC/C. J. Biol. Chem. 283:23039-23047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shah, J. V., and D. W. Cleveland. 2000. Waiting for anaphase: Mad2 and the spindle assembly checkpoint. Cell 103:997-1000. [DOI] [PubMed] [Google Scholar]
- 48.Shannon, K. B., J. C. Canman, and E. D. Salmon. 2002. Mad2 and BubR1 function in a single checkpoint pathway that responds to a loss of tension. Mol. Biol. Cell 13:3706-3719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sironi, L., M. Mapelli, S. Knapp, A. De Antoni, K. T. Jeang, and A. Musacchio. 2002. Crystal structure of the tetrameric Mad1-Mad2 core complex: implications of a ‘safety belt’ binding mechanism for the spindle checkpoint. EMBO J. 21:2496-2506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sudakin, V., G. K. Chan, and T. J. Yen. 2001. Checkpoint inhibition of the APC/C in HeLa cells is mediated by a complex of BUBR1, BUB3, CDC20, and MAD2. J. Cell Biol. 154:925-936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tang, Z., R. Bharadwaj, B. Li, and H. Yu. 2001. Mad2-Independent inhibition of APCCdc20 by the mitotic checkpoint protein BubR1. Dev. Cell 1:227-237. [DOI] [PubMed] [Google Scholar]
- 52.Vigneron, S., S. Prieto, C. Bernis, J. C. Labbe, A. Castro, and T. Lorca. 2004. Kinetochore localization of spindle checkpoint proteins: who controls whom? Mol. Biol. Cell 15:4584-4596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Waters, J. C., R. H. Chen, A. W. Murray, and E. D. Salmon. 1998. Localization of Mad2 to kinetochores depends on microtubule attachment, not tension. J. Cell Biol. 141:1181-1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xu, L., H. X. Deng, Y. Yang, J. H. Xia, W. Y. Hung, and T. Siddque. 1997. Assignment of mitotic arrest deficient protein 2 (MAD2L1) to human chromosome band 5q23.3 by in situ hybridization. Cytogenet. Cell Genet. 78:63-64. [DOI] [PubMed] [Google Scholar]
- 55.Yang, M., B. Li, C. J. Liu, D. R. Tomchick, M. Machius, J. Rizo, H. Yu, and X. Luo. 2008. Insights into Mad2 regulation in the spindle checkpoint revealed by the crystal structure of the symmetric Mad2 dimer. PLoS Biol. 6:e50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yu, H. 2002. Regulation of APC-Cdc20 by the spindle checkpoint. Curr. Opin. Cell Biol. 14:706-714. [DOI] [PubMed] [Google Scholar]
- 57.Yu, H. 2006. Structural activation of Mad2 in the mitotic spindle checkpoint: the two-state Mad2 model versus the Mad2 template model. J. Cell Biol. 173:153-157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang, J., R. Neisa, and Y. Mao. 2009. Oncogenic adenomatous polyposis coli mutants impair the mitotic checkpoint through direct interaction with Mad2. Mol. Biol. Cell 20:2381-2388. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.