Abstract
Extracellular amino acids induce the yeast SPS sensor to endoproteolytically cleave transcription factors Stp1 and Stp2 in a process termed receptor-activated proteolysis (RAP). Ssy5, the activating endoprotease, is synthesized with a large N-terminal prodomain and a C-terminal chymotrypsin-like catalytic (Cat) domain. During biogenesis, Ssy5 cleaves itself and the prodomain and Cat domain remain associated, forming an inactive primed protease. Here we show that the prodomain is a potent inhibitor of Cat domain activity and that its inactivation is a requisite for RAP. Accordingly, amino acid-induced signals trigger proteasome-dependent degradation of the prodomain. A mutation that stabilizes the prodomain prevents Stp1 processing, whereas destabilizing mutations lead to constitutive RAP-independent Stp1 processing. We fused a conditional degron to the prodomain to synthetically reprogram the amino acid-responsive SPS signaling pathway, placing it under temperature control. Our results define a regulatory mechanism that is novel for eukaryotic proteases functioning within cells.
Proteolytic processing is a mechanism used by eukaryotes to control the activity of latent transcription factors (9). For example, regulated intramembrane proteolysis (RIP) and ubiquitin/proteasome-dependent processing (RUP) are two mechanisms that employ proteolysis-dependent activation of latent membrane-anchored transcription factors (24). RIP is catalyzed by membrane-bound proteases that cleave inactive precursor molecules to release active transcription factors from their membrane anchor (10). In the RUP mechanism, a polyubiquitylated membrane-anchored precursor is partially degraded by the 26S proteasome, resulting in the release of an active transcription factor (23). Both RIP and RUP rely on constitutively active proteases that process substrate transcription factors when they become available and bind the protease. Control of substrate availability is achieved by altering the intracellular compartmentalization of the substrate (RIP) or by covalent modification to target the substrate to an enclosed protease compartment (RUP). Consequently, in these pathways, the regulation of signal transduction occurs at the level of substrate availability and not by direct regulation of the proteolytic activity of the processing protease.
By studying the mechanism responsible for amino acid-induced gene expression in the yeast Saccharomyces cerevisiae, we have found that transcription factor activity can be regulated by directly controlling the enzymatic activity of an intracellular protease (3). We have termed this mechanism receptor-activated proteolysis (RAP) to signify the critical involvement of a plasma membrane receptor. RAP is distinguished from the previously described protease-based signaling mechanisms, RIP and RUP, in that it does not rely on the modulation of substrate accessibility.
RAP enables yeast cells to respond to amino acids in the growth medium by increasing the expression of amino acid permeases, leading to an enhanced capacity to take up these nutrients. The signal transduction pathway governing this transcriptional response is known as the SPS sensing pathway. Signaling requires a functional Ssy1-Ptr3-Ssy5 (SPS) sensor, the SCFGrr1 ubiquitin ligase complex, casein kinase-dependent phosphorylation, and two homologous transcription factors, Stp1 and Stp2 (1, 17, 19, 27, 29, 34, 35). In the absence of SPS sensor signaling, Stp1 and Stp2 exhibit latent characteristics and are excluded from the nucleus by the presence of N-terminal negative regulatory domains (4, 5). Upon induction by extracellular amino acids, the primary amino acid receptor Ssy1 initiates signals (48) that, via hyperphosphorylation of Ptr3 (28), result in activation of the Ssy5 protease, leading to the endoproteolytic processing of Stp1 and Stp2, which enter the nucleus and activate the transcription of amino acid permease genes (4).
The core SPS sensor component Ssy5 is the RAP-controlled protease (1, 3, 34). Ssy5 exhibits homology to chymotrypsin-like serine proteases and is expressed as a zymogen that cleaves itself into an N-terminal prodomain and a C-terminal catalytic (Cat) domain. The Cat domain contains a conserved catalytic triad with a predicted active-site serine (S640) that is required for function (1, 2). N-terminal sequencing of the Cat domain identified alanine 382 as the first residue, consistent with the notion that autoprocessing takes place between prodomain residue 381 and Cat domain residue 382 (34).
The mechanism of RAP-dependent activation of Stp1 and Stp2 is not well understood, but several observations point to an important regulatory role for the Ssy5 Pro domain. First, in comparison to the size of characterized serine proteases, the prodomain of Ssy5 is unusually large, suggesting a more complex regulatory function. Further, after autolytic processing, the prodomain and Cat domain remain associated, forming a primed protease that associates with Stp1 and, although catalytically competent, does not process its substrate (3). Last, Stp1 cleavage correlates in time with the downregulation of prodomain levels. These observations led us to propose that after assembling into a primed protease, the prodomain inhibits the Cat domain and dissociates before substrate processing (3).
In this study, we examined the role of the Ssy5 prodomain in controlling the endoproteolytic Stp1-processing activity of the Cat domain. We found that the prodomain undergoes proteasomal degradation as a consequence of SPS sensor signaling. Mutations in SSY5 that induce prodomain degradation result in constitutive protease activity, whereas deletion of the N-terminal 90 amino acids abolishes Stp1 processing without affecting autolysis or Stp1 binding. These results strongly suggest that the prodomain functions as a potent inhibitor of the Cat domain. We have critically tested this model by synthetically targeting the prodomain for regulated proteasomal degradation and found that proteasomal degradation of the prodomain is sufficient to fully induce Cat domain-dependent Stp1 processing. The regulatory mechanism described here represents a novel way of controlling an intracellular eukaryotic protease.
MATERIALS AND METHODS
Media.
Standard media, including yeast extract-peptone-dextrose (YPD) medium, ammonia-based synthetic minimal dextrose (SD) medium, supplemented as required to enable the growth of auxotrophic strains, and ammonia-based synthetic complete dextrose (SC) were prepared as described previously (4). When needed, l-leucine was added at a concentration of 1.3 mM to induce SPS sensor signaling. Sensitivity to 2-{[({[(4-methoxy-6-methyl)-1,3,5-triazin-2-yl]-amino}carbonyl)amino-]-sulfonyl}-benzoic acid (MM; 450 μg/ml) on complex medium is described elsewhere (5). Sensitivity to 1 mM azetidine-2-carboxylic acid (AzC) was tested on solid SD medium.
Strains and plasmids.
The S. cerevisiae strains used in this study (Table 1) are isogenic descendants of S288C-derived strain AA255/PLY115 (6) or W303-1B (43), as indicated. The plasmids used in this work are listed in Table 2. Plasmids pSH105 and pSH106 were obtained by recombination of a Pro-HA-SSY541-326 PCR fragment amplified from pFL005 and AatII-restricted pCA174. Plasmids pSH119 and pSH120 were created identically to pSH105 and pSH106 but with different primer pairs. A fragment containing a temperature-regulated degron was amplified from plasmid pKL187 (36) and recombined into BamHI-linearized pHK48 (19), after which the 1,791-bp AatII/NotI fragment from this plasmid was recombined into PmeI-linearized pSH120, creating plasmid pTP110. A PCR-generated fragment with the critical arginine (Rds) replaced with alanine (As) was recombined into SacII-linearized pTP110, creating pTP111. Plasmids pTP112, pTP113, pTP114, pTP115, pTP116, pTP117, pTP118, pTP119, pTP120, and pTP121, encoding mutant alleles affecting Pro domain function, were obtained by recombination of PCR fragments carrying the indicated mismatch mutations and a silent mutation eliminating a diagnostic BamHI site. The sequences of the mutagenic oligonucleotides and PCR primers used are available upon request.
TABLE 1.
Yeast strains used in this study
| Strain | Genotype | Reference |
|---|---|---|
| AA255/PLY115-derived strains | ||
| CAY265 | MATaura3-52 ssy5Δ2::hisG gap1Δ::PAGP1-lacZ | 3 |
| CAY276 | MATalys2Δ201 ura3-52 ssy5Δ2::hisG grr1Δ50::hphMX4 | This work |
| CAY285 | MATalys2Δ201 ura3-52 ptr3Δ15::hisG ssy1Δ13::hisG ssy5Δ2::hisG | 3 |
| HKY77 | MATalys2Δ201 ura3-52 ssy5Δ2::hisG | 19 |
| HKY84 | MATalys2Δ201 ura3-52 ssy1Δ13::hisG ssy5Δ2::hisG | 19 |
| HKY85 | MATalys2Δ201 ura3-52 ptr3Δ15::hisG ssy5Δ2::hisG | 19 |
| TPY101 | MATalys2Δ201 ura3-52 ssy1Δ13::hisG ssy5Δ2::hisG ubr1Δ::natMX | This work |
| W303-1B-derived strains | ||
| CAY220 | MATα ura3-52 leu2Δ1 ssd1 RPT6 (CIM3) | 4 |
| CMY763 | MATα ura3-52 leu2Δ1 ssd1 rpt6 (cim3-1) | 21 |
TABLE 2.
Plasmids used in this study
| Plasmid | Description | Reference |
|---|---|---|
| pCA030 | pRS317 (LYS2) containing PAGP1-lacZ | 5 |
| pCA047 | pRS316 (URA3) containing STP1-3HA | 4 |
| pCA122 | pRS317 (LYS2) containing STP1-3HA | 5 |
| pCA177 | pRS316 (URA3) containing SSY5-HA-KlTRP1 | 3 |
| pCA195 | pRS316 (URA3) containing HA-SSY5 | 3 |
| pCA204 | pRS317 (LYS2) containing STP1-MYC-kanMX | 3 |
| pFL001 | pRS316 (URA3) containing SSY5 | 3 |
| pSH105 | pRS316 (URA3) containing HAi-SSY541-699-GST | This work |
| pSH106 | pRS316 (URA3) containing HAi-SSY561-699-GST | This work |
| pSH119 | pRS316 (URA3) containing HAi-SSY591-699-GST | This work |
| pSH120 | pRS316 (URA3) containing HAi-SSY51-699-GST | This work |
| pHK048 | pRS316 (URA3) containing SSY5-myc | 19 |
| pTP110 | pRS316 (URA3) containing Ubi-Rds-tsDHFR-HAi-SSY51-699-GST | This work |
| pTP111 | pRS316 (URA3) containing Ubi-As-tsDHFR-HAi-SSY51-699-GST | This work |
| pTP112 | pRS316 (URA3) containing HAi-SSY51-699-GST (L126S V129A E131A) | This work |
| pTP113 | pRS316 (URA3) containing HAi-SSY51-699-GST (L126S E131A) | This work |
| pTP114 | pRS316 (URA3) containing HAi-SSY51-699-GST (L126S V129A) | This work |
| pTP115 | pRS316 (URA3) containing HAi-SSY51-699-GST (E131A) | This work |
| pTP116 | pRS316 (URA3) containing HAi-SSY51-699-GST (E131V) | This work |
| pTP117 | pRS316 (URA3) containing HAi-SSY51-699-GST (V129A) | This work |
| pTP118 | pRS316 (URA3) containing HAi-SSY51-699-GST (V129D) | This work |
| pTP119 | pRS316 (URA3) containing HAi-SSY51-699-GST (V129G) | This work |
| pTP120 | pRS316 (URA3) containing HAi-SSY51-699-GST (L126W) | This work |
| pTP121 | pRS316 (URA3) containing HAi-SSY51-699-GST (L126S) | This work |
β-Galactosidase activity assays.
Semiquantitative β-galactosidase activity was measured in N-lauroylsarcosine-permeabilized cells as described elsewhere (5).
Immunoblot analysis.
Whole-cell extracts were prepared under denaturing conditions using NaOH and trichloroacetic acid as described previously (39). Primary antibodies were diluted as follows: 12CA5 ascites fluid (antihemagglutinin [anti-HA] monoclonal antibody), 1:1,000; purified rabbit polyclonal anti-glutathione S-transferase (anti-GST) antibody, 1:5,000; anti-myc monoclonal antibody (Roche Applied Science), 1:1,000; horseradish peroxidase (HRP)-conjugated 3F10 anti-HA monoclonal antibody (Roche Applied Science), 1:5,000; anti-GST 3-4C monoclonal antibody (Zymed Laboratories Inc.), 1:500; anti-myc-HRP 9E10 monoclonal antibody (Roche Applied Science), 1:1,000; anti-FLAG polyclonal antibody (Sigma), 1:5,000; anti-Pgk1 antibody (Molecular Probes), 1:5,000. Immunoreactive bands were visualized by chemiluminescence detection (SuperSignal West Dura Extended-Duration Substrate; Pierce) and quantified using a LAS1000 system (Fuji Photo Film Co., Ltd.).
CHX chase.
Cells were inoculated at an optical density at 600 nm of 0.5 and grown at 30°C for 3 h. Cycloheximide (CHX) was added to the cells at a concentration of 100 μg/ml, and where indicated, leucine was added to a concentration of 1.3 mM (+ leu). Extracts were prepared from samples taken at the time points indicated and analyzed by immunoblotting. Cells harboring temperature-sensitive alleles were treated similarly but were pregrown at 25°C for 4 h, after which the culture was shifted to 37°C for 30 min prior to CHX addition.
PCR mutagenesis.
Prodomain-encoding sequences were amplified using error-prone PCR (47) in the presence of 1.25 mM dCTP and TTP, 0.025 mM dATP and dGTP, 3.5 mM MgCl2, and 0.125 mM MnCl2. The resulting PCR products and AatII-linearized pSH120 were introduced together into strain HKY84 (ssy5Δ ssy1Δ), and Ura+ transformants with recombinant plasmids were selected on SC medium lacking uracil. Cells carrying plasmids expressing constitutively active alleles of SSY5 were selected on YPD containing MM.
RESULTS
SSY5* encodes a constitutively active protease that functions independently of other SPS sensor components.
We set out to critically test whether the prodomain is responsible for the proper regulation of the Ssy5 protease. Previously, we reported that fusion of a 6HA tag to the N terminus of Ssy5 results in constitutive Stp1 processing in the absence of inducing amino acids. Also, we found that N-terminally HA-tagged Ssy5 cleaves Stp1 when these proteins are heterologously expressed in Schizosaccharomyces pombe, a yeast that lacks an identifiable SPS sensing pathway (3). In contrast to our results, Liu et al. (28) recently reported that the insertion of a similar HA tag at the N terminus of Ssy5 did not result in constitutive Stp1 processing. This apparent discrepancy prompted us to fully characterize the functional properties of N- and C-terminally HA-tagged Ssy5 alleles.
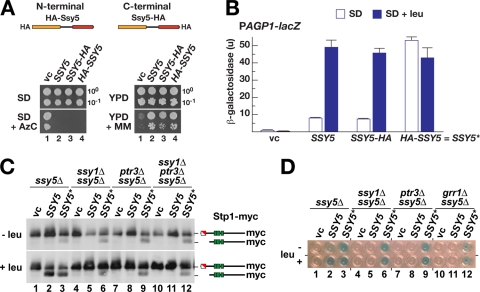
SSY5 alleles encoding HA tags at either the N or the C terminus are functional and complement ssy5Δ-encoded phenotypes (Fig. 1 A); i.e., they complement SPS sensor-dependent uptake of a toxic proline analogue (AzC) (5) and branched-chain amino acid uptake in the presence of MM (MM is a sulfonylurea analogue that inhibits branched-chain amino acid synthesis) (25). In order to quantify the extent of SPS sensor-induced transcriptional activation and to detect possible constitutive behavior of HA-tagged Ssy5, we monitored the ability of epitope-tagged Ssy5 to restore SPS sensor-induced gene expression in an ssy5Δ strain by monitoring AGP1 promoter activity of a PAGP1-lacZ reporter gene integrated chromosomally (3). Interestingly, while cells carrying wild-type or C-terminally HA-tagged Ssy5 responded normally to SPS sensor signaling (Fig. 1B, SSY5 and SSY5-HA), cells with N-terminally HA-tagged Ssy5 exhibited constitutively high levels of β-galactosidase activity, even in medium lacking the SPS sensor inducer leucine (Fig. 1B, HA-SSY5). Hence, the insertion of the 6HA tag at the N terminus of Ssy5 clearly creates an artificially activated species, here designated Ssy5*.
FIG. 1.
Stp1 is constitutively processed in strains expressing N-terminally HA-tagged Ssy5 (Ssy5*). (A) Schematic diagram and functional analysis of N- and C-terminally HA-tagged Ssy5. The empty vector pRS316 (vc), pFL001 (Ssy5), pCA177 (Ssy5-HA), and pCA195 (HA-Ssy5) were individually introduced into ssy5Δ mutant strain CAY265. Dilutions of cultures grown in SD were spotted onto SD, SD with AzC (SD + AzC), YPD, and YPD with MM (YPD + MM). Plates were incubated for 2 to 3 days at 30°C. (B) β-Galactosidase activity in ssy5Δ mutant strain HKY77 [pCA030 (PAGP1-lacZ)] carrying pRS316 (vc), pFL001 (SSY5), pCA177 (SSY5-HA), or pCA195 (HA-SSY5) and grown in SD or SD supplemented with leucine (SD + leu). Error bars indicate 95% confidence intervals based on assays carried out using four independent transformants. The constitutive HA-SSY5 allele is designated SSY5*. (C) Immunoblot analysis of extracts prepared from strains HKY77 (ssy5Δ), HKY84 (ssy5Δ ssy1Δ), HKY85 (ssy5Δ ptr3Δ), and CAY285 (ssy5Δ ptr3Δ ssy1Δ) carrying pCA204 (Stp1-myc) and pRS316 (vc), pFL001 (SSY5), or pCA195 (SSY5*) and grown in SD (− leu) and 30 min after induction with leucine (+ leu). (D) β-Galactosidase activity from PAGP1-lacZ (pCA030) expressed in HKY77 (ssy5Δ), HKY84 (ssy5Δ ssy1Δ), HKY85 (ssy5Δ ptr3Δ), and CAY276 (ssy5Δ grr1Δ) carrying pRS316 (vc), pFL001 (SSY5), pCA177 (SSY5-HA), or pCA195 (SSY5*). The strains were grown in SD or SD supplemented with leucine (+ leu) as indicated. β-Galactosidase activity was detected by 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) staining (blue precipitate).
Ssy5* may represent a protease that is hyperresponsive to basal (noninduced) SPS sensor signaling, retaining its dependency on the presence of the other SPS sensor components. Alternatively, Ssy5* may function in a fully uncontrolled manner that is independent of the other signaling components. We tested these possibilities by monitoring Stp1 processing in ssy1Δ, ptr3Δ, and ssy1Δ ptr3Δ mutant strains harboring the SSY5* allele (Fig. 1C). SSY5*-induced Stp1 processing was observed in all of the strains, both in the absence and in the presence of leucine in the growth medium, although processing was more effective in leucine-induced cells with a functional SPS sensor (Fig. 1C, lane 3). To test if processing of Stp1 resulted in promoter activation in these experiments, we monitored β-galactosidase expression from the AGP1 promoter. Indeed, SSY5* facilitated expression from the AGP1 promoter in all SPS sensor-deficient strains, including a grr1Δ mutant strain (Fig. 1D). Together, these observations indicate that Ssy5* is able to process Stp1 without the participation of the other SPS sensor components. It is likely that the fusion of an HA tag to the N terminus affects the conformation of the Pro domain in a manner that favors Cat domain-mediated Stp1 processing.
26S proteasome-dependent prodomain degradation correlates with Stp1 processing.
We have previously reported that prodomain levels rapidly decrease upon SPS sensor induction by amino acids in a manner that correlates in time with Stp1 processing (3, 19). These findings led us to propose that the dissociation of the prodomain from the Cat domain, or a conformational change in the prodomain, changes its stability and may be the key regulatory step in Ssy5 activation (3). However, the relevance of reduced prodomain levels and the correlation with Ssy5 activity have recently been questioned (28), which required us to examine the amino acid-induced downregulation of the prodomain in more detail.
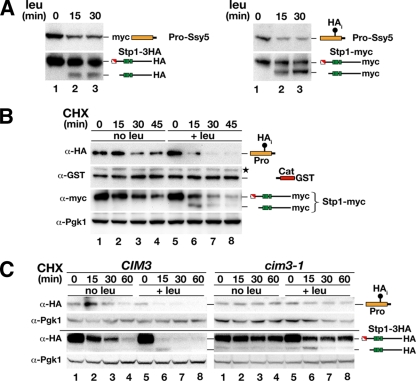
First, we addressed the potential experimental artifact that prodomain downregulation was a consequence of the insertion of an N-terminal HA tag. Leucine-induced downregulation was apparent when either an N-terminal myc tag (19) or an internal HA tag (HAi) inserted after amino acid position 216 (3) was used to tag the prodomain (Fig. 2 A, upper panels). Importantly, both of these epitope-tagged Ssy5 constructs exhibit fully regulated activity, judging by their ability to rapidly induce Stp1 processing in response to leucine addition (Fig. 2A, lower panels). These results indicate that prodomain downregulation is independent of the epitope tag used to detect it.
FIG. 2.
Amino acid-induced proteasomal degradation of the Ssy5 prodomain correlates with Stp1 processing. (A) SD-grown cultures of ssy5Δ mutant strain HKY77 carrying plasmids pHK048 (myc-Ssy5) and pCA122 (Stp1-HA) (left panel) or pSH120 (HAi-Ssy5-GST) and pCA204 (Stp1-myc) (right panel) were induced with leucine. Immunoblot analysis of cell extracts prepared from subsamples of each culture removed at the indicated time after leucine addition. (B) Immunoblot analysis of cell extracts from strain HKY77 carrying pSH120 (HAi-Ssy5-GST) and pCA204 (Stp1-myc) treated with CHX. Cells were pregrown in SD, the culture was split into two equal volumes at t = 0, and the subcultures received an aliquot of CHX (final concentration equal to 100 mg/ml) and leucine as indicated. (C) Immunoblot analysis of cell extracts from CIM3 (CAY220) and cim3-1 (CMY763) mutant strains carrying pRS315 (LEU2) and pSH120 (HAi-Ssy5-GST) or pCA047 (Stp1-HA). Cells were pregrown in SD at 25°C, and cultures were split into two equal volumes and incubated at 37°C for 30 min prior to the addition of CHX and leucine as indicated. Samples were taken at the indicated time points after CHX addition. The immunoreactive forms of the Ssy5 Pro domain and Stp1 are schematically represented at their corresponding positions of migration. Levels of Pgk1 served as an internal control for protein loading.
Next, we directly tested whether downregulation of the prodomain is the result of altered rates of degradation or synthesis. We repeated the time course experiments in the presence of the translation inhibitor CHX and extended the analysis by simultaneously monitoring Cat domain levels using a C-terminal GST tag (Fig. 2B). In the absence of amino acid induction, the prodomain is relatively stable and was readily detected over the entire 45-min time course (Fig. 2B, lanes 1 to 4). In contrast, leucine induction rapidly reduced prodomain levels (Fig. 2B, lanes 5 to 8); levels were already significantly reduced 15 min postinduction and were barely detectable by the 30-min time point. Consequently, amino acid induction enhances prodomain degradation. Again, the reduced prodomain levels correlated with Stp1 processing. Both full-length and processed forms of Stp1 have a high turnover rate (half-life, ≤10 min) (5); consequently, the processed form of Stp1 is most abundant at the 15-min time point. The levels of the Cat domain were stable, as were the levels of the protein loading control Pgk1 (38).
Finally, to investigate whether the 26S proteasome is involved in prodomain downregulation, we assayed amino acid-induced degradation in strains carrying a temperature-sensitive mutation in RPT6 (cim3-1), which encodes one of the ATPases of the 19S regulatory particle of the proteasome. Proteasomal substrates are stabilized at nonpermissive temperatures in strains carrying cim3-1 (21, 37). We found that the prodomain was stabilized under both noninducing (no leu) and inducing (+leu) conditions in cells harboring the cim3-1 mutation relative to the isogenic wild-type strain (Fig. 2C, upper panels). Apparently, both basal and SPS sensor-induced degradation of the prodomain is mediated by the 26S proteasome. Similarly, we found cim3-1-induced stabilization of full-length Stp1 under both noninducing (no leu) and inducing (+leu) conditions compared to the isogenic wild-type strain (Fig. 2C, lower panel). As shown previously, Stp1 is processed even when proteasomal activity is impaired (1, 4).
Progressive deletions within the first 90 amino acids of the Ssy5 prodomain define negative- and positive-acting sequences required for proper Stp1 processing.
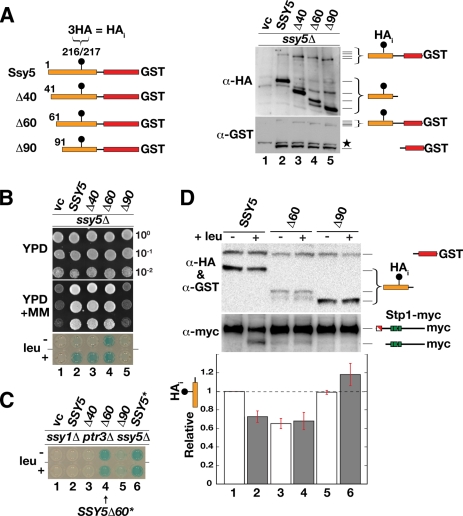
Based on the observation that manipulation of the prodomain with an N-terminal HA epitope results in constitutively active Ssy5 (Fig. 1), we focused on the possibility that the prodomain possesses autoregulatory activity. We created a series of N-terminal deletion mutations in the context of the functional SSY5 allele with an internal HA tag (HAi) and a C-terminal GST tag (Fig. 3 A, left panel). This HAi-Ssy5-GST allele (SSY5) fully complements the ssy5Δ mutation and correctly responds to amino acid induction (Fig. 3B). Consistently, the N- and C-terminal tags do not affect autolytic cleavage into separate pro- and Cat domains (Fig. 3A, right panel, lane 2) or proteasome-dependent degradation (Fig. 2B).
FIG. 3.
Deletion analysis of the prodomain yields constitutive and nonresponsive Ssy5 alleles. (A) Schematic representation of the N-terminal deletion constructs analyzed. The positions of the N-terminal residues, each preceded by an initiator methionine, are indicated (left panel). Each construct is expressed with an internal 3 × HA tag (HAi) and a C-terminal GST fusion protein. Shown is an immunoblot analysis of protein extracts from an ssy5Δ mutant strain (CAY265) carrying pRS316 (vc), pSH120 (SSY5), pSH105 (Δ40), pSH106 (Δ60), or pSH119 (Δ90) (right panel); extracts were prepared from cells grown in SD. The immunoreactive forms of Ssy5 are schematically depicted at their corresponding positions of migration. A star marks the position of an unrelated cross-reacting protein. (B) Dilution series of cell suspensions from strains as in panel A carrying pCA030 (PAGP1-lacZ). Cells were pregrown in SD and spotted onto YPD and YPD plus MM. Expression of β-galactosidase from PAGP1-lacZ was detected by X-Gal staining of cells grown in SD without or with leucine as indicated. (C) Strain CAY285 (ssy1Δ ptr3Δ ssy5Δ) carrying pCA030 (PAGP1-lacZ) and pRS316 (vc), pSH120 (SSY5), pSH105 (Δ40), pSH106 (Δ60), pSH119 (Δ90), or pCA205 (6HA-SSY5) was grown with or without leucine as indicated, and the levels of X-Gal staining were assessed in permeabilized cells (lower panel). (D) Immunoblot analysis of protein extracts from an ssy5Δ mutant strain (HKY77) carrying pCA204 (Stp1-myc) and pSH120 (SSY5), pSH106 (Δ60), or pSH119 (Δ90). Extracts were prepared from cells grown in SD and from cells harvested 30 min after induction by leucine as indicated. The immunoreactive forms of Ssy5 and Stp1 are schematically depicted at their corresponding positions of migration. The signal intensity of the immunoreactive bands corresponding to the HA-tagged prodomain and the GST-tagged Cat domain were quantified; the prodomain/Cat domain ratios were determined and normalized to the ratio of Ssy5 in uninduced cells (lane 1), and mean values were plotted. Error bars show standard errors (n = 3).
Three N-terminal truncation mutant proteins in which the first 40, 60, and 90 amino acids (Δ40, Δ60 and Δ90), respectively, were removed were evaluated for their effects on Ssy5 maturation into processed pro- and Cat domains. All of the truncated proteins were processed, indicating that deletion of up to the first 90 amino acids does not impair the catalytic activity of the Cat domain (Fig. 3A, right panels). Next, we assessed the functionality of the truncated proteins using a growth-based assay and analyzed their ability to induce the PAGP1-lacZ reporter (Fig. 3B). Whereas the Δ40 allele behaved like wild-type SSY5 (Fig. 3B, dilution 3), the Δ60 allele induced constitutive activation of the PAGP1-lacZ reporter (Fig. 3B, dilution 4). The latter result prompted us to test whether the Δ60 allele, like the SSY5* allele, bypassed the requirement of the other SPS sensor components (Fig. 3C). When the Δ60 allele was introduced into an ssy1Δ ptr3Δ ssy5Δ mutant strain, PAGP1-lacZ was expressed independently of amino acid induction, indicating that the allele encodes a constitutively active species. In contrast, the Δ90 allele lacked Stp1-processing activity since the ssy5Δ mutant host strain remained MM sensitive and PAGP1-lacZ expression was not induced by leucine (Fig. 3B, dilution 5). Thus, the Δ90 allele encodes a protein that is catalytically competent (i.e., prodomain and Cat domain autolysis occurs normally) but is unable to process Stp1.
Based on the previously observed correlation between prodomain degradation and Stp1 processing (Fig. 2), we postulated that cells with the constitutive Δ60 allele would have lower, constitutively downregulated levels of the prodomain. Similarly, we predicted that cells with the nonresponsive Δ90 allele would fail to downregulate the prodomain in response to amino acid induction. These predictions were tested by measuring the levels of the prodomain and Cat domain and the state of Stp1 processing in cells before and after leucine induction. Cells with wild-type SSY5 (HAi-SSY5-GST) responded to leucine induction with a 30% (±10%) reduction of the prodomain signal and accompanying Stp1 processing (Fig. 3D, lanes 1 and 2). Uninduced cells harboring a Δ60 allele also exhibited a 35% (±10%) reduction in prodomain levels relative to uninduced wild-type SSY5, and no further reduction was observed upon leucine induction (Fig. 3D, lanes 3 and 4). Cells harboring the Δ90 allele exhibited constant prodomain levels and no Stp1 processing (Fig. 3D, lanes 5 and 6).
Taken together, these results show that, in all instances, the prodomain levels inversely correlate with Stp1-processing activity; i.e., at high prodomain levels, no Stp1 processing is observed, and at low prodomain levels, Stp1 processing is observed. Presumably, deletion of the first 60 residues of the prodomain results in the constitutive activation of Ssy5 as a consequence of increased prodomain turnover. The larger deletion that removes the N-terminal 90 residues results in a protein that is unable to respond to amino acid induction, the likely consequence of the removal of sequences required for signal-induced degradation of the prodomain.
Constitutive activating mutations cluster in a conserved region of the Ssy5 prodomain.
To critically test whether constitutive activity is linked to prodomain degradation, we screened for point mutations in prodomain-encoding sequences of SSY5 that result in constitutive protease activity (see Materials and Methods). Briefly, a plasmid library with mutations specifically localized to sequences encoding the prodomain was generated by error-prone PCR. This library was introduced into a ssy5Δ ssy1Δ double-mutant yeast strain, and out of 2 × 106 transformants, 100 reproducibly scored positive for growth on YPD plus MM, a phenotype that is diagnostic for constitutive SPS sensor-regulated gene expression. Sixteen plasmids carrying constitutive alleles were further analyzed, and their inserts were sequenced; see Table 3 for a summary of the genetic selection and resulting mutations. Most of the inserts contained multiple mutations; however, we noted that several mutations affected three specific codons clustered within a conserved region (amino acids 126 to 131). Specifically, nine inserts carried mutations in codon 131, coding for glutamate; the mutations resulted in the substitution of alanine, valine, lysine, or glycine at this residue. Of the remaining inserts, five carried mutations in valine codon 129; here, the mutations resulted in the substitution of aspartate, alanine, or glycine at this residue. The last two inserts carried mutations resulting in the replacement of leucine residue 126 with either tryptophan or serine.
TABLE 3.
Summary of PCR-generated SSY5 constitutive allelesa
| Amino acid codon affected in plasmid insert and resulting change | No. of times isolated |
|---|---|
| E131 | |
| V | 4 |
| A | 3 |
| G | 1 |
| K | 1 |
| V129 | |
| D | 3 |
| G | 1 |
| A | 1 |
| L126 | |
| S | 1 |
| W | 1 |
Analyzed were 2 × 106 transformants, 300 MMr colonies in a primary selection, and 100 MMr colonies in a rescreening, and the sequences of inserts of 16 plasmids were analyzed.
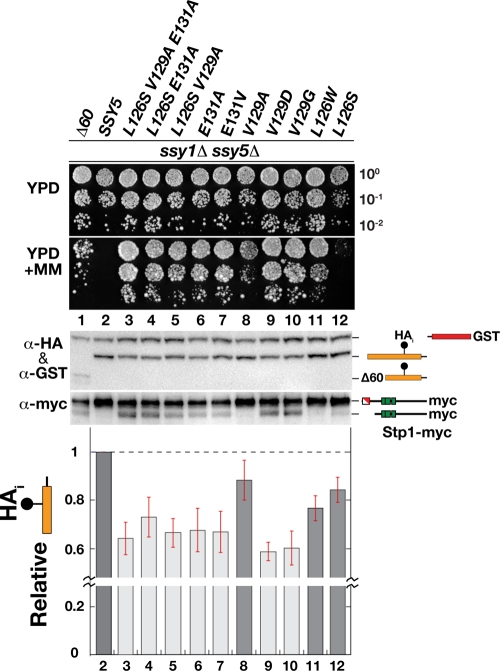
Since the inserts contained several mutations in addition to the ones just described, we introduced each of the mutations affecting these three conserved amino acid residues, both individually and in combination, into the fully functional and doubly tagged HAi-SSY5-GST allele. These new mutant alleles, presumably affecting Pro domain function, were subjected to further analysis. First, the mutant alleles were tested for the ability to complement a ssy5Δ mutation, and as expected, all fully complemented growth phenotypes in a manner indistinguishable from that of wild-type SSY5 (data not shown). Next, the constitutive activity of the mutant alleles was examined in an ssy5Δ ssy1Δ double-mutant strain using a growth-based assay on YPD plus MM (Fig. 4, upper panels). The growth characteristics enabled the single mutations to be classified into three groups. Group I mutations, E131A/V (dilutions 6 and 7) and V129D/G (dilutions 9 and 10), supported robust growth indicative of a high degree of constitutive activity. Group II mutations, V129A and L126W (dilutions 8 and 11), resulted in visibly weaker growth than group I mutations in the presence of MM. The single group III mutation, L126S, conferred poor growth on both YPD plus MM and YPD, which made it difficult to assess its constitutive activity. However, in combination with the weak group II V129A mutation, the L126S mutation had an enhancing effect that markedly improved growth on MM (Fig. 4, compare dilution 5 with dilution 8). The latter finding likely explains our ability to isolate mutations affecting this residue.
FIG. 4.
Mutations decreasing prodomain levels lead to constitutive Stp1 processing. Growth of HKY84 (ssy5Δ ssy1Δ) carrying pCA204 (Stp1-myc) and plasmid pSH106 (Δ60), pSH120 (HAi-Ssy5-GST), or pTP112-pTP121 expressing the indicated SSY5 alleles. Dilutions prepared from cells pregrown in SD were spotted onto YPD and YPD plus MM. Shown is an immunoblot analysis of protein extracts prepared from the strains grown in SD. The immunoreactive forms of Ssy5 and Stp1 are schematically depicted at their corresponding positions of migration. The signal intensities of the immunoreactive bands corresponding to the HA-tagged prodomain and the GST-tagged Cat domain were quantified; the prodomain/Cat domain ratios were determined and normalized to the ratio in Ssy5 (lane 2), and mean values were plotted. Error bars show standard errors (n = 5). Note the break in the y axis.
The status of Stp1 processing was examined in the cells carrying the constitutive alleles (Fig. 4, lower middle panel). Consistent with the growth phenotypes, the expression of group I alleles induced readily detectable levels of Stp1 processing, whereas Stp1 processing was barely or not detectable in cells expressing group II and group III mutations. Hence, the YPD-plus-MM growth phenotype provides a more sensitive assay of constitutive Ssy5 activity than immunoblot analysis of Stp1 processing. The higher sensitivity of growth-based assays is expected since both chromosomal copies of wild-type STP1 and STP2 are present in our strains; our analysis does not detect the processing of these native factors. Also, it has been reported that Ssy5-dependent processing of Stp2 occurs at lower levels of SPS sensor signaling (46). Thus, the weaker constitutive group II alleles may preferentially result in Stp2 processing.
Finally, we directly tested the possibility that the mutant alleles possess constitutively reduced prodomain levels. Indeed, we found a striking correlation between constitutive activity and prodomain downregulation (Fig. 4, upper middle and lower panels). The strong group I mutations reduced prodomain levels to between 30 and 40% of the wild-type level, while the weaker group II mutations reduced levels between 10 and 20%. Taken together, these results show that all constitutive mutations induce signal-independent degradation of the prodomain, and importantly, the endoproteolytic Stp1-processing activity of the Ssy5 protease is detected when low prodomain levels are observed.
Synthetically induced degradation of the prodomain is sufficient to activate Ssy5.
The results of all previous experiments are consistent with the prodomain functioning as an inhibitor of Stp1-processing activity of the Cat domain after assisting its correct folding. We directly tested this notion by asking if posttranslational removal of the prodomain would suffice to activate Ssy5 protease activity. To accomplish this, we fused a temperature-regulated N-degron (15, 16, 26, 36) to the N terminus of the Ssy5 prodomain (Fig. 5 A, left panel, Ssy5-Rds). This N-degron is latent at 25°C but induces rapid, proteasome-dependent degradation at elevated temperatures (35 to 37°C). Importantly, the N-degron relies on the presence of a destabilizing N-terminal arginine residue (Rds) that is created by cotranslational processing of appropriately fused ubiquitin. At elevated temperatures, this residue is exposed due to a temperature-induced conformational change in the linking temperature-sensitive dihydrofolate reductase moiety. The residue is then recognized by the N-end rule ubiquitin ligase Ubr1, which leads to polyubiquitylation and proteasomal degradation (44, 49). The replacement of the N-terminal arginine with a stabilizing alanine residue (As), which is not recognized by Ubr1, is expected to yield a stable protein (44, 49).
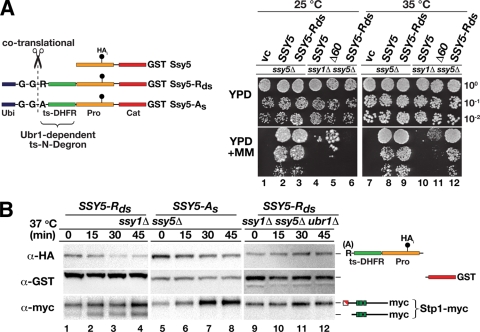
FIG. 5.
Synthetic N-degron-regulated prodomain degradation results in temperature-controlled Stp1 processing. (A) Ssy5 N-degron constructs are schematically represented. The constructs have an N-terminal ubiquitin moiety (Ubi), a destabilizing arginine (R) or a stabilizing alanine (A) residue, and a temperature-sensitive dihydrofolate reductase (ts-DHFR) linker fused at the N terminus of Ssy5 with an internal prodomain HA epitope (HAi) and a C-terminal GST tag. The ubiquitin is cotranslationally cleaved (scissors) following the diglycine (G-G) residues, resulting in proteins with an N-terminal arginine or alanine residue. Arginine, but not alanine, is an N-end rule substrate recognized by Ubr1. Growth of HKY77 (ssy5Δ) and HKY84 (ssy5Δ ssy1Δ) carrying a vector control (vc), pSH120 (SSY5), pSH106 (Δ60), or pTP110 (SSY5-Rds) is shown. Cells were pregrown in SD, dilutions were prepared and spotted onto YPD and YPD plus MM, and the plates were incubated at 25 or 35°C for 6 or 3 days, respectively. pSH106 (Δ60) was used as a positive constitutive control (right panel). (B) Immunoblot analysis of protein extracts from HKY84 (ssy5Δ ssy1Δ) carrying pCA204 (Stp1-myc) and pTP110 (SSY5-Rds, lanes 1 to 4) or pTP111 (SSY5-As, lanes 5 to 8) and from TPY101 (ssy5Δ ssy1Δ ubr1Δ) carrying pCA204 (Stp1-myc) and pTP110 (SSY5-Rds, lanes 9 to 12). Cells were grown in SD at 25°C, cultures were shifted to 37°C at t = 0, and extracts were prepared from equivalent numbers of cells harvested at the indicated time points.
When the SSY5-Rds allele was introduced into an ssy5Δ mutant strain, it fully complemented the MM mutant growth phenotype at both 25 and 35°C (Fig. 5A, compare dilution 2 with dilution 3 and dilution 8 with dilution 9). Thus, the N-degron-containing prodomain is functional and capable of operating in the context of the other SPS sensor components. Importantly, when the SSY5-Rds allele was introduced into an ssy1Δ ssy5Δ double-mutant strain, the cells did not grow at 25°C, indicating that the presence of the N-degron does not constitutively activate Stp1 processing. However, when these cells were incubated at 35°C, a temperature predicted to trigger N-degron-dependent prodomain degradation, robust growth was observed (Fig. 5A, compare dilution 6 with dilution 12). Clearly, the activity of this novel SSY5 allele is temperature regulated and functions independently of a functional SPS sensor.
To directly test if the SSY5-Rds growth phenotype was a result of temperature-induced prodomain degradation, we measured the stability of the prodomain encoded by SSY5-Rds in ssy1Δ ssy5Δ double-mutant cells after shifting the temperature from 25 to 37°C by immunoblot analysis (Fig. 5B). The stability of the prodomain was severely reduced at the elevated temperature, and the prodomain was rapidly degraded, whereas the Cat domain remained stable (Fig. 5B, lanes 1 to 4). The reduced level of the prodomain correlated precisely with Stp1 processing. The temperature-induced degradation and Cat domain activation were fully dependent on the regulated N-degron since substitution of the destabilizing N-terminal arginine residue with the stabilizing alanine residue (Fig. 5B, lanes 5 to 8) or deletion of UBR1 (Fig. 5B, lanes 9 to 12) prevented both prodomain degradation and Stp1 processing. These experiments unambiguously demonstrate that the prodomain functions as a potent inhibitor of the Cat domain and its degradation is sufficient for the endoproteolytic cleavage of Stp1.
DISCUSSION
According to our current understanding of RAP, the Ssy5 endoprotease is expressed as a zymogen. During biogenesis, the prodomain assists the folding of the Cat domain. Once active, the Cat domain autolytically cleaves Ssy5 between the two domains and the noncovalently associated domains remain attached, forming an inactive but primed protease that binds its substrates, Stp1 and Stp2 (3). After amino acid induction of the SPS sensor, the prodomain is degraded by the ubiquitin proteasome system, presumably as a consequence of a conformational change. Once released from its inhibitory interaction with the prodomain, the Cat domain endoproteolytically processes Stp1 and Stp2. Hence, the prodomain is required for maturation of the protease and subsequently functions as a potent inhibitor that prevents Ssy5 proteolytic activity in the absence of signaling.
Consistent with an inhibitory function, we found a tight correlation between reduced prodomain levels and Stp1 processing. This observation can be accounted for by a model in which the prodomain attains two alternative conformations, a stable inhibitory conformation and an unstable noninhibitory conformation. According to this model, the conformational switch from the inhibitory to the noninhibitory conformation is triggered by amino acid-induced signals and leads to the degradation of the prodomain by the 26S proteasome. This model predicts that the constitutive mutant alleles that result in unregulated processing of Stp1 (SSY5*, SSY5Δ60, and substitution mutations in codons 126, 129, and 131) encode prodomains with properties that mimic or favor the unstable noninhibitory conformation. Indeed, the constitutive nature of these alleles was traced to one common attribute; i.e., they all exhibited decreased prodomain levels (Fig. 3 and 4). In contrast, the enhanced stability of the nonsignaling ssy5Δ90 truncated prodomain (Fig. 3) can be explained by its being trapped in the inhibitory conformation. Hence, sequence elements responsible for the induced conformational switch of the prodomain reside N terminally of residue 90, while the basic inhibitory function of the Pro domain resides C terminally of this residue. This mapping is consistent with the observation that our isolated constitutive mutations cluster at positions 126, 129, and 131. In summary, the proposed conformational model of the activation of primed Ssy5 accounts for both the normal and the mutant behavior of this regulated protease.
The mechanisms underlying the amino acid-induced switch of the Ssy5 prodomain from its stable inhibitory to unstable noninhibiting conformation remains elusive. Posttranslational modifications, including phosphorylation and ubiquitylation, are given candidates to function as the trigger. Notably, the casein kinase Yck1 or Yck2 (Yck1/2) is required for SPS sensor signaling. These rather promiscuous kinases have been shown to constitutively phosphorylate Stp1 and to phosphorylate Ptr3 in an amino acid-induced manner (1, 28). Hence, given the involvement of these kinases in both upstream and downstream signaling events, Ssy5 might also be a substrate during SPS sensor signaling. In several instances, Yck1/2-catalyzed phosphorylation has been shown to lead to Grr1-dependent polyubiquitylation and subsequent degradation of the modified substrates by the 26S proteasome (30, 40). Grr1 is an F-box protein of the SCF E3 ubiquitin ligase complex, which is required for transducing SPS sensor-mediated signals (5, 8). Consequently, the possibility that Yck1/2-dependent phosphorylation and Grr1-mediated ubiquitylation govern the conformation of the Pro domain, and hence its inhibitory activity, needs to be investigated.
To unambiguously test the inhibitory role of the prodomain, we synthetically reprogrammed the SPS sensing pathway by placing the degradation of the prodomain under temperature control (Fig. 5). Two experimental aspects require comment. First, at 25°C, the N-degron-containing prodomain is fully functional and capable of operating in the context of the other SPS sensor components. Thus, the presence of the N-degron does not interfere with SPS sensor function, but more importantly, the stability of the N-degron-containing prodomain remains controlled by amino acid-induced and SPS sensor-mediated signaling events. Presumably, Stp1 processing at 25°C is the result of the same conformational switch affecting the stability of the wild-type prodomain. Second, at the elevated inducing temperature, the presence of the synthetic N-degron leads to efficient Stp1 processing in the absence of a functional SPS sensor, a situation normally incompatible with Stp1 processing. Thus, similar to the constitutive active mutations that affect Pro domain stability, direct targeting of the prodomain for degradation, independently of upstream signaling events, suffices to liberate a fully competent Cat domain that processes its substrate. This result demonstrates that the critical event in the activation of primed Ssy5 is liberation of the Cat domain from prodomain inhibition.
Although N-degron-induced prodomain degradation suffices to activate the Cat domain, it remains to be elucidated whether prodomain degradation is required for Stp1 processing in the context of SPS sensor signaling. We raise this issue based on our finding that amino acid-induced processing occurs in the temperature-sensitive proteasomal cim3-1 mutant under conditions where the prodomain exhibits clearly enhanced stability (Fig. 2C). This result, consistent with previous reports (1, 4), opens the possibility that the unstable noninhibitory Pro domain conformation has a decreased affinity for the Cat domain; its dissociation may relieve inhibition. However, rigorous interpretation of the apparent Cat domain activation in the absence of prodomain degradation is confounded by the experimental setup. Specifically, in no instance do the experimental conditions used here or in previously published work (1, 4) result in complete inhibition of the proteasome. Thus, residual proteasomal activity may still account for the observed Stp1 processing. Consequently, at this time it is impossible to evaluate the importance of direct proteasomal involvement in signaling. Regardless, in cells with a fully active ubiquitin proteasome system, amino acid-induced signaling results in reduced prodomain levels, indicating that signaling and degradation are normally coupled events.
An important characteristic of RAP is that Ssy5 autolysis, which occurs concomitantly with its biogenesis, is not sufficient to trigger Stp1 processing (3). In this respect, Ssy5 differs from classical chymotrypsin-like proteases that are most commonly activated by intermolecular proteolytic processing of an inactive zymogen precursor (32). The processing of the prodomain within the zymogen is frequently associated with a conformational change that suffices to fully activate the protease (18, 41, 45). In contrast to this established concept, Ssy5 requires a second signal-mediated activation step.
Although, to our knowledge, the mode of Ssy5 activation that we document here is unique among eukaryotic proteases, similar two-step mechanisms have been described for secreted bacterial serine proteases, e.g., the secreted bacterial protease alpha-lytic protease, a distant relative of Ssy5 (2, 34). Alpha-lytic protease is expressed with a prodomain that functions as an effective catalyst of Cat domain folding (13, 14). Similar to Ssy5, alpha-lytic protease undergoes autolysis upon Cat domain maturation. The cleaved prodomain and Cat domain of alpha-lytic protease remain strongly associated, forming a complex in which the C-shaped prodomain surrounds the beta barrel of the folded Cat domain, sterically blocking the active site. The active Cat domain is liberated from the prodomain by proteolysis in the extracellular milieu (7, 12, 20). Despite the obvious similarities between the two-step activation of Ssy5 and that of alpha-lytic protease, there is one striking difference; i.e., the Ssy5 prodomain requires a signal transduced by a plasma membrane-localized receptor complex to initiate a conformational change that triggers its degradation.
Serine proteases are involved in a variety of important physiological processes in metazoan organisms, e.g., food digestion, blood coagulation, immune response, and signal transduction. Dysregulation of the proteolytic activity of several of these proteases is linked to disease states in humans (11, 22, 31, 33, 45). A thorough understanding of the full repertoire of regulatory processes governing the function of this class of proteases, including the RAP mechanism described here, is a requisite for the development of new approaches to the beneficial modulation of protease functions. Interestingly, with respect to the central regulatory role of the prodomain of Ssy5 in RAP, the potential of targeting prodomains to engineer proteases with unique catalytic properties has been recognized (42). In line with this, we emphasize that we were able to successfully reprogram Ssy5 to control its activity by temperature. In light of the modularity and transferability of the Ssy5 recognition and cleavage site (5), such a protease is an ideal signal input device to engineer synthetic temperature-regulated signaling pathways.
Acknowledgments
We thank members of the Ljungdahl laboratory for constructive comments throughout the course of this work.
This research was supported by the Swedish Research Council (P.O.L.). The Ludwig Institute for Cancer Research is gratefully acknowledged for support during the initial stages of this work.
Footnotes
Published ahead of print on 26 April 2010.
REFERENCES
- 1.Abdel-Sater, F., M. El Bakkoury, A. Urrestarazu, S. Vissers, and B. André. 2004. Amino acid signaling in yeast: casein kinase I and the Ssy5 endoprotease are key determinants of endoproteolytic activation of the membrane-bound Stp1 transcription factor. Mol. Cell. Biol. 24:9771-9785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andréasson, C. 2004. Ligand-activated proteolysis in nutrient signaling. Ph.D. thesis. Karolinska Institute, Stockholm, Sweden.
- 3.Andréasson, C., S. Heessen, and P. O. Ljungdahl. 2006. Regulation of transcription factor latency by receptor-activated proteolysis. Genes Dev. 20:1563-1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Andréasson, C., and P. O. Ljungdahl. 2002. Receptor-mediated endoproteolytic activation of two transcription factors in yeast. Genes Dev. 16:3158-3172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Andréasson, C., and P. O. Ljungdahl. 2004. The N-terminal regulatory domain of Stp1p is modular and, fused to an artificial transcription factor, confers full Ssy1p-Ptr3p-Ssy5p sensor control. Mol. Cell. Biol. 24:7503-7513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Antebi, A., and G. R. Fink. 1992. The yeast Ca(2+)-ATPase homologue, PMR1, is required for normal Golgi function and localizes in a novel Golgi-like distribution. Mol. Biol. Cell 3:633-654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baker, D., J. L. Silen, and D. A. Agard. 1992. Protease pro region required for folding is a potent inhibitor of the mature enzyme. Proteins 12:339-344. [DOI] [PubMed] [Google Scholar]
- 8.Bernard, F., and B. André. 2001. Ubiquitin and the SCF(Grr1) ubiquitin ligase complex are involved in the signalling pathway activated by external amino acids in Saccharomyces cerevisiae. FEBS Lett. 496:81-85. [DOI] [PubMed] [Google Scholar]
- 9.Brivanlou, A. H., and J. E. Darnell, Jr. 2002. Signal transduction and the control of gene expression. Science 295:813-818. [DOI] [PubMed] [Google Scholar]
- 10.Brown, M. S., J. Ye, R. B. Rawson, and J. L. Goldstein. 2000. Regulated intramembrane proteolysis: a control mechanism conserved from bacteria to humans. Cell 100:391-398. [DOI] [PubMed] [Google Scholar]
- 11.Coughlin, S. R. 2000. Thrombin signalling and protease-activated receptors. Nature 407:258-264. [DOI] [PubMed] [Google Scholar]
- 12.Cunningham, E. L., and D. A. Agard. 2004. Disabling the folding catalyst is the last critical step in alpha-lytic protease folding. Protein Sci. 13:325-331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cunningham, E. L., and D. A. Agard. 2003. Interdependent folding of the N- and C-terminal domains defines the cooperative folding of alpha-lytic protease. Biochemistry 42:13212-13219. [DOI] [PubMed] [Google Scholar]
- 14.Cunningham, E. L., T. Mau, S. M. Truhlar, and D. A. Agard. 2002. The pro region N-terminal domain provides specific interactions required for catalysis of alpha-lytic protease folding. Biochemistry 41:8860-8867. [DOI] [PubMed] [Google Scholar]
- 15.Dohmen, R. J., and A. Varshavsky. 2005. Heat-inducible degron and the making of conditional mutants. Methods Enzymol. 399:799-822. [DOI] [PubMed] [Google Scholar]
- 16.Dohmen, R. J., P. Wu, and A. Varshavsky. 1994. Heat-inducible degron: a method for constructing temperature-sensitive mutants. Science 263:1273-1276. [DOI] [PubMed] [Google Scholar]
- 17.Eckert-Boulet, N., B. Regenberg, and J. Nielsen. 2005. Grr1p is required for transcriptional induction of amino acid permease genes and proper transcriptional regulation of genes in carbon metabolism of Saccharomyces cerevisiae. Curr. Genet. 47:139-149. [DOI] [PubMed] [Google Scholar]
- 18.Eder, J., and A. R. Fersht. 1995. Pro-sequence-assisted protein folding. Mol. Microbiol. 16:609-614. [DOI] [PubMed] [Google Scholar]
- 19.Forsberg, H., and P. O. Ljungdahl. 2001. Genetic and biochemical analysis of the yeast plasma membrane Ssy1p-Ptr3p-Ssy5p sensor of extracellular amino acids. Mol. Cell. Biol. 21:814-826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fuhrmann, C. N., B. A. Kelch, N. Ota, and D. A. Agard. 2004. The 0.83 A resolution crystal structure of alpha-lytic protease reveals the detailed structure of the active site and identifies a source of conformational strain. J. Mol. Biol. 338:999-1013. [DOI] [PubMed] [Google Scholar]
- 21.Ghislain, M., A. Udvardy, and C. Mann. 1993. S. cerevisiae 26S protease mutants arrest cell division in G2/metaphase. Nature 366:358-362. [DOI] [PubMed] [Google Scholar]
- 22.Hedstrom, L. 2002. An overview of serine proteases. Curr. Protoc. Protein Sci. Chapter 21:Unit 21.10. [DOI] [PubMed] [Google Scholar]
- 23.Hoppe, T., K. Matuschewski, M. Rape, S. Schlenker, H. D. Ulrich, and S. Jentsch. 2000. Activation of a membrane-bound transcription factor by regulated ubiquitin/proteasome-dependent processing. Cell 102:577-586. [DOI] [PubMed] [Google Scholar]
- 24.Hoppe, T., M. Rape, and S. Jentsch. 2001. Membrane-bound transcription factors: regulated release by RIP or RUP. Curr. Opin. Cell Biol. 13:344-348. [DOI] [PubMed] [Google Scholar]
- 25.Jørgensen, M. U., M. B. Bruun, T. Didion, and M. C. Kielland-Brandt. 1998. Mutations in five loci affecting GAP1-independent uptake of neutral amino acids in yeast. Yeast 14:103-114. [DOI] [PubMed] [Google Scholar]
- 26.Kanemaki, M., A. Sanchez-Diaz, A. Gambus, and K. Labib. 2003. Functional proteomic identification of DNA replication proteins by induced proteolysis in vivo. Nature 423:720-724. [DOI] [PubMed] [Google Scholar]
- 27.Klasson, H., G. R. Fink, and P. O. Ljungdahl. 1999. Ssy1p and Ptr3p are plasma membrane components of a yeast system that senses extracellular amino acids. Mol. Cell. Biol. 19:5405-5416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu, Z., J. Thornton, M. Spirek, and R. A. Butow. 2008. Activation of the SPS amino acid-sensing pathway in Saccharomyces cerevisiae correlates with the phosphorylation state of a sensor component, Ptr3. Mol. Cell. Biol. 28:551-563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ljungdahl, P. O. 2009. Amino-acid-induced signalling via the SPS-sensing pathway in yeast. Biochem. Soc. Trans. 37:242-247. [DOI] [PubMed] [Google Scholar]
- 30.Moriya, H., and M. Johnston. 2004. Glucose sensing and signaling in Saccharomyces cerevisiae through the Rgt2 glucose sensor and casein kinase I. Proc. Natl. Acad. Sci. U. S. A. 101:1572-1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Neurath, H. 1984. Evolution of proteolytic enzymes. Science 224:350-357. [DOI] [PubMed] [Google Scholar]
- 32.Neurath, H., and G. H. Dixon. 1957. Structure and activation of trypsinogen and chymotrypsinogen. Fed. Proc. 16:791-801. [PubMed] [Google Scholar]
- 33.Page, M. J., R. T. Macgillivray, and E. Di Cera. 2005. Determinants of specificity in coagulation proteases. J. Thromb. Haemost. 3:2401-2408. [DOI] [PubMed] [Google Scholar]
- 34.Poulsen, P., L. Lo Leggio, and M. C. Kielland-Brandt. 2006. Mapping of an internal protease cleavage site in the Ssy5p component of the amino acid sensor of Saccharomyces cerevisiae and functional characterization of the resulting pro- and protease domains by gain-of-function genetics. Eukaryot. Cell 5:601-608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Poulsen, P., B. Wu, R. F. Gaber, K. Ottow, H. A. Andersen, and M. C. Kielland-Brandt. 2005. Amino acid sensing by Ssy1. Biochem. Soc. Trans. 33:261-264. [DOI] [PubMed] [Google Scholar]
- 36.Sanchez-Diaz, A., M. Kanemaki, V. Marchesi, and K. Labib. 2004. Rapid depletion of budding yeast proteins by fusion to a heat-inducible degron. Sci. STKE 2004:PL8. [DOI] [PubMed] [Google Scholar]
- 37.Schork, S. M., M. Thumm, and D. H. Wolf. 1995. Catabolite inactivation of fructose-1,6-bisphosphatase of Saccharomyces cerevisiae. Degradation occurs via the ubiquitin pathway. J. Biol. Chem. 270:26446-26450. [DOI] [PubMed] [Google Scholar]
- 38.Schüle, T., M. Rose, K. D. Entian, M. Thumm, and D. H. Wolf. 2000. Ubc8p functions in catabolite degradation of fructose-1,6-bisphosphatase in yeast. EMBO J. 19:2161-2167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Silve, S., C. Volland, C. Garnier, R. Jund, M. R. Chevallier, and R. Haguenauer-Tsapis. 1991. Membrane insertion of uracil permease, a polytopic yeast plasma membrane protein. Mol. Cell. Biol. 11:1114-1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Spielewoy, N., K. Flick, T. I. Kalashnikova, J. R. Walker, and C. Wittenberg. 2004. Regulation and recognition of SCFGrr1 targets in the glucose and amino acid signaling pathways. Mol. Cell. Biol. 24:8994-9005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stroud, R. M., A. A. Kossiakoff, and J. L. Chambers. 1977. Mechanisms of zymogen activation. Annu. Rev. Biophys. Bioeng. 6:177-193. [DOI] [PubMed] [Google Scholar]
- 42.Takagi, H., and M. Takahashi. 2003. A new approach for alteration of protease functions: pro-sequence engineering. Appl. Microbiol. Biotechnol. 63:1-9. [DOI] [PubMed] [Google Scholar]
- 43.Thomas, B. J., and R. Rothstein. 1989. Elevated recombination rates in transcriptionally active DNA. Cell 56:619-630. [DOI] [PubMed] [Google Scholar]
- 44.Varshavsky, A. 2008. The N-end rule at atomic resolution. Nat. Struct. Mol. Biol. 15:1238-1240. [DOI] [PubMed] [Google Scholar]
- 45.Whitcomb, D. C., and M. E. Lowe. 2007. Human pancreatic digestive enzymes. Dig. Dis. Sci. 52:1-17. [DOI] [PubMed] [Google Scholar]
- 46.Wielemans, K., C. Jean, S. Vissers, and B. André. 2010. Amino acid signaling in yeast: post-genome duplication divergence of the Stp1 and Stp2 transcription factors. J. Biol. Chem. 285:855-865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wilson, D. S., and A. D. Keefe. 2001. Random mutagenesis by PCR. Curr. Protoc. Mol. Biol. Chapter 8:Unit 8.3. [DOI] [PubMed] [Google Scholar]
- 48.Wu, B., K. Ottow, P. Poulsen, R. F. Gaber, E. Albers, and M. C. Kielland-Brandt. 2006. Competitive intra- and extracellular nutrient sensing by the transporter homologue Ssy1p. J. Cell Biol. 173:327-331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Xia, Z., A. Webster, F. Du, K. Piatkov, M. Ghislain, and A. Varshavsky. 2008. Substrate-binding sites of UBR1, the ubiquitin ligase of the N-end rule pathway. J. Biol. Chem. 283:24011-24028. [DOI] [PMC free article] [PubMed] [Google Scholar]