Abstract
The non-long terminal repeat (non-LTR) retrotransposon R2 is inserted into the 28S rRNA genes of many animals. Expression of the element appears to be by cotranscription with the rRNA gene unit. We show here that processing of the rRNA cotranscript at the 5′ end of the R2 element in Drosophila simulans is rapid and utilizes an unexpected mechanism. Using RNA synthesized in vitro, the 5′ untranslated region of R2 was shown capable of rapid and efficient self-cleavage of the 28S-R2 cotranscript. The 5′ end generated in vitro by the R2 ribozyme was at the position identical to that found for in vivo R2 transcripts. The RNA segment corresponding to the R2 ribozyme could be folded into a double pseudoknot structure similar to that of the hepatitis delta virus (HDV) ribozyme. Remarkably, 21 of the nucleotide positions in and around the active site of the HDV ribozyme were identical in R2. R2 elements from other Drosophila species were also shown to encode HDV-like ribozymes capable of self-cleavage. Tracing their sequence evolution in the Drosophila lineage suggests that the extensive similarity of the R2 ribozyme from D. simulans to that of HDV was a result of convergent evolution, not common descent.
A critical step in the life cycle of a retrotransposable element is the generation of an RNA transcript that serves as a substrate for both protein synthesis and reverse transcription. For those elements with long terminal repeats (LTRs), the sequences necessary for starting and stopping transcription are located within the repeats, and the result is a terminally redundant RNA. Most elements without LTRs (i.e., the non-LTR retrotransposons or LINES) appear to encode internal promoters that initiate transcription at the first base of the element (7, 25, 26, 35). However, other non-LTR retrotransposons have been suggested to rely on their cotranscription with active transcription units already present within the genome (9). This cotranscription approach appears to be an attractive option for those families of elements that have evolved a preference for insertion into sites within specific genes. Examples of these site-specific elements include multiple families of R elements that are inserted into rRNA genes and the families of elements that are inserted into spliced leader exons (9, 11, 15, 24, 36).
Best studied among the site-specific non-LTR retrotransposons are the R2 elements that are inserted into the 28S rRNA genes (Fig. 1 A) (9). R2 elements have existed since the origin of most animal taxa, and all active copies reported to date are found in the same, highly conserved site of the 28S rRNA gene (2, 16, 17, 34). The exceptional sequence specificity of R2 has enabled detailed studies of its retrotransposition mechanism (5); however, few specifics about the generation of the R2 transcript were known. In Drosophila species, full-length R2 transcripts are readily detected in animals undergoing R2 retrotransposition (8). Nuclear run-on transcription experiments indicate that R2 RNA abundance is controlled at the level of transcription rather than at the posttranscriptional level (8, 38). Attempts to identify a promoter at the 5′ ends of R2 elements have proven unsuccessful (12). Instead, R2 transcriptional regulation appears to be accomplished by whether ribosomal DNA (rDNA) units with R2 insertions are themselves transcribed (8, 38, 40). An important question thus becomes that of how R2 transcripts are processed from 28S-R2 cotranscripts.
FIG. 1.
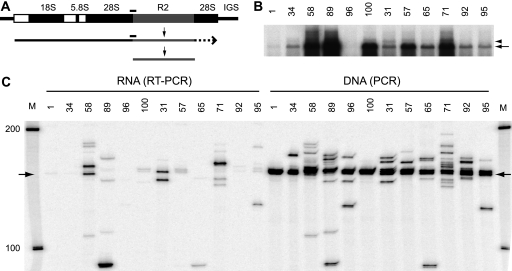
Analysis of full-length R2 elements and their transcripts in D. simulans. (A) Diagram of an rRNA transcription unit with 18S, 5.8S, and 28S genes (black boxes), transcribed spacers (white boxes), and R2 insertion (gray box). R2 RNA transcripts are diagramed as processed from a cotranscript with the rRNA unit. Black bars above the DNA unit and the RNA cotranscript represent the products of PCR and RT-PCR assays, respectively, as shown in panel C. (B) Northern blot of total RNA from 12 D. simulans stocks probed with a fragment from the 5′ end of the R2 element. The arrow points to the full-length, 3,600-nt R2 transcript. The arrowhead points to the presumed 28S cotranscript discussed in the text. (C) A primer complementary to sequences within the R2 element was annealed to total RNA from the 12 stocks and used to prime reverse transcription by the M-MLV reverse transcriptase. The resulting cDNA was PCR amplified using an end-labeled 28S primer (80 nt upstream of the junction) and an R2 primer (108 nt downstream), and the products were separated on an 8 M urea, 7.5% polyacrylamide gel (left half). Genomic DNAs from each stock were directly amplified using the same PCR primers to reveal the lengths of R2 junctions present in each stock (right half). Arrows point to product lengths from the common R2 junction found in all stocks.
Two possible pathways exist for the processing of R2 sequences from a 28S rRNA cotranscript. First, the R2 junctions could mimic processing sites that are involved in the generation of mature 18S, 5.8S, and 28S rRNAs from the single RNA precursor (21). However, previously noted conserved sequences and secondary structures identified at the 5′ and 3′ ends of the R2 elements appear unrelated to the normal processing sites in rRNA precursors (12, 13, 32). Second, the R2 RNA itself could be autocatalytic. Examples of such autocatalysis include self-splicing required for the propagation of group I and group II introns as well as for the expression of the inserted gene (3). Indeed, several group I introns are inserted into the large rRNA genes at positions near the R2 insertion site (18, 27). In addition, group II introns utilize a retrotransposition mechanism with many similarities to the mechanism described for R2 retrotransposition (19). However, the conserved sequences and secondary structures at the 5′ and 3′ ends of the R2 elements noted above also appeared to be unrelated to group I and group II introns (12, 13, 32). Furthermore, the R2 target sequences make up part of the catalytic site of the large subunit. Therefore, the frequent deletions and insertions observed at the junctions of R2 elements with the 28S gene suggest that even if splicing did occur, the resulting 28S rRNA would not be functional (9).
Alternatively, a class of smaller autocatalytic RNAs is characterized by self-cleavage (6). Most of these RNAs have been found associated with RNA viruses and viroids and function to process the RNA from longer replication precursors (6). In this report, we show that the 5′ end of the R2 transcript forms a ribozyme capable of efficient self-cleavage at the junction between the 28S rRNA and the R2 element. The R2 ribozyme has remarkable structural and sequence identity to the ribozyme encoded by the hepatitis delta virus (HDV).
MATERIALS AND METHODS
Nucleic acid isolation.
Total RNA and genomic DNA were isolated from adult flies as previously described (8, 20). Initial nucleic acid pellets were treated with either DNase I (0.1 unit/μl; Promega) or RNase A (0.2 μg/μl; Sigma), extracted with phenol, and ethanol precipitated. The integrity of the DNA and RNA on 1% agarose gels was monitored and their concentrations estimated by optical density at 260 nm.
Northern blot analysis.
Total RNA (10 μg) from each Drosophila simulans stock was separated on 2.2 M formaldehyde-0.8% agarose gels as previously described (8). RNAs were transferred to a nylon membrane and probed with a 250-nucleotide (nt) α-32P-labeled antisense RNA generated from the 5′ end of the R2 element (8).
PCR and RT-PCR amplifications.
Procedures were largely as previously described (8). Total RNAs (0.1 μg) were incubated with a primer annealed to the R2 sequence from 505 to 525 nucleotides from the 5′ end [R2(525); 5′-CAACCTCCTTTCTCGCCATC-3′] in buffer (50 mM Tris-HCl [pH 8.3], 75 mM KCl, 3 mM MgCl2, 20 mM dithiothreitol [DTT]) at 65°C for 5 min. The annealed product was then incubated with 1 mM deoxynucleoside triphosphates (dNTPs), 20 mM DTT, 20 units of Moloney murine leukemia virus (M-MLV) reverse transcriptase (RT; Invitrogen) in the same buffer for 50 min at 37°C. Two microliters of the RT reaction mixture was then used in a standard PCR amplification using a primer annealed to R2 sequences from 87 to 108 nt from the 5′ end [R2(108); 5′-AGAGACTTGTGAGTTACAGAG-3′] paired with a γ-32P-end-labeled primer annealed to the 28S gene 81 to 61 nucleotides upstream of the R2 insertion site [28S(−81); 5′-TGCCCAGTCCTCTGAATGTC-3′]. Parallel PCR amplifications using genomic DNA from each of the stocks were done using the same primers. The amplified DNAs were denatured and separated on an 8 M urea, 7.5% polyacrylamide gel.
DNA templates for the T7 transcription reactions.
For D. simulans, the DNA segment from position −170 of the 28S rRNA gene (5′-CAATCAATTCAGAACTGGCACG-3′) to position 342 of the R2 element (5′-CGCTGCGTTTGGTTCATATTGGTC-3′) was PCR amplified from genomic DNA and cloned into the pCR2.1-TOPO cloning vector (Invitrogen). Multiple clones were sequenced, and an insert with a standard R2 junction was removed from the cloning vector by digestion with EcoRI and gel purified. DNA templates of the desired size and with an upstream T7 promoter for RNA synthesis were then generated by PCR amplification. The primers used were 28S(−95) (5′-TAATACGACTCACTATAGGGCACAATGTGATTTCTGCCCAGT-3′), 28S(−24) (5′-TAATACGACTCACTATAGGGGGAGTAACTATGACTCTCTTAAGG-3′), 28S(−7) (5′-TAATACGACTCACTATAGGGGGTACCTAATAAGCTTAAGGGGATCTGGGGTAATTGCGAG), R2(250) (5′GTGTTCGTAGTTCCAATATGAGTA-3′), R2(225) (5′-ATTTCCTCTGGGTAACCAGC-3′), R2(200) (5′-GGAAAAGTTTTTGCCGCTCA-3′), R2(184) (5′-CTCAAATTAGCTGCTAAACA-3′), R2(177) (5′-TAGCTGCTAAACAAGTTTAG-3′), R2(165) (5′-AAGTTTAGCATTACCGGGGA-3′), and R2(155) (5′-TTACCGGGGACCACCACGAGG-3′). Unincorporated primers and nucleotides were removed with a Qiaquick PCR purification kit (Qiagen). A plasmid in which the 116-nt J1/2 segment of D. simulans was replaced with a 10-nt linker (see Fig. 3, construct l) was generated by separate PCR amplification of the two flanking regions from a standard R2 junction with primers containing a BglII site. Primer 28S(−170) was paired with primer 5′-CTAGAGATCTGATCCCCTTAAGAGAGTCATA-3′, and primer R2(342) was paired with primer 5′-CTAGAGATCTCTCAAAACCTCCTCGTGGTGG-3′. The amplified products were digested with BglII, ligated with T4 ligase, reamplified using primers 28S(−170) and R2(342), and cloned as described above in pCR2.1-TOPO. DNA templates containing 28S/R2 junctions from four other Drosophila species were generated using the PCR primers 28S(−24) and either R2D. yakuba (142) (5′-CTCAAATTAGCTGCTAAACA-3′), R2D. ananassae (132) (5′-CTCAAATCAGCTACCAGTAA-3′), R2D. pseudoobscura (154) (5′-TTCAAGTTAGTTACTATTAG-3′), or R2D. falleni (185) (5′-TCGCTCGAATTAGCTGCTAATCAG-3′). PCR products were cloned and sequenced and DNA templates generated as described above.
FIG. 3.
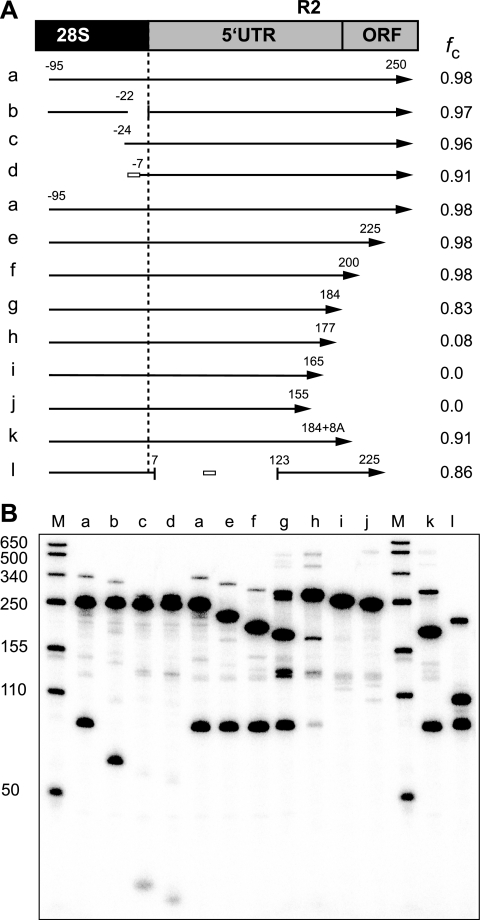
The 5′ end of R2 RNA can function as a self-cleaving ribozyme. (A) Diagram of the 5′ junction of the R2 element with the 28S gene. Only the region corresponding to the R2 5′ untranslated region (5′ UTR) and beginning of the open reading frame (ORF) are shown. Arrows below the diagram represent the in vitro-generated RNAs tested for self-cleavage (fc). Negative numbers indicate the position within the upstream 28S sequences and positive numbers the R2 sequences. White boxes represent substituted sequences for some regions. To the right of each arrow is the average fraction of RNA cleaved in two separate cotranscription/cleavage assays. (B) A 5% acrylamide denaturing gel showing the products for each of the tested RNAs in cotranscription/cleavage assays. Lane M, RNA length markers with sizes indicated.
Cotranscription/cleavage assay.
Approximately 0.1 μg of PCR template was incubated in transcription buffer (40 mM Tris-HCl [pH 7.9]; 6 mM MgCl2; 10 mM NaCl; 10 mM DTT; 1 mM each of ATP, CTP, and GTP; 0.5 mM UTP; 20 units RNaseOUT [Invitrogen]; 20 units T7 RNA polymerase [Fermentas]; and trace amounts of [α-32P]UTP) for 1 h at 42°C. Reactions were stopped by the addition of 4 volumes of 95% formamide, 10 mM EDTA on ice. RNA products were denatured at 92°C for 3 min and separated on 8 M urea, 5% polyacrylamide gels and the dried gels exposed to a phosphorimager screen and analyzed using QuantityOne (Bio-Rad).
Isolation of uncleaved RNA.
RNAs were generated by T7 RNA polymerase as described above, except the reactions were done at 25°C. Gel fragments containing the full-length RNAs were excised and soaked overnight in 300 mM Na acetate, 10 mM EDTA, 0.1% SDS, 1 mg/ml yeast tRNA. The solution was then extracted with phenol equilibrated with 10 mM Tris-HCl (pH 7.9), 5 mM EDTA-chloroform-isoamyl alcohol (1:1:0.01) and then extracted with chloroform-isoamyl alcohol (1:0.01) and ethanol precipitated. RNA was resuspended on in 10 mM Tris-HCl (pH 7.9) on ice.
Primer extensions.
A γ-32P-5′-end-labeled primer ending at position 108 of the R2 element (described above) was annealed to either 10 μg of total RNA or 30 ng of in vitro-generated RNA with or without 5 μg of nonspecific RNA in buffer (50 mM Tris-HCl [pH 8.3], 75 mM KCl, 3 mM MgCl2, 20 mM DTT) at 65°C for 20 min and then cooled at 25°C for 10 min. The reactions were then continued as described above for RT amplification. The cDNAs were denatured and separated on 8 M urea, 7.5% polyacrylamide gel next to a 35S-labeled sequencing ladder.
RESULTS
Analysis of R2 transcripts.
Individual stocks of D. simulans have significantly different levels of R2 transcripts (8). A Northern blot of RNA isolated from 12 stocks probed with sequences from the 5′ end of the R2 element is shown in Fig. 1B. The five stocks previously found to support active R2 retrotransposition, stocks 58, 89, 100, 57, and 71 (8, 39), had the highest levels of a 3,600-nt RNA, with 3,600 nt being the expected size of a full-length R2 transcript (Fig. 1B, arrow). Longer transcripts, corresponding to potential R2-28S rRNA cotranscripts, could also be detected (arrowhead); however, their abundance did not correlate with the level of the full-length R2 transcripts (e.g., compare stocks 31 and 57). In a more sensitive assay, 28S-R2 cotranscripts were scored by RT-PCR. A primer annealing to sequences within the R2 element was first used to prime reverse transcription of total RNA. The resulting cDNA was then used for PCR amplification with one primer located within the 28S gene a short distance upstream of the R2 insertion site and the second primer within the R2 element (Fig. 1C, left side). There was little correlation between the levels of these RT-PCR products and the levels of full-length R2 transcripts seen in Fig. 1B. More importantly, the RT-PCR products differed in length between stocks, suggesting that they did not correspond to transcripts derived from the standard (i.e., the most common) R2 junction found in all D. simulans stocks (12, 34).
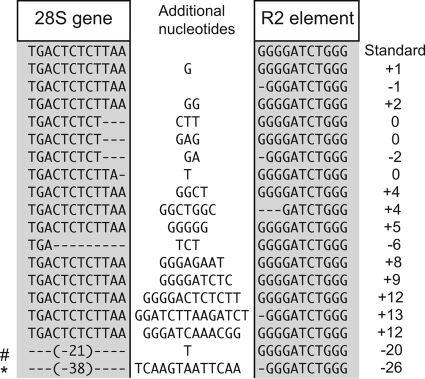
To survey the 5′ junctions of R2 copies within each stock, the same PCR primers used in the RT-PCR were also used to directly amplify genomic DNA (Fig. 1C, right side). From 50 to 98% of the R2 copies in each stock gave rise to products that were within a few base pairs of the standard length (Fig. 1C, intense band indicated with arrow). To describe the origin of the many bands that differed from this common length, the 5′ junctions of R2 copies from several stocks were cloned and sequenced. A summary of the different R2 5′ junction sequences obtained is shown in Fig. 2. The variant junctions contained from 1 to 38 nucleotide deletions of the 28S gene, 1 to 3 nucleotide deletions of the R2 element, and 1 to 14 additional nucleotides located between the 28S gene and R2 sequences. The additional nucleotides corresponded to short duplications of the terminal R2 sequences as well as apparently random sequences. This 5′ sequence variation, seen at different levels with the R2 elements in all species, was previously suggested to be a result of the inefficiency of the R2 reverse transcriptase (polymerase) in using the upstream target sequence to prime synthesis of the second DNA strand during retrotransposition (5).
FIG. 2.
5′ junctions of full-length R2 elements. Sequences were derived by PCR from several D. simulans stocks shown in Fig. 1 (34, 58, and 89) or obtained from the GenBank trace archives of the D. simulans genome. Shaded boxes indicate the sequences corresponding to the upstream 28S gene or the R2 element. Nucleotides added during the retrotransposition process are shown in the middle unshaded region. Many of the full-length R2 junctions in the D. simulans stocks studied here are identical and define the standard (most common) junction (top sequence). All other junctions are present at low frequencies. To the right of each sequence is indicated the difference in length, compared to the standard junction length, of PCR or RT-PCR products in assays similar to those represented in Fig. 1C. #, the junction used as construct b in Fig. 3. *, the junction tested for self-cleavage in Fig. 5.
The insertions and deletions of sequences at the 5′ end of the D. simulans R2 elements readily explain the different lengths of the PCR products seen in Fig. 1C. Comparison of these direct PCR products with the RT-PCR products also seen in this figure reveals that most of the 28S-R2 cotranscripts that could be detected corresponded to R2 elements with 5′ ends that differed from the common sequence. The paucity of 28S-R2 cotranscripts corresponding to the many common junctions present in all stocks suggested that if the R2 transcripts seen on a Northern blot were derived by cotranscription with the 28S gene, then their cleavage from the 28S gene must be extremely rapid.
Identification of the R2 ribozyme.
The apparent speed of the R2 5′ end processing suggested that this step might be autocatalytic rather than dependent upon cellular RNases. This model was tested by using T7 RNA polymerase to generate in vitro RNA comprising 170 nt of the upstream 28S rRNA sequences and up to 525 nt of the R2 sequences. Depending on the length of the DNA template and the conditions of the transcription reaction, from 15% to over 90% of the RNA products detected by electrophoresis after T7 transcription corresponded to the sizes predicted from self-cleavage near the 28S-R2 junction (data not shown).
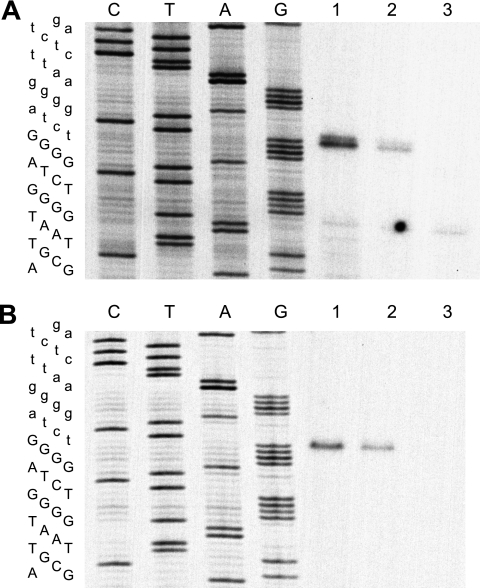
A series of templates were tested in cotranscription/cleavage assays to determine the minimum length of the 28S and R2 RNA regions responsible for self-cleavage (Fig. 3). RNA from a construct which would initially give rise to a 345-nt product (Fig. 3A, construct a) was observed to be 98% cleaved into 95-nt and 250-nt fragments during the transcription reaction (Fig. 3B, lane a). Further analysis of the 28S deletions revealed that RNA retaining only 7 nucleotides of the 28S sequence could support cleavage (lane d), as did RNA in which the 28S sequences from position −21 to position −1 had been deleted (lane b). Thus, upstream 28S sequences were not required for the formation or activity of the R2 ribozyme. Analysis of the series of R2 sequence deletions revealed that deletion after position 200 maintained maximal cleavage (lanes a, e, and f), deletion after position 177 reduced cleavage significantly (lane h), and deletion beyond position 165 eliminated cleavage (lanes i and j).
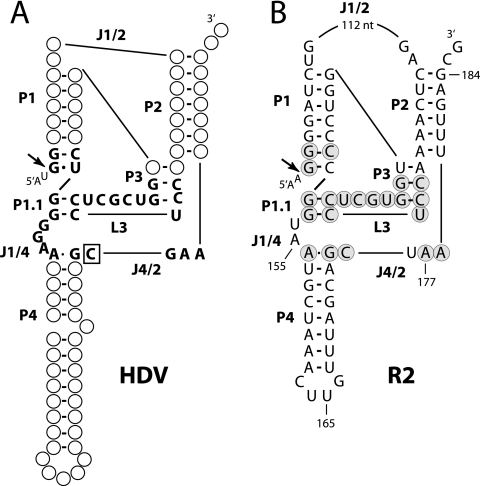
With the ribozyme pinpointed within the first 200 nucleotides of the R2 element, possible secondary structures of this region were compared to the five known classes of self-cleaving RNAs (6). R2 RNA could be readily folded into a structure similar to those of the genomic and antigenomic ribozymes of hepatitis delta virus (HDV) (1, 10). Comparison of the R2 structure to that of the genomic HDV ribozyme is shown in Fig. 4. The HDV ribozyme (Fig. 4A) is composed of a double pseudoknot with five base-paired regions (P1 to P4 and P1.1). The residues involved in catalysis are located in loop region 3 (L3) and in the nucleotides joining P1 and P4 (J1/4) and P4 and P2 (J4/2). Two C nucleotides in L3 anneal to G nucleotides in J1/4 to form P1.1. On the basis of mutagenesis experiments, with the exception of the catalytic nucleotide (Fig. 4A, boxed C in segment J4/2), none of the HDV nucleotides are absolutely necessary for cleavage (1, 28). The similarly folded R2 ribozyme (Fig. 4B) differs from the HDV structure only in that the joining segment J1/2 is over 100 nt longer in R2, and the length of L3 is 1 nucleotide shorter in R2. Remarkably, of 27 positions in and around the catalytic core of HDV, 21 are the same as those in the R2 structure (Fig. 4B, shaded R2 nucleotides).
FIG. 4.
R2 RNA sequences can be folded into a structure similar to that of the HDV ribozyme. (A) Diagram of the HDV genomic ribozyme (1, 5, 10). P, base-paired region; L, loop at the end of a P region; J, nucleotides joining paired regions. Only nucleotides within and near the active site are presented with the catalytic C in J4/2 boxed. (B) Folded structure of the R2 standard sequence given the same nomenclature as HDV. Nucleotides in and around the putative active site that are in common with those of HDV are enclosed in gray circles. Endpoints of constructs g, h, i, and j in Fig. 3 are indicated. Arrows indicate the HDV and R2 self-cleavage sites (Fig. 6). Sequences upstream of the cleavage site are indicated with a smaller font.
The model for the R2 ribozyme shown in Fig. 4 suggested that additional RNA constructs should be analyzed. First, the 116-nucleotide J1/2 region of R2 was replaced with a 10-nucleotide linker (Fig. 3, construct l). This RNA cleaved efficiently, suggesting that the long J1/2 segment in R2 had minimal effect on the formation or activity of the ribozyme. Second, deletion of the R2 sequences beyond position 184 (construct g) reduced cleavage from 98% to 83% even though this RNA should contain all the components of the ribozyme. An RNA which contained 184 nt of the R2 sequence as well as eight A nucleotides (construct k) was therefore tested. The presence of the poly(A) tail increased self-cleavage, suggesting that R2 sequences downstream of the structure in Fig. 4 may be involved in folding of the ribozyme, perhaps by allowing the correct structure to form before the T7 RNA polymerase releases the RNA.
Finally, the finding that no upstream 28S sequences were needed for the formation or activity of the R2 ribozyme suggested that many of the R2 junctions shown in Fig. 2 that contained deletions of 28S sequences or additional nucleotides would be able to self-cleave from a 28S cotranscript. Indeed, construct b used in Fig. 3 was just such a naturally occurring R2 variant junction (Fig. 2, sequence identified by “#”). Another frequent R2 variation found in D. simulans was the deletion of the terminal G nucleotide from the R2 element itself. In 3 of the 4 examples shown in Fig. 2, the nucleotide immediately upstream of the deletion is an A residue. In these cases, the presumed folded RNAs have an A-C wobble at the base of the P1 stem (Fig. 4B). Because the HDV ribozyme has been found to self-cleave with the same A-C wobble (30), one of these R2 5′ variants (Fig. 2, junction labeled with an asterisk) was tested for self-cleavage. Under the same transcription/cleavage conditions used for Fig. 3, 84% of the RNA generated with the variant A-C junction underwent self-cleavage, compared to 98% of the RNA with the standard R2 5′ junction (Fig. 5 A).
FIG. 5.
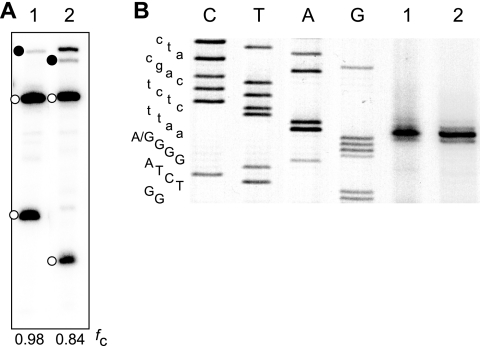
R2 self-cleavage can occur with an A-C wobble at the base of the P1 stem. RNAs were tested in coupled T7 transcription/cleavage reactions as described for Fig. 3. (A) RNA 1 has the standard R2 junction, while RNA 2 has the junction indicated with an asterisk in Fig. 2. Because of a deletion, this junction has an A at position 1 of the R2 element rather than a G and thus would contain an A-C wobble at the base of the P1 segment (Fig. 4B). A 5% polyacrylamide denaturing gel showing the self-cleavage products for the two RNAs is shown (intervening lanes were removed). Solid circles indicate uncleaved RNA, while open circles indicate the self-cleaved products. The fraction of each RNA undergoing self-cleavage is indicated at the bottom of the gel. (B) The cleavage site for each RNA was determined by primer extension with the products run on an 8 M urea, 7.5% polyacrylamide gel as described for Fig. 6. The sequence ladder is indicated to the left of the gel with R2 sequences in capital letters and upstream sequences in lower case. The G-to-A difference at position 1 of the sequence in RNAs 1 and 2 is indicated.
Comparison of the 5′ ends of in vivo- and in vitro-generated R2 transcripts.
To determine whether self-cleavage of the R2 ribozyme occurred at the site predicted by its similarity to HDV, primer extension experiments with M-MLV reverse transcriptase were conducted with the in vitro cleavage products (Fig. 6 A). The R2 cleavage site was 5′ to the G residue at the base of P1, identical to the cleavage position in HDV (Fig. 4A and B, arrows). Primer extension experiments conducted with total RNA isolated from two R2 active stocks of D. simulans revealed the same 5′ junction (Fig. 6B). The cleavage location of the RNA containing an A-C wobble at the base of region P1 was also tested (Fig. 5B). As was found with the HDV ribozyme (30), substitution of the A-C wobble did not change the location of the cleavage site. The similarity of the in vivo R2 5′ ends to that generated in vitro strongly suggests that the 5′ end of endogenous R2 transcripts detected in flies are generated by self-cleavage of the R2 RNA from a 28S rRNA-R2 cotranscript. Furthermore, in spite of the considerable sequence variation at the R2 5′ junctions, self-cleaved R2 transcripts are identical in length (Fig. 6B).
FIG. 6.
Location of the R2 self-cleavage site. (A) RNA was made as in the standard cotranscription/cleavage assay, using constructs g (lane 1), h (lane 2), and j (lane 3), as described for Fig. 3. M-MLV reverse transcriptase was used to generate cDNA from each cleavage product using an annealed primer ending at position 108 in the R2 element. The cDNA was run on an 8 M urea, 7.5% polyacrylamide gel next to a sequencing ladder generated from a plasmid containing an R2 junction using the “108” primer. The R2 sequence is presented to the left of the gel in capital letters and upstream sequences in lowercase. (B) Similar to that in panel A, except 10 μg of total RNA from three D. simulans stocks was used for the primer extension reactions. Lane 1, stock 89; lane 2, stock 100; and lane 3, stock 96 (see R2 transcript levels in Fig. 1B). The extension products 1 nucleotide longer than this position, which are readily detected for the in vitro cleavage but blurred by the necessity of using an intensifying screen for the in vivo cleavage, arose by the reverse transcriptase, adding an additional base as it ran off the RNA template (29).
Temperature and ionic requirements for R2 RNA self-cleavage.
The fraction of the RNA undergoing cleavage during the T7 transcription reaction was found to decrease as the temperature of the reaction was decreased. For example, with the use of construct a in Fig. 3, RNA synthesis by T7 RNA polymerase at 25°C resulted in only 30% of the RNA recovered as self-cleaved products, RNA synthesis at 15°C resulted in 15% self-cleaved products, and RNA synthesis at 5°C resulted in less than 5% self-cleaved products. Therefore, the T7 transcription reaction was conducted at 25°C, the full-length (noncleaved) RNA band isolated from denaturing gels at low ionic strength in the presence of EDTA, and the cleavage reaction examined under different conditions.
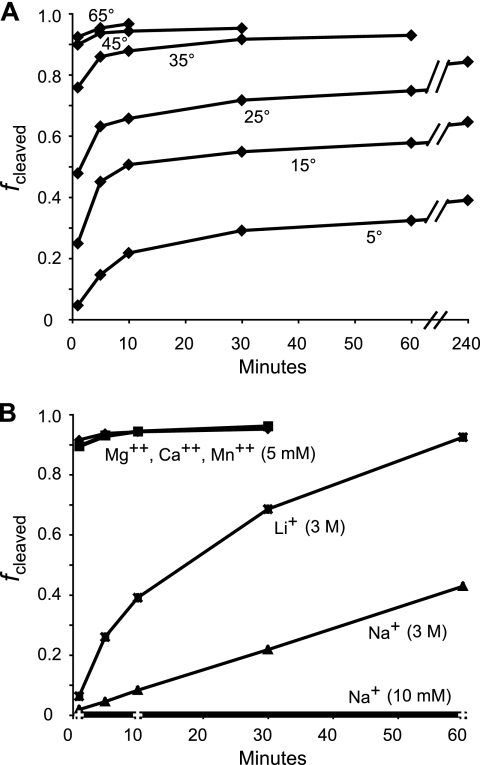
As in the coupled transcription/cleavage reaction, temperature was a critical factor in the rate of R2 self-cleavage. As shown in Fig. 7 A, in 6 mM MgCl2, 10 mM NaCl most RNA was cleaved in the first few minutes of the incubations at 35°C and above. Incubation at 25°C, 15°C, and 5°C resulted in lower levels of cleavage and revealed what appears to be two phases to the cleavage reaction. A significant fraction of the total cleavage occurs in the first 5 min, with further cleavage occurring only slowly over time. This suggests that the addition of Mg2+ to start the reaction induces the rapid assembly of the RNA into active and inactive conformations. Cleavage of the properly folded RNA occurs rapidly while misfolded RNA refolds to the active ribozyme conformation with subsequent cleavage at a rate dependent on the temperature.
FIG. 7.
Temperature and ionic requirements of the R2 self-cleavage reaction. (A) Uncleaved RNA comprising 95 nt of upstream 28S and 200 nt of R2 was synthesized with T7 RNA polymerase at 25°C, isolated from a 5% acrylamide denaturing gel, purified in the presence of 10 mM EDTA, and resuspended in 10 mM Tris-HCl (pH 7.9) on ice. The RNA was equilibrated at the indicated temperature and the reaction started by making the solution 6 mM MgCl2, 10 mM NaCl, and 40 mM Tris-HCl (pH 7.9). (B) Uncleaved RNA isolated as for panel A was incubated in 10 mM NaCl, 40 mM Tris at 40°C. The reaction was started by making the solution 5 mM MgCl2, 5 mM CaCl2, 5 mM MnCl2, 3 M LiCl with 10 mM EDTA or 3 M NaCl with 10 mM EDTA. Data for an incubation of the RNA in 10 mM NaCl and 40 mM Tris-HCl (pH 7.9) are also shown.
When the ionic conditions were varied (Fig. 7B), cleavage was observed to be highly dependent upon the presence of divalent cations. Either 5 mM Mg2+, Ca2+, or Mn2+ supported over 90% cleavage within a few minutes. Interestingly, incubation in Mn2+ (and prolonged incubations in Ca2+ and Mg2+) supported secondary cleavage at a site within J1/2 where another GGGGA sequence is capable of forming the first five base pairs of the P1 stem (Fig. 4). Similar to what was found for HDV, high levels of Na+ and Li+ (3 M) also supported cleavage, but at much lower rates, giving rise to uncertainty as to whether divalent cations are involved only in the formation of the correct structures or also in the catalytic reaction itself (4, 31).
Evolution of the R2 ribozyme within the Drosophila genus.
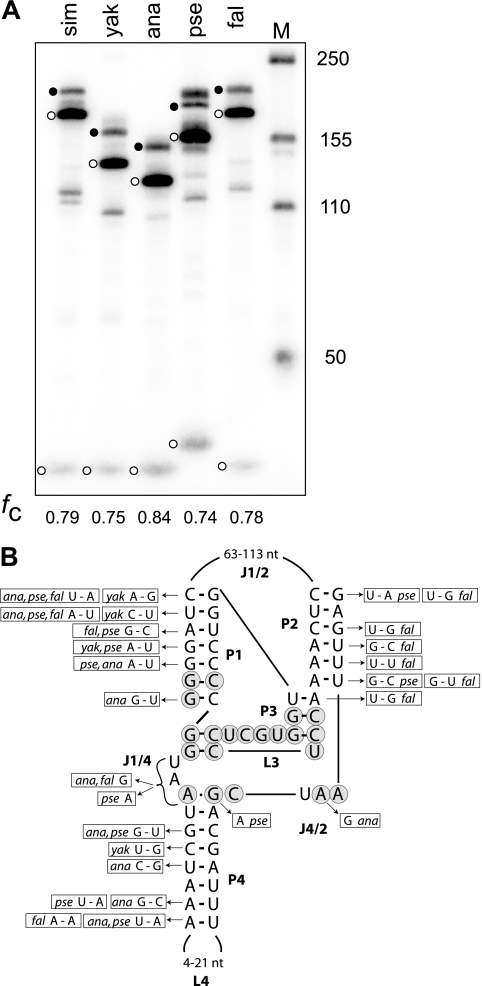
Because R2 elements have been active in the rRNA genes of most Drosophila lineages since the origin of this genus (estimated 50 million years) (20, 34), the evolution of the R2 ribozyme could be followed over this time period. RNA sequences from the R2 elements of other Drosophila species could be folded into a double pseudoknot structure similar to those of D. simulans and HDV. To test if these RNAs could self-cleave, R2 5′ junctions from four species that spanned much of the evolutionary history of Drosophila (Drosophila yakuba, D. ananassae, D. pseudoobscura, and D. falleni) were cloned and their cotranscripts tested in transcription/cleavage assays as described for Fig. 3. The in vitro-generated RNA started 24 bp upstream of the R2 insertion site (the D. pseudoobscura construct had three “additional nucleotides” at the 28S/R2 junction) and ended at the last paired base of the P2 helix. While termination of the RNA at this position in D. simulans did not give the maximum level of RNA cleavage (Fig. 3B, lane g), the elimination of downstream R2 sequence enabled a more direct comparison among the presumptive ribozymes. As shown in Fig. 8 A, R2 transcripts from all four species exhibited 74% to 84% self-cleavage in the cotranscription/cleavage assays similar to that observed for D. simulans.
FIG. 8.
The 5′ ends of R2 elements from other Drosophila species are also ribozymes. (A) RNA corresponding to the sequence from 24 nucleotides upstream in the 28S gene (the D. pseudoobscura construct contained 3 additional nucleotides) to the last nucleotide of the double pseudoknot structure from four species and D. simulans was synthesized and assayed for self-cleavage in the T7 transcription reaction. The uncleaved RNA (solid circle) and self-cleaved products (open circles) for each species are indicated. Lengths varied predominantly because of the different numbers of nucleotides in segments J1/2 and L4 among the species. sim, D. simulans; yak, D. yakuba; ana, D. ananassae; pse, D. pseudoobscura; and fal, D. falleni. The fraction of the synthesized RNA undergoing self-cleavage (fc) is shown at the bottom. (B) Summary of the nucleotide differences in the R2 elements from other species compared to the D. simulans sequence.
Shown in Fig. 8B are the nucleotide differences in the four species compared to the D. simulans sequence. The length and sequence of the J1/2 segment and the loop of P4 were highly variable, and virtually all positions within the P1, P2, and P4 helices had undergone covariation. Interestingly, the C residue that is paired to the G nucleotide at the base of P1 was a wobble U in D. ananassae and D. pseudoobscura, similar to the predominant wobble G-U in HDV. Of those nucleotides putatively involved in the active site, the only rigidly conserved sequences were the 13 nucleotides corresponding to most of P3 and all of L3. The J4/2 segment was conserved in length, but there were single G-to-A and A-to-G transitions in two of the species. Finally, considerable variation was detected in the J1/4 segment, with this sequence being UAA in D. simulans and D. yakuba, a single A in D. pseudoobscura, and a single G in both D. ananassae and D. falleni. It is interesting to note that the antigenomic HDV ribozyme also has a single G for its J1/4 segment (10). Thus, the R2 ribozyme had undergone considerable sequence change; however, the most conserved R2 sequences (shaded nucleotides) overlap extensively with the catalytic domain of HDV.
DISCUSSION
As found for other elements and viruses, the 5′ and 3′ untranslated regions of R2 contain conserved sequences and form RNA secondary structures which are critical to its life cycle. RNA corresponding to the R2 3′ untranslated region assembles into a conformation that is recognized by the R2 protein (32). This interaction positions the 3′ end of the RNA transcript in the proper orientation to enable the protein subunit to perform the first half of an R2 retrotransposition reaction: cleavage of one strand of the DNA target site and the use of the newly generated DNA end to prime reverse transcription of the R2 transcript (22, 23). More recently, RNA from the 5′ untranslated region was found to fold into a structure that can also be bound by the R2 protein (5, 14). This interaction is used to correctly position a second R2 protein subunit on the DNA target to perform the remaining steps of the retrotransposition reaction: cleavage of the second target DNA strand and second-strand synthesis of the element. In Drosophila species, the RNA sequences involved in the latter steps are predominately located within the large J1/2 segment of the structure shown in Fig. 4B (W. Moss and D. Turner, unpublished data).
Establishing that the 5′ untranslated region of R2 also encodes an autocatalytic ribozyme further improves our understanding of the R2 life cycle. First, it is our strongest evidence to date that R2 transcripts are processed from a 28S cotranscript. Previously, processing of the putative 28S cotranscript seemed likely to involve a mechanism that mimicked one of the many processing or modification steps associated with the maturation of rRNA. Instead, the evolution of a self-cleaving ribozyme suggests that it is advantageous for the element to rapidly separate itself from the complex series of events associated with the assembly of the large ribosomal subunit. Self-cleavage also enables this step of the retrotransposition pathway to be independent of possible regulation controlling the cellular RNases.
Second, autocatalysis reconciles the apparent contradiction between R2's inefficient 5′ integration mechanism and its widely successful history in many animals. While the frequent deletions of the upstream 28S gene and additional nucleotides inserted between the 28S gene and the full-length R2 element were suggested to indicate that there was little selective pressure on the R2 retrotransposition mechanism to maintain the integrity of the upstream target site, they also raised the question as to whether a large fraction of the insertions were “dead” copies. This is an important point because only a limited number of the rDNA units within the locus, possibly only 30 to 40 units, are transcriptionally active (8, 38, 40) and thus only a small percentage of the R2 copies are transcribed. The finding that R2 ribozyme activity is independent of nucleotide insertions or deletions of the 28S gene provides support for the model in which most copies of the element which retain all R2 sequences can provide transcripts capable of supporting new retrotransposition events.
Third, analysis of the R2 ribozyme structure provides insights into possible models for the translation of the R2 transcript (12). Because the R2 transcript is processed from an RNA polymerase I transcript, it will not contain a 5′ methyl cap (13). While mRNAs without such caps are usually unstable, the 5′ end of the R2 RNA generated by the ribozyme is likely to be a stable structure that could be resistant to 5′ exonuclease-initiated degradation. Uncapped mRNAs are also typically not translated. The single open reading frame (ORF) of Drosophila R2 elements begins near the end of the ribozyme structure (Fig. 3A). Indeed, the UAA (or UGA) termination codon that represents the amino-terminal boundary of the ORF in all Drosophila R2 elements corresponds to the conserved sequences in the J4/2 segment of the R2 ribozyme. There is no conserved Met initiation codon downstream of this termination codon. Many viral RNAs are able to overcome the lack of a 5′ methyl cap by encoding an internal ribosomal entry site (IRES) responsible for the initiation of protein synthesis at non-Met codons (13). It will be interesting to determine if the 5′ untranslated region of the R2 elements, in addition to serving as a site for binding by the R2 protein and as a ribozyme, is also able to initiate translation by forming an IRES.
The most striking feature of the R2 ribozyme is its similarity to the HDV ribozyme. Not only can R2 RNA be folded into a similar secondary structure (Fig. 4), but of the 27 nucleotides in and around the catalytic core of the HDV enzyme, 21 are identical in sequence in R2. This identity is all the more remarkable because changes in many of these nucleotide positions in HDV can be compensated for by changes elsewhere in the structure; thus, with the exception of the catalytic nucleotide, none are absolutely necessary for cleavage (1, 28). However, rather than common descent, the HDV and R2 ribozymes appear to represent a striking example of convergent evolution. For example, we have scanned the sequences at the 5′ ends of R2 elements from more-distant insects. While sequence identity to the catalytic core of the Drosophila R2 ribozymes could usually be detected in these non-Drosophila R2s, the R2 ribozyme from D. simulans has greater similarity to HDV than it does to the putative ribozymes of these distant R2 elements. In addition, following the discovery of an HDV-like ribozyme in an intron of the human CPEB3 gene (33), Webb and coworkers (37) have recently identified HDV-like self-cleaving enzymes in a wide variety of organisms. The catalytic core of the R2 ribozymes shows striking similarity to these other HDV-like ribozymes, but not in any consistent phylogenic manner. For example, the catalytic core of the R2 ribozyme from D. simulans is more similar in sequence to the drz-Bflo-1 ribozyme of the lancelet Branchiostoma floridae, while the R2 ribozyme from D. pseudoobscura is more similar to the dzr-Agam1-3 ribozyme of the mosquito Anopheles gambiae. These results suggest a rapid evolution of the HDV-like ribozyme sequences. However, due to the limited parameter space available to a ribozyme, multiple, independent evolution of highly similar catalytic cores have arisen. It will be interesting to determine whether R2 elements from more-diverse animals (e.g., those in vertebrates) continue to utilize this same parameter space or have evolved entirely new ribozyme structures.
Finally, the discovery that the 5′ end of the R2 transcript is generated by a ribozyme suggests that other non-LTR retrotransposons will also encode ribozymes. Indeed, several of the HDV-like ribozymes found by Webb et al. (37) were near reverse transcriptase-like sequences. The ability to self-cleave would enable any non-LTR retrotransposon to generate a precise 5′ end to its RNA transcript from any transcription unit. While this serves as an advantage to the element, the presence of an efficient ribozyme could effectively block expression of the inserted gene. In the case of R2, the organism is not adversely affected, because many other rDNA units can make the abundant 28S rRNA needed for development. However, the presence of a self-cleaving ribozyme on a non-LTR retrotransposon that is inserted more randomly in the genome would require the autocatalytic step to be either less efficient or regulated.
Acknowledgments
We thank members of the laboratory for their discussions. We especially thank Gloria Culver and Doug Turner for discussions and comments on the manuscript.
This work was supported by National Institutes of Health grant R01GM42790.
Footnotes
Published ahead of print on 26 April 2010.
REFERENCES
- 1.Been, M. D., and G. S. Wickham. 1997. Self-cleaving ribozymes of hepatitis delta virus RNA. Eur. J. Biochem. 247:741-753. [DOI] [PubMed] [Google Scholar]
- 2.Burke, W. D., H. S. Malik, W. C. Lathe III, and T. H. Eickbush. 1998. Are retrotransposons long-term hitchhikers? Nature 392:141-142. [DOI] [PubMed] [Google Scholar]
- 3.Cech, T. R. 1993. Structure and mechanism of the large catalytic RNAs: group I and group II introns and ribonuclease P, p. 239-269. In R. F. Gesteland and J. F. Atkins (ed.), The RNA world. Cold Spring Harbor Laboratory Press, Plainview, NY.
- 4.Chen, J. H., B. Gong, P. C. Bevilacqua, P. R. Carey, and B. L. Golden. 2009. A catalytic metal ion interacts with the cleavage site G.U wobble in the HDV ribozyme. Biochemistry 48:1498-1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Christensen, S. M., J. Ye, and T. H. Eickbush. 2006. RNA from the 5′ end of the R2 retrotransposon controls R2 protein binding to and cleavage of its DNA target site. Proc. Natl. Acad. Sci. U. S. A. 103:17602-17607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cochrane, J. C., and S. A. Strobel. 2008. Catalytic strategies of self-cleaving ribozymes. Acc. Chem. Res. 41:1027-1035. [DOI] [PubMed] [Google Scholar]
- 7.DeBerardinis, R. J., and H. H. Kazazian, Jr. 1999. Analysis of the promoter from an expanding mouse retrotransposon subfamily. Genomics 56:317-323. [DOI] [PubMed] [Google Scholar]
- 8.Eickbush, D. G., J. Ye, X. Zhang, W. D. Burke, and T. H. Eickbush. 2008. Epigenetic regulation of retrotransposons within the nucleolus of Drosophila. Mol. Cell. Biol. 28:6452-6461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eickbush, T. H. 2002. R2 and Related site-specific non-long terminal repeat retrotransposons, p. 813-835. In N. L. Craig, R. Craigie, M. Gellert, and A. M. Lambowitz (ed.), Mobile DNA II. ASM Press, Washington, DC.
- 10.Ferré-D'Amaré, A. R., K. Zhou, and J. A. Doudna. 1998. Crystal structure of a hepatitis delta virus ribozyme. Nature 395:567-574. [DOI] [PubMed] [Google Scholar]
- 11.Gabriel, A., T. J. Yen, D. C. Schwartz, C. L. Smith, J. D. Boeke, B. Sollner-Webb, and D. W. Cleveland. 1990. A rapidly rearranging retrotransposon within the miniexon gene locus of Crithidia fasciculate. Mol. Cell. Biol. 10:615-624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.George, J. A., and T. H. Eickbush. 1999. Conserved features at the 5′ end of Drosophila R2 retrotransposable elements: implications for transcription and translation. Insect Mol. Biol. 8:3-10. [DOI] [PubMed] [Google Scholar]
- 13.Kieft, J. S. 2008. Viral IRES RNA structures and ribosome interactions. Trends Biochem. Sci. 33:274-283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kierzek, E., S. M. Christensen, T. H. Eickbush, R. Kierzek, D. H. Turner, and W. N. Moss. 2009. Secondary structures for 5′ regions of R2 retrotransposon RNAs reveal a novel conserved pseudoknot and regions that evolve under different constraints. J. Mol. Biol. 390:428-432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kojima, K. K., and H. Fujiwara. 2003. Evolution of target specificity in the R1 clade non-LTR retrotransposons. Mol. Biol. Evol. 20:351-361. [DOI] [PubMed] [Google Scholar]
- 16.Kojima, K. K., and H. Fujiwara. 2005. Long-term inheritance of the 28S rDNA-specific retrotransposon R2. Mol. Biol. Evol. 22:2157-2165. [DOI] [PubMed] [Google Scholar]
- 17.Kojima, K. K., K. Kuma, H. Toh, and H. Fujiwara. 2006. Identification of rDNA-specific non-LTR retrotransposons in Cnidaria. Mol. Biol. Evol. 23:1984-1993. [DOI] [PubMed] [Google Scholar]
- 18.Kruger, K., P. J. Grabowski, A. J. Zaug, J. Sands, D. E. Gottschling, and T. R. Cech. 1982. Self-spicing RNA: autoexcision and autocyclization of the ribosomal RNA intervening sequence of Tetrahymena. Cell 31:147-157. [DOI] [PubMed] [Google Scholar]
- 19.Lambowitz, A. M., and S. Zimmerly. 2004. Mobile group II introns. Annu. Rev. Genet. 38:1-35. [DOI] [PubMed] [Google Scholar]
- 20.Lathe III, W. C., and T. H. Eickbush. 1997. A single lineage of R2 retrotransposable elements is an active, evolutionarily stable component of the Drosophila rDNA locus. Mol. Biol. Evol. 14:1232-1241. [DOI] [PubMed] [Google Scholar]
- 21.Long, E. O., and I. B. Dawid. 1980. Alternative pathways in the processing of ribosomal RNA precursor in Drosophila melanogaster. J. Mol. Biol. 138:873-878. [DOI] [PubMed] [Google Scholar]
- 22.Luan, D. D., and T. H. Eickbush. 1995. RNA template requirements for target DNA-primed reverse transcription by the R2 retrotransposable element. Mol. Cell. Biol. 15:3882-3891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Luan, D. D., M. H. Korman, J. L. Jakubczak, and T. H. Eickbush. 1993. Reverse transcription of R2Bm RNA is primed by a nick at the chromosomal target site: a mechanism for non-LTR retrotransposition. Cell 72:595-605. [DOI] [PubMed] [Google Scholar]
- 24.Malik, H. S., and T. H. Eickbush. 2000. NeSL-1, an ancient lineage of site-specific non-LTR retrotransposons from Caenorhabditis elegans. Genetics 154:193-203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McLean, C., A. Bucheton, and D. J. Finnegan. 1993. The 5′ untranslated region of the I factor, a long interspersed element-like retrotransposon of Drosophila melanogaster, contains an internal promoter and sequences that regulate expression. Mol. Cell. Biol. 13:1042-1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mizrokhi, L. J., S. G. Gorgieva, and Y. V. Ilyin. 1988. Jockey, a mobile Drosophila element similar to mammalian LINEs, is transcribed from the internal promoter by RNA polymerase II. Cell 54:685-691. [DOI] [PubMed] [Google Scholar]
- 27.Muscarella, D. E., and V. M. Vogt. 1989. A mobile group I intron in the nuclear rDNA of Physarum polycephalum. Cell 56:443-454. [DOI] [PubMed] [Google Scholar]
- 28.Nehdi, A., and J.-P. Perreault. 2006. Unbiased in vitro selection reveals the unique character of the self-cleaving antigenomic HDV RNA sequence. Nucleic Acids Res. 34:584-592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Patel, P. H., and B. D. Preston. 1994. Marked infidelity of human immunodeficiency virus type 1 reverse transcriptase at RNA and DNA template ends. Proc. Natl. Acad. Sci. U. S. A. 91:549-553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Perrotta, A. T., and M. D. Been. 1996. Core sequences and a cleavage site wobble pair required for HDV antigenomic ribozyme self-cleavage. Nucleic Acids Res. 24:1314-1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Perrotta, A. T., and M. D. Been. 2006. HDV ribozyme activity in monovalent cations. Biochemistry 45:11357-11365. [DOI] [PubMed] [Google Scholar]
- 32.Ruschak, A. M., D. H. Mathews, A. Bibillo, S. L. Spinelli, J. L. Childs, T. H. Eickbush, and D. H. Turner. 2004. Secondary Structure models of the 3′ untranslated regions of diverse R2 RNAs. RNA 10:978-987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Salehi-Ashtiani, K., A. Lupták, A. Litovchick, and J. W. Szostak. 2006. A genomewide search for ribozymes reveals an HDV-like sequence in the human CPEB3 gene. Science 313:1788-1792. [DOI] [PubMed] [Google Scholar]
- 34.Stage, D. E., and T. H. Eickbush. 2009. Origin of nascent lineages and the mechanisms used to prime second-strand DNA synthesis in the R1 and R2 retrotransposons of Drosophila. Genome Biol. 10:R49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Swergold, G. D. 1990. Identification, characterization, and cell specificity of a human LINE-1 promoter. Mol. Cell. Biol. 10:6718-6729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Villanueva, M. S., S. P. Williams, C. B. Beard, F. F. Richards, and S. Aksoy. 1991. A new member of a family of site-specific retrotransposons is present in the spliced leader RNA genes of Trypanosoma cruzi. Mol. Cell. Biol. 11:6139-6148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Webb, C.-H., N. J. Riccitelli, D. J. Ruminski, and A. Lupták. 2009. Widespread occurrence of self-cleaving ribozymes. Science 326:953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ye, J., and T. H. Eickbush. 2006. Chromatin structure and transcription of the R1- and R2-inserted rRNA genes of Drosophila melanogaster. Mol. Cell. Biol. 26:8781-8790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang, X., and T. H. Eickbush. 2005. Characterization of active R2 retrotransposition in the rDNA locus of Drosophila simulans. Genetics 170:195-205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhou, J., and T. H. Eickbush. 2009. The pattern of R2 retrotransposon activity in natural populations of Drosophila simulans reflects the dynamic nature of the rDNA locus. PLoS Genet. 5:e1000386. [DOI] [PMC free article] [PubMed] [Google Scholar]