Abstract
The mTOR signaling complex integrates signals from growth factors and nutrient availability to control cell growth and proliferation, in part through effects on the protein-synthetic machinery. Protein synthesis rates fluctuate throughout the cell cycle but diminish significantly during the G2/M transition. The fate of the mTOR complex and its role in coordinating cell growth and proliferation signals with protein synthesis during mitosis remain unknown. Here we demonstrate that the mTOR complex 1 (mTORC1) pathway, which stimulates protein synthesis, is actually hyperactive during mitosis despite decreased protein synthesis and reduced activity of mTORC1 upstream activators. We describe previously unknown G2/M-specific phosphorylation of a component of mTORC1, the protein raptor, and demonstrate that mitotic raptor phosphorylation alters mTORC1 function during mitosis. Phosphopeptide mapping and mutational analysis demonstrate that mitotic phosphorylation of raptor facilitates cell cycle transit through G2/M. Phosphorylation-deficient mutants of raptor cause cells to delay in G2/M, whereas depletion of raptor causes cells to accumulate in G1. We identify cyclin-dependent kinase 1 (cdk1 [cdc2]) and glycogen synthase kinase 3 (GSK3) pathways as two probable mitosis-regulated protein kinase pathways involved in mitosis-specific raptor phosphorylation and altered mTORC1 activity. In addition, mitotic raptor promotes translation by internal ribosome entry sites (IRES) on mRNA during mitosis and is demonstrated to be associated with rapamycin resistance. These data suggest that this pathway may play a role in increased IRES-dependent mRNA translation during mitosis and in rapamycin insensitivity.
Cell growth and cell division are tightly coordinated processes required for cells to remain equal in size after division. In unicellular organisms, cell growth and proliferation are coordinated by nutrient availability, whereas their multicellular counterparts must also respond to growth factor input. Both processes lead to organismal growth as well as to increased cell number and cell mass. Cell growth and cell proliferation are also linked via the mTOR signaling pathway (16, 17). The mTOR kinase forms a distinct signaling complex (mTORC1) that participates in the coordination of nutrient and growth factor signaling. mTORC1 is composed of the kinase mTOR, the adaptor protein raptor, and the regulatory protein LST8 (25, 33, 34, 72).
Accumulation of cellular proteins leads to cell growth and cell division. However, cell growth occurs only during certain phases of the cell cycle, necessitating that protein synthesis rates oscillate during cell cycling (40). In addition, in quiescent cells in G0, protein synthesis rates are significantly reduced, whereas a select group of mRNAs maintain active translation (20, 68). During the G1 phase, overall protein synthesis rates increase through S phase to allow cells to grow and enter another round of cell division while maintaining cell size (2, 3, 42, 45). As with G0, entrance into mitosis (G2/M phase) results in a global downregulation by as much as 60 to 80% of cap-dependent mRNA translation in primary, immortalized, and some transformed cells (5, 14, 29).
Studies report several possible mechanisms for inhibition of protein synthesis during mitosis. Translation initiation requires the formation of an initiation factor complex known as eukaryotic translation initiation factor 4F (eIF4F), which consists of cap binding protein eIF4E, molecular scaffold protein eIF4G, and RNA helicase eIF4A. Together, they recruit ribosomes to mRNAs via bridging interactions between the 7-methyl-GTP (m7GTP) 5′ cap and the small 40S ribosomal subunit. Downregulation of protein synthesis during G2/M was first ascribed to hypophosphorylation of eIF4E and the eIF4E binding proteins (4E-BPs) (5, 46). 4E-BPs are activated by hypophosphorylation, which allows them to bind and sequester eIF4E, preventing it from binding eIF4G and thereby blocking cap-dependent mRNA translation. More recently, several studies suggest that 4E-BP1, the major 4E-BP and a key target of mTORC1, is actually hyperphosphorylated (inactivated) during mitosis (26, 49). It is puzzling, then, that the phosphatidylinositol 3-kinase (PI3K)/AKT network and AKT itself (which modulate mTORC1 activity) are reportedly inactivated during late mitosis (1, 9, 22). In addition, phosphorylation of another mTORC1 target, ribosomal S6 kinase 1 (S6K1), and its activity are actually highest during G2/M phase, consistent with elevated mTORC1 activity during mitosis (6).
In this study we show that, despite repression of AKT and other activators of mTORC1 activity in mitosis, mTORC1 remains active and phosphorylates 4E-BP1 and S6K1 during G2/M. We describe the multisite phosphorylation of raptor during mitosis, and we identify seven mitosis-specific raptor phosphorylation sites. By developing phosphomimetic and phosphorylation-deficient mutants of raptor, we show that hyperphosphorylated raptor promotes cell cycle transit through G2/M, whereas hypophosphorylated raptor promotes transit through G1. Raptor phosphorylation is shown to involve kinase pathways that are known to be active during mitosis, including cyclin-dependent kinase 1 (cdk1 [cdc2]) and glycogen synthase kinase 3 (GSK3) pathways that are also upregulated in certain human cancers, including breast cancers. These and other findings disclose a novel regulatory network for mTORC1 that is active during mitosis, important for G2/M progression and increased internal ribosome entry site (IRES)-dependent translation during mitosis, and indirectly associated with rapamycin resistance.
MATERIALS AND METHODS
Cell culture and inhibitors.
All inhibitors were obtained from Calbiochem unless otherwise noted. The PP242 inhibitor was a gift from K. M. Shokat of the University of California, San Francisco. Human embryonic kidney (HEK) 293T and 293GP cells (expressing the gag-pol gene from HIV) were cultured in Dulbecco's modified Eagle medium (DMEM; Cellgro) containing 10% bovine calf serum (HyClone). HaCaT and 4T1 cells and neonatal human dermal fibroblasts (NHDF) were maintained in DMEM (Cellgro) with 10% fetal bovine serum (FBS; Gemini). MCF10A cells were cultured in Ham's F-12-DMEM (Cellgro) supplemented with 5% horse serum (Gibco), 2 mM glutamine (BioWhittaker), 20 ng/ml epidermal growth factor (EGF; Invitrogen), 100 ng/ml cholera toxin (Calbiochem), 0.01 mg/ml bovine insulin (Gibco), and 500 ng/ml hydrocortisone. All experiments were performed with exponentially growing cells plated at approximately 40% cell density.
Metabolic labeling and preparation of extracts.
Cells were pulse-labeled for 30 min with 10 μCi of [35S]methionine and [35S]cysteine mix per ml (Easytag Express protein labeling mix; Perkin Elmer) in DMEM without methionine and cysteine and containing 2% dialyzed FBS. Cells were washed twice in ice-cold 1× phosphate-buffered saline (PBS) and lysed in ice-cold buffer A (0.5% [vol/vol] Igepal, 150 mM NaCl, 50 mM HEPES, pH 7.0, 2 mM Na3VO4, 25 mM β-glycerophosphate, complete protease inhibitor mix [Roche]). Debris was removed by centrifugation at 13,000 × g and 4°C for 10 min, and the supernatants were stored at −80°C. Protein content was determined by the Bradford method (Bio-Rad). Immunoprecipitations (IPs) involving mTORC1 were performed as described previously (51).
Mitotic arrest and G1 synchronization.
HaCaT, MCF10A, NHDF, and 4T1 cells were arrested in mitosis by incubation of cells with 5 mM thymidine for 16 h and released for 8 h, followed by addition of 40 ng/ml nocodazole for 6 h (76). At the end of this procedure, mitotic cells were dislodged by mechanical shake-off, resulting in significant (>80%) enrichment of the mitotic cell fraction. 293T cells were partially arrested by 12 h of incubation with 20 ng/ml nocodazole. G1/S synchronization was achieved by a double thymidine block. Cells were treated with 5 mM thymidine for 16 h, released for 10 h, and blocked again with 5 mM thymidine for an additional 16 h before release. Cell cycle distribution was determined by fluorescence-activated cell sorter (FACS) analysis after release from the double block. For treatment with inhibitors, chemicals were added at the same time as nocodazole. LY294002 was used at 10 μM, rapamycin at 100 nM, CGP74514A at 1.5 μM, SB218078 (SB) at 2.5 μM, TBB (4,5,6,7-tetrabromobenzotriazole) at 25 μM, PD98059 at 10 μM, and 6-bromoindirubin-3′-oxime (BIO) at 2 μM. AICAR (aminoimidazole carboxamide ribonucleotide; Toronto Research Chemicals) was used at 2 mM for 1 h.
Measurement of cell proliferation and cell cycle distribution.
Cell proliferation was measured by a standard MTT [3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide] cell proliferation assay. Cells were plated in quadruplicate (per time point) in 96-well format and subsequently quantified four times over a course of 96 h. Wells were treated with 20% sterile-filtered 5-mg/ml MTT (Sigma) in DMEM over 6 h at 37°C. The reduced formazan was then solubilized overnight in a solution of 50% N,N-dimethylformamide (DMF) and 20% SDS. Colorimetric quantification was performed with a Tecan Sunrise microplate reader at 570 nm, with a negative reference at 450 nm. Relative proliferation rates were determined from the first-order logarithmic regression analysis of the colorimetric change over time per condition. Results were normalized to control cells. For cell cycle distribution analysis, HaCaT cells at similar densities were trypsinized, ethanol fixed, and labeled with 200 mg/ml propidium iodide in 38 mM sodium citrate in the presence of RNase A. DNA histograms were acquired using a FACSCalibur flow cytometer (Becton Dickinson) and analyzed using ModFit LT (Ventry Software) or FlowJo (Tree Star).
See the supplemental material for experimental procedures related to antibodies, immunoprecipitations, the S6K kinase assay, transfection and retrovirus production, tryptic phosphopeptide mapping, RNA interference (RNAi) and lentivirus transduction, and DNA constructs.
RESULTS
mTORC1 is active during mitosis despite decreased PI3K/AKT activity.
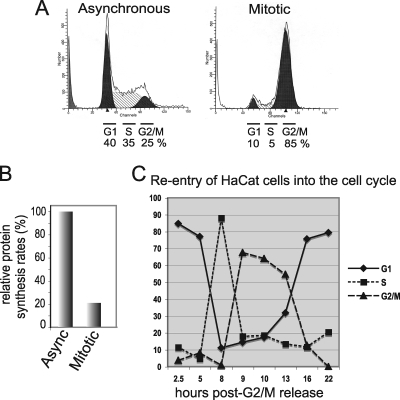
To address the role of PI3K/AKT/mTORC1 signaling during mitosis, HaCaT cells were arrested at prophase/prometaphase by sequential incubation with thymidine and nocodazole followed by mechanical shake-off. Immortalized keratinocytes (HaCaT cells) were utilized because they arrest well and demonstrate significant downregulation of protein synthesis during mitosis, unlike most highly transformed cell lines (11). Furthermore, mitotic arrest of HaCaT cells by this approach does not induce apoptosis or terminal withdrawal from the cell cycle, unlike other methods (50). FACS analysis of the cell cycle distribution demonstrated a >80% accumulation of HaCaT cells in G2/M due to thymidine-nocodazole treatment compared to untreated asynchronous cells (Fig. 1A). Translation rates for mitotic cells were reduced by 80% compared to those for untreated asynchronous cells (Fig. 1B). Importantly, with release from G2/M cell cycle blockade, almost 90% of HaCaT cells rapidly reentered the cell cycle (Fig. 1C). This is evidenced by quantitative movement of cells from G2/M into G1 (80 to 90%) by 2.5 h postrelease, by movement into S phase by 8 h, and by reentry into G1 by 16 to 20 h.
FIG. 1.
Mitotic arrest of HaCaT cells. (A) Thymidine and nocodazole double-drug treatment achieves more than 80% arrest of HaCaT cells at G2/M. FACS analysis shown is representative of typical results for untreated asynchronous and treated mitotically arrested cells. (B) Mitotic HaCaT cells are translationally arrested. Cells were labeled with [35S]methionine as asynchronous or mitotically arrested cultures, and the specific activity of incorporation into equal amounts of protein was determined by trichloroacetic acid (TCA) precipitation and scintillation counting. Standard errors of the means (SEMs) were determined from three independent experiments. (C) The majority of HaCaT cells reenter the cell cycle following G2/M arrest. Following thymidine-nocodazole block and accumulation of >80% cells in G2/M (A), cells were released from drug blockade and examined by FACS analysis for cell cycle distribution as a function of time postrelease. Results are an averages of 3 independent experiments.
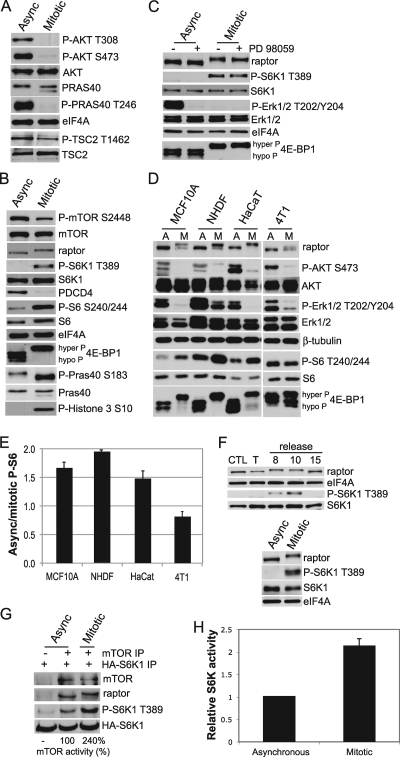
AKT is phosphorylated at position S473 by the mTOR-rictor (rapamycin-insensitive) complex known as mTORC2, followed by activation by phosphoinositide-dependent kinase 1 (PDK1) of phosphorylation at position T308 (19, 28, 55). Phosphorylation of AKT at T308 and S473 and phosphorylation of its immediate substrates PRAS40 (which migrates as a doublet during mitosis) at T246 and TSC2 at T1462 were strongly reduced during mitosis, whereas total protein levels of AKT, PRAS40, and TSC2 were unchanged (Fig. 2A) (10, 28, 31, 35, 55). Interestingly, the mTORC1 targets S6K1 (and in turn ribosome protein S6) and 4E-BP1 were strongly hyperphosphorylated at their mTORC1-specific sites during mitosis, and PDCD4 levels were reduced, consistent with S6K phosphorylation and activation (Fig. 2B). PRAS40 has been shown to be a target of mTOR (43). Accordingly, PRAS40 phosphorylation at S183 by mTORC1, which leads to an active mTOR/raptor complex, was increased during mitosis (Fig. 2B) (65). Surprisingly, phosphorylation of mTOR at S2448 was decreased in mitosis, which correlates with decreased phosphorylation of TSC2 at T1462 and phosphorylation of S6K1 at T389 (Fig. 2A and B). It has also been proposed that AKT phosphorylates mTOR at S2448 (13, 58). Because phosphorylation of AKT was decreased in mitotic cells, this could potentially explain the decrease in mTOR phosphorylation. However, replacement of serine with alanine at this site has been shown to have no effect on mTOR activity (58). These data therefore suggest that mTOR remains strongly active despite downregulation of AKT activity in mitosis. As shown later, analysis of mTORC1 kinase activity during mitosis substantiates this conclusion.
FIG. 2.
Immunoblot and phosphoimmunoblot analysis of mTORC1-associated proteins obtained from asynchronous and mitotic HaCaT cells. (A and B) Amino acid position corresponds to specific phosphoprotein sequences. Note that in G2/M-arrested cells raptor mobility is retarded. Phosphorylation of histone 3 at position S10 is shown as a marker of mitosis. Equal amounts of protein lysates were used for analyses, which are typical of at least three independent experiments. eIF4A is provided as a loading control. (C) Inhibition of the Erk1/2 pathway with MEK inhibitor PD98059 does not prevent mTORC1 activation during mitosis. Asynchronous and mitotic HaCaT cells were treated with 10 μM PD98059, and equal amounts of protein lysates were analyzed by immunoblotting. Slowly migrating (hyperphosphorylated) and fast-migrating (hypophosphorylated) 4E-BP1 proteins are shown. (D) Lysates of asynchronous and mitotic cell lines shown were assessed for the indicated phosphoproteins and proteins, demonstrating that raptor mobility retardation and mTORC1-target phosphorylation during mitosis are not cell line-specific events. (E) P-S6/S6 ratios were determined by densitometry and averaged from three independent experiments, with background fluorescence subtracted. SEMs are shown. (F) HaCaT cells were grown asynchronously (CTL) or arrested at G1 using thymidine (T) and released for the indicated times. (Top) Cell lysates were blotted for the proteins shown. (Bottom) Lysates from asynchronous or mitotic cells dislodged by mechanical shake-off without other treatments were resolved and immunoblotted for the indicated proteins. (G) mTOR binding to raptor and S6K1 complexes in asynchronous and mitotic HaCaT cells and mTORC1 activity. HA-S6K1 was isolated from LY294002-rapamycin-treated HEK 293T cells to prevent its phosphorylation, mixed with mTORC1 isolated from asynchronous or mitotic HaCaT cells, and then subjected to an in vitro kinase assay. Proteins were resolved by SDS-PAGE and detected by immunoblot analysis. All results shown are representative of at least three independent experiments using equal amounts of protein lysates. Mitotic S6K activity was normalized by comparing the ratio of band density of phospho-S6K1 to total HA-S6K1 for mitotic cells to that for asynchronous cells. Values were determined from three independent experiments. (H) S6K1 activity from equal amounts of asynchronous or mitotic cell lysates was determined using in vitro kinase assays. SEMs were calculated from three independent experiments.
Both S6K1 and 4E-BP1 are reportedly also phosphorylated by cdc2/cdk1 during mitosis (26, 59). 4E-BP1 has been shown to be phosphorylated by cdc2 at position T70, which is only one of several phosphorylation sites required for inactivation of 4E-BP1 (22, 26, 57). Thus, activity of mTOR is likely required for additional 4E-BP1 phosphorylation during mitosis. S6K1 is phosphorylated by cdc2 at several sites but not at the mTOR phosphorylation site, T389 (59). The S6K1 target, S6 protein, was also highly phosphorylated during mitosis; this was expected if mTORC1 is active, as was downregulation of PDCD4 levels (Fig. 2B) (6, 12). Although the Ras/Erk signaling network reportedly activates mTORC1 during mitosis (39), we could not detect Erk1/2 phosphorylation at its activation sites in mitotically arrested HaCaT cells (Fig. 2C). Proof that Erk1/2 is not required for mTORC1 activity during mitosis was obtained by blocking this pathway with MEK inhibitor PD98059, which also did not prevent 4E-BP1 or S6K1 phosphorylation (Fig. 2C). To exclude cell-type-specific effects of mitosis on mTORC1, we compared primary cells (neonatal human dermal fibroblasts [NHDF]), immortalized cells (HaCaT and immortalized breast epithelial cells [MCF10A]), and highly transformed cells (4T1 mouse mammary tumor cells). We found that all cell types displayed diminished AKT phosphorylation and increased 4E-BP1 phosphorylation in mitosis (Fig. 2D), shown by slower mobility for 4E-BP1, indicative of increased mTOR activity during mitosis. In addition, we also observed that S6 phosphorylation was increased in mitosis in primary and immortalized cells but not transformed cells (Fig. 2D and E), despite equal S6 protein levels (Fig. 2D).
mTORC1 phosphorylation of its distinct targets has recently been shown to be differentially regulated in a cell-specific manner, which may be relevant to our observation of uncoupled 4E-BP1 and S6K1 regulation in response to diverse treatments, as noted below (10). The differential regulation of mTORC1 targets among different cell types may help explain why some investigators have reported decreased S6K1 activity during mitosis while others have reported increased signaling (6, 59, 64). In this regard, we note that not all mTORC1 targets are phosphorylated during mitosis. For instance, eIF4GI is phosphorylated downstream of mTORC1 in response to various signals but was hypophosphorylated during mitosis in HaCaT and other cells (see Fig. S1 in the supplemental material; data not shown) (6, 51). Further, S6K1 itself is differentially regulated during mitosis. For instance, in HaCaT cells, we observed the hyperphosphorylation of ribosome protein S6, indicative of increased S6K1 activity (Fig. 2B). However, another well-known target of S6K1, eEF2K, was slightly hypophosphorylated during mitosis, while eEF2 was hyperphosphorylated (see Fig. S1 in the supplemental material) (8, 67). In mitotic HeLa cells, eEF2 is dephosphorylated (62), although we have found that this cell line is deregulated in mitotic signaling pathways, making it a poor model for this research (our unpublished studies). Decreased translational elongation in the growth cone during mitosis, which could result from hypophosphorylation of eEF2K, has been reported as a possible mechanism of translational inhibition during mitosis (61).
It was evident that the electrophoretic migration of raptor in mitotic HaCaT cell extracts was slower than that in asynchronous cells (Fig. 2B to D). To determine whether a nocodazole-specific modification could account for altered raptor migration, we accumulated cells in mitosis through release from thymidine block or by mechanical shake-off in the absence of drugs. Enrichment of mitotic cells occurred 8 to 10 h after thymidine release (data not shown) (61). Both of these techniques revealed slowly migrating raptor and increased mTORC1 activity in mitotic cells (Fig. 2F). We therefore investigated whether asynchronous and mitotic mTORC1 displays altered kinase activity. HEK 293 cells were transfected with a hemagglutinin (HA)-tagged-S6K1 expression vector and treated with LY294002 and rapamycin to block PI3K and mTOR, respectively, and dephosphorylated S6K1 was recovered by IP. mTORC1 was recovered by mTOR IP from asynchronous or G2/M-arrested HaCaT cells. An in vitro kinase assay was then performed to examine the activity of mTORC1 isolated from asynchronous and mitotic HaCaT cells based on its ability to phosphorylate dephosphorylated S6K1. mTORC1 isolated from mitotic HaCaT cells exhibited an almost 2.5-fold increase in activity, determined by measuring in vitro T389 phosphorylation of the immunoprecipitated, hypophosphorylated S6K1 protein, compared to mTORC1 obtained from asynchronous cells (Fig. 2G). Thus, mTORC1 purified from mitotic cells is more than twice as active as that in asynchronous cells. Moreover, the form of raptor that copurifies with mTOR as a component of mTORC1 in mitotic cells corresponds entirely to the slower-mobility raptor protein (Fig. 2G). In vitro kinase activity of mitotic mTORC1 was also found to be increased almost 2-fold using a commercial in vitro kinase assay (data not shown). Other groups have also reported increased mTORC1 activity during mitosis (37). We tested the vitro kinase activity of S6K1, and as expected from increased mTORC1 signaling, mitotic lysates displayed an ∼2-fold increase in S6K1 activity compared to asynchronous cells (Fig. 2H).
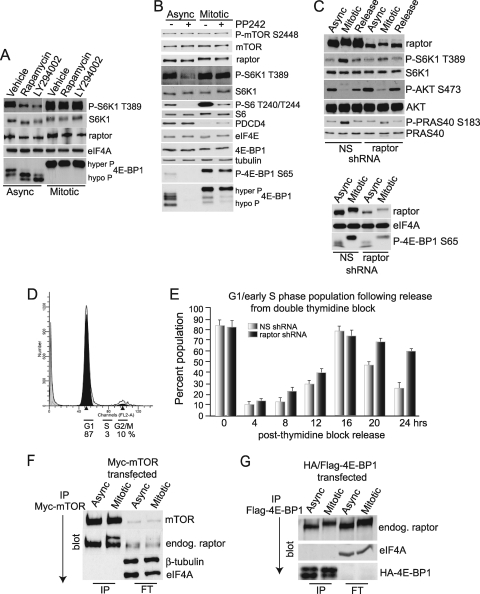
Mitotic cells differ from asynchronous cells in response to inhibition of mTORC1 upstream pathways but not formation of mTORC1.
As greater than 50% silencing of mTOR could not be achieved (data not shown), to further investigate the mTOR pathway, asynchronously growing or mitosis-arrested cells were treated with established inhibitors of mTOR signaling. Both rapamycin (an mTORC1-allosteric inhibitor) and LY294002 (a PI3K and mTOR inhibitor) reduced mTORC1 activity in asynchronous cells, as measured by decreased phosphorylation of S6K1 at T389 and hypophosphorylation of 4E-BP1 in cells treated with the inhibitors (Fig. 3A). However, neither treatment had any effect on S6K1 or 4E-BP1 phosphorylation in mitotic cells. To confirm that mTORC1 is the kinase responsible for phosphorylation of these targets during mitosis, an ATP-competitive inhibitor of the mTOR active site (PP242 [15]) was added to asynchronous and mitotic cells and phosphorylation of downstream targets was analyzed. This small-molecule inhibitor differs from rapamycin in that it strongly blocks both mTORC1 and mTORC2 activities, whereas rapamycin complexes with the mTOR binding protein FKBP12, partially inhibiting mTORC1 via a poorly described mechanism (15, 27). mTOR kinase activity inhibition by PP242 blocked phosphorylation of S6K1 (T389), S6 (T240/T244), and 4E-BP1 (S65) in asynchronous and, to a lesser extent, mitotic cells (Fig. 3B). The level of PDCD4 also increased in mitotic cells treated with PP242, providing further evidence for mTORC1 inhibition. Furthermore, silencing of rictor, an essential mTORC2 component (53), did not decrease phosphorylation of these targets during mitosis, eliminating the possibility of mTORC2 involvement (see Fig. S2 in the supplemental material). Collectively, these data indicate that mTORC1 has increased activity and is responsible for phosphorylation of its downstream targets during mitosis. The inability of rapamycin to inhibit mitotic mTORC1 suggests differences in the activity of the complex during different phases of the cell cycle and possibly rapamycin sensitivity (53).
FIG. 3.
Raptor is modified during mitosis and required to prevent G1 cell cycle arrest. (A) Activity of mTORC1 in HaCaT cells is not prevented during mitosis by mTOR inhibitors rapamycin and LY294002. Equal amounts of protein lysates from asynchronous and mitotic HaCaT cells were examined by SDS-PAGE and immunoblot analysis as shown. 4E-BP1 was resolved into hyper- and hypophosphorylated forms. Cells were treated with vehicle alone as a control. (B) Phosphorylation of mTORC1 targets is prevented by mTOR-specific kinase inhibitor PP242 during mitosis. Lysates were treated as for panel A. (C) shRNA-mediated downregulation of raptor decreases phosphorylation at the mTORC1-specific sites of S6K1 (T389) and PRAS40 (S183) (top) and decreases mTOR-dependent phosphorylation of 4E-BP1 (S65) (bottom). Equal amounts of protein were examined by immunoblotting as shown. Blots are representative of at least three independent experiments. (D) Double thymidine block of HaCaT cells causes accumulation of cells in G1. Cells were treated as described in Materials and Methods and analyzed by FACS. Typically, 85 to 90% of cells accumulate in G1. (E) Raptor silencing impairs cell cycle progression into S phase. HaCaT cells with and without raptor silencing were accumulated in G1 by double thymidine block and then released, and reaccumulation in G1/early S phase was assessed by FACS. (F and G) Both mitotic and asynchronous raptor binds mTOR and mTORC1 substrate 4E-BP1. (F) Asynchronous and partially mitotically arrested 293 cells were transfected with a Myc-mTOR expression vector. Cells were approximately equally distributed between G1 and G2/M, with a smaller fraction in S phase. Cells were immunoprecipitated with specific antibodies, and associated proteins were identified by immunoblot analysis, as indicated. Typical results of three independent studies are shown. IP, immunoprecipitate; FT, flowthrough. (G) HaCaT cells were transfected with a Flag-HA-4E-BP1 expression vector and either left asynchronous or mitotically arrested by thymidine-nocodazole block. Equal amounts of protein lysates were immunoprecipitated for Flag-4E-BP1, and associated proteins were identified by immunoblot analysis. FT indicates the flowthrough fraction not retained in the immunoprecipitate.
We assessed the effect of raptor depletion on mitotic mTORC1 signaling. Asynchronous and mitotic HaCaT cells were silenced with control nonsilencing (NS) or raptor short hairpin RNAs (shRNAs), achieving raptor silencing of >70% (Fig. 3C). Raptor silencing in mitotic cells partially blocked phosphorylation of S6K1 at position T389, PRAS40 at position S183, and 4E-BP1 at S65 (Fig. 3C). Silencing also slightly elevated the levels of S473-phosphorylated AKT independently of the cell cycle (Fig. 3C), possibly through decreased mTORC1 feedback inhibition (31). While raptor silencing was insufficient to fully prevent mTORC1 phosphorylation of its targets, it clearly slowed progression through the cell cycle (Fig. 3D and E). After release of HaCaT cells from a double thymidine block, which synchronizes approximately 85% of the cells at G1 despite raptor silencing (Fig. 3D), both NS control and raptor-silenced cells exited G1 (Fig. 3E). However, whereas most NS control and raptor-silenced cells continued to cycle back to G1 by 16 h following release, raptor-silenced cells transited more slowly through G1, demonstrating a significant delay in progression through the cell cycle and accumulation back in G1 (Fig. 3E). Similarly, inhibition of mTORC1 with rapamycin also causes G1 arrest (27, 69). We therefore explored whether raptor is modified in mitosis, possibly mediating differential association with known binding partners required for efficient cell cycle progression.
For these studies we used HEK 293 cells because 12 h of treatment with nocodazole only partially arrests the population of cells in G1, providing almost equal numbers of cells in prometaphase and G1/early S (see Fig. 4B [FACS analysis]). 293 cells were first transfected with an expression vector for Myc-tagged mTOR, which was then immunoprecipitated from asynchronous and mitotic cell lysates. Under these conditions, mTOR was associated with both electrophoretically faster (lower band) and slower (upper band) forms of raptor only in the partially arrested population (Fig. 3F). We also examined asynchronous and mitotic cells for raptor binding to 4E-BP1, with which it interacts through a target of rapamycin signaling (TOS) motif (56). HaCaT cells were transduced with a Flag-HA-tagged 4E-BP1 construct and arrested at mitosis by thymidine-nocodazole block or grown asynchronously, followed by immunoprecipitation of Flag-4E-BP1 and analysis of endogenous raptor association. Similar to the mTOR interaction, raptor from both asynchronous and mitotic cells bound equally well to 4E-BP1 (Fig. 3G). We therefore conclude that mitotic raptor associates equally well with at least two of its established binding targets. mTORC1 responds differently to upstream signaling pathways and to inhibitors in mitotic cells than it does in cycling cells, and it exhibits differential activity toward its substrates compared to mTORC1 of asynchronous cells. As described above, we excluded the possibility that mTORC2 phosphorylates mTORC1 targets during mitosis, because silencing of the mTORC2-specific rictor protein did not alter phosphorylation of 4E-BP1, PRAS40, or S6K1 during mitosis, nor did it affect cell cycle progression (see Fig. S2 in the supplemental material) (54).
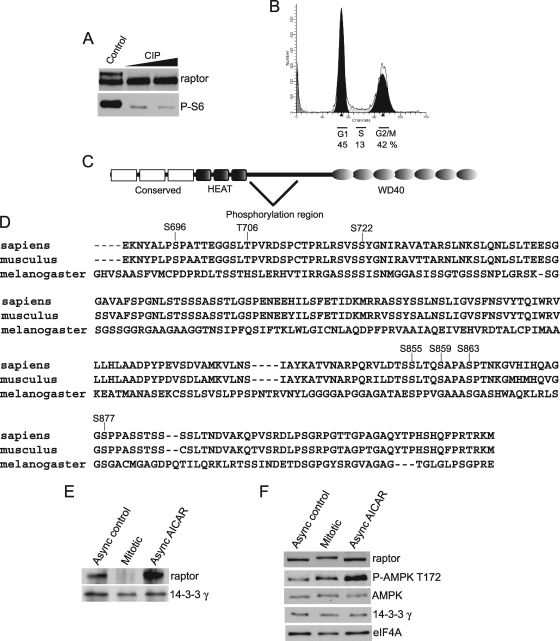
FIG. 4.
Raptor is hyperphosphorylated during mitosis. (A) Mixed mitotic 293 cell lysates produced by partial block with nocodazole were incubated with increasing amounts of calf intestinal phosphatase (CIP) at 37°C. (B) 293 cells were partially blocked in G2/M by 12 h of nocodazole treatment. FACS analysis demonstrates an almost equal distribution of cells in G1 and G2/M. (C) Diagram of raptor showing conserved motifs and the identified mitosis-specific phosphorylation region. (D) Alignment of human, mouse, and Drosophila phosphorylated regions of raptor. Sites identified by this work as ones where phosphorylation is increased during mitosis are indicated. (E and F) Raptor does not interact with 14-3-3γ during mitosis. 14-3-3γ was immunoprecipitated from lysates of cells treated as indicated and probed for interaction with raptor by immunoblotting. (F) Input lysates were resolved by electrophoresis, and levels of phospho-AMPK and 14-3-3γ during mitosis and after activation of AMPK with AICAR were assessed by immunoblotting.
Raptor is phosphorylated at multiple sites during mitosis.
Since the expression of raptor protein from a transfected plasmid carrying raptor cDNA without its 5′ and 3′ regulatory elements also demonstrated altered electrophoretic mobility during mitosis (data not shown), the modification was most likely a result of mitosis-specific phosphorylation. We therefore used 293 cells arrested at mitosis with a single nocodazole block for 12 h, producing both fast- and slow-mobility raptor forms, and subjected lysates to phosphatase treatment (Fig. 4A). Under these conditions, roughly equal fractions of the cell population were found in G2/M and G1/early S, with a smaller fraction in authentic S phase (Fig. 4B). Calf intestinal phosphatase (CIP) treatment reverted the slower-migrating mitotic raptor band to the faster form found in asynchronous cells, confirming phosphorylation as the alteration responsible for raptor mobility (Fig. 4A). Both fast- and slowly migrating raptor protein forms were isolated by gel electrophoresis from mitotically arrested 293 cells subjected to nocodazole block and compared to raptor isolated from asynchronous cells. Phosphopeptide mapping was carried out by mass spectrometry (MS) of the raptor proteins. Slow-mobility raptor protein isolated from mitotic cells contained seven phosphorylation sites; these sites, which were not strongly present or detectable in high-mobility raptor or raptor isolated from asynchronous cells, were mapped to and identified in individual amino acids (Fig. 4C and D). Raptor is composed of a central region that contains no known sequence motifs, a C terminus containing 7 WD40 elements (25, 33), and mTOR binding sites in both the N and C termini (Fig. 4C). All of the identified phosphorylation sites that were strongly represented in mitotically isolated raptor mapped to the central region. All phosphorylation sites are also conserved between human and mouse raptor proteins as shown, and four are found in Drosophila (S722, S855, S863, and S877). Raptor is therefore multiply phosphorylated during mitosis at seven sites. Although we cannot formally exclude the possibility of additional mitosis-specific phosphorylation sites not detected by MS, while the manuscript was under review, five of these raptor phosphorylation sites, all except S722 and S721, were also identified by others, although not with respect to mTORC1 activity in mitosis. Insulin was shown to stimulate phosphorylation of S863, and other identified sites were shown to be associated with mTORC1 activity (18, 66).
Phosphorylation of raptor by AMPK was recently reported to be associated with the sequestration of raptor by 14-3-3γ protein, effectively preventing mTORC1 function during AMPK activation (24). We found that one of the reported AMPK raptor sites is also phosphorylated during mitosis, as determined by MS. We therefore sought to determine whether mitosis-specific phosphorylation of raptor is also associated with raptor/14-3-3γ interaction. Asynchronous, mitotic, or AICAR-treated HaCaT cells were subjected to IP with 14-3-3γ antibody, and raptor was detected by immunoblotting. As shown in Fig. 4E, 14-3-3γ associates with raptor in asynchronous cells and this association is increased by AMPK activation with AICAR treatment, as expected, but mitotic raptor does not co-IP with 14-3-3γ. 14-3-3γ levels are unaltered during mitosis (Fig. 4F). These findings therefore indicate that the mitosis-specific phosphorylation of raptor is independent of 14-3-3γ binding and may have distinct functions.
Phosphorylation-deficient mutants of raptor associate with mTORC1 but delay cell transit through G2/M.
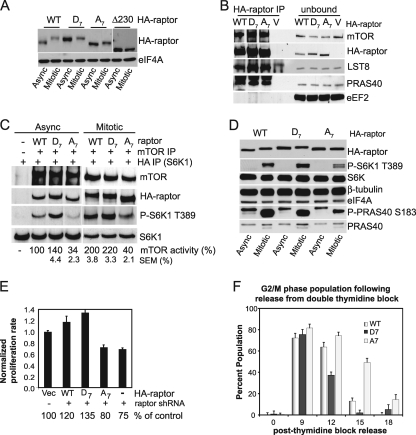
To determine the function of mitosis-mediated phosphorylation of raptor, the seven identified phosphorylation sites were mutated or deleted and the proteins were expressed as Flag- and HA-tagged proteins from retrovirus vectors in HaCaT cells. Exogenously expressed proteins consisted of wild-type raptor, mutants with all seven phosphorylation sites mutated to aspartic acid (D7) to mimic phosphorylation or to nonphosphorylatable alanines (A7), and a mutant containing a 230-amino-acid (aa) deletion encompassing the entire phosphorylated region of raptor (Δ230) (Fig. 5A). When expressed in HaCaT cells, wild-type raptor migrated more slowly during mitosis. The shift, already present in asynchronous cells, was much less pronounced for raptor D7 during mitosis and only slight for raptor A7. Raptor Δ230 was poorly expressed (darker exposure for Δ230 in Fig. 5A), and it completely failed to shift to slower mobility during mitosis. While raptor Δ230 was capable of forming complexes with mTOR and its expression in HaCaT cells slowed their growth, its expression was quickly lost from cells despite continued selection, suggesting that it was unstable, toxic, or both (see Fig. S3A in the supplemental material; data not shown). We therefore focused on the D7 and A7 raptor mutants. Wild-type, D7, and A7 raptor proteins were all recovered in complexes of mTORC1 with mTOR, LST8, and PRAS40 (Fig. 5B). Because raptor phosphorylation mutants interact with mTOR similarly to wild-type raptor protein, studies were carried out to determine whether mTORC1 that contains raptor phosphorylation mutants has functional kinase activity.
FIG. 5.
Effect of overexpression of raptor phosphorylation mutants in HaCaT cells. (A) Expression of phosphoraptor mutants. Asynchronous and mitotic HaCaT cells were transduced with HA-tagged raptor constructs, and equal amounts of lysates from asynchronous or mitotic cells were immunoblotted for the indicated proteins: wild-type (WT) raptor, phosphomimetic (aspartic acid) raptor (D7) and phosphorylation-deficient-alanine raptor (A7) (both mutated in the seven identified mitosis-phosphorylation sites), and Δ230 raptor, deleted of the phosphorylation region containing all seven identified phosphorylation sites. Lighter and darker exposures of raptor protein blots are shown. Typical results of multiple independent experiments are shown. (B) WT, D7, and A7 raptor proteins all form mTORC1 complexes. HA-raptor proteins were immunoprecipitated from equal amounts of transfected HaCaT cell lysates, and associated proteins were detected by immunoblot analysis. Unbound (flowthrough) proteins of the immunoprecipitate are shown. eEF2, which does not associate with mTORC1, is shown. Control vector (V) does not express a raptor protein. (C) In vitro kinase assays of mTORC1 activity. mTOR was immunoprecipitated using conditions that preserve mTORC1 integrity and mixed with dephosphorylated, immunoprecipitated HA-S6K1 in an in vitro kinase assay, as described for Fig. 2E. Results were normalized to asynchronous wild-type raptor control and are expressed as the means of the density ratios of P-S6K1 T389 to S6K1 ± SEMs from three independent experiments (P < 0.05). (D) Expression of raptor A7 decreases mTORC1 activity during mitosis. Asynchronous and mitotic HaCaT cells were transduced with HA-tagged raptor constructs, and equal amounts of lysates from asynchronous or mitotic cells were immunoblotted for the indicated proteins. (E) Proliferation rates of cells overexpressing raptor constructs or silenced for endogenous raptor. Proliferation rates were normalized to the vector control (Vec). Standard deviations of the means are shown for three experiments. P was <0.05 by paired t test. (F) Cells expressing raptor A7 accumulate in G2. Raptor silencing alone results in G1 accumulation. Control HaCaT cells express nonsilencing (NS) shRNA and vector alone rather than HA-raptor or vector alone and raptor shRNA. Cells were analyzed by FACS. Typical results are shown.
We assessed the effect of the different raptor mutants on mTORC1 in vitro kinase activity. Compared to wild-type raptor (mTORC1 expression normalized to 100% activity), phosphomimetic raptor D7 increased the ability of mTORC1 to phosphorylate S6K1 by ∼40%, whereas phosphorylation-deficient raptor A7 decreased it to about one-third in asynchronous cells (Fig. 5C). In contrast, in mitotic cells, D7 raptor activity did not strongly further increase S6K1 phosphorylation above that of wild-type raptor, whereas A7 raptor remained poorly phosphorylated (Fig. 5C). These data are consistent with “activation” of raptor by aspartic acid substitutions that cannot be significantly further activated in mitosis. Significantly decreased signaling by mTORC1 containing the raptor A7 mutant was also found in vivo, evidenced by reduced P-S6K1 T389 and P-PRAS40 S183 in mitotic raptor A7-expressing cells (Fig. 5D). Nevertheless, cells overexpressing mutant raptor proteins did not differ in their responses to rapamycin treatment (see Fig. S3B in the supplemental material), suggesting that unknown mitotic raptor modifications may play a role in the rapamycin insensitivity of mitotic mTORC1 or that mitotic alteration of other components of mTORC1 do so. We note that it was not possible to test all permutations of the identified raptor phosphorylation sites (over 5,000) and that there is no means for prioritizing them at this time.
Cell proliferation assays were performed to determine whether raptor mutants influence rates of cell cycling. Raptor depletion by RNAi decreased cell proliferation rates (∼25%), and overexpression of wild-type raptor increased cell proliferation rates (20%) (Fig. 5E). The D7 raptor phosphomimetic mutant increased cell proliferation rates by almost 40%, even more than wild-type raptor, while phosphorylation-deficient raptor A7 reproducibly decreased cell proliferation by ∼20% (Fig. 5E) (P values for all results are <0.05 by paired t test). Changes in cell proliferation were consistent with the cell cycle transit results (Fig. 5F). In cells overexpressing raptor mutants, D7-expressing cells traversed G2/M faster than both wild-type raptor and A7-expressing cells. The A7-overexpressing cells traversed G2/M at a slower rate than wild-type and D7 raptor-expressing cells, consistent with their slower proliferation rate (Fig. 5F). The cell cycle distribution of cells expressing the different raptor proteins in a raptor-silenced background was therefore analyzed. As noted previously, raptor silencing delays cell cycling, retaining cells at G1 (Fig. 3E). Interestingly, expression of wild-type raptor, mutant raptor D7, or raptor A7 all rescued the G1 accumulation phenotype resulting from raptor silencing (Table 1). Expression of the phosphorylation-deficient raptor A7 mutant (but not the phosphomimetic D7 mutant) also caused increased cell accumulation at G2/M (Fig. 5F; Table 1). The D7 mutant cells, as expected, showed the opposite result, with faster transit through G2/M (Fig. 5F) and faster transit through the G1 and S phases of the cell cycle (Table 1; data not shown). This result explains the lack of change in cell cycle distribution with the raptor phosphomimetic D7 mutant. Therefore, while overall raptor levels are important for G1 progression, as shown by the delay in G1 transit in raptor-silenced cells, raptor phosphorylation is associated with facilitated progression through G2/M.
TABLE 1.
Raptor phosphorylation alters cell cycle distributiona
| HA-raptor expressed | Presence of raptor shRNA | % of cells in: |
||
|---|---|---|---|---|
| G1 | S | G2/M | ||
| None | − | 45.6 | 19.3 | 35.1 |
| WT | + | 46.0 | 11.0 | 43.0 |
| D7 | + | 46.4 | 10.7 | 42.9 |
| A7 | + | 34.8 | 9.1 | 56.1 |
| None | + | 72.9 | 13.0 | 13.6 |
HaCaT cells were transfected with nonsilencing shRNA or raptor silencing shRNAs and HA-raptor expression vector or vector alone (none). Cells were analyzed by FACS. Results of three independent experiments were averaged and are shown.
Raptor phosphorylation in G2/M stimulates cap-independent IRES-mediated mRNA translation.
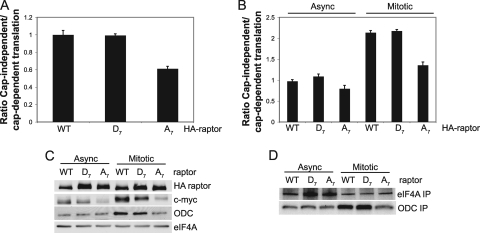
A major target of mTORC1 activity is regulation of translation. We therefore assessed the effect of overexpression of wild-type, D7, or A7 raptor proteins on overall protein synthesis in HaCaT cells. Interestingly, overexpression of wild-type, D7, or A7 raptor did not affect overall protein synthesis rates in asynchronous cells, nor did it alter the well-established downregulation of protein synthesis in mitosis (data not shown). Cap-independent, IRES-mediated translation of specific mRNAs plays an important role in mitotic progression (47, 70). Consequently, we addressed the possibility that mTORC1 signaling during mitosis could affect IRES-mediated mRNA translation. Translation directed by the encephalomyocarditis virus (EMCV) IRES in HaCaT cells overexpressing wild-type or mutant raptor proteins was analyzed by transfection of a combined cap-dependent and IRES-dependent bicistronic luciferase mRNA. EMCV IRES-directed translation was decreased by ∼45% in asynchronous cells overexpressing raptor A7 (Fig. 6A; see Fig. S4A and B in the supplemental material). To specifically measure cap-independent translation during mitosis, 293 cells (which transfect at much higher efficiency than HaCaT cells) were cotransfected with raptor-expressing vectors, a cap-dependent β-galactosidase expression vector (73), and an EMCV IRES enhanced green fluorescent protein (EGFP) expression vector and cell lysates were subjected to IP after [35S]methionine and [35S]cysteine radiolabeling. Cap-independent translation (the ratio of green fluorescent protein [GFP] to β-galactosidase) increased by almost 2.5-fold during mitosis but was blocked by overexpression of raptor A7 (Fig. 6B; see Fig. S4C in the supplemental material). Similar results were obtained with the vascular endothelial growth factor A (VEGF-A) IRES reporter (data not shown). In addition, a small number of cellular mRNAs contain IRES elements and translate via these elements during mitosis, including ornithine decarboxylase (ODC) and c-myc mRNAs (47). Immunoblot analysis of mitotic and interphase HaCaT cells indicated that, while both ODC and c-myc protein levels increased during mitosis in wild-type and D7 raptor-expressing cells, raptor A7 overexpression largely blocked this effect (Fig. 6C), again consistent with a requirement for phosphorylated raptor in IRES-dependent mRNA translation during mitosis. To determine whether A7 raptor expression specifically impaired ODC and c-myc mRNA translation during mitosis, lysates of [35S]methionine- and [35S]cysteine-radiolabeled HaCaT cells were immunoprecipitated with antibodies against eIF4A (cap-dependent) and ODC (cap-independent) proteins. ODC translation increased during mitosis by almost 3-fold in wild-type and D7 raptor-expressing cells but not in cells overexpressing raptor A7 (Fig. 6D). Thus, raptor phosphorylation and likely mTORC1 activity promote IRES-dependent mRNA translation during mitosis.
FIG. 6.
Phosphorylation of raptor during mitosis is required for cap-independent translation. (A) HaCaT cells were transduced with the indicated HA-raptor constructs and transfected with a bicistronic reporter expressing Renilla luciferase (EMCV IRES) and firefly luciferase (cap-independent translation). The ratio of firefly to Renilla luciferase activity was assessed 24 h later. WT, wild type. (B) 293 cells were cotransfected with the indicated HA-raptor constructs and pCR3-βgal and pIRES-EGFP constructs, subcultured, and grown asynchronously or arrested with nocodazole. Cells were then radiolabeled, and lysates were subjected to immunoprecipitation with anti-β-galactosidase (cap-dependent) or anti-GFP (cap-independent) antibodies. Shown are the average ratios of GFP/β-galactosidase levels from three experiments with SEMs. (C) The indicated transduced HaCaT cell lysates were immunoblotted for the proteins shown and normalized to the asynchronous wild-type raptor control. (D) HaCaT cells transduced with HA-raptor constructs as shown were grown asynchronously or arrested at G2/M and radiolabeled, and lysates were subjected to immunoprecipitation with the indicated antibodies and normalized to the asynchronous wild-type raptor control. SEMs are indicated.
Mitosis-mediated raptor phosphorylation involves cdc2 and GSK3 pathways.
Four of the identified phosphorylated amino acids in raptor (S696, T706, S863, and S877) are potential targets (based on consensus motifs) of cdc2 and GSK3, which are active kinases during mitosis (63). Two residues (S696 and S859) are also potential targets of Chk1, which, apart from its role in the DNA damage response, is also involved in normal mitotic entry and spindle checkpoint function (38, 74).
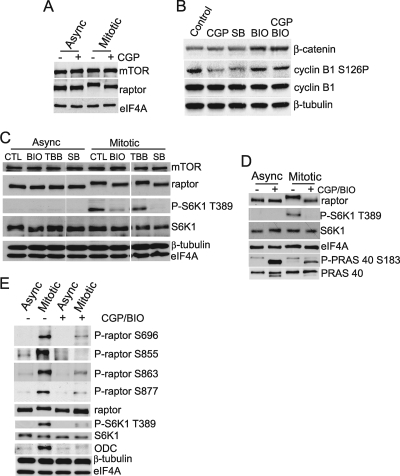
To explore the possible control of mTORC1 phosphorylation during mitosis, asynchronous and mitosis-arrested HaCaT cells were treated with the cdc2-specific inhibitor CGP74514A (CGP) and raptor phosphorylation was analyzed by determining mobility shift (Fig. 7A). Inhibitors were added after mitotic arrest by nocodazole treatment, and then mitotic cells were removed by the mechanical shake-off method, ensuring that the cells acquired were almost entirely mitotic. CGP74514A had no effect on the migration or abundance of raptor in asynchronous HaCaT cells but incompletely restored a faster-mobility form of raptor in mitotic cells (Fig. 7A). It is therefore likely that the cdc2 pathway is necessary for phosphorylation of some residues that alter raptor mobility during mitosis. Control studies in asynchronous cells demonstrated reduction of cyclin B1 S126 phosphorylation by CGP (Fig. 7B). Asynchronous and mitotic HaCaT cells were treated with inhibitors of GSK3, Chk1, and CK2, a kinase also involved in cell cycle transit that is not predicted to recognize any of the 7 identified raptor phosphorylation sites. Inhibitors of GSK3 (BIO) and Chk1 (SB), but not CK2 (TBB), partially reduced raptor electrophoretic mobility during mitosis compared to that of control untreated raptor (Fig. 7C), similar to the effect of CGP74514A. All inhibitors behaved as expected on control targets (Fig. 7B), including reduced S126 phosphorylation on cyclin B by CGP and SB and increased β-catenin levels by BIO or BIO/CGP. Alteration of raptor mobility by BIO or SB was accompanied by decreased phosphorylation of S6K1 at the mTORC1 site (T389) (Fig. 7C). SB (which inhibits both cdc2 and Chk1 [32]) increased raptor mobility during mitosis and decreased S6K1 phosphorylation, again suggesting that cdc2 is involved in mitotic mTORC1 phosphorylation (Fig. 7C). Since Chk1 silencing did not alter raptor mobility during mitosis (data not shown), the effect of SB on raptor phosphorylation was most likely due to cdc2 inhibition. Although CK2 is active during mitosis, its inhibition also did not alter raptor mobility (Fig. 7C).
FIG. 7.
Inhibitors of cdc2 and GSK3 kinase pathways prevent mitosis-specific phosphorylation of raptor. (A) Asynchronous and mitotic HaCaT cells were treated with cdc2 kinase inhibitor CGP74514A or left untreated, and equal amounts of protein lysates were analyzed by immunoblotting for the indicated proteins. eIF4A is shown as a control for protein loading. (B) Control data for inhibitor action. Inhibitor CGP was used at 1.5 μM, SB218078 (SB) was used at 2.5 μM, and BIO was used at 2 μM, as described in Materials and Methods. Equal amounts of HaCaT protein lysates were examined by immunoblot analysis as shown for total and phosphorylated forms of the proteins. Typical results of three independent experiments are shown. β-Tubulin is shown as a loading control. (C) Asynchronous or mitotic HaCaT cells were treated with the indicated inhibitors: BIO, GSK3; TBB, CK2; SB218078 (SB), Chk1 and cdc2. Equal amounts of lysates were analyzed by immunoblotting. β-Tubulin and eIF4A are shown as loading controls. (D) As in panel C, but cells were either left untreated or treated with a combination of BIO (GSK3 inhibitor) and CGP74514A (cdc2 inhibitor). Proteins were subjected to immunoblotting as shown. (E) As for panel D, cells were immunoblotted for the raptor phospho-specific sites or for total proteins, as indicated.
To determine whether raptor phosphorylation involves both cdc2 and GSK3 pathways, both were inhibited upon HaCaT cell entrance into mitosis. Combined cdc2 and GSK3 inhibition largely eliminated slow-mobility mitotic raptor and strongly blocked S6K1 phosphorylation at position T389 and PRAS40 phosphorylation at position S183 (Fig. 7D). Inhibition of other kinases active in mitosis, including Plk1, Aurora A, and Aurora B, had no detectable effect on raptor mobility in mitosis or S6K1 phosphorylation (data not shown), nor did Erk1/2 inhibition (Fig. 2C). Moreover, immunoprecipitated mTORC1 that was obtained from mitotically arrested cells treated with CGP and BIO displayed ∼2-fold-diminished in vitro kinase activity with dephosphorylated S6K1 as a target, compared to mTORC1 isolated from control mitotic cells (Table 2). There was no effect of other inhibitors on mTORC1 in vitro kinase activity when mTORC1 was isolated from asynchronous cells (data not shown). To further corroborate the site-specific modifications of raptor during mitosis and their control by the cdc2/GSK3 pathways, we utilized phospho-specific antibodies to probe endogenous raptor in asynchronous and mitotic cells untreated or treated with BIO/CGP. As demonstrated in Fig. 7E, using phospho-site-specific antibodies, phosphorylation of raptor sites S696, S855, S863, and S877 was strongly increased in untreated mitotic cells compared to asynchronous cells. However, phosphorylation of these sites was significantly blocked during mitosis by inhibition of cdc2/GSK3, but not Erk1/2 (Fig. 7E; data not shown). Moreover, expression of ODC was severely decreased in mitotic cells treated with CGP/BIO, as previously seen with raptor A7 expression (Fig. 7E). These data indicate that raptor is phosphorylated during mitosis through signaling pathways involved in mitotic functions and that phosphorylation is important for both G2/M progression and IRES-dependent mRNA translation.
TABLE 2.
In vitro mTORC1 activitya
| Cell condition | Treatment | mTORC1 activity (%) ± SEM |
|---|---|---|
| Asynchronous | None | 100 |
| Asynchronous | CGP + BIO | 97 ± 6 |
| Mitotic | None | 220 ± 20 |
| Mitotic | CGP + BIO | 139 ± 5 |
Assay of mTORC1 in vitro kinase activity was performed by determining S6K1 phosphorylation at T389 compared to total HA-S6K1, as described for Fig. 2F. Activity was normalized to that of asynchronous control cells. mTORC1 activity is diminished when mTOR is isolated from mitotic cells treated with CGP74514A and BIO compared to that for untreated mitotic cells. Values are averages from three independent sets of data.
DISCUSSION
In complex metazoans, mTOR assimilates signals from growth factors and nutrient availability to control cell size and proliferation (reviewed in reference 72). Complex metazoans coordinate cell size and proliferation to maintain organ and organismal size (reviewed in reference 44). For example, mutations that cause an increase in cell proliferation in the Drosophila wing are countered by reduced cell size, producing mutant wings of normal size (41). In contrast, mutations in the mTORC1 pathway are distinct because they alter cell growth, organ growth, and cellular proliferation. For example, mutation of the TSC2 ortholog gigas in Drosophila alters the cell cycle, resulting in endoduplication and enlarged cells, as well as enlarged organ size (30). In addition, S6K1 knockout mice are smaller in size and smaller in cell size, demonstrating an uncoupling of cell size from cell proliferation (60). How the insulin-stimulated PI3K/AKT/mTORC1 nexus regulates cell size and cell proliferation is still poorly understood. Activation of the mTORC1 pathway increases cell proliferation in a multifaceted manner. mTORC1 activation inhibits the cell cycle inhibitor p27 and blocks increased levels of cyclin D1, CDK4, and other cell cycle-stimulatory factors (21, 52). In addition, mTORC1 stimulates cell growth by increasing macromolecular synthesis, including increased ribosome biogenesis, increased overall protein synthesis, and increased nutrient import, and by antagonizing catabolic processes such as autophagy and fatty acid β-oxidation (72).
Available evidence indicates that mTORC1 activation is associated with anabolic processes, particularly increased protein synthesis. Our data show that established upstream activating networks of mTORC1, such as PI3K and Ras/Erk, are downregulated during mitosis, yet mTORC1 remains active. Indeed, inhibitors of the PI3K/AKT/mTORC1 pathway, including LY294002 and rapamycin, did not prevent mTORC1 activity during mitosis. Mitotic mTORC1 is therefore distinct from the asynchronous complex, a key difference being mitosis-specific raptor phosphorylation. The phosphorylated form of raptor establishes intact, active mTORC1 complexes with mTOR and LST8, it can bind mTORC1 targets such as PRAS40 and 4E-BP1, and it displays moderately higher in vitro and in vivo kinase activities. Our results also suggest that PRAS40 is not simply a competitive inhibitor of mTORC1 since it is associated with mTORC1 despite increased activity of the complex in mitosis. It is paradoxical that mTORC1 is active in mitosis, a period during which global protein synthesis is impaired. This is likely because translation of certain mRNAs must continue for production of factors involved in mitotic progression (47, 48, 70). Our results indicate that phosphorylation of raptor, which is associated with mTORC1 activity in mitosis, stimulates cap-independent translation. However, our experiments with raptor phosphorylation mutants show that ablation of raptor phosphorylation, while decreasing mTORC1 activity in asynchronous cells, has no effect on rapamycin sensitivity. Other factors may therefore play a role in promoting rapamycin resistance during mitosis. Our data also add to the growing evidence that mTORC1 output to its targets is likely distinctly regulated, suggestive of another layer of complexity in the mTOR regulatory network that is temporally controlled and possibly cell type specific (10).
Several of the mitotic raptor phosphorylation sites are conserved in multiple higher eukaryotes, and all map to a middle region of raptor lacking established motifs or structural elements. The phosphomimetic raptor D7 mutant, in which aspartate residues are substituted for serine and threonine phosphorylation sites, binds mTOR and is associated with higher in vitro mTORC1 kinase activity and increased cell proliferation. Conversely, phosphorylation-deficient raptor A7 protein decreases endogenous raptor-silenced cell accumulation in G1, decreases mTORC1 activity, and instead promotes increased accumulation of cells in G2. These data suggest that raptor also has a role in progression through the G2/M phase of the cell cycle. These results help to explain several previous observations. For example, a high-throughput screen for factors involved in maintaining cell size and cell cycle progression identified raptor as a protein involved in both G1 arrest (as expected) and decreased cytokinesis (4), which agrees with our findings that raptor plays a role in mitosis. Moreover, insulin-mediated activation of the canonical dTOR/dRaptor pathway in Drosophila was found to inhibit G2/M cell cycle progression (71). Our results suggest that, during G2/M, upstream signaling by PI3K/AKT is decreased (possibly to disable other PI3K/AKT targets) but cdc2/GSK3 pathways increase mitotic mTORC1 activity, cap-independent (IRES) mRNA translation, and progression through G2/M. A recent report has shown that S6K1 can phosphorylate and inhibit GSK3 by AKT feedback inhibition (75). How then is GSK3 active during mitosis to phosphorylate raptor? One possibility is that, as noted above, S6K1 output is compartmentalized to only signal to certain factors during mitosis. Accordingly, we do not observe an increase in S6K1-mediated IRS-1 phosphorylation (data not shown). In another high-throughput screen, dRaptor was found to be required for proper mitotic spindle assembly in Drosophila S2 cells (23). Phosphorylated raptor might therefore play a role in mitosis by adapting mTOR to target proteins in addition to S6K1 and 4E-BP1. It is important to note that phosphorylation of raptor during mitosis has consequences distinct from those of AMPK-mediated raptor phosphorylation, In this regard, our data have determined that the mitotic modification of raptor prevents its association with 14-3-3γ during mitosis.
mTORC1 containing phosphorylated raptor displays characteristics distinct from those of mTORC1 isolated from asynchronous cells and akin to those found for human cytomegalovirus (HCMV)-infected cells (36). HCMV infection, similar to mitosis, alters raptor electrophoretic mobility and prevents it from being inhibited by rapamycin (36). Our results reveal a novel region in raptor that is specifically hyperphosphorylated during mitosis. This phosphorylation is associated with cell cycle-dependent alteration of mTORC1 activity and increased phosphorylation of some of its known targets and affects cap-independent translation during mitosis. Furthermore, mitosis-specific raptor phosphorylation is required for normal cell growth and progression through G2/M. It is important to note that a recent study demonstrated that p90 ribosomal S6 kinases (Rsks) 1 and 2, which are activated by the Ras/mitogen-activated protein kinase (MAPK) pathway, also phosphorylate raptor at position 722, which stimulates its activity (7). These findings are distinct from the work reported here for two reasons: the Rsks phosphorylate raptor at position 722, which is only one of the mitosis-specific phosphorylation sites we identified, and inhibition of Erk/MAPK activity did not prevent mTORC1 activity or raptor phosphorylation-dependent electrophoretic mobility shift during mitosis. Our findings therefore disclose a new mechanism of cell cycle-specific mTORC1 regulation and function and an important new mechanism by which rapamycin resistance of tumor cells may arise.
Supplementary Material
Acknowledgments
We declare no conflict of interest.
Footnotes
Published ahead of print on 3 May 2010.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Alvarez, B., A. C. Martinez, B. M. Burgering, and A. C. Carrera. 2001. Forkhead transcription factors contribute to execution of the mitotic programme in mammals. Nature 413:744-747. [DOI] [PubMed] [Google Scholar]
- 2.Baserga, R. 1962. A radioautographic study of the uptake of 14C leucine by tumor cells in deoxyribonucleic acid synthesis. Biochim. Biophys. Acta 61:445-450. [Google Scholar]
- 3.Baserga, R., and W. E. Kisieleski. 1962. Recent observations on cell proliferation and metabolism by radioautography with tritiated compounds. Atompraxis 8:386-391. [PubMed] [Google Scholar]
- 4.Bjorklund, M., M. Taipale, M. Varjosalo, J. Saharinen, J. Lahdenpera, and J. Taipale. 2006. Identification of pathways regulating cell size and cell-cycle progression by RNAi. Nature 439:1009-1013. [DOI] [PubMed] [Google Scholar]
- 5.Bonneau, A. M., and N. Sonenberg. 1987. Involvement of the 24-kDa cap-binding protein in regulation of protein synthesis in mitosis. J. Biol. Chem. 262:11134-11139. [PubMed] [Google Scholar]
- 6.Boyer, D., R. Quintanilla, and K. K. Lee-Fruman. 2008. Regulation of catalytic activity of S6 kinase 2 during cell cycle. Mol. Cell. Biochem. 307:59-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carriere, A., M. Cargnello, L. A. Julien, H. Gao, E. Bonneil, P. Thibault, and P. P. Roux. 2008. Oncogenic MAPK signaling stimulates mTORC1 activity by promoting RSK-mediated raptor phosphorylation. Curr. Biol. 18:1269-1277. [DOI] [PubMed] [Google Scholar]
- 8.Celis, J. E., P. Madsen, and A. G. Ryazanov. 1990. Increased phosphorylation of elongation factor 2 during mitosis in transformed human amnion cells correlates with a decreased rate of protein synthesis. Proc. Natl. Acad. Sci. U. S. A. 87:4231-4235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chaussepied, M., and D. Ginsberg. 2004. Transcriptional regulation of AKT activation by E2F. Mol. Cell 16:831-837. [DOI] [PubMed] [Google Scholar]
- 10.Choo, A. Y., S. O. Yoon, S. G. Kim, P. P. Roux, and J. Blenis. 2008. Rapamycin differentially inhibits S6Ks and 4E-BP1 to mediate cell-type-specific repression of mRNA translation. Proc. Natl. Acad. Sci. U. S. A. 105:17414-17419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Connolly, E., S. Braunstein, S. Formenti, and R. J. Schneider. 2006. Hypoxia inhibits protein synthesis through a 4E-BP1 and elongation factor 2 kinase pathway controlled by mTOR and uncoupled in breast cancer cells. Mol. Cell. Biol. 26:3955-3965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dorrello, N. V., A. Peschiaroli, D. Guardavaccaro, N. H. Colburn, N. E. Sherman, and M. Pagano. 2006. S6K1- and betaTRCP-mediated degradation of PDCD4 promotes protein translation and cell growth. Science 314:467-471. [DOI] [PubMed] [Google Scholar]
- 13.Edinger, A. L., and C. B. Thompson. 2004. An activated mTOR mutant supports growth factor-independent, nutrient-dependent cell survival. Oncogene 23:5654-5663. [DOI] [PubMed] [Google Scholar]
- 14.Fan, H., and S. Penman. 1970. Regulation of protein synthesis in mammalian cells. II. Inhibition of protein synthesis at the level of initiation during mitosis. J. Mol. Biol. 50:655-670. [DOI] [PubMed] [Google Scholar]
- 15.Feldman, M. E., B. Apsel, A. Uotila, R. Loewith, Z. A. Knight, D. Ruggero, and K. M. Shokat. 2009. Active-site inhibitors of mTOR target rapamycin-resistant outputs of mTORC1 and mTORC2. PLoS Biol. 7:e38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fingar, D. C., and J. Blenis. 2004. Target of rapamycin (TOR): an integrator of nutrient and growth factor signals and coordinator of cell growth and cell cycle progression. Oncogene 23:3151-3171. [DOI] [PubMed] [Google Scholar]
- 17.Fingar, D. C., S. Salama, C. Tsou, E. Harlow, and J. Blenis. 2002. Mammalian cell size is controlled by mTOR and its downstream targets S6K1 and 4EBP1/eIF4E. Genes Dev. 16:1472-1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Foster, K. G., H. A. Acosta-Jaquez, Y. Romeo, B. Ekim, G. A. Soliman, A. Carriere, P. P. Roux, B. A. Ballif, and D. C. Fingar. 2010. Regulation of mTOR complex 1 (mTORC1) by raptor S863 and multi-site phosphorylation. J. Biol. Chem. 285:80-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Frodin, M., T. L. Antal, B. A. Dummler, C. J. Jensen, M. Deak, S. Gammeltoft, and R. M. Biondi. 2002. A phosphoserine/threonine-binding pocket in AGC kinases and PDK1 mediates activation by hydrophobic motif phosphorylation. EMBO J. 21:5396-5407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fuge, E. K., E. L. Braun, and M. Werner-Washburne. 1994. Protein synthesis in long-term stationary-phase cultures of Saccharomyces cerevisiae. J. Bacteriol. 176:5802-5813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gao, N., D. C. Flynn, Z. Zhang, X. S. Zhong, V. Walker, K. J. Liu, X. Shi, and B. H. Jiang. 2004. G1 cell cycle progression and the expression of G1 cyclins are regulated by PI3K/AKT/mTOR/p70S6K1 signaling in human ovarian cancer cells. Am. J. Physiol. Cell Physiol. 287:C281-C291. [DOI] [PubMed] [Google Scholar]
- 22.Gingras, A. C., B. Raught, S. P. Gygi, A. Niedzwiecka, M. Miron, S. K. Burley, R. D. Polakiewicz, A. Wyslouch-Cieszynska, R. Aebersold, and N. Sonenberg. 2001. Hierarchical phosphorylation of the translation inhibitor 4E-BP1. Genes Dev. 15:2852-2864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Goshima, G., R. Wollman, S. S. Goodwin, N. Zhang, J. M. Scholey, R. D. Vale, and N. Stuurman. 2007. Genes required for mitotic spindle assembly in Drosophila S2 cells. Science 316:417-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gwinn, D. M., D. B. Shackelford, D. F. Egan, M. M. Mihaylova, A. Mery, D. S. Vasquez, B. E. Turk, and R. J. Shaw. 2008. AMPK phosphorylation of raptor mediates a metabolic checkpoint. Mol. Cell 30:214-226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hara, K., Y. Maruki, X. Long, K. Yoshino, N. Oshiro, S. Hidayat, C. Tokunaga, J. Avruch, and K. Yonezawa. 2002. Raptor, a binding partner of target of rapamycin (TOR), mediates TOR action. Cell 110:177-189. [DOI] [PubMed] [Google Scholar]
- 26.Heesom, K. J., A. Gampel, H. Mellor, and R. M. Denton. 2001. Cell cycle-dependent phosphorylation of the translational repressor eIF-4E binding protein-1 (4E-BP1). Curr. Biol. 11:1374-1379. [DOI] [PubMed] [Google Scholar]
- 27.Heitman, J., N. R. Movva, and M. N. Hall. 1991. Targets for cell cycle arrest by the immunosuppressant rapamycin in yeast. Science 253:905-909. [DOI] [PubMed] [Google Scholar]
- 28.Hresko, R. C., and M. Mueckler. 2005. mTOR.RICTOR is the Ser473 kinase for Akt/protein kinase B in 3T3-L1 adipocytes. J. Biol. Chem. 280:40406-40416. [DOI] [PubMed] [Google Scholar]
- 29.Huang, J. T., and R. J. Schneider. 1991. Adenovirus inhibition of cellular protein synthesis involves inactivation of cap-binding protein. Cell 65:271-280. [DOI] [PubMed] [Google Scholar]
- 30.Ito, N., and G. M. Rubin. 1999. gigas, a Drosophila homolog of tuberous sclerosis gene product-2, regulates the cell cycle. Cell 96:529-539. [DOI] [PubMed] [Google Scholar]
- 31.Jacinto, E., V. Facchinetti, D. Liu, N. Soto, S. Wei, S. Y. Jung, Q. Huang, J. Qin, and B. Su. 2006. SIN1/MIP1 maintains rictor-mTOR complex integrity and regulates Akt phosphorylation and substrate specificity. Cell 127:125-137. [DOI] [PubMed] [Google Scholar]
- 32.Jackson, J. R., A. Gilmartin, C. Imburgia, J. D. Winkler, L. A. Marshall, and A. Roshak. 2000. An indolocarbazole inhibitor of human checkpoint kinase (Chk1) abrogates cell cycle arrest caused by DNA damage. Cancer Res. 60:566-572. [PubMed] [Google Scholar]
- 33.Kim, D. H., D. D. Sarbassov, S. M. Ali, J. E. King, R. R. Latek, H. Erdjument-Bromage, P. Tempst, and D. M. Sabatini. 2002. mTOR interacts with raptor to form a nutrient-sensitive complex that signals to the cell growth machinery. Cell 110:163-175. [DOI] [PubMed] [Google Scholar]
- 34.Kim, D. H., D. D. Sarbassov, S. M. Ali, R. R. Latek, K. V. Guntur, H. Erdjument-Bromage, P. Tempst, and D. M. Sabatini. 2003. GbetaL, a positive regulator of the rapamycin-sensitive pathway required for the nutrient-sensitive interaction between raptor and mTOR. Mol. Cell 11:895-904. [DOI] [PubMed] [Google Scholar]
- 35.Kovacina, K. S., G. Y. Park, S. S. Bae, A. W. Guzzetta, E. Schaefer, M. J. Birnbaum, and R. A. Roth. 2003. Identification of a proline-rich Akt substrate as a 14-3-3 binding partner. J. Biol. Chem. 278:10189-10194. [DOI] [PubMed] [Google Scholar]
- 36.Kudchodkar, S. B., Y. Yu, T. G. Maguire, and J. C. Alwine. 2006. Human cytomegalovirus infection alters the substrate specificities and rapamycin sensitivities of raptor- and rictor-containing complexes. Proc. Natl. Acad. Sci. U. S. A. 103:14182-14187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu, X., and X. F. Zheng. 2007. Endoplasmic reticulum and Golgi localization sequences for mammalian target of rapamycin. Mol. Biol. Cell 18:1073-1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Loffler, H., B. Rebacz, A. D. Ho, J. Lukas, J. Bartek, and A. Kramer. 2006. Chk1-dependent regulation of Cdc25B functions to coordinate mitotic events. Cell Cycle 5:2543-2547. [DOI] [PubMed] [Google Scholar]
- 39.Ma, L., Z. Chen, H. Erdjument-Bromage, P. Tempst, and P. P. Pandolfi. 2005. Phosphorylation and functional inactivation of TSC2 by Erk implications for tuberous sclerosis and cancer pathogenesis. Cell 121:179-193. [DOI] [PubMed] [Google Scholar]
- 40.Mathews, M. B., N. Sonenberg, and J. W. B. Hershey. 1996. Origins and principles of translational control, p. 1-31. In J. W. B. Hershey, M. B. Mathews, and N. Sonenberg (ed.), Translational control. Cold Spring Harbor Press, Cold Spring Harbor, NY. [DOI] [PMC free article] [PubMed]
- 41.Neufeld, T. P., A. F. de la Cruz, L. A. Johnston, and B. A. Edgar. 1998. Coordination of growth and cell division in the Drosophila wing. Cell 93:1183-1193. [DOI] [PubMed] [Google Scholar]
- 42.Olszewska, M. J. 1964. Double autoradiography of nuclei in the interphase. Exp. Cell Res. 33:571-574. [DOI] [PubMed] [Google Scholar]
- 43.Oshiro, N., R. Takahashi, K. Yoshino, K. Tanimura, A. Nakashima, S. Eguchi, T. Miyamoto, K. Hara, K. Takehana, J. Avruch, U. Kikkawa, and K. Yonezawa. 2007. The proline-rich Akt substrate of 40 kDa (PRAS40) is a physiological substrate of mammalian target of rapamycin complex 1. J. Biol. Chem. 282:20329-20339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Potter, C. J., and T. Xu. 2001. Mechanisms of size control. Curr. Opin. Genet. Dev. 11:279-286. [DOI] [PubMed] [Google Scholar]
- 45.Prescott, D. M. 1962. Nucleic acid and protein metabolism in the macnonuclei of two ciliated protozoa. J. Histochem. Cytochem. 10:145-153. [Google Scholar]
- 46.Pyronnet, S., J. Dostie, and N. Sonenberg. 2001. Suppression of cap-dependent translation in mitosis. Genes Dev. 15:2083-2093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pyronnet, S., L. Pradayrol, and N. Sonenberg. 2000. A cell cycle-dependent internal ribosome entry site. Mol. Cell 5:607-616. [DOI] [PubMed] [Google Scholar]
- 48.Pyronnet, S., and N. Sonenberg. 2001. Cell-cycle-dependent translational control. Curr. Opin. Genet. Dev. 11:13-18. [DOI] [PubMed] [Google Scholar]
- 49.Qin, H., B. Raught, N. Sonenberg, E. G. Goldstein, and A. M. Edelman. 2003. Phosphorylation screening identifies translational initiation factor 4GII as an intracellular target of Ca(2+)/calmodulin-dependent protein kinase I. J. Biol. Chem. 278:48570-48579. [DOI] [PubMed] [Google Scholar]
- 50.Qin, X., and P. Sarnow. 2004. Preferential translation of internal ribosome entry site-containing mRNAs during the mitotic cycle in mammalian cells. J. Biol. Chem. 279:13721-13728. [DOI] [PubMed] [Google Scholar]
- 51.Ramirez-Valle, F., S. Braunstein, J. Zavadil, S. C. Formenti, and R. J. Schneider. 2008. eIF4GI links nutrient sensing by mTOR to cell proliferation and inhibition of autophagy. J. Cell Biol. 181:293-307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sabatini, D. M. 2006. mTOR and cancer: insights into a complex relationship. Nat. Rev. Cancer 6:729-734. [DOI] [PubMed] [Google Scholar]
- 53.Sarbassov, D. D., S. M. Ali, D. H. Kim, D. A. Guertin, R. R. Latek, H. Erdjument-Bromage, P. Tempst, and D. M. Sabatini. 2004. Rictor, a novel binding partner of mTOR, defines a rapamycin-insensitive and raptor-independent pathway that regulates the cytoskeleton. Curr. Biol. 14:1296-1302. [DOI] [PubMed] [Google Scholar]
- 54.Sarbassov, D. D., D. A. Guertin, S. M. Ali, and D. M. Sabatini. 2005. Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science 307:1098-1101. [DOI] [PubMed] [Google Scholar]
- 55.Sarbassov, D. D., S. M. Ali, and D. M. Sabatini. 2005. Growing roles for the mTOR pathway. Curr. Opin. Cell Biol. 17:596-603. [DOI] [PubMed] [Google Scholar]
- 56.Schalm, S. S., and J. Blenis. 2002. Identification of a conserved motif required for mTOR signaling. Curr. Biol. 12:632-639. [DOI] [PubMed] [Google Scholar]
- 57.Schalm, S. S., D. C. Fingar, D. M. Sabatini, and J. Blenis. 2003. TOS motif-mediated raptor binding regulates 4E-BP1 multisite phosphorylation and function. Curr. Biol. 13:797-806. [DOI] [PubMed] [Google Scholar]
- 58.Sekulic, A., C. C. Hudson, J. L. Homme, P. Yin, D. M. Otterness, L. M. Karnitz, and R. T. Abraham. 2000. A direct linkage between the phosphoinositide 3-kinase-AKT signaling pathway and the mammalian target of rapamycin in mitogen-stimulated and transformed cells. Cancer Res. 60:3504-3513. [PubMed] [Google Scholar]
- 59.Shah, O. J., S. Ghosh, and T. Hunter. 2003. Mitotic regulation of ribosomal S6 kinase 1 involves Ser/Thr, Pro phosphorylation of consensus and non-consensus sites by Cdc2. J. Biol. Chem. 278:16433-16442. [DOI] [PubMed] [Google Scholar]
- 60.Shima, H., M. Pende, Y. Chen, S. Fumagalli, G. Thomas, and S. C. Kozma. 1998. Disruption of the p70(s6k)/p85(s6k) gene reveals a small mouse phenotype and a new functional S6 kinase. EMBO J. 17:6649-6659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sivan, G., N. Kedersha, and O. Elroy-Stein. 2007. Ribosomal slowdown mediates translational arrest during cellular division. Mol. Cell. Biol. 27:6639-6646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Smith, E. M., and C. G. Proud. 2008. cdc2-cyclin B regulates eEF2 kinase activity in a cell cycle- and amino acid-dependent manner. EMBO J. 27:1005-1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Stark, G. R., and W. R. Taylor. 2006. Control of the G2/M transition. Mol. Biotechnol. 32:227-248. [DOI] [PubMed] [Google Scholar]
- 64.Stolovich, M., T. Lerer, Y. Bolkier, H. Cohen, and O. Meyuhas. 2005. Lithium can relieve translational repression of TOP mRNAs elicited by various blocks along the cell cycle in a glycogen synthase kinase-3- and S6-kinase-independent manner. J. Biol. Chem. 280:5336-5342. [DOI] [PubMed] [Google Scholar]
- 65.Wang, L., T. E. Harris, R. A. Roth, and J. C. Lawrence, Jr. 2007. PRAS40 regulates mTORC1 kinase activity by functioning as a direct inhibitor of substrate binding. J. Biol. Chem. 282:20036-20044. [DOI] [PubMed] [Google Scholar]
- 66.Wang, L., J. C. Lawrence, Jr., T. W. Sturgill, and T. E. Harris. 2009. Mammalian target of rapamycin complex 1 (mTORC1) activity is associated with phosphorylation of raptor by mTOR. J. Biol. Chem. 284:14693-14697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wang, X., W. Li, M. Williams, N. Terada, D. R. Alessi, and C. G. Proud. 2001. Regulation of elongation factor 2 kinase by p90(RSK1) and p70 S6 kinase. EMBO J. 20:4370-4379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Werner-Washburne, M., E. L. Braun, M. E. Crawford, and V. M. Peck. 1996. Stationary phase in Saccharomyces cerevisiae. Mol. Microbiol. 19:1159-1166. [DOI] [PubMed] [Google Scholar]
- 69.Wicker, L. S., R. C. Boltz, Jr., V. Matt, E. A. Nichols, L. B. Peterson, and N. H. Sigal. 1990. Suppression of B cell activation by cyclosporin A, FK506 and rapamycin. Eur. J. Immunol. 20:2277-2283. [DOI] [PubMed] [Google Scholar]
- 70.Wilker, E. W., M. A. van Vugt, S. A. Artim, P. H. Huang, C. P. Petersen, H. C. Reinhardt, Y. Feng, P. A. Sharp, N. Sonenberg, F. M. White, and M. B. Yaffe. 2007. 14-3-3σ controls mitotic translation to facilitate cytokinesis. Nature 446:329-332. [DOI] [PubMed] [Google Scholar]
- 71.Wu, M. Y., M. Cully, D. Andersen, and S. J. Leevers. 2007. Insulin delays the progression of Drosophila cells through G2/M by activating the dTOR/dRaptor complex. EMBO J. 26:371-379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wullschleger, S., R. Loewith, and M. N. Hall. 2006. TOR signaling in growth and metabolism. Cell 124:471-484. [DOI] [PubMed] [Google Scholar]
- 73.Xi, Q., R. Cuesta, and R. J. Schneider. 2004. Tethering of eIF4G to adenoviral mRNAs by viral 100k protein drives ribosome shunting. Genes Dev. 18:1997-2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zachos, G., E. J. Black, M. Walker, M. T. Scott, P. Vagnarelli, W. C. Earnshaw, and D. A. Gillespie. 2007. Chk1 is required for spindle checkpoint function. Dev. Cell 12:247-260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhang, H. H., A. I. Lipovsky, C. C. Dibble, M. Sahin, and B. D. Manning. 2006. S6K1 regulates GSK3 under conditions of mTOR-dependent feedback inhibition of Akt. Mol. Cell 24:185-197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zieve, G. W., D. Turnbull, J. M. Mullins, and J. R. McIntosh. 1980. Production of large numbers of mitotic mammalian cells by use of the reversible microtubule inhibitor nocodazole. Nocodazole accumulated mitotic cells. Exp. Cell Res. 126:397-405. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.