Abstract
Abasic (AP) sites are potent blocks to DNA and RNA polymerases, and their repair is essential for maintaining genome integrity. Although AP sites are efficiently dealt with through the base excision repair (BER) pathway, genetic studies suggest that repair also can occur via nucleotide excision repair (NER). The involvement of NER in AP-site removal has been puzzling, however, as this pathway is thought to target only bulky lesions. Here, we examine the repair of AP sites generated when uracil is removed from a highly transcribed gene in yeast. Because uracil is incorporated instead of thymine under these conditions, the position of the resulting AP site is known. Results demonstrate that only AP sites on the transcribed strand are efficient substrates for NER, suggesting the recruitment of the NER machinery by an AP-blocked RNA polymerase. Such transcription-coupled NER of AP sites may explain previously suggested links between the BER pathway and transcription.
High levels of transcription affect genome stability by stimulating mutagenesis and homologous recombination, both of which are well-known consequences of DNA damage (for reviews, see references 2 and 3). Consistently with a link between transcription and DNA damage accumulation, highly transcribed sequences in the yeast Saccharomyces cerevisiae are more susceptible to exogenous mutagens (19), and spontaneous transcription-associated mutagenesis (TAM) is further elevated when DNA repair is compromised (11, 30, 38). While there likely are multiple causes for and types of damage that accumulate in transcriptionally active DNA, we recently demonstrated that apurinic/apyrimidinic (AP) sites can be a significant source of TAM (30). AP sites are one of the most common DNA lesions and can be a potent block to replicative DNA polymerases (reviewed in reference 15). Blocked DNA synthesis can be rescued either by a recombination/template switch mechanism or through the recruitment of specialized translesion synthesis (TLS) DNA polymerases (14). In yeast, TLS polymerase ζ (Polζ), together with the deoxycytidyl transferase Rev1, is required for most AP-site bypass, with a C nucleotide usually being inserted across from the template lesion (20, 31, 39). Depending on the base lost, AP-site bypass via TLS has the potential to be highly mutagenic.
AP sites can be generated by the spontaneous hydrolysis of the sugar base glycosidic linkage or are formed when a specialized DNA N-glycosylase releases its cognate damaged base from the sugar phosphate backbone (reviewed in references 6 and 15). In yeast, genetic studies have demonstrated that most spontaneous AP sites originate from the incorporation of dUTP in place of dTTP during DNA synthesis (22). Subsequent uracil removal by Ung1, the sole uracil DNA glycosylase in S. cerevisiae, creates a potentially toxic/mutagenic AP site. As a part of the cellular defense mechanism against uracil incorporation into DNA, dUTPase, an essential enzyme in yeast (16) as well as in Escherichia coli (13), catalyzes the conversion of dUTP to dUMP. Thymidylate synthase then uses a folate-derived cofactor to convert dUMP to dTMP, a requisite intermediate in dTTP synthesis. A reduction in either dUTPase or thymidylate synthase activity is associated with an increase in the cellular dUTP/dTTP ratio and enhanced replacement of thymine in DNA with uracil. It should be noted that thymidylate synthase is a common target of chemotherapeutics, which underscores the importance of maintaining a low dUTP/dTTP ratio for genome stability in actively dividing cells (reviewed in reference 5). Additionally, a dUTPase is sometimes packaged into retroviral virions, suggesting that maintaining a low dUTP/dTTP ratio is important for the production and/or stability of the DNA intermediate required for the viral life cycle (9).
The formation of an AP site initiates the base excision repair (BER) pathway, which uses the undamaged cDNA strand to restore the lost base. During BER, an AP site usually is processed by an AP endonuclease, which cleaves the phosphodiester bond on the 5′ side of the lesion. In yeast, the major AP endonuclease is encoded by APN1 (40), with APN2 specifying a minor activity (27). AP-site repair also can be initiated in an Apn1-independent manner by a glycosylase-associated AP lyase activity, which nicks the sugar phosphate backbone on the 3′ side of the AP site. Three of the five yeast glycosylases, Ntg1, Ntg2, and Ogg1, have such an AP lyase activity, but only Ntg1 and Ntg2 have been implicated in general AP-site repair (37, 46). Following the cleavage of the DNA backbone, blocked ends are removed to allow gap filling by DNA polymerase and the ligation of the remaining nick.
The nucleotide excision repair (NER) pathway removes bulky, helix-distorting lesions from DNA and is best known for its role in repairing UV-induced pyrimidine dimers (15). In humans, defects in NER result in the disease xeroderma pigmentosum, which is characterized by an extreme sensitivity to sunlight and a predisposition to skin cancer. Although NER and BER generally are considered to remove different types of damage, the elimination of both pathways can have synergistic effects on damage sensitivity and genome stability in yeast, suggesting a functional overlap between these pathways (46, 48). Given the specificity of the BER pathway for AP sites, it has been suggested that the functional redundancy of BER and NER reflects a role for NER in AP-site repair. Such a role is supported by the ability of the human and bacterial NER machineries to initiate the repair of AP sites in vitro (26, 33).
There are two major subclasses of NER: global genomic and transcription-coupled NER (GG-NER and TC-NER, respectively; reviewed in reference 23). GG-NER repairs lesions throughout the genome without regard to the transcriptional status of the corresponding DNA. TC-NER specifically repairs lesions in the transcribed strand of active genes, which leads to the faster removal of damage and fewer mutations on the transcribed than on the nontranscribed strand. What distinguishes the two subclasses of NER is how the initiating lesion is recognized; the subsequent repair steps are identical. In GG-NER, a lesion-associated distortion in the DNA helix is recognized directly by dedicated proteins that then recruit downstream NER components. In TC-NER, an RNA polymerase (RNAP) II complex stalled at a lesion signals for the recruitment of the NER machinery. Subsequent steps in NER include localized DNA unwinding around the lesion by a helicase complex, the excision of a lesion-containing oligonucleotide by endonucleases that nick the backbone at defined distances 5′ and 3′ of the lesion, the filling of the resulting gap by DNA polymerase, and the ligation of the remaining nick.
In the current study, we examine the role of NER in the repair of uracil-derived AP sites that lead to the reversion of highly transcribed reporter alleles in yeast. Because uracil specifically replaces thymine under high-transcription conditions, we can infer which strand of the duplex contained the mutagenic AP site in individual revertants. While Apn1 appears to repair AP sites on both strands of the reporters, results suggest that Ntg1/Ngt2 and NER have complementary, strand-specific repair activities. The lyases efficiently repair only AP sites that are on the nontranscribed strand of the reporters, while NER has the reverse specificity, repairing only AP sites located on the transcribed strand. These results provide a novel framework for linking lesions that are substrates for BER-specific glycosylases to the process of TC-NER.
MATERIALS AND METHODS
Strain construction.
All yeast strains were derived from YPH45 (MATα ura3-52 ade2-101oc trp1Δ1). Wild-type (WT) strains containing the his4Δ::pTET-lys2ΔA746 allele in both orientations relative to ARS306 were described previously (30). Strains containing the his4Δ::pTET-lys2-TAA allele in both orientations were derived from his4Δ::pTET-LYS2 strains by two-step allele replacement using BglII-digested pSR982. pSR982 contains an internal portion of the lys2-TAA allele and was constructed by inserting an XbaI/XhoI fragment of pSR835 (1) into XbaI/XhoI-digested pRS306 (42). Mutant derivatives of pTET-lys2 strains were constructed by one-step gene disruption using PCR-generated deletion cassettes containing a selectable marker. The subsequent Cre/loxP-mediated deletion of the marker gene was carried out as appropriate (21, 35). A complete strain list is given in Table S1 in the supplemental material.
Mutation rates and spectra.
Mutation rates were determined using the method of the median, and 95% confidence intervals were calculated as described previously (43). Each rate was based on data obtained from 10 to 24 independent cultures. With the exception of Dut1 overexpression experiments, 1 ml nonselective yeast extract-peptone (YEP) medium supplemented with 2% glycerol plus 2% ethanol (YEPGE) was inoculated with 250,000 cells from an overnight culture grown in the same medium. To suppress transcription from pTET, doxycycline hyclate (Sigma) was added to the growth medium to a concentration of 2 μg/ml. After 3 days of growth at 30°C, cells were washed with H2O and appropriate dilutions were plated either on synthetic, lysine-deficient medium containing 2% dextrose (SCD-Lys) to select Lys+ revertants or on YEP medium supplemented with 2% dextrose to determine the total number of cells in each culture. CAN1 forward mutation rates were determined by plating cells on SCD-Arg medium supplemented with 60 μg/ml l-canavanine sulfate (SCD-Arg+Can; Sigma). In Dut1 overexpression experiments, strains were transformed with either pRS426 (10) or p426-GAL1-DUT1 (30) and selectively plated on SCD-Ura medium. Five to seven colonies were pooled to inoculate a 5-ml SC-Ura culture supplemented with 2% glycerol, 2% ethanol, and 2% galactose (SCGGE-Ura). The following day, multiple 1-ml SCGGE-Ura cultures were started using 500,000 cells of the overnight culture. After 4 days of growth at 30°C, appropriate dilutions were plated on SCD-Ura to determine total cell numbers, on SCD-Ura-Lys to determine the number of Lys+ colonies, or on SCD-Ura-Arg+Can to determine the number of canavanine-resistant colonies in each culture.
To isolate independent Lys+ revertants, individual colonies were used to inoculate 1-ml YEPGE cultures (or SCGGE-Ura cultures in Dut1 experiments). Following 2 to 3 days of growth at 30°C, an appropriate fraction of each culture was plated on SCD-Lys (or SCD-Ura-Lys in Dut1 experiments) medium. A single Lys+ colony from each culture was purified, and genomic DNA was prepared using a 96-well format in microtiter plates (http://jinks-robertsonlab.duhs.duke.edu/protocols/yeast_prep.html). The lys2ΔA746 reversion window was amplified using primers 5′-GCCTCATGATAGTTTTTCTAACAAATACG and 5′-CCCATCACACATACCATCAAATCCAC, and the PCR product was sequenced by the Duke University DNA Analysis Facility using primer 5′-GTAACCGGTGACGATGAT.
RESULTS
We previously used a tetracycline-regulated (pTET) frameshift reversion assay to examine TAM in BER-deficient yeast strains. In the pTET-lys2ΔA746 system, activated transcription selectively increases complex mutations in which the compensatory +1 mutation is accompanied by a nearby base substitution (29). Complex mutations are a hallmark of Polζ-dependent TLS activity, and we have argued that the base substitution marks the site of the underlying DNA damage (25). In an apn1 background, complex mutations were highly enriched at a 6A hot spot (mutations are reported in the context of the DNA strand that has the same sequence as the mRNA), so named because the selected frameshift occurs in a mononucleotide run of six adenines (30) (Fig. 1). Upon the additional elimination of the Ntg1 and Ntg2 lyases, which further disables BER, there was a synergistic increase in the reversion rate and further enrichment of complex mutations at the 6A hot spot. Not only did the 6A hot spot complex mutations require high transcription from pTET but they also were dependent on the presence of Ung1 and Polζ and were reduced upon the overexpression of the yeast dUTPase. These unique genetic requirements indicate that highly activated transcription leads to elevated dUTP incorporation into the transcribed target gene. The subsequent Ung1-mediated excision of misincorporated uracil and the mutagenic bypass of the unrepaired AP site by Polζ elevates the mutation rate, especially that of complex events.
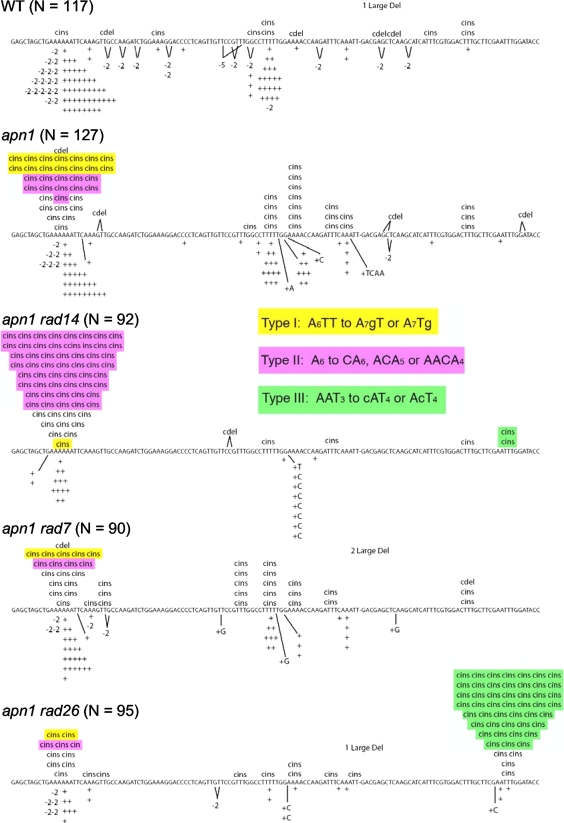
FIG. 1.
pTET-lys2ΔA746 reversion spectra. Only the first 130 nucleotides (nt) of the reversion window are shown, as no mutations were outside of this region. The position of the adenine deleted to create the frameshift allele is indicated by a dash within the sequence. Simple insertions and deletions are indicated below the sequence by a plus sign (1-nt addition), −2, −5, etc.; complex insertions and deletions (cins and cdel, respectively) are indicated above the sequence. Complex mutations at the 6A hot spot associated with a T→G transversion (type I events) are highlighted in yellow, and those associated with an A→C transversion (type II events) are highlighted in pink. Type III events are highlighted in green. N is the number of independent revertants sequenced.
Synergistic increase in TAM in an apn1 rad14 double mutant.
The effect of disrupting both the BER and NER pathwayson the reversion of the highly transcribed pTET-lys2ΔA746 allele was examined in an apn1 rad14 double mutant background. Rad14 is the yeast homolog of the human XPA protein, which is required for early steps in both the GG-NER and TC-NER subclasses of the NER pathway (4). In an apn1 background, the loss of Rad14 is associated with reduced viability and increased mutagenesis upon MMS treatment, leading to the suggestion that NER is important in AP-site processing (48). We previously reported 2.6-fold and 3.4-fold increases in the reversion rates of the pTET-lys2ΔA746 allele in rad14 and apn1 single mutant backgrounds, respectively (29, 30). In an apn1 rad14 double mutant, the reversion rate under high-transcription conditions was increased an astounding 230-fold relative to that of the WT (Table 1). As in the BER-defective (apn1 ntg1 ntg2 triple mutant) background, the increased mutagenesis in an apn1 rad14 double mutant was abrogated when transcription was repressed by the addition of doxycycline (+DOX) to the growth medium or when the gene encoding either Polζ (REV3) or uracil DNA glycosylase (UNG1) was deleted (Table 1). The overexpression of the dUTPase-encoding DUT1 gene, which lowers the cellular dUTP/dTTP ratio, also reduced the reversion rate of the pTET-lys2ΔA746 allele by 30-fold in the apn1 rad14 background (from 7.4 × 10−6 to 0.24 × 10−6), while the forward mutation rate of the moderately transcribed CAN1 gene was reduced only 2-fold (from 2.3 × 10−6 to 1.2 × 10−6). The distinctive genetic requirements of TAM in the apn1 rad14 double mutant indicate that, as documented previously for the apn1 ntg1 ntg2 triple mutant (30), most mutations in the pTET-lys2ΔA746 system arise during the bypass of AP sites generated when Ung1 removes uracil that is selectively incorporated into highly transcribed DNA.
TABLE 1.
Reversion rates of pTET-lys2 alleles under high-transcription conditions
| Genotype | Lys+ ratea (×10−8) for: |
|
|---|---|---|
| pTET-lys2ΔA746 | pTET-lys2-TAA | |
| WT | 4.28b (3.35-6.74) | 2.49 (1.22-3.43) |
| apn1 | 14.5b (12.4-18.1) | 184 (49-197) |
| rad14 | 11.2c (9.15-14.2) | 5.73 (4.29-6.45) |
| apn1 rad14 | 963 (798-1,120) | 1,460 (1,350-1,590) |
| apn1 rad14 rev3 | 6.5 (4.56-8.61) | 1.16 (0.87-2.04) |
| apn1 rad14 ung1 | 17 (15.1-20.7) | 18.3 (11.9-23.4) |
| apn1 rad14 + DOX | 0.66 (0.41-0.88) | 1.70 (1.39-2.44) |
Transcription-associated mutations in an apn1 rad14 mutant are distinctly different from those in an apn1 ntg1 ntg2 mutant.
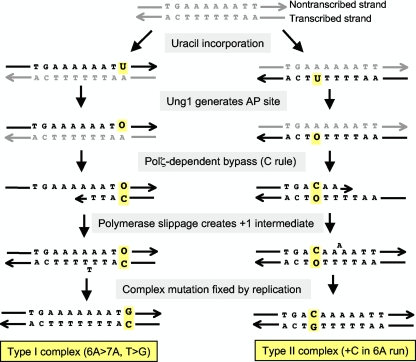
As in the apn1 ntg1 ntg2 triple mutant, there was a striking enrichment of complex mutations at the 6A hot spot in the apn1 rad14 double mutant, with these events comprising more than 60% of the pTET-lys2ΔA746 reversion spectrum (Fig. 1). The patterns of complex mutations, however, were distinctly different in the double and triple mutants (Table 2). In the apn1 ntg1 ntg2 strain, ∼95% of the complex mutations converted a 6A run to a 7A run, and this change was accompanied by a T-to-G transversion immediately downstream of the run (5′-AAAAAATT to AAAAAAAgT or AAAAAAATg) (bases added to the 6A run are underlined, and base substitutions are in lowercase). We will refer to these mutations as type I complex insertions and have argued that they are initiated when one of the two mutated thymines is replaced with uracil (30). The subsequent removal of the uracil by Ung1 generates an AP site that is bypassed by the Polζ-dependent insertion of dCTP (the C rule), followed by slippage to generate the selected +1 frameshift in the 6A run (Fig. 2). In the apn1 rad14 strain, the complex mutations at the 6A run consisted of the insertion of a single cytosine into the 5′ portion of the run (5′-AAAAAA to ACAAAAA, AACAAAA, or AAACAAA). We refer to this second class of mutations as type II insertions. Because type II events are completely dependent on the presence of Polζ, we suggest that they arise through a lesion-initiated misincorporation-slippage mechanism that is similar to that proposed for type I events. As shown in Fig. 2, we postulate that type II complex mutations are initiated when dCTP is inserted opposite an Ung1-generated AP site located within the 6T run that is complementary to the 6A run. AP-site bypass then would be followed by slippage within the remaining portion of the run to produce the requisite +1 frameshift. It should be noted that the C insertion always occurs before one of the first three adenines of the 6A run, the polar pattern that is expected if misincorporation precedes slippage.
TABLE 2.
Complex mutations at the 6A hot spota (5′-AGCTGAAAAAATTC)
| Type | Sequence | No. of events in mutant background: |
||
|---|---|---|---|---|
| apn1 | apn1 ntg1 ntg2 | apn1 rad14 | ||
| I | AGCTGAAAAAAAATgC | 10 | 26 | 0 |
| AGCTGAAAAAAAAgTC | 4 | 16 | 1 | |
| II | AGCTGCAAAAAATTC | 4 | 0 | 20 |
| AGCTGACAAAAATTC | 5 | 0 | 22 | |
| AGCTGAACAAAATTC | 2 | 0 | 10 | |
| Other | AGCTcAAAAAAATTC | 3 | 0 | 2 |
| AGCTaAAAAAAATTC | 2 | 0 | 0 | |
| AGCaGAAAAAAATTC | 1 | 0 | 0 | |
| AGCTGAAAAAAAaTC | 1 | 0 | 0 | |
| cGCTGAAAAAAATTC | 0 | 0 | 3 | |
| AGCcGAAAAAAATTC | 0 | 0 | 2 | |
| AGCTGCAAAAAAaaa | 0 | 0 | 1 | |
| AGCTGAAAAAACTTC | 0 | 0 | 1 | |
| AGCTGAAAAgTC | 1 | 0 | 0 | |
| AGCTGAAAAAAAAAATgC | 0 | 1 | 0 | |
| Total no. of complex events at 6A run | 33 | 43 | 62 | |
| Total no. of revertants sequenced | 127 | 94 | 92 | |
Bases added to the 6A run are underlined, and base substitutions are in lowercase. Spectra for apn1 single and apn1 ntg1 ntg2 triple mutants are from reference 30.
FIG. 2.
Models for generating type I and type II complex mutations in the pTET-lys2DA746 reversion assay. The starting duplex is in gray, and all newly synthesized DNA is in black. Relevant bases are highlighted in yellow. O indicates the AP site. See the text for details.
TAM in a nonsense reversion assay.
In the pTET-lys2ΔA746 system, the majority of the transcription-associated complex mutations in both the apn1 ntg1 ntg2 and apn1 rad14 backgrounds were confined to a single hot spot (the 6A run), making it difficult to generalize the results and exclude sequence-specific effects. We thus repeated analyses using the pTET-lys2-TAA allele, which contains a TAA stop codon (1). The selection of Lys+ revertants in this system allows the detection of any base substitution that destroys the stop codon and removes the demand for a coincident +1 mutation. In a WT background, transcription stimulated the reversion of the pTET-lys2-TAA allele approximately 10-fold (from 2.8 × 10−9 to 2.5 × 10−8), and all seven possible base changes that can revert the stop codon were observed in approximately equal numbers (Table 3).
TABLE 3.
Rates of pTET-lys2-TAA revertant types under high-transcription conditions
| Genotypea (n) | Total Lys+ rate (×10−8) (95% CIc) | Rate of mutation typeb (×10−8) at: |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| Position 1 |
Position 2 |
Position 3 |
Other | ||||||
| T→A | T→C | T→G | A→C | A→T | A→C | A→T | |||
| WT (84) | 2.49 (1.22-3.43) | 0.24 (8/84) | 0.42 (14/84) | 0.18 (6/84) | 0.53 (18/84) | 0.33 (11/84) | 0.21 (7/84) | 0.12 (4/84) | 0.44 (16/84) |
| apn1 (86) | 184 (49-197) | 6.4 (3/86) | 4.3 (2/86) | 21 (10/86) | 140 (66/86) | 2.1 (1/86) | 8.6 (4/86) | — (0/86) | — (0/86) |
| apn1 ntg1 ntg2 (94) | 468 (367-1,150) | 5.0 (1/94) | 15 (3/94) | 390 (78/94) | 55 (11/94) | — (0/94) | 5.0 (1/94) | — (0/94) | — (0/94) |
| apn1 rad14 (85) | 1,460 (1,350-1,590) | — (0/85) | 17 (1/85) | 34 (2/85) | 1,300 (75/85) | — (0/85) | 86 (5/85) | 17 (1/85) | 17 (1/85) |
The numbers in parentheses are the numbers of revertants sequenced.
Rates of mutation types were calculated by multiplying the proportion of the event (in parentheses) by the total Lys+ rate. With the exception of one mutation in the WT background, “others” contained multiple mutations, one of which reverted the stop codon. —, rate not calculated.
CI, confidence interval.
When APN1 was deleted, there was a 74-fold increase in the reversion rate of the highly transcribed pTET-lys2-TAA allele, and this was accompanied by a shift in the proportions of mutation types (Table 3). Most notably, there was an increase in A→C mutations from 30% of the total mutations in the WT to 80% in the apn1 mutant. Although the additional deletion of NTG1 and NTG2 from the apn1 background elevated the pTET-lys2-TAA reversion rate only 2.5-fold, the increase was almost entirely due to the accumulation of T→G mutations (Table 3). Their proportion increased from 12% in the apn1 single mutant to 77% in the apn1 ntg1 ntg2 triple mutant, corresponding to an ∼20-fold rate increase in the rate of this specific class. It should be noted that there was no increase in the reciprocal A→C mutation rate in the apn1 ntg1 ntg2 triple relative to the apn1 single mutant. In the apn1 rad14 background, the reversion rate of the pTET-lys2-TAA allele was 8-fold higher than in the apn1 single mutant background. This rate increase reflects a specific increase in A→C mutations; there was no significant increase in the T→G class (Table 3).
As observed with the pTET-lys2ΔA746 allele, the elevated reversion rates of the pTET-lys2-TAA allele in both the apn1 rad14 double and apn1 ntg1 ntg2 triple mutant backgrounds were dependent on a high level of transcription and on the presence of both Rev3 and Ung1 (Table 1 and data not shown). Because of these genetic dependencies, we suggest that the selective increase in T→G transversions in the apn1 ntg1 ntg2 triple mutant is initiated when uracil replaces thymine at the first position of the TAA stop codon. In a similar manner, the selective increase in A→C mutations in the apn1 rad14 double mutant would reflect the incorporation of uracil in place of one of the thymines opposite the second and third positions of the TAA stop codon. In both cases, the Ung1-dependent removal of uracil incorporated into highly transcribed DNA would create an AP site. The Polζ-dependent bypass of the AP site using the C rule then would generate the observed T→G and A→C transversion patterns, which are precisely the same as those observed among complex mutations in the frameshift reversion assay.
Orientation of the pTET-lys2 reporter does not affect strand specificity of AP-associated mutations.
In both the frameshift and base substitution assays, T→G transversions (type I mutations in the frameshift reversion assay) were selectively enhanced in the apn1 ntg1 ntg2 triple mutant, while only A→C transversions (type II mutations in the frameshift reversion assay) were elevated in the apn1 rad14 double mutant background. These data indicate that the Ntg1/Ntg2 lyase activities efficiently process uracil-derived AP sites that are on one strand of the duplex, while the NER machinery only processes those AP sites that are on the complementary strand. There are two possible sources of this repair bias: (i) the location of the AP site on the leading versus lagging strand of DNA replication, or (ii) the location of the AP site on the transcribed versus nontranscribed strand of the pTET-lys2 alleles.
To determine the source of the AP-site repair bias, we inverted the pTET-lys2 reporter alleles with respect to the nearby ARS306 origin of replication. This reverses the identities of the leading and lagging strands during DNA replication but does not alter the identities of the transcribed and nontranscribed strands (TS and NTS, respectively). As described in our earlier studies (29), the transcription complex and the replication fork move in the same direction in the SAME constructs; these constructs were used to generate the data reported above. In the SAME constructs, the NTS strand serves as the lagging-strand template and the TS strand is the leading-strand template. In the configuration denoted OPPO, where the transcription machinery and replication fork converge, the NTS and TS strands are the leading- and the lagging-strand templates, respectively.
In the apn1 single, apn1 rad14 double, and apn1 ntg1 ntg2 triple mutant OPPO strains, the reversion rates and spectra obtained with the pTET-lys2ΔA746 reporter were indistinguishable from those obtained from the SAME strains of comparable genotype (Table 4 ). Thus, type I complex events, which contain a T→G transversion, were selectively elevated in both the SAME and OPPO apn1 ntg1 ntg2 triple mutants. Conversely, only type II complex insertions, which contain an A→C transversion, were increased in the SAME and OPPO apn1 rad14 double mutants. The mutation rates and base substitution patterns also were similar for the SAME and OPPO strains containing the pTET-lys2-TAA reporter: A→C mutations were much more prevalent than T→G mutations in the apn1 SAME and OPPO constructs, T→G mutations were selectively elevated upon the additional deletion of NTG1 and NTG2 in both construct types, and only A→C mutations were increased in the SAME and OPPO apn1 rad14 double mutants. Based on the very similar behaviors of the SAME and OPPO constructs, we conclude that the direction of replication fork movement, and hence the position of an AP site on the leading versus lagging strand of replication, does not significantly affect the mechanism of AP-site processing.
TABLE 4.
Effect of replication direction on base substitution patterns in highly transcribed lys2 allelesa
| Allele | Genotype | Lys+ rates (×10−8) with SAME allele |
Lys+ rates (×10−8) with OPPO allele |
||||
|---|---|---|---|---|---|---|---|
| Total (95% CI) | T→G (fraction) | A→C (fraction) | Total (95% CI) | T→G (fraction) | A→C (fraction) | ||
| pTET-lys2ΔA746 | apn1 | 14.5 (12.4-18.1) | 1.6 (14/127) | 1.3 (11/127) | 10.1 (8.9-12.1) | 1.0 (14/136) | 0.5 (6/136) |
| apn1 ntg1 ntg2 | 150 (125-192) | 67 (42/94) | — (0/94) | 178 (130-196) | 93 (42/93) | — (0/93) | |
| apn1 rad14 | 963 (798-1,120) | 10 (1/92) | 540 (52/92) | 890 (809-1,030) | — (0/94) | 550 (58/94) | |
| pTET-lys2-TAA | apn1 | 184 (49-197) | 21 (10/86) | 150 (70/86) | 130 (100-283) | — (0/90) | 120 (82/90) |
| apn1 ntg1 ntg2 | 468 (367-1,150) | 390 (78/94) | 55 (11/94) | 422 (226-769) | 190 (43/95) | 200 (45/95) | |
| apn1 rad14 | 1,460 (1,350-1,590) | 34 (2/85) | 1,400 (80/85) | 2,780 (2,160-3,980) | — (0/89) | 2,700 (87/89) | |
T→G and A→C rates were calculated by multiplying the total Lys+ rate by the proportion of the relevant mutation type in the corresponding spectrum. In the frameshift reversion assay, T→G and A→C mutations are characteristic of the type I and type II complex mutations, respectively, at the 6A run. CI, confidence interval. —, rate not calculated.
The GG-NER subpathway does not repair transcription-associated AP sites.
Reversing the direction of DNA replication through the lys2 reporters did not alter reversion rates or spectra, suggesting that the observed A→C and T→G biases reflect a repair-related asymmetry associated with transcription. Of particular significance, the A→C mutation signature in apn1 rad14 mutants reflects the bypass of AP sites that are specifically located on the TS, suggesting that the repair of these sites via NER is limited to the TC-NER subpathway. In the experiments described above, NER was completely disabled by eliminating Rad14, which is required for the formation of an active preincision complex during both GG-NER and TC-NER. To demonstrate that an NER-related asymmetry during AP-site repair reflects TC-NER, it is necessary to individually disrupt the GG-NER and TC-NER subpathways.
Yeast strains defective in Rad7 or Rad16, as well as cells from XP complementation group C and E patients, retain the ability to repair UV-induced pyrimidine dimers on the TS but are defective in the repair of UV dimers on the NTS (44, 50). We thus deleted RAD7 from an apn1 strain containing either the pTET-lys2ΔA746 or pTET-lys2-TAA reporter, creating a situation in which GG-NER is specifically disabled but TC-NER still functions. With both reporters, the reversion rates and spectra in the apn1 rad7 double mutants were indistinguishable from those of the parent apn1 strain, confirming that the loss of GG-NER does not compromise AP-site repair (Table 5). We also examined the effect of deleting RAD7 in an apn1 ntg1 ntg2 background. According to our mutagenesis model, the AP sites underlying A→C base substitutions (type II complex mutations in the pTET-lys2ΔA746 assay) are actively repaired by NER in an apn1 ntg1 ntg2 mutant, leading to the elevation of only T→G transversions (type I complex mutations in the frameshift assay). If AP sites on the TS are recognized and repaired by GG-NER, the deletion of the RAD7 gene in the apn1 ntg1 ntg2 background should result in the elevation of A→C transversions. The mutation rate and spectra in the apn1 ntg1 ntg2 rad7 quadruple mutant, however, were indistinguishable from those in the apn1 ntg1 ntg2 triple mutant (Table 5), providing additional support for a model in which AP sites are targeted for removal only by the transcription-coupled subpathway of NER.
TABLE 5.
Reversion of lys2 alleles in the absence of GG-NER factor Rad7 and TC-NER factor Rad26
| Genotype |
pTET-lys2ΔA746 reversion rate (×10−8) |
pTET-lys2-TAA reversion rate (×10−8) |
|||||
|---|---|---|---|---|---|---|---|
| Total (95% CIa) | Type I, T→G (fraction) | Type II, A→C (fraction) | Type III, A>C (fraction) | Total (95% CI) | T→G (fraction) | A→C (fraction) | |
| apn1 | 14.5 (12.4-18.1) | 1.6 (14/127) | 1.3 (11/127) | —b (0/127) | 184 (49-197) | 21 (10/86) | 150 (70/86) |
| apn1 rad14 | 963 (798-1,120) | 10 (1/92) | 540 (52/92) | 21 (2/92) | 1,460 (1,350-1,590) | 34 (2/85) | 1,400 (8/85) |
| apn1 rad7 | 19.7 (13.9-35.2) | 1.1 (5/90) | 0.9 (4/90) | — (0/90) | 135 (86-282) | 19 (13/91) | 100 (68/91) |
| apn1 rad26 | 49.8 (36.9-70.6) | 1.0 (2/95) | 1.6 (3/95) | 25 (47/95) | 522 (256-546) | 5.9 (1/88) | 490 (83/88) |
| apn1 rad7 rad26 | 42.4 (36.8-59) | 2.0 (4/86) | 2.5 (5/86) | 8.9 (19/86) | 416 (312-507) | 4.4 (1/94) | 400 (90/94) |
| apn1 ntg1 ntg2 | 150 (125-192) | 67 (42/94) | — (0/94) | — (0/94) | 468 (367-1,150) | 390 (78/94) | 55 (11/94) |
| apn1 ntg1 ntg2 rad7 | 81.7 (28.3-130) | 22 (24/91) | — (0/91) | — (0/91) | 440 (197-583) | 370 (118/142) | 59 (19/142) |
| apn1 ntg1 ntg2 rad26 | 195 (156-261) | 68 (33/95) | 6.2 (3/95) | 2.1 (1/95) | 841 (673-917) | 280 (31/92) | 520 (57/92) |
CI, confidence interval.
—, rate not calculated.
AP-derived mutations are elevated in the absence of the TC-NER factor Rad26.
Rad26 is a member of the Swi/Snf family of chromatin remodeling factors and is the yeast homolog of the human CSB protein. CSB initiates the recruitment of the NER machinery to the sites of stalled RNA polymerase complexes (49); in its absence, the preferential repair of UV lesions on the TS of active genes is abolished (reviewed in reference 24). In contrast, the disruption of RAD26 in yeast only partially disables TC-NER, suggesting that there are additional transcription repair coupling factors (TRCFs) that function independently of Rad26 (51). To further examine whether the NER-directed processing of AP sites is limited to the TS, we examined the effect of Rad26 loss on the rates of the A→C and T→G mutations that are diagnostic of AP-site bypass on the TS and NTS, respectively.
In an apn1 single or apn1 rad7 double mutant strain containing the pTET-lys2ΔA746 allele, the further deletion of the RAD26 gene led to only a modest 2- to 3-fold increase in the overall reversion rate (Table 5). There may have been a very slight increase in type II complex mutations, but it was negligible relative to the 400-fold increase observed upon the complete loss of NER (apn1 rad14 double mutant). Although we did not observe the expected increase in type II events, a novel and very strong complex insertion hot spot was evident in the apn1 rad26 double mutant (Fig. 1). Most notably, almost all of the mutations at the new hot spot contained an A→C transversion in addition to the selected +1 frameshift mutation (5′-AATTT was changed to cATTTT or AcTTTT), which is identical to the base substitution pattern associated with type II complex events. We refer to events at the new hot spot as type III complex insertions and suggest that they, like the type II events, are generated during the bypass of an AP site located specifically on the TS. Finally, in an apn1 ntg1 ntg2 background, the deletion of RAD26 elevated neither the overall reversion rate of the pTET-lys2ΔA746 allele nor the rate of the type II complex mutations. In contrast to the dramatic increase in type III events observed in the apn1 rad26 double and apn1 rad7 rad26 triple mutants, these events were only slightly elevated in the apn1 ntg1 ntg2 rad26 quadruple mutant. We currently have no explanation for the rarity of type III events in the quadruple mutant strain.
With the pTET-lys2-TAA allele, the deletion of RAD26 resulted in an increase only in A→C mutations. This was evident upon the deletion of RAD26 in the apn1 single, the apn1 rad7 double, or the apn1 ntg1 ntg2 triple mutant background (Table 5). We note that the increase in A→C mutations in the apn1 rad26 mutant was less than that seen in the apn1 rad14 mutant, which is consistent with only a partial defect in TC-NER upon Rad26 loss. Taken together, the results obtained from the various rad26 mutants support the hypothesis that the NER machinery processes only those AP sites that are located on the TS of active genes. We suggest, however, that the role of Rad26 in TC-NER is very context dependent, with Rad26 being the major TRCF at some positions, such as that defined by type III complex insertions. At other positions, such as the 6A hot spot, we speculate that other candidate TRCFs are more important than or can compensate for the loss of Rad26.
DISCUSSION
We recently used the pTET-lys2ΔA746 frameshift reversion assay to document a novel source of TAM that is specifically linked to increased dUTP incorporation under high-transcription conditions (30). The subsequent release of uracil by the Ung1 glycosylase generates an AP site that, if not repaired, is bypassed in a mutagenic, Polζ-dependent manner. Upon the loss of Apn1, the major AP endonuclease in yeast, there was a striking enrichment of two distinct types of complex frameshift mutations (type I and type II) at a 6A hot spot. In the additional absence of the Ntg1 and Ntg2 lyases, a condition that more severely disables AP-site repair, there was a synergistic increase in type I complex mutations but no apparent increase in type II mutations. The current study expands these analyses in two ways. First, the pTET-lys2ΔA746 frameshift reversion assay was used to examine the role of the yeast NER pathway in preventing TAM due to unrepaired AP sites. The elimination of NER by the deletion of RAD14 in an apn1 background resulted in a synergistic increase in complex mutations at the previously identified 6A hot spot, but these were strikingly limited to type II events. Second, we developed a nonsense reversion assay (the pTET-lys2-TAA allele) and used this system to demonstrate that the distinctive occurrence of T→G versus A→C mutations associated with AP-site bypass in apn1 ntg1 ntg1 triple versus apn1 rad14 double mutants, respectively, is not limited to a specific hot spot or to frameshift-associated complex mutations.
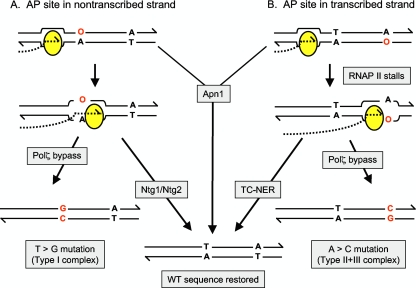
The AP-associated mutagenesis in the highly transcribed pTET-lys2 reporters is the result of dUTP incorporation in place of dTTP, allowing one to infer which strand of the DNA contained the underlying, Ung1-generated AP site. We use the convention of reporting mutations in the context of the DNA strand that has the same sequence as that of the mRNA and hence corresponds to the nontranscribed strand (NTS) of the gene. T→G and A→C mutations thus reflect AP-site bypass on the NTS and TS strands, respectively. With both reporters, T→G and A→C mutations (type I and type II complex events, respectively, in the frameshift assay) were elevated in an apn1 single mutant. In the apn1 ntg1 ntg2 triple mutants, however, only type I frameshifts and T→G base substitutions were elevated. This indicates that the Ntg1 and Ntg2 lyases are functionally redundant with Apn1 for AP-site repair on the NTS, and also that the remaining NER pathway has little, if any, activity against these AP sites. In striking contrast, only those mutations characteristic of AP-site bypass on the TS strand (type II complex mutations and A→C base substitutions in the pTET-lys2ΔA746 and pTET-lys2-TAA assays, respectively) were elevated in an apn1 rad14 background. This demonstrates that the NER pathway is able to efficiently repair AP sites when they are on the TS strand and, conversely, that the Ntg1/Ntg2 lyases have little activity against these AP sites. As illustrated in Fig. 3, these analyses suggest a strict division of labor between the Ntg1/Ntg2 lyases and the NER pathway with regard to repairing AP sites on the NTS and TS strands, respectively, of actively transcribed DNA.
FIG. 3.
Model for the differential repair of AP sites on the TS and NTS of active genes. See the text for details.
Although the NER pathway generally is thought to repair only bulky, helix-distorting lesions, several genetic studies have implicated the yeast NER machinery in the repair of AP sites (46, 48). The results presented here provide insight into how AP sites engage the NER machinery and how, through an AP intermediate, the BER pathway can be linked to TC-NER. Our results indicate that AP sites normally are not detected by the damage recognition proteins/complexes that initiate NER and instead trigger the pathway via their ability to block RNAP II. It should be noted that an AP site is indeed a potent block to mammalian RNAP II in vitro (47). This model makes the specific prediction that the disruption of the GG-NER pathway should have no effect on AP-site repair, while the loss of TC-NER should be equivalent to completely disabling NER. Consistently with this prediction, we observed that the loss of Rad7, which is required for GG-NER in yeast, did not elevate the mutagenesis associated with Ung1-derived AP sites in either of the pTET-lys2 reporters. Although it is not possible to do the complementary experiment and completely disable TC-NER in yeast, the efficiency of this subpathway can be reduced by the deletion of the RAD26 gene (51). The deletion of RAD26 in an apn1 background led to a small but significant increase in the reversion rate of both pTET-lys2 reporters. The observation that reversion rates were elevated to a lesser extent in the apn1 rad26 than in the apn1 rad14 strains is consistent with Rad26 being only one of several factors that can mediate TC-NER in yeast. Additional TRCFs that might be relevant in this system include the RNAP II subunit Rpb9 (32), transcription elongation factors (17), and factors involved with mRNP biogenesis and export (18). Def1, which forms a complex with Rad26, also may provide an alternative mechanism for TC-NER by promoting the ubiquitination and degradation of a stalled RNAP II complex (45). The pTET-lys2 reversion assays described here will be useful for clarifying the in vivo contributions of these factors to the efficient repair of lesions on the transcribed strand of active genes.
In the base substitution assay, the slight reversion rate increase in the apn1 rad26 double mutant relative to that of the apn1 single mutant was accompanied by the predicted enrichment of A→C mutations relative to that of T→G mutations (TS and NTS mutations, respectively). In the case of the frameshift reversion assay, a differential effect on type I versus type II complex mutations was difficult to assess, because the spectrum in the apn1 rad26 double mutant was dominated by complex mutations at a novel hot spot (type III mutations). Like type II mutations, however, the type III mutations contained an A→C mutation and hence can be explained by the Polζ-dependent bypass of a persistent AP site on the TS. The appearance of the novel type III events was unexpected and indicates that the relative roles of specific TRCFs in TC-NER are likely to be context dependent in yeast. Finally, our observations provide novel insight into how lesions that normally are substrates for the BER pathway also can engage TC-NER, a connection that has been tenuous and highly controversial (see reference 24). While a lesion such as 8-oxoguanine may be able to directly pause or block RNAP in some sequence contexts, it presents no impediment and can be a source of transcriptional mutagenesis in other contexts (7, 8). The data reported here demonstrate that uracil in DNA, which would not be expected to impede RNA polymerase under any circumstances, nevertheless can trigger TC-NER in an Ung1-dependent manner. While we make the simplifying assumption that the resulting AP site is directly responsible for the observed TC-NER, a protein-bound AP site may be the relevant block in some or all cases. We suggest that the transcription-coupled repair of glycosylase-generated AP sites is likely a conserved mechanism that links BER to TC-NER in a wide variety of systems (7, 28).
The demonstration that the repair of AP sites via the NER pathway is limited to the TS of active genes is not totally unexpected (see reference 47); a more unanticipated result is the apparent limitation of the lyase activities of Ntg1 and Ntg2 to the NTS of the pTET-lys2 reporters. One can think about this NTS-associated specificity in two ways: either a stalled RNAP II complex sterically prevents the access of these lyases to AP sites on the TS, or the lyases are able to efficiently access AP sites only within a transient-transcription bubble or single-stranded region (e.g., the displaced strand in an R-loop) on the NTS. The former scenario makes the prediction that Ntg1/Ntg2 should more efficiently repair AP sites in nontranscribed DNA; in the latter case, only AP sites in transcriptionally active DNA should be efficient substrates for Ntg1/Ntg2. With regard to the possibility that Ntg1/Ntg2 has a preference for nonduplex DNA, it has been reported that the glycosylase activities of mammalian NEIL1, NEIL2, and NEIL3 are most active against some lesions when they are in single-stranded DNA or bubbles (12, 34). With respect to the physical inhibition of repair by a stalled RNAP II complex, it generally is assumed that the complex needs to be either backtracked away from the lesion or removed/degraded for the appropriate repair factors to gain access to the lesion during TC-NER (45). While yeast appears to utilize both mechanisms, it is interesting that mammalian cells seem to rely only on CSB-dependent backtracking and bacterial cells only on the Mfd-dependent removal of RNA polymerase (24).
In summary, the data reported here demonstrate that the NER pathway functions as an alternative mechanism to the Apn1-initiated repair of the Ung1-generated AP sites that accumulate in highly transcribed DNA. Importantly, mutagenesis patterns indicate that only those AP sites that are on the TS of an active gene are repaired by NER in yeast. We suggest that the observed TC-NER of AP sites, which are obligate intermediates during BER, is a general mechanism for linking the repair of nondistorting base damages, which do not themselves block RNAP II, to the process of TC-NER. Although we previously reported that the lyase activities of the Ntg1 and Ntg2 glycosylases likewise provide an alternative to Apn1 for AP-site processing, the data presented here demonstrate that these proteins mediate the efficient repair of only those AP sites that are on the NTS of a highly transcribed gene. The mechanism of how this novel strand bias is achieved awaits further investigation. Finally, the strand-related repair biases reported here may be relevant to the strand-related mutational biases that have been linked to gene expression levels in mammalian germ cells (36) and cancer genomes (41).
Supplementary Material
Acknowledgments
This work was supported by grant GM038464 from the National Institutes of Health.
We thank Phil Hanawalt, Graciela Spiva, Tom Petes, and Dennis Grogan for insightful comments on the manuscript and members of the S.J.R. laboratory for discussions throughout the course of this work.
Footnotes
Published ahead of print on 26 April 2010.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Abdulovic, A. L., B. K. Minesinger, and S. Jinks-Robertson. 2008. The effect of sequence context on spontaneous Polζ-dependent mutagenesis in Saccharomyces cerevisiae. Nucleic Acids Res. 36:2082-2093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aguilera, A. 2002. The connection between transcription and genomic instability. EMBO J. 21:195-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aguilera, A., and B. Gomez-Gonzalez. 2008. Genome instability: a mechanistic view of its causes and consequences. Nat. Rev. Genet. 9:204-217. [DOI] [PubMed] [Google Scholar]
- 4.Bankmann, M., L. Prakash, and S. Prakash. 1992. Yeast RAD14 and human xeroderma pigmentosum group A DNA-repair genes encode homologous proteins. Nature 355:555-558. [DOI] [PubMed] [Google Scholar]
- 5.Berger, S. H., D. L. Pittman, and M. D. Wyatt. 2008. Uracil in DNA: consequences for carcinogenesis and chemotherapy. Biochem. Pharmacol. 76:697-706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boiteux, S., and M. Guillet. 2004. Abasic sites in DNA: repair and biological consequences in Saccharomyces cerevisiae. DNA Repair 3:1-12. [DOI] [PubMed] [Google Scholar]
- 7.Brégeon, D., Z. A. Doddridge, H. J. You, B. Weiss, and P. W. Doetsch. 2003. Transcriptional mutagenesis induced by uracil and 8-oxoguanine in Escherichia coli. Mol. Cell 12:959-970. [DOI] [PubMed] [Google Scholar]
- 8.Brégeon, D., P. A. Peignon, and A. Sarasin. 2009. Transcriptional mutagenesis induced by 8-oxoguanine in mammalian cells. PLoS Genet. 5:e1000577. [DOI] [PMC free article] [PubMed]
- 9.Chen, R., H. Wang, and L. M. Mansky. 2002. Roles of uracil-DNA glycosylase and dUTPase in virus replication. J. Gen. Virol. 83:2339-2345. [DOI] [PubMed] [Google Scholar]
- 10.Christianson, T. W., R. S. Sikorski, M. Dante, J. H. Shero, and P. Hieter. 1992. Multifunctional yeast high-copy number vectors. Gene 110:119-122. [DOI] [PubMed] [Google Scholar]
- 11.Datta, A., and S. Jinks-Robertson. 1995. Association of increased spontaneous mutation rates with high levels of transcription in yeast. Science 268:1616-1619. [DOI] [PubMed] [Google Scholar]
- 12.Dou, H., S. Mitra, and T. K. Hazra. 2003. Repair of oxidized bases in DNA bubble structures by human DNA glycosylases NEIL1 and NEIL2. J. Biol. Chem. 278:49679-49684. [DOI] [PubMed] [Google Scholar]
- 13.el-Hajj, H. H., H. Zhang, and B. Weiss. 1988. Lethality of a dut (deoxyuridine triphosphatase) mutation in Escherichia coli. J. Bacteriol. 170:1069-1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Friedberg, E. C. 2005. Suffering in silence: the tolerance of DNA damage. Nat. Rev. Mol. Cell Biol. 6:943-953. [DOI] [PubMed] [Google Scholar]
- 15.Friedberg, E. C., G. C. Walker, W. Siede, R. D. Wood, R. A. Schultz, and T. Ellenberger. 2006. DNA repair and mutagenesis, 2nd ed. ASM Press, Washington, DC.
- 16.Gadsden, M. H., E. M. McIntosh, J. C. Game, P. J. Wilson, and R. H. Haynes. 1993. dUTP pyrophosphatase is an essential enzyme in Saccharomyces cerevisiae. EMBO J. 12:4425-4431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gaillard, H., C. Tous, J. Botet, C. Gonzalez-Aguilera, M. J. Quintero, L. Viladevall, M. L. Garcia-Rubio, A. Rodriguez-Gil, A. Marin, J. Arino, J. L. Revuelta, S. Chavez, and A. Aguilera. 2009. Genome-wide analysis of factors affecting transcription elongation and DNA repair: a new role for PAF and Ccr4-not in transcription-coupled repair. PLoS Genet. 5:e1000364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gaillard, H., R. E. Wellinger, and A. Aguilera. 2007. A new connection of mRNP biogenesis and export with transcription-coupled repair. Nucleic Acids Res. 35:3893-3906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.García-Rubio, M., P. Huertas, S. Gonzalez-Barrera, and A. Aguilera. 2003. Recombinogenic effects of DNA-damaging agents are synergistically increased by transcription in Saccharomyces cerevisiae: new insights into transcription-associated recombination. Genetics 165:457-466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gibbs, P. E., J. McDonald, R. Woodgate, and C. W. Lawrence. 2005. The relative roles in vivo of Saccharomyces cerevisiae Polη, Polζ, Rev1 protein and Pol32 in the bypass and mutation induction of an abasic site, T-T (6-4) photoadduct and T-T cis-syn cyclobutane dimer. Genetics 169:575-582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gueldener, U., J. Heinisch, G. J. Koehler, D. Voss, and J. H. Hegemann. 2002. A second set of loxP marker cassettes for Cre-mediated multiple gene knockouts in budding yeast. Nucleic Acids Res. 30:e23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guillet, M., and S. Boiteux. 2003. Origin of endogenous DNA abasic sites in Saccharomyces cerevisiae. Mol. Cell. Biol. 23:8386-8394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hanawalt, P. C. 2002. Subpathways of nucleotide excision repair and their regulation. Oncogene 21:8949-8956. [DOI] [PubMed] [Google Scholar]
- 24.Hanawalt, P. C., and G. Spivak. 2008. Transcription-coupled DNA repair: two decades of progress and surprises. Nat. Rev. Mol. Cell Biol. 9:958-970. [DOI] [PubMed] [Google Scholar]
- 25.Harfe, B. D., and S. Jinks-Robertson. 2000. DNA polymerase ζ introduces multiple mutations when bypassing spontaneous DNA damage in Saccharomyces cerevisiae. Mol. Cell 6:1491-1499. [DOI] [PubMed] [Google Scholar]
- 26.Huang, J. C., D. S. Hsu, A. Kazantsev, and A. Sancar. 1994. Substrate spectrum of human excinuclease: repair of abasic sites, methylated bases, mismatches, and bulky adducts. Proc. Natl. Acad. Sci. U. S. A. 91:12213-12217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Johnson, R. E., C. A. Torres-Ramos, T. Izumi, S. Mitra, S. Prakash, and L. Prakash. 1998. Identification of APN2, the Saccharomyces cerevisiae homolog of the major human AP endonuclease HAP1, and its role in the repair of abasic sites. Genes Dev. 12:3137-3143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Khobta, A., N. Kitsera, B. Speckmann, and B. Epe. 2009. 8-Oxoguanine DNA glycosylase (Ogg1) causes a transcriptional inactivation of damaged DNA in the absence of functional Cockayne syndrome B (Csb) protein. DNA Repair 8:309-317. [DOI] [PubMed] [Google Scholar]
- 29.Kim, N., A. L. Abdulovic, R. Gealy, M. J. Lippert, and S. Jinks-Robertson. 2007. Transcription-associated mutagenesis in yeast is directly proportional to the level of gene expression and influenced by the direction of DNA replication. DNA Repair 6:1285-1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim, N., and S. Jinks-Robertson. 2009. dUTP incorporation into genomic DNA is linked to transcription in yeast. Nature 459:1150-1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kow, Y. W., G. Bao, B. Minesinger, S. Jinks-Robertson, W. Siede, Y. L. Jiang, and M. M. Greenberg. 2005. Mutagenic effects of abasic and oxidized abasic lesions in Saccharomyces cerevisiae. Nucleic Acids Res. 33:6196-6202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li, S., and M. J. Smerdon. 2004. Dissecting transcription-coupled and global genomic repair in the chromatin of yeast GAL1-10 genes. J. Biol. Chem. 279:14418-14426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lin, J. J., and A. Sancar. 1989. A new mechanism for repairing oxidative damage to DNA: (A)BC excinuclease removes AP sites and thymine glycols from DNA. Biochemistry 28:7979-7984. [DOI] [PubMed] [Google Scholar]
- 34.Liu, M., V. Bandaru, J. P. Bond, P. Jaruga, X. Zhao, P. P. Christov, C. J. Burrows, C. J. Rizzo, M. Dizdaroglu, and S. S. Wallace. 2010. The mouse ortholog of NEIL3 is a functional DNA glycosylase in vitro and in vivo. Proc. Natl. Acad. Sci. U. S. A. 107:4925-4930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Longtine, M. S., A. McKenzie III, D. J. Demarini, N. G. Shah, A. Wach, A. Brachat, P. Philippsen, and J. R. Pringle. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14:953-961. [DOI] [PubMed] [Google Scholar]
- 36.Majewski, J. 2003. Dependence of mutational asymmetry on gene-expression levels in the human genome. Am. J. Hum. Genet. 73:688-692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Meadows, K. L., B. Song, and P. W. Doetsch. 2003. Characterization of AP lyase activities of Saccharomyces cerevisiae Ntg1p and Ntg2p: implications for biological function. Nucleic Acids Res. 31:5560-5567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Morey, N. J., C. N. Greene, and S. Jinks-Robertson. 2000. Genetic analysis of transcription-associated mutation in Saccharomyces cerevisiae. Genetics 154:109-120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Otsuka, C., S. Sanadai, Y. Hata, H. Okuto, V. N. Noskov, D. Loakes, and K. Negishi. 2002. Difference between deoxyribose- and tetrahydrofuran-type abasic sites in the in vivo mutagenic responses in yeast. Nucleic Acids Res. 30:5129-5135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Popoff, S. C., A. S. Spira, A. W. Johnson, and B. Demple. 1990. The yeast structural gene (APN1) for the major apurinic endonuclease: homology to E. coli endonucelase IV. Proc. Natl. Acad. Sci. U. S. A. 87:4193-4197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rubin, A. F., and P. Green. 2009. Mutation patterns in cancer genomes. Proc. Natl. Acad. Sci. U. S. A. 106:21766-21770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sikorski, R. S., and P. Hieter. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122:19-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Spell, R. M., and S. Jinks-Robertson. 2004. Determination of mitotic recombination rates by fluctuation analysis in Saccharomyces cerevisiae, p. 3-12. In A. S. Waldman (ed.), Genetic recombination: reviews and protocols, vol. 262. Humana Press, Totowa, NJ. [DOI] [PubMed] [Google Scholar]
- 44.Sugasawa, K. 2009. UV-DDB: a molecular machine linking DNA repair with ubiquitination. DNA Repair 8:969-972. [DOI] [PubMed] [Google Scholar]
- 45.Svejstrup, J. Q. 2007. Contending with transcriptional arrest during RNAPII transcript elongation. Trends Biochem. Sci. 32:165-171. [DOI] [PubMed] [Google Scholar]
- 46.Swanson, R. L., N. J. Morey, P. W. Doetsch, and S. Jinks-Robertson. 1999. Overlapping specificities of base excision repair, nucleotide excision repair, recombination, and translesion synthesis pathways for DNA base damage in Saccharomyces cerevisiae. Mol. Cell. Biol. 19:2929-2935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tornaletti, S., L. S. Maeda, and P. C. Hanawalt. 2006. Transcription arrest at an abasic site in the transcribed strand of template DNA. Chem. Res. Toxicol. 19:1215-1220. [DOI] [PubMed] [Google Scholar]
- 48.Torres-Ramos, C. A., R. E. Johnson, L. Prakash, and S. Prakash. 2000. Evidence for the involvement of nucleotide excision repair in the removal of abasic sites in yeast. Mol. Cell. Biol. 20:3522-3528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Van Gool, A. J., R. Verhage, S. M. A. Swagemakers, P. van de Putte, J. Brouwer, C. Toelstra, D. Bootsma, and J. H. J. Hoeijmakers. 1994. RAD26, the functional S. cerevisiae homolog of the Cockayne syndrome B gene ERCC6. EMBO J. 13:5361-5369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Verhage, R., A.-M. Zeeman, N. de Groot, F. Gleig, D. D. Bang, P. van de Putte, and J. Brouwer. 1994. The RAD7 and RAD16 genes, which are essential for pyrimidine dimer removal from the silent mating type loci, are also required for repair of the nontranscribed strand of an active gene in Saccharomyces cerevisiae. Mol. Cell. Biol. 14:6135-6142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Verhage, R. A., A. J. van Gool, N. de Groot, J. H. Hoeijmakers, P. van de Putte, and J. Brouwer. 1996. Double mutants of Saccharomyces cerevisiae with alterations in global genome and transcription-coupled repair. Mol. Cell. Biol. 16:496-502. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.