Abstract
In response to stress, cells can utilize several cellular processes, such as autophagy, which is a bulk-lysosomal degradation pathway, to mitigate damages and increase the chances of cell survival. Deregulation of autophagy causes upregulation of p62 and the formation of p62-containing aggregates, which are associated with neurodegenerative diseases and cancer. The Nrf2-Keap1 pathway functions as a critical regulator of the cell's defense mechanism against oxidative stress by controlling the expression of many cellular protective proteins. Under basal conditions, Nrf2 is ubiquitinated by the Keap1-Cul3-E3 ubiquitin ligase complex and targeted to the 26S proteasome for degradation. Upon induction, the activity of the E3 ubiquitin ligase is inhibited through the modification of cysteine residues in Keap1, resulting in the stabilization and activation of Nrf2. In this current study, we identified the direct interaction between p62 and Keap1 and the residues required for the interaction have been mapped to 349-DPSTGE-354 in p62 and three arginines in the Kelch domain of Keap1. Accumulation of endogenous p62 or ectopic expression of p62 sequesters Keap1 into aggregates, resulting in the inhibition of Keap1-mediated Nrf2 ubiquitination and its subsequent degradation by the proteasome. In contrast, overexpression of mutated p62, which loses its ability to interact with Keap1, had no effect on Nrf2 stability, demonstrating that p62-mediated Nrf2 upregulation is Keap1 dependent. These findings demonstrate that autophagy deficiency activates the Nrf2 pathway in a noncanonical cysteine-independent mechanism.
p62, also known as sequestosome 1 (SQSTM-1) and A170, has emerged as a multifaceted adaptor protein that exhibits diverse biological functions through interacting with numerous proteins (16). It was originally characterized as an interacting partner of atypical protein kinase C (aPKC), which is mediated through the Phox and Bem1p (PB1) domain, a domain conserved on the N terminus of p62 that can cause p62 to either homodimerize or heterodimerize with the PB1 domain of other proteins (6). Other domains encoded in p62 are the ZZ-type zinc finger domain that binds receptor-interacting protein-1 (RIP1) and the TRAF6 binding site (TBS), which both have been shown to activate the NF-κB signaling pathway (21). At the C terminus of p62 there is a ubiquitin-associated domain (UBA), which has been shown to bind monoubiquitin- or polyubiquitin-conjugated proteins in a noncovalent manner (6). The PB1 domain of p62 also interacts with the S5a and Rpt1 subunits of the 26S proteasome, which supports its function as a shuttling protein that brings ubiquitinated proteins to the proteasome to be degraded (20).
p62 is also found in protein aggregates that are positive for both ubiquitin and microtubule-associated protein 1 light chain 3 (LC3), a well-characterized marker of a cellular process known as autophagy. Subsequent experiments demonstrated the direct interaction of p62 and LC3 through a 22-amino acid sequence known as the LC3-interacting region (LIR) (6). Therefore, p62 is thought to be the link between polyubiquitinated proteins and autophagy (6). Autophagy, which means “self-eating,” is a ubiquitous process that occurs in all eukaryotic cells. It is a complex catabolic process in which double-membrane vesicles (autophagosomes) engulf large intracellular components, such as organelles or proteins, and then fuse with lysosomes to degrade its contents (25). Autophagy plays a major role in maintaining cellular homeostasis during nutrient deprivation and in eliminating damaged proteins and organelles that accumulate during stress (25). It has been shown that allelic loss of the essential autophagy gene encoding Beclin1 not only is frequently found in ovarian, prostate, and breast cancer but also renders mice prone to hepatocellular carcinoma, lung adenocarcinoma, mammary hyperplasia, and lymphoma (19, 24, 26). Additionally, mice with a deficiency in autophagy have tissues with signs of metabolic impairment, accumulate abnormal organelles and lipid droplets, and have both p62- and polyubiquitin-containing protein aggregates (24). Protein accumulation, particularly of p62, has been shown to be cytotoxic, which can cause cell death, neurodegeneration, and inflammation (8, 24). Recently, it has been shown that excessive accumulation of p62 due to a defect in autophagy causes an increase in oxidative stress, which can potentially lead to deregulation of several signal transduction pathways and altered gene expression (12). However, the pathways that are affected by p62 accumulation and the physiological significance of these effects have yet to be fully elucidated.
One of the main cellular defense mechanisms is the Nrf2-Keap1 signaling pathway. Nrf2 is a critical transcription factor that neutralizes reactive oxygen species (ROS) to restore cellular redox balance (27). It regulates the antioxidant response by activating a battery of genes bearing an antioxidant response element (ARE), including antioxidant enzymes, phase II detoxifying enzymes, xenobiotic transporters, and other stress response proteins (1, 4, 5, 9, 11, 15, 17). The Nrf2 signaling pathway is negatively regulated by Keap1, a substrate adaptor protein of a Cullin 3 (Cul3)-Rbx1-E3 ubiquitin ligase complex that is responsible for the ubiquitination and proteasomal degradation of Nrf2 (3, 13, 27, 29). Under basal conditions, Nrf2 levels remain relatively low; however, a plethora of inducers, which include phytoantioxidants and oxidative stress conditions, can induce Nrf2 protein levels and activate the pathway (9, 27). Upon activation, the activity of the ligase is inhibited through modification of cysteine residues in Keap1, resulting in stabilization of Nrf2 and the activation of Nrf2-dependent cytoprotective genes (27, 28).
Here, we report that upregulation of endogenous p62 by autophagy deficiency, or ectopic expression of p62, sequesters Keap1 into aggregates through direct interaction between these two proteins. Sequestration of Keap1 into aggregates results in a decrease in Nrf2 ubiquitination, an increase in Nrf2 stability, and the activation of ARE-bearing genes. This newly identified mechanism of Nrf2 upregulation by autophagy deficiency adds another dimension to the complex regulation of the Nrf2-Keap1 pathway. This noncanonical mechanism of Nrf2 activation may explain the previously reported cysteine-independent activation of Nrf2 by certain Nrf2 inducers (7, 23).
MATERIALS AND METHODS
Recombinant DNA.
p62 was initially purchased from Open Biosystems and was cloned into the pcDNA3.1 expression vector (Invitrogen) using standard recombinant DNA technology. The p62 and Keap1 mutants were generated by site-directed mutagenesis using the PCR and DpnI method that has previously been described (28). Plasmids for hemagglutinin (HA)-Ub, Keap1, Keap1-chitin binding domain (CBD), and Nrf2 and generation of the human NQO1-ARE TATA-Inr luciferase reporter plasmid have also been described previously (28). The glutathione S-transferase (GST) fusion proteins were expressed in Escherichia coli and purified using GST beads (Amersham Biosciences). The 35S-labeled proteins were generated using an in vitro transcription and translation kit according to the manufacturer's instructions (Promega). To construct the fluorescently tagged proteins for live-cell imaging, p62-WT, p62-M, Keap1, Nrf2, and Cul3 were cloned into the cyan fluorescent protein (CFP), yellow fluorescent protein (YFP), or red fluorescent protein (RFP) vector, each a generous gift from Alexander Sorkin. The following restriction enzyme cutting sites were utilized to generate the fluorescently tagged proteins: EcoRI/XhoI (p62-CFP and p62M-CFP), XhoI/BamHI (Keap1-YFP, Nrf2-YFP, and Nrf2-RFP), and XhoI/KpnI (Cul3-RFP).
Cell culture, transfection, and chemicals.
All cell culture dishes and coverslips used for HEK293 cells were coated with 0.1 mg/ml poly-d-lysine (Sigma). MDA-MB-231 and HEK293T cells were purchased from the American Type Culture Collection (ATCC). The Keap1−/− mouse embryonic fibroblast (MEF) cells were a generous gift from Masayuki Yamamoto. The Atg5+/+, Atg5−/−, Beclin1+/+, and Beclin1+/− immortalized baby mouse kidney (iBMK) cells were a generous gift from Eileen White. Cells were maintained in either Eagle's minimal essential medium (MEM) or Dulbecco's modified Eagle's medium (DMEM) (Invitrogen) in the presence of 10% fetal bovine serum (FBS), 1% glutamine, and 0.1% gentamicin. All cells were incubated at 37°C in a humidified incubator containing 5% CO2. Transfection of cDNA was performed using either Lipofectamine Plus (Invitrogen) or TurboFect (Fermentas), according to the manufacturer's instructions.
Immunoprecipitation and antibodies.
For immunoprecipitation, cell lysates were collected at 48 h posttransfection in radio immunoprecipitation assay (RIPA) buffer containing 10 mM sodium phosphate (pH 8.0), 150 mM NaCl, 1% Triton X-100, 1% sodium deoxycholate, 0.1% sodium dodecyl sulfate (SDS). A total of 1 mM dithiothreitol (DTT), 1 mM phenylmethylsulfonyl fluoride (PMSF), and a protease inhibitor cocktail (PIC) (Sigma) were also added to the RIPA buffer. Cell lysates were incubated with chitin beads (New England Biolabs), myc beads (Santa Cruz Biotechnology), or 1 μg of antibody with 10 μl of protein A-agarose beads on a rotator at 4°C overnight. To detect protein expression in the total cell lysates, 10 μl of the cell lysates in RIPA buffer was mixed with 10 μl of 2× sample buffer (50 mM Tris-HCl [pH 6.8], 2% SDS, 10% glycerol, 100 mM DTT, 0.1% bromophenol blue) and boiled for 5 min. The immunoprecipitated complexes were washed with RIPA buffer plus DTT plus PMSF plus PIC three times and eluted in sample buffer by boiling for 5 min. Samples were then resolved by SDS-polyacrylamide gel electrophoresis (PAGE) and transferred onto nitrocellulose membranes for immunoblot analysis. The antibody against the chitin-binding domain (CBD) was purchased from New England Biolabs. Additionally, antibodies against HA, Nrf2, Keap1, myc, p62, NQO1, and β-actin and both mouse and rabbit antibody-horseradish peroxidase (HRP) were all purchased from a commercial source (Santa Cruz Biotechnology).
Luciferase reporter gene assay.
For the dual-luciferase reporter gene assay, HEK293 cells were transfected with the NQO1 ARE-luciferase plasmid along with the Renilla luciferase expression plasmid pGL4.74 (hRluc/TK) (Promega) and different amounts of either wild-type p62 (p62-WT) or mutant p62 (p62-M) cDNA. As a positive control, cells were treated with a known inducer of Nrf2, tert-butylhydroquinone (tBHQ) (Sigma) or sulforaphane (SF) (Sigma). At 48 h posttransfection, the transfected cells were lysed with passive lysis buffer (Promega) and both firefly and Renilla luciferase activities were measured with the dual-luciferase reporter assay system purchased from Promega. Firefly luciferase activity was normalized to Renilla luciferase activity. The experiment was carried out in triplicate and expressed as the mean ± the standard deviation (SD).
mRNA extraction and real-time qRT-PCR.
Total mRNA was extracted using TRIzol (Invitrogen) according to the manufacturer's instructions. Using equal amounts of mRNA and the Transcriptor First Strand cDNA synthesis kit (Roche), cDNA was generated and used for real-time quantitative reverse transcription-PCR (qRT-PCR). The following TaqMan probes were obtained from the universal probe library (Roche): human Nrf2 (hNrf2), no. 70; hKeap1, no. 10; hHO-1, no. 25; hNQO1, no. 87; hGCLM, no. 18; hMRP2, no. 25; and hGAPDH (glyceraldehyde-3-phosphate dehydrogenase), no. 25. Both the forward and reverse primers for hNrf2, hKeap1, hHO-1, hNQO1, hGCLM, hMRP2, and hGAPDH were synthesized by Integrated DNA Technologies, and the sequences are as follows: hNrf2, ACACGGTCCACAGCTCATC (forward) and TGTCAATCAAATCCATGTCCTG (reverse); hKeap1, ATTGGCTGTGTGGAGTTGC (forward) and CAGGTTGAAGAACTCCTCTTGC (reverse); hHO-1, AACTTTCAGAAGGGCCAGGT (forward) and CTGGGCTCTCCTTGTTGC (reverse); hNQO1, ATGTATGACAAAGGACCCTTCC (forward) and TCCCTTGCAGAGAGTACATGG (reverse); hGCLM, GACAAAACACAGTTGGAACAGC (forward) and CAGTCAAATCTGGTGGCATC (reverse); hMRP2, TGAGCATGCTTCCCATGAT (forward) and CTTCTCTAGCCGCTCTGTGG (reverse); and hGAPDH, CTGACTTCAACAGCGACACC (forward) and TGCTGTAGCCAAATTCGTTGT (reverse).
The real-time PCR was performed as followed: one cycle of predenaturation (95°C for 5 min), 45 cycles of amplification (95°C for 10 s and 60°C for 20 s), and a cooling program of 50°C for 30 s. Reactions for each sample were done in triplicate, and the experiment was repeated three times. The data were expressed as relative mRNA levels and were normalized to GAPDH.
Ubiquitination assay.
HEK293 cells transfected with expression plasmids for HA-Ub and the indicated proteins for 48 h were treated for 4 h with 10 μM MG132 (Sigma) to block proteasome degradation. Cells were then lysed in a buffer containing 2% SDS, 150 mM NaCl, 10 mM Tris-HCl and 1 mM DTT. The cell lysates were boiled immediately for 10 min to inactivate cellular ubiquitin hydrolases to preserve ubiquitin-protein conjugates. The heated lysates were then cooled and diluted five times with a Tris-buffered salt (TBS) solution without SDS and used for immunoprecipitation with an antibody against Nrf2 (C-20) or Keap1. Immunoprecipitated proteins were subjected to immunoblot analysis with an antibody against HA.
Protein half-life analysis.
HEK293 cells transfected with either p62-WT or p62-M for 24 h were treated with 25 μM cycloheximide (CHX) for the indicated time points. Endogenous Nrf2 was detected by immunoblot analysis. The relative intensities of the Nrf2 bands were quantified using the ChemiDoc CRS gel documentation system with the Quantity One software (Bio-Rad). The data were then plotted on a log graph, with the amount of Nrf2 before CHX treatment set as 1.
Fluorescently tagged proteins and immunofluorescence.
Cells were grown on 35-mm glass-bottom dishes (In Vivo Scientific) for live-cell imaging. Cells were transfected with the indicated fluorescently labeled proteins. Twenty-four h posttransfection, cells were gently washed once with 1× phosphate-buffered saline (PBS), and phenol red-free DMEM supplemented with 10% FBS was added.
For indirect immunofluorescence, cells were grown on glass coverslips (Fisher Scientific) in 35-mm dishes (BD Biosciences). Cells were fixed in prechilled methanol. Colocalization of stably integrated p62 or endogenous p62 and Keap1 was detected using double-label indirect immunofluorescence with primary antibodies against Keap1, p62, and/or green fluorescent protein (GFP) (Santa Cruz Biotechnology) and mouse 488 and rabbit 594 secondary antibodies (Invitrogen-Molecular Probes). All images were taken with the Zeiss Observer.Z1 microscope using the Slidebook 4.2.0.11 computer program (Intelligent Imaging Innovations, Inc.).
RESULTS
The interaction between p62 and Keap1 and the domains that are required for the interaction.
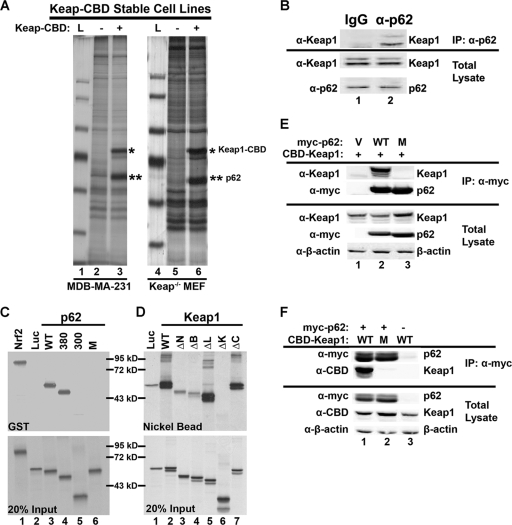
In an attempt to identify Keap1-interacting proteins, several stable cell lines harboring Keap1-CBD were generated. Pulldown experiments were performed using the stable cell lines containing vector control (−) or Keap1-CBD (+) to identify Keap1-CBD-interacting proteins. One protein was significantly enriched in the Keap1-CBD stable cell lines, compared to the control cell line (Fig. 1A, compare lanes 2 and 3 and lanes 5 and 6). Copurification of this protein with Keap1-CBD was observed with all cell lines tested (data not shown). Mass spectrometry analysis identified this protein as p62 (Fig. 1A, **). The silver-stained gels derived from MDA-MB-231 cell lysate or Keap1-null MEF cell lysate are shown in Fig. 1A. Next, interaction of endogenous p62 with endogenous Keap1 was confirmed using immunoprecipitation analysis. An antibody against Keap1 precipitated p62, and the IgG negative control did not (Fig. 1B).
FIG. 1.
The interaction between p62 and Keap1 and the domains that are required for the interaction. (A) Identification of the Keap1-interacting protein p62. Two stable cell lines, MDA-MB-231 and Keap1−/−, that expressed either vector control (−) or CBD-tagged Keap1 (+) were used in a pulldown assay using chitin beads. Samples were run on an SDS-PAGE gel and silver stained. The distinct band was excised and identified as p62. L, protein ladder. (B) Interaction of endogenous p62 and Keap1. Cell lysates from HEK293 cells were immunoprecipitated (IP) using an antibody against p62 or IgG (negative control). The resulting immunoprecipitates were subjected to SDS-PAGE and analyzed by immunoblotting with a Keap1 antibody. (C) The Keap1-interacting domain of p62 is 349-DPSTGE-354. p62 deletion mutants (p62-380 [deletion of aa 381 to the C terminus] and p62-300 [aa 301 to the C terminus]), along with p62 wild-type (p62-WT) and a mutant in which amino acids 349 to 354 were changed to alanines (p62-M), were constructed into an expression vector containing a T7 promoter. In vitro transcription and translation were performed to generate 35S-labeled full-length p62-WT, p62-M, and its deletion proteins. Each of these proteins was incubated with GST-Keap1, which was produced and purified from E. coli bacteria. Keap1-associated proteins were pulled down using glutathione beads. Equivalent amounts of p62 proteins were used in the GST pulldown assay. (D) The Kelch domain of Keap1 is the binding domain of p62. Full-length Keap1, its deletion proteins (N-terminal [ΔN], BTB domain [ΔB], linker domain [ΔL], Kelch domain [ΔK], and the C terminus [ΔC]), and p62-WT were generated by in vitro transcription and translation methods. Each of the 35S-labeled Keap1 proteins were incubated with p62-WT followed by nickel affinity chromatography. Equal amounts of Keap1 proteins were used for this pulldown assay. (E) Immunoprecipitation analysis was performed using cell lysates of HEK293 cells cotransfected with an expression vector for either p62-WT or p62-M, along with an expression vector for Keap1. Expression of each protein was detected using anti-Keap1, anti-myc (for p62 proteins), and anti-β-actin (for loading control) antibodies. (F) Three arginines in the Kelch domain of Keap1 are responsible for the binding of p62. The three arginine residues at positions 380, 415, and 483 were replaced with alanine residues using site-directed mutagenesis. The indicated constructs were coexpressed with myc-p62 in HEK293 cells by transfection, and lysates were immunoprecipitated using an anti-myc antibody. Precipitates were subjected to SDS-PAGE and immunoblotted with the indicated antibodies.
To identify the domain in p62 that is required for association with Keap1, p62 deletion mutants were constructed in an expression vector containing a T7 promoter. The deletion mutant constructs used in the following experiments were p62-380, a mutant containing a deletion from amino acid 381 to the C terminus, and p62-300, a mutant containing a deletion from amino acid 301 to the C terminus. In vitro transcription and translation were performed to generate 35S-labeled full-length p62 (WT) and its deletion proteins. These proteins were then incubated with GST-Keap1, which was produced and purified from E. coli bacteria. Keap1-associated proteins were pulled down using glutathione beads. Equivalent amounts of p62 proteins were used in the GST pulldown assay (Fig. 1C, bottom). Keap1 associated with p62-WT and p62-380; however, p62 did not associate with p62-300 (Fig. 1C, top, compare lanes 3 and 4 to lane 5), indicating that the region of p62 containing amino acids 300 to 380 is important for the interaction. Nrf2 and luciferase (Luc) were included in this experiment as positive and negative controls, respectively (Fig. 1C, lanes 1 and 2). After careful examination of the region of amino acids 300 to 380, a cluster of negatively charged amino acids was revealed (349-DPSTGE-354). This primary sequence resembles the ETGE motif of Nrf2 that is required for the interaction with the Kelch domain of Keap1. Thus, a p62 mutant (p62-M), in which all six amino acids (349-DPSTGE-354) were replaced with alanine residues, was constructed. Indeed, p62-M lost its binding with GST-Keap1 (Fig. 1C, top, lane 6), demonstrating that 349-DPSTGE-354 in p62 is required for the interaction between p62 and Keap1. To further confirm this, immunoprecipitation analysis was performed using cell lysates of HEK293 cells cotransfected with an expression vector for either p62-WT or p62-M, along with an expression vector for Keap1. Expression of each protein was detected using antibodies against Keap1, myc (for p62 proteins), and β-actin (for loading control) (Fig. 1E, bottom). Immunoprecipitation analysis indicates that Keap1 associated with p62-WT but not with p62-M (Fig. 1E, top, compare lanes 2 and 3), demonstrating that 349-DPSTGE-354 is essential for the interaction between Keap1 and p62.
In a similar experiment, the domain in Keap1 that interacts with p62 was identified. Full-length Keap1 and its deletion proteins were generated and 35S labeled using an in vitro transcription and translation method. p62-WT was also generated by in vitro transcription and translation but was not radiolabeled. Each of the 35S-labeled Keap1 proteins were incubated with p62-WT followed by nickel affinity chromatography. Equal amount of Keap1 proteins were used for this pulldown assay (Fig. 1D, bottom). Keap1-ΔK, a mutant with the Kelch domain deleted, completely lost association with p62 (Fig. 1D, top, lane 6), indicating that the Kelch domain contains the p62-interacting residues. The other mutants, with deletions of the N terminus (ΔN), BTB domain (ΔB), linker domain (ΔL), and the C terminus (ΔC), were still able to be pulled down by p62 (Fig. 1D, top, lanes 3 to 5 and 7). Three positively charged amino acids (R380, R415, and R483) in the Kelch domain of Keap1 are necessary for interacting with Nrf2 (18). Therefore, it is highly possible that the same amino acids may be needed for the p62-Keap1 interaction. Thus, a Keap1 mutant (Keap1-M), in which the three arginine residues at positions 380, 415, and 483 were replaced with alanine residues, was constructed. Interaction of Keap1 or Keap1-M with p62-WT was tested in HEK293 cells using immunoprecipitation analysis. Keap1, and not Keap1-M, associated with p62 (Fig. 1F). Taken together, these results clearly demonstrate that there is a direct interaction between p62 and Keap1 and that the amino acids 349-DPSTGE-354 in p62 and the three arginine residues (at positions 380, 415, and 483) in Keap1 are essential for the interaction of these two proteins.
p62 upregulated the Nrf2 signaling pathway.
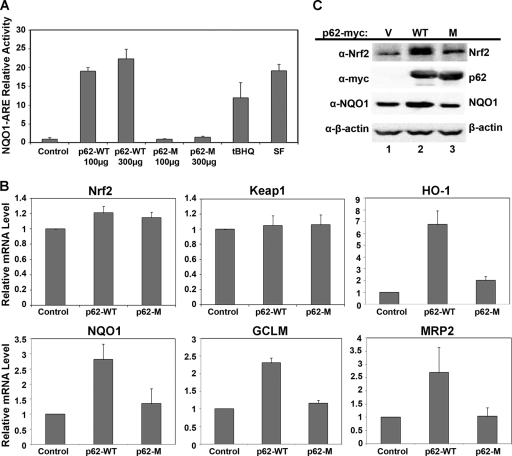
Because the same three arginine residues in Keap1 are required for interaction with either Nrf2 or p62, it is likely that p62 competes with Nrf2 for binding to Keap1. Next, the functional role of p62 in regulating Nrf2 and its antioxidant response was tested. Relative firefly luciferase activity in HEK293 cells transfected with an expression vector for either p62-WT or p62-M was measured, along with vectors for ARE-firefly luciferase and Renilla luciferase reporter genes driven by the NQO1 and TK promoters, respectively. p62-WT, but not p62-M, enhanced the ARE-mediated luciferase activity in a dose-dependent manner (Fig. 2A). tBHQ and SF, known inducers of Nrf2, were used as positive controls (Fig. 2A). To confirm that p62 is able to activate the Nrf2-mediated antioxidant response, qRT-PCR was used to measure the transcription of several Nrf2 target genes. In accordance with the results from the reporter gene assay, p62-WT, but not p62-M, increased mRNA expression of Nrf2 target genes, such as HO-1, NQO1, GCLM, and MRP2, without significantly affecting the transcription of Nrf2 or Keap1 (Fig. 2B). These results indicate that p62 most likely activates Nrf2 target genes through upregulation of Nrf2 at the protein level. Next, immunoblot analysis was performed in HEK293 cells transfected with an expression vector for vector alone, p62-WT, or p62-M. Ectopic expression of p62-WT significantly enhanced the levels of Nrf2 and NQO1, whereas p62-M did not (Fig. 2C, compare lanes 1 and 2 and lanes 1 and 3). Collectively, these results demonstrate that overexpression of p62 is able to upregulate the Nrf2 signaling pathway by enhancing the protein level of Nrf2.
FIG. 2.
p62 upregulated the Nrf2 signaling pathway. (A) p62 regulates the transcriptional activity of Nrf2. Different amounts of the expression plasmids of p62-WT or p62-M were transfected into HEK293 cells along with the NQO1-ARE promoter firefly luciferase and Renilla luciferase as an internal control. As a positive control, cells were treated with known inducers of Nrf2, tBHQ, or SF overnight. Thirty-six hours posttransfection, both firefly and Renilla luciferase activities were measured. (B) Upregulation of Nrf2 downstream genes by p62. HEK293 cells were transfected with either the expression plasmid for vector, p62-WT, or p62-M. Forty-eight hours posttransfection, total mRNA was extracted using TRIzol, and qRT-PCR was performed to measure the mRNA levels of Nrf2, Keap1, HO-1, NQO1, GLCM, and MRP2. Values were normalized to GAPDH, and samples were done in triplicates. Data represented are the mean ± standard deviation (SD). (C) An increase in p62 caused an increase in Nrf2 protein levels. HEK293 cells were transfected with an expression vector for vector, p62-WT, or p62-M. Cell lysates were collected and subjected to immunoblot analysis using the following antibodies: anti-Nrf2, anti-myc (p62), anti-NQO1, and anti-β-actin.
p62 sequestered Keap1 into aggregates.
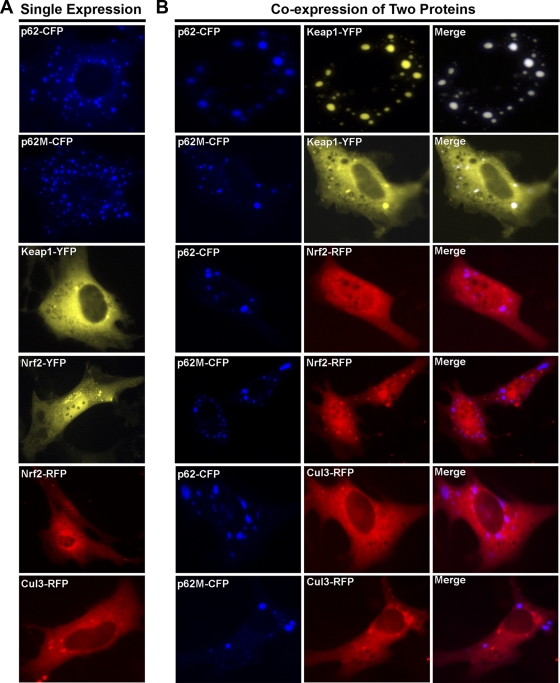
Subcellular localization of p62, Keap1, Cul3, and Nrf2 and p62-positive aggregate formation were investigated with live cells. p62, Keap1, Cul3, and Nrf2 were individually tagged with CFP, YFP, or RFP. To ensure that the function of these proteins was not affected by the addition of the fluorescent tag, all of the constructs were first tested for their expression and regulation. As expected, all constructs had high expression levels and normal function (data not shown). These proteins were then expressed singly or in combination in HEK293 cells, and cellular localization of these proteins was visualized by real-time live imaging under a fluorescent microscope. As shown in Fig. 3A, overexpression of p62-CFP or p62M-CFP alone formed aggregates in the cytoplasm, which are typically characterized by punctate dots (Fig. 3A, first and second panels from top). Ectopically expressed Keap1-YFP was localized predominantly in the cytoplasm (Fig. 3A, third panel from top), while Nrf2-YFP and Nrf2-RFP were present in both compartments and the majority of the cells had more nuclear localization (Fig. 3A, fourth and fifth panels from top). Similarly to Keap1-YFP, Cul3-RFP was localized mainly in the cytoplasm (Fig. 3A, bottom panel).
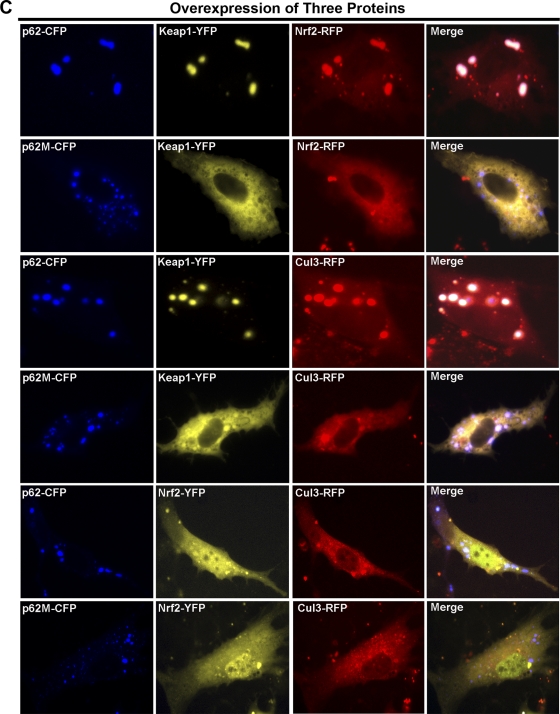
FIG. 3.
p62 sequestered Keap1 into aggregates. (A) The cellular localization of p62, Keap1, Nrf2, and Cul3. HEK293 cells were singly transfected with an expression vector for the fluorescently tagged protein. The subcellular localization of the proteins was monitored live. (B and C) The colocalization of proteins. Live imaging of the proteins was monitored with HEK293 cells that were cotransfected with expression vectors for the indicated fluorescently tagged proteins.
We have previously reported that Keap1 functions as the substrate adaptor protein that brings Nrf2 into the core Cul3-Rbx1 E3 ubiquitin ligase complex for ubiquitination (29). The fact that p62 interacts directly with Keap1 led us to examine the colocalization of p62 with Keap1, Cul3, or Nrf2 (Fig. 3B). Coexpression of Keap1-YFP with p62-CFP caused aggregation of both proteins (Fig. 3B, first row from top). In contrast, p62M-CFP lost its ability to recruit Keap1 into aggregates (Fig. 3B, second row from top). Neither p62-CFP nor p62M-CFP was able to recruit Nrf2 into aggregates (Fig. 3B, third and fourth rows from top). Similarly, Cul3 was not recruited into aggregates as well (Fig. 3B, fifth and sixth rows from top).
Next, HEK293 cells were simultaneously transfected with three different proteins to test localization of the indicated proteins (Fig. 3C). In contrast to the diffused whole-cell localization of Nrf2 seen when p62-CFP and Nrf2-RFP were expressed in the absence of Keap1 (Fig. 3B, third row from top), overexpression of both p62 and Keap1 was able to bring a certain amount of Nrf2 into aggregates (Fig. 3C, first row from top). Nonetheless, Nrf2 was still localized diffusely in the entire cell (Fig. 3C, first row from top). This can be attributed to Nrf2 having the ability to bind to a Kelch domain that is not occupied by p62 in a Keap1 homodimer. As expected, Nrf2 was not detected in the aggregates when p62M-CFP was transfected in combination with Nrf2 and Keap1 (Fig. 3C, second row from top). Similarly to Nrf2, some of Cul3 was colocalized into aggregates when Keap1 along with p62-CFP, but not p62M-CFP, was coexpressed (Fig. 3C, compare third and fourth rows from top). Cul3 still localized diffusely in the cytoplasm; however, Cul3 also colocalized with Keap1 and p62 aggregates (Fig. 3C, third row from top). This was not observed with cells with Keap1, p62M, and Cul3 (Fig. 3C, fourth row from top). Lastly, when p62-CFP or p62M-CFP, along with Nrf2-YFP and Cul3-RFP, was coexpressed in the absence of Keap1, neither p62 nor p62M was able to recruit Nrf2 or Cul3 into aggregates (Fig. 3C, fifth and sixth rows from top). Collectively, these results indicate the necessity of Keap1 in p62-dependent Nrf2 and Cul3 aggregation.
p62 decreased ubiquitination of Nrf2, leading to an increase in Nrf2 stability.
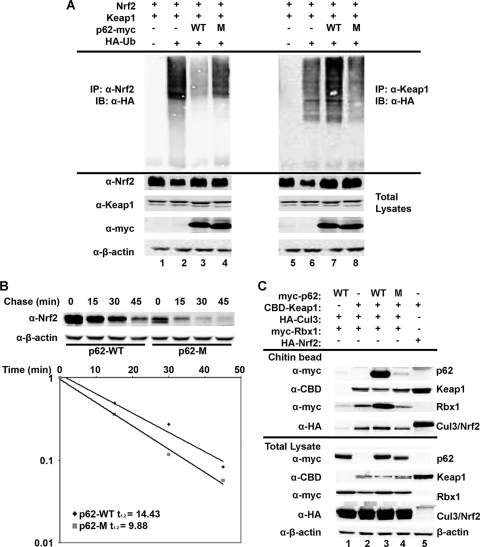
To test the possibility that p62 upregulates the Nrf2 signaling pathway by decreasing the active E3 ubiquitin ligase for Nrf2, ubiquitination of Nrf2 and Keap1 in the presence of p62-WT or p62-M was tested using an in vivo ubiquitination assay. Indeed, overexpression of p62-WT significantly decreased ubiquitination of Nrf2, while it enhanced ubiquitination of Keap1 (Fig. 4A, lanes 3 and 7). However, overexpression of p62-M had no effect on ubiquitination of either Nrf2 or Keap1 (Fig. 4A, lanes 4 and 8). We demonstrate, in combination with the imagining data presented in Fig. 3C, that p62 is able to sequester Cul3 into aggregates through Keap1, which may explain the increase in Keap1 autoubiquitination. Next, the half-life of Nrf2 was determined in the presence of p62-WT or p62-M by using CHX and immunoblot analysis. The half-life of Nrf2 was 14.43 or 9.93 min in the presence of p62-WT or p62-M, respectively (Fig. 4B). Thus, taken together, these results suggest that overexpressed p62 is able to sequester the Keap1-Cul3-E3 ubiquitin ligase complexes into aggregates, which prevents Nrf2 from being ubiquitinated. Therefore, under p62-overexpressed conditions, Nrf2 is stabilized and the Nrf2-mediated antioxidant response is activated.
FIG. 4.
p62 decreased ubiquitination of Nrf2, leading to an increase in Nrf2 stability. (A) p62 decreased the ubiquitination of Nrf2 and increased the ubiquitination of Keap1. HEK293 cells were cotransfected with an expression vector for Nrf2, Keap1, HA-ubiquitin, and either p62-WT or p62-M. In vivo ubiquitination analysis was performed 48 h posttransfection. IB, immunoblotting. (B) p62 increased the half-life of Nrf2. HEK293 cells transfected with either p62-WT or p62-M were treated with 25 μM CHX for the indicated time periods (chase). Endogenous Nrf2 levels were detected by immunoblotting, and the intensity of the Nrf2 bands was quantified and plotted on a semilog graph. (C) p62-WT increased the association of Keap1, Cul3, and Rbx1. Immunoprecipitation analysis was conducted with HEK293 cells transfected with the indicated proteins. Nrf2 was included as a positive control.
Next, Keap1-Cul3-Rbx1 complex formation in the presence of p62-WT or p62-M was compared. Cells were transfected with CBD-Keap1, HA-Cul3, and myc-Rbx1 along with either p62-WT or p62-M. Keap1-containing complexes were pulled down using chitin beads and subjected to immunoblot analysis (Fig. 4C). p62-WT increased the association of Keap1, Cul3, and Rbx1 compared to the lane with no p62 or p62-M (Fig. 4C, compare Rbx1 and Cul3 rows of lane 3 to those of lanes 2 and 4). As a negative control, no Keap1-CBD was transfected (Fig. 4C, lane 1), and as a positive control, pulldown of exogenous Nrf2 was done (Fig. 4C, lane 5). Collectively, these results not only recapitulate the interaction between p62 and Keap1 but also suggest that p62 enhances the association of Keap1 with Cul3 and Rbx1, which offers a possible explanation for the shift in ubiquitination from Nrf2 to Keap1.
Autophagy-defective cells sequestered Keap1 into aggregates.
To further confirm that the Nrf2 pathway is upregulated during deregulation of autophagy, we used primary immortalized baby mouse kidney (iBMK) cells to monitor the localization of Keap1. Localization of Keap1 was conducted in both autophagy-competent (Beclin1+/+ and Atg5+/+) and autophagy-deficient (Beclin1+/− and Atg5−/−) iBMK cells either stably expressing GFP alone (control) or GFP-tagged p62. In one set of experiments, the iBMK cell lines were transiently transfected with Keap1-CFP. Localization of the proteins was visualized in live cells using a fluorescent microscope. Both the cytoplasm and nucleus were uniformly green in the GFP control for all four cell lines (Beclin1+/+, Beclin1+/−, Atg5+/+, and Atg5−/−), with Keap1-CFP remaining evenly distributed in the cytoplasm (Fig. 5 A and B, first and third rows from top). The localization of p62-GFP and Keap1-CFP in the Beclin1+/+ and Atg5+/+ cells was also primarily in the cytoplasm, with a few small protein aggregates (Fig. 5A and B, second rows from top). In the autophagy-deficient cell lines, Beclin1+/− and Atg5−/−, p62-GFP formed large aggregates, with which Keap1 completely colocalized (Fig. 5A and B, fourth rows from top). Additionally, indirect immunofluorescence using antibodies against Keap1 and GFP demonstrated that endogenous Keap1 indeed colocalized with the p62-GFP aggregates in the autophagy-deficient iBMK cells (Fig. 5C and D, fourth rows from top), confirming our observations of iBMK cells ectopically expressing CFP-tagged Keap1 (Fig. 5A and B, fourth rows from top). As expected, in Atg5+/+ and Beclin1+/+ cells, p62-GFP and endogenous Keap1 localized mainly in the cytoplasm (Fig. 5C and D, first and third rows from top). With the control cells of Atg5+/+, Atg5−/−, Beclin1+/+, and Beclin1+/− stably expressing GFP, whole-cell GFP staining was observed, while endogenous Keap1 was localized primarily in the cytoplasm (Fig. 5C and D, second rows from top). Collectively, these results indicate that autophagy deficiency activates the Nrf2 pathway through recruitment of Keap1 into aggregates due to the excessive accumulation of p62.
FIG. 5.
Autophagy-defective cells sequestered Keap1 into aggregates. (A and B) Keap1 was sequestered into aggregates in primary autophagy-deficient cells. Atg5+/+, Atg5−/−, Beclin1+/+, and Beclin+/− iBMK cells stably expressing GFP alone or p62-GFP were transfected with CFP-tagged Keap1. Live imaging was taken 24 h posttransfection. (C and D) Endogenous Keap1 was sequestered into aggregates in primary autophagy-deficient cells. The cells were grown on coverslips and fixed in methanol. Indirect immunofluorescence staining was conducted using an antibody against GFP and Keap1.
DISCUSSION
Classically, the Nrf2-Keap1 pathway is induced by oxidative stress through modifications of one or more cysteine residues in Keap1. It was suggested that the modifications of cysteine residues caused a conformational change of the Keap1-Cul3-Rbx1 E3 ubiquitin ligase complex, resulting in a decrease in ubiquitination and degradation of Nrf2 (28). In this report, we demonstrate a noncanonical mechanism of Nrf2 activation that is caused by deregulation of autophagy, which is different from the canonical pathway. Autophagy deficiency causes the formation of aggregates and the excessive accumulation of p62, which recruits Keap1 through direct binding. Normally, Keap1 functions as the substrate adaptor protein that brings Nrf2 into the core complex of the E3 ubiquitin ligase, consisting of Cul3 and Rbx1, and facilitates the ubiquitination of Nrf2 (27). Thus, by sequestering Keap1 into aggregates, the ability of the E3 ubiquitin ligase complex to ubiquitinate Nrf2 is hindered, which prevents its degradation. This renders Nrf2 more stable, leading to the activation of its target genes. Our results provide mechanistic insight into the previous findings by Liu et al., who demonstrated that overexpression of p62 increased nuclear Nrf2, ARE-luciferase, and NQO1 protein levels (10). We believe that many endogenous proteins exist that can upregulate the Nrf2 pathway by disrupting the Keap1-Nrf2 interaction. These proteins can cause different stress signaling pathways to converge to activate Nrf2 to maintain cellular redox homeostasis. One example is p21, which is known to bind to the DLG motif of Nrf2. We showed that the interaction of p21 with Nrf2 disrupted the binding of Nrf2 to the E3 ubiquitin ligase complex and prevented Keap1-dependent ubiquitination and degradation of Nrf2 (2). Therefore, it is important to identify other proteins that disrupt the interaction between Nrf2 and Keap1.
The binding of Nrf2 to Keap1 has been well characterized and has been mapped to the Neh2 domain of Nrf2 and the Kelch domain of Keap1 (27). The binding of Keap1 to Nrf2 is through the “hinge-and-latch” two-site binding model, in which each Kelch domain from a Keap1 homodimer binds to Nrf2 through two sites in the Neh2 domain (22). The “hinge” is the high-affinity binding site ETGE, and the low-affinity binding site DLG is the “latch” (22). We found that p62 has a similar ETGE motif between amino acids 349 and 354. When the amino acids were mutated to alanine residues, p62 lost binding to Keap1, indicating that these are indeed the residues required for the direct interaction of p62 and Keap1. Previously, it has been shown that there are three pertinent arginine residues (Arg380, -415, and -483) in the Kelch domain of Keap1 that are responsible for Nrf2 binding (18). Similarly, upon mutation of the arginine residues to alanines, Keap1 lost binding to p62, implying that p62 may be competing with Nrf2 for Keap1 binding. Binding of both the DLG and ETGE motifs of Nrf2 to the Kelch domain of a Keap1 homodimer is essential in presenting the lysine residues of Nrf2 in the correct orientation to accept ubiquitin (14, 22, 30). Therefore, when the Kelch domain of Keap1 is occupied by p62, this prevents the correct positioning of Nrf2 and prohibits the Keap1-Cul3-E3 ubiquitin ligase complex from ubiquitinating Nrf2. Additionally, p62-WT enhanced the ubiquitination of Keap1, indicating that the core complex (Cul3-Rbx1) is still functional. In conjunction with this finding, we observed colocalization of p62-WT, Keap1, Cul3, and Nrf2 in aggregates (Fig. 3C). Based on the presence of Nrf2 in the aggregates (Fig. 3C), we speculate that one Kelch domain of the Keap1 homodimer binds to p62 and the other binds to Nrf2, which prevents Nrf2 ubiquitination. Additionally, the core Cul3-Rbx1 complex is still able to bind to Keap1 because the binding domain of Cul3 to Keap1 is different from that of p62 and Nrf2. Furthermore, we observed an increase in association of Keap1-Cul3-Rbx1 in the presence of p62-WT, but not of p62-M (Fig. 4C). It is well established that Keap1-Cul3-Rbx1 undergoes dynamic association and dissociation, which is a highly regulated process. Therefore, recruitment of this complex into aggregates by p62 may disrupt dissociation of Keap1 from the core complex. These data explain why p62 increased the autoubiquitination of Keap1 (Fig. 4A), which is consistent with our previous findings that two Keap1 mutants with enhanced association to Cul3 had an increase in Keap1 autoubiquitination and a decrease in Nrf2 ubiquitination (29).
Autophagy deficiency has been shown to cause p62 to accumulate because it cannot be properly degraded (12). Consequently, this leads to the formation of protein aggregates, which enhances the production of ROS and damage to DNA, protein, and organelles (12). Since the Nrf2-Keap1 pathway is an important cellular defense mechanism that copes with oxidative stress (9), it is possible that the ROS generated from autophagy deficiency activates Nrf2 indirectly. To test this indirect mechanism, autophagy-deficient cells were treated with ROS scavengers, such as N-acetylcysteine (NAC) or glutathione (GSH). Treatment with NAC or GSH did not affect the protein level of Nrf2 or the interaction between p62 and Keap1 (data not shown). This was consistent with a recent paper showing that NAC treatment had no effect on Atg7-deficient hepatocytes (7). Therefore, the upregulation of Nrf2 is not due to the indirect mechanism of enhanced ROS production caused by a deficiency in autophagy, but rather due to the accumulation of p62 and the direct interaction of p62 and Keap1. This indicates that this mechanism of Nrf2 activation is dominant over the canonical ROS-mediated activation of Nrf2 in autophagy deficiency.
During the preparation of the manuscript, a recent article demonstrated that p62 disrupts the interaction between Nrf2 and Keap1, which is consistent with our results. The authors showed that autophagy deficiency not only caused p62 accumulation and hepatic injury but also upregulated the Nrf2 pathway, which exacerbated hepatotoxicity. However, the protective role of Nrf2 against many human diseases has been well documented. Although Komatsu et al. observed improved liver injury in double-knockout (Atg7 and Nrf2) mice, we believe that p62 overexpression, and not Nrf2, is the primary culprit in hepatic injury. As shown in their Fig. 5a, the double-knockout mice had milder p62 accumulation than the excessive accumulation of p62 in Atg7 single-knockout mice (7). The functional significance of Nrf2 activation by deregulation of autophagy in human diseases is still unclear and warrants further investigation.
Acknowledgments
We thank R. Mathew for his intellectual contribution.
This study was supported by the following grants: RO1ES015010 (NIEHS) and RSG-07-154 (ACS), which were awarded to D.D.Z., and ES006694 (NIH), a center grant.
Footnotes
Published ahead of print on 26 April 2010.
REFERENCES
- 1.Banning, A., S. Deubel, D. Kluth, Z. Zhou, and R. Brigelius-Flohe. 2005. The GI-GPx gene is a target for Nrf2. Mol. Cell. Biol. 25:4914-4923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen, W., Z. Sun, X. J. Wang, T. Jiang, Z. Huang, D. Fang, and D. D. Zhang. 2009. Direct interaction between Nrf2 and p21(Cip1/WAF1) upregulates the Nrf2-mediated antioxidant response. Mol. Cell 34:663-673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Furukawa, M., and Y. Xiong. 2005. BTB protein Keap1 targets antioxidant transcription factor Nrf2 for ubiquitination by the Cullin 3-Roc1 ligase. Mol. Cell. Biol. 25:162-171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ishii, T., K. Itoh, S. Takahashi, H. Sato, T. Yanagawa, Y. Katoh, S. Bannai, and M. Yamamoto. 2000. Transcription factor Nrf2 coordinately regulates a group of oxidative stress-inducible genes in macrophages. J. Biol. Chem. 275:16023-16029. [DOI] [PubMed] [Google Scholar]
- 5.Kim, Y. C., H. Masutani, Y. Yamaguchi, K. Itoh, M. Yamamoto, and J. Yodoi. 2001. Hemin-induced activation of the thioredoxin gene by Nrf2. A differential regulation of the antioxidant responsive element by a switch of its binding factors. J. Biol. Chem. 276:18399-18406. [DOI] [PubMed] [Google Scholar]
- 6.Kirkin, V., D. G. McEwan, I. Novak, and I. Dikic. 2009. A role for ubiquitin in selective autophagy. Mol. Cell 34:259-269. [DOI] [PubMed] [Google Scholar]
- 7.Komatsu, M., H. Kurokawa, S. Waguri, K. Taguchi, A. Kobayashi, Y. Ichimura, Y. S. Sou, I. Ueno, A. Sakamoto, K. I. Tong, M. Kim, Y. Nishito, S. I. Iemura, T. Natsume, T. Ueno, E. Kominami, H. Motohashi, K. Tanaka, and M. Yamamoto. 2010. The selective autophagy substrate p62 activates the stress responsive transcription factor Nrf2 through inactivation of Keap1. Nat. Cell Biol. 12:213-223. [DOI] [PubMed] [Google Scholar]
- 8.Komatsu, M., S. Waguri, M. Koike, Y. S. Sou, T. Ueno, T. Hara, N. Mizushima, J. Iwata, J. Ezaki, S. Murata, J. Hamazaki, Y. Nishito, S. Iemura, T. Natsume, T. Yanagawa, J. Uwayama, E. Warabi, H. Yoshida, T. Ishii, A. Kobayashi, M. Yamamoto, Z. Yue, Y. Uchiyama, E. Kominami, and K. Tanaka. 2007. Homeostatic levels of p62 control cytoplasmic inclusion body formation in autophagy-deficient mice. Cell 131:1149-1163. [DOI] [PubMed] [Google Scholar]
- 9.Lau, A., N. F. Villeneuve, Z. Sun, P. K. Wong, and D. D. Zhang. 2008. Dual roles of Nrf2 in cancer. Pharmacol. Res. 58:262-270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu, Y., J. T. Kern, J. R. Walker, J. A. Johnson, P. G. Schultz, and H. Luesch. 2007. A genomic screen for activators of the antioxidant response element. Proc. Natl. Acad. Sci. U. S. A. 104:5205-5210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Maher, J. M., X. Cheng, A. L. Slitt, M. Z. Dieter, and C. D. Klaassen. 2005. Induction of the multidrug resistance-associated protein family of transporters by chemical activators of receptor-mediated pathways in mouse liver. Drug Metab. Dispos. 33:956-962. [DOI] [PubMed] [Google Scholar]
- 12.Mathew, R., C. M. Karp, B. Beaudoin, N. Vuong, G. Chen, H. Y. Chen, K. Bray, A. Reddy, G. Bhanot, C. Gelinas, R. S. Dipaola, V. Karantza-Wadsworth, and E. White. 2009. Autophagy suppresses tumorigenesis through elimination of p62. Cell 137:1062-1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McMahon, M., K. Itoh, M. Yamamoto, and J. D. Hayes. 2003. Keap1-dependent proteasomal degradation of transcription factor Nrf2 contributes to the negative regulation of antioxidant response element-driven gene expression. J. Biol. Chem. 278:21592-21600. [DOI] [PubMed] [Google Scholar]
- 14.McMahon, M., N. Thomas, K. Itoh, M. Yamamoto, and J. D. Hayes. 2006. Dimerization of substrate adaptors can facilitate cullin-mediated ubiquitylation of proteins by a “tethering” mechanism: a two-site interaction model for the Nrf2-Keap1 complex. J. Biol. Chem. 281:24756-24768. [DOI] [PubMed] [Google Scholar]
- 15.Moinova, H. R., and R. T. Mulcahy. 1999. Up-regulation of the human gamma-glutamylcysteine synthetase regulatory subunit gene involves binding of Nrf-2 to an electrophile responsive element. Biochem. Biophys. Res. Commun. 261:661-668. [DOI] [PubMed] [Google Scholar]
- 16.Moscat, J., and M. T. Diaz-Meco. 2009. p62 at the crossroads of autophagy, apoptosis, and cancer. Cell 137:1001-1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nioi, P., M. McMahon, K. Itoh, M. Yamamoto, and J. D. Hayes. 2003. Identification of a novel Nrf2-regulated antioxidant response element (ARE) in the mouse NAD(P)H:quinone oxidoreductase 1 gene: reassessment of the ARE consensus sequence. Biochem. J. 374:337-348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Padmanabhan, B., K. I. Tong, T. Ohta, Y. Nakamura, M. Scharlock, M. Ohtsuji, M. I. Kang, A. Kobayashi, S. Yokoyama, and M. Yamamoto. 2006. Structural basis for defects of Keap1 activity provoked by its point mutations in lung cancer. Mol. Cell 21:689-700. [DOI] [PubMed] [Google Scholar]
- 19.Qu, X., J. Yu, G. Bhagat, N. Furuya, H. Hibshoosh, A. Troxel, J. Rosen, E. L. Eskelinen, N. Mizushima, Y. Ohsumi, G. Cattoretti, and B. Levine. 2003. Promotion of tumorigenesis by heterozygous disruption of the beclin 1 autophagy gene. J. Clin. Invest. 112:1809-1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Seibenhener, M. L., J. R. Babu, T. Geetha, H. C. Wong, N. R. Krishna, and M. W. Wooten. 2004. Sequestosome 1/p62 is a polyubiquitin chain binding protein involved in ubiquitin proteasome degradation. Mol. Cell. Biol. 24:8055-8068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Seibenhener, M. L., T. Geetha, and M. W. Wooten. 2007. Sequestosome 1/p62—more than just a scaffold. FEBS Lett. 581:175-179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tong, K. I., Y. Katoh, H. Kusunoki, K. Itoh, T. Tanaka, and M. Yamamoto. 2006. Keap1 recruits Neh2 through binding to ETGE and DLG motifs: characterization of the two-site molecular recognition model. Mol. Cell. Biol. 26:2887-2900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang, X. J., Z. Sun, W. Chen, Y. Li, N. F. Villeneuve, and D. D. Zhang. 2008. Activation of Nrf2 by arsenite and monomethylarsonous acid is independent of Keap1-C151: enhanced Keap1-Cul3 interaction. Toxicol. Appl. Pharmacol. 230:383-389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.White, E., C. Karp, A. M. Strohecker, Y. Guo, and R. Mathew. 2010. Role of autophagy in suppression of inflammation and cancer. Curr. Opin. Cell Biol. 22:212-217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yorimitsu, T., and D. J. Klionsky. 2005. Autophagy: molecular machinery for self-eating. Cell Death Differ. 12(Suppl. 2):1542-1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yue, Z., S. Jin, C. Yang, A. J. Levine, and N. Heintz. 2003. Beclin 1, an autophagy gene essential for early embryonic development, is a haploinsufficient tumor suppressor. Proc. Natl. Acad. Sci. U. S. A. 100:15077-15082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang, D. D. 2006. Mechanistic studies of the Nrf2-Keap1 signaling pathway. Drug Metab. Rev. 38:769-789. [DOI] [PubMed] [Google Scholar]
- 28.Zhang, D. D., and M. Hannink. 2003. Distinct cysteine residues in Keap1 are required for Keap1-dependent ubiquitination of Nrf2 and for stabilization of Nrf2 by chemopreventive agents and oxidative stress. Mol. Cell. Biol. 23:8137-8151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang, D. D., S. C. Lo, J. V. Cross, D. J. Templeton, and M. Hannink. 2004. Keap1 is a redox-regulated substrate adaptor protein for a Cul3-dependent ubiquitin ligase complex. Mol. Cell. Biol. 24:10941-10953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zipper, L. M., and R. T. Mulcahy. 2002. The Keap1 BTB/POZ dimerization function is required to sequester Nrf2 in1 cytoplasm. J. Biol. Chem. 277:36544-36552. [DOI] [PubMed] [Google Scholar]