Abstract
Severe acute respiratory syndrome coronavirus (SARS-CoV) was identified to be the causative agent of SARS with atypical pneumonia. Angiotensin-converting enzyme 2 (ACE2) is the major receptor for SARS-CoV. It is not clear whether ACE2 conveys signals from the cell surface to the nucleus and regulates expression of cellular genes upon SARS-CoV infection. To understand the pathogenesis of SARS-CoV, human type II pneumocyte (A549) cells were incubated with the viral spike protein or with SARS-CoV virus-like particles containing the viral spike protein to examine cytokine modulation in lung cells. Results from oligonucleotide-based microarray, real-time PCR, and enzyme-linked immunosorbent assays indicated an upregulation of the fibrosis-associated chemokine (C-C motif) ligand 2 (CCL2) by the viral spike protein and the virus-like particles. The upregulation of CCL2 by SARS-CoV spike protein was mainly mediated by extracellular signal-regulated kinase 1 and 2 (ERK1/2) and AP-1 but not the IκBα-NF-κB signaling pathway. In addition, Ras and Raf upstream of the ERK1/2 signaling pathway were involved in the upregulation of CCL2. Furthermore, ACE2 receptor was activated by casein kinase II-mediated phosphorylation in cells pretreated with the virus-like particles containing spike protein. These results indicate that SARS-CoV spike protein triggers ACE2 signaling and activates fibrosis-associated CCL2 expression through the Ras-ERK-AP-1 pathway.
Severe acute respiratory syndrome (SARS) is an atypical pneumonia that occurred in several countries during late 2002 and the first half of 2003. A novel coronavirus, SARS-coronavirus (SARS-CoV), isolated from SARS patients was identified to be the causative agent of SARS. SARS-CoV infected more than 8,000 people, with a worldwide mortality rate of 9.6% (8, 20). The virus contains a positive-sense single-stranded RNA genome of approximately 30,000 nucleotides. Four major structural proteins including spike (S), membrane (M), envelope (E), and nucleocapsid (N) make up the SARS-CoV particles (31, 36). Angiotensin (Ang)-converting enzyme 2 (ACE2) and CD209L (L-SIGN) have been identified to be the receptors for SARS-CoV (15, 27). SARS-CoV spike protein induced ACE2-mediated interleukin-8 (IL-8) release from lung cells via activation of activation protein 1 (AP-1) (4). Nevertheless, involvement of ACE2 in virus pathogenesis is not fully understood.
Dysregulation of inflammatory cytokines and adhesion molecules may be involved in lung injury that causes acute respiratory distress syndrome. High levels of proinflammatory cytokines such as interleukin-6, transforming growth factor β (TGF-β), and tumor necrosis factor alpha (TNF-α) were detected in the sera and ACE2+ cells of SARS patients (12, 45). Elevated levels of cytokines, including alpha interferon (IFN-α), IFN-β, IFN-γ, CCL3, CCL5, and CXCL10, were also detected in SARS-CoV-infected macrophages, dendritic cells, and a colon carcinoma cell line (1, 5, 25). It is possible that the high fatality rate of SARS results from a severe immune response caused by cytokines and chemokines.
CCL2 [chemokine (C-C motif) ligand 2; monocyte chemoattractant protein-1, (MCP-1)] is a CC chemokine that attracts monocytes, memory T lymphocytes, and basophils. CCL2 and its receptor CCR2 are involved in inflammatory reactions, including monocyte/macrophage migration, Th2 cell polarization, and the production of TGF-β and procollagen in fibroblast cells (9, 10). CCL2 is thus associated with several lung inflammatory disorders including acute respiratory distress syndrome, asthma, and pulmonary fibrosis (35). These inflammatory disorders and pulmonary infiltration are known to account for the progressive respiratory failure and death of SARS patients. In addition, upregulation of CCL2 was detected in the sera of SARS patients and the supernatant of a SARS-CoV-infected culture system (5, 16). However, mechanisms by which SARS-CoV is involved in the upregulation of CCL2 are not known.
In this study, we have taken a step forward in understanding the pathogenesis of SARS-CoV by examining SARS-CoV-mediated cytokine modulation in human type II pneumocyte (A549) cells and monkey kidney Vero E6 cells. Both pretreatment of A549 cells with SARS-CoV virus-like particles (VLPs) and preincubation of the cells with the viral spike protein upregulate the expression of fibrosis-associated CCL2. SARS-CoV may interact with ACE2 receptor and activate casein kinase II-mediated ACE2 phosphorylation, which is critical for SARS-CoV-induced CCL2 upregulation. In addition, Ras, Raf, MEK, extracellular signal-regulated kinase 1 and 2 (ERK1/2), and AP-1 are directly involved in SARS-CoV-induced CCL2 upregulation. These data suggest that the intracellular ACE2 signaling pathway in the pneumocytes of SARS-CoV-infected patients confers risks of lung fibrosis leading to respiratory failure.
MATERIALS AND METHODS
Cell lines.
Human alveolar basal epithelial cells (A549; type II pneumocytes) and African green monkey kidney cells (Vero E6) were maintained at 37°C with 5% CO2 in RPMI 1640 medium (Gibco) and Dulbecco's modified Eagle's medium (Gibco), respectively, supplemented with 10% heat-inactivated fetal bovine serum, 100 U/ml penicillin, and 100 μg/ml streptomycin. Sf9 (Spodoptera frugiperda) insect cells were maintained at 27°C in serum-free Sf-900 II SFM medium (Gibco).
Plasmids. (i) pGLM-PRM, pGLM-PRM-mT1, pGLM-PRM-mT1T2, and pGLM-PRM-mκB.
pGLM-PRM (kindly provided by T. Yoshimura, Laboratory of Immunobiology, National Cancer Institute, Frederick, MD) represents a firefly luciferase reporter plasmid containing the proximal regulatory region of CCL2 promoter from −107 to +60 (41). pGLM-PRM-mT1 represents a derivative of pGLM-PRM in which the AP-1 binding site from −97 to −91 (−97/−91) was mutated. For generation of the plasmid pGLM-PRM-mT1, DNA fragment PRM-mT1 amplified from pGLM-PRM with the primer set X107mT1 and H60CCL2 (see Table S1 at http://homepage.ntu.edu.tw/∼f90442001) was digested with XhoI and HindIII and subcloned into the XhoI-HindIII sites of pGLM-PRM. Plasmid pGLM-PRM-mT1T2 bears mutations at both AP-1 binding sites (−97/−91 and −69/−63) and was derived from plasmid pGLM-PRM-mT1 by site-directed mutagenesis with the primer set mT2CCL2F and mT2CCL2R (see Table S1 at http://homepage.ntu.edu.tw/∼f90442001). Plasmid pGLM-PRM-mκB, in which the NF-κB binding site in the CCL2 promoter from −89 to −80 was mutated, was generated from the wild-type plasmid pGLM-PRM by site-directed mutagenesis with the primer set mκBCCL2F and mκBCCL2R (see Table S1 at http://homepage.ntu.edu.tw/∼f90442001). Site-directed mutagenesis was performed with a QuikChange II XL site-directed mutagenesis kit followed the protocol provided by the manufacturer (Stratagene).
(ii) Plasmid pNFκB-Luc.
Reporter plasmid pNFκB-Luc consists of tandem repeats of the NF-κB binding site and was kindly provided by Zee-Fen Chang (National Taiwan University College of Medicine, Taipei, Taiwan).
Generation of recombinant baculoviruses expressing SARS-CoV viral proteins.
To generate recombinant baculoviruses expressing SARS-CoV proteins S and E, BaculoDirect Linear DNA was cotransfected into Sf9 cells with plasmids pENTR-S and pENTR-E encoding the C-terminal V5- and His-tagged full-length SARS-CoV S and E protein (TW1 strain; GenBank accession number AY291451) (14), respectively, with Cellfectin Reagent (Invitrogen) according to the manufacturer's instructions. Recombinant baculoviruses expressing M protein were generated by cotransfecting BacVector virus DNA with plasmid pBac-1-M encoding the C-terminal V5- and His-tagged full-length SARS-CoV M protein via the BacVector Transfection System (Novagen). The culture medium was collected at 3 days posttransfection and subjected to plaque assay for determining the titers of the recombinant baculoviruses.
Production and purification of SARS-CoV VLPs.
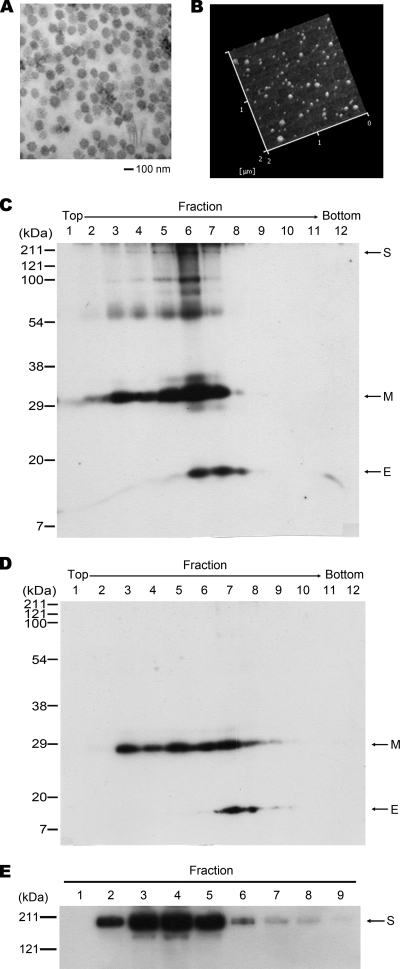
SARS-CoV VLPs were generated by coinfecting Sf9 cells with recombinant baculoviruses expressing the viral structural proteins S, E, and M. Successful production of SARS-CoV VLPs in Sf9 cells was confirmed by electron microscopy (Fig. 1A). SARS-CoV VLPs secreted into the culture medium were harvested and purified by sucrose gradient centrifugation as described previously (40). Characteristics of the purified SARS-CoV VLP were examined by atomic force microscopy (AFM) (Fig. 1B), as described previously (29), and by Western blot analysis (Fig. 1C). In addition, SARS-CoV VLPs composed of the viral structural proteins E and M but not the spike protein [SARS-CoV(EM)] were produced by coinfecting Sf9 cells with recombinant baculoviruses expressing the E protein and baculoviruses expressing the M protein (Fig. 1D). The SARS-CoV(EM) VLP was used as a negative control in this study.
FIG. 1.
Purification of SARS-CoV VLPs and the viral spike protein. The production of SARS-CoV VLPs in Sf9 cells was analyzed by electron microscopy (A) and AFM (B; the height image in liquid is shown). Western blot analysis was performed for identification of the SARS-CoV VLPs (C) and SARS-CoV(EM) VLPs (D) following purification by sucrose gradient centrifugation and for identification of the viral spike protein following Ni2+ Sepharose purification (E). Antibodies against the His and V5 tags of the recombinant viral proteins were used in Western blot analysis. Fractions 6 in panel C, 7 in panel D, and 4 in panel E were used in this study.
Purification of SARS-CoV spike protein.
Purification of the C-terminal V5-and His-tagged full-length SARS-CoV spike protein was carried out from the culture medium of Sf9 cells infected with the recombinant baculovirus encoding the viral spike protein at 4 days postinfection by using an Ni2+ Sepharose purification system, as described by the manufacturer (GE Healthcare). Fraction 4, which contains the majority of the spike protein as detected by Western blot analysis (Fig. 1E), was collected and used in this study.
RNA isolation and reverse transcription (RT)-quantitative real-time PCR.
Total RNA was isolated from culture cells by acid guanidinium thiocyanate-phenol-chloroform extraction as described previously (7) or isolated by TRIzol reagent (Invitrogen) extraction.
To detect the expression of CCL2 mRNA in cultured cells, a two-step quantitative reverse transcription-PCR was applied. In the first step, 5 μg of the total RNA was reverse transcribed with avian myeloblastosis virus reverse transcriptase (Roche Applied Science) in the presence of oligo(dT) primer. The products were then subjected to real-time PCR amplification with the CCL2-specific primer set, CCL2F and CCL2R (see Table S1 at http://homepage.ntu.edu.tw/∼f90442001) and iQ SYBR green Supermix (Bio-Rad). Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA was quantified in parallel as an internal control. Quantification was performed with an iCycler iQ real-time PCR detection system (Bio-Rad).
RNA amplification and microarray analysis.
For microarray analysis, A549 cells incubated with approximately 5 × 106 SARS-CoV VLPs were subjected to RNA isolation by using a PicoPure RNA Isolation Kit and amplification by using a RiboAmp RNA amplification kit (Arcturus), as described previously (43). Gene expression profiles were analyzed with an Affymetrix high-density oligonucleotide microarray (Human Genome U133 Plus 2.0 Array) using the Affymetrix microarray system protocol as described by the manufacturer. GeneChip operating software (GCOS) and robust multichip average (RMA) statistics were applied for data analysis.
DNA transfection and luciferase reporter assay.
The promoter activity of CCL2 was examined by luciferase reporter assay following a cotransfection of A549 cells with the firefly luciferase reporter plasmid pGLM-PRM (or its mutated forms) and the Renilla luciferase-expressing control plasmid phRL-TK (Promega). To examine the effect of SARS-CoV on the promoter activity of CCL2, a further incubation with the SARS-CoV VLP was performed at 48 h posttransfection. Transfection of the reporter plasmid was performed with Lipofectamine 2000 reagent (Invitrogen) as described by the manufacturer. The cells were lysed with radioimmunoprecipitation assay (RIPA) buffer (50 mM Tris-HCl, 1% NP-40, 0.05% sodium deoxycholate, 150 mM NaCl, and 1 mM EDTA), supplemented with protease inhibitor cocktail (Roche) and phenylmethylsulfonyl fluoride (PMSF), and subjected to the analysis of firefly luciferase activity and Renilla luciferase activity according to the manufacturer's protocol (Dual-Glo Luciferase Assay System; Promega). The firefly luciferase activity was normalized against the control Renilla luciferase activity.
Western blot analysis, immunoprecipitation, and enzyme-linked immunosorbent assay (ELISA).
For detection of proteins expressed in cultured cells, cells were lysed in RIPA buffer supplemented with phosphatase inhibitor (1 mM NaF and 1 mM NaVO3), protease inhibitor cocktail, and PMSF. The protein lysates were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis, Western blot analysis (3), and immunoprecipitation (26), as described previously. For quantification of CCL2 protein, the culture medium was subjected to analysis with an MCP1/CCL2 ELISA kit (Biosource) following the procedures described by the manufacturer.
Antibodies and kinase inhibitors.
Antibodies specific to phospho-ERK1/2, ERK1/2, phospho-Ser/Thr (MPM-2), and phosphotyrosine (clone 4G10) were purchased from Upstate. Antibodies to phospho-Jun N-terminal protein kinase (JNK), JNK, phospho-p38, p38, and IκBα were purchased from Cell Signaling. Antibody to ACE2 was purchased from R&D Systems, and antibody specific to tubulin was purchased from Sigma. Antibodies to c-Fos and the His epitope were purchased from Santa Cruz, and antibody to c-Jun was purchased from Calbiochem. Antibody to the V5 epitope was purchased from Invitrogen. MEK inhibitor PD98059 and Raf inhibitor GW5074 were purchased from Tocris. JNK inhibitor SP600125, p38 inhibitor SB203580, and Ras inhibitor manumycin A were purchased from Calbiochem. The casein kinase II inhibitor 5,6-dichlorobenzimidazole ribose (DRB) was purchased from Sigma.
RESULTS
Upregulation of CCL2 in SARS-CoV VLP-pretreated A549 and Vero E6 cells.
Previous studies have demonstrated the presence of SARS-CoV RNAs and antigens in the type II pneumocytes of SARS patients (21, 39). To mimic SARS-CoV infection, SARS-CoV VLPs were generated and incubated with human type II pneumocyte (A549) cells. Total RNA was isolated from the A549 cells 24 h after incubation with SARS-CoV VLPs and subjected to microarray analysis. Gene expression profiles were analyzed by GCOS and RMA statistics analysis. This revealed upregulation of 124 genes and downregulation of 128 genes by more than 2-fold in A549 cells treated with SARS-CoV VLPs at VLP-to-cell ratios of 10:1 and 100:1 compared to genes expressed in control cells. (For well-established genes, see Table S2 at http://homepage.ntu.edu.tw/∼f90442001.) The differentially expressed genes are involved in humoral immune responses, apoptosis, cell cycle, and transcriptional regulation. Among those involved in humoral immune responses, CCL2 had mRNA levels elevated by nearly 3-fold whether the cells were treated at a VLP-to-cell ratio of 10:1 or 100:1. The upregulation of CCL2 mRNA was confirmed by RT-quantitative real-time PCR in A549 and Vero E6 cells pretreated with SARS-CoV VLPs (Fig. 2A). In addition, the upregulation is likely to be mediated by the viral spike protein. Preincubation of both A549 and Vero E6 cells with purified SARS-CoV spike protein upregulated the levels of CCL2 mRNA, consistent with the results in cells pretreated with the SARS-CoV VLPs consisting of the viral S, M, and E proteins. In contrast, there was no significant difference in expression levels of CCL2 between control cells and cells pretreated with SARS-CoV(EM) VLPs that lack the viral spike protein (Fig. 2A). Furthermore, upregulation of CCL2 mRNA was detected in A549 cells 4 h after the addition of the SARS-CoV VLPs or purified spike protein, and the elevated CCL2 mRNA level lasted for at least 20 h (Fig. 2B). When an ELISA was performed, upregulation of CCL2 protein was detected in culture media of both the SARS-CoV VLP-treated and the spike protein-pretreated A549 cells (Fig. 2C). This upregulation is consistent with the pulmonary fibrosis-associated CCL2 elevation detected in the sera of SARS patients (16) and in the SARS-CoV-infected A549 cells (44). Taken together, these results indicate an important role of CCL2 in the pathogenesis of SARS-CoV. Cellular signaling pathways involved in the SARS-CoV spike protein-induced CCL2 upregulation were further examined.
FIG. 2.
The elevation of CCL2 expression in SARS-CoV VLP-treated and SARS-CoV spike protein-treated A549 and Vero E6 cells. RNA was isolated from A549 cells at 24 h (A) or at indicated time points (B) after the treatment of purified SARS-CoV VLPs or SARS-CoV(EM) VLPs at a VLP-to-cell ratio of 10:1 or of the viral spike protein (1 μg/106 cells) as indicated. The level of CCL2 mRNA was determined by quantitative real-time PCR. In panel A, the CCL2 mRNA in Vero E6 cells was analyzed as a comparison. In addition, culture medium of the A549 cells was collected at 24 h after the treatment and subjected to ELISA to determine the level of secreted CCL2 (C). Cells without any of the treatments were analyzed in parallel as the controls. The results shown represent the means plus standard deviations (error bars) from four independent experiments. ***, P < 0.001; **, P < 0.01 (for comparisons between the treated and untreated control cells).
Transcriptional regulation of CCL2 in A549 cells pretreated with SARS-CoV VLPs or with the viral spike protein.
To understand the mechanisms by which CCL2 is upregulated in SARS patients, we first examined whether SARS-CoV infection was directly involved in the transcriptional regulation of CCL2.
The CCL2 promoter has distal and proximal regulatory regions. In addition to two distal NF-κB binding sites, there are one NF-κB binding site, two AP-1 binding sites, and one Sp1 binding site in the proximal regulatory region (33). The Sp1 binding site is critical for the basal transcriptional activity of CCL2. The proximal NF-κB and AP-1 binding sites are essential for the transcriptional activities of the CCL2 promoter induced in response to cytokine and tetradecanoyl phorbol acetate (TPA), respectively (28, 33).
To identify cis elements involved in the SARS-CoV-induced upregulation of CCL2, A549 cells were transfected with luciferase reporter plasmid pGLM-PRM which contains the wild-type proximal regulatory region of the human CCL2 promoter from −107 to +60 (Fig. 3A) and then incubated with SARS-CoV VLPs. Results from the luciferase assay demonstrated induction of CCL2 promoter activity by more than 2-fold compared with expression in control cells without incubation with SARS-CoV VLPs (Fig. 3B). Interestingly, the induction of CCL2 promoter activity by SARS-CoV VLPs was abolished when transfection was performed with plasmid pGLM-PRM-mT1T2 in which both AP-1 binding sites in the proximal regulatory region have been mutated. Furthermore, the CCL2 promoter with a mutated NF-κB binding site retained the ability to be induced by SARS-CoV VLPs. On the other hand, when A549 cells were transfected with the reporter plasmid pNFκB-Luc that carries repeated NF-κB binding sites in the promoter, no difference in the luciferase activity was detected whether the cells were treated with SARS-CoV VLPs or not. Western blot analysis further demonstrated SARS-CoV VLP-induced upregulation of both c-Fos and c-Jun, which constitute AP-1 (Fig. 4). These results support the role of AP-1 in trans-activating the CCL2 promoter upon SARS-CoV infection.
FIG. 3.
The involvement of AP-1 in the SARS-CoV VLP-induced upregulation of CCL2. (A) Illustration of the luciferase reporter plasmids containing the proximal regulatory region of the human CCL2 gene. pGLM-PRM-mT1T2 and pGLM-PRM-mκB plasmids that contain mutated (m) AP-1 and NF-κB binding sites are indicated. (B) Luciferase reporter assay. A549 cells were incubated for 24 h with SARS-CoV VLPs at 2 days posttransfection of the luciferase reporter plasmids, as indicated. Cell lysates were harvested and subjected to a luciferase reporter assay. The results shown represent the means plus standard deviations (error bars) from four independent experiments.
FIG. 4.
The increased expression of c-Fos and c-Jun in SARS-CoV VLP-treated A549 cells. Cell lysates prepared from A549 cells at various time points after treatment of SARS-CoV VLPs were subjected to Western blot analysis with antibodies against c-Fos, c-Jun, and tubulin as indicated. Samples indicated as 0 min represent controls in which cells were washed with fresh medium right after treatment with the SARS-CoV VLPs. Relative intensity (RI) shown was calculated by normalization of the intensities from the controls.
MAPK pathway is involved in the AP-1-mediated, SARS-CoV VLP-induced upregulation of CCL2 expression.
The transcriptional activity of AP-1 can be regulated by mitogen-activated protein kinases (MAPKs) (17). To further examine the mechanism involved in the AP-1-mediated, SARS-CoV VLP-induced upregulation of CCL2, A549 cells pretreated with SARS-CoV VLPs or the viral spike protein were analyzed for the activity of MAPKs. As shown in Fig. 5A, ERK1/2 was highly phosphorylated in A549 cells at 15 min postincubation with SARS-CoV VLPs. On the other hand, JNK was activated to a lesser extent, and no significant difference was detected in the phosphorylation status of p38 with or without incubation with the SARS-CoV VLPs. Similar results were observed in A549 cells pretreated with the viral spike protein (Fig. 5B). In addition, phosphorylation of ERK1/2 and JNK, but not p38, was elevated in Vero E6 cells at 15 min postincubation with SARS-CoV VLPs (Fig. 6). These data strongly support the role of specific MAPKs in the SARS-CoV-induced upregulation of the CCL2 signaling pathway.
FIG. 5.
The role of MAPK pathway in the SARS-CoV-induced upregulation of CCL2 in A549 cells. A549 cells were treated with SARS-CoV VLPs (A) or with the viral spike protein (B) for various time periods. Cell lysates were then prepared and subjected to Western blot analysis with antibodies against phospho-ERK1/2 (p-ERK), ERK1/2 (ERK), phospho-JNK (p-JNK), JNK, phospho-p38 (p-p38), and p38 as indicated. Samples indicated as 0 min represent controls in which cells were washed with fresh medium right after treatment with the SARS-CoV VLPs or the viral spike protein as indicated. Relative phosphorylated levels of ERK, JNK, and p38 are shown.
FIG. 6.
The role of the MAPK pathway in SARS-CoV-induced upregulation of CCL2 in Vero E6 cells. Vero E6 cells were treated with SARS-CoV VLPs for various time periods. Cell lysates were then prepared and subjected to Western blot analysis with antibodies against phospho-ERK1/2 (p-ERK) and ERK1/2 (ERK) (A), phospho-JNK (p-JNK) and JNK (B), and phospho-p38 (p-p38) and p38 (C) as indicated. Samples marked time zero represent controls as described in the legend of Fig. 5.
The roles of ERK and JNK in SARS-CoV-induced upregulation of CCL2 were confirmed by MAPK inhibitors. Pretreatment of A549 cells with the MAPK inhibitors PD98059 and SP600125 abolished, respectively, the phosphorylation of ERK1/2 and JNK induced by SARS-CoV VLPs and spike protein (Fig. 7). In addition, neither SARS-CoV VLPs nor the viral spike protein had a significant effect on IκB degradation (Fig. 8). Specific MAPKs that confer the SARS-CoV-induced upregulation of CCL2 were further examined by reporter assay following a transfection of A549 cells with luciferase reporter plasmids and pretreatment of MAPK inhibitors prior to the incubation with SARS-CoV VLPs. The results demonstrated that MEK inhibitor PD98059 completely abolished the VLP-induced CCL2 promoter activity, but the JNK inhibitor had only a slight effect (Fig. 9), indicating that SARS-CoV upregulates expression of CCL2 mainly via the ERK1/2-AP-1 signaling pathway.
FIG. 7.
The effects of ERK and JNK inhibitors on SARS-CoV-induced activation of MAPK. A549 cells were pretreated with MEK inhibitor PD98059 (20 μM; PD) or JNK inhibitor SP600125 (30 μM; SP) for 1 h before incubation with SARS-CoV VLPs (A) or with the viral spike protein (B) for 2 h. Cell lysates were prepared and subjected to Western blot analysis with antibodies against phospho-ERK1/2 (p-ERK), ERK1/2 (ERK), phospho-JNK (p-JNK), and JNK as indicated.
FIG. 8.
IκB-NF-κB signaling pathway is not involved in SARS-CoV-induced CCL2 expression. A549 cells were treated with SARS-CoV VLPs (A) or with the viral spike protein (B) for various time periods as indicated. Cell lysates were prepared and subjected to Western blot analysis with specific antibodies against IκBα and tubulin as indicated. Samples marked time zero represent controls as described in the legend to Fig. 5.
FIG. 9.
ERK1/2 regulate the promoter activity of CCL2 induced by SARS-CoV VLPs. At 2 days posttransfection of A549 cells with pGLM-PRM plasmid, the cells were treated with the inhibitor PD98059 (PD) or SP600125 (SP) for 1 h prior to a further incubation with SARS-CoV VLPs for 24 h. A luciferase reporter assay was performed. The results shown represent the means plus standard deviations (error bars) from four independent experiments. DMSO, dimethyl sulfoxide.
The Ras-Raf-MEK-ERK signaling cascade mediates signal transduction from cell surface to nucleus and regulates specific gene expression in response to extracellular stimuli. This signaling cascade plays a part in biological processes including cell growth, apoptosis, and cell cycle progression (2, 22). To identify the key player that activates ERK1/2 and mediates the SARS-CoV VLP-induced CCL2 expression, A549 cells were treated with Ras or Raf inhibitor prior to incubation with SARS-CoV VLPs. Western blot analysis demonstrated that both the Ras inhibitor manumycin A and the Raf inhibitor GW5074 diminished the phosphorylation of ERK1/2 induced by SARS-CoV VLPs (Fig. 10A) or the viral spike protein (Fig. 10B). Correspondingly, the SARS-CoV-induced upregulation of CCL2 mRNA was diminished when A549 cells were pretreated with MEK, Ras, or Raf inhibitor (Fig. 10C), but JNK and p38 inhibitors had no obvious effect on SARS-CoV-induced upregulation of CCL2. These results suggest that Ras and Raf are the key players involved in the SARS-CoV-induced activation of ERK1/2 and the upregulation of CCL2.
FIG. 10.
The effects of Ras and Raf inhibitors on the SARS-CoV-induced activation of ERK. A549 cells were pretreated with the Ras inhibitor manumycin A (MA; 3 μM), or Raf inhibitor GW5074 (GW; 1 μM) for 30 min prior to incubation with SARS-CoV VLPs (A) or with the viral spike protein (B) for 2 h. Cell lysates were prepared and subjected to Western blot analysis with antibodies against phospho-ERK1/2 (p-ERK) and ERK1/2 (ERK) as indicated. In addition, total RNA was isolated from A549 cells that had been treated with MAPK cascade inhibitors: MA and GW for 30 min and PD, SP, and p38 inhibitor SB203580 (SB) for 1 h, prior to the 6-h incubation with SARS-CoV VLPs or the viral spike protein. The level of CCL2 mRNA was determined by quantitative real-time PCR (C). The results shown represent the means plus standard deviations (error bars) from three independent experiments.
Role of ACE2 involved in SARS-CoV VLP- and spike protein-induced CCL2 upregulation.
ACE2 has been identified as the major receptor for SARS-CoV (27). Here, we examined the role of ACE2 involved in the SARS-CoV-induced upregulation of CCL2 with anti-ACE2-specific antibodies. As shown in Fig. 11A, upregulation of CCL2 mRNA was blocked in A549 cells pretreated with anti-ACE2 antibodies but not with the control IgG. The anti-ACE2 antibodies also diminished the SARS-CoV VLP- and spike protein-induced phosphorylation of ERK1/2 (Fig. 11B). Taken together, these results suggest that an interaction between SARS-CoV spike protein and the ACE2 receptor mediates the phosphorylation of ERK1/2 and results in the upregulation of CCL2 expression.
FIG. 11.
ACE2 is involved in the SARS-CoV-induced activation of ERK and upregulation of CCL2. (A) SARS-CoV spike protein upregulates the expression of CCL2 via the ACE2 signaling pathway. A549 cells were pretreated with anti-ACE2 antibody (Ab) or control IgG for 20 min prior to the incubation with SARS-CoV VLPs or the viral spike protein for 24 h. Total RNA was isolated for determination of the level of CCL2 mRNA by quantitative real-time PCR. The results shown represent the means plus standard deviations (error bars) from three independent experiments. (B) SARS-CoV spike protein activates ERK via the ACE2 signaling pathway. A549 cells were pretreated with anti-ACE2 antibody for 20 min prior to incubation with SARS-CoV VLPs or the viral spike protein for 2 h. Cell lysates were prepared and subjected to Western blot analysis with antibodies against phospho-ERK1/2 (p-ERK) and ERK1/2 (ERK) as indicated.
ACE2 is a homologue of ACE (38) and is known to be a type I transmembrane protein that plays an important role in the renin-angiotensin system (23). Sequence analysis revealed a potential tyrosine kinase phosphorylation site in ACE2 from amino acid residues 775 to 781 and a casein kinase II phosphorylation site from amino acid residues 787 to 790 (Fig. 12A), but the physiological significance of ACE2 phosphorylation remains unclear. Previous studies demonstrated that, in the presence of small interfering RNA (siRNA) targeted to ACE2, multiplication of SARS-CoV in the permissive Vero E6 cells was abolished (30). In this study, Vero E6 cells pretreated with SARS-CoV VLPs were subjected to immunoprecipitation with anti-phospho-Ser/Thr antibodies and immunodetected with specific anti-ACE2 antibodies. As shown in Fig. 12B, the phosphorylated form of ACE2 was dramatically increased with the treatment of SARS-CoV VLPs. However, in the presence of the casein kinase II inhibitor DRB (37), the VLP-induced ACE2 phosphorylation was significantly reduced. These results indicate that the SARS-CoV VLP induces the casein kinase II-mediated phosphorylation of SARS-CoV receptor ACE2 at Ser-787.
FIG. 12.
Phosphorylation of ACE2 in SARS-CoV VLP-treated Vero E6 cells. (A) The domain structure of ACE2. ACE2 is a type I integral membrane protein with a signal peptide (amino acid residues 1 to 17), an ectodomain (amino acid residues 18 to 740), a transmembrane domain (amino acid residues 741 to 763), and a cytoplasmic domain (amino acid residues 764 to 805). Potential phosphorylation sites for tyrosine kinase (TK; amino acid residues 775 to 781) and casein kinase II (CK II; amino acid residues 787 to 790) in the cytoplasmic domain are indicated. (B) Phosphorylated form of ACE2 in the SARS-CoV-permissive Vero E6 cells. Vero E6 cells were pretreated with the casein kinase II inhibitor DRB (100 μM) for 1 h prior to incubation with SARS-CoV VLPs for various times, as indicated. Cell lysates were prepared and subjected to immunoprecipitation (IP) with anti-phospho-Ser/Thr antibodies, followed by Western blot (WB) analysis with anti-ACE2 antibodies. Cell lysates without the process of immunoprecipitation (10% of input) were analyzed in parallel as loading controls. Lane C represents protein lysate prepared from untreated Vero E6 cells as an ACE2 positive control.
The role of casein kinase II in the ACE2 signaling pathway induced by SARS-CoV was further examined by pretreating A549 cells with the casein kinase II inhibitor DRB before incubation with SARS-CoV VLPs or the viral spike protein. As shown in Fig. 13, DRB blocked the upregulation of CCL2 (panel A) and inhibited the phosphorylation of ERK1/2 induced by SARS-CoV VLPs or by the viral spike protein (panel B). These results suggest that casein kinase II-mediated phosphorylation of ACE2 at Ser-787 is critical for the activation of ERK1/2 and upregulation of CCL2 mediated by the SARS-CoV VLPs.
FIG. 13.
Critical role of casein kinase II in the SARS-CoV-induced ERK activation and upregulation of CCL2 in A549 cells. A549 cells were pretreated with the casein kinase II inhibitor DRB for 1 h. Following a 6-h incubation of the DRB-treated cells with SARS-CoV VLPs or the viral spike protein, total RNA was isolated to determine the level of CCL2 mRNA by quantitative real-time PCR (A). The results shown represent the means plus standard deviations (error bars) from four independent experiments. Meanwhile, cell lysates were prepared from the DRB-treated cells following a 2-h incubation with SARS-CoV VLPs or the viral spike protein and subjected to Western blot analysis with antibodies against phospho-ERK1/2 (p-ERK) and ERK1/2 (ERK) as indicated (B).
DISCUSSION
In this study, we have demonstrated the upregulation of CCL2 in both SARS-CoV VLP-treated and the viral spike protein-treated human pneumocyte (A549) cells and monkey kidney (Vero E6) cells. These results are consistent with the observation that an elevated CCL2 level was detected in the sera of SARS patients (16) and in the SARS-CoV-infected A549 cells (44). The AP-1 cis elements, but not the NF-κB binding site, are involved in the regulation of CCL2 expression. SARS-CoV VLPs induced ERK1/2 activation mediated by the Ras, Raf, and MEK cascade through interacting with the SARS-CoV receptor ACE2. ACE2 could be activated by casein kinase II when cells were incubated with SARS-CoV VLPs. These data reveal a novel ACE2 signaling pathway with physiological and pathological significance.
AP-1, NF-κB, and Sp1, which interact with the cis elements on the proximal regulatory region of CCL2 promoter, have been predicted to be responsible for the regulation of CCL2 expression. But AP-1 was suggested to be the major transcription activator of CCL2 (6, 42). In this study, we have demonstrated that AP-1 is involved in SARS-CoV-induced CCL2 upregulation. SARS-CoV VLPs increased the transcriptional activity of AP-1 via upregulation of c-Fos and c-Jun. Previous studies indicated that the expression and activity of c-Fos and c-Jun are regulated by the MAPK family in response to environmental stimuli. Phosphorylation of c-Fos at Thr-232, -325, and -331 and Ser-374 and of c-Jun at Ser-63 and -73 increases the transcriptional activity of c-Fos and c-Jun (32, 34). In addition, ERK1/2 involves in the upregulation of c-Fos via transactivation of the ternary complex factor (TCF) (17). Here, we have demonstrated the activation of AP-1 and ERK1/2 by SARS-CoV VLPs and by the viral spike protein. Phosphorylated sites of c-Fos and c-Jun critical for the SARS-CoV-mediated upregulation of CCL2 need to be determined. Interestingly, previous studies also demonstrated the involvement of SARS-CoV nucleocapsid protein in the activation of the AP-1 signaling pathway (13). These data indicate that the viral spike and nucleocapsid proteins may coordinately regulate the expression of CCL2 and that its upregulation by the viral proteins results in severe lung inflammatory disorders in SARS patients.
ACE2 is a homologue of ACE but plays an opposite role to ACE in the renin-angiotensin system (23). ACE converts angiotensin I (Ang I) to Ang II and induces vasoconstriction, whereas ACE2 cleaves Ang II to Ang-(1-7) and induces vasodilation and apoptosis. Amino acid sequence analysis revealed potential phosphorylation sites for protein kinase C (PKC), protein kinase A, and casein kinase II in the short cytoplasmic domain of ACE, but only a casein kinase II-mediated phosphorylation at Ser-1270 was evident (19). The phosphorylation triggers retention of ACE on the plasma membrane. On the other hand, earlier studies have demonstrated that tumor necrosis factor alpha convertase (ADAM17, or TACE) can mediate ACE2 shedding. In addition, SARS-CoV spike protein can induce ACE2 shedding in the presence of the convertase activity of ADAM17 (11, 24). In this study, we have identified a SARS-CoV-induced, casein kinase II-mediated phosphorylation of ACE2 at Ser-787. Whether the phosphorylation of ACE2 initiates shedding and thus decreases the level of ACE2 needs to be further elucidated.
ACE is not just a C-terminal peptidyl dipeptidase in the renin-angiotensin system; it can also initiate outside-in signaling. ACE interacts with JNK and MKK7 (18). Phosphorylation of ACE at Ser-1270 activated JNK and phosphorylated c-Jun. The present study has demonstrated that SARS-CoV could induce CCL2 upregulation through the activation of ERK1/2 and AP-1. In addition, phosphorylation of ACE2 by casein kinase II is involved in ERK1/2 activation. A possible mechanism by which SARS-CoV induces CCL2 expression was proposed (Fig. 14). Casein kinase II phosphorylates ACE2 at Ser-787 upon binding of SARS-CoV to its ACE2 receptor. The binding causes a conformational change of ACE2 and activates the ACE2 downstream signal transduction pathways with which ERK1/2 and AP-1 are involved. ACE2 may be involved in an intracellular signaling pathway to activate ERK1/2 via interacting with upstream factors such as Ras, phosphatidylinositol 3-kinase (PI3K), and PKC. Detailed mechanisms would need further investigation.
FIG. 14.
A current model of the ACE2 signaling pathway involved in SARS-CoV-induced CCL2 expression. Infection of lung epithelial cells with SARS-CoV induces casein kinase II (CK II)-mediated phosphorylation of the ACE2 receptor, leading to the activation of ERK1/2 and AP-1 and the upregulation of CCL2. Ras, Raf, and MEK may contribute to the SARS-CoV-induced ERK1/2 activation and CCL2 upregulation. The elevated level of CCL2 protein detected in the sera of SARS-CoV-infected patients may account for the development of lung fibrosis.
The current study is the first report demonstrating a potential role of the phosphorylated form of ACE2 as a signal transducer on the plasma membrane. Our findings also suggest a possible mechanism of SARS-CoV-induced lung fibrosis via ACE2-mediated intracellular signal transduction. These results may provide new targets for drug development against SARS-CoV infection.
Acknowledgments
We thank C.-F. Lo (National Taiwan University) for providing the Sf9 cell line and baculoviruses and Z.-F. Chang (National Taiwan University) and T. Yoshimura (National Cancer Institute) for providing luciferase reporter plasmids. We are grateful to Z.-F. Chang, T.-L. Tseng (National Taiwan University), and Jeffrey J.-Y. Yen (Academia Sinica, Taiwan) for helpful comments on this paper.
This work was supported by research grants NSC92-3112-B-002-009 and NSC93-2751-B-002-003-Y from the National Science Council of the Republic of China.
Footnotes
Published ahead of print on 19 May 2010.
REFERENCES
- 1.Castilletti, C., L. Bordi, E. Lalle, G. Rozera, F. Poccia, C. Agrati, I. Abbate, and M. R. Capobianchi. 2005. Coordinate induction of IFN-alpha and -gamma by SARS-CoV also in the absence of virus replication. Virology 341:163-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chang, L., and M. Karin. 2001. Mammalian MAP kinase signalling cascades. Nature 410:37-40. [DOI] [PubMed] [Google Scholar]
- 3.Chang, M. F., C. Y. Sun, C. J. Chen, and S. C. Chang. 1993. Functional motifs of delta antigen essential for RNA binding and replication of hepatitis delta virus. J. Virol. 67:2529-2536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chang, Y. J., C. Y. Liu, B. L. Chiang, Y. C. Chao, and C. C. Chen. 2004. Induction of IL-8 release in lung cells via activator protein-1 by recombinant baculovirus displaying severe acute respiratory syndrome-coronavirus spike proteins: identification of two functional regions. J. Immunol. 173:7602-7614. [DOI] [PubMed] [Google Scholar]
- 5.Cheung, C. Y., L. L. Poon, I. H. Ng, W. Luk, S. F. Sia, M. H. Wu, K. H. Chan, K. Y. Yuen, S. Gordon, Y. Guan, and J. S. Peiris. 2005. Cytokine responses in severe acute respiratory syndrome coronavirus-infected macrophages in vitro: possible relevance to pathogenesis. J. Virol. 79:7819-7826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cho, N. H., S. Y. Seong, M. S. Huh, N. H. Kim, M. S. Choi, and I. S. Kim. 2002. Induction of the gene encoding macrophage chemoattractant protein 1 by Orientia tsutsugamushi in human endothelial cells involves activation of transcription factor activator protein 1. Infect. Immun. 70:4841-4850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chomczynski, P., and N. Sacchi. 1987. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 162:156-159. [DOI] [PubMed] [Google Scholar]
- 8.Drosten, C., S. Gunther, W. Preiser, S. van der Werf, H. R. Brodt, S. Becker, H. Rabenau, M. Panning, L. Kolesnikova, R. A. Fouchier, A. Berger, A. M. Burguiere, J. Cinatl, M. Eickmann, N. Escriou, K. Grywna, S. Kramme, J. C. Manuguerra, S. Muller, V. Rickerts, M. Sturmer, S. Vieth, H. D. Klenk, A. D. Osterhaus, H. Schmitz, and H. W. Doerr. 2003. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N. Engl. J. Med. 348:1967-1976. [DOI] [PubMed] [Google Scholar]
- 9.Gharaee-Kermani, M., E. M. Denholm, and S. H. Phan. 1996. Costimulation of fibroblast collagen and transforming growth factor beta1 gene expression by monocyte chemoattractant protein-1 via specific receptors. J. Biol. Chem. 271:17779-17784. [DOI] [PubMed] [Google Scholar]
- 10.Gu, L., S. Tseng, R. M. Horner, C. Tam, M. Loda, and B. J. Rollins. 2000. Control of TH2 polarization by the chemokine monocyte chemoattractant protein-1. Nature 404:407-411. [DOI] [PubMed] [Google Scholar]
- 11.Haga, S., N. Yamamoto, C. Nakai-Murakami, Y. Osawa, K. Tokunaga, T. Sata, T. Sasazuki, and Y. Ishizaka. 2008. Modulation of TNF-alpha-converting enzyme by the spike protein of SARS-CoV and ACE2 induces TNF-alpha production and facilitates viral entry. Proc. Natl. Acad. Sci. U. S. A. 105:7809-7814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.He, L., Y. Ding, Q. Zhang, X. Che, Y. He, H. Shen, H. Wang, Z. Li, L. Zhao, J. Geng, Y. Deng, L. Yang, J. Li, J. Cai, L. Qiu, K. Wen, X. Xu, and S. Jiang. 2006. Expression of elevated levels of pro-inflammatory cytokines in SARS-CoV-infected ACE2+ cells in SARS patients: relation to the acute lung injury and pathogenesis of SARS. J. Pathol. 210:288-297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.He, R., A. Leeson, A. Andonov, Y. Li, N. Bastien, J. Cao, C. Osiowy, F. Dobie, T. Cutts, M. Ballantine, and X. Li. 2003. Activation of AP-1 signal transduction pathway by SARS coronavirus nucleocapsid protein. Biochem. Biophys. Res. Commun. 311:870-876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hsueh, P. R., C. H. Hsiao, S. H. Yeh, W. K. Wang, P. J. Chen, J. T. Wang, S. C. Chang, C. L. Kao, and P. C. Yang. 2003. Microbiologic characteristics, serologic responses, and clinical manifestations in severe acute respiratory syndrome, Taiwan. Emerg. Infect. Dis. 9:1163-1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jeffers, S. A., S. M. Tusell, L. Gillim-Ross, E. M. Hemmila, J. E. Achenbach, G. J. Babcock, W. D. Thomas, Jr., L. B. Thackray, M. D. Young, R. J. Mason, D. M. Ambrosino, D. E. Wentworth, J. C. Demartini, and K. V. Holmes. 2004. CD209L (L-SIGN) is a receptor for severe acute respiratory syndrome coronavirus. Proc. Natl. Acad. Sci. U. S. A. 101:15748-15753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jiang, Y., J. Xu, C. Zhou, Z. Wu, S. Zhong, J. Liu, W. Luo, T. Chen, Q. Qin, and P. Deng. 2005. Characterization of cytokine/chemokine profiles of severe acute respiratory syndrome. Am. J. Respir. Crit. Care. Med. 171:850-857. [DOI] [PubMed] [Google Scholar]
- 17.Karin, M. 1995. The regulation of AP-1 activity by mitogen-activated protein kinases. J. Biol. Chem. 270:16483-16486. [DOI] [PubMed] [Google Scholar]
- 18.Kohlstedt, K., R. P. Brandes, W. Muller-Esterl, R. Busse, and I. Fleming. 2004. Angiotensin-converting enzyme is involved in outside-in signaling in endothelial cells. Circ. Res. 94:60-67. [DOI] [PubMed] [Google Scholar]
- 19.Kohlstedt, K., F. Shoghi, W. Muller-Esterl, R. Busse, and I. Fleming. 2002. CK2 phosphorylates the angiotensin-converting enzyme and regulates its retention in the endothelial cell plasma membrane. Circ. Res. 91:749-756. [DOI] [PubMed] [Google Scholar]
- 20.Ksiazek, T. G., D. Erdman, C. S. Goldsmith, S. R. Zaki, T. Peret, S. Emery, S. Tong, C. Urbani, J. A. Comer, W. Lim, P. E. Rollin, S. F. Dowell, A. E. Ling, C. D. Humphrey, W. J. Shieh, J. Guarner, C. D. Paddock, P. Rota, B. Fields, J. DeRisi, J. Y. Yang, N. Cox, J. M. Hughes, J. W. LeDuc, W. J. Bellini, and L. J. Anderson. 2003. A novel coronavirus associated with severe acute respiratory syndrome. N. Engl. J. Med. 348:1953-1966. [DOI] [PubMed] [Google Scholar]
- 21.Kuiken, T., R. A. Fouchier, M. Schutten, G. F. Rimmelzwaan, G. van Amerongen, D. van Riel, J. D. Laman, T. de Jong, G. van Doornum, W. Lim, A. E. Ling, P. K. Chan, J. S. Tam, M. C. Zambon, R. Gopal, C. Drosten, S. van der Werf, N. Escriou, J. C. Manuguerra, K. Stohr, J. S. Peiris, and A. D. Osterhaus. 2003. Newly discovered coronavirus as the primary cause of severe acute respiratory syndrome. Lancet 362:263-270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kyriakis, J. M., and J. Avruch. 2001. Mammalian mitogen-activated protein kinase signal transduction pathways activated by stress and inflammation. Physiol. Rev. 81:807-869. [DOI] [PubMed] [Google Scholar]
- 23.Lambert, D. W., N. M. Hooper, and A. J. Turner. 2008. Angiotensin-converting enzyme 2 and new insights into the renin-angiotensin system. Biochem. Pharmacol. 75:781-786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lambert, D. W., M. Yarski, F. J. Warner, P. Thornhill, E. T. Parkin, A. I. Smith, N. M. Hooper, and A. J. Turner. 2005. Tumor necrosis factor-alpha convertase (ADAM17) mediates regulated ectodomain shedding of the severe-acute respiratory syndrome-coronavirus (SARS-CoV) receptor, angiotensin-converting enzyme-2 (ACE2). J. Biol. Chem. 280:30113-30119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Law, H. K., C. Y. Cheung, H. Y. Ng, S. F. Sia, Y. O. Chan, W. Luk, J. M. Nicholls, J. S. Peiris, and Y. L. Lau. 2005. Chemokine up-regulation in SARS-coronavirus-infected, monocyte-derived human dendritic cells. Blood 106:2366-2374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee, C. H., S. C. Chang, C. J. Chen, and M. F. Chang. 1998. The nucleolin binding activity of hepatitis delta antigen is associated with nucleolus targeting. J. Biol. Chem. 273:7650-7656. [DOI] [PubMed] [Google Scholar]
- 27.Li, W., M. J. Moore, N. Vasilieva, J. Sui, S. K. Wong, M. A. Berne, M. Somasundaran, J. L. Sullivan, K. Luzuriaga, T. C. Greenough, H. Choe, and M. Farzan. 2003. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature 426:450-454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li, Y. S., and P. E. Kolattukudy. 1994. Functional role of the cis-acting elements in human monocyte chemotactic protein-1 gene in the regulation of its expression by phorbol ester in human glioblastoma cells. Mol. Cell. Biochem. 141:121-128. [DOI] [PubMed] [Google Scholar]
- 29.Lin, S., C. K. Lee, S. Y. Lee, C. L. Kao, C. W. Lin, A. B. Wang, S. M. Hsu, and L. S. Huang. 2005. Surface ultrastructure of SARS coronavirus revealed by atomic force microscopy. Cell. Microbiol. 7:1763-1770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lu, C. Y., H. Y. Huang, T. H. Yang, L. Y. Chang, C. Y. Lee, and L. M. Huang. 2008. siRNA silencing of angiotensin-converting enzyme 2 reduced severe acute respiratory syndrome-associated coronavirus replications in Vero E6 cells. Eur. J. Clin. Microbiol. Infect. Dis. 27:709-715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marra, M. A., S. J. Jones, C. R. Astell, R. A. Holt, A. Brooks-Wilson, Y. S. Butterfield, J. Khattra, J. K. Asano, S. A. Barber, S. Y. Chan, A. Cloutier, S. M. Coughlin, D. Freeman, N. Girn, O. L. Griffith, S. R. Leach, M. Mayo, H. McDonald, S. B. Montgomery, P. K. Pandoh, A. S. Petrescu, A. G. Robertson, J. E. Schein, A. Siddiqui, D. E. Smailus, J. M. Stott, G. S. Yang, F. Plummer, A. Andonov, H. Artsob, N. Bastien, K. Bernard, T. F. Booth, D. Bowness, M. Czub, M. Drebot, L. Fernando, R. Flick, M. Garbutt, M. Gray, A. Grolla, S. Jones, H. Feldmann, A. Meyers, A. Kabani, Y. Li, S. Normand, U. Stroher, G. A. Tipples, S. Tyler, R. Vogrig, D. Ward, B. Watson, R. C. Brunham, M. Krajden, M. Petric, D. M. Skowronski, C. Upton, and R. L. Roper. 2003. The genome sequence of the SARS-associated coronavirus. Science 300:1399-1404. [DOI] [PubMed] [Google Scholar]
- 32.Monje, P., M. J. Marinissen, and J. S. Gutkind. 2003. Phosphorylation of the carboxyl-terminal transactivation domain of c-Fos by extracellular signal-regulated kinase mediates the transcriptional activation of AP-1 and cellular transformation induced by platelet-derived growth factor. Mol. Cell. Biol. 23:7030-7043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ping, D., P. L. Jones, and J. M. Boss. 1996. TNF regulates the in vivo occupancy of both distal and proximal regulatory regions of the MCP-1/JE gene. Immunity 4:455-469. [DOI] [PubMed] [Google Scholar]
- 34.Pulverer, B. J., J. M. Kyriakis, J. Avruch, E. Nikolakaki, and J. R. Woodgett. 1991. Phosphorylation of c-Jun mediated by MAP kinases. Nature 353:670-674. [DOI] [PubMed] [Google Scholar]
- 35.Rose, C. E., Jr., S. S. Sung, and S. M. Fu. 2003. Significant involvement of CCL2 (MCP-1) in inflammatory disorders of the lung. Microcirculation 10:273-288. [DOI] [PubMed] [Google Scholar]
- 36.Rota, P. A., M. S. Oberste, S. S. Monroe, W. A. Nix, R. Campagnoli, J. P. Icenogle, S. Penaranda, B. Bankamp, K. Maher, M. H. Chen, S. Tong, A. Tamin, L. Lowe, M. Frace, J. L. DeRisi, Q. Chen, D. Wang, D. D. Erdman, T. C. Peret, C. Burns, T. G. Ksiazek, P. E. Rollin, A. Sanchez, S. Liffick, B. Holloway, J. Limor, K. McCaustland, M. Olsen-Rasmussen, R. Fouchier, S. Gunther, A. D. Osterhaus, C. Drosten, M. A. Pallansch, L. J. Anderson, and W. J. Bellini. 2003. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science 300:1394-1399. [DOI] [PubMed] [Google Scholar]
- 37.Schubert, U., T. Schneider, P. Henklein, K. Hoffmann, E. Berthold, H. Hauser, G. Pauli, and T. Porstmann. 1992. Human-immunodeficiency-virus-type-1-encoded Vpu protein is phosphorylated by casein kinase II. Eur. J. Biochem. 204:875-883. [DOI] [PubMed] [Google Scholar]
- 38.Tipnis, S. R., N. M. Hooper, R. Hyde, E. Karran, G. Christie, and A. J. Turner. 2000. A human homolog of angiotensin-converting enzyme. Cloning and functional expression as a captopril-insensitive carboxypeptidase. J. Biol. Chem. 275:33238-33243. [DOI] [PubMed] [Google Scholar]
- 39.To, K. F., J. H. Tong, P. K. Chan, F. W. Au, S. S. Chim, K. C. Chan, J. L. Cheung, E. Y. Liu, G. M. Tse, A. W. Lo, Y. M. Lo, and H. K. Ng. 2004. Tissue and cellular tropism of the coronavirus associated with severe acute respiratory syndrome: an in-situ hybridization study of fatal cases. J. Pathol. 202:157-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tsai, C. W., S. C. Chang, and M. F. Chang. 1999. A 12-amino acid stretch in the hypervariable region of the spike protein S1 subunit is critical for cell fusion activity of mouse hepatitis virus. J. Biol. Chem. 274:26085-26090. [DOI] [PubMed] [Google Scholar]
- 41.Ueda, A., Y. Ishigatsubo, T. Okubo, and T. Yoshimura. 1997. Transcriptional regulation of the human monocyte chemoattractant protein-1 gene. Cooperation of two NF-κB sites and NF-κB/Rel subunit specificity. J. Biol. Chem. 272:31092-31099. [DOI] [PubMed] [Google Scholar]
- 42.Wang, N., L. Verna, S. Hardy, J. Forsayeth, Y. Zhu, and M. B. Stemerman. 1999. Adenovirus-mediated overexpression of c-Jun and c-Fos induces intercellular adhesion molecule-1 and monocyte chemoattractant protein-1 in human endothelial cells. Arterioscler. Thromb. Vasc. Biol. 19:2078-2084. [DOI] [PubMed] [Google Scholar]
- 43.Wu, S. C., S. C. Chang, H. Y. Wu, P. J. Liao, and M. F. Chang. 2008. Hepatitis C virus NS5A protein down-regulates the expression of spindle gene Aspm through PKR-p38 signaling pathway. J. Biol. Chem. 283:29396-29404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yen, Y. T., F. Liao, C. H. Hsiao, C. L. Kao, Y. C. Chen, and B. A. Wu-Hsieh. 2006. Modeling the early events of severe acute respiratory syndrome coronavirus infection in vitro. J. Virol. 80:2684-2693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang, Y., J. Li, Y. Zhan, L. Wu, X. Yu, W. Zhang, L. Ye, S. Xu, R. Sun, Y. Wang, and J. Lou. 2004. Analysis of serum cytokines in patients with severe acute respiratory syndrome. Infect. Immun. 72:4410-4415. [DOI] [PMC free article] [PubMed] [Google Scholar]