Abstract
The ubiquitin-proteasome system (UPS) is a key player in regulating the intracellular sorting and degradation of proteins. In this study we investigated the role of the UPS in different steps of the coronavirus (CoV) infection cycle. Inhibition of the proteasome by different chemical compounds (i.e., MG132, epoxomicin, and Velcade) appeared to not only impair entry but also RNA synthesis and subsequent protein expression of different CoVs (i.e., mouse hepatitis virus [MHV], feline infectious peritonitis virus, and severe acute respiratory syndrome CoV). MHV assembly and release were, however, not appreciably affected by these compounds. The inhibitory effect on CoV protein expression did not appear to result from a general inhibition of translation due to induction of a cellular stress response by the inhibitors. Stress-induced phosphorylation of eukaryotic translation initiation factor 2α (eIF2α) generally results in impaired initiation of protein synthesis, but the sensitivity of MHV infection to proteasome inhibitors was unchanged in cells lacking a phosphorylatable eIF2α. MHV infection was affected not only by inhibition of the proteasome but also by interfering with protein ubiquitination. Viral protein expression was reduced in cells expressing a temperature-sensitive ubiquitin-activating enzyme E1 at the restrictive temperature, as well as in cells in which ubiquitin was depleted by using small interfering RNAs. Under these conditions, the susceptibility of the cells to virus infection was, however, not affected, excluding an important role of ubiquitination in virus entry. Our observations reveal an important role of the UPS in multiple steps of the CoV infection cycle and identify the UPS as a potential drug target to modulate the impact of CoV infection.
The cellular ubiquitin-proteasome system (UPS), which is important for intracellular protein degradation in eukaryotic cells, plays a central role in cellular protein homeostasis (59, 64). Since all viruses exploit and manipulate the infrastructure and metabolism of their host cell to their own advantage, it is not surprising that the UPS has also been implicated in the infection cycle and virus-host interplay of several viruses (7, 14, 48, 52, 70).
The UPS controls many different processes, including the regulation of cell cycle progression, apoptosis, and antigen presentation (17). Proteins destined for proteasomal degradation are conjugated with chains of the small protein ubiquitin, which constitute the recognition motif for the proteasome (21). In addition to targeting proteins for degradation, conjugation with ubiquitin can also regulate intracellular protein sorting, as has been described for numerous membrane proteins (22). Attachment of ubiquitin moieties to protein substrates occurs by the sequential action of three enzymes. First, the ubiquitin-activating enzyme E1 forms a high-energy thiolester bond with ubiquitin, after which ubiquitin is transferred to the ubiquitin-conjugating enzyme E2. Subsequently, the ubiquitin is conjugated to a lysine side chain or to the N terminus of the substrate by the corporate action of E2 and an E3 ubiquitin ligase, with the latter enzyme determining the substrate specificity of the process. Subsequently, the UPS targets these polyubiquitinated substrates to the catalytic 20S core complex of the proteasome, which subsequently cleaves them into smaller peptides. The proteasome controls not only hydrolysis of functionally active proteins but also the degradation of misfolded polypeptides.
Coronaviruses (CoVs) are enveloped, positive-strand RNA viruses and are common pathogens in many animal species. With a size of 28 to 32 kb, CoVs have the largest genome among RNA viruses known to date. Several CoVs cause severe disease in animals, including porcine transmissible gastroenteritis virus, bovine coronavirus, avian infectious bronchitis viruses, and feline infectious peritonitis virus (FIPV). With the discovery of new human CoVs (HCoVs), such as the severe acute respiratory syndrome (SARS)-CoV (15), HCoV-NL63 (62), and HCoV-HKU1 (67), interest in CoV research has significantly increased. The well-studied mouse hepatitis virus (MHV) is often used as a model CoV.
The CoV infection cycle starts with the attachment of the virus to a specific cellular receptor. The spike (S) protein, a class I fusion protein, is responsible for virus entry by mediating both receptor binding and the subsequent fusion of the viral envelope with a host membrane (6, 10). After virus entry, the viral genome is released into the cytosol of the cell, where it is translated into two large replicase polyproteins. These are autoproteolytically processed to produce 15 or 16 mature nonstructural proteins (nsp's), which assemble into viral replication-transcription complexes that are thought to be associated with a virus-induced network of modified endoplasmic reticulum (ER) membranes, which includes double membrane vesicles and other unusual membrane structures (18, 31, 54, 63). Subsequently, a nested set of (sub)genomic mRNAs is produced (42), which are translated into the viral structural and accessory proteins. Together with the newly synthesized genomic RNA, the structural proteins assemble into progeny virions by budding through membranes of the ER-to-Golgi intermediate compartment (ERGIC) (32). The newly synthesized virions are subsequently released by exocytosis.
In the present study we investigated the importance of the UPS during CoV infection. Besides a previous study reporting that inhibition of the proteasome affected MHV entry (68), no comprehensive analysis of the involvement of the UPS in the CoV replicative cycle has been performed until now. Here, we interfered with the UPS either by treating cells with chemical inhibitors of the proteasome, by using cells that express a temperature-sensitive form of the ubiquitin-activating enzyme E1, or by knockdown of ubiquitin synthesis with small interfering RNAs (siRNAs). Whereas CoV RNA synthesis and subsequent protein expression were severely reduced under all experimental conditions tested, virus entry appeared only to be affected by the chemical inhibitors of the proteasome.
MATERIALS AND METHODS
Cells and viruses.
Murine LR7 (34), feline FCWF, and Vero-E6 cells were used to propagate the viruses (i.e., recombinant MHV, FIPV, and SARS-CoV, respectively) and for infection experiments. All work with live SARS-CoV was performed inside biosafety cabinets in the biosafety level 3 facility at Leiden University Medical Center. Chinese Hamster-E36 and -ts20 cells (33) were maintained at 31°C in α-minimal essential medium supplemented with 10% (vol/vol) fetal calf serum (Bodinco B.V.), 100 U of penicillin/ml, and 100 μg of streptomycin/ml. The mouse embryonic fibroblasts (MEFs) expressing wild-type or mutant (S51A) eIF2α (50) and HeLa-CEACAM1a (63) cells were maintained in complete Dulbecco modified Eagle medium (Cambrex BioScience) containing 10% (vol/vol) fetal calf serum (Bodinco B.V.), 100 U of penicillin/ml, and 100 μg of streptomycin/ml, supplemented with 1× nonessential amino acids (Invitrogen). MHV-EFLM (13), MHV-nsp2EGFP (63), FIPV-Δ3abcFL (13), and SARS-CoV-GFP (53) were used for the infection experiments.
Chemicals.
Stocks of 10 mM MG132, 1 mM epoxomicin, and 5 mM lactacystin (all obtained from Sigma-Aldrich) were prepared in dimethyl sulfoxide (DMSO). A stock of 1 mM Velcade (Millennium Pharmaceuticals, Inc.) was prepared in phosphate-buffered saline (PBS). All stocks were stored at −20°C.
Luciferase assays.
Cell monolayers infected with FL-expressing viruses were lysed at the indicated times postinfection using the appropriate buffer provided with the firefly luciferase (FL) assay system (Promega). Intracellular luciferase expression was measured according to the manufacturer's instructions, and the relative light units (RLU) were determined with a Berthold Centro LB 960 plate luminometer.
Confocal immunofluorescence microscopy.
Cells were fixed with 4% paraformaldehyde in PBS and subsequently permeabilized with 0.1% Triton X-100 in PBS. When indicated, the MHV-infected cells were incubated for 1 h with the first antibody directed against nsp2-3 (kindly provided by Susan Baker) (25) or against double-stranded RNA (dsRNA; English and Scientific Consulting Bt. [K1]) (51) diluted in PBS containing 10% normal goat serum. After several washing steps, the cells were incubated with an appropriate dilution of secondary antibody in the same buffer for 1 h. After three subsequent washing steps, the coverslips were mounted in FluorSave (Calbiochem). The immunofluorescence staining was analyzed by using a confocal laser-scanning microscope (Leica). Green fluorescent protein (GFP) was excited at 488 nm, Cy3 at 568 nm, and Cy5 at 633 nm.
Metabolic labeling and immunoprecipitation.
At 4.5 h postinfection, the MHV-infected cells were starved for 30 min in cysteine- and methionine-free modified Eagle medium containing 10 mM HEPES (pH 7.2) and 5% dialyzed fetal calf serum. The medium was then replaced by the same medium containing 100 μCi of 35S in vitro cell-labeling mixture (Amersham Biosciences), after which the cells were further incubated for 30 min. Subsequently, the cells were either lysed or incubation was continued with culture medium (chase). The cells were lysed, and cell lysates were subjected to immunoprecipitation as described previously (40), using polyclonal antisera directed against MHV (k135) and the M protein (anti-Mc) (35). Culture supernatants were subjected to immunoprecipitation in the absence of detergents using the A3.10 monoclonal antibody directed against the S protein (66). The immunoprecipitates were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and autoradiography.
Reporter RNA synthesis and transfection.
The reporter plasmid pM5f-RL-M3 (63) was linearized by using a PacI restriction site directly downstream of the poly(A) sequence. Subsequently, RNA transcripts were produced by using the T7 MessageMachine kit (Ambion) according to the manufacturer's instructions. Next, 0.5 pmol of RNA was transfected into cells by using Lipofectamine 2000 (Invitrogen). Cells were treated with 10 μg of MG132/ml or mock treated for 4 h, after which the cells were lysed and Renilla luciferase activity was measured with a Renilla luciferase assay kit (Promega) according to the manufacturer's protocol.
Immunocytochemistry.
Cell monolayers were fixed, permeabilized, and processed for immunocytochemistry as described previously (12). Peroxidase was visualized by using an AEC substrate kit from Vector Laboratories. MHV-positive cells were detected and counted by using bright-field light microscopy.
SARS-CoV nsp5 protease assay.
To obtain recombinant SARS-CoV main protease (Mpro), nsp5 was expressed in Escherichia coli BL21(DE3) from expression vector pMal-SARS-CoV-Mpro-His as the C-terminal domain of a maltose-binding protein (MBP) fusion protein (24). Autocatalytic cleavage of the fusion protein during expression liberated the authentic nsp5 N terminus. At its C terminus, recombinant nsp5 was extended with a His6 tag to allow for affinity chromatography purification of the protein. As a negative control for enzymatic activity, a C145A mutant nsp5 (1) was expressed from the same plasmid (wild-type and mutant plasmids were kindly provided by John Ziebuhr and Tanja Schirmeister). Expression of SARS-CoV wild-type nsp5 and the C145A mutant was induced by using autoinduction medium ZYM-5052 (57), and recombinant proteins were purified by using Talon metal affinity resin beads (Clontech) as previously described (24). Fluorimetric in vitro activity assays were performed at 30°C using a synthetic 8-amino-acid peptide substrate labeled with a fluorescence resonance energy transfer pair (24) and containing a consensus CoV Mpro cleavage site, Val-X-Leu-Asn↓Ser. In a 100-μl reaction volume, 0.43 μg of purified nsp5 and 50 μM the Dabcyl-VRLQSGTC-fluorescein peptide substrate were incubated in a 20 mM Tris-HCl buffer (pH 7.5) containing 0.1 mM EDTA, 1 mM dithiothreitol, 200 mM NaCl, and 12.5% DMSO. The increase in fluorescence (485-nm excitation, 535-nm emission) resulting from cleavage of the substrate was measured for 60 min in a Mithras LB940 fluorimeter (Berthold), in the presence or absence of Velcade. Cleavage rates were calculated by fitting a linear curve through data obtained during the first 10 min of the assay. The cleavage rates are expressed in dF/min (i.e., the change in fluorescence per minute).
Ubiquitin knockdown.
siRNA duplexes targeting different sites within the coding sequences of UBA52 and RPS27A were designed by and obtained from Ambion, Inc. (three siRNAs per gene). Scrambled siRNAs or siRNAs targeting FL (GL2+GL3) (all from Ambion) were taken along as controls in each experiment. One day after seeding, the HeLa-CEACAM1a cells were transfected with a final concentration of 10 nM siRNA using Oligofectamine (Invitrogen). At 72 h after transfection, the cells were inoculated with MHV-EFLM. At 6 h postinfection, the cell number and viability was measured by Wst-1 assay according to the manufacturer's protocol (Roche Diagnostics GmbH). Subsequently, intracellular luciferase expression was determined as described above. Each siRNA experiment was performed in triplicate. For each well, RLU values were corrected for cell number and viability as determined by the Wst-1 assay.
Western blotting.
Depletion of ubiquitin after siRNA transfection was confirmed by Western blotting. To this end, cells were lysed in ice-cold lysis buffer (20 mM morpholinepropanesulfonic acid [pH 7.2], 5 mM EDTA, 2 mM EGTA, and 0.5% [wt/vol] Nonidet P-40, containing 30 mM NaF, 40 mM β-glycerophosphate, 20 mM sodium pyrophosphate, 1 mM sodium orthovanadate, 1 mM phenylmethylsulfonyl fluoride, 3 mM benzamidine, 1.5 μM pepstatin A, and 10 μM leupeptin). Cell lysates were cleared by centrifugation at 100,000 × g at 4°C for 30 min. Proteins present in the cell lysates were separated by SDS-PAGE and transferred to a nitrocellulose membrane (0.1 μM; Schleicher & Schuell). Subsequently, the membrane was incubated overnight in blocking buffer PBS containing 0.05% Tween 20 and 0.5% cold water fish skin gelatin (Sigma-Aldrich). Next, the membrane was washed three times with PBS (containing 0.05% Tween 20) and incubated for 16 h at 4°C with a peroxidase-labeled mouse polyclonal antibody against ubiquitin (P4D1; Santa Cruz Biotechnology, Inc.). After extensive washing of the membrane, the amount of protein was visualized and quantitated by using an Enhanced ChemoLuminescence Plus kit, a Typhoon imager, and ImageQuant TL software (all from Amersham Biosciences). Immunoblotting for endogenous levels of eIF2α and eIF2αP was performed as described previously (43).
RESULTS
MG132 inhibits MHV at multiple steps of the infection cycle.
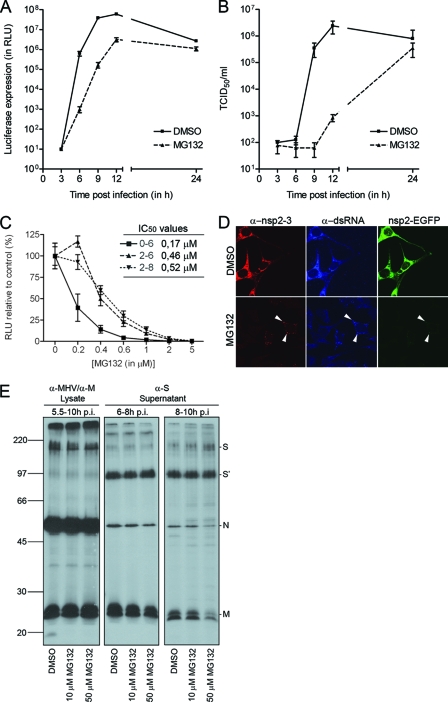
We started our analysis of the role of the UPS in the CoV replicative cycle by studying the one-step growth of MHV strain A59 (MHV-A59) in LR7 cells in the absence or presence of the well-known proteasome inhibitor MG132. In this assay the effect of inhibition of proteasome activity on different steps of the infection cycle was investigated. To this end, LR7 cells were inoculated with MHV-EFLM in the presence or absence of 10 μM MG132. MHV-EFLM is a recombinant virus that expresses the FL reporter gene from a subgenomic mRNA (13). The intracellular FL level, which is an indirect measure for viral RNA synthesis (43, 63), was significantly delayed in the presence of MG132 at 6, 9, and 12 h postinfection (Fig. 1 A). As a likely consequence, the extracellular accumulation of progeny virions at these time points was also dramatically affected (Fig. 1B).
FIG. 1.
MHV infection is reduced by treatment with MG132. (A) LR7 cells were inoculated with MHV-EFLM (multiplicity of infection of 1) in the absence (DMSO control) or presence of 10 μM MG132. After 2 h, the inoculum was replaced with fresh medium containing either DMSO or 10 μM MG132. The luciferase expression levels at the indicated time points were determined. Standard deviations (n = 3) are indicated. (B) In parallel, viral infectivity in the culture media was determined by a quantal assay on LR7 cells. The 50% tissue culture infectious dose (TCID50) values are indicated. (C) LR7 cells were infected with MHV-EFLM in the absence (DMSO control) or presence of different concentrations of MG132. The drug was present from 0 to 6, from 2 to 6, or from 2 to 8 h postinfection. The intracellular luciferase expression levels were determined at the end of the treatment. Luciferase expression is indicated as a percentage relative to the DMSO control. The 50% inhibitory concentrations (IC50) for each condition are indicated in the graph. (D) MHV-nsp2EGFP-infected LR7 cells were mock treated or treated with 10 μM MG132 from 2 to 7 h postinfection. At 7 h postinfection, the cells were fixed and processed for immunofluorescence. The white arrowheads indicate the putative replication sites. (E) MHV-infected cells were labeled with 35S-labeled amino acids and subsequently chased. Progeny virions released into the culture medium from 6 to 8 and from 8 to 10 h postinfection (p.i.) were affinity purified using antibodies against the S protein. At 10 h postinfection, the cells were lysed and processed for immunoprecipitation with a mixture of anti-MHV and anti-M protein serum. Immunoprecipitates were analyzed by SDS-PAGE. Cells were mock treated or treated with 10 or 50 μM MG132 from 5.5 h postinfection onward. Molecular mass markers are indicated at the left in kilodaltons, while the positions of the MHV structural proteins (i.e., M, N, and S) in the gel are depicted at the right (S′ refers to the furin-cleaved forms of the S proteins).
To investigate which specific step(s) of the MHV infection cycle might be affected by the lack of proteasome activity, we evaluated the effects of adding MG132 to the culture medium at different time points. First, we determined the dose-response curves when the drug was applied either already during (i.e., from 0 to 6 h postinfection) or after the virus inoculation period (i.e., from 2 to 6 h or from 2 to 8 h postinfection). As shown in Fig. 1C, virus replication (i.e., the ability to reproduce in cells) was much more affected when the drug was already present during virus inoculation (50% inhibitory concentration [IC50] = 0.17 μM) than upon addition of the drug at 2 h postinfection (IC50 ∼ 0.5 μM). The duration of the MG132 postincubation period did not appear to affect the IC50 (compare the periods from 2 to 6 and from 2 to 8 h). These results suggest that early steps in the virus life cycle, as well as later steps, which include viral RNA synthesis and subsequent protein expression, are affected by the inhibition of proteasome activity.
Confocal microscopy was next used to confirm the inhibition of MHV RNA synthesis and subsequent protein expression by MG132. To this end, an infection experiment was performed with a recombinant virus that expresses nsp2 fused to enhanced GFP (EGFP) from a subgenomic mRNA (19). This fusion protein is recruited to the virus-induced membrane network with which viral RNA synthesis is thought to be associated. In this experiment, LR7 cells infected with MHV-nsp2EGFP were mock treated or treated with MG132 from 2 to 7 h postinfection. Subsequently, the cells were fixed and stained for two additional markers of the putative replication sites by using antibodies directed either against nsp2 and nsp3 or against dsRNA. In agreement with our previous results, in the presence of MG132 the staining for nsp2-3, dsRNA, and nsp2EGFP was dramatically reduced in comparison with the untreated control cells (Fig. 1D). Interestingly, it appeared that the expression of nsp2EGFP was more affected than that of nsp2-3 by treatment with MG132, since barely any EGFP fluorescence could be detected. Although nsp2-3 is expressed directly from the viral genome, the nsp2EGFP fusion protein is expressed from a subgenomic viral mRNA, and hence transcription is required for its expression. These results therefore strengthen the conclusion that MG132 affects MHV RNA synthesis.
The UPS has been demonstrated to play an important role in the assembly and release of some viruses (20, 56, 60, 65). The reduction of extracellular infectivity shown in Fig. 1B may indicate a similar effect of MG132 on assembly of MHV. Therefore, we studied the effect of the drug on the synthesis and release of MHV particles by using an assembly assay. In this assay, cells infected with MHV were metabolically labeled from 5 to 5.5 h postinfection in the absence of MG132. Subsequently, the cells were chased in the presence of cycloheximide to inhibit protein synthesis and, in addition, they were mock treated or treated with MG132. After 30 min (i.e., at 6 h postinfection), the chase medium was refreshed, and the release of newly assembled viral particles was monitored from 6 to 8 h and from 8 to 10 h postinfection. Virus particles were affinity isolated from the culture media harvested at 8 or 10 h postinfection using antibodies to the S protein. At 10 h postinfection, cells were lysed, and cell lysates were monitored for viral protein production. As shown in Fig. 1E, no appreciable effect of MG132 on virus assembly could be observed. Only in the presence of 50 μM MG132 was a slight decrease in the amount of affinity-isolated N and M proteins detected. From these results we conclude that the proteasome inhibitor MG132 affects multiple steps in the MHV replicative cycle but that assembly of the virus was not significantly affected by the drug.
The inhibitory effect of MG132 does not result from induction of a cellular stress response.
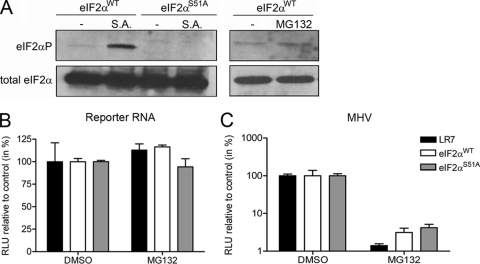
Previously, Neznanov et al. showed that the inhibitory effect of proteasome inhibitors on vesicular stomatitis virus infection was partially due to the induction of a cellular stress response by proteasome inhibitors, the initiation of which required the protein kinase GCN2 (39). Upon activation, this kinase phosphorylates serine 51 of eukaryotic initiation factor 2α (eIF2α), which results in global translational repression (3, 4). To test whether the inhibition of the proteasome affected MHV RNA synthesis via the induction of a cellular stress response, we used eIF2α wild-type and nonphophorylatable eIF2αS51A mutant MEFs. The phenotype of the MEFs was verified by treatment with sodium arsenite, which causes phosphorylation of eIF2α in wild-type MEFs, but not in MEFs with mutated eIF2α (Fig. 2 A). In addition, treatment of the wild-type MEFs for 4 h with 10 μM MG132 did not significantly induce phosphorylation of eIF2α. Next, the effect of inhibition of proteasome activity on mRNA translation was investigated by transfection of synthetic luciferase reporter mRNAs into MEF-, as well as LR7 cells, followed by a treatment for 4 h with 10 μM MG132. Subsequently, the cells were lysed, and the intracellular luciferase levels were determined. As shown in Fig. 2B, treatment with MG132 did not inhibit mRNA translation in any of the cells tested. Likewise, differences in the level of protein synthesis could not be detected by performing metabolic labeling of LR7 cells that were either treated with MG132 or mock treated for 4 h (data not shown). However, when the different cells were infected with MHV-EFLM and subsequently treated with MG132 or mock treated from 2 to 6 h postinfection, the luciferase expression was severely affected by the treatment with MG132 (Fig. 2C). From these results, we conclude that the inhibitory effect of MG132 on MHV infection is not caused by the induction of a cellular stress response. Therefore, we suggest that the inhibitory effect of MG132 on reporter gene expression does result from an inhibitory effect of the compound on viral RNA synthesis rather than from inhibition of translation of viral mRNAs.
FIG. 2.
The inhibitory effect of MG132 on MHV replication does not result from induction of a cellular stress response. (A) The phosphorylation status of eIF2α in the eIF2WT and eIF2S51A was determined by Western blotting with eIF2αP-specific antibodies (top) and related to total eIF2α levels by stripping and reprobing of the membrane (bottom). As a control, the cells were treated with 0.5 mM sodium arsenite (S.A.) for 30 min, which is known to induce the phosphorylation of eIF2α at serine 51 (36). In addition, the phosphorylation status of eIF2α in the eIF2WT MEFs was determined after treatment with 10 μM MG132 for 4 h. (B) The indicated cells (i.e., LR7, eIF2WT, and eIF2S51A) were transfected with a reporter RNA and subsequently cultured in the absence or presence of 10 μM MG132 (from 2 to 6 h posttransfection). The luciferase levels at 6 h posttransfection are indicated as a percentage relative to the DMSO control. (C) The different cells were infected with MHV-EFLM and subsequently cultured in the absence or presence of 10 μM MG132 (from 2 to 6 h postinfection). The intracellular luciferase expression levels at 6 h postinfection are indicated as a percentage relative to the DMSO control of each individual cell line. The standard deviations (n = 6) are indicated.
CoV infection is affected by different proteasome inhibitors.
Next, we studied whether the inhibition of virus infection caused by MG132 is specific for this proteasome inhibitor or whether it is also observed when other inhibitors of the proteasome are used. Although MG132 has been shown to also inhibit proteases other than the proteasome (i.e., cathepsin A and tripeptidyl peptidase II) (30), epoxomicin, lactacystin, and Velcade target the proteasome more specifically (8, 37, 41). The used concentrations of all drugs and of their solvent were nontoxic to the cells, as determined by cell viability assays (data not shown). Epoxomicin and Velcade reduced luciferase expression driven by MHV in LR7 cells approximately to the same extent as MG132, both when present already during virus inoculation and when added directly thereafter (Fig. 3 A). Lactacystin, however, was not very potent in its antiviral activity under these experimental conditions. In agreement with the results obtained for MG132, both drugs affected MHV infection more extensively when already applied during the inoculation period than thereafter.
FIG. 3.
MHV infection is reduced by treatment with different proteasome inhibitors. (A) LR7 cells were infected with MHV-EFLM in the absence or presence of different concentrations of the inhibitors MG132, epoxomicin, lactacystin, and Velcade. The compounds were added either from 0 to 6 or from 2 to 6 h after virus inoculation (p.i.). The intracellular luciferase expression levels at 6 h postinfection are indicated as a percentage relative to the DMSO control. (B) LR7 cells transfected with the pEGFP-degron plasmid were treated with the indicated concentrations of the inhibitors for 6 h. Subsequently, the cells were fixed and processed for microscopic analysis. Representative images for each condition are shown. (C) LR7 cells infected with MHV-EFLM, FCWF cells infected with FIPV-Δ3abcFL, and Vero cells infected with SARS-CoV-GFP were treated with the indicated concentrations of Velcade. Velcade was applied to the cells 1 h prior to infection, after which the intracellular luciferase activity was measured at 7 h postinfection for MHV-EFLM and FIPV-Δ3abcFL. Quantification of GFP expression in Vero cells infected with SARS-CoV-GFP (53) was performed at 18 h postinfection by using a molecular light imager (Berthold Technologies). The reporter gene expression levels (i.e., FL for MHV and FIPV; GFP for SARS-CoV) are indicated as a percentage relative to the PBS control. The standard deviations are indicated (i.e., n = 3 for MHV and FIPV and n = 4 for SARS-CoV).
To confirm that these drugs indeed block proteasome activity, we next applied them to LR7 cells expressing a GFP containing a degron sequence. Under normal conditions this protein is not stable because it is rapidly degraded by the proteasome and thus not detected (23). However, when cells were treated with MG132, epoxomicin, or Velcade, GFP expression was stabilized in a dose-dependent manner, which correlated with the inhibitory effect of the drugs on MHV infection (Fig. 3B). Consistently, lactacystin was much less effective in this assay, in agreement with its limited antiviral activity.
Next, we evaluated whether the antiviral activity of the proteasome inhibitors also exists for other CoV family members. To this end, Velcade was applied to FCWF and Vero cells infected with recombinant FIPV or SARS-CoV, respectively, each of which expressed a reporter gene allowing the straightforward measurement of viral RNA synthesis. Velcade (also known as Bortezomib) is a proteasome inhibitor approved for clinical use against multiple myelomas (46). The drug was already applied to the cells 1 h prior to virus inoculation. As a control, the inhibitor was applied to LR7 cells infected with MHV-EFLM. Velcade exhibited a dose-dependent antiviral effect for all three CoVs (Fig. 3C). Overall, these results demonstrate that inhibition of virus infection by proteasome inhibitors is a general feature of CoVs.
Velcade does not inhibit SARS-CoV main protease activity.
Velcade is known to block the multicatalytic activity of the proteasome, which includes chymotrypsinlike and trypsinlike protease subunits that are distant relatives of the main protease (Mpro) domain in CoV nsp5 (71). Given its critical role in replicase polyprotein cleavage, inhibition of the activity of nsp5, rather than inhibition of proteasome function, would explain a deleterious effect of Velcade on CoV RNA synthesis. To investigate whether the drug indeed interferes with Mpro activity, recombinant nsp5 and a fluorescently labeled peptide substrate were used to perform in vitro protease assays in the presence or absence of Velcade (Fig. 4). Cleavage of the peptide substrate by nsp5 results in an increase of fluorescence due to the fact that the C-terminal fluorescein group of the substrate is no longer quenched by the N-terminal Dabcyl group. As anticipated, the C145A active site mutant protein that was used as negative control was unable to cleave the substrate (Fig. 4A). Different concentrations of Velcade were tested, but even at 50 μM the drug did not inhibit the activity of nsp5. As shown in Fig. 4B, the relative cleavage rates were similar in the presence of Velcade (4.3 dF/min). From this analysis, we conclude that Velcade does not inhibit the proteolytic activity of the CoV Mpro.
FIG. 4.
Velcade does not inhibit SARS-CoV main protease activity. A fluorimetric SARS-CoV nsp5 protease activity assay (see Materials and Methods) was performed. (A) Change in fluorescence as a result of cleavage of a Dabcyl-VRLQSGTC-fluorescein peptide substrate by wild-type nsp5 or an active site mutant (nsp5C145A) in the presence or absence of 50 μM Velcade. Symbols: □, wild-type nsp5; ○, wild-type nsp5 + Velcade; ▾, nsp5C145A. (B) Cleavage rate for wild-type nsp5 in the absence or presence of different concentrations of Velcade.
RNA synthesis but not entry of MHV is affected in cells deficient for ubiquitin conjugation.
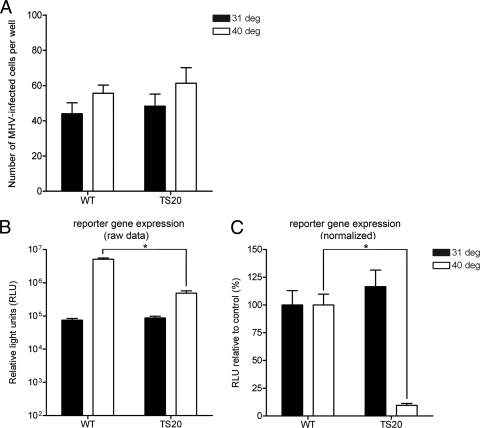
We next determined the role of the host ubiquitination machinery in MHV infection, since proteins destined for proteasomal degradation are often, but not always, modified with ubiquitin. To investigate the effect of ubiquitination on MHV infection we first made use of Chinese Hamster lung cells (ts20) that exhibit a temperature-sensitive defect in ubiquitin conjugation, resulting from their expressing a thermolabile ubiquitin-activating enzyme E1 (33). Wild-type cells (E36) were taken along as controls. The cells were transfected with a plasmid encoding the MHV receptor (i.e., CEACAM-1a [13]) to render them susceptible to MHV infection and grown for 24 h at the permissive temperature. The E36 and ts20 cells were transfected to the same extent as controlled by the cotransfection of a Renilla luciferase encoding expression plasmid (data not shown). Subsequently, 1 h prior to inoculation with MHV-EFLM, the cells were placed at either the permissive (31°C) or the nonpermissive (40°C) temperature. At 8 h postinfection, the cells were fixed and stained for viral antigen, after which the virus-positive cells were counted. Comparable numbers of MHV-infected wild-type and ts20 cells were observed at both temperatures. (Fig. 5 A). The temperature shift for 8 h did not significantly affect cell viability in both cell lines, as determined by a cell proliferation assay (data not shown). In parallel, cells were lysed, and the FL activity was measured. In the E36 (control) cells, viral luciferase levels were much higher at 40°C than at 31°C (Fig. 5B), a finding consistent with the known enhancement of CoV RNA synthesis at this higher temperature (49). In the ts20 cells, similar FL levels could be measured at 31°C compared to the control cells. At 40°C, however, although the number of MHV-infected cells were similar to that of the control cells, the FL levels were significantly lower in the ts20 cells (Fig. 5B). Overall, these results show that RNA synthesis but not entry of MHV (as determined by luciferase expression and number of infected cells, respectively) is significantly reduced in cells that are affected in ubiquitin conjugation.
FIG. 5.
Replication but not entry of MHV is affected in cells deficient for ubiquitin conjugation. E36 (indicated as wild-type [WT]) or ts20 (indicated as TS20) Chinese hamster cells grown in 24-wells clusters were infected with MHV-EFLM at the indicated temperatures (i.e., 31 and 40°C). Incubation was continued at the same temperatures for 8 h. (A) Indicated are the total numbers of virus-infected cells per well for each condition, which were determined by staining for viral antigen with a polyclonal anti-MHV serum. (B and C) In parallel, the intracellular luciferase expression levels at 8 h postinfection were determined. The raw RLU values are shown in panel B, whereas in panel C the data are expressed as a percentage relative to the expression levels at 31 and 40°C in the wild-type cells (i.e., normalized data). The standard deviations are indicated (n = 6). *, P < 0.05 as determined by statistical analysis using the Student t test.
To confirm the results obtained with the cells expressing a thermolabile ubiquitin-activating enzyme E1, we also studied MHV entry and RNA synthesis after ubiquitin depletion. Ubiquitin is synthesized in cells either in the form of polyubiquitin polypeptide chains or as ubiquitin-ribosomal protein fusions. siRNA-mediated depletion of UBA52 and Rps27A, which encode ubiquitin-ribosomal fusion proteins, resulted in significantly reduced levels of polyubiquitinated proteins and free ubiquitin (Fig. 6 A). Next, siRNA-treated cells were infected with MHV-EFLM, after which entry was assessed by determining the number of MHV-positive cells by immunocytochemistry. As shown in Fig. 6B, entry of MHV was hardly affected by depletion of ubiquitin. In a parallel experiment, MHV RNA synthesis was analyzed by determining the luciferase expression levels. The results show that reporter gene expression was significantly reduced in the UBA52- and Rps27A-depleted cells compared to the siRNA control (Fig. 6C), which correlated with the observed effects of ubiquitin depletion. These results, which are in agreement with those obtained with the ts20 cells, indicate that protein ubiquitination facilitates MHV infection but does not appear to be important for entry into host cells.
FIG. 6.
MHV replication is reduced in cells depleted of ubiquitin. HeLa-CEACAM1a cells grown in 96-wells clusters were transfected with 10 nM siRNAs targeting the indicated ubiquitin genes (i.e., UBA52 and Rps27A). Mock- and scrambled siRNA-transfected cells were used as controls. (A) At 72 h after transfection, the cells were lysed and processed for Western blotting with antibodies against ubiquitin and β-actin (loading control). The levels of free ubiquitin and ubiquitin conjugates were quantified and are expressed as a percentages relative to mock-transfected cells. Note that the values were corrected for the loading control. (B) The siRNA-transfected cells were inoculated with MHV-EFLM. At 6 h postinfection, the cells were fixed and stained for viral antigen using the polyclonal anti-MHV serum. The total numbers of MHV-infected cells per well are indicated, as well as the standard deviations (n = 3). (C) In parallel, the intracellular luciferase expression levels measured at 6 h postinfection are shown as a percentage relative to the control (scrambled siRNA-transfected cells). Note that the RLU values are corrected for cell viability. *, P < 0.05 as determined by statistical analysis using the Student t test.
DISCUSSION
In the present study we have shown that a properly functioning UPS is required for optimal CoV infection. Interfering with the UPS by treating cells with chemical inhibitors of the proteasome, by using cells expressing a temperature-sensitive ubiquitin-activating enzyme E1, or by depletion of ubiquitin with siRNAs severely affected MHV RNA synthesis and subsequent protein expression. In addition, the proteasome inhibitors appeared to interfere with an early step of the MHV infectious cycle. However, virus entry seemed not to be affected after interference with protein ubiquitination. The effects of the proteasome inhibitors were not limited to MHV but also apply other CoVs such as SARS-CoV and FIPV.
Inhibition of CoV infection by inhibitors of proteasome activity most likely results from impairment of the proteasome rather than from inhibition of a viral factor as the effect was observed for different CoVs and with several (highly specific) inhibitors. In agreement herewith, the drug Velcade (Bortezomib), which inhibited infection of all CoVs tested and which is known to block the chymotrypsinlike activity of the proteasome, did not inhibit the proteolytic activity of the chymotrypsinlike proteinase residing in SARS-CoV nsp5, as established by using an in vitro peptide cleavage assay. The negative effect of the proteasome inhibitors on CoV RNA synthesis and subsequent protein expression might be directly linked to proteasome inhibition but might also be caused be an indirect effect. MG132 did, however, not induce significant attenuation of cellular translation. In concordance with this, the effect of MG132 on MHV RNA synthesis was still observed in cells lacking a phosphorylatable eIF2α. A detailed mechanistic understanding of the inhibition of proteasome inhibitors on CoV infection requires the identification of specific cellular processes that are affected by inhibition of proteasome activity and required for efficient virus replication.
Our results indicate that an early step in the coronavirus infection cycle is affected by different inhibitors of proteasome activity since a more profound effect on MHV infection was observed when the drugs were already present during virus inoculation. Although we cannot exclude a possible negative effect of these inhibitors on an early step in viral RNA synthesis, it might well be that it is the entry of MHV that is affected by different inhibitors of the proteasome, which would be in agreement with a previous study reporting an inhibitory effect of MG132 on the entry of MHV (68). For several other viruses, including influenza virus, herpes simplex virus, and minute virus of mice, proteasome inhibitors have also been shown to affect virus entry (14, 28, 47).
What could be the mechanism of CoV entry inhibition? Proteasome inhibitors are known to reduce the levels of free ubiquitin in the cell (38). This effect is, however, unlikely to explain the inhibitory action of the proteasome inhibitors on coronavirus entry. Depletion of free ubiquitin by using RNA interference did not affect virus entry, nor was entry reduced in cells expressing a thermolabile ubiquitin-activating enzyme E1 at the restrictive temperature. Consistently, the MHV receptor appeared not to be ubiquitinated (68). Possibly, the proteasome is directly involved in the coronavirus entry process, perhaps during particle disassembly. In agreement with this, Yu and Lai found MHV particles to be trapped within endosomes upon treatment with MG132, which may indicate that proteasome activity is somehow important for the release of virus from the endosome into the cytosol (68). As with MHV, the entry of herpes simplex virus into the cell, a process that is also facilitated by the proteasome, appeared to be independent of protein ubiquitination (14). The authors of that study hypothesized that a proteasomal degradation process not involving ubiquitin conjugation may play a role in herpes simplex virus entry (26, 27). The same explanation may hold true for CoVs.
Ubiquitination was found to be critical for efficient RNA synthesis by MHV. Similar results were recently obtained for coxsackievirus and vaccinia virus (48, 52). However, the mechanism(s) by which ubiquitination facilitates virus infection remains unclear. For coxsackievirus, the viral RNA polymerase was found to be modified by ubiquitin moieties, which might be critical for its functioning. Infection with this virus was shown to induce the accumulation of protein-ubiquitin conjugates, with a concomitant decrease in the levels of free ubiquitin (52). During MHV infection, however, such an increase in protein-ubiquitin conjugates was not observed (data not shown). Interestingly, several studies have recently shown that the papainlike protease (PLpro) domain of CoVs contains deubiquitinating activity (2, 9, 45, 58). Hence, the requirement of ubiquitination for efficient CoV replication and the deubiquitinating activity of a nonstructural protein that is localized to the replication-transcription complexes may somehow be connected.
In contrast to MHV entry and RNA synthesis, assembly of MHV appeared to be independent of proteasome functioning. For several other enveloped viruses, however, the importance of the UPS for the assembly and release of progeny virions has been clearly demonstrated (5, 11, 29). This dependence appears to relate to these viruses exploiting the cellular vacuolar protein sorting (VPS) machinery for their budding. This machinery is involved in the formation of multivesicular bodies (MVBs), which depends on monoubiquitinated cargo proteins (44). Viruses shown to be dependent for their assembly on the VPS4 protein, an essential cellular component of the MVB sorting pathway, were also sensitive to proteasome inhibitors in their assembly. The insensitivity of MHV assembly to proteasome inhibitors suggests that this process is not dependent on the VPS machinery. Indeed, the production of progeny virus particles appeared not to be affected in cells expressing a nonfunctional VPS4 (unpublished results).
This study has demonstrated important roles for the UPS in multiple steps of the CoV infection cycle. Although further studies are needed to elucidate the precise mechanism(s) by which the UPS facilitates CoV infection, these results appear to indicate that proteasome inhibitors present a new class of anticoronaviral drugs. Several other studies have also suggested proteasome inhibitors as attractive antiviral compounds (48, 61). In agreement with this, proteasome inhibitors have already been shown to protect against coxsackievirus-induced myocarditis in a mouse model (16) and to prolong survival of mice inoculated with Epstein-Barr virus-transformed cells (72). Of note, the proteasome inhibitor Velcade, which was also used in the present study, has already been approved for the treatment of multiple myelomas (46, 55, 69). The major advantage of drugs targeting host, rather than viral components, may be the lower probability of generating viral drug-resistant variants. Evolutionary escape possibilities for the virus are expected to be limited since direct mutation of the drug target is not possible. Therefore, the appearance of drug-resistant variants may be restricted compared to conventional antiviral drug approaches. In an accompanying study (42a), we examined the feasibility for inhibiting proteasome activity as an effective means to treat CoV infections in vivo. Interestingly, however, rather than having a protective effect, treatment of MHV-infected mice with Velcade resulted in enhanced disease.
Acknowledgments
This study was supported by grants from the M. W. Beijerinck Virology Fund (Royal Netherlands Academy of Arts and Sciences) and The Netherlands Organization for Scientific Research (NWO-VIDI-700.54.421) to C. A. M. de Haan.
We thank Mikihiko Naito (Institute of Molecular and Cellular Biosciences, The University of Tokyo, Tokyo, Japan) for providing the pEGFP-degron plasmid, Costas Koumenis (Radiation Oncology, University of Pennsylvania, Philadelphia, PA) for providing the eIF2α wild-type and eIF2αS51A MEFs, and Susan Baker (Department of Microbiology and Immunology, Loyola University, Maywood, IL) for the nsp2-3 antibody. We also thank Amy Sims and Ralph Baric (University of North Carolina, Chapel Hill, NC) for providing GFP-expressing recombinant SARS-CoV, John Ziebuhr (The Queen's University of Belfast, Belfast, United Kingdom) and Tanja Schirmeister (University of Würzburg, Würzburg, Germany) for providing the SARS-CoV nsp5 expression constructs, Linda Boomaars-van der Zanden for technical assistance, and Marne Hagemeijer and Mijke Vogels for stimulating discussions.
Footnotes
Published ahead of print on 19 May 2010.
REFERENCES
- 1.Anand, K., J. Ziebuhr, P. Wadhwani, J. R. Mesters, and R. Hilgenfeld. 2003. Coronavirus main proteinase (3CLpro) structure: basis for design of anti-SARS drugs. Science 300:1763-1767. [DOI] [PubMed] [Google Scholar]
- 2.Barretto, N., D. Jukneliene, K. Ratia, Z. Chen, A. D. Mesecar, and S. C. Baker. 2005. The papain-like protease of severe acute respiratory syndrome coronavirus has deubiquitinating activity. J. Virol. 79:15189-15198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berlanga, J. J., J. Santoyo, and C. De Haro. 1999. Characterization of a mammalian homolog of the GCN2 eukaryotic initiation factor 2α kinase. Eur. J. Biochem. 265:754-762. [DOI] [PubMed] [Google Scholar]
- 4.Berlanga, J. J., I. Ventoso, H. P. Harding, J. Deng, D. Ron, N. Sonenberg, L. Carrasco, and C. de Haro. 2006. Antiviral effect of the mammalian translation initiation factor 2α kinase GCN2 against RNA viruses. EMBO J. 25:1730-1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bieniasz, P. D. 2006. Late budding domains and host proteins in enveloped virus release. Virology 344:55-63. [DOI] [PubMed] [Google Scholar]
- 6.Bosch, B. J., R. van der Zee, C. A. de Haan, and P. J. Rottier. 2003. The coronavirus spike protein is a class I virus fusion protein: structural and functional characterization of the fusion core complex. J. Virol. 77:8801-8811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burch, A. D., and S. K. Weller. 2004. Nuclear sequestration of cellular chaperone and proteasomal machinery during herpes simplex virus type 1 infection. J. Virol. 78:7175-7185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burger, A. M., and A. K. Seth. 2004. The ubiquitin-mediated protein degradation pathway in cancer: therapeutic implications. Eur. J. Cancer 40:2217-2229. [DOI] [PubMed] [Google Scholar]
- 9.Chen, Z., Y. Wang, K. Ratia, A. D. Mesecar, K. D. Wilkinson, and S. C. Baker. 2007. Proteolytic processing and deubiquitinating activity of papain-like proteases of human coronavirus NL63. J. Virol. 81:6007-6018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Compton, S. R., S. W. Barthold, and A. L. Smith. 1993. The cellular and molecular pathogenesis of coronaviruses. Lab. Anim. Sci. 43:15-28. [PubMed] [Google Scholar]
- 11.Crump, C. M., C. Yates, and T. Minson. 2007. Herpes simplex virus type 1 cytoplasmic envelopment requires functional Vps4. J. Virol. 81:7380-7387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.de Haan, C. A., Z. Li, E. te Lintelo, B. J. Bosch, B. J. Haijema, and P. J. Rottier. 2005. Murine coronavirus with an extended host range uses heparan sulfate as an entry receptor. J. Virol. 79:14451-14456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.de Haan, C. A., L. van Genne, J. N. Stoop, H. Volders, and P. J. Rottier. 2003. Coronaviruses as vectors: position dependence of foreign gene expression. J. Virol. 77:11312-11323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Delboy, M. G., D. G. Roller, and A. V. Nicola. 2008. Cellular proteasome activity facilitates herpes simplex virus entry at a postpenetration step. J. Virol. 82:3381-3390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Drosten, C., S. Gunther, W. Preiser, S. van der Werf, H. R. Brodt, S. Becker, H. Rabenau, M. Panning, L. Kolesnikova, R. A. Fouchier, A. Berger, A. M. Burguiere, J. Cinatl, M. Eickmann, N. Escriou, K. Grywna, S. Kramme, J. C. Manuguerra, S. Muller, V. Rickerts, M. Sturmer, S. Vieth, H. D. Klenk, A. D. Osterhaus, H. Schmitz, and H. W. Doerr. 2003. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N. Engl. J. Med. 348:1967-1976. [DOI] [PubMed] [Google Scholar]
- 16.Gao, G., J. Zhang, X. Si, J. Wong, C. Cheung, B. McManus, and H. Luo. 2008. Proteasome inhibition attenuates coxsackievirus-induced myocardial damage in mice. Am. J. Physiol. Heart Circ. Physiol. 295:H401-H408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goldberg, A. L. 2007. Functions of the proteasome: from protein degradation and immune surveillance to cancer therapy. Biochem. Soc. Trans. 35:12-17. [DOI] [PubMed] [Google Scholar]
- 18.Gosert, R., A. Kanjanahaluethai, D. Egger, K. Bienz, and S. C. Baker. 2002. RNA replication of mouse hepatitis virus takes place at double-membrane vesicles. J. Virol. 76:3697-3708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hagemeijer, M. C., M. H. Verheije, M. Ulasli, I. A. Shaltiel, L. A. de Vries, F. Reggiori, P. J. Rottier, and C. A. de Haan. 2010. Dynamics of coronavirus replication-transcription complexes. J. Virol. 84:2134-2149. [DOI] [PMC free article] [PubMed]
- 20.Harty, R. N., M. E. Brown, J. P. McGettigan, G. Wang, H. R. Jayakar, J. M. Huibregtse, M. A. Whitt, and M. J. Schnell. 2001. Rhabdoviruses and the cellular ubiquitin-proteasome system: a budding interaction. J. Virol. 75:10623-10629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hershko, A., and A. Ciechanover. 1998. The ubiquitin system. Annu. Rev. Biochem. 67:425-479. [DOI] [PubMed] [Google Scholar]
- 22.Hicke, L., and R. Dunn. 2003. Regulation of membrane protein transport by ubiquitin and ubiquitin-binding proteins. Annu. Rev. Cell Dev. Biol. 19:141-172. [DOI] [PubMed] [Google Scholar]
- 23.Ishioka, T., R. Katayama, R. Kikuchi, M. Nishimoto, S. Takada, R. Takada, S. Matsuzawa, J. C. Reed, T. Tsuruo, and M. Naito. 2007. Impairment of the ubiquitin-proteasome system by cellular FLIP. Genes Cells 12:735-744. [DOI] [PubMed] [Google Scholar]
- 24.Kaeppler, U., N. Stiefl, M. Schiller, R. Vicik, A. Breuning, W. Schmitz, D. Rupprecht, C. Schmuck, K. Baumann, J. Ziebuhr, and T. Schirmeister. 2005. A new lead for nonpeptidic active-site-directed inhibitors of the severe acute respiratory syndrome coronavirus main protease discovered by a combination of screening and docking methods. J. Med. Chem. 48:6832-6842. [DOI] [PubMed] [Google Scholar]
- 25.Kanjanahaluethai, A., and S. C. Baker. 2000. Identification of mouse hepatitis virus papain-like proteinase 2 activity. J. Virol. 74:7911-7921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kerscher, O. 2007. SUMO junction: what's your function? New insights through SUMO-interacting motifs. EMBO Rep. 8:550-555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kerscher, O., R. Felberbaum, and M. Hochstrasser. 2006. Modification of proteins by ubiquitin and ubiquitin-like proteins. Annu. Rev. Cell Dev. Biol. 22:159-180. [DOI] [PubMed] [Google Scholar]
- 28.Khor, R., L. J. McElroy, and G. R. Whittaker. 2003. The ubiquitin-vacuolar protein sorting system is selectively required during entry of influenza virus into host cells. Traffic 4:857-868. [DOI] [PubMed] [Google Scholar]
- 29.Kian Chua, P., M. H. Lin, and C. Shih. 2006. Potent inhibition of human hepatitis B virus replication by a host factor Vps4. Virology 354:1-6. [DOI] [PubMed] [Google Scholar]
- 30.Kisselev, A. F., and A. L. Goldberg. 2001. Proteasome inhibitors: from research tools to drug candidates. Chem. Biol. 8:739-758. [DOI] [PubMed] [Google Scholar]
- 31.Knoops, K., M. Kikkert, S. H. Worm, J. C. Zevenhoven-Dobbe, Y. van der Meer, A. J. Koster, A. M. Mommaas, and E. J. Snijder. 2008. SARS-coronavirus replication is supported by a reticulovesicular network of modified endoplasmic reticulum. PLoS Biol. 6:e226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Krijnse-Locker, J., M. Ericsson, P. J. Rottier, and G. Griffiths. 1994. Characterization of the budding compartment of mouse hepatitis virus: evidence that transport from the RER to the Golgi complex requires only one vesicular transport step. J. Cell Biol. 124:55-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kulka, R. G., B. Raboy, R. Schuster, H. A. Parag, G. Diamond, A. Ciechanover, and M. Marcus. 1988. A Chinese hamster cell cycle mutant arrested at G2 phase has a temperature-sensitive ubiquitin-activating enzyme, E1. J. Biol. Chem. 263:15726-15731. [PubMed] [Google Scholar]
- 34.Kuo, L., G. J. Godeke, M. J. Raamsman, P. S. Masters, and P. J. Rottier. 2000. Retargeting of coronavirus by substitution of the spike glycoprotein ectodomain: crossing the host cell species barrier. J. Virol. 74:1393-1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Locker, J. K., J. K. Rose, M. C. Horzinek, and P. J. Rottier. 1992. Membrane assembly of the triple-spanning coronavirus M protein. Individual transmembrane domains show preferred orientation. J. Biol. Chem. 267:21911-21918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McEwen, E., N. Kedersha, B. Song, D. Scheuner, N. Gilks, A. Han, J. J. Chen, P. Anderson, and R. J. Kaufman. 2005. Heme-regulated inhibitor kinase-mediated phosphorylation of eukaryotic translation initiation factor 2 inhibits translation, induces stress granule formation, and mediates survival upon arsenite exposure. J. Biol. Chem. 280:16925-16933. [DOI] [PubMed] [Google Scholar]
- 37.Meng, L., R. Mohan, B. H. Kwok, M. Elofsson, N. Sin, and C. M. Crews. 1999. Epoxomicin, a potent and selective proteasome inhibitor, exhibits in vivo anti-inflammatory activity. Proc. Natl. Acad. Sci. U. S. A. 96:10403-10408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mimnaugh, E. G., H. Y. Chen, J. R. Davie, J. E. Celis, and L. Neckers. 1997. Rapid deubiquitination of nucleosomal histones in human tumor cells caused by proteasome inhibitors and stress response inducers: effects on replication, transcription, translation, and the cellular stress response. Biochemistry 36:14418-14429. [DOI] [PubMed] [Google Scholar]
- 39.Neznanov, N., E. M. Dragunsky, K. M. Chumakov, L. Neznanova, R. C. Wek, A. V. Gudkov, and A. K. Banerjee. 2008. Different effect of proteasome inhibition on vesicular stomatitis virus and poliovirus replication. PLoS One 3:e1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Oostra, M., C. A. de Haan, R. J. de Groot, and P. J. Rottier. 2006. Glycosylation of the severe acute respiratory syndrome coronavirus triple-spanning membrane proteins 3a and M. J. Virol. 80:2326-2336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ostrowska, H., C. Wojcik, S. Omura, and K. Worowski. 1997. Lactacystin, a specific inhibitor of the proteasome, inhibits human platelet lysosomal cathepsin A-like enzyme. Biochem. Biophys. Res. Commun. 234:729-732. [DOI] [PubMed] [Google Scholar]
- 42.Pasternak, A. O., W. J. Spaan, and E. J. Snijder. 2006. Nidovirus transcription: how to make sense? J. Gen. Virol. 87:1403-1421. [DOI] [PubMed] [Google Scholar]
- 42a.Raaben, M., G. C. M. Grinwis, P. J. M. Rottier, and C. A. M. de Haan. 2010. The proteasome inhibitor Velcade enhances rather than reduces disease in mouse hepatitis coronavirus-infected mice. J. Virol. 84:7880-7885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Raaben, M., M. J. Groot Koerkamp, P. J. Rottier, and C. A. de Haan. 2007. Mouse hepatitis coronavirus replication induces host translational shutoff and mRNA decay, with concomitant formation of stress granules and processing bodies. Cell Microbiol. 9:2218-2229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Raiborg, C., T. E. Rusten, and H. Stenmark. 2003. Protein sorting into multivesicular endosomes. Curr. Opin. Cell Biol. 15:446-455. [DOI] [PubMed] [Google Scholar]
- 45.Ratia, K., K. S. Saikatendu, B. D. Santarsiero, N. Barretto, S. C. Baker, R. C. Stevens, and A. D. Mesecar. 2006. Severe acute respiratory syndrome coronavirus papain-like protease: structure of a viral deubiquitinating enzyme. Proc. Natl. Acad. Sci. U. S. A. 103:5717-5722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Richardson, P. G., C. Mitsiades, T. Hideshima, and K. C. Anderson. 2006. Bortezomib: proteasome inhibition as an effective anticancer therapy. Annu. Rev. Med. 57:33-47. [DOI] [PubMed] [Google Scholar]
- 47.Ros, C., C. J. Burckhardt, and C. Kempf. 2002. Cytoplasmic trafficking of minute virus of mice: low-pH requirement, routing to late endosomes, and proteasome interaction. J. Virol. 76:12634-12645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Satheshkumar, P. S., L. C. Anton, P. Sanz, and B. Moss. 2009. Inhibition of the ubiquitin-proteasome system prevents vaccinia virus DNA replication and expression of intermediate and late genes. J. Virol. 83:2469-2479. [DOI] [PMC free article] [PubMed]
- 49.Sawicki, S. G., D. L. Sawicki, D. Younker, Y. Meyer, V. Thiel, H. Stokes, and S. G. Siddell. 2005. Functional and genetic analysis of coronavirus replicase-transcriptase proteins. PLoS Pathog. 1:e39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Scheuner, D., B. Song, E. McEwen, C. Liu, R. Laybutt, P. Gillespie, T. Saunders, S. Bonner-Weir, and R. J. Kaufman. 2001. Translational control is required for the unfolded protein response and in vivo glucose homeostasis. Mol. Cell 7:1165-1176. [DOI] [PubMed] [Google Scholar]
- 51.Schonborn, J., J. Oberstrass, E. Breyel, J. Tittgen, J. Schumacher, and N. Lukacs. 1991. Monoclonal antibodies to double-stranded RNA as probes of RNA structure in crude nucleic acid extracts. Nucleic Acids Res. 19:2993-3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Si, X., G. Gao, J. Wong, Y. Wang, J. Zhang, and H. Luo. 2008. Ubiquitination is required for effective replication of coxsackievirus B3. PLoS One 3:e2585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sims, A. C., R. S. Baric, B. Yount, S. E. Burkett, P. L. Collins, and R. J. Pickles. 2005. Severe acute respiratory syndrome coronavirus infection of human ciliated airway epithelia: role of ciliated cells in viral spread in the conducting airways of the lungs. J. Virol. 79:15511-15524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sims, A. C., J. Ostermann, and M. R. Denison. 2000. Mouse hepatitis virus replicase proteins associate with two distinct populations of intracellular membranes. J. Virol. 74:5647-5654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sterz, J., I. von Metzler, J. C. Hahne, B. Lamottke, J. Rademacher, U. Heider, E. Terpos, and O. Sezer. 2008. The potential of proteasome inhibitors in cancer therapy. Expert Opin. Invest. Drugs. 17:879-895. [DOI] [PubMed] [Google Scholar]
- 56.Strack, B., A. Calistri, M. A. Accola, G. Palu, and H. G. Gottlinger. 2000. A role for ubiquitin ligase recruitment in retrovirus release. Proc. Natl. Acad. Sci. U. S. A. 97:13063-13068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Studier, F. W. 2005. Protein production by auto-induction in high density shaking cultures. Protein Expr. Purif. 41:207-234. [DOI] [PubMed] [Google Scholar]
- 58.Sulea, T., H. A. Lindner, E. O. Purisima, and R. Menard. 2005. Deubiquitination, a new function of the severe acute respiratory syndrome coronavirus papain-like protease? J. Virol. 79:4550-4551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tanahashi, N., H. Kawahara, Y. Murakami, and K. Tanaka. 1999. The proteasome-dependent proteolytic system. Mol. Biol. Rep. 26:3-9. [DOI] [PubMed] [Google Scholar]
- 60.Taylor, G. M., P. I. Hanson, and M. Kielian. 2007. Ubiquitin depletion and dominant-negative VPS4 inhibit rhabdovirus budding without affecting alphavirus budding. J. Virol. 81:13631-13639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Teale, A., S. Campbell, N. Van Buuren, W. C. Magee, K. Watmough, B. Couturier, R. Shipclark, and M. Barry. 2009. Orthopoxviruses require a functional ubiquitin-proteasome system for productive replication. J. Virol. 83:2099-2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.van der Hoek, L., K. Pyrc, M. F. Jebbink, W. Vermeulen-Oost, R. J. Berkhout, K. C. Wolthers, P. M. Wertheim-van Dillen, J. Kaandorp, J. Spaargaren, and B. Berkhout. 2004. Identification of a new human coronavirus. Nat. Med. 10:368-373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Verheije, M. H., M. Raaben, M. Mari, E. G. Te Lintelo, F. Reggiori, F. J. van Kuppeveld, P. J. Rottier, and C. A. de Haan. 2008. Mouse hepatitis coronavirus RNA replication depends on GBF1-mediated ARF1 activation. PLoS Pathog. 4:e1000088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Voges, D., P. Zwickl, and W. Baumeister. 1999. The 26S proteasome: a molecular machine designed for controlled proteolysis. Annu. Rev. Biochem. 68:1015-1068. [DOI] [PubMed] [Google Scholar]
- 65.Watanabe, H., Y. Tanaka, Y. Shimazu, F. Sugahara, M. Kuwayama, A. Hiramatsu, K. Kiyotani, T. Yoshida, and T. Sakaguchi. 2005. Cell-specific inhibition of paramyxovirus maturation by proteasome inhibitors. Microbiol. Immunol. 49:835-844. [DOI] [PubMed] [Google Scholar]
- 66.Weismiller, D. G., L. S. Sturman, M. J. Buchmeier, J. O. Fleming, and K. V. Holmes. 1990. Monoclonal antibodies to the peplomer glycoprotein of coronavirus mouse hepatitis virus identify two subunits and detect a conformational change in the subunit released under mild alkaline conditions. J. Virol. 64:3051-3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Woo, P. C., S. K. Lau, C. M. Chu, K. H. Chan, H. W. Tsoi, Y. Huang, B. H. Wong, R. W. Poon, J. J. Cai, W. K. Luk, L. L. Poon, S. S. Wong, Y. Guan, J. S. Peiris, and K. Y. Yuen. 2005. Characterization and complete genome sequence of a novel coronavirus, coronavirus HKU1, from patients with pneumonia. J. Virol. 79:884-895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yu, G. Y., and M. M. Lai. 2005. The ubiquitin-proteasome system facilitates the transfer of murine coronavirus from endosome to cytoplasm during virus entry. J. Virol. 79:644-648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zavrski, I., L. Kleeberg, M. Kaiser, C. Fleissner, U. Heider, J. Sterz, C. Jakob, and O. Sezer. 2007. Proteasome as an emerging therapeutic target in cancer. Curr. Pharm. Des. 13:471-485. [DOI] [PubMed] [Google Scholar]
- 70.Zhang, Z., N. Torii, A. Furusaka, N. Malayaman, Z. Hu, and T. J. Liang. 2000. Structural and functional characterization of interaction between hepatitis B virus X protein and the proteasome complex. J. Biol. Chem. 275:15157-15165. [DOI] [PubMed] [Google Scholar]
- 71.Ziebuhr, J., E. J. Snijder, and A. E. Gorbalenya. 2000. Virus-encoded proteinases and proteolytic processing in the Nidovirales. J. Gen. Virol. 81:853-879. [DOI] [PubMed] [Google Scholar]
- 72.Zou, P., J. Kawada, L. Pesnicak, and J. I. Cohen. 2007. Bortezomib induces apoptosis of Epstein-Barr virus (EBV)-transformed B cells and prolongs survival of mice inoculated with EBV-transformed B cells. J. Virol. 81:10029-10036. [DOI] [PMC free article] [PubMed] [Google Scholar]