Abstract
N-(4-Chlorophenyl)-N′-(2,2,6,6-tetramethyl-piperidin-4-yl)-oxalamide (NBD-556) is a low-molecular-weight compound that reportedly blocks the interaction between human immunodeficiency virus type 1 (HIV-1) gp120 and its receptor CD4. We investigated whether the enhancement of binding of anti-gp120 monoclonal antibodies (MAbs) toward envelope (Env) protein with NBD-556 are similar to those of soluble CD4 (sCD4) by comparing the binding profiles of the individual MAbs to Env-expressing cell surfaces. In flow cytometric analyses, the binding profiles of anti-CD4-induced epitope (CD4i) MAbs toward NBD-556-pretreated Env-expressing cell surfaces were similar to the binding profiles toward sCD4-pretreated cell surfaces. To investigate the binding position of NBD-556 on gp120, we induced HIV-1 variants that were resistant to NBD-556 and sCD4 in vitro. At passage 21 in the presence of 50 μM NBD-556, two amino acid substitutions (S375N in C3 and A433T in C4) were identified. On the other hand, in the selection with sCD4, seven mutations (E211G, P212L, V255E, N280K, S375N, G380R, and G431E) appeared during the passages. The profiles of the mutations after the selections with NBD-556 and sCD4 were very similar in their three-dimensional positions. Moreover, combinations of NBD-556 with anti-gp120 MAbs showed highly synergistic interactions against HIV-1. We further found that after enhancing the neutralizing activity by adding NBD-556, the contemporaneous virus became highly sensitive to antibodies in the patient's plasma. These findings suggest that small compounds such as NBDs may enhance the neutralizing activities of CD4i and anti-V3 antibodies in vivo.
Human immunodeficiency virus type 1 (HIV-1) replicates continuously in the face of a strong antibody (Ab) response, although Abs effectively control many viral infections (3). Neutralizing Abs (NAbs) are directed against the HIV-1 envelope (Env) protein, which is a heterodimer comprising an extensively glycosylated CD4-binding subunit (gp120) and an associated transmembrane protein (gp41). Env proteins are present on the virion surface as “spikes” composed of trimers of three gp120-gp41 complexes (20, 21, 29). These spikes resist neutralization through epitope occlusion within the oligomer, extensive glycosylation, extension of variable loops from the surface of the complex, and steric and conformational blocking of receptor binding sites (16, 18, 20).
Ab access to conserved regions is further limited because viral entry is a stepwise process involving conformational changes that lead to only transient exposure of conserved domains such as the coreceptor binding site (4, 5). However, some early strains of HIV-1 appear to be highly susceptible to neutralization by Abs (1, 10). For instance, subtype A HIV-1 envelopes from the early stage of infection exhibit a broad range of neutralization sensitivities to both autologous and heterologous plasma (1), suggesting that at least a subset of the envelopes have some preserved and/or exposed neutralization epitopes. It is well known that the potential for neutralizing properties of particular Abs is enhanced after binding of soluble CD4 (sCD4), especially NAbs against CD4-induced epitopes (CD4i Abs) (27) and some anti-V3 Abs (22). CD4i Abs are detected in plasma samples from many patients at an early stage of HIV-1 infection (9). Consequently, we hypothesize that small compounds such as sCD4 can enhance the neutralizing activities of CD4i Abs and some anti-V3 Abs not only in vitro but also in vivo.
In a previous report, two low-molecular-weight compounds that presumably interfere with viral entry of HIV-1 into cells were described (35). These two N-phenyl-N′-(2,2,6,6-tetramethyl-piperidin-4-yl)-oxalamide analogs, NBD-556 and NBD-557, comprise a novel class of HIV-1 entry inhibitors that block the interaction between gp120 and CD4. These compounds were found to be equally potent inhibitors of both X4 and R5 viruses in CXCR4- and CCR5-expressing cell lines, respectively (35). Schön et al. (25) also reported that NBD-556 binds to gp120 in a process characterized by a large favorable change in enthalpy that is partially compensated for by a large unfavorable entropy change, representing a thermodynamic signature similar to that observed for binding of sCD4 to gp120. In a recent study, Madani et al. (23) reported the following findings: (i) NBD-556 binds within the Phe43 cavity, a highly conserved and functionally important pocket formed as gp120 assumes the CD4-bound conformation; (ii) the NBD-556 phenyl ring projects into the Phe43 cavity; (iii) the enhancement of CD4-independent infection by NBD-556 requires the induction of conformational changes in gp120; and (iv) increased affinities of NBD-556 analogs toward gp120 improve the antiviral potency during infection of CD4-expressing cells. The latter two studies demonstrated that low-molecular-weight compounds such as NBDs can induce conformational changes in the HIV-1 gp120 glycoprotein similar to those observed upon sCD4 binding (23, 25). The authors of these studies concluded that their data supported the importance of gp120 residues near the Phe43 cavity in binding to NBD-556 and lent credence to the docked binding mode.
In the present study, we investigated the binding position of NBD-556 on gp120 by inducing HIV-1 variants that were resistant to NBD-556 by exposing HIV-1IIIB to increasing concentrations of the compound in vitro. We also induced sCD4-resistant HIV-1IIIB variants and compared the profile of the sCD4-resistant mutations to that of the NBD-556-resistant mutations. We subsequently examined the virological properties of pseudotyped HIV-1 clones carrying the NBD-556 and sCD4 resistance-associated env gene mutations. Our findings provide a foundation for understanding the interaction of NBD-556 with the CD4-binding site of HIV-1 gp120. We also evaluated the anti-HIV-1 interactions between plasma NAbs and NBD-556 in vitro and considered the possibility of using the data as a key to opening the shield covering the conserved epitopes targeted by NAbs.
(This study was presented in part at the 15th Conference on Retroviruses and Opportunistic Infection, Boston, MA, 3 to 6 February 2008 [Abstract 736].)
MATERIALS AND METHODS
Cells, culture conditions, and reagents.
The CD4-positive T-cell line PM1 was maintained in RPMI 1640 (Sigma, St. Louis, MO) supplemented with 10% heat-inactivated fetal calf serum (FCS; HyClone Laboratories, Logan, UT), 50 U of penicillin/ml, and 50 μg of streptomycin/ml. PM1/CCR5 cells were generated by standard retrovirus-mediated transduction of PM1 cells with pBABE-CCR5 provided by the National Institutes of Health AIDS Research and Preference Reagent Program (NIH ARRRP) (24, 34). PM1/CCR5 cells were maintained in RPMI 1640 supplemented with 10% heat-inactivated FCS, 50 U of penicillin/ml, 50 μg of streptomycin/ml, and 0.1 mg of G418 (Invitrogen, Carlsbad, CA)/ml. The TZM-bl cell line was obtained from the NIH ARRRP and maintained in Dulbecco modified Eagle medium (Sigma) supplemented with 10% FCS.
NBD-556 (molecular weight, 337.84) and YYA-004 (molecular weight, 303.4), which has the same structure as JRC-I-300 (23), were synthesized as previously described (23, 25, 30). KD-247 (12), 3E4, and 0.5γ (unpublished) are anti-gp120-V3 monoclonal Abs (MAbs). 17b (27), 4C11, and 4E9C (unpublished) are MAbs against CD4-induced epitopes (CD4i Abs). 17b, 2G12 (a MAb against the gp120 glycan), and b12 (a MAb against the CD4-binding site [CD4bs] epitope) were provided by the NIH ARRRP. The 0.5δ antibody established in our laboratory is an anti-CD4bs MAb (unpublished results). RPA-T4 (an anti-CD4 MAb) was purchased from BD Biosciences Pharmingen (San Jose, CA). Recombinant human sCD4 was purchased from R&D Systems, Inc. (Minneapolis, MN).
MAbs 3E4, 0.5γ, 0.5δ, 4C11, and 4E9C were human MAbs established from a patient with long-term nonprogressive illness. B cells from the patient's peripheral blood mononuclear cells (PBMC) were transformed by Epstein-Barr virus, followed by cloning. Culture supernatant from an individual clone was screened for reactivity to gp120SF2 by enzyme-linked immunosorbent assay (ELISA). The specificity of the antibodies was determined by gp120 capture ELISA and fluorescence-activated cell sorting analysis of HIV-1JR-FL-infected PM1 cells in the presence or absence of sCD4. The binding specificity was further assessed by an ELISA using peptides corresponding to the V3 sequence of various isolates. Based on these binding data, we classified them as follows: V3 MAbs, 3E4 and 0.5γ; CD4bs MAb, 0.5δ; and CD4i MAbs, 4C11 and 4E9C.
The laboratory-adapted HIV-1 strains HIV-189.6, HIV-1BaL, HIV-1SF162, HIV-1JR-FL, and HIV-1YU2 were propagated in phytohemagglutinin-activated PBMC. These viruses were then passaged in PM1/CCR5 cells, and the culture supernatants were stored at −150°C prior to use. R5 primary HIV-1 isolates (HIV-1Pt.1, HIV-1Pt.2, HIV-1Pt.3, and HIV-1Pt.4) were isolated from four Japanese patients in our laboratory. All patients were at a stage of chronic infection. HIV-1Pt.1, HIV-1Pt.3, and HIV-1Pt.4 were isolated from drug-naive patients, and HIV-1Pt.2 was isolated from a drug-experienced patient and passaged in phytohemagglutinin-activated PBMC. Infected PBMC were cocultured with PM1/CCR5 cells for 4 to 5 days, and the culture supernatants were stored at −150°C until used. Nucleotide sequences of the gp120 from the four primary isolates were deposited in the DNA Data Bank of Japan under accession numbers AB553911 to AB553914.
Susceptibility assay.
The sensitivities of six laboratory-adapted viruses, four primary isolates, and HIV-1IIIB viruses passaged in the presence of sCD4 or NBD-556 were determined by the MTT [3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide] assay as previously described with minor modifications (31). Briefly, PM1/CCR5 cells (2 × 103 cells/well) were exposed to 100 times the 50% tissue culture infective dose (TCID50) of the viruses in the presence of various concentrations of sCD4 or NBD-556 in 96-well round-bottom microculture plates, followed by incubation at 37°C for 7 days. After removal of 100 μl of the medium, 10 μl of MTT solution (7.5 mg/ml) in phosphate-buffered saline (PBS) was added to each well. The plate was then incubated at 37°C for 3 h. Subsequently, the produced formazan crystals were dissolved by adding 100 μl of acidified isopropanol containing 4% (vol/vol) Triton X-100 to each well. The optical densities at a wavelength of 570 nm were measured in a microplate reader. All assays were performed in duplicate or triplicate. We also determined the concentration for 50% cytotoxicity (CC50) by using the MTT assay.
The sensitivities of the HIV-1Pt.3 primary isolate to KD-247 (anti-V3 MAb), 4E9C (CD4i MAb), and autologous plasma IgG in the presence or absence of NBD-556 were also determined by using the MTT assay. To exclude any influence of plasma factors, such as antiviral drugs, cytokines, and chemokines, on the neutralization activities, we used IgG from the patient's plasma, which was purified using protein A-Sepharose (Affi-gel Protein A; Bio-Rad, Hercules, CA) (19).
Flow cytometric analysis.
HIV-1JR-FL chronically infected PM1 cells were preincubated with or without sCD4 (0.5 μg/ml) and NBD-556 (1, 3, 10, 30, 90, and 100 μM) for 15 min and then incubated with various anti-HIV-1 MAbs (17b, 4C11, KD-247, 3E4, and 0.5γ) at 4°C for 30 min. The cells were washed with PBS, and a fluorescein isothiocyanate-conjugated goat anti-human IgG Ab was used for Ab detection. Flow cytometry was performed with a FACSCalibur flow cytometer (BD Biosciences), and the data were analyzed by using the BD CellQuest version 3.1 software (BD Biosciences).
Data analysis and evaluation of synergy.
Analyses of the synergistic, additive, and antagonistic effects of the antiviral agents were initially performed according to the median effect principle using the CalcuSyn version 2 computer program (6) to provide estimates of the 50% inhibitory concentration (IC50) values of the antiviral agents in combination. Combination indices (CIs) were estimated from the data and reflected the nature of the interactions between KD-247 and sCD4 or NBD-556 and between NBD-556 and CD4i MAb 4C11 or anti-CD4bs MAb 0.5δ against HIV-1JR-FL or HIV-1IIIB on PM1/CCR5 cells as determined by the MTT assay. A CI of <0.9 indicated synergy, a CI between 0.9 and 1.1 indicated additivity, and a CI of >1.1 indicated antagonism. The CI value was directly proportional to the amount of synergy for the combination regimen. For example, values of <0.5 represented a high degree of synergy, while values of >1.5 represented significant antagonism. This approach has been widely used in analyses of antiviral interactions and was chosen to allow comparability with published literature.
Docking simulation.
The structure for NBD-556 was built in SYBYL 7.1 (Tripos, St. Louis, MO) and minimized with the MMFF94 force field and partial charges (15). Using FlexSIS through its SYBYL module, docking of NBD-556 was performed into the crystal structure of gp120 obtained from the Protein Data Bank (PDB; entry 1RZJ). The binding site was defined as residues Val255, Asp368, Glu370, Ser375, Ile424, Trp427, Val430, and Val475, including residues located within a radius of 4.4 Å. The structure of the ligand was treated flexibly, and all other options were set to their default values. Figures were generated using SwissPdb Viewer version 3.9 (SPdbViewer) (13) and ViewerLite version 5.0 (Accelrys, Inc., San Diego, CA). We also generated a simian immunodeficiency virus (SIV) gp120 figure (PDB entry 2BF1) to compare the sites of the mutations in HIV-1 gp120 using the same software programs.
Isolation of NBD-556- and sCD4-resistant mutants from HIV-1IIIB in vitro.
To select NBD-556 and sCD4 escape viruses, HIV-1IIIB was treated with various concentrations of NBD-556 or sCD4 and then infected into PM1/CCR5 cells as previously described with minor modifications (32). Viral replication was monitored by observation of any cytopathic effects in PM1/CCR5 cells. The culture supernatants were harvested on day 7 and used to infect fresh PM1/CCR5 cells for the next round of culture in the presence of increasing concentrations of NBD-556 or sCD4. When the viruses began to propagate in the presence of NBD-556 or sCD4, the concentration was further increased. After the viruses were passaged using up to 50 μM NBD-556 or 20 μg of sCD4/ml in PM1/CCR5 cells, the resulting viruses, designated NBD-556(20)14p, NBD-556(50)17p, and sCD4(20)5p, were recovered from the passaged cell culture supernatants.
Proviral DNA extracts from cells cultured with several concentrations of NBD-556 and sCD4 were subjected to PCR amplification using Taq polymerase (Takara, Shiga, Japan). The amplified products were cloned into pCR2.1 (Invitrogen), and the env regions in both the passaged and selected viruses were sequenced by using an ABI Prism 3110 automated DNA sequencer (ABI, Foster City, CA).
Construction of mutant Env expression vectors.
Proviral DNA was extracted from the passaged HIV-1IIIB-infected PM1/CCR5 cells by using a QIAamp DNA blood minikit (Qiagen, Valencia, CA). For the construction of Env expression vectors, we used pCXN2, which contains a chicken actin promoter. Briefly, we amplified the passaged HIV-1IIIB gp160 regions using LA Taq (Takara) with the primers ENVA (5′-GGCTTAGGCATCTCCTATGGCAGGAAGAA-3′) and ENVN (5′-CTGCCAATCAGGGAAGTAGCCTTGTGT-3′). The PCR products were inserted into pCR-XL-TOPO (Invitrogen). The EcoRI fragment of pCR-XL-IIIB containing the entire env region was ligated into pCXN2 to give pCXN-IIIBwt. pCXN-IIIB(S375N), pCXN-IIIB(V255E), and pCXN-IIIB(A433T) were generated by site-directed mutagenesis using a QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA) in accordance with the manufacturer's instructions with the primer pairs S375Nfw (5′-AAATTGTAACGCACAATTTTAATTGTGGAGG-3′) and S375Nrv (5′-CCTCCACAATTAAAATTGTGCGTTACAATTT-3′), V255Efw (5′-GAATTAGGCCAGTAGAATCAACTCAACTGCT-3′) and V255Erv (5′-AGCAGTTGAGTTGATTCTACTGGCCTAATTC-3′), and A433Tfw (5′-CAGGAAGTAGGAAAAACAATGTATGCCCCTC-3′) and A433Trv (5′-GAGGGGCATACATTGTTTTTCCTACTTCCTG-3′), respectively.
Pseudovirus preparation.
Portions, 5 μg of pSG3ΔEnv and 0.5 μg of pRSV-Rev (17), supplied by the NIH ARRRP, and a 4.5-μg portion of HIV-1IIIB Env-expressing pCXN2 were cotransfected into 293T cells using the Effectene transfection reagent (Qiagen). At 48 h after transfection, the pseudovirus-containing supernatants were harvested, filtered through a 0.2-μm-pore-size filter, and stored at −80°C. The pseudovirus activities were measured with a luminescence assay using TZM-bl cells as previously described (28).
Single-round virus infection assay.
A single-cycle infectivity assay was used to measure the neutralization of HIV-1IIIB pseudoviruses as described previously (26, 28). Briefly, NBD-556, YYA-004, sCD4, 2G12, b12, RPA-T4, or 4C11 at various concentrations and a pseudovirus suspension corresponding to 100 TCID50 were preincubated in the absence or presence of 1 μM NBD-556 for 15 min on ice. The virus-compound mixtures were added to TZM-bl cells, which had been seeded in a 96-well plate (1.5 × 104 cells/well) on the previous day. The cultures were incubated for 2 days at 37°C, washed with PBS, and lysed with lysis solution (Galacto-Star mammalian reporter gene assay system; ABI). After transfer of the cell lysates to luminometer plates, the β-galactosidase activity (in relative light units) in each well was measured by using 50-fold-diluted Galacto-Star substrate in a reaction buffer diluent (100 μl/well; ABI) in a TR717 microplate luminometer (ABI). The reduction in infectivity was determined by comparing the relative light units in the presence or absence of each compound and expressed as the percentage of neutralization. Each assay was repeated two to three times.
RESULTS
Anti-HIV-1 activities of sCD4, NBD-556, and YYA-004 for laboratory strains and primary HIV-1 isolates.
Initially, we determined the inhibitory activities of sCD4, NBD-556, and YYA-004, which has a phenyl group instead of the p-chlorophenyl group of NBD-556 (Fig. 1), on the infection of PM1/CCR5 cells by different laboratory-adapted HIV-1 strains and different HIV-1 primary isolates of subtype B, including both X4 and R5 viruses, by using a previously reported method (33). sCD4 inhibited the laboratory-adapted HIV-1 strains HIV-1IIIB, HIV-189.6, HIV-1BaL, HIV-1SF162, HIV-1JR-FL, and HIV-1YU2 with IC50s ranging from 0.26 to 6.1 μg/ml (Table 1). NBD-556 inhibited the X4 virus HIV-1IIIB and dualtropic virus HIV-189.6 with IC50s of 7.8 and 11.4 μM, respectively, but did not inhibit the R5 viruses HIV-1BaL, HIV-1SF162, HIV-1JR-FL, and HIV-1YU2 with IC50s of >30 μM. We also tested sCD4 and NBD-556 against the R5 primary isolates HIV-1Pt.1, HIV-1Pt.2, HIV-1Pt.3, and HIV-1Pt.4. sCD4 effectively inhibited all of the primary isolates at concentrations of 0.2 to 7.4 μg/ml. On the other hand, NBD-556 inhibited two of the four primary isolates, HIV-1Pt.1 and HIV-1Pt.3, with IC50s of 3.6 and 11.8 μM, respectively (Table 1). YYA-004 did not show significant anti-HIV activity against any of the strains tested up to a concentration of 100 μM. The in vitro cytotoxicities of NBD-556 and YYA-004 toward PM1/CCR5 cells used for the anti-HIV-1 infectivity studies were determined by using the MTT assay. The CC50 values of NBD-556 and YYA-004 toward PM1/CCR5 cells were 140 and 350 μM, respectively (Table 1).
FIG. 1.
Structures of NBD-556 and YYA-004.
TABLE 1.
Inhibitory activities of sCD4 and NBD-556 toward infection by laboratory and primary strains of HIV-1
| Virus | Subtype | Cell | Mean IC50a ± SD |
||
|---|---|---|---|---|---|
| sCD4 (μg/ml) | NBD-556 (μM) | YYA-004 (μM) | |||
| Laboratory-adapted viruses | |||||
| X4 | |||||
| HIV-1IIIB | B | PM1/CCR5 | 0.26 ± 0.17 | 7.8 ± 2.6 | >100 |
| Dual | |||||
| HIV-189.6 | B | PM1/CCR5 | 0.87 ± 0.09 | 11.4 ± 2.4 | >100 |
| R5 | |||||
| HIV-1BaL | B | PM1/CCR5 | 1.7 ± 0.28 | >30 | >100 |
| HIV-1SF162 | B | PM1/CCR5 | 3.6 ± 0.64 | >30 | >100 |
| HIV-1JR-FL | B | PM1/CCR5 | 3.6 ± 0.71 | >30 | >100 |
| HIV-1YU2 | B | PM1/CCR5 | 6.1 ± 2.00 | >30 | >100 |
| Primary isolates | |||||
| R5 | |||||
| HIV-1Pt.1 | B | PM1/CCR5 | 0.2 ± 0.04 | 3.6 ± 0.67 | >100 |
| HIV-1Pt.2 | B | PM1/CCR5 | 1.6 ± 0.21 | >30 | >100 |
| HIV-1Pt.3 | B | PM1/CCR5 | 3.7 ± 0.42 | 11.8 ± 1.6 | >100 |
| HIV-1Pt.4 | B | PM1/CCR5 | 7.4 ± 1.30 | >30 | >100 |
PM1/CCR5 cells (2 × 103) were exposed to 100 TCID50 of each virus and then cultured in the presence of various concentrations of sCD4, NBD-556, or YYA-004 as indicated. The IC50s were determined by using the MTT assay on day 7 of culture. All assays were conducted in duplicate, and the data shown represent the means derived from the results of two to three independent experiments. For NBD-556, CC50 = 140 μM; for YYA-004, CC50 = 350 μM. (The CC50 is the concentration for 50% cytotoxicity.)
Comparison of Ab binding to cell surface-expressed HIV-1JR-FL Env with NBD-556 and sCD4.
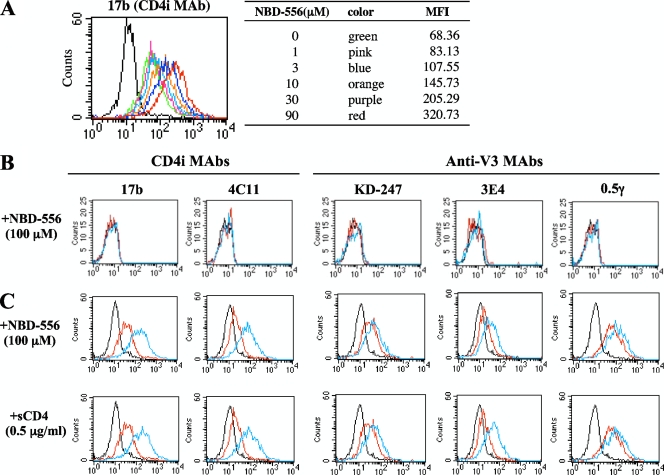
To compare the effect of NBD-556 with that of sCD4 with respect to the induction of conformational change in the trimeric gp120, the binding of CD4i MAbs (17b and 4C11) and anti-V3 MAbs (KD-247, 3E4, and 0.5γ) to cell surface-expressed Env proteins on HIV-1JR-FL chronically infected PM1 cells was analyzed by fluorescence-activated cell sorting. Comparisons of the binding profiles of the Abs to the cell surfaces were carried out using the mean fluorescence intensity (MFI). The binding of CD4i MAb 17b increased gradually as the amount of the CD4-mimicking small compound NBD-556 increased from 0 to 90 μM (Fig. 2A, the MFI increased from 68.36 to 320.73). As shown in Fig. 2C, the binding of both CD4i MAbs 17b and 4C11 increased remarkably after pretreatment with 100 μM NBD-556 (the MFIs increased from 43.3 to 201.57 and from 24.43 to 96.06, respectively). Moreover, the binding of all of the anti-V3 MAbs—KD-247, 3E4, and 0.5γ—was enhanced by pretreatment with NBD-556 (the MFIs increased from 34.59 to 51.9, from 22.97 to 39.07, and from 86.61 to 145.08, respectively). sCD4 pretreatment of the Env-expressing cell surface also caused remarkable enhancement of the binding for not only the CD4i MAbs but also the three anti-V3 MAbs, similar to pretreatment with NBD-556. These results indicate that the CD4-mimicking compound NBD-556 can induce the conformational changes in gp120 that are caused by binding of sCD4.
FIG. 2.
Comparisons of MAb binding to cell surface-expressed gp120 with sCD4 and NBD-556. HIV-1JR-FL chronically infected PM1 cells were preincubated with or without NBD-556 (A, 1 to 90 μM; C, 100 μM) or sCD4 (C, 0.5 μg/ml), and uninfected PM1 cells were also preincubated with or without NBD-556 (B, 100 μM) for 15 min, and then incubated with various anti-HIV-1 MAbs (17b, 4C11, KD-247, 3E4, and 0.5γ) at 4°C for 30 min. The cells were washed, and a fluorescein isothiocyanate-conjugated anti-human IgG was used for detection. (A) Color lines show the concentrations of NBD-556: green, 0 μM; pink, 1 μM; blue, 3 μM; orange, 10 μM; purple, 30 μM; and red, 90 μM. (B and C) Red line shows no preincubated with NBD-556 or sCD4. Blue line shows preincubated with NBD-556 or sCD4. Black line shows using a control IgG MAb.
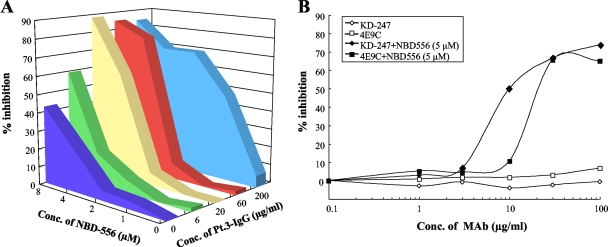
Highly synergistic interactions of KD-247 combined with NBD-556.
Both neutralizing anti-V3 MAb KD-247 and NBD-556 block the viral entry process, especially at the stage of the interaction between CD4 and gp120 (CD4-binding site). Each of these agents binds to either the V3 loop or the CD4 cavity. Furthermore, our previous observations suggested that neutralizing MAb KD-247 selects escape variants with greater sensitivities to sCD4 (33). Based on this notion, we examined the synergy of this MAb with sCD4 or the CD4-mimicking compound NBD-556 against wild-type HIV-1JR-FL. The multiple-drug effect analysis of Chou et al. (6) was used to analyze the effects of combining KD-247 with sCD4 or NBD-556. As shown in Table 2, all of the CI values for KD-247 with the two CD4-gp120 interaction inhibitors (sCD4 and NBD-556) were <0.5 against HIV-1JR-FL at all of the inhibitory concentrations tested. In particular, the CI values for the combinations of KD-247 with NBD-556 were <0.1 for IC75 and IC90. These results suggest that combinations of KD-247 with the CD4-gp120 binding inhibitors sCD4 and NBD-556 produce very highly synergistic effects. We further examined the synergy of CD4i MAb 4C11 or anti-CD4bs MAb 0.5δ with NBD-556 against wild-type HIV-1IIIB. The combination of 4C11 and NBD-556 showed synergy against HIV-1IIIB for IC50 and IC75. As expected, the IC values for NBD-556 and anti-CD4 binding site MAb, 0.5δ, which may compete with the CD4 mimetic for the CD4-binding site, were >5 against HIV-1IIIB at all of the inhibitory concentrations tested. However, at lower concentrations, additive effects were observed between NBD-556 and anti-CD4bs MAb 0.5δ (data not shown). These results indicate that NBD-556 may bind within or near the epitope of the anti-CD4bs MAb and then induce the conformational changes in Env.
TABLE 2.
Combination indices for KD-247, 4C11, or 0.5δ and for sCD4 or NBD-556 against HIV-1JR-FL and HIV-1IIIB
| Combination | Virus | CI values at different ICsa |
||
|---|---|---|---|---|
| IC50 | IC75 | IC90 | ||
| KD-247+sCD4 | HIV-1JR-FL | 0.313 | 0.266 | 0.277 |
| KD-247+NBD-556 | HIV-1JR-FL | 0.174 | 0.043 | 0.011 |
| 4C11+NBD-556 | HIV-1IIIB | 0.473 | 0.445 | 0.860 |
| 0.5δ+NBD-556 | HIV-1IIIB | 47.8 | 20.1 | 8.56 |
The multiple-drug effect analysis of Chou et al. (6) was used to analyze the effects of the drugs in combination. IC, inhibitory concentration. CI < 0.9, synergy; CI = 0.9 to 1.1, additivity; CI > 1.1, antagonism. The data shown are representative of two or three separate experiments.
Selection of NBD-556 and sCD4 escape variants.
To select NBD-556- and sCD4-resistant HIV-1 variants in vitro, we exposed PM1/CCR5 cells to HIV-1IIIB and serially passaged the viruses in the presence of increasing concentrations of NBD-556 or sCD4. As a control, HIV-1IIIB was passaged under the same conditions without the antiviral agents to allow us to monitor the spontaneous changes that occurred in the virus during prolonged PM1/CCR5 cell passages (designated the passage control). The selected viruses were initially propagated in the presence of 1 μM NBD-556 or 0.5 μg of sCD4/ml and, during the course of the selection procedure, the concentrations of the NBD-556 and sCD4 were increased to 50 μM and 20 μg/ml, respectively. At passages 14 and 17 for NBD-556 and passage 5 for sCD4, the supernatants containing the viruses, which were designated HIV-1NBD-R(20)14p, HIV-1NBD-R(50)17p, and HIV-1sCD4-R(20)5p, respectively, were harvested, and the sensitivities of the viruses to NBD-556 and sCD4 were determined by the MTT assay (Table 3). The IC50s for NBD-556 against HIV-1IIIB, HIV-1NBD-R(20)14p, and HIV-1NBD-R(50)17p were 12, >30, and >30 μM, respectively. The IC50s of sCD4 against HIV-1IIIB and HIV-1sCD4-R(20)5p were 0.52 and >10 μg/ml, respectively. HIV-1NBD-R(20)14p, HIV-1NBD-R(50)17p, and HIV-1sCD4-R(20)5p were also examined for their cross-resistance with one another. Each resistant variant was found to be cross-resistant to NBD-556 and sCD4 (Table 3). These results indicate that the HIV-1IIIB virus acquired resistant phenotypes against NBD-556 and sCD4 during the distinct in vitro selection processes.
TABLE 3.
Inhibitory activities of NBD-556 and sCD4 toward infection of HIV-1IIIB escape variants from NBD-556 and sCD4
| Virus | IC50a |
|
|---|---|---|
| NBD-556 (μM) | sCD4 (μg/ml) | |
| HIV-1IIIB | 12 | 0.52 |
| HIV-1NBD-R(20)14p | >30 | 5.7 |
| HIV-1NBD-R(50)17p | >30 | >10 |
| HIV-1sCD4-R(20)5p | >30 | >10 |
PM1/CCR5 cells (2 × 103) were exposed to 100 TCID50 of each passaged virus and then cultured in the presence of various concentrations of sCD4 or NBD-556. The IC50s were determined by using the MTT assay on day 7 of culture. All assays were conducted in duplicate. The data shown are representative of two or three separate experiments.
Sequences of the envelope region of the NBD-556 and sCD4 mutants.
To determine the genetic basis of the resistance in the variant HIV-1IIIB strains, the C1 to C4 region of the env gene was amplified from genomic DNA extracted from the infected cells and cloned, and the PCR-amplified products were sequenced (Fig. 3). At passage 8 for 6 μM NBD-556, five mutations (A281D, E370A, S375N, A433T, and A436T) were observed. At passage 21 in the culture where HIV-1IIIB was propagating in the presence of 50 μM NBD-556, four amino acid substitutions of Ser to Asn at position 375 (S375N, 11 of 11 clones) in C3, Ala to Lys at position 342 (A432K, 1 of 11 clones) in C4, Ala to Thr at position 433 (A433T, 4 of 11 clones) in C4, and Ala to Thr at position 436 (A436T, 1 of 11 clones) in C4 were observed (Fig. 3A). These results did not contradict a previous study in which gp120 mutants (S375W, I424A, W427A, and M475A) with changes in residues that contacted the Phe43 cavity did not detectably bind NBD-556 by isothermal titration calorimetry (23). On the other hand, in the selection with sCD4, seven mutations (E211G, P212L, V255E, N280K, S375N, G380R, and G431E) appeared during the passages. At passage 5 in the culture where HIV-1IIIB was propagating in the presence of sCD4 (20 μg/ml), four substitutions of E211G (1 of 10 clones), V255E (5 of 10 clones), G380R (1 of 10 clones), and G431E (2 of 10 clones) were detected for sCD4 at 20 μg/ml (Fig. 3B).
FIG. 3.
Alignment of the gp120 amino acid sequences from the indicated passages in the NBD-556 and sCD4 escape processes. The amino acid sequences were deduced from the nucleotide sequences of the env-encoding regions of proviral DNA isolated from cells infected with the HIV-1IIIB variants selected in the presence of NBD-556 (A) or sCD4 (B) and the passage control. The amino acid sequences of the envelope proteins of the baseline HIV-1IIIB are shown at the top as a reference. The identity of the sequences at the individual amino acid positions is indicated by dots. The numbers of clones with the given amino acid substitutions among a total of 8 to 12 clones are listed. The number in parentheses denotes the concentrations of NBD-556 or sCD4. The major mutations of NBD-556 and sCD4-resistant variants at final passage are boxed.
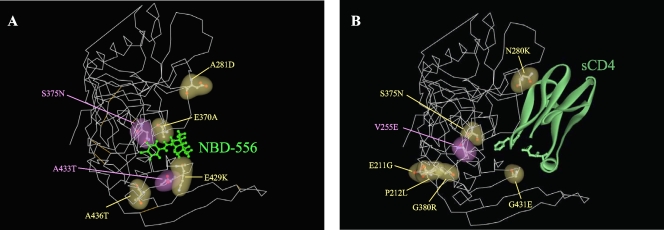
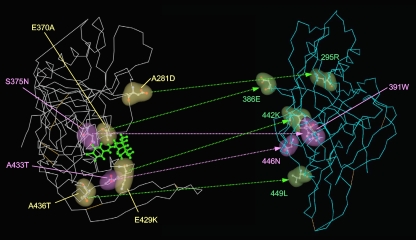
To compare the two mutation profiles obtained during the in vitro selection with NBD-556 and sCD4, molecular modeling of NBD-556 docked into gp120 was performed by docking simulations using the FlexSIS module of SYBYL 7.1 (Fig. 4). The atomic coordinates of the crystal structure of gp120 with sCD4 were retrieved from the PDB (entry 1RZJ). As shown in Fig. 4, almost all of the mutations lay along the inside of the CD4 cavity in the selection of NBD-556, with similar three-dimensional positions to the mutations induced by sCD4. These findings demonstrate that NBD-556 binds to the CD4 cavity or in the vicinity of the CD4-binding site.
FIG. 4.
Locations of substitutions in HIV-1IIIB gp120 induced by in vitro selection with NBD-556 or sCD4. The side chains of the mutated residues that appeared during the in vitro selection with NBD-556 (A) or sCD4 (B) are shown in yellow and purple. The amino acid substitutions that confer resistance in HIV-1 are indicated in purple. The crystal structure of gp120 with sCD4 was retrieved from the PDB (entry 1RZJ). The structure of compound NBD-556 docked into gp120 was created by using the FlexSIS module of SYBYL 7.1.
Sensitivities of β-galactosidase reporter HIV strains pseudotyped with the sCD4- and NBD-556-resistant envelope mutations to NBD-556, sCD4, and MAbs.
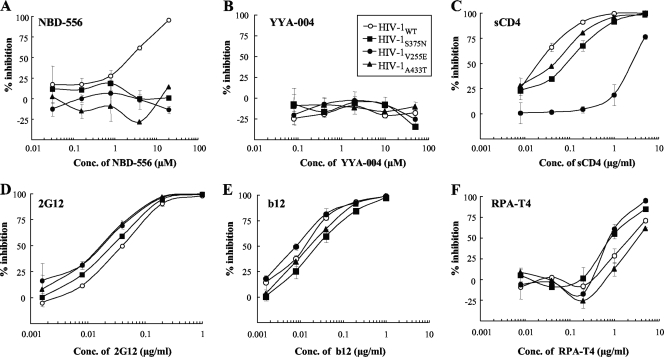
To confirm whether the mutations were responsible for the reduced sensitivities to NBD-556 and sCD4, a single-round replication assay was performed. The β-galactosidase reporter viruses were pseudotyped with wild-type Env (HIV-1WT) or Env singly mutated with V255E in C2 (HIV-1V255E), S375N in C3 (HIV-1S375N), and A433T in C4 (HIV-1A433T). The mutations that arose in the absence of NBD-556 (the passage control) are not related to resistance because the control passage did not show any significant increase in IC50 (data not shown). With respect to the mutations in the presence of NBD-556 three mutations, S375N, V255E, and A433T were consistently and increasingly observed during the process of selection. Additional mutations in “escape variants” other than S375N, V255E, and A433T were observed; however, these mutations were not consistently detected in passages and did not accumulate during selection. Thus, we considered the three mutations—S375N, V255E, and A433T—related to the development of resistance to both NBD-556 and sCD4, although some involvement of additional mutations in the development of a resistant phenotype is undeniable. As shown in Fig. 5A, all of the mutant clones were completely resistant to NBD-556 at concentrations of up to 20 μM. YYA-004 without the p-chlorophenyl group was unable to inhibit infection of all of the clones tested (Fig. 5B). The clone with V255E, which was induced by in vitro selection with sCD4, was highly resistant to sCD4 compared to the wild-type virus (114-fold-higher IC50) (Fig. 5C). However, the other pseudotyped viruses, HIV-1S375N and HIV-1A433T, were slightly resistant compared to HIV-1WT (4- and 2-fold-higher IC50s, respectively). We also examined the sensitivities of the pseudotyped clones containing Env mutations to anti-gp120 glycan MAb 2G12, anti-CD4bs MAb b12, and anti-CD4 MAb RPA-T4 by a single-round replication assay (Fig. 5D to F). All of the mutant viruses showed almost the same neutralization sensitivities as the wild-type virus to the 2G12, b12, and RPA-T4 MAbs. These results indicate that the three mutations induced by in vitro selection with NBD-556 and sCD4 were responsible for the resistance to NBD-556, whereas the NBD-selected variants containing S375N in C3 and A433T in C4 of gp120 had moderately resistant phenotypes against sCD4, as shown by the sensitivities of the NBD-556-passaged viruses to sCD4 determined by the multiround assay (Table 3).
FIG. 5.
Sensitivities of β-galactosidase reporter HIV strains pseudotyped with the sCD4 and NBD-556 resistance envelope mutations to NBD-556, YYA-004, sCD4, and MAbs. The sensitivities of β-galactosidase reporter HIV strains pseudotyped with the sCD4 and NBD-556 resistance envelope mutations to NBD-556 (A), YYA-004 (B), sCD4 (C), 2G12 (D), b12 (E), and RPA-T4 (F) are shown. NBD-556, YYA-004, sCD4, and MAbs at various concentrations and a pseudovirus suspension corresponding to 100 TCID50 were preincubated for 15 min on ice and then added to the target cells (TZM-bl). The inhibitory effects were determined by measuring the β-galactosidase activities on day 2 of culture. All assays were conducted in triplicate, and the data shown represent the means ± the standard deviations (SD) derived from the results of two to three independent experiments.
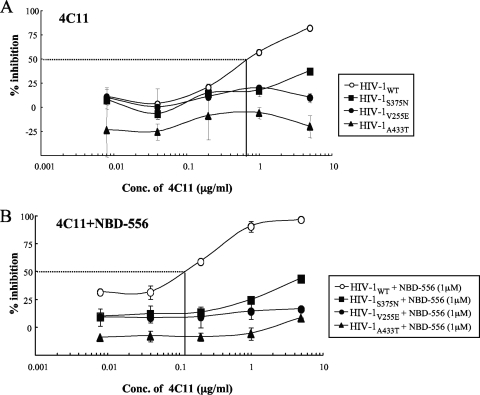
To examine whether the resistance mutations affected the sensitivity of a CD4i MAb against HIV-1, we determined the sensitivities of HIV strains pseudotyped with the sCD4- and NBD-556-resistant envelope mutations to CD4i MAb 4C11 with or without the CD4-mimicking compound. As expected, NBD-556-pretreated HIV-1WT was more sensitive to 4C11 than the untreated virus (IC50s, 0.12 versus 0.72 μg/ml) (Fig. 6). On the other hand, all of the mutant viruses were completely resistant to 4C11 with or without NBD-556 pretreatment. These results suggest that the CD4 and NBD-556 resistance mutations in gp120 hide the epitope for a particular Ab against a CD4-induced epitope, similar to primary R5 viruses.
FIG. 6.
Sensitivities of β-galactosidase reporter HIV strains pseudotyped with the sCD4 and NBD-556 resistance envelope mutations to CD4i MAb 4C11 with or without NBD-556. The sensitivities of β-galactosidase reporter HIV strains pseudotyped with the sCD4 and NBD-556 resistance envelope mutations to CD4i MAb 4C11 in the absence (A) or presence (B) of NBD-556 are shown. 4C11 at various concentrations and a pseudovirus suspension corresponding to 100 TCID50 were preincubated with or without NBD-556 (1 μM) for 15 min on ice and then added to the target cells (TZM-bl). The inhibitory effects were determined by measuring the β-galactosidase activities on day 2 of culture. All assays were conducted in triplicate, and the data shown represent the means ± the SD derived from the results of two to three independent experiments.
NBD-556-mediated enhancement of the neutralization activities of plasma Abs against an autologous isolate.
Neutralization escape has been documented in HIV-1 subtype B viruses, with contemporaneous viruses showing less sensitivity to autologous neutralization than earlier viruses (2). For one patient (patient 3 [Pt.3]) infected with a subtype B virus, the autologous neutralizing activities in plasma IgG obtained close to the time of the virus isolation were measured in the presence or absence of NBD-556 (0, 1, 2, 4, and 8 μM) by the MTT assay. As shown in Fig. 7A, the plasma IgG neutralizing activity was much less potent against the variant (HIV-1Pt.3) from the same time point (IC50 of >200 μg/ml for IgG). However, HIV-1Pt.3 pretreated with at least 1 μM NBD-556 became sensitive to the contemporaneous plasma IgG compared to the untreated virus. To examine which kinds of NAbs are enhanced by NBD-556, we determined the susceptibilities of HIV-1Pt.3 to anti-V3 MAb KD-247 and CD4i MAb 4E9C with or without NBD-556. The virus was completely resistant to both MAbs (IC50s of >100 μg/ml) in the absence of NBD-556, while NBD-556-pretreated HIV-1Pt.3 became sensitive to KD-247 and 4E9C (IC50s of 10.0 and 20.8 μg/ml, respectively) (Fig. 7B). These results indicate that CD4-mimicking small compounds such as NBDs have potent NAb-enhancing activities toward plasma Abs that cannot access the neutralizing epitopes hidden within the trimeric Env, such as CD4i and anti-V3 Abs.
FIG. 7.
NBD-556-mediated enhancement of the neutralization activity of plasma IgG against the autologous isolate. (A) The sensitivities of the HIV-1Pt.3 primary isolate to the autologous plasma IgG (Pt.3-IgG) in the absence or presence of NBD-556 (1, 2, 4, and 8 μM) were determined by the MTT assay. (B) The sensitivities of the HIV-1Pt.3 primary isolate to KD-247 (anti-V3 MAb; diamonds) and 4E9C (anti-CD4i MAb; squares) in the absence (open symbols) or presence (filled symbols) of 5 μM NBD-556 were determined by the same assay. The data shown are representative of two or three separate experiments.
DISCUSSION
In this study, we observed that NBD-556 could bind to a CD4-binding site, followed by the induction of conformational changes in gp120 similar to those observed upon sCD4 binding. Although we used a limited number of viruses and plasma IgG preparations obtained from an HIV-1-positive patient for testing the synergistic effects between NBD-556 and neutralizing antibody, we also found highly synergistic interactions between NBD-556 and not only CD4i MAbs but also anti-V3 MAbs. Moreover, our data indicated that small compounds such as NBDs can enhance the potency of NAbs in HIV-1-infected patients against the contemporaneous viruses, which are resistant to neutralization by Abs in the plasma.
We illustrated the sites of the mutations induced by NBD-556 on the structure of unliganded gp120 of SIV obtained from the PDB (entry 2BF1) to compare the sites before and after binding of the CD4-mimicking compound. As shown in Fig. 8, the mutations lay in front of the outer domain in gp120, which was near to or within the CD4-binding site. These findings indicate that NBD-556 attaches to the CD4-binding site or the surrounding residues in the unliganded form of gp120 and that, after the conformational changes of the envelope glycoproteins, probably the CD4-liganded form induced by the attack by NBD-556, the compound could penetrate and be held for a while in the CD4 cavity. In a recent study, Haim et al. (14) showed that sCD4-mimicking compounds have the ability to inactivate HIV-1 by prematurely triggering active but transient intermediate states of the envelope glycoproteins. In the transient intermediate states, several neutralizing epitopes in gp120 may be accessible to the neutralizing Abs. These data and our present results suggest that some NBD analogs, which bind to the cavity tightly and for a longer time, as well as cell surface CD4 inducing a more stable envelope glycoprotein intermediate state, show highly potent NAb-enhancing activities.
FIG. 8.
Comparisons of the locations of the mutations induced by NBD-556 between the structures of unliganded and liganded gp120. The side chains of the mutated residues that appeared during in vitro selection with NBD-556 are shown in yellow, green and purple in the liganded (left) or unliganded (right) structures. The amino acid substitutions that confer resistance in HIV-1 are indicated in purple (S375N and A433T). The crystal structures of liganded and unliganded gp120 were retrieved from the PDB (entries 1RZJ and 2BF1, respectively). The corresponding sites of the NBD-resistant mutations are also shown on the unliganded gp120.
Madani et al. (23) reported that replacement of gp120 Ser375 with a glycine residue dramatically reduced the HIV-1 sensitivity to enhancement by any of the NBD-556 analogs, suggesting that a certain element of the Ser375 side chain contributes to the NBD-556 efficacy. They also reported that viruses bearing envelope glycoproteins with Ser375 mutated to alanine exhibited greater enhancement by NBD-556 and some NBD-556 analogs than the viruses with wild-type envelope glycoproteins, suggesting that the hydroxyl group of Ser375 is detrimental to the binding and/or activity of some NBD-556 analogs that contain large para-phenyl substituents. Mutations of other gp120 residues lining the Phe43 cavity or vestibule (Val255, Thr257, Glu429, and Val430) significantly decreased the enhancement of virus infection by the NBD-556 analogs. Our in vitro selection study showed that the key mutations for NBD-556 resistance were S375N and A433T and that minor mutations related to NBD-556 resistance were A281D, E370A, E429K, and A436T (Fig. 4). Thus, alterations to several gp120 residues, namely, S375N, A433T, and V255E, that line the Phe43 pocket or reside around and inside the cavity can negatively affect the entry inhibitory effect of NBD-556 on HIV-1 infection (Fig. 5).
Decker et al. (9) reported that the chemokine coreceptor binding sites of HIV-1 from subtypes A, B, C, D, F, G, and H and circulating recombinant form (CRF) 01, CRF02, and CRF11 elicit high titers of CD4i Abs during natural human infection and that these Abs bind and neutralize viruses as divergent as HIV-2 in the presence of sCD4. Recently, Davis et al. (7) showed that transplantation of HIV-1 V3 epitopes into an HIV-2 envelope scaffold provides a sensitive and specific means to detect and quantify HIV-1 V3 epitope-specific NAbs in human sera. They used this HIV-2/HIV-1 V3 scaffolding strategy to study the kinetics of the development and breadth of V3-specific NAbs in longitudinal sera from individuals acutely infected with subtype C or subtype B HIV-1. Their results indicated that high-titer broadly reactive V3-specific Abs are among the first to be elicited during acute and early HIV-1 infection, although these Abs lack neutralizing potency against primary HIV-1 viruses, which effectively shield V3 from Ab binding to the functional Env trimer (8). These observations strongly support the idea that the major problem facing the development of CD4i-based or V3-based immunogens is not sequence variation within the epitopes, but rather that access of most CD4i and anti-V3 Abs to their epitopes in functional Env complexes is blocked. As shown in Fig. 7A, plasma IgG from a seropositive patient exhibited strongly enhanced neutralizing activity against the contemporaneous virus after treatment with NBD-556. Therefore, we consider that small compounds such as NBDs can enhance the neutralizing activities of CD4i and certain anti-V3 Abs in vivo at the acute stage of HIV-1 infection or in combination with anti-V3 NAbs as a passive immunization.
In general, small molecules have certain advantages from a therapeutic standpoint because of their low propensity for immunogenicity, high metabolic stability, easy large-scale production, and relatively low cost. Small molecule Ab-enhancing therapeutics such as NBD compounds would have additional benefits over available treatment approaches to HIV. Since CD4i and anti-cryptic V3 Abs are already present in a large number of HIV-1-infected patients, no prevaccination would be necessary for the induction of NAbs. Moreover, the use of bifunctional small molecules, such as an entry inhibitor and a NAb enhancer, should be effective for passive immunization of the anti-HIV NAbs enhanced by the accessibility of epitopes after binding of sCD4, such as 17b (27) and KD-247 (11, 12). Elucidation of the molecular details governing the interactions between gp120 and NBD compounds will assist in optimization efforts, as well as in the evaluation of this strategy in more complex biological models for HIV infection. Consequently, we will continue to synthesize such NBD analogs to search for drugs with more potent power to change the tertiary structure of the envelope glycoproteins and lower toxicity toward the host cells.
Acknowledgments
We thank The Chemo-Sero-Therapeutic Research Institute for kindly providing MAb KD-247. We are grateful to Yosuke Maeda for providing the PM1/CCR5 cells. We also thank Akiko Honda-Shibata, Yoko Kawanami, and Syoko Yamashita for excellent technical assistance.
This study was supported in part by the Ministry of Health, Labor, and Welfare of Japan (H20-AIDS-002 and H21-AIDS-010); a Grant-in-Aid for Scientific Research (C-20591206) from the Ministry of Education, Science, and Culture of Japan; and the Cooperative Research Project on Clinical and Epidemiological Studies of Emerging and Re-emerging Infectious Diseases.
Footnotes
Published ahead of print on 26 May 2010.
REFERENCES
- 1.Blish, C. A., R. Nedellec, K. Mandaliya, D. E. Mosier, and J. Overbaugh. 2007. HIV-1 subtype A envelope variants from early in infection have variable sensitivity to neutralization and to inhibitors of viral entry. AIDS 21:693-702. [DOI] [PubMed] [Google Scholar]
- 2.Bunnik, E. M., L. Pisas, A. C. van Nuenen, and H. Schuitemaker. 2008. Autologous neutralizing humoral immunity and evolution of the viral envelope in the course of subtype B human immunodeficiency virus type 1 infection. J. Virol. 82:7932-7941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Burton, D. R. 2002. Antibodies, viruses, and vaccines. Nat. Rev. Immunol. 2:706-713. [DOI] [PubMed] [Google Scholar]
- 4.Burton, D. R., R. C. Desrosiers, R. W. Doms, W. C. Koff, P. D. Kwong, J. P. Moore, G. J. Nabel, J. Sodroski, I. A. Wilson, and R. T. Wyatt. 2004. HIV vaccine design and the neutralizing antibody problem. Nat. Immunol. 5:233-236. [DOI] [PubMed] [Google Scholar]
- 5.Burton, D. R., R. L. Stanfield, and I. A. Wilson. 2005. Antibody versus HIV in a clash of evolutionary titans. Proc. Natl. Acad. Sci. U. S. A. 102:14943-14948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chou, T. C., and P. Talaly. 1977. A simple generalized equation for the analysis of multiple inhibitions of Michaelis-Menten kinetic systems. J. Biol. Chem. 252:6438-6442. [PubMed] [Google Scholar]
- 7.Davis, K. L., F. Bibollet-Ruche, H. Li, J. M. Decker, O. Kutsch, L. Morris, A. Salomon, A. Pinter, J. A. Hoxie, B. H. Hahn, P. D. Kwong, and G. M. Shaw. 2009. Human immunodeficiency virus type 2 (HIV-2)/HIV-1 envelope chimeras detect high titers of broadly reactive HIV-1 V3-specific antibodies in human plasma. J. Virol. 83:1240-1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Davis, K. L., E. S. Gray, P. L. Moore, J. M. Decker, A. Salomon, D. C. Montefiori, B. S. Graham, M. C. Keefer, A. Pinter, L. Morris, B. H. Hahn, and G. M. Shaw. 2009. High titer HIV-1 V3-specific antibodies with broad reactivity but low neutralizing potency in acute infection and following vaccination. Virology 387:414-426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Decker, J. M., F. Bibollet-Ruche, X. Wei, S. Wang, D. N. Levy, W. Wang, E. Delaporte, M. Peeters, C. A. Derdeyn, S. Allen, E. Hunter, M. S. Saag, J. A. Hoxie, B. H. Hahn, P. D. Kwong, J. E. Robinson, and G. M. Shaw. 2005. Antigenic conservation and immunogenicity of the HIV coreceptor binding site. J. Exp. Med. 201:1407-1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Derdeyn, C. A., J. M. Decker, F. Bibollet-Ruche, J. L. Mokili, M. Muldoon, S. A. Denham, M. L. Heil, F. Kasolo, R. Musonda, B. H. Hahn, G. M. Shaw, B. T. Korber, S. Allen, and E. Hunter. 2004. Envelope-constrained neutralization-sensitive HIV-1 after heterosexual transmission. Science 303:2019-2022. [DOI] [PubMed] [Google Scholar]
- 11.Eda, Y., T. Murakami, Y. Ami, T. Nakasone, M. Takizawa, K. Someya, M. Kaizu, Y. Izumi, N. Yoshino, S. Matsushita, H. Higuchi, H. Matsui, K. Shinohara, H. Takeuchi, Y. Koyanagi, N. Yamamoto, and M. Honda. 2006. Anti-V3 humanized antibody KD-247 effectively suppresses ex vivo generation of human immunodeficiency virus type 1 and affords sterile protection of monkeys against a heterologous simian/human immunodeficiency virus infection. J. Virol. 80:5563-5570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eda, Y., M. Takizawa, T. Murakami, H. Maeda, K. Kimachi, H. Yonemura, S. Koyanagi, K. Shiosaki, H. Higuchi, K. Makizumi, T. Nakashima, K. Osatomi, S. Tokiyoshi, S. Matsushita, N. Yamamoto, and M. Honda. 2006. Sequential immunization with V3 peptides from primary human immunodeficiency virus type 1 produces cross-neutralizing antibodies against primary isolates with a matching narrow-neutralization sequence motif. J. Virol. 80:5552-5562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guex, N., A. Diemand, and M. C. Peitsch. 1999. Protein modeling for all. Trends Biochem. Sci. 24:364-367. [DOI] [PubMed] [Google Scholar]
- 14.Haim, H., Z. Si, N. Madani, L. Wang, J. R. Courter, A. Princiotto, A. Kassa, M. DeGrace, K. McGee-Estrada, M. Mefford, D. Gabuzda, A. B. Smith III, and J. Sodroski. 2009. Soluble CD4 and CD4-mimetic compounds inhibit HIV-1 infection by induction of a short-lived activated state. PLoS Pathog. 5:e1000360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Halgren, T. A. 1996. Merck molecular force field. I. Basis, form, scope, parameterization, and performance of MMFF94. J. Comput. Chem. 17:490-519. [Google Scholar]
- 16.Hatada, M., K. Yoshimura, S. Harada, J. Shibata, and S. Matsushita. 2010. HIV-1 evasion of a neutralizing anti-V3 antibody involves acquisition of a potential glycosylation site in V2. J. Gen. Virol. 91:1335-1345. [DOI] [PubMed]
- 17.Hope, T. J., X. J. Huang, D. McDonald, and T. G. Parslow. 1990. Steroid-receptor fusion of the human immunodeficiency virus type 1 Rev transactivator: mapping cryptic functions of the arginine-rich motif. Proc. Natl. Acad. Sci. U. S. A. 87:7787-7791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Johnson, W. E., and R. C. Desrosiers. 2002. Viral persistence: HIV's strategies of immune system evasion. Annu. Rev. Med. 53:499-518. [DOI] [PubMed] [Google Scholar]
- 19.Kimura, T., K. Yoshimura, K. Nishihara, Y. Maeda, S. Matsumi, A. Koito, and S. Matsushita. 2002. Reconstitution of spontaneous neutralizing antibody response against autologous human immunodeficiency virus during highly active antiretroviral therapy. J. Infect. Dis. 185:53-60. [DOI] [PubMed] [Google Scholar]
- 20.Kwong, P. D., R. Wyatt, J. Robinson, R. W. Sweet, J. Sodroski, and W. A. Hendrickson. 1998. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature 393:648-659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lu, M., S. C. Blacklow, and P. S. Kim. 1995. A trimeric structural domain of the HIV-1 transmembrane glycoprotein. Nat. Struct. Biol. 2:1075-1082. [DOI] [PubMed] [Google Scholar]
- 22.Lusso, P., P. L. Earl, F. Sironi, F. Santoro, C. Ripamonti, G. Scarlatti, R. Longhi, E. A. Berger, and S. E. Burastero. 2005. Cryptic nature of a conserved, CD4-inducible V3 loop neutralization epitope in the native envelope glycoprotein oligomer of CCR5-restricted, but not CXCR4-using, primary human immunodeficiency virus type 1 strains. J. Virol. 79:6957-6968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Madani, N., A. Schon, A. M. Princiotto, J. M. Lalonde, J. R. Courter, T. Soeta, D. Ng, L. Wang, E. T. Brower, S. H. Xiang, Y. D. Kwon, C. C. Huang, R. Wyatt, P. D. Kwong, E. Freire, A. B. Smith III, and J. Sodroski. 2008. Small-molecule CD4 mimics interact with a highly conserved pocket on HIV-1 gp120. Structure 16:1689-1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maeda, Y., K. Yusa, and S. Harada. 2008. Altered sensitivity of an R5X4 HIV-1 strain 89.6 to coreceptor inhibitors by a single amino acid substitution in the V3 region of gp120. Antivir. Res. 77:128-135. [DOI] [PubMed] [Google Scholar]
- 25.Schön, A., N. Madani, J. C. Klein, A. Hubicki, D. Ng, X. Yang, A. B. Smith III, J. Sodroski, and E. Freire. 2006. Thermodynamics of binding of a low-molecular-weight CD4 mimetic to HIV-1 gp120. Biochemistry 45:10973-10980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shibata, J., K. Yoshimura, A. Honda, A. Koito, T. Murakami, and S. Matsushita. 2007. Impact of V2 mutations on escape from a potent neutralizing anti-V3 monoclonal antibody during in vitro selection of a primary human immunodeficiency virus type 1 isolate. J. Virol. 81:3757-3768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thali, M., J. P. Moore, C. Furman, M. Charles, D. D. Ho, J. Robinson, and J. Sodroski. 1993. Characterization of conserved human immunodeficiency virus type 1 gp120 neutralization epitopes exposed upon gp120-CD4 binding. J. Virol. 67:3978-3988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang, F. X., T. Kimura, K. Nishihara, K. Yoshimura, A. Koito, and S. Matsushita. 2002. Emergence of autologous neutralization-resistant variants from preexisting human immunodeficiency virus (HIV) quasispecies during virus rebound in HIV type 1-infected patients undergoing highly active antiretroviral therapy. J. Infect. Dis. 185:608-617. [DOI] [PubMed] [Google Scholar]
- 29.Wyatt, R., P. D. Kwong, E. Desjardins, R. W. Sweet, J. Robinson, W. A. Hendrickson, and J. G. Sodroski. 1998. The antigenic structure of the HIV gp120 envelope glycoprotein. Nature 393:705-711. [DOI] [PubMed] [Google Scholar]
- 30.Yamada, Y., C. Ochiai, K. Yoshimura, T. Tanaka, N. Ohashi, T. Narumi, W. Nomura, S. Harada, S. Matsushita, and H. Tamamura. 2010. CD4 mimics targeting the mechanism of HIV entry. Bioorg. Med. Chem. Lett. 20:354-358. [DOI] [PubMed] [Google Scholar]
- 31.Yoshimura, K., R. Kato, M. F. Kavlick, A. Nguyen, V. Maroun, K. Maeda, K. A. Hussain, A. K. Ghosh, S. V. Gulnik, J. W. Erickson, and H. Mitsuya. 2002. A potent human immunodeficiency virus type 1 protease inhibitor, UIC-94003 (TMC-126), and selection of a novel (A28S) mutation in the protease active site. J. Virol. 76:1349-1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yoshimura, K., R. Kato, K. Yusa, M. F. Kavlick, V. Maroun, A. Nguyen, T. Mimoto, T. Ueno, M. Shintani, J. Falloon, H. Masur, H. Hayashi, J. Erickson, and H. Mitsuya. 1999. JE-2147: a dipeptide protease inhibitor (PI) that potently inhibits multi-PI-resistant HIV-1. Proc. Natl. Acad. Sci. U. S. A. 96:8675-8680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yoshimura, K., J. Shibata, T. Kimura, A. Honda, Y. Maeda, A. Koito, T. Murakami, H. Mitsuya, and S. Matsushita. 2006. Resistance profile of a neutralizing anti-HIV monoclonal antibody, KD-247, that shows favourable synergism with anti-CCR5 inhibitors. AIDS 20:2065-2073. [DOI] [PubMed] [Google Scholar]
- 34.Yusa, K., Y. Maeda, A. Fujioka, K. Monde, and S. Harada. 2005. Isolation of TAK-779-resistant HIV-1 from an R5 HIV-1 GP120 V3 loop library. J. Biol. Chem. 280:30083-30090. [DOI] [PubMed] [Google Scholar]
- 35.Zhao, Q., L. Ma, S. Jiang, H. Lu, S. Liu, Y. He, N. Strick, N. Neamati, and A. K. Debnath. 2005. Identification of N-phenyl-N′-(2,2,6,6-tetramethyl-piperidin-4-yl)-oxalamides as a new class of HIV-1 entry inhibitors that prevent gp120 binding to CD4. Virology 339:213-225. [DOI] [PubMed] [Google Scholar]