Abstract
Simian retroviruses are precursors of all human retroviral pathogens. However, little is known about the prevalence and coinfection rates or the genetic diversity of major retroviruses—simian immunodeficiency virus (SIV), simian T-cell lymphotropic virus type 1 (STLV-1), and simian foamy virus (SFV)—in wild populations of nonhuman primates. Such information would contribute to the understanding of the natural history of retroviruses in various host species. Here, we estimate these parameters for wild West African red colobus monkeys (Piliocolobus badius badius) in the Taï National Park, Côte d'Ivoire. We collected samples from a total of 54 red colobus monkeys; samples consisted of blood and/or internal organs from 22 monkeys and additionally muscle and other tissue samples from another 32 monkeys. PCR analyses revealed a high prevalence of SIV, STLV-1, and SFV in this population, with rates of 82%, 50%, and 86%, respectively. Forty-five percent of the monkeys were coinfected with all three viruses while another 32% were coinfected with SIV in combination with either STLV or SFV. As expected, phylogenetic analyses showed a host-specific pattern for SIV and SFV strains. In contrast, STLV-1 strains appeared to be distributed in genetically distinct and distant clades, which are unique to the Taï forest and include strains previously described from wild chimpanzees in the same area. The high prevalence of all three retroviral infections in P. b. badius represents a source of infection to chimpanzees and possibly to humans, who hunt them.
Lentiviruses and deltaretroviruses that infect African nonhuman primates have received considerable attention as they are the precursors of all pathogenic human retroviruses: human immunodeficiency virus types 1 and 2 (HIV-1/HIV-2) and human T-cell lymphotropic virus type 1 (HTLV-1). These human infections are the results of past zoonotic transfers of simian immunodeficiency virus (SIV) and simian T-cell lymphotropic viruses type 1 (STLV-1) from wild monkeys and apes into local human populations, presumably through primate hunting and handling of primate bushmeat (13, 19, 43, 46, 55, 58, 59). Via the same route, zoonotic transmission of simian foamy virus (SFV), a spumaretrovirus whose exact pathogenicity in human hosts is still unknown, has also been shown (64). The increasing contact between humans and wild primates implies that further zoonotic transmission of retroviruses is likely to happen (42, 63). Studying the occurrence and circulation of simian retroviruses such as SIV, STLV-1, and SFV in wild primate populations enables us to better understand retrovirus evolution in primates and also provides tools for monitoring possible future retroviral zoonotic events.
Systematic studies of SIV, STLV-1, and SFV in wild primates are relatively rare. Many use bushmeat samples, which can vary in their quality and are prone to cross-contamination from butchering and storage with other carcasses. Confiscated primates are also not representative of the situation in the wild since the animals are caught at a young age when the occurrence of different retroviruses may be extremely low (24). The technical possibilities for the detection of various pathogens in noninvasive samples such as urine and feces have greatly improved and are frequently used; however, in general, the sensitivity of detection methods is higher when blood and tissue samples are used (25, 32, 47). Such samples can be collected if fresh carcasses are found, or they can be collected by anesthetizing live primates for sampling purpose, animal translocation, or medical intervention, such as snare removal. The practical and ethical issues of each of the sampling methods have been discussed elsewhere (12, 14).
Red colobus monkeys [Procolobus (Piliocolobus)] are interesting subjects for retroviral infection studies for a number of reasons. First, they are widely distributed (yet in a fragmented manner) from East to West Africa, which suggests that red colobus species and subspecies, or more likely ancestor(s) of these, could have been key hosts in transmitting retroviruses across tropical Africa (4, 54). Second, as they are herbivore primates, the hunting of other primates can be excluded as a route of infection. Finally, these monkeys are frequently hunted by humans and chimpanzees and represent a possibly large reservoir for retroviruses and other pathogens that ought to be investigated further (2, 45).
Very little information is available about the prevalence and coinfection of SIV, STLV-1, and SFV in wild red colobus monkeys across Africa. In other colobine monkeys only SIV has been documented: in olive colobus (Procolobus verus) in Côte d'Ivoire and in black and white colobus (Colobus guereza) in Cameroon (7, 8). Based on fecal samples from habituated adult individuals, the prevalence of SIV in West African red colobus monkeys (SIVwrc; local subspecies, Piliocolobus badius badius) has been estimated to a minimum of 26% in the Taï National Park, Côte d'Ivoire, but the authors recognized the low sensitivity of viral RNA detection in fecal samples (34). Another study conducted on the same population revealed that 5 out of 10 blood samples were SIV positive (7). These results highlight that the most reliable prevalence data are based on analyses of blood/tissue samples although such sampling is not always feasible for reasons discussed above. Published prevalence information concerning STLV-1 and SFV in wild red colobus monkeys (STLV-1wrc and SFVwrc) in the same area is restricted to results obtained from analyses of a limited number of blood and necropsy samples collected as a part of studies whose focus was on cross-species transmission of these two viruses to chimpanzees (27, 28). However, these samples indicated a high prevalence of STLV-1wrc and SFVwrc in the red colobus monkey population (56% and 90%, respectively). A recent study from Uganda, East Africa, estimated the prevalence of SIV, STLV-1, and SFV in another red colobus species (Piliocolobus rufomitratus tephrosceles) to be 22.6%, 6.4%, and 97%, respectively (15). The study was performed using blood samples collected from anesthetized wild red colobus monkeys living in their natural habitat, which allowed reliable assessment of the prevalence and genetic diversity of these three retroviruses.
The preliminary data from the Taï National Park indicate that there might be great variation in the prevalence of retroviruses across the African continent, even in closely related species of wild primates. Here, we aimed at generating reliable prevalence and coinfection data for SIVwrc, STLV-1wrc, and SFVwrc based on the analysis of blood and tissue samples from wild Western red colobus monkeys. We expected that this would allow for proper comparison of retroviral prevalence in the allied species P. b. badius and P. r. tephrosceles.
MATERIALS AND METHODS
Study site and animals.
Field work was conducted in the evergreen rainforest of the Taï National Park in Côte d'Ivoire, West Africa (5°15′ to 6°07′N, 7°25′ to 7°54′W). The West African red colobus monkey (P. b. badius) is the most abundant primate species in this forest and shares its habitat with eight other diurnal primates: chimpanzee (Pan troglodytes verus), sooty mangabey (Cercocebus atys), black-and-white colobus (Colobus polykomos), olive colobus (P. verus), Diana monkey (Cercopithecus diana), lesser spot-nosed monkey (Cercopithecus petaurista), Campbell's monkey (Cercopithecus campbelli), and greater spot-nosed monkey (Cercopithecus nictitans) (36). Samples were collected from wild nonhabituated West African red colobus monkeys within an area of approximately 100 km2.
Sample collection.
Blood samples were collected from 10 adult red colobus monkeys under general anesthesia (28). At the same time other biological samples and anatomical measurements were collected. The samples were centrifuged shortly after collection, and the cell-rich layer (buffy coat) was frozen immediately in liquid nitrogen. As part of a chimpanzee health monitoring project, veterinarians have been performing necropsies on all carcasses of any species found in the forest. Since the year 2001 necropsy samples have been collected from 12 adult red colobus monkey carcasses, and these were transported on ice directly to the camp. Chimpanzees in the area regularly hunt red colobus (2) and sometimes leave behind pieces of muscle or other tissues from their prey. The veterinary project collected such tissue samples from observed hunts of 32 individual red colobus monkeys as soon as the chimpanzees had left the site. These samples were collected using single-use gloves, transported at ambient temperature, and preserved at camp a maximum of 12 h after the death of the monkey. For the necropsies and chimpanzee meal remains, multiple samples were collected if possible, and up to three samples per individual were analyzed. All samples were stored in liquid nitrogen at the field site and later transported on dry ice to Robert Koch Institute, Berlin, Germany, where samples were stored at −80°C until analysis. All parts of the study were performed under permission of the Ministry of Research and the National Park authorities of Côte d'Ivoire.
DNA extraction, PCR, and sequencing.
DNA was extracted using either a DNA tissue kit or DNA blood kit (Qiagen, Hilden, Germany).
Samples were tested for SIV with a seminested PCR with primers specifically designed for the detection of pol regions of SIV from the Western red colobus/olive colobus (SIVwrc/SIVolc) group (SIVwrc S1 [CAT GGC AAA TGG ATT GTA CTC A], SIVwrc R2 [GTG CCA TTG CTA ATG CTG TTT C], SIVwrc S3 [CCA AAT TCT TGT TCT ATC CCT AAC C], and SIVwrc R3 [AGC AAA AAT CAT ATC AGC AGA AGA T]). These primers were based on SIVwrc and SIVolc sequences published by Courgnaud and colleagues (7). We used the primers SIVwrc S1 and SIVwrc R2 in the first round PCR, and the primer pair SIVwrc S1 and SIVwrc R3 (expected amplicon size approximately 250 bp) and the pair SIVwrc S3 and SIVwrc R2 (expected amplicon size approximately 300 bp) were used in two parallel seminested PCRs. The cycler conditions were 94°C for 5 min and 30 cycles of 94°C for 15 s, 55°C for 30 s, 72°C for 30 s, with a final step of 72°C for 10 min and then cooling to 4°C. SIVwrc-negative samples and one sample from each SIVwrc-positive individual were also tested with a generic SIV PCR known to detect most primate lentiviruses to determine if the monkeys also carried other types of SIV (6). We used the primers DR1 (TRC AYA CAG GRG CWG AYG A) and DR2 (AIA DRT CAT CCA TRT AYT G) in the first round PCR and primers DR4 (GGI ATW CCI CAY CCD GCA GG) and DR5 (GGI GAY CCY TTC CAY CCY TGH GG) in a nested PCR. The cycler conditions were 94°C for 2 min and 30 cycles of 94°C for 15 s, 50°C decreasing by 0.5°C each cycle to 35°C for 30 s, and 72°C for 1 min; this was followed by 15 cycles of 94°C for 15 s, 50°C for 30 s, and 72°C for 1 min, with a final step at 72°C for 5 min and then cooling to 4°C. The expected amplicon size was 194 bp.
Samples were tested for proviral DNA of STLV-1 by a tax-specific real-time PCR (23). We used the primers SK43 (CGG ATA CCC AGT CTA CGT GT) and SK44 (GAG CCG ATA ACG CGT CCA TCG) and the probe HTLV TAX TM (6FAM-CGC CCT ATG GCC ACC TGT CCA GA XT P; 6FAM is 6-carboxyfluorescein), and the cycler conditions were 95°C for 10 min and 45 cycles of 95°C for 15 s and 60°C for 35 s. The expected amplicon size was approximately 190 bp. A fragment of the long terminal repeat (LTR) region was then sequenced from positive samples as this region of the primate T-cell lymphotropic virus type 1 (PTLV-1) genome evolves more rapidly and is frequently used for phylogenetic analyses. We used the primers S10, H, and X (26, 27) derived from nucleotides 7929 to 7948, 8756 to 8735, and 8296 to 8316, respectively, from the prototype HTLV-1 sequence ATK (accession number J02029) (48). We used primer S10 (GGC CCT AAT AAT TCT ACC CG) and primer H (AGT TCA GGA GGC ACC ACA GGC G) for the first round and primer X (GAG CTC GAG CAG ATG ACA ATG ACC ATG AG) and primer H in a seminested PCR. The cycler conditions were 94°C for 5 min and 35 cycles of 94°C for 30 s, 58°C for 30 s, and 72°C for 1 min (30 s for seminested PCR), with a final step at 72°C for 10 min and then cooling to 4°C. The expected amplicon size for the seminested PCR was 450 bp.
Samples were tested for SFV with a PCR specifically designed to amplify a fragment of SFVwrc pol (28). We used the primers SFVwrc 1s (CAT ACA ATT ACC ACT CCA AGC CT), SFVwrc 2as (CAG ACA AAT CCA GTC ATA CCA TC), SFVwrc 3s (CTC AGT ACT GGT GGC CAA ATC TTA GA), and SFVwrc 4as (CCA GTC ATA CCA TCG ACT ACT ACA AGG). In the first-round PCR we used primers SFVwrc 1s and SFVwrc 2as, and then for two parallel seminested PCRs we used primers SFVwrc 1s and SFVwrc 4as (expected amplicon size of approximately 430 bp) and primers SFVwrc 3s and SFVwrc 2as (expected amplicon size of approximately 270 bp); the cycler conditions were 96°C for 5 min and 40 cycles of 96°C for 1 min, 56°C for 30 s, and 72°C for 1 min, with a final step at 72°C for 10 min and then cooling to 4°C. One sample from each individual was also tested with a generic SFV PCR (15) to check for the presence of other non-red colobus strains of SFV. We used the primers SIF2 (TAG CWG AYA ARC TTG CCA CCC AAG G) and SIR1 (GTC GTT TWA TIT CAC TAT TTT TCC TTT CCA C) in the first round and the primers SIF3 (CCA ARC CTG GAT GCA GAG YTG GAT CA) and SIR3 (ACT TTG GGG RTG RTA AGG AGT ACT G) in a nested PCR. The cycler conditions were 95°C for 5 min and 40 cycles of 95°C for 30 s, 45°C for 45 s, and 72°C for 1 min, with a final step at 72°C for 10 min and then cooling to 4°C. The expected amplicon size for was approximately 630 bp.
PCR products were visualized with gel electrophoresis before being purified. Gel extraction was performed when necessary. Sequencing was performed in both directions using the Sanger method, with all PCR products being sequenced on both strands. Comparison to the public database using NCBI BLAST (1) always confirmed that the expected proviral sequences had been amplified.
Prevalence and coinfection of SIV, STLV-1, and SFV and correlation between infections.
The prevalence of the three retroviruses was calculated in Stata (Stata/SE, version 10.0, for Windows; Stata Corp., College Station TX), as well as the corresponding 95% confidence interval (CI) for proportions (normal approximation). We calculated the percentage of monkeys with single infections of the individual viruses, and in order to investigate if infections were linked to each other, we also calculated the percentage of individual monkeys with dual (for all possible viral combinations) or triple infections. Kendall tau-b test, including Fisher's exact test, was used to determine the degree of correlation between the infections. Prevalence and coinfection of SIV, STLV-1, and SFV and the correlation between infections were calculated on basis of the results obtained from buffy coat samples and necropsy samples only. The samples collected after chimpanzee meals were considered not to be of sufficient quality to be included in these analyses. These samples, however, were used to obtain additional nucleotide sequences for the phylogenetic analyses.
Sequence analyses.
Newly generated sequences were added to data sets considered to encompass the overall genetic diversity of SIV, STLV-1, and SFV. Alignments were edited manually using SeaView, version 4 (16). To allow detection of saturation, the number of transitions and transversions versus divergence was first plotted in DAMBE (65). If distances calculated under the global time reversible (GTR) model are chosen as a measure of divergence, then both transitions (ts) and transversions (tv) should increase in a linear manner with GTR distance. In general, ts should also accumulate faster than tv. However, with GTR distance increasing, multiple substitutions are expected to occur at the same sites, ultimately leading to the loss of the initial correlation. Saturation should thus translate into plateauing plots, with tv outnumbering ts (30). While the PTLV-1 data set was apparently exempt from saturation, the SIV and SFV data sets exhibited clear patterns of ts saturation at the third positions of codons. Accordingly, we performed all following analyses on SIV and SFV data sets stripped of this position (a conservative approach). All three data sets were haplotyped (reduced to unique sequences) using FaBox (60). The overall process resulted in our starting data sets being composed as follows: (i) SIV pol, 75 taxa and 152 bp; (ii) PTLV-1 LTR, 47 taxa and 422 bp; and (iii) SFV pol, 51 taxa and 252 bp.
Nucleotide substitution models to obtain the best fit for the data were then selected using jModeltest, version 0.1.1 (17, 44). According to the Akaike information criterion (AIC), comparisons of model likelihoods were most favorable to GTR+I+G (GTR with a proportion of invariant sites [I] and gamma-distributed [G] rate heterogeneity) (SIV), Hasegawa Kishino and Yano (HKY)+I+G (PTLV-1), and GTR+G (SFV). Phylogenetic analyses were performed in both maximum-likelihood (ML) and Bayesian frameworks, under the appropriate model of nucleotide substitution. ML analyses were performed on the PhyML webserver (http://www.atgc-montpellier.fr/phyml/) (17, 18). Equilibrium frequencies, topology, and branch lengths were optimized; the starting tree was determined using BioNJ and both nearest-neighbor interchange (NNI) and subtree pruning and regrafting (SPR) algorithms of the tree search were used (keeping the best outcome). Branch robustness was assessed by performing nonparametric bootstrapping (500 replicates). Bayesian analyses were performed using BEAST, version 1.5.3 (11). Besides allowing modeling nucleotide substitution processes, BEAST also allows for modeling rate variation among tree branches and tree shape. All analyses were run under the assumption of a relaxed, uncorrelated log-normal clock. For SIV and SFV data sets, analyses were performed assuming two different tree shape speciation models (Yule and birth-death processes). Given the expected relatively shallow depth of the PTLV-1 phylogenetic tree, a speciation model (birth-death process) and a coalescent model (constant population size) were employed. Two runs of 10,000,000 generations were run per data set per tree shape model (i.e., four runs total for each data set). Trees and numerical values taken were sampled every 1,000 generations. Tracer, version 1.5, was used to check that individual runs had reached convergence, that independent runs converged, and that chain mixing was satisfactory (effective sample size values of >200) (11). Trees sampled in duplicate runs were then gathered into a single file (after removal of a visually conservative 10% burn-in period) using LogCombiner, version 1.5.3 (distributed with BEAST), and the information of 18,000 trees per data set per tree shape model was summarized onto the maximum clade credibility tree using TreeAnnotator, version 1.5.3 (distributed with BEAST). Posterior probabilities (pps) were taken as a measure of branch robustness.
For Bayesian analyses, no major discrepancy in topology or branch support was detectable using different tree shape models. ML and Bayesian methods globally supported congruent topologies with consistent branch supports (even though bootstrap and posterior probability are not directly comparable [10]). All xml files (including sequence alignments) used for Bayesian analyses are available at http://sebastiencalvignac.fr/emergingzoonoses/index.html. Figures summarizing phylogenetic analyses were drawn using FigTree, version 1.3.1 (http://tree.bio.ed.ac.uk/software/figtree/).
Nucleotide sequence accession numbers.
All sequences generated in this study were deposited in GenBank under the accession numbers FN825787 to FN825803 and FN859997 to FN860025.
RESULTS
Prevalence of SIV, STLV-1, and SFV.
All three viruses were detected by specific PCR and confirmed with sequencing and BLAST. Further, all newly sequenced SIV, STLV-1, and SFV strains could be linked to strains previously found in red colobus monkeys. Results are summarized in Table 1. SIV1wrc was detected in 8 out of 10 anesthetized monkeys and in 10 out of 12 carcasses. The overall prevalence was 82% (95% CI, 66 to 98%). STLV-1wrc was detected in six anesthetized monkeys and in five carcasses. The overall prevalence was 50% (95% CI, 29 to 71%). SFVwrc was detected in all anesthetized monkeys and in nine carcasses. The overall prevalence was 86% (95% CI, 72 to 100%). In the samples from chimpanzee meal remains (attributed to 32 red colobus monkeys), SIV, STLV-1, and SFV were detected in five, nine, and six individuals, respectively.
TABLE 1.
Overview of red colobus PCR results for SIV, STLV-1, and SFVa
| Red colobus animal no. | Origin of sample(s)b | Sample(s) tested | PCR result |
||
|---|---|---|---|---|---|
| SIV (wrc/generic) | STLV | SFV (wrc/generic)c | |||
| 124 | Darted | Buffy coat | + | + | + |
| 125 | Darted | Buffy coat | + | + | + |
| 126 | Darted | Buffy coat | + | + | + |
| 127 | Darted | Buffy coat | + | − | + |
| 128 | Darted | Buffy coat | + | + | + |
| 129 | Darted | Buffy coat | + | + | + |
| 130 | Darted | Buffy coat | + | + | + |
| 131 | Darted | Buffy coat | − | − | + |
| 132 | Darted | Buffy coat | + | − | −/+ |
| 133 | Darted | Buffy coat | − | − | + |
| 3 | Necropsy | Spleen, lymph node | + | − | −/+ |
| 12 | Necropsy | Spleen, lymph node | + | + | + |
| 23 | Necropsy | Lung, heart | − | − | − |
| 28 | Necropsy | Liver | + | − | +/− |
| 43 | Necropsy | Spleen, lung, muscle | + | + | − |
| 66 | Necropsy | Lung, unidentified tissue | + | + | + |
| 71 | Necropsy | Lung | − | − | + |
| 72 | Necropsy | Liver, spleen | + | + | + |
| 213 | Necropsy | Spleen, kidney, muscle | + | − | + |
| 236 | Necropsy | Lymph node | + | − | + |
| 268 | Necropsy | Lymph node, intestine | −/+ | − | − |
| 276 | Necropsy | Liver, lung | + | + | + |
| 15 | CMR | Muscle, bone marrow | +/− | + | − |
| 29 | CMR | Blood | − | + | − |
| 30 | CMR | Blood in RNA later | − | + | − |
| 45 | CMR | Blood | − | − | +/− |
| 68 | CMR | Trachea, lymph node | − | + | + |
| 204 | CMR | Blood | +/no material | − | −/no material |
| 210 | CMR | Muscle, blood | − | + | + |
| 211 | CMR | Muscle | − | + | − |
| 212 | CMR | Muscle | +/− | + | − |
| 269 | CMR | Muscle, brain | −/+ | − | − |
| 278 | CMR | Muscle, bone marrow | − | + | +/no material |
| 279 | CMR | Muscle, bone marrow | − | − | + |
| 280 | CMR | Muscle | +/− | + | +/no material |
| Remaining 19 animals | CMR | Muscle (7), blood (7), other tissue (7)d | −e | − | −f |
For SIV and SFV two different primer sets were used; where results agree only one result appears in the table.
CMR, chimpanzee meal remain.
Samples were tested with specific primers for red colobus virus (wrc) and with generic primers for virus.
Numbers in parentheses are numbers of samples tested.
Partial result; no material was available to test four individuals with generic primers for SIV.
Partial result; no material was available to test five individuals with generic primers for SFV.
SIV, SFV, and STLV-1 coinfections and correlation between infections.
Forty-five percent (n = 10) of the monkeys were coinfected with all three viruses, 27% (n = 6) were infected with SIV in combination with SFV, and 5% (n = 1) were infected with SIV in combination with STLV-1. Fourteen percent (n = 3) were infected with SFV only, and 5% (n = 1) were infected with SIV only. Five percent (n = 1) were negative for all viruses. Of note, no monkey was infected with STLV-1 only or with the virus combination STLV-1 and SFV. There was no statistically significant correlation between any of the infections (Kendall tau-b coefficient and Fisher exact P value of 0.47 and 0.09 for SIV/STLV-1, 0.16 and 0.47 for SIV/SFV, and 0.13 and 1.00 for STLV-1/SFV, respectively).
Phylogenetic analyses.
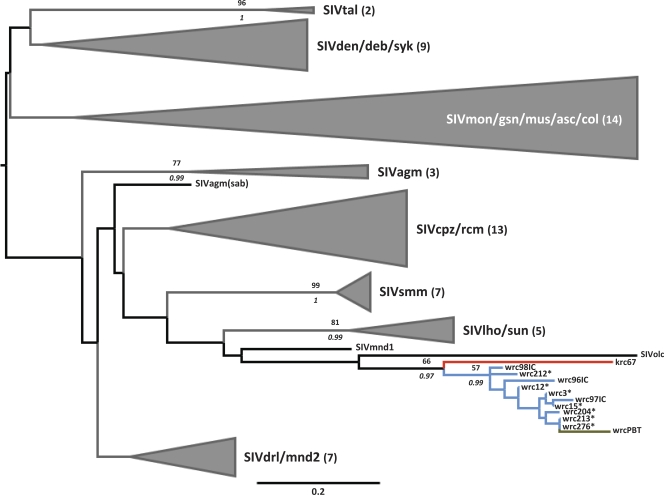
To infer the phylogenetic relationships of the newly described SIV, STLV-1, and SFV strains with previously characterized strains, we used ML and Bayesian methods. The main pattern of SIV host specificity was retrieved by these analyses (Fig. 1). SIVwrc sequences from P. b. badius were found to form a clade with SIVwrc strains previously identified from P. b. badius from the same area and Piliocolobus badius temminckii from Gambia, the branch defining the bipartition receiving reasonable statistical support (bootstrap value [Bp], 57; pp, 0.99) (Fig. 1). The sister taxon to this group appeared to be the strain identified from Kibale P. r. tephrosceles, which also grouped with other SIVwrc with reasonable statistical support (Bp of 66; pp of 0.97) (Fig. 1). SIVolc, a strain identified from an olive colobus monkey [Procolobus (Procolobus) verus] appeared as completing a big colobine clade in the ML analysis (Bp of 58) but was found to group with the clade comprised of SIV in L'Hoest's monkey (Cercopithecus lhoesti) and the sun-tailed guenon (Cercopithecus solatus) (SIVlho/sun) in Bayesian analyses, though with very low branch support (pp of 0.57).
FIG. 1.
Maximum-likelihood tree based on the analysis of SIV partial pol sequences (152 bp). The topologies of Bayesian maximum clade credibility trees obtained under two different tree priors were similar when shallow evolutionary depths were considered (deep branching patterns were not similar). Branches leading to strains isolated from red colobus monkeys in Côte d'Ivoire are blue, those leading to strains isolated from Ugandan red colobus monkeys are red, and the one branch leading to a Gambian strain is green. Major SIV groups are represented graphically to improve readability: in every case the number of strains represented is indicated in parentheses after group names. Numbers above branches represent bootstrap values (Bp); italicized numbers below branches represent posterior probability values (pp) obtained using the birth-death model. Bp and pp are indicated only where Bp is ≥50 and pp is ≥0.95. Asterisks indicate strains identified in the present study. Note that this tree is mid-point rooted due to the lack of information regarding the position of the root in SIV phylogeny. Tal, talapoin monkey (Miopithecus talapoin); den, Dent's monkey (Cercopithecus denti); deb, De Brazza's guenon (Cercopithcus neglectus); syk, Sykes'monkey (Cercopithecus mitis); mon, mona monkey (Cercopithecus mona); gsn, greater spot-nosed guenon (Cercopithecus nictitans); mus, mustached guenon (Cercopithecus cephus); asc, red-tailed guenon (Cercopithecus ascanius); col, mantled guereza (Colobus guereza); agm, African green monkey (Chlorocebus aethiops); cpz, chimpanzee (Pan troglodytes); rcm, red-capped mangabey (Cercocebus torquatus); smm, sooty mangabey monkey (Cercocebus atys); lho, L'Hoest's monkey (Cercopithecus lhoesti); sun, sun-tailed guenon (Cercopithecus solatus); mnd, mandrill (Mandrillus sphinx); drl, drill (Mandrillus leucophaeus); olc, olive colobus monkey (Procolobus verus); wrc, Western red colobus (P. b. badius); krc, Eastern red colobus (P. r. tephrosceles); wrcPBT, Western red colobus (P. b. temminckii).
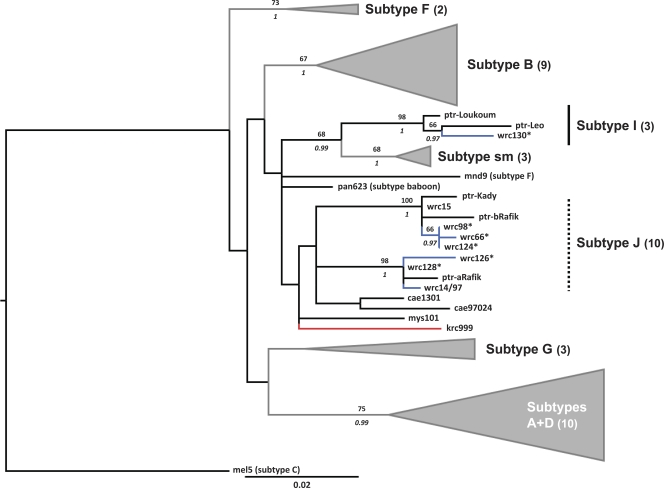
The general design of PTLV-1 strains clustering into geographical subtypes was retrieved by the analyses whose results are summarized in Fig. 2. Into that scheme, our STLV-1wrc sequences from P. b. badius did not form one monophyletic group. Sequences were distributed in three distinct clades, corresponding to subtype I or nested into subtype J (Fig. 2), as defined by Junglen et al. (21). In all cases, these clades were well supported and comprised chimpanzee strains (Bp of 98 to 99; pp of 1) (Fig. 2). The P. r. tephrosceles strain did not exhibit a close relationship to any of the STLV-1wrc strains found in the Taï forest, nor did it show particular affinity to any of the previously described PTLV-1 subtypes (Fig. 2).
FIG. 2.
Maximum-likelihood tree based on the analysis of PTLV-1 partial LTR sequences (422 bp). The topologies of Bayesian maximum clade credibility trees obtained under two different tree priors were similar when shallow evolutionary depths were considered (deep branching patterns were not similar). Branches leading to strains isolated from red colobus monkeys in Côte d'Ivoire are blue, and those leading to strains isolated from Ugandan red colobus monkeys are red. Major PTLV-1 groups are represented graphically to improve readability: in every case the number of strains represented is indicated in parentheses after group names. Numbers above branches represent Bp values, italicized numbers below branches represent pp values obtained using the birth-death model. Bp and pp are indicated only where Bp is ≥50 and pp is ≥0.95. Asterisks indicate strains identified in the present study. Three of these strains were actually identified in more than one individual: wrc15 (published as STLVwrc [27]) is identical to wrc129* and wrc212* (plus ptr-Dorry); wrc66* is identical to wrc72*; and wrc126* is identical to wrc211*. Sm, sooty mangabey monkey (Cercocebus atys); ptr, chimpanzee (Pan troglodytes); mnd, mandrill (Mandrillus sphinx); cae, African green monkey (Chlorocebus aethiops); msy, Barbary macaque (Macaca sylvanus); wrc, Western red colobus (P. b. badius); krc, Eastern red colobus P. r. tephrosceles).
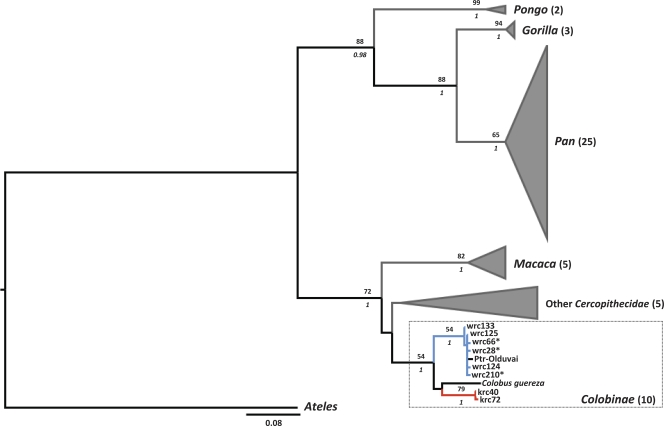
SFV phylogeny exhibited the expected pattern of marked host specificity together with plausible long-term cospeciation (Fig. 3). SFVwrc from P. b. badius respected that rule, forming a monophyletic group supported by reasonable Bp and pp values (Bp of 54; pp of 1) (Fig. 3). P. r. tephrosceles strains clustered together with high support (Bp of 79; pp of 1) (Fig. 3). The existence of a colobine clade was also reasonably supported (Bp of 54: pp of 1) though its inner branching order could not be determined (Fig. 3).
FIG. 3.
Maximum-likelihood tree based on the analysis of SFV partial pol sequences (252 bp). The topologies of Bayesian maximum clade credibility trees obtained under two different tree priors were similar. Branches leading to strains isolated from red colobus monkeys in Côte d'Ivoire are blue, those leading to strains isolated from Ugandan red colobus monkeys are red. Noncolobine tips are represented graphically to improve readability: in every case the number of strains represented is indicated between parentheses after family, subfamily or genus names. Numbers above branches represent bootstrap values (Bp), italicized numbers below branches represent posterior probability values (pp) obtained using the birth-death model. Bp and pp are indicated only where Bp is ≥50 and pp is ≥0.95. Asterisks indicate strains identified in the present study. Two of these strains were actually identified in more than one individual: wrc125 is identical to wrc3*, wrc12, wrc45*, wrc68*, wrc71*, wrc126, wrc127, wrc128, wrc129, wrc130, wrc131, wrc213*, wrc236*, and wrc276* (15 wrc sequences plus ptr-Leo); wrc133 is identical to wrc132*. Ptr, chimpanzee (P. troglodytes); krc, Eastern red colobus (P. r. tephrosceles); wrc, Western red colobus (P. b. badius).
DISCUSSION
Prevalence of SIV, STLV-1, and SFV.
Most data on retroviruses in wild primates are derived from studies based on bushmeat samples or confiscated and captive populations and might therefore not be representative for the situation in the wild. Therefore, the main data we use here for comparison are based on a study by Goldberg et al. (15) using samples obtained from wild primates under anesthesia.
We estimate that the prevalence of SIVwrc in P. b. badius in Taï National Park in Cote d'Ivoire is 82%. This is somewhat higher than previously estimated for this population (26% based on noninvasive samples from 53 individuals and 50% based on 10 blood samples), which could be due to differences in test material and sample sizes (7, 35). However, our results confirm that this population has one of the highest prevalences of this virus found in wild nonhuman primates to date. Further, the prevalence in P. r. badius is more than three times higher than that in the closely related species of red colobus monkey, P. r. tephrosceles, living in Kibale, Uganda. The fact that there is no overlap in the 95% CI of the estimated SIV prevalence in the Kibale study and that in the present study (8 to 37% and 66 to 98% for Kibale and Taï, respectively) shows that there is a significant difference between these two populations/species (15). Previous studies have shown that SIV prevalence varies greatly between species and that some populations, such as mandrills in Cameroon and sooty mangabeys in Côte d'Ivoire, have a high frequency of SIV infection (estimated prevalence of 79% [95% CI, 54 to 99%] and 59% [95% CI, 35 to 88], respectively) (46, 51, 56). The effect that the high rate of SIV occurrence might have on the P. b. badius population is unknown. In general, natural SIV infections have been considered nonpathogenic and even asymptomatic throughout the course of infection (49). However, it has recently been discovered that SIV in wild chimpanzees (SIVcpz) has a negative effect on health and fertility and causes AIDS-like immunopathology in wild chimpanzees from Gombe National Park, Tanzania (22). It is possible that a similar effect will be found in other primate species when additional prospective, long-term follow-up studies of SIV-infected wild primates become available. It should be underlined that this type of study requires continuous investigation, and the discovery of SIV pathogenicity in the Gombe chimpanzees was only possible because good demographic data had been obtained from a long-term field study.
Also for STLV-1 we found that the prevalence is much higher in P. b. badius than in P. r. tephrosceles monkeys, with no overlap found in the 95% CI: 50% in P. b. badius (95% CI, 29 to 71%) and 6.4% in P. r. tephrosceles (95% CI, 0 to 15%) (15). It appears that there is a great deal of variation in the estimated prevalence of STLV-1 in wild primates, ranging from 0 to 89%, depending on species and region (9, 25, 37, 50). Interestingly, with the comparisons possible from our study, it is striking to observe such extreme differences also between closely related primate species. This probably means that both genetic diversity and retroviral prevalence are determined by geographical location for STLV-1s. However, further studies on additional populations are needed to adequately assess if the prevalence in P. b. badius is unusually high or if that of P. r. tephrosceles is extraordinarily low.
In contrast to SIV and STLV-1, there was not much difference in the prevalences of SFV in our study (86%; 95% CI, 72 to 100%) and in P. r. tephrosceles (97%; 95% CI, 90 to 100%) (15). High SFV prevalence has also been found in other wild primate populations, where infections can reach 100% (3, 33). The fact that this generally high prevalence is unique to SFV (compared to SIV and STLV-1) might be explained by a lesser sensitivity to the behavioral differences that exist between species and even populations.
The results from our study, as well as those from the red colobus study in Uganda, are based on relatively small sample sizes (15). However, samples from wild red colobus monkeys and primates in general are difficult to obtain, and one should not refrain from discussing possible reasons for differences in viral prevalence between populations, based on the data we have to date. Virus biology can be one possible reason for the observed difference in SIV and STLV-1 prevalences since these viruses in the Taï and Kibale populations are all distinct. Behavioral differences should also be considered although no further demographic data are available for our study animals, and the routes of retroviral transmission in wild primates are not fully understood (20, 38). In general, the intense social behavior of red colobus monkeys could give the opportunity for frequent retrovirus transmission. The monkeys live in promiscuous multimale groups of about 50 individuals, the males fight each other to mate receptive females throughout their adult lives, and frequent aggressive harassment of mating couples occurs (36, 52). There are, however, behavioral differences between the red colobus monkey populations/species found in Taï and Kibale that might explain, at least partly, the difference in the prevalences of SIV and STLV-1 at these sites. First, the Taï P. b. badius population has a defined breeding season of 5 to 6 months every year, whereas the Kibale P. r. tephrosceles population breeds all year round (A. Korstjens, personal communication). During the intense breeding season in Taï, the receptive females mate with virtually all males available. In Kibale, there are receptive females available all year round, and the average number of males in the group is lower than in Taï (3.5 versus 10), which makes monopoly by dominating males easier (36, 40, 52). This means that the P. b. badius females in Taï overall mate with a larger number of partners, which is a risk factor in the spread of sexually transmitted diseases (41). Second, although a precise comparison is difficult, it appears that there is more aggression associated with breeding in Taï than in Kibale because of the intense male competition over access to females during a restricted breeding period (A. Korstjens, personal communication). Frequent fighting facilitates close contact between individuals and hence represents a risk of viral transmission. Finally, the P. b. badius males in Taï were also more frequently seen with red, ulcerated penises than the males in Kibale (A. Korstjens, personal communication). This could be a sign of other sexually transmitted diseases which could ultimately make retroviral transmission easier. Further studies are required to diagnose and determine the extent of sexually transmitted diseases in this red colobus population as well as to investigate their possible effect on retrovirus transmission.
Coinfection and correlation of viruses.
There was a high degree of coinfection of the retroviruses in the P. b. badius monkeys, as nearly half of the individuals were triple infected and nearly one-third were dually infected with SIV and one of the other retroviruses. This is not surprising considering the relatively high prevalences of all the individual viruses. In comparison, the level of coinfections in the P. r. tephrosceles population was substantially lower (3% of the monkeys had triple infections, and 23% had dual infections, all of which included SFV), as would be expected with the relatively lower prevalences of both SIV and STLV-1 (15). Interestingly, in the Taï P. b. badius population, no individual included in the coinfection analysis was positive for STLV-1 alone or in combination with SFV; all the STLV-1-positive individuals were at the same time infected with SIV. It is possible that STLV-1 is frequently transmitted together with SIV; however, there was no significant correlation among any of the viruses in the Taï P. b. badius population. Also in the Kibale P. r. tephrosceles population, no correlation was found among these viruses (15).
Phylogeny.
With data accumulating, the mechanisms of retrovirus evolution have taken more precise and distinctive shapes. SFV and SIV mostly show species-specific distribution, either as a result of host-parasite cospeciation or preferential host switching (5, 53, 61). In contrast to SFV and SIV, the distribution of the genetic diversity of STLV-1 seems to be more complex, which most likely reflects more frequent cross-species transmission of this virus between different primate species than for lenti- and spumaretroviruses (21).
Also in our study, SIV and SFV phylogenies both exhibited the pattern of host-specific association of these retroviruses. This was especially true for SIVwrc and SFVwrc sequences determined from P. b. badius. In the SIV tree, SIVwrc sequences from P. b. badius are clustered together with those determined from habituated P. b. badius in the same area and in P. b. temminckii in Gambia (34, 35). Of note, these red colobus populations are both called Western red colobus but are believed to belong to different subspecies (54). In the phylogeny of mitochondrial DNA, P. b. temminckii appears as being nested into the genetic diversity of P. b. badius. SIVwrc therefore conforms to the same pattern. In the SFV tree, SFVwrc strains cluster into one clade but are at this time associated with one strain previously identified as coming from a chimpanzee, most probably as the result of a cross-species transmission event linked to chimpanzee hunting behavior (28). For both SIV and SFV, strains identified from P. r. tephrosceles in Kibale grouped with strains of P. b. badius in Taï but were never interspersed with them, suggesting reciprocal monophyly. As this conforms to the host phylogeny (54), this might be explained by host-parasite cospeciation for SFVwrc, a common process for retroviruses of this genus (53). For SIVwrc, preferential host switching (5) would seem a much more likely explanation given the presumed overall short time scale of primate lentivirus evolution (62).
In contrast to SIV and SFV, the phylogeny of STLV-1 is thought to be linked to geography rather than to host species (57). Given this geographical component and the slow evolutionary rate of STLV-1 (29), one could expect relatively low genetic variation on small geographical scales. However, we found that the novel strains of STLV-1wrc in P. b. badius living in a relatively small area of the Taï rainforest showed high genetic variability, being distributed into three distinct lineages (of which one shows some affinity to STLV-1sm described from sooty mangabeys from Sierra Leone, whereas STLV-1krc from Kibale P. r. tephrosceles show no affinity to any of these lineages). Thus, Taï strains were found in the two main lineages constituting the recently described subtype J as well as in the group formed by subtype I sequences (21). This is a considerable extension of their known genetic diversity as only two Taï STLV-1wrc strains have been described so far (27). Importantly, all new STLV-1wrc strains were interspersed with those found from P. t. verus living in the area. This confirms previous findings of cross-species transmission of STLV-1 from the red colobus monkeys to the chimpanzees and underlines that the major part of the diversity of STLV-1 strains found in those chimpanzees possibly stems from hunting and eating P. b. badius (27). To the authors' knowledge a comparable pattern of genetic diversity has so far been found only in two other herbivore primate species (C. nictitans and Cercopithecus cephus) (31). Under the hypothesis that geographical proximity rather than host specificity is the main determinant of the presence of a given strain in a given species, it is tempting to make the assumption that Taï P. b. badius monkeys are infected with a wide variety of STLV-1 strains because the Taï rainforest is a place of high endemicity for STLV-1. This might in turn indicate that STLV-1 has been circulating in that forest zone for a longer period than in other regions or that it has not been submitted to similar degrees of a bottleneck effect. It would therefore be interesting to investigate in more detail the overall STLV-1 diversity in Taï and also other areas of tropical Africa since, at the moment, only a two-point comparison is possible. Increasing the sampling of P. b. badius as well as getting samples from other Taï primate species, some of which live in close contact with red colobus monkeys (36, 39), could be a first step into the process.
Conclusion.
This study shows that retroviral infections with SIV, STLV-1, and SFV are common in red colobus monkeys (P. b. badius) in the Taï National Park, Côte d'Ivoire. Comparing our results with those obtained from a study of a sister species (P. r. tephrosceles) in Uganda shows that the prevalence of these retroviruses in wild primates can vary dramatically, even between closely related species. We further demonstrate a high genetic variability of STLV-1 in this herbivore monkey species, which might be taken as an indication that Taï is a hot spot of diversity for this retrovirus.
Acknowledgments
We thank the Ivorian authorities for long-term support, especially the Ministry of Environment and Forests as well as the Ministry of Research, the directorship of the Taï National Park, the Office Ivoirien des Parcs et Réserves, and the Swiss Research Centre in Abidjan. We also thank S. Metzger, W. Rietschel, and field assistants and students of the Taï Chimpanzee Project for assistance in sample collection and U. Thiesen, A. Blasse, A. Kopp, S. Handrick, J. Hinzmann, and A. Hübner for assistance in the laboratory. For sequencing we thank J. Tesch. We thank A. Korstjens, K. Zuberbühler, and P. Beziers for useful discussions on red colobus monkey behavior.
The study was funded by the Max Planck Institute for Evolutionary Anthropology, Leipzig, Germany, and the Robert Koch Institute, Berlin, Germany.
Footnotes
Published ahead of print on 19 May 2010.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Boesch, C., and H. Boesch-Achermann. 2000. The chimpanzees of the Taï forest: behavioural ecology and evolution. Oxford University Press, Oxford, United Kingdom.
- 3.Calattini, S., E. Nerrienet, P. Mauclere, M. C. Georges-Courbot, A. Saib, and A. Gessain. 2004. Natural simian foamy virus infection in wild-caught gorillas, mandrills and drills from Cameroon and Gabon. J. Gen. Virol. 85:3313-3317. [DOI] [PubMed] [Google Scholar]
- 4.Cardini, A., and S. Elton. 2009. The radiation of red colobus monkeys (Primates, Colobinae): morphological evolution in a clade of endangered African primates. Zool. J. Linn. Soc. 157:197-224. [Google Scholar]
- 5.Charleston, M. A., and D. L. Robertson. 2002. Preferential host switching by primate lentiviruses can account for phylogenetic similarity with the primate phylogeny. Syst. Biol. 51:528-535. [DOI] [PubMed] [Google Scholar]
- 6.Clewley, J. P., J. C. Lewis, D. W. Brown, and E. L. Gadsby. 1998. A novel simian immunodeficiency virus (SIVdrl) pol sequence from the drill monkey, Mandrillus leucophaeus. J. Virol. 72:10305-10309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Courgnaud, V., P. Formenty, C. Akoua-Koffi, R. Noe, C. Boesch, E. Delaporte, and M. Peeters. 2003. Partial molecular characterization of two simian immunodeficiency viruses (SIV) from African colobids: SIVwrc from Western red colobus (Piliocolobus badius) and SIVolc from olive colobus (Procolobus verus). J. Virol. 77:744-748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Courgnaud, V., X. Pourrut, F. Bibollet-Ruche, E. Mpoudi-Ngole, A. Bourgeois, E. Delaporte, and M. Peeters. 2001. Characterization of a novel simian immunodeficiency virus from guereza colobus monkeys (Colobus guereza) in Cameroon: a new lineage in the nonhuman primate lentivirus family. J. Virol. 75:857-866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Courgnaud, V., S. Van Dooren, F. Liegeois, X. Pourrut, B. Abela, S. Loul, E. Mpoudi-Ngole, A. Vandamme, E. Delaporte, and M. Peeters. 2004. Simian T-cell leukemia virus (STLV) infection in wild primate populations in Cameroon: evidence for dual STLV type 1 and type 3 infection in agile mangabeys (Cercocebus agilis). J. Virol. 78:4700-4709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Douady, C. J., F. Delsuc, Y. Boucher, W. F. Doolittle, and E. J. Douzery. 2003. Comparison of Bayesian and maximum likelihood bootstrap measures of phylogenetic reliability. Mol. Biol. Evol. 20:248-254. [DOI] [PubMed] [Google Scholar]
- 11.Drummond, A. J., and A. Rambaut. 2007. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol. Biol. 7:214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fedigan, L. M. 8 March 2010, posting date. Ethical issues faced by field primatologists: asking the relevant questions. Am. J. Primatol. doi: 10.1002/ajp.20814. [DOI] [PubMed]
- 13.Gao, F., E. Bailes, D. L. Robertson, Y. Chen, C. M. Rodenburg, S. F. Michael, L. B. Cummins, L. O. Arthur, M. Peeters, G. M. Shaw, P. M. Sharp, and B. H. Hahn. 1999. Origin of HIV-1 in the chimpanzee Pan troglodytes troglodytes. Nature 397:436-441. [DOI] [PubMed] [Google Scholar]
- 14.Gillespie, T. R., C. L. Nunn, and F. H. Leendertz. 2008. Integrative approaches to the study of primate infectious disease: implications for biodiversity conservation and global health. Am. J. Phys. Anthropol. Suppl. 47:53-69. [DOI] [PubMed] [Google Scholar]
- 15.Goldberg, T. L., D. M. Sintasath, C. A. Chapman, K. M. Cameron, W. B. Karesh, S. Tang, N. D. Wolfe, I. B. Rwego, N. Ting, and W. M. Switzer. 2009. Coinfection of Ugandan red colobus (Procolobus [Piliocolobus] rufomitratus tephrosceles) with novel, divergent delta-, lenti-, and spumaretroviruses. J. Virol. 83:11318-11329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gouy, M., S. Guindon, and O. Gascuel. 2010. SeaView version 4: a multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol. Biol. Evol. 27:221-224. [DOI] [PubMed] [Google Scholar]
- 17.Guindon, S., and O. Gascuel. 2003. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 52:696-704. [DOI] [PubMed] [Google Scholar]
- 18.Guindon, S., F. Lethiec, P. Duroux, and O. Gascuel. 2005. PHYML online—a web server for fast maximum likelihood-based phylogenetic inference. Nucleic Acids Res. 33:W557-W559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hahn, B. H., G. M. Shaw, K. M. De Cock, and P. M. Sharp. 2000. AIDS as a zoonosis: scientific and public health implications. Science 287:607-614. [DOI] [PubMed] [Google Scholar]
- 20.Heeney, J. L., A. G. Dalgleish, and R. A. Weiss. 2006. Origins of HIV and the evolution of resistance to AIDS. Science 313:462-466. [DOI] [PubMed] [Google Scholar]
- 21.Junglen, S., C. Hedemann, H. Ellerbrok, G. Pauli, C. Boesch, and F. H. Leendertz. 2010. Diversity of STLV-1 strains in wild chimpanzees (Pan troglodytes verus) from Cote d'Ivoire. Virus Res. 150:143-147. [DOI] [PubMed] [Google Scholar]
- 22.Keele, B. F., J. H. Jones, K. A. Terio, J. D. Estes, R. S. Rudicell, M. L. Wilson, Y. Li, G. H. Learn, T. M. Beasley, J. Schumacher-Stankey, E. Wroblewski, A. Mosser, J. Raphael, S. Kamenya, E. V. Lonsdorf, D. A. Travis, T. Mlengeya, M. J. Kinsel, J. G. Else, G. Silvestri, J. Goodall, P. M. Sharp, G. M. Shaw, A. E. Pusey, and B. H. Hahn. 2009. Increased mortality and AIDS-like immunopathology in wild chimpanzees infected with SIVcpz. Nature 460:515-519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kwok, S., G. Ehrlich, B. Poiesz, R. Kalish, and J. J. Sninsky. 1988. Enzymatic amplification of HTLV-I viral sequences from peripheral blood mononuclear cells and infected tissues. Blood 72:1117-1123. [PubMed] [Google Scholar]
- 24.Mugisha, L., C. Kücherer, H. Ellerbrok, S. Junglen, J. Opuda-Asibo, O. O. Joseph, G. Pauli, and F. H. Leendertz. Retroviruses in wild-born semi-captive East African sanctuary chimpanzees (Pan troglodytes schweinfurthii) Open Vet. Sci. J., in press.
- 25.Leendertz, F. H., C. Boesch, H. Ellerbrok, W. Rietschel, E. Couacy-Hymann, and G. Pauli. 2004. Non-invasive testing reveals a high prevalence of simian T-lymphotropic virus type 1 antibodies in wild adult chimpanzees of the Tai National Park, Cote d'Ivoire. J. Gen. Virol. 85:3305-3312. [DOI] [PubMed] [Google Scholar]
- 26.Leendertz, F. H., C. Boesch, S. Junglen, G. Pauli, and H. Ellerbrok. 2003. Characterization of a new simian T-lymphocyte virus type 1 (STLV-1) in a wild living chimpanzee (Pan troglodytes verus) from Ivory Coast: evidence of a new STLV-1 group? AIDS Res. Hum. Retroviruses 19:255-258. [DOI] [PubMed] [Google Scholar]
- 27.Leendertz, F. H., S. Junglen, C. Boesch, P. Formenty, E. Couacy-Hymann, V. Courgnaud, G. Pauli, and H. Ellerbrok. 2004. High variety of different simian T-cell leukemia virus type 1 strains in chimpanzees (Pan troglodytes verus) of the Tai National Park, Cote d'Ivoire. J. Virol. 78:4352-4356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Leendertz, F. H., F. Zirkel, E. Couacy-Hymann, H. Ellerbrok, V. A. Morozov, G. Pauli, C. Hedemann, P. Formenty, S. A. Jensen, C. Boesch, and S. Junglen. 2008. Interspecies transmission of simian foamy virus in a natural predator-prey system. J. Virol. 82:7741-7744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lemey, P., O. G. Pybus, S. Van Dooren, and A. M. Vandamme. 2005. A Bayesian statistical analysis of human T-cell lymphotropic virus evolutionary rates. Infect. Genet. Evol. 5:291-298. [DOI] [PubMed] [Google Scholar]
- 30.Lemey, P., M. Salemi, L. Bassit, and A. M. Vandamme. 2002. Phylogenetic classification of TT virus groups based on the N22 region is unreliable. Virus Res. 85:47-59. [DOI] [PubMed] [Google Scholar]
- 31.Liegeois, F., B. Lafay, W. M. Switzer, S. Locatelli, E. Mpoudi-Ngole, S. Loul, W. Heneine, E. Delaporte, and M. Peeters. 2008. Identification and molecular characterization of new STLV-1 and STLV-3 strains in wild-caught nonhuman primates in Cameroon. Virology 371:405-417. [DOI] [PubMed] [Google Scholar]
- 32.Ling, B., M. L. Santiago, S. Meleth, B. Gormus, H. M. McClure, C. Apetrei, B. H. Hahn, and P. A. Marx. 2003. Noninvasive detection of new simian immunodeficiency virus lineages in captive sooty mangabeys: ability to amplify virion RNA from fecal samples correlates with viral load in plasma. J. Virol. 77:2214-2226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu, W., M. Worobey, Y. Li, B. F. Keele, F. Bibollet-Ruche, Y. Guo, P. A. Goepfert, M. L. Santiago, J. B. Ndjango, C. Neel, S. L. Clifford, C. Sanz, S. Kamenya, M. L. Wilson, A. E. Pusey, N. Gross-Camp, C. Boesch, V. Smith, K. Zamma, M. A. Huffman, J. C. Mitani, D. P. Watts, M. Peeters, G. M. Shaw, W. M. Switzer, P. M. Sharp, and B. H. Hahn. 2008. Molecular ecology and natural history of simian foamy virus infection in wild-living chimpanzees. PLoS Pathog. 4:e1000097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Locatelli, S., B. Lafay, F. Liegeois, N. Ting, E. Delaporte, and M. Peeters. 2008. Full molecular characterization of a simian immunodeficiency virus, SIVwrcpbt from Temminck's red colobus (Piliocolobus badius temminckii) from Abuko Nature Reserve, The Gambia. Virology 376:90-100. [DOI] [PubMed] [Google Scholar]
- 35.Locatelli, S., F. Liegeois, B. Lafay, A. D. Roeder, M. W. Bruford, P. Formenty, R. Noe, E. Delaporte, and M. Peeters. 2008. Prevalence and genetic diversity of simian immunodeficiency virus infection in wild-living red colobus monkeys (Piliocolobus badius badius) from the Tai forest, Cote d'Ivoire SIVwrc in wild-living Western red colobus monkeys. Infect. Genet. Evol. 8:1-14. [DOI] [PubMed] [Google Scholar]
- 36.McGraw, W. S., K. Zuberbühler, and R. Noe. 2007. Monkeys of the Taï Forest: an African primate community. Cambridge University Press, New York, NY.
- 37.Meertens, L., J. Rigoulet, P. Mauclere, M. Van Beveren, G. M. Chen, O. Diop, G. Dubreuil, M. C. Georges-Goubot, J. L. Berthier, J. Lewis, and A. Gessain. 2001. Molecular and phylogenetic analyses of 16 novel simian T cell leukemia virus type 1 from Africa: close relationship of STLV-1 from Allenopithecus nigroviridis to HTLV-1 subtype B strains. Virology 287:275-285. [DOI] [PubMed] [Google Scholar]
- 38.Nerrienet, E., X. Amouretti, M. C. Muller-Trutwin, V. Poaty-Mavoungou, I. Bedjebaga, H. T. Nguyen, G. Dubreuil, S. Corbet, E. J. Wickings, F. Barre-Sinoussi, A. J. Georges, and M. C. Georges-Courbot. 1998. Phylogenetic analysis of SIV and STLV type I in mandrills (Mandrillus sphinx): indications that intracolony transmissions are predominantly the result of male-to-male aggressive contacts. AIDS Res. Hum. Retroviruses 14:785-796. [DOI] [PubMed] [Google Scholar]
- 39.Noe, R., and R. Bshary. 1997. The formation of red colobus-diana monkey associations under predation pressure from chimpanzees. Proc. Biol. Sci. 264:253-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nunn, C. L. 1999. The number of males in primate social groups: a comparative test of the socioecological model. Behav. Ecol. Sociobiol. 46:1-13. [Google Scholar]
- 41.Nunn, C. L., J. L. Gittleman, and J. Antonovics. 2000. Promiscuity and the primate immune system. Science 290:1168-1170. [DOI] [PubMed] [Google Scholar]
- 42.Peeters, M., V. Courgnaud, B. Abela, P. Auzel, X. Pourrut, F. Bibollet-Ruche, S. Loul, F. Liegeois, C. Butel, D. Koulagna, E. Mpoudi-Ngole, G. M. Shaw, B. H. Hahn, and E. Delaporte. 2002. Risk to human health from a plethora of simian immunodeficiency viruses in primate bushmeat. Emerg. Infect. Dis. 8:451-457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Plantier, J. C., M. Leoz, J. E. Dickerson, F. De Oliveira, F. Cordonnier, V. Lemee, F. Damond, D. L. Robertson, and F. Simon. 2009. A new human immunodeficiency virus derived from gorillas. Nat. Med. 15:871-872. [DOI] [PubMed] [Google Scholar]
- 44.Posada, D. 2008. jModelTest: phylogenetic model averaging. Mol. Biol. Evol. 25:1253-1256. [DOI] [PubMed] [Google Scholar]
- 45.Refisch, J., and I. Kone. 2005. Impact of commercial hunting on monkey populations in the Tai region, Cote d'Ivoire. Biotropica 37:136-144. [Google Scholar]
- 46.Santiago, M. L., F. Range, B. F. Keele, Y. Li, E. Bailes, F. Bibollet-Ruche, C. Fruteau, R. Noe, M. Peeters, J. F. Brookfield, G. M. Shaw, P. M. Sharp, and B. H. Hahn. 2005. Simian immunodeficiency virus infection in free-ranging sooty mangabeys (Cercocebus atys atys) from the Tai Forest, Cote d'Ivoire: implications for the origin of epidemic human immunodeficiency virus type 2. J. Virol. 79:12515-12527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Santiago, M. L., C. M. Rodenburg, S. Kamenya, F. Bibollet-Ruche, F. Gao, E. Bailes, S. Meleth, S. J. Soong, J. M. Kilby, Z. Moldoveanu, B. Fahey, M. N. Muller, A. Ayouba, E. Nerrienet, H. M. McClure, J. L. Heeney, A. E. Pusey, D. A. Collins, C. Boesch, R. W. Wrangham, J. Goodall, P. M. Sharp, G. M. Shaw, and B. H. Hahn. 2002. SIVcpz in wild chimpanzees. Science 295:465. [DOI] [PubMed] [Google Scholar]
- 48.Seiki, M., S. Hattori, Y. Hirayama, and M. Yoshida. 1983. Human adult T-cell leukemia virus: complete nucleotide sequence of the provirus genome integrated in leukemia cell DNA. Proc. Natl. Acad. Sci. U. S. A. 80:3618-3622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Silvestri, G. 2009. Immunity in natural SIV infections. J. Intern. Med. 265:97-109. [DOI] [PubMed] [Google Scholar]
- 50.Sintasath, D. M., N. D. Wolfe, M. Lebreton, H. Jia, A. D. Garcia, J. Le Doux-Diffo, U. Tamoufe, J. K. Carr, T. M. Folks, E. Mpoudi-Ngole, D. S. Burke, W. Heneine, and W. M. Switzer. 2009. Simian T-lymphotropic virus diversity among nonhuman primates, Cameroon. Emerg. Infect. Dis. 15:175-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Souquiere, S., F. Bibollet-Ruche, D. L. Robertson, M. Makuwa, C. Apetrei, R. Onanga, C. Kornfeld, J. C. Plantier, F. Gao, K. Abernethy, L. J. White, W. Karesh, P. Telfer, E. J. Wickings, P. Mauclere, P. A. Marx, F. Barre-Sinoussi, B. H. Hahn, M. C. Muller-Trutwin, and F. Simon. 2001. Wild Mandrillus sphinx are carriers of two types of lentivirus. J. Virol. 75:7086-7096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Struhsaker, T. T. 1975. The red colobus monkey. University of Chicago Press, Chicago, IL.
- 53.Switzer, W. M., M. Salemi, V. Shanmugam, F. Gao, M. E. Cong, C. Kuiken, V. Bhullar, B. E. Beer, D. Vallet, A. Gautier-Hion, Z. Tooze, F. Villinger, E. C. Holmes, and W. Heneine. 2005. Ancient co-speciation of simian foamy viruses and primates. Nature 434:376-380. [DOI] [PubMed] [Google Scholar]
- 54.Ting, N. 2008. Mitochondrial relationships and divergence dates of the African colobines: evidence of Miocene origins for the living colobus monkeys. J. Hum. Evol. 55:312-325. [DOI] [PubMed] [Google Scholar]
- 55.Vandamme, A. M., M. Salemi, M. Van Brussel, H. F. Liu, K. Van Laethem, M. Van Ranst, L. Michels, J. Desmyter, and P. Goubau. 1998. African origin of human T-lymphotropic virus type 2 (HTLV-2) supported by a potential new HTLV-2d subtype in Congolese Bambuti Efe Pygmies. J. Virol. 72:4327-4340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.VandeWoude, S., and C. Apetrei. 2006. Going wild: lessons from naturally occurring T-lymphotropic lentiviruses. Clin. Microbiol. Rev. 19:728-762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Van Dooren, S., E. J. Verschoor, Z. Fagrouch, and A. M. Vandamme. 2007. Phylogeny of primate T lymphotropic virus type 1 (PTLV-1) including various new Asian and African non-human primate strains. Infect. Genet. Evol. 7:374-381. [DOI] [PubMed] [Google Scholar]
- 58.Van Heuverswyn, F., Y. Li, C. Neel, E. Bailes, B. F. Keele, W. Liu, S. Loul, C. Butel, F. Liegeois, Y. Bienvenue, E. M. Ngolle, P. M. Sharp, G. M. Shaw, E. Delaporte, B. H. Hahn, and M. Peeters. 2006. Human immunodeficiency viruses: SIV infection in wild gorillas. Nature 444:164. [DOI] [PubMed] [Google Scholar]
- 59.Verdonck, K., E. Gonzalez, S. Van Dooren, A. M. Vandamme, G. Vanham, and E. Gotuzzo. 2007. Human T-lymphotropic virus 1: recent knowledge about an ancient infection. Lancet Infect. Dis. 7:266-281. [DOI] [PubMed] [Google Scholar]
- 60.Villesen, P. 2007. FaBox: an online toolbox for fasta sequences. Mol. Ecol. Notes 7:965-968. [Google Scholar]
- 61.Wertheim, J. O., and M. Worobey. 2007. A challenge to the ancient origin of SIVagm based on African green monkey mitochondrial genomes. PLoS Pathog. 3:e95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wertheim, J. O., and M. Worobey. 2009. Dating the age of the SIV lineages that gave rise to HIV-1 and HIV-2. PLoS Comput. Biol. 5:e1000377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wolfe, N. D., P. Daszak, A. M. Kilpatrick, and D. S. Burke. 2005. Bushmeat hunting, deforestation, and prediction of zoonoses emergence. Emerg. Infect. Dis. 11:1822-1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wolfe, N. D., W. M. Switzer, J. K. Carr, V. B. Bhullar, V. Shanmugam, U. Tamoufe, A. T. Prosser, J. N. Torimiro, A. Wright, E. Mpoudi-Ngole, F. E. McCutchan, D. L. Birx, T. M. Folks, D. S. Burke, and W. Heneine. 2004. Naturally acquired simian retrovirus infections in central African hunters. Lancet 363:932-937. [DOI] [PubMed] [Google Scholar]
- 65.Xia, X., and Z. Xie. 2001. DAMBE: software package for data analysis in molecular biology and evolution. J. Hered. 92:371-373. [DOI] [PubMed] [Google Scholar]