Abstract
The influenza A virus genome consists of 8 negative-stranded RNA segments. NS1 is a nonstructural protein that participates in different steps of the virus infectious cycle, including transcription, replication, and morphogenesis, and acts as a virulence factor. Human Staufen1 (hStau1), a protein involved in the transport and regulated translation of cellular mRNAs, was previously identified as a NS1-interacting factor. To investigate the possible role of hStau1 in the influenza virus infection, we characterized the composition of hStau1-containing granules isolated from virus-infected cells. Viral NS1 protein and ribonucleoproteins (RNPs) were identified in these complexes by Western blotting, and viral mRNAs and viral RNAs (vRNAs) were detected by reverse transcription (RT)-PCR. Also, colocalization of hStau1 with NS1, nucleoprotein (NP), and PA in the cytosol of virus-infected cells was shown by immunofluorescence. To analyze the role of hStau1 in the infection, we downregulated its expression by gene silencing. Human HEK293T cells or A549 cells were silenced using either short hairpin RNAs (shRNAs) or small interfering RNAs (siRNAs) targeting four independent sites in the hStau1 mRNA. The yield of influenza virus was reduced 5 to 10 times in the various hStau1-silenced cells compared to that in control silenced cells. The expression levels of viral proteins and their nucleocytoplasmic localization were not affected upon hStau1 silencing, but virus particle production, as determined by purification of virions from supernatants, was reduced. These results indicate a role for hStau1 in late events of the influenza virus infection, possibly during virus morphogenesis.
The influenza A virus genome is formed by eight segments of negative-sense, single-stranded RNA that encode 12 different proteins, nine of which are present in the virion (43, 57). Genomic RNAs form viral ribonucleoprotein (RNP) complexes (vRNPs) by association with viral RNA (vRNA) polymerase and nucleoprotein (NP). The polymerase complex is formed by the PA, PB1, and PB2 proteins and carries out both viral transcription and replication events in the cell nucleus (28, 29). The influenza virus genome encodes two nonstructural proteins, NS1 and the more recently identified PB1-F2 (11). NS1 accumulates in the nucleus at early times postinfection and in both the nucleus and cytoplasm at later times (6). The existence of mutant viruses lacking NS1 (22, 33) suggests that it is not the product of an essential gene, although the phenotypes of NS1 point and deletion mutants indicate that its function may be related to transcription and replication events (18), late-viral synthesis (27), modulation of the innate immune response (15), and viral morphogenesis (20) (reviewed in reference 26). Such a variety of roles may be related to the capacity of NS1 to interact with viral RNPs (39) and also with cellular factors, such as proteins involved in posttranslational processing of mRNAs, such as cleavage and polyadenylation specificity factor (CPSF) (41), NS1-BP (58), proteins of the nuclear pore complex (47), proteins involved in interferon signaling (such as PKR and RIG-I) (36, 40) or involved in translation (PABP, eIF4G, and Staufen1) (1, 7, 17).
Human Staufen1 (hStau1) was first identified in a yeast two-hybrid screen using NS1 as bait (17). It is the human homologue to Drosophila melanogaster Staufen (dmStau), a protein essential for the proper localization of certain mRNAs during the formation of the anteroposterior axis of the embryo of D. melanogaster and for the asymmetric division of neuroblasts (19). The hStau1 protein is associated with polysomes and localizes in dendrites of cultured neurons in structures called RNA granules (32, 54). The size of these granules is about 10 MDa, and their composition, including cytoskeleton proteins such as tubulin and actin, motor proteins, such as kinesin and dynein, ribosomal proteins, and proteins involved in the regulation of translation, suggests a role for hStau1 in the transport and localized translation of mRNAs (54). Previous data have shown that hStau1 and NS1 proteins are associated to the polysome fraction of influenza virus-infected cells and coimmunoprecipitate both in infected and cotransfected cells. Furthermore, the overexpression of both proteins from cDNA induces the redistribution of hStau1 from the cytoplasm to the nucleus (17). On the other hand, hStau1 has been shown to participate in HIV virion assembly, forming a complex with HIV genomic RNA and pr55gag (10) in the membrane of infected cells. Both the overexpression and the depletion of hStau1 affect the multimerization of pr55gag (8). In this report we have analyzed the possible function of the hStau1 protein during influenza virus infection. We describe the association of hStau1 not only with the NS1 protein but also with the viral RNP. Both mRNAs and vRNAs were found associated to hStau1 complexes in vivo. In addition, by using cells in which the expression levels of hStau1 have been reduced by silencing, we show that the viral yields and the formation of extracellular virions are reduced, although the expression and the nucleocytoplasmic transport of the viral proteins are not affected.
MATERIALS AND METHODS
Biological materials.
The HEK293T (13), A549 (24), BHK21, and MDCK (18, 23) cell lines were cultured as previously described. Plasmid pChStaufen-TAP, expressing the recombinant Staufen-TAP protein, and the control plasmid pC-TAP have been described previously (54). The plasmids pCMV-PB1, pCMV-PB2, pCMV-PA, pCMV-NP, pHHNS1, and pHHDelNS1, used to reconstitute functional RNPs, have been described previously (18, 20). The influenza virus WSN and delNS1 strains were used for low-multiplicity-of-infection (MOI) and high-MOI assays, respectively. Vesicular stomatitis virus (VSV) was used as an unrelated control virus. Antibodies specific for hStau1 (38), NP and NS1 (37), M1 and M2 (4, 46), PB1 (45), and PA and PB2 (3, 42) have been described previously. The commercial antibody against human β-actin was purchased from Sigma.
Plasmid construction.
The plasmid pRMut1 containing the mutant hStau1 protein was obtained by site-directed mutagenesis of the plasmid pRHST (38) using oligonucleotides 5′-GGCATTTTTCTTTGAAATCGCCGCGCTTTTCCCTTCACCTTCCCC-3′ and 5′-GGGGAAGGTGAAGGGAAAAGCGCGGCGATTTCAAAGAAAAATGCC-3′. Lysines 129 and 130 in the third RNA binding domain (RBD) were mutated to alanines using the site-directed mutagenesis kit from Stratagene, as recommended by the manufacturer. To generate the pChStaufen-Mut1-TAP, the fragment containing the coding sequence for hStau1-Mut1 was amplified by PCR using the oligonucleotides 5′-ATGAAACTTGGAAAAAAACCAATGTATAAGCCTGTTGACCCTTACTCTCGGATGCAGTC-3′ and 5′-TAGCCCTCTCGAGTCAGCACCTCCCACACACAGACATT-3′. The purified fragment was further cloned into the EcoRV site of the pC-TAP plasmid (54). The plasmid pSR-puro-iStau1 was generated by cloning the sequence 5′-TTTGCCTTGAACGGACACTTAAA-3′, specific for hStau1, into the HindIII/EcoRI sites of the pSR-puro plasmid (Oligoengine). Plasmid pSR-puro-Tm, which contains the sequence 5′-CAATTCTCCGAACGTGTCACGT-3′ from Thermotoga maritima (Qiagen), which is not expressed in HEK293T cells, was a gift from R. Alfonso-Dunn.
Generation of iStau cell lines.
HEK293T cells were transfected with pSR-puro-iStau1 or pSR-puro-Tm plasmids. One day posttransfection, cells were diluted and grown in the presence of 2 μg/ml of puromycin until independent clones were available for isolation. The individual clones were amplified until the hStau1 protein levels could be determined by Western blotting and an immunofluorescence assay. Two of the analyzed clones presented levels of hStau1 protein lower than that in the parental HEK293T cell line: clone 1-4 and clone 1-8. Cultured cells were further maintained with 4 μg/ml puromycin.
Transfection-infection.
HEK293T cells were used to perform transfection-infection experiments. Cells were transfected with 20 μg of pChStau-TAP, pChStaufen-Mut1TAP, or pC-TAP in p150 dishes using the calcium phosphate method (56). Infection was carried out 16 h after transfection, using the influenza virus WSN or delNS1 strain at a multiplicity of 3 to 5 PFU/cell for 6 h.
Viral growth in the HEK293T, iStau, or A549 cell lines was analyzed by low-MOI (10−3 PFU/cell) experiments that progressed for 60 h.
siRNA transfection.
Cultured A549 cells were transfected independently with 5 nM each small interfering RNA (siRNA): (S1 (s13547), S2 (s13546), and S3 (s13548) specific for hStau1 or an irrelevant siRNA (AM4611) from Ambion, using Lipofectamine (Invitrogen) as recommended by the manufacturer. Transfection was carried out for two consecutive days to increase the silencing efficiency before infection.
Protein expression and purification.
The expression of His-thSTL and His-thSTL-Mut1 proteins in Escherichia coli was induced by isopropyl-β-d-thiogalactopyranoside, and the recombinant proteins were purified by chromatography on Ni-nitrilotriacetic acid (NTA)-agarose as previously described (37).
Protein analyses.
For protein labeling in vivo, cells were washed, incubated for 1 h in Dulbecco's minimal essential medium (DMEM) lacking methionine-cysteine (Met-Cys) and labeled for 1 h at various times postinfection with [35S]Met-Cys (200 μCi/ml) (59). Total protein was recovered in Laemmli sample buffer and analyzed by polyacrylamide gel electrophoresis and autoradiography. For TAP purification, cell extracts were obtained by lysis in a buffer containing 50 mM Tris-HCl, 100 mM NaCl, and 5 mM EDTA, pH 7.5 (TNE), 0.5% NP-40, 1 mM dithiothreitol (DTT), human placental RNase inhibitor (HPRI) (40 U/ml), and the complete protease inhibitor cocktail (Roche) for 30 min at 4°C. After centrifugation at 2,500 rpm and 4°C, the supernatant was further centrifuged for 10 min at 10,000 rpm and 4°C. The lysates were incubated with IgG-Sepharose (Stratagene) for 12 h at 4°C (approximately 5-μl-bed volume/mg protein). The resin was washed 10 times with 10 resin volumes of IPP-150 buffer (54) and five times in 50 mM Tris-HCl, pH 8, 0.5 mM EDTA, 1 mM DTT. The complexes bound to the resin were digested with 1 U of tobacco etch virus protease per 107 cells for 3 h at room temperature. The supernatant was mixed with five washes of the resin with IPP150-CBB buffer (54) and incubated with calmodulin-agarose resin for 12 h at 4°C. The resin was washed 10 times in IPP150-CBB buffer and eluted in a buffer containing 10 mM Tris-HCl, pH 8, 0.1% NP-40, 10 mM β-mercaptoethanol, 1 mM imidazol, and 3 mM EGTA. In the indicated experiments, 20 μg of RNase A (Fermentas, Ontario, Canada) was added during cell lysis. The purified proteins were analyzed by polyacrylamide gel electrophoresis and Western blotting.
For immunofluorescence experiments, cells were fixed for 20 min in PBS containing 4% paraformaldehyde and permeabilized for 5 min in PBS containing 0.5% Triton X-100. After blocking in PBS containing 20% fetal bovine serum (FBS) and 0.5% Tween 20, cells were incubated with the corresponding primary antibodies diluted in PBS-0.2% BSA for 1 h. After washing with PBS, the preparations were further incubated with goat anti-rabbit, goat anti-mouse, or goat anti-rat antibodies bound to Alexa 488 and/or Alexa 594 fluorochromes. The preparations were mounted in Prolong reagent and analyzed by confocal microscopy using a Bio-Rad Radiance 2100 laser scanning system on a Zeiss Axiovert 200 microscope. Images of 1,024 by 1,024 pixels and an eight-bit gray-scale depth were acquired sequentially every 0.2 to 0.3 μm employing LaserSharp v5.0 software (Bio-Rad) and analyzed using LaserPix v.4 image program (Bio-Rad). The colocalization mask was used for colocalization analysis. All experiments were repeated at least three times, and a representative experiment is shown.
RNA binding activity assay.
To determine the RNA binding activity of wild-type hStau1 and Mut1 proteins, a Northwestern assay was performed with purified proteins separated on 10% polyacrylamide gels and transferred to a nitrocellulose filter. The filter was hybridized using a probe corresponding to the 3′ UTR of bicoid mRNA (32), as described previously (25).
RNA analyses.
The RNA associated to hStau1-TAP was obtained from the purified complexes by proteinase K (0.2 mg/ml) and 0.5% SDS treatment in TNE buffer for 30 min at 37°C. After phenol extraction, RNAs were precipitated with 2 ethanol volumes and 20 μg of glycogen. Semiquantitative RT-PCR for NP and M segments was performed to determine the presence of vRNA or mRNAs. To detect the NP segment, the oligonucleotides 5′-TGGAATTGGACGATTCTACAT-3′ and 5′-TCTTAGGATCTTTCCCCGC-3′ were used, respectively. To detect the M segment, the primers used were 5′-CTCATGGAATGGCTAAAGACA-3′ and 5′-GCATTTTGGACAAAGCGTCTA-3′, respectively. The reverse transcription (RT) reaction was performed in the presence of the corresponding oligonucleotide for 30 min at 42°C. After that time, the reaction mix was incubated 2 min at 94°C to inactivate the AMV enzyme, and the second oligonucleotide was included in the mixture for 30 PCR rounds of 94°C for 30 s, 53°C (for NP segment) or 50°C (for M segment) for 30 s, and 68°C for 30 s and a final extension time of 7 min at 68°C. Serial dilutions of total RNA and the associated RNAs were used to ensure amplification in the lineal range.
Purification of virions.
Low (10−3-PFU/ml) multiplicities of infection were performed for 60 h in each cell line, and the culture supernatants were centrifuged over a 33% to 50% sucrose step gradient in TNE for 60 min at 40,000 rpm and 4°C in an SW41 rotor. The 50% to 33% interface was collected, diluted in the same buffer, and further centrifuged through a 33% sucrose cushion for 60 min at 40,000 rpm and 4°C in an SW41 rotor, as previously described (20). To determine the protein composition, the pellet was resuspended in Laemmli sample buffer and analyzed by Western blotting. To analyze RNA content, pellets were resuspended in 1 ml of TRIzol reagent (Invitrogen), and the RNA extraction procedure was followed as suggested by the manufacturer. RNA was analyzed by denaturing polyacrylamide gel electrophoresis and silver staining as previously described (30).
RESULTS AND DISCUSSION
Human Stau1 interacts with NS1 and viral RNPs.
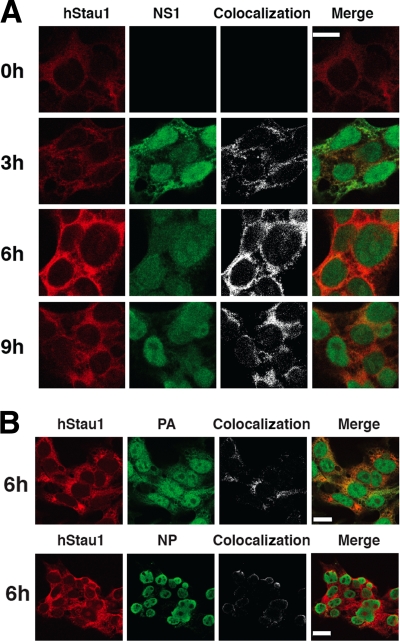
Previous results in our laboratory have shown the interaction of endogenous hStau1 with NS1 protein during influenza virus infection (17). In order to verify this interaction, protein localization analyses were performed using antibodies specific for NS1 and hStau1 in HEK293T cells. As shown in Fig. 1A, both proteins colocalized from early times in the cytoplasm of the infected cells. Although the overexpression of NS1 and hStau1 was shown to cause the relocalization of the latter to the nucleus (17), we did not observe such an effect in the infection, probably due to the lower concentration of free soluble endogenous hStau1 protein in the cytosol. Furthermore, both proteins colocalized in the cytosol when cells were transfected with plasmids expressing NS1 and hStau1 under an RNA polymerase II promoter (data not shown). In addition, both NP and PA proteins were also found to colocalize with hStau1 in the cytoplasm (Fig. 1B). This result strongly suggests that hStau1 is interacting with viral RNPs, as most of the NP found in the cytosol later in the infection is assumed to be in the form of RNPs.
FIG. 1.
Colocalization of hStau1 protein with influenza viral proteins in infected cells. Cultures of HEK293T cells were infected with WSN virus and fixed at the times after infection indicated. The cells were processed for immunofluorescence with antibodies specific for the hStau1, NS1, NP, or PA proteins as indicated in Materials and Methods. The left columns of panels show the localization pattern of each protein. The right columns of panels show the colocalization sites of hStau1 and the viral markers used, as well as the merged images. (A) hStau1 protein is shown in red, and NS1 protein in green. The scale bar represents 10 μm. (B) hStau1 is shown in red, and either PA or NP is shown in green. The scale bar represents 20 μm.
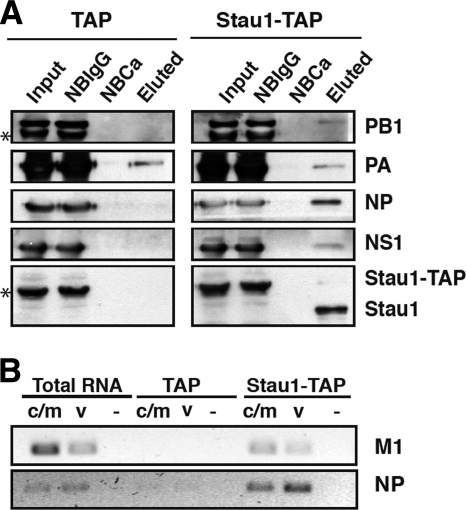
To verify the interaction of hStau1 with viral RNPs during the infection, HEK293T cells were transfected with the pChStaufen-TAP or the pCTAP constructs using the calcium phosphate protocol (56). Sixteen hours later, the cells were infected at a high multiplicity with the WSN strain of influenza virus for 6 h, and total cell extracts were used for TAP purification. In agreement with previous results (17), NS1 protein was found in the final TAP elutions obtained from cells transfected with the pChStaufen-TAP plasmid, while there was no detectable NS1 protein in the elutions obtained from cells transfected with control pCTAP plasmid (Fig. 2A). Furthermore, NP, PB1, and PA proteins were also found associated to purified hStau1 derived from cells expressing hStau1-TAP (Fig. 2A), although a substantial background signal of PA appears in the corresponding TAP elution due to unspecific binding to the resin.
FIG. 2.
Association of viral proteins and RNAs with the recombinant hStau1-TAP protein in infected cells. Cultures of HEK293T cells were transfected with a plasmid expressing the hStau1-TAP protein or the TAP tag as a control and subsequently infected with WSN virus at an MOI of 5 PFU/cell. At 6 h postinfection, total cell extracts were prepared and used for TAP purification as indicated in Materials and Methods. (A) Aliquots of the total extract (Input), material not bound to IgG-Sepharose resin (NBIgG), material not bound to calmodulin resin (NBCa), and material eluted from the calmodulin resin (Eluted) were analyzed by Western blotting with antibodies specific for PB1, PA, NP, NS1, and hStau1, as indicated on the right. The asterisks on the left indicate the band of hStau1-TAP, which cross-reacts with rabbit antisera. (B) Total RNA was isolated from the cell extracts of the eluted fractions, and RT-PCR was performed with primers specific for M1 and NP RNAs of positive (c/m) or negative (v) polarity. Tenfold serial dilutions of the RNAs were analyzed until the PCR signal for the TAP control was negligible. As a negative control, the primers were omitted in the RT reaction step (−) but were included in the PCR step.
Binding of viral RNAs to hStau1 is not direct and does not depend on NS1 protein.
The association of NP and polymerase subunits to purified hStau1 suggested its interaction with progeny viral RNPs and hence an association to vRNA would be predicted. Nevertheless, the presence of viral mRNAs in hStau1 complexes could not be excluded, as viral mRNAs are structurally similar to cellular ones, and hStau1 complexes are involved in transport and translation of cellular mRNAs (34, 54). To clarify this question, RNA was obtained from TAP-purified hStau1 or control complexes and used for the detection of both viral mRNAs and vRNAs corresponding to the early-expressed NP segment and the late-expressed M segment. As presented in Fig. 2B, both mRNAs and vRNAs specific for these viral RNA segments were found in hStau1 complexes, suggesting a dual role of hStau1 in influenza virus infection.
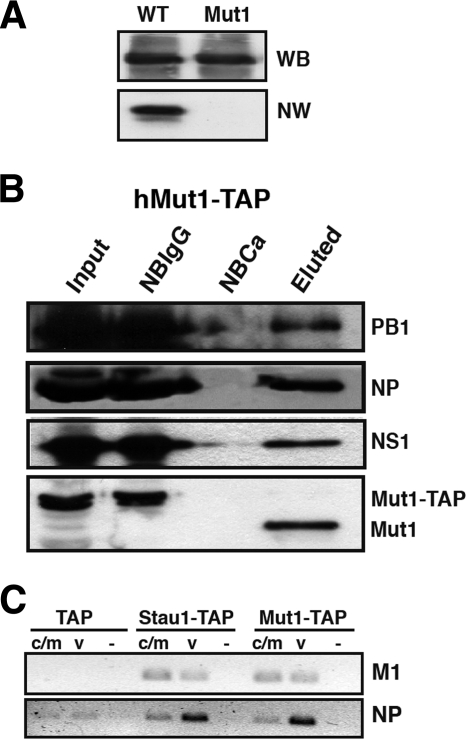
It is known that D. melanogaster Staufen protein (dmStau) binds specifically, but not necessarily directly, certain cellular mRNAs (50), and double-stranded RBD3 (dsRBD3) and dsRBD4 have been shown to bind RNAs with the highest affinity (55). Mutagenesis experiments with dmStau and the determination of the dsRNA-RBD3 atomic structure (44) revealed the importance of the lysine residues at positions 129 and 130 in dsRNA-RBD3 to preserve the RNA binding activity. To test whether viral RNAs bind directly to hStau1, we generated a mutant protein in which lys129 and lys130 have been mutated to alanines (hStau1-Mut1). The RNA binding activity of wild-type (wt) and mutant proteins was determined by Northwestern assays using wt and hStau1-Mut1 proteins purified after expression in E. coli (Fig. 3A). These results showed that hStau1-Mut1 binds RNA much less efficiently than wt hStau1. Direct in vitro RNA binding assays indicated that the affinity of hStau1-Mut1 binding for RNA was around 10 times lower than wt hStau1 (data not shown). Therefore, we used the pChStau-Mut1-TAP construct to perform transfection-infection and TAP purification experiments similar to those described above. Western blot analyses of purified hStau1-Mut1 complexes showed the association of the NS1 protein (Fig. 3B), corroborating that the interaction of NS1 and hStau1 is not mediated by binding to the same RNA molecule (17). In addition, the NP and polymerase subunits were also found associated to hStau1-Mut1 protein (Fig. 3B). Consistent with these observations, vRNAs specific for NP and M1 RNA segments were found associated to purified hStau1-Mut1 complexes (Fig. 3C). In addition, the association of viral mRNAs was also maintained for the hStau1-Mut1 protein, suggesting that neither viral mRNAs nor genomic vRNAs are directly bound to hStau1, but are to another component of the complex.
FIG. 3.
Role of hStau1 RBD in the interaction with viral RNPs and RNAs. (A) RNA binding activity of wild-type and Mut1 hStau1 proteins. Equal amounts of either protein, as shown by the Western blot (WB) panel, were used to perform a Northwestern (NW) assay using a probe corresponding to the 3′ UTR of bicoid mRNA. (B) 293T cells were transfected with the plasmid expressing a mutant hStau1-TAP protein (Mut1-TAP) and infected with the influenza virus WSN strain at 24 h posttransfection. Cell extracts were used to perform TAP purification. Aliquots of the total extract (Input), material not bound to IgG-Sepharose resin (NBIgG), material not bound to calmodulin resin (NBCa), and material eluted from the calmodulin resin (Eluted) were analyzed by Western blotting with antibodies specific for PB1, NP, NS1, and hStau1, as indicated on the right. (C) Total RNA was isolated from the eluted fractions shown in panel B, and RT-PCR was performed with primers specific for M1 and NP RNAs of positive (c/m) or negative (v) polarity. Tenfold serial dilutions of the RNAs were analyzed until the PCR signal for the TAP control was negligible. As a negative control, the primers were omitted in the RT reaction step (−) but were included in the PCR step.
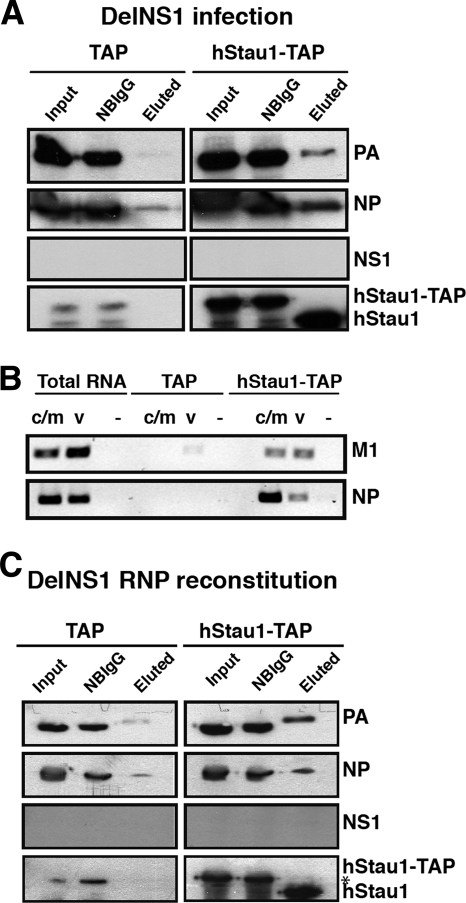
It has been shown previously that in influenza virus-infected cells, the NS1 protein interacts with viral translation complexes (7) and viral transcription-and-replication complexes (39). Since the results mentioned above suggest that hStau1 interacts with the viral RNP, it is conceivable that NS1 could be responsible for this association. To explore this possibility, we have used delNS1-defective virus (22) to infect cells previously transfected with the pChStaufen-TAP or TAP control constructs. After IgG pulldown and TEV digestion of the intracellular complexes containing hStau1-TAP proteins, the presence of RNP proteins was ascertained. As shown in Fig. 4, NP and PB1 proteins and also the vRNAs were found associated to hStau1, suggesting that NS1 is not necessary for hStau1-RNP interaction. Furthermore, the presence of viral mRNAs in hStau1 complexes suggests that the NS1 protein is not required for the transport of these in hStau1 complexes (Fig. 4B).
FIG. 4.
NS1 protein is not necessary for the interaction of vRNPs and viral mRNAs with hStau1-TAP. Cultures of HEK293T cells were transfected with a plasmid expressing hStau1-TAP protein or the TAP tag as a control and subsequently infected with the delNS1 virus strain at an MOI of 2 PFU/cell. At 6 h postinfection, total cell extracts were prepared and used for TAP purification as indicated in Materials and Methods. (A) Aliquots of the total extract (Input), material not bound to IgG-Sepharose resin (NBIgG), and material eluted from the IgG-Sepharose resin (Eluted) were analyzed by Western blotting with antibodies specific for PA, NP, NS1, and hStau1, as indicated on the right. (B) Total RNA was isolated from the cell extracts or the eluted fractions, and RT-PCR was performed with primers specific for M1 and NP RNAs of positive (c/m) or negative (v) polarity. Tenfold serial dilutions of the RNAs were analyzed until the PCR signal for the TAP control was negligible. As a negative control, the primers were omitted in the RT reaction step (−) but were included in the PCR step. (C) Cultured 293T cells were cotransfected with the plasmid expressing hStau1-TAP protein or the TAP tag and the plasmids necessary to reconstitute an active vRNP in the absence of NS1 protein. TAP purification was performed 24 h posttransfection as described in Materials and Methods. Aliquots of the total extract (Input), material not bound to IgG-Sepharose resin (NBIgG), and material eluted after TEV digestion (Eluted) were analyzed by Western blotting with antibodies specific for PA, NP, NS1, and hStau1, as indicated on the right. The asterisk indicates a cross-reaction of the anti-hStau1 antibody.
This result was corroborated by cotransfection of plasmids expressing PA, PB1, PB2, NP, and plasmid pHHdelNS, which expresses a NS-virus-like RNA with an almost complete deletion of NS1 (21), and the plasmid expressing hStau1-TAP or the TAP tag alone (Fig. 4C). Cell extracts were used to perform TAP purification, and the proteins were analyzed by Western blotting. The results indicated that these in vivo reconstituted RNPs are able to interact with hStau1-TAP independently of the NS1 protein.
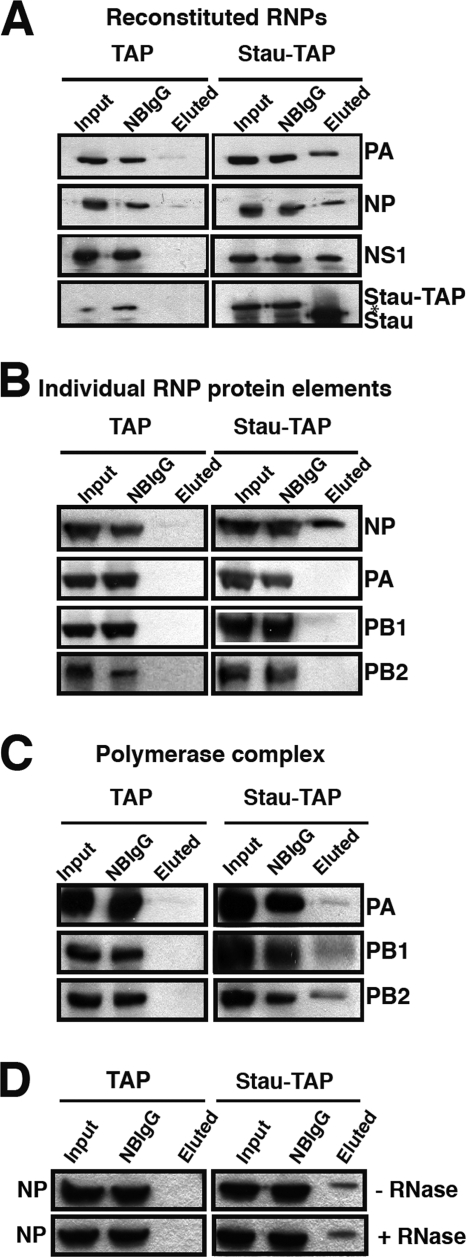
Both NP and the polymerase complex are able to interact with Staufen complexes.
So far, we have shown the interaction of hStau1 with the genomic vRNPs. However, these experiments could not identify which element in the RNP is responsible for the interaction. To analyze this issue, we reconstituted wt NS RNPs in HEK293T cells by cotransfection of plasmids expressing PB1, PB2, PA, NP, and pHHNS, as well as the plasmid expressing hStau1-TAP (or the TAP tag as a control). After 24 h, cell extracts were obtained and used to perform TAP purification. Western blot analysis revealed the interaction of the in vivo reconstituted RNP with hStau1-TAP (Fig. 5A). Functional activity of the RNP reconstituted in vivo is shown by the expression of the NS1 protein (Fig. 5A). This result verified the interaction of hStau1 with the RNPs during the infection (Fig. 1 and 2A). To determine whether this interaction requires the functional RNP complex or may occur with any independent element of the RNP, HEK293T cells were cotransfected with plasmids expressing hStau1-TAP (or the TAP tag alone) and the individual RNP components. The results after TAP purification and Western blot analysis are shown in Fig. 5B. NP is able to interact with hStau1-TAP, whereas the individual polymerase subunits are not. However, when all the subunits of the polymerase were transfected together with hStau1-TAP to allow the formation of the heterotrimeric complex, all subunits were found to associate to hStau1 (Fig. 5C). To analyze whether the interaction of NP and hStau1-TAP was mediated by their binding to the same RNA, 24 h after cotransfection of the hStau1-TAP and NP plasmids, cell extracts were obtained, divided in two portions, and used to perform TAP purification in the presence or absence of RNase A. As shown in Fig. 5D, this interaction is RNA independent. These results suggest that the association of hStau1 with viral RNPs is mediated by independent interactions with the polymerase complex and the NP monomers.
FIG. 5.
Influenza virus NP and polymerase complex interact with hStau1 complexes. To determine which viral RNP proteins interact with hStau1, HEK293T cells were cotransfected with the plasmid expressing hStau1-TAP (or the TAP tag as a control) and all plasmids required to generate an active RNP (pCMV-PB1, pCMV-PB2, pCMV-PA, pCMV-NP and pHH-NS1) (A), each of the plasmids individually expressing the protein components of the RNP (B), the plasmids expressing the viral polymerase (C), or a plasmid expressing NP (D) (the cell lysis was performed in the presence or absence of RNase A). In every set of experiments, after 24 h posttransfection, cell extracts were used to carry out TAP purification. Aliquots of the total extract (Input), material not bound to IgG-Sepharose resin (NBIgG), and material eluted from the IgG-Sepharose resin (eluted) were analyzed by Western blotting with antibodies specific for PA, PB1, PB2, NP, NS1, and hStau1, as indicated to the right. The asterisk in panel A indicates a cross-reaction of the anti-hStau1 antibody.
Relevance of hStau1 in influenza virus infection.
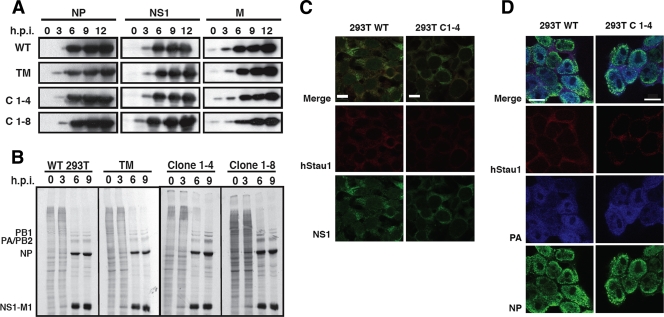
To analyze the possible role of hStau1 protein in influenza virus infection, we used gene silencing to generate HEK293T-based cell lines in which hStau1 expression is reduced. The cells were transfected with the plasmid pSR-puro-iStau, which encodes a short hairpin (shRNA) directed to hStau1 mRNA and a puromycin resistance marker. Multiple cell clones were selected by incubation with puromycin, and the levels of endogenous hStau1 were analyzed by Western blot and immunofluorescence assays. Two of them, clones 1-4 and 1-8, showed diminished levels of hStau1 compared to the parental cell line (iStau cells) and were further used for infection experiments (Fig. 6A). As a control, HEK293T cells silenced with an irrelevant sequence derived from Thermotoga maritima (iTm cells) were used. Both hStau1 isoforms were silenced (see Fig. 6A, arrows), and the downregulation was around 50% for clone 1-4.
FIG. 6.
The yield of influenza virus is reduced in hStau1-silenced HEK293T cells. (A) The levels of hStau1 in WT HEK293T cells, TM clone cells, and the hStau1-silenced cell clones 1-4 and 1-8 were determined by Western blotting using actin as a standard. The two hStau1 isoforms are indicated by arrows, while the asterisk denotes a cross-reaction of the anti-hStau1 antibody. (B) Cultures of HEK293T cells (WT), cells silenced with the irrelevant TM sequence (TM), and the hStau1-silenced clones 1-4 and 1-8 were infected with WSN virus at an MOI of 10−3 PFU/cell, and aliquots of the supernatants were sampled at the indicated times after infection. Virus titers and standard deviations are presented, as determined by plaque assay with MDCK cells.
To test whether the presence of hStau1 is important for virus replication, the growth kinetics of the WSN strain of influenza virus was analyzed by low-multiplicity-infection assays, performed in independent but parallel triplicates, with the iStau and control HEK293T cell lines. Titration of the supernatants obtained at various times postinfection showed a close to 10-fold decrease in viral production in iStau clone 1-4 cells (compared to parental HEK293T or iTm-silenced cells) (Fig. 6B). To check whether these reduced virus yields were indeed due to downregulation of hStau1 and could be generalized to any human cell line, we used three different siRNAs specific for hStau1 to transfect A549 cells and tested the virus production in triplicate low-multiplicity infections. The yield of virus paralleled the hStau1 silencing induced by all three specific siRNAs, compared to the control siRNA (Fig. 7 A and B). As a specificity control, we carried out similar experiments but using VSV as an unrelated negative-stranded RNA virus. As presented in Fig. 7C and D, no reduction in virus titers was detected upon silencing of hStau1.
FIG. 7.
The yield of influenza virus is specifically reduced in A549 cells. (A) The level of hStau1 in A549 cells transfected with an irrelevant siRNA (control) or with 3 different siRNAs specific for hStau1 (S1, S2, and S3) was determined by Western blotting using actin as standard. (B) Cultures of A549 cells transfected with the siRNAs were infected with WSN virus at an MOI of 10−3 PFU/cell, and aliquots of the supernatants were sampled at the indicated times after infection. Virus titers and standard deviations are presented, as determined by plaque assay in MDCK cells. (C) The level of hStau1 was determined as described in the legend for panel A. (D) Cultures of A549 cells transfected with the siRNAs were infected with VSV at an MOI of 10−3 PFU/cell, and aliquots of the supernatants were sampled at the indicated times after infection. Virus titers and standard deviations are presented, as determined by plaque assay in BHK21 cells.
hStau1 silencing does not affect viral protein expression or localization, but production of viral particles is reduced.
To determine which step in the viral infection cycle is affected by the reduction in hStau1 levels, silenced or control cells were infected with the WSN strain at a high multiplicity, and the time course of virus proteins NS1, NP, and M1 was determined by Western blotting. The results showed that there are no differences in the accumulation of the viral proteins produced in the iStau cell lines compared to the iTm cell line or the parental HEK293T cell line (Fig. 8A). In addition, similar time courses of virus protein synthesis were examined. At various time points after infection, cells were pulse-labeled with [35S]Met-Cys, and total protein extracts were analyzed by gel electrophoresis. As shown in Fig. 8B, the viral protein synthesis was not affected by the reduction in hStau1 levels.
FIG. 8.
Protein accumulation, synthesis, and localization after infection of wt and hStau1-silenced cells. Cultures of HEK293T and the hStau1-silenced cells were infected at a high MOI with WSN virus, and total cell extracts were prepared at different times postinfection. (A) The accumulation of viral proteins was determined by Western blotting with antibodies specific for the NP, NS1, and M proteins. (B) The cells were pulse-labeled with [35S]Met-Cys at the indicated times after infection. The labeled total cell extracts were analyzed by polyacrylamide-gel electrophoresis and autoradiography. The positions of specific virus proteins are indicated on the left. Cultures of HEK293T cells (WT) and the hStau1-silenced clone 1-4 were infected with WSN virus at an MOI of 5 PFU/cell, fixed at 6 h after infection, and processed for immunofluorescence with antibodies specific for hStau1 (in red) and either NS1 (in green) (C) or NP (in green) and PA (in blue) (D). The scale bar represents 10 μm.
As hStau1 colocalizes with both NS1 and viral RNPs from early times in the infection (Fig. 1A and B), it is possible that the reduction in hStau1 levels could affect the nucleocytosolic transport of viral proteins, and although their expression is not affected, they could not get out from the nucleus. Localization experiments were performed by using immunofluorescence to determine the expression pattern of NS1, NP, and PA in the iStau cell lines. As shown in Fig. 8C and D, there were no changes in the nucleocytosolic distribution of the viral proteins.
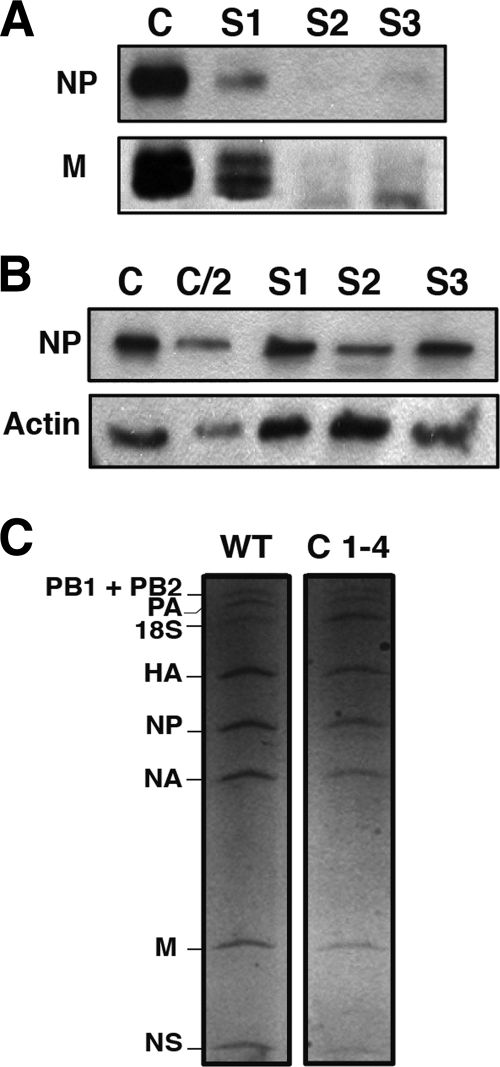
The analyses carried out so far did not reveal any differences in the virus infections of iStau versus iTm or parental HEK293T cells. To test whether the production of virus particles was affected, the supernatants obtained from low-MOI infections of different cell lines were used to purify virions, and these were quantified by Western blotting using antibodies specific for NP and M1 proteins. As presented in Fig. 9A, the yield of viral particles was reduced in the supernatant of A549 cells transfected with siRNAs specific for hStau1 compared to that of the control siRNA-silenced cells. In contrast, the relative accumulations of NP protein in the extracts obtained from the infected cells at the time that the virus was harvested were not substantially different (Fig. 9B), suggesting that fewer virions are generated in hStau1-silenced cells compared to the control-silenced ones. In addition, a comparison of the vRNA contents of virions purified from normal or hStau1-silenced cells did not reveal any misbalance among the different genomic RNA segments (Fig. 9C). In single-cycle experiments performed with the various silenced or control cells, no differences in the shapes of particles obtained were detectable by electron microscopy of stained cellular sections of the infected cells (data not shown). All together, these results suggest that the reduction of virus infectivity described above (Fig. 6 and 7) probably reflects a diminished capacity of hStau1-silenced cells to generate virus particles.
FIG. 9.
Production of virus particles in hStau1-silenced cells. Cultures of A549 cells were transfected with 3 different siRNAs specific for hStau1 (S1, S2, and S3) or an irrelevant siRNA as a control (C) and used for low-multiplicity infections with WSN influenza virus. Virus particles were purified from the supernatant media, and total cell extracts were prepared in Laemmli buffer. (A) Equal aliquots of the purified virions were analyzed by PAGE and Western blotting with anti-NP or anti-M1 antibodies. (B) Samples from the cell extracts were analyzed by PAGE and Western blotting with anti-NP (NP) or anti-actin antibodies (Actin). C/2 denotes loading one half the amount than in the control (C). (C) HEK293T cells (WT) and iStau clone 1-4 (C 1-4) cells were infected at a high MOI, and after 7 h, the supernatant was used to purify viral particles. Virion RNAs were purified and analyzed by denaturing polyacrylamide-urea gel electrophoresis and silver staining. The positions of the various RNA segments are indicated on the left.
Possible roles of hStau1 during influenza virus infection.
A number of features make influenza virus unusual among RNA-containing viruses infecting animal cells. Its genome is segmented, has negative polarity, and is composed of RNPs rather than naked RNAs. In addition, the replication and transcription of these RNPs occur in the nucleus of the infected cells, but final virion morphogenesis takes place at the plasma membrane (43). All together, these features require the virus to find ways to export the progeny RNPs from the nucleus to the cytoplasm, transport them to the vicinity of the cell membrane, and package a complete set of RNPs into single virions. The results presented above indicate that the hStau1 protein, which was first identified as a human protein associated to the influenza virus NS1 protein (17, 38), also associates to viral RNPs and virus mRNAs in influenza virus-infected cells (Fig. 2A and B). Several recent reports used genomic silencing to identify host genes involved in influenza virus multiplication (5, 31, 35), but the hStau1 gene was not identified as a factor essential for virus replication. In contrast, a general analysis of influenza virus interactors in human cells verified the interaction of hStau1 with NS1 protein (48). This general analysis failed to reveal the interactions of hStau1 with the virus NP and polymerase showed here but revealed an additional interaction with the M1 protein.
hStau1 is involved in the formation of cytoplasmic RNA granules that mediate the controlled localization and translation of specific cellular mRNAs during differentiation and development (50, 51) but is also present in P bodies and stress granules (2, 52, 53). Therefore, it is possible that hStau1 association to viral mRNAs is connected to their expression and turnover. In fact, it has been reported that hStau1 is involved in the efficient translation of mRNAs containing a structured 5′ UTR (14), and NS1 has been described to enhance the translation of viral mRNAs (1, 12, 16). However, no change in the pattern of virus protein synthesis was observed when hStau1 accumulation was downregulated by gene silencing (Fig. 8), consistent with the low structural complexity of influenza virus mRNAs. Hence, we have to conclude that hStau1 is not essential for virus protein expression, at least as detected by the levels of downregulation used.
The association of hStau1 with viral RNPs, and hence with vRNAs, probably represents an additional virus-host interaction, not related to virus RNA replication or transcription, as hStau1 colocalization with RNP markers occurs in the cytoplasm (Fig. 1). Furthermore, this association was found to be independent of the RNA-binding activity of hStau1 and the presence of NS1. Hence, we propose that the association occurs mainly by direct or indirect protein-protein interaction with the polymerase complex and NP (Fig. 5) and could be involved in the localization of the progeny RNPs under the plasma membrane or in the clustering of the various RNP species for efficient packaging, although the analysis of the vRNAs present in virions produced in normal cells or hStau1 knockdown cells failed to show any misbalance among the viral RNA segments (Fig. 9C). It is noteworthy that hStau1 interacts both with NS1 and elements of the virus RNP. It is conceivable that the virus subverts the role of hStau1 to improve the RNP packaging into infectious virus while it also uses hStau1-NS1 interaction for proper mRNA localization and expression.
A precedent of such a role for the hStau1 protein has been reported for HIV-infected cells, as it was shown to become incorporated into HIV virions by interaction with the Pr55gag protein and to regulate the assembly of virus particles (8-10). We have been unable to detect the presence of the hStau1 protein in purified influenza virions by Western blotting (data not shown). Such a negative result is consistent with the data recently published by Shaw et al. (49) on the proteomic characterization of host proteins present in influenza virions.
Acknowledgments
We thank Adolfo García-Sastre for providing the delNS1 virus strain and anti-M antibodies. The technical assistance of Yolanda Fernández, Noelia Zamarreño, and Marcela Benavides is gratefully acknowledged.
S.D.L. is a postdoctoral fellow from FISS, Ministerio de Ciencia e Innovación. J.P. is a fellow from Ministerio de Ciencia e Innovación. R.M.M. was a fellow from Ministerio de Educación y Ciencia. This work was funded by Ministerio de Educación y Ciencia (BFU2007-60046), Comunidad de Madrid (S-SAL-0815-2006) (VIRHOST), and Fundación Marcelino Botín.
Footnotes
Published ahead of print on 26 May 2010.
REFERENCES
- 1.Aragón, T., S. de la Luna, I. Novoa, L. Carrasco, J. Ortín, and A. Nieto. 2000. Translation factor eIF4GI is a cellular target for NS1 protein, a translational activator of influenza virus. Mol. Cell. Biol. 20:6259-6268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barbee, S. A., P. S. Estes, A. M. Cziko, J. Hillebrand, R. A. Luedeman, J. M. Coller, N. Johnson, I. C. Howlett, C. Geng, R. Ueda, A. H. Brand, S. F. Newbury, J. E. Wilhelm, R. B. Levine, A. Nakamura, R. Parker, and M. Ramaswami. 2006. Staufen- and FMRP-containing neuronal RNPs are structurally and functionally related to somatic P bodies. Neuron 52:997-1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bárcena, J., M. Ochoa, S. de la Luna, J. A. Melero, A. Nieto, J. Ortín, and A. Portela. 1994. Monoclonal antibodies against influenza virus PB2 and NP polypeptides interfere with the initiation step of viral mRNA synthesis in vitro. J. Virol. 68:6900-6909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bourmakina, S. V., and A. Garcia-Sastre. 2005. The morphology and composition of influenza A virus particles are not affected by low levels of M1 and M2 proteins in infected cells. J. Virol. 79:7926-7932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brass, A. L., I. C. Huang, Y. Benita, S. P. John, M. N. Krishnan, E. M. Feeley, B. J. Ryan, J. L. Weyer, L. van der Weyden, E. Fikrig, D. J. Adams, R. J. Xavier, M. Farzan, and S. J. Elledge. 2009. The IFITM proteins mediate cellular resistance to influenza A H1N1 virus, West Nile virus, and dengue virus. Cell 139:1243-1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Briedis, D. J., G. Conti, E. A. Munn, and B. W. J. Mahy. 1981. Migration of influenza virus-specific polypeptides from cytoplasm to nucleus of infected cells. Virology 111:154-164. [DOI] [PubMed] [Google Scholar]
- 7.Burgui, I., T. Aragón, J. Ortín, and A. Nieto. 2003. PABP1 and eIF4GI associate to influenza virus NS1 protein in viral mRNA translation initiation complexes. J. Gen. Virol. 84:3263-3274. [DOI] [PubMed] [Google Scholar]
- 8.Chatel-Chaix, L., L. Abrahamyan, C. Frechina, A. J. Mouland, and L. DesGroseillers. 2007. The host protein Staufen1 participates in human immunodeficiency virus type 1 assembly in live cells by influencing pr55Gag multimerization. J. Virol. 81:6216-6230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chatel-Chaix, L., K. Boulay, A. J. Mouland, and L. Desgroseillers. 2008. The host protein Staufen1 interacts with the Pr55Gag zinc fingers and regulates HIV-1 assembly via its N-terminus. Retrovirology 5:41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chatel-Chaix, L., J. F. Clement, C. Martel, V. Beriault, A. Gatignol, L. DesGroseillers, and A. J. Mouland. 2004. Identification of Staufen in the human immunodeficiency virus type 1 Gag ribonucleoprotein complex and a role in generating infectious viral particles. Mol. Cell. Biol. 24:2637-2648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen, W., P. A. Calvo, D. Malide, J. Gibbs, U. Schubert, I. Bacik, S. Basta, R. O'Neill, J. Schickli, P. Palese, P. Henklein, J. R. Bennink, and J. W. Yewdell. 2001. A novel influenza A virus mitochondrial protein that induces cell death. Nat. Med. 7:1306-1312. [DOI] [PubMed] [Google Scholar]
- 12.de la Luna, S., P. Fortes, A. Beloso, and J. Ortín. 1995. Influenza virus NS1 protein enhances the rate of translation initiation of viral mRNAs. J. Virol. 69:2427-2433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.DuBridge, R. B., P. Tang, H. C. Hsia, P. M. Leong, J. H. Miller, and M. P. Calos. 1987. Analysis of mutation in human cells by using an Epstein-Barr virus shuttle system. Mol. Cell. Biol. 7:379-387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dugré-Brisson, S., G. Elvira, K. Boulay, L. Chatel-Chaix, A. J. Mouland, and L. DesGroseillers. 2005. Interaction of Staufen1 with the 5′ end of mRNA facilitates translation of these RNAs. Nucleic Acids Res. 33:4797-4812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Egorov, A., S. Brandt, S. Sereinig, J. Romanova, B. Ferko, D. Katinger, A. Grassauer, G. Alexandrova, H. Katinger, and T. Muster. 1998. Transfectant influenza A viruses with long deletions in the NS1 protein grow efficiently in Vero cells. J. Virol. 72:6437-6441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Enami, K., T. A. Sato, S. Nakada, and M. Enami. 1994. Influenza virus NS1 protein stimulates translation of the M1 protein. J. Virol. 68:1432-1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Falcón, A. M., P. Fortes, R. M. Marión, A. Beloso, and J. Ortín. 1999. Interaction of influenza virus NS1 protein and the human homologue of Staufen in vivo and in vitro. Nucleic Acids Res. 27:2241-2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Falcón, A. M., R. M. Marión, T. Zürcher, P. Gómez, A. Portela, A. Nieto, and J. Ortín. 2004. Defective RNA replication and late gene expression in temperature-sensitive (A/Victoria/3/75) influenza viruses expressing deleted forms of NS1 protein. J. Virol. 78:3880-3888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ferrandon, D., L. Elphick, V. C. Nusslein, and D. St. Johnston. 1994. Staufen protein associates with the 3′UTR of bicoid mRNA to form particles that move in a microtubule-dependent manner. Cell 79:1221-1232. [DOI] [PubMed] [Google Scholar]
- 20.Garaigorta, U., A. M. Falcon, and J. Ortin. 2005. Genetic analysis of influenza virus NS1 gene: a temperature-sensitive mutant shows defective formation of virus particles. J. Virol. 79:15246-15257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Garaigorta, U., and J. Ortín. 2007. Mutation analysis of a recombinant NS replicon shows that influenza virus NS1 protein blocks the splicing and nucleo-cytoplasmic transport of its own viral mRNA. Nucleic Acids Res. 35:4573-4582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.García-Sastre, A., A. Egorov, D. Matassov, S. Brandt, D. E. Levy, J. E. Durbin, P. Palese, and T. Muster. 1998. Influenza A virus lacking the NS1 gene replicates in interferon-deficient systems. Virology 252:324-330. [DOI] [PubMed] [Google Scholar]
- 23.Gaush, C. R., W. L. Hard, and T. F. Smith. 1966. Characterization of an established line of canine kidney cells (MDCK). Proc. Soc. Exp. Biol. Med. 122:931-935. [DOI] [PubMed] [Google Scholar]
- 24.Giard, D. J., S. A. Aaronson, G. J. Todaro, P. Arnstein, J. H. Kersey, H. Dosik, and W. P. Parks. 1973. In vitro cultivation of human tumors: establishment of cell lines derived from a series of solid tumors. J. Natl. Cancer Inst. 51:1417-1423. [DOI] [PubMed] [Google Scholar]
- 25.González, S., and J. Ortín. 1999. Distinct regions of influenza virus PB1 polymerase subunit recognize vRNA and cRNA templates. EMBO J. 18:3767-3775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hale, B. G., R. E. Randall, J. Ortin, and D. Jackson. 2008. The multifunctional NS1 protein of influenza A viruses. J. Gen. Virol. 89:2359-2376. [DOI] [PubMed] [Google Scholar]
- 27.Hatada, E., M. Hasegawa, K. Shimizu, M. Hatanaka, and R. Fukuda. 1990. Analysis of influenza A virus temperature-sensitive mutants with mutations in RNA segment 8. J. Gen. Virol. 71:1283-1292. [DOI] [PubMed] [Google Scholar]
- 28.Herz, C., E. Stavnezer, R. M. Krug, and T. Gurney. 1981. Influenza virus, an RNA virus, synthesizes its messenger RNA in the nucleus of infected cells. Cell 26:391-400. [DOI] [PubMed] [Google Scholar]
- 29.Honda, A., J. Mukaigawa, A. Yokoiyama, A. Kato, S. Ueda, K. Nagata, M. Krystal, D. P. Nayak, and A. Ishihama. 1990. Purification and molecular structure of RNA polymerase from influenza virus A/PR8. J. Biochem. 107:624-628. [DOI] [PubMed] [Google Scholar]
- 30.Jorba, N., S. Juarez, E. Torreira, P. Gastaminza, N. Zamarreno, J. P. Albar, and J. Ortin. 2008. Analysis of the interaction of influenza virus polymerase complex with human cell factors. Proteomics 8:2077-2088. [DOI] [PubMed] [Google Scholar]
- 31.Karlas, A., N. Machuy, Y. Shin, K. P. Pleissner, A. Artarini, D. Heuer, D. Becker, H. Khalil, L. A. Ogilvie, S. Hess, A. P. Maurer, E. Muller, T. Wolff, T. Rudel, and T. F. Meyer. 2010. Genome-wide RNAi screen identifies human host factors crucial for influenza virus replication. Nature 463:818-822. [DOI] [PubMed] [Google Scholar]
- 32.Kiebler, M. A., I. Hemraj, P. Verkade, M. Kohrmann, P. Fortes, R. M. Marion, J. Ortin, and C. G. Dotti. 1999. The mammalian staufen protein localizes to the somatodendritic domain of cultured hippocampal neurons: implications for its involvement in mRNA transport. J. Neurosci. 19:288-297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kochs, G., I. Koerner, L. Thiel, S. Kothlow, B. Kaspers, N. Ruggli, A. Summerfield, J. Pavlovic, J. Stech, and P. Staeheli. 2007. Properties of H7N7 influenza A virus strain SC35M lacking interferon antagonist NS1 in mice and chickens. J. Gen. Virol. 88:1403-1409. [DOI] [PubMed] [Google Scholar]
- 34.Köhrmann, M., M. Luo, C. Kaether, L. DesGroseillers, C. G. Dotti, and M. A. Kiebler. 1999. Microtubule-dependent recruitment of Staufen-green fluorescent protein into large RNA-containing granules and subsequent dendritic transport in living hippocampal neurons. Mol. Biol. Cell 10:2945-2953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.König, R., S. Stertz, Y. Zhou, A. Inoue, H. H. Hoffmann, S. Bhattacharyya, J. G. Alamares, D. M. Tscherne, M. B. Ortigoza, Y. Liang, Q. Gao, S. E. Andrews, S. Bandyopadhyay, P. De Jesus, B. P. Tu, L. Pache, C. Shih, A. Orth, G. Bonamy, L. Miraglia, T. Ideker, A. Garcia-Sastre, J. A. Young, P. Palese, M. L. Shaw, and S. K. Chanda. 2010. Human host factors required for influenza virus replication. Nature 463:813-817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lu, Y., M. Wambach, M. G. Katze, and R. M. Krug. 1995. Binding of the influenza virus NS1 protein to double-stranded RNA inhibits the activation of the protein kinase that phosphorylates the elF-2 translation initiation factor. Virology 214:222-228. [DOI] [PubMed] [Google Scholar]
- 37.Marión, R. M., T. Aragón, A. Beloso, A. Nieto, and J. Ortín. 1997. The N-terminal half of the influenza virus NS1 protein is sufficient for nuclear retention of mRNA and enhancement of viral mRNA translation. Nucleic Acids Res. 25:4271-4277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Marión, R. M., P. Fortes, A. Beloso, C. Dotti, and J. Ortín. 1999. A human sequence homologue of staufen is an RNA-binding protein that localizes to the polysomes of the rough endoplasmic reticulum. Mol. Cell. Biol. 19:2212-2219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Marión, R. M., T. Zürcher, S. de la Luna, and J. Ortín. 1997. Influenza virus NS1 protein interacts with viral transcription-replication complexes in vivo. J. Gen. Virol. 78:2447-2451. [DOI] [PubMed] [Google Scholar]
- 40.Mibayashi, M., L. Martinez-Sobrido, Y. M. Loo, W. B. Cardenas, M. Gale, Jr., and A. Garcia-Sastre. 2007. Inhibition of retinoic acid-inducible gene I-mediated induction of beta interferon by the NS1 protein of influenza A virus. J. Virol. 81:514-524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nemeroff, M. E., S. M. Barabino, Y. Li, W. Keller, and R. M. Krug. 1998. Influenza virus NS1 protein interacts with the cellular 30 kDa subunit of CPSF and inhibits 3′ end formation of cellular pre-mRNAs. Mol. Cell 1:991-1000. [DOI] [PubMed] [Google Scholar]
- 42.Ochoa, M., J. Bárcena, S. de la Luna, J. A. Melero, A. R. Douglas, A. Nieto, J. Ortín, J. J. Skehel, and A. Portela. 1995. Epitope mapping of cross-reactive monoclonal antibodies specific for the influenza A virus PA and PB2 polypeptides. Virus Res. 37:305-315. [DOI] [PubMed] [Google Scholar]
- 43.Palese, P., and M. Shaw. 2006. Orthomyxoviridae: the viruses and their replication, p. 1647-1689. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 5th ed., vol. 1. Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 44.Ramos, A., P. Bayer, and G. Varani. 1999. Determination of the structure of the RNA complex of a double-stranded RNA-binding domain from Drosophila Staufen protein. Biopolymers 52:181-196. [DOI] [PubMed] [Google Scholar]
- 45.Resa-Infante, P., N. Jorba, N. Zamarreno, Y. Fernandez, S. Juarez, and J. Ortin. 2008. The host-dependent interaction of alpha-importins with influenza PB2 polymerase subunit is required for virus RNA replication. PLoS One 3:e3904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Salvatore, M., C. F. Basler, J. P. Parisien, C. M. Horvarth, S. Bourmakina, H. Zheng, T. Muster, P. Palese, and A. García-Sastre. 2002. Effects of influenza A virus NS1 protein on protein expression: the NS1 protein enhances translation and is not required for shutoff of host protein synthesis. J. Virol. 76:1206-1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Satterly, N., P. L. Tsai, J. van Deursen, D. R. Nussenzveig, Y. Wang, P. A. Faria, A. Levay, D. E. Levy, and B. M. Fontoura. 2007. Influenza virus targets the mRNA export machinery and the nuclear pore complex. Proc. Natl. Acad. Sci. U. S. A. 104:1853-1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shapira, S. D., I. Gat-Viks, B. O. Shum, A. Dricot, M. M. de Grace, L. Wu, P. B. Gupta, T. Hao, S. J. Silver, D. E. Root, D. E. Hill, A. Regev, and N. Hacohen. 2009. A physical and regulatory map of host-influenza interactions reveals pathways in H1N1 infection. Cell 139:1255-1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shaw, M. L., K. L. Stone, C. M. Colangelo, E. E. Gulcicek, and P. Palese. 2008. Cellular proteins in influenza virus particles. PLoS Pathog. 4:e1000085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.St. Johnston, D., D. Beuchle, and C. Nusslein-Volhard. 1991. staufen, a gene required to localize maternal RNAs in the Drosophila egg. Cell 66:51-63. [DOI] [PubMed] [Google Scholar]
- 51.St. Johnston, D., and C. Nusslein-Volhard. 1992. The origin of pattern and polarity in the Drosophila embryo. Cell 68:201-219. [DOI] [PubMed] [Google Scholar]
- 52.Thomas, M. G., L. J. Martinez Tosar, M. Loschi, J. M. Pasquini, J. Correale, S. Kindler, and G. L. Boccaccio. 2005. Staufen recruitment into stress granules does not affect early mRNA transport in oligodendrocytes. Mol. Biol. Cell 16:405-420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Thomas, M. G., L. J. Tosar, M. A. Desbats, C. C. Leishman, and G. L. Boccaccio. 2009. Mammalian Staufen 1 is recruited to stress granules and impairs their assembly. J. Cell Sci. 122:563-573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Villacé, P., R. M. Marión, and J. Ortín. 2004. The composition of Staufen-containing RNA granules from human cells indicate a role in the regulated transport and translation of messenger RNAs. Nucleic Acids Res. 32:2411-2420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wickham, L., T. Duchaine, M. Luo, I. R. Nabi, and L. DesGroseillers. 1999. Mammalian Staufen is a double-stranded RNA- and tubulin-binding protein which localizes to the rough endoplasmic reticulum. Mol. Cell. Biol. 19:2220-2227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wigler, M., A. Pellicer, S. Silverstein, R. Axel, G. Urlaub, and L. Chasin. 1979. DNA-mediated transfer of the adenine phosphoribosyltransferase locus into mammalian cells. Proc. Natl. Acad. Sci. U. S. A. 76:1373-1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wise, H. M., A. Foeglein, J. Sun, R. M. Dalton, S. Patel, W. Howard, E. C. Anderson, W. S. Barclay, and P. Digard. 2009. A complicated message: identification of a novel PB1-related protein translated from influenza A virus segment 2 mRNA. J. Virol. 83:8021-8031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wolff, T., R. E. O'Neill, and P. Palese. 1998. NS1-binding protein (NS1-BP): a novel human protein that interacts with the influenza A virus nonstructural NS1 protein is relocalized in the nuclei of infected cells. J. Virol. 72:7170-7180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zürcher, T., R. M. Marion, and J. Ortin. 2000. Protein synthesis shut-off induced by influenza virus infection is independent of PKR activity. J. Virol. 74:8781-8784. [DOI] [PMC free article] [PubMed] [Google Scholar]