Abstract
Porcine parvovirus (PPV) is a major cause of reproductive failure in swine. The mechanisms implicated in the first steps of infection that lead to the delivery of the PPV genome to the nucleus are poorly understood. In the present work, a panel of chemical inhibitors was used to dissect the cellular mechanisms involved in establishing a PPV infection. The results demonstrated that following binding to sialic acids on cell surface glycoproteins, the virus used both clathrin-mediated endocytosis and macropinocytosis pathways to gain access into cells. Virus obtained from infected cells was present either as isolated particles or as aggregates, and these two forms could be separated by low-speed centrifugation. Isolated and purified particles strongly preferred entry by clathrin-mediated endocytosis, whereas aggregates clearly favored macropinocytosis. Subsequent endosomal acidification and traffic to the late endosomes were also shown to be essential for infection. The microtubule network was found to be important during the first 10 h of infection, whereas an intact actin network was required for almost the whole viral cycle. Proteasome processing was found to be essential, and capsid proteins were ubiquitinated relatively early during infection. Taken together, these results provided new insights into the first steps of PPV infection, including the use of alternative entry pathways, unique among members of this viral family.
Porcine parvovirus (PPV) is a major causative agent of reproductive failure in swine, a syndrome that includes infertility, early embryonic death, mummified fetuses, and stillbirth (54). PPV belongs to the Parvovirus genus in the Parvovirinae subfamily of the Parvoviridae family (55). This family is characterized by small nonenveloped, icosahedral viruses with a diameter of about 26 nm. The genome of these viruses is a linear, negative single-stranded DNA of about 5 kb featuring distinct hairpin termini (3, 4). Transcript mapping revealed promoters for both the nonstructural and structural protein gene cassettes, and intricate splicing mechanisms generate several proteins from each promoter (4). The 3-dimensional (3D) structure of this virus has been determined by X-ray crystallography (49). The compact structure of the capsid confers great stability under different conditions, including wide ranges of pH and high temperatures (11). Infectious particles contain a total of 60 VP1/VP2 proteins arranged in a T=1 capsid (49). The VP1 protein consists of the VP2 sequence with an N-terminal extension that is normally folded within the particle (49). During entry, about 22 to 25 amino acids of the N termini of the majority of the VP2 proteins are cleaved off, forming VP3 (11) and allowing the N terminus of VP1 to be externalized during passage in the endosomes (8). The unique N-terminal part of the VP1 protein contains a viral phospholipase A2 (PLA2) motif. This protein is not crucial for the assembly of progeny virions but is essential for the infectivity of the virions (57). The enzyme's activity provides the virus with the means to breach the endosomal barrier (16, 68).
Parvoviruses deploy a plethora of strategies to deliver the genome to their site of replication, the nucleus (10, 11, 61). The sturdy, extracellular viral particles undergo multistep conformational changes that are locally and temporally regulated by specific intracellular signals after interaction of the capsid with cell surface receptor (11, 64). Particle-to-infectivity ratios are at least 250:1 (68). Therefore, productive and nonproductive pathways are difficult to distinguish, making it challenging to understand the specific trafficking of parvoviruses. Nevertheless, several discrete steps have been recognized (27, 64): (i) initial interaction with cell surface receptors (17, 19-23, 36), (ii) trafficking through the endosomal pathway (32, 41, 52, 60, 68), (iii) escape from the endosomes through the newly exposed viral PLA2 (16, 39, 41, 52), and (iv) cytoskeleton-driven transport to the nucleus (38, 50, 60). Although most parvoviruses use equivalent routes for gaining access to the cell, there are considerable differences among species. The mechanisms involved in these early steps are poorly understood for PPV.
Some viruses use complicated multistep attachment and binding to specific receptors, while others bind more common structures, such as sialic acids (9, 58). These structures are located at the ends of glycans; they are fairly accessible for protein binding and for virus docking; and their density may increase avidity (2). Several parvoviruses bind specifically to the transferrin receptor, including feline parvovirus (FPV) (40) and canine parvovirus (CPV) (41). Minute virus of mice (MVM) and bovine parvovirus (BPV) bind the cells via sialic acids (24, 31), whereas the human parvovirus B19 binds to the blood group P antigen and integrin α5β1 on erythroid progenitor cells (7, 63). In the case of PPV, the specific receptor remains unknown, but the transferrin receptor is not essential, since the virus is able to enter quail cells lacking this receptor (unpublished data).
Binding to specific receptors can trigger entry of the virus via the ubiquitous and constitutive clathrin-coated pit endocytosis mechanism (45). This well-studied pathway requires specific receptor attachment to promote cell membrane invagination and assembly of the clathrin cage (42). At the early-endosome stage, a sorting step determines if the vesicle is recycled back to the cell membrane or proceeds further in the endosomal pathway toward the late endosomes and lysosomes (5). Another well-known endocytosis mechanism takes place in cholesterol-rich lipid rafts (29). Caveolar endocytosis is not constitutive and needs to be triggered. This entry mode has been shown to be responsible for simian virus 40 (SV40) infection (37), and among the parvoviruses, only adeno-associated virus 5 (AAV5) is known to use it (1). A third mechanism for virus entry into a cell is macropinocytosis (53). Although this actin-driven endocytosis displays a low rate in several cell types, it can be upregulated after the interaction of a virus with the cell (15). In contrast to the two endocytosis pathways described above, actin-driven endocytosis does not require specific receptor binding (25). After their formation, the macropinosome vesicles are acidified as in the endosomal pathway. Other, less common “nonclathrin-noncaveolar” endocytosis mechanisms are now emerging (33, 35, 48).
Most of the entry pathways described above lead to endosomal or endosome-like pathways, characterized by acidification and exposure to several proteases meant to destroy the vesicle contents. However, several viruses take advantage of these changes in the environment to trigger conformational changes in their capsids or fusion with the endosome's membrane (50, 51). For parvoviruses, externalization of the unique part of the VP1 capsid protein is essential to infection (16, 39, 41, 52), since it exposes the catalytically active viral PLA2 domain, which is essential for establishing a productive infection (68). The transport of endosomal vesicles is mediated by microtubules (MTs) and proceeds toward the microtubule organization center (MTOC), located in the perinuclear region (18, 62). Accordingly, trafficking to the late endosomes/lysosomes is beneficial not only for conformational changes but also for transport to the nucleus. Once it has escaped to the cytoplasm, the capsid itself could interact with the MT motors or with the actin network components (44, 46, 59).
Important cellular processes involved in different virus infections may include the proteasome (12, 47, 56, 67). This component degrades targeted proteins into small peptides, which are then processed by cellular proteases, yielding amino acids (28). In the case of parvoviruses, the role of the proteasome can be beneficial or detrimental to the infection cycle, depending on the virus. AAV virions are degraded by the proteasome, aborting the infection (13). On the other hand, proteasome processing is required for productive MVM infection, although the mechanism remains unclear (46, 47).
In this study, a panel of chemical inhibitors was used to investigate the cellular entry and transport of PPV. To minimize the side effects of these pharmacological inhibitors, two drugs, known to be the most selective, were used to target cellular components at concentrations that least compromised their specificity while remaining effective. The infection could be inhibited by removal of sialic acid moieties from cell surface glycoproteins. Virus entry could also be partially inhibited by both clathrin and macropinocytosis inhibitors, while isolated/purified or aggregated PPV particles were shown to preferentially use alternative entry modes. After the virus gained entry into the cells, inhibition of endosomal acidification, traffic to the late endosomes, or destruction of either microtubules or actin networks greatly reduced the infection. Finally, capsid proteins were ubiquitinated early during infection, as shown by coimmunoprecipitation, and inhibition of the proteolytic activities of the proteasome almost completely abolished the infection.
MATERIALS AND METHODS
Chemicals and antibodies.
Neuraminidase (Neura), amiloride (Ami), chlorpromazine (Chl), nystatin (Ny), methyl-β-cyclodextrin (MBC), and MG-132 were purchased from Sigma. Cytochalasin D (CyD), nocodazole (Noc), paclitaxel (Pac), latrunculin A (LatA), and lactacystin (Lac) were purchased from Calbiochem. Bafilomycin A1 (Baf) was purchased from LC Labs and brefeldin A (BFA) from BioLegend. Protease inhibitors (Complete; EDTA-free tablets) and the colorimetric substrate nitroblue tetrazolium (NBT)-5-bromo-4-chloro-3-indolylphosphate (BCIP) were purchased from Roche Applied Science. The 3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2H-tetrazolium bromide (MTT) viability assay kit was purchased from Sigma. Anti-PPV antibodies included mouse monoclonal antibody 3C9 (ATCC CRL-1745), specific to the PPV capsid, and a polyclonal rabbit anti-VP2 (α-VP2) antibody obtained via rabbit immunization (8). The mouse monoclonal anti-ubiquitin (α-Ub) antibody (P4D1) was purchased from Santa Cruz. Secondary antibodies included alkaline phosphatase (AP)-conjugated goat anti-mouse and anti-rabbit antibodies (Bio-Rad) and Alexa Fluor 488-conjugated goat anti-mouse and anti-rabbit antibodies (Invitrogen).
Cells and viruses.
Porcine testis (PT) fibroblasts, derived from ST cells (ATCC CRL-1746) (4), were grown at 37°C in Dulbecco's modified Eagle's medium (GIBCO-Invitrogen) containing d-glucose and l-glutamine and supplemented with 7% heat-inactivated fetal bovine serum (Wisent) and antibiotics (penicillin/streptomycin; Invitrogen). The NADL-2 vaccine strain of PPV was used for infection (ATCC VR-742), and viral stocks were obtained by propagation in cell culture. PPV was collected in the supernatant after cell lysis and was used directly in the experiments unless otherwise indicated. Viral stocks were titrated by immunofluorescence (IF) in 96-well plates at 20 h postinfection (p.i.) with the capsid-specific mouse monoclonal antibody 3C9, together with an Alexa Fluor 488-conjugated anti-mouse antibody as a secondary antibody. Fluorescent nuclei were scored, and virus titers were expressed as fluorescent focus-forming units (FFU) per milliliter.
From the crude preparation, aggregates and isolated particles of PPV were separated by centrifugation at 15,000 × g for 10 min. Isolated particles were obtained directly in the supernatant, and aggregates were found in the pellet, reconstituted with the same volume of cell culture medium. Electron micrographs showed that there were no or very few isolated particles in the aggregate preparations and very few aggregates in the isolated stock (data not shown).
PPV purification.
Purified virus was obtained from PPV-infected PT cell supernatants. Crude supernatants were first cleared by centrifugation at 10,000 × g for 45 min at 4°C. PPV was precipitated from the supernatant by using 7.5% polyethylene glycol 8000 (PEG 8000)-1.5 M NaCl (final concentrations) for 16 h at 4°C and was collected by centrifugation at 15,000 × g for 45 min at 4°C. The pellet was dissolved in 5 ml of 10 mM Tris-HCl (pH 7.5) and was dialyzed overnight against the same buffer. Virus was also extracted from the cell debris fraction from the first centrifugation. The pellet was resuspended in 10 ml of 10 mM Tris-HCl (pH 7.5) and was treated with 2 ml of 1× trypsin and 50 U of DNase I for 4 h at 37°C. Then PPV was extracted using 1 volume of chloroform and centrifugation at 3,500 × g for 10 min. These two PPV preparations were then purified using ultracentrifugation over sucrose cushions (2 ml of 50% sucrose plus 2 ml of 20% sucrose) at 200,000 × g for 2 h. Fractions were collected and subjected to 10% sodium dodecyl sulfide-polyacrylamide gel electrophoresis (SDS-PAGE) and Coomassie staining to detect PPV proteins. Positive fractions were pooled and dialyzed against 100 mM sodium phosphate buffer (pH 7.0). A second round of purification by ultracentrifugation was performed as described above. The final purified PPV was titrated by IF as described in the previous section.
Drug treatments and PPV infection.
For IF experiments, cells were plated onto glass coverslips at 5 × 104 per well in 24-well plates. For quantitative PCR (qPCR) experiments, cells were plated directly onto the plates, as described for IF experiments. For inhibitors, dose-response curves were obtained by treating cells with increasing concentrations of each inhibitor, and toxic doses were determined by the MTT assay according to the manufacturer's instructions (Sigma). The optimal dose of each inhibitor was determined to be the concentration displaying the best inhibition of infection without detectable toxic effects on the cells. One day after plating, cells were treated with the optimal dose of inhibitor for the times indicated below and were infected with NADL-2 at a multiplicity of infection (MOI) of 2 for IF and 0.1 for qPCR. Cells were either fixed (IF) or harvested (qPCR) 20 h p.i. As a control, the impact of the inhibitors on cell growth was also evaluated by qPCR with specific c-myc primers as described below, after treatment of the cells with optimal doses of inhibitors for 20 h.
For neuraminidase treatments, cells were treated for 1 h with increasing concentrations of neuraminidase and were washed three times with 1× phosphate-buffered saline (PBS, comprising 2.7 mM KCl, 1.5 mM KH2PO4, 144.3 mM NaCl, and 8.1 mM Na2HPO4) to remove cleaved sialic acids. Cells were then infected, in the presence of neuraminidase, for 2 h and were washed again. Infection was allowed to proceed for a total of 20 h in the presence of a low dose of neuraminidase (2 mU/ml).
For entry experiments, cells were treated for 1 h with either inhibitor, infected with NADL-2 in the presence of the inhibitors, and washed 2 h p.i. to remove all viruses in the supernatant. Cells were then treated with neuraminidase (2 mU/ml) to remove all bound virus that failed to enter the cells. Binding experiments were also performed with entry inhibitors. Cells were treated for 1 h at 37°C and were subsequently chilled to 4°C in order to completely inhibit PPV entry in the cells. PPV was then added, and cells were incubated at 4°C for 2 h. Cells were gently washed with cold 1× PBS to remove unbound virus and were transferred to 37°C for 2 h to allow entry. Cells were washed again and were incubated for a total of 20 h at 37°C.
For cytoplasmic trafficking experiments and proteasome processing, cells were treated 1 h prior to infection, and the inhibitors remained present for the duration of the infection. However, since the inhibitor for the actin network (latrunculin A) can block cytoplasmic trafficking and macropinocytosis, it was added 2 h p.i. and was kept for the rest of the infection. Pulse experiments were also performed by adding the inhibitors (Noc, LatA, Baf, BFA, and MG-132) at different times throughout the infection.
Immunofluorescence.
At designated times, or at 20 h p.i., cells were fixed with 3% formaldehyde in IF buffer (1× PBS, 0.02% sodium azide, and 0.1% bovine serum albumin [BSA]) for 30 min and were washed three times with 1× PBS. Cells were permeabilized with 3% Triton X-100 in IF buffer for 30 min and were washed three times with 1× PBS. PPV was detected with the mouse monoclonal anti-capsid antibody 3C9 as a primary antibody for 1 h (diluted 1:50 in IF buffer). Cells were washed with 1× PBS and were incubated with a goat anti-mouse secondary antibody, conjugated with Alexa Fluor 488, for 1 h (1:2,000 in IF buffer). Finally, DNA was stained with Hoechst 33258 (2 μg/ml) for 30 min, and coverslips were fixed on slides that were kept at 4°C in the dark until they were read. Percentages of infected cells were obtained by scoring the virus-positive nuclei as a proportion of total nuclei. For each experiment, the infection level of untreated cells was arbitrarily set at 100%. For each inhibitor, at least 300 cells were scored for each coverslip (samples in triplicate) in at least three independent experiments.
qPCR.
At 20 h p.i., cells were washed with 1× PBS. Cells were scraped from the plate in 150 μl of STE buffer (1 mM EDTA, 150 mM NaCl, 20 mM Tris-HCl [pH 7.5]) and were harvested in tubes. Then 500 μl of heated lysis buffer was added (0.75% SDS, 1.25 M NaCl, 20 mM Tris [pH 7.5], 10 mM EDTA, 100 μg/ml proteinase K, 100 μg/ml RNase). Tubes were incubated at 37°C for 4 h and were chilled at −20°C for at least 30 min. The supernatants were collected after a 20-min centrifugation at 20,000 × g and 4°C. DNA was then extracted with 500 μl Miniprep Express matrix (MP Biomedicals) according to the manufacturer's instructions. DNA was eluted with 100 μl of PCR water. Each sample was diluted 1:10 for PCR purposes, and qPCR was performed as described previously (66) to determine the number of PPV genome copies. Primers specific to the VP2 region (forward primer, 5′-GGG GGA GGG CTT GGT TAG AAT CAC-3′; reverse primer, 5′-ACC ACA CTC CCC ATG CGT TAG C-3′; based on GenBank accession no. NC_001718.1) were used. The cellular DNA content was used to normalize the results with qPCR targeting the c-myc gene (forward primer, 5′-CTC CCT GAG ACT CTG CCA TC-3′; reverse primer, 5′-GCT GCC TCT TTT CCA CAG AA-3′; based on GenBank accession no. X97040.1). Cycling conditions were as follows: 95°C for 10 min, followed by 40 cycles of 95°C for 15 s and 60°C for 60 s. Fluorescence was acquired after each cycle. Amplifications were carried out in a RotorGene 3000 system (Corbett) with 2× Sybr green master mix (ABSciences). Standard curves were performed using a plasmid containing PPV or c-myc DNA. A myc DNA fragment was cloned into the pSmartHC AmpR vector (GenBank accession no. AF399742; Lucigen) by the addition of HindIII and BamHI restriction sites (forward primer, 5′-CGTAAGCTTTCGGACTCTCTGCTCTCCTC-3′; reverse primer, 5′-CTGTCTAGAGCTGCCTCTTTTCCACAGAA-3′). PPV curves were made using the infectious clone in the pSmartHC AmpR vector. Melting curve analysis was performed from 60°C to 95°C, with a temperature increase of 1°C at each step of 30 s, for a specific product amplification control.
Coimmunoprecipitation and Western blotting.
Cells were plated in 6-well plates (3 × 105 cells/well) 1 day before the experiment. Cells either were mock treated or were treated with MG-132 at 5 μM. Cells were lysed at different times postinfection using lysis buffer (50 mM Tris-HCl [pH 7.5], 150 mM NaCl, 1% NP-40, 1× Complete protease inhibitor cocktail [Roche Applied Science]) and were incubated for 2 h on ice. Lysates were precleared with 20 μl of protein G-agarose beads for 30 min on ice with gentle shaking and were centrifuged for 30 s at 10,000 × g (quick spin). Supernatants were incubated with a specific antibody, either an anti-capsid (3C9) or an α-Ub (P4D1; Santa Cruz) antibody, for 1 h on ice, with gentle shaking. A 50-μl volume of protein G-agarose beads was then added, and the mixture was further incubated for at least 2 h. Beads were pelleted by quick-spin centrifugation and were then washed three times with lysis buffer, twice with washing buffer 1 (50 mM Tris-HCl [pH 7.5], 500 mM NaCl, 0.1% NP-40), and once with washing buffer 2 (50 mM Tris-HCl [pH 7.5], 0.1% NP-40). For each wash, 500 μl of buffer was added and incubated 5 min on ice with agitation before a quick spin. Proteins were eluted with 2× SDS-PAGE sample buffer (125 mM Tris-HCl [pH 6.8], 4% SDS, 0.2% bromophenol blue, 5% β-mercaptoethanol) in boiling water for 5 min. Samples were separated by 10% SDS-PAGE and were transferred to nitrocellulose membranes. Primary antibodies (rabbit α-VP2 at 1:1,000 or mouse α-Ub at 1:200) were diluted in 5% milk in 1× TBS buffer (10 mM Tris-Cl [pH 8.0], 150 mM NaCl) and were incubated with the membrane for 1 h with agitation. Anti-mouse or anti-rabbit antibodies conjugated with AP were used as secondary antibodies (diluted 1:1,000 in 5% milk in 1× TBS buffer). Detection was performed directly on the membrane by using the colorimetric AP substrate NBT-BCIP, according to the manufacturer's recommendations (Roche).
Confocal microscopy.
Cells were plated on glass coverslips (5 × 10 4 cells/well in a 24-well plate) 1 day prior to the experiment. Cells were then treated or mock treated with 5 μm MG132. Cells were infected with NADL-2 and fixed at the indicated times. Indirect IF was performed as described above. Coverslips were incubated with the mouse anti-capsid antibody 3C9. Goat anti-mouse antibodies coupled with Alexa Fluor 488 were used as secondary antibodies. Images were collected on a Bio-Rad Radiance 2000 confocal system, with an Ar-Kr laser and a 60× oil objective (numerical aperture [N.A.], 1.4). Fluorescence was detected at 515 nm.
Statistical analysis.
Data were analyzed by the unpaired two-tailed t test using 99% confidence intervals. Analyses were performed with GraphPad Prism software, version 5.
RESULTS
Overall PPV replication kinetics.
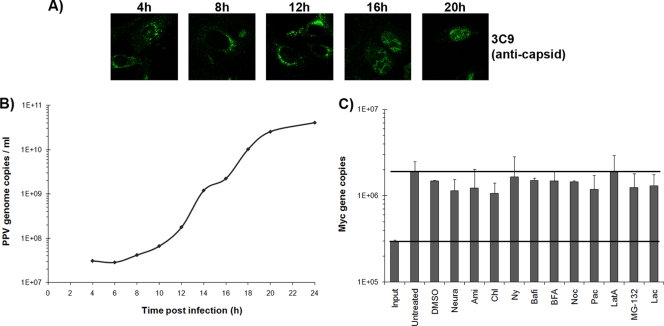
The general replication kinetics of the NADL-2 strain of PPV on PT cells is shown in Fig. 1. The first steps of the high-MOI (50 FFU/cell) infection were followed by the detection of viral capsids by indirect immunofluorescence (with monoclonal antibody 3C9). After cell entry, capsids accumulated on one side of the nucleus by 4 to 8 h postinfection (p.i). Newly synthesized virions were found in the nucleus by 16 to 20 h p.i., before cell lysis (Fig. 1A). DNA replication, monitored by quantitative PCR (qPCR) on cell lysates after infection at a low MOI (0.1 FFU/cell), started at 8 to 12 h p.i. (Fig. 1B).
FIG. 1.
Kinetics of PPV infection. (A) Detection of viral capsids at different times p.i. in PT cells by immunofluorescence using the capsid-specific antibody 3C9 and an Alexa Fluor 488-coupled secondary antibody. (B) Amplification of the viral genome. qPCR specific to PPV was performed on cell lysates collected at different times p.i. (C) Cell DNA content in the presence of inhibitors, as an indication of side effects of the inhibitors on cell growth, determined by qPCR with c-myc gene-specific primers after 20 h of incubation with the optimal dose of each inhibitor.
Inhibitor optimization.
All inhibitors were tested for toxic effects on the cells used in this study (PT cells) by the MTT assay (Sigma). The toxic doses, which are the first concentrations that have an effect on cell viability, are shown in Table 1. Then a dose-response curve was obtained in order to determine the concentration that had the most effect on viral infection (data not shown). The optimal dose of each inhibitor could be defined as the concentration displaying the greatest inhibition of viral infection without being toxic to the cells (Table 1). The effects of the inhibitors on cell growth were also evaluated by incubating cells with the optimal dose of each inhibitor for 20 h. qPCR using specific c-myc gene primers was performed to evaluate cell density. As demonstrated in Fig. 1C, the inhibitors used in this study failed to have any significant short-term effect on cell growth. This result correlated with that of the MTT assay.
TABLE 1.
Chemical inhibitors
| Cellular component | Inhibitor | Effect on cells | Optimal concna | Toxicityb |
|---|---|---|---|---|
| Sialic acids | Neuraminidase | Catalyzes the hydrolysis of N-acetylneuraminic acids on cell surface glycoproteins | 5 mU/ml | 20 mU/ml |
| Clathrin | Chlorpromazine | Inhibits pit formation by clathrin relocation at the endosomes | 10 μM | 100 μM |
| K+ depletion | Aggregates clathrin in empty small cages | Buffer | ||
| Caveolae | Nystatin | Inhibits caveolin pit formation; sequesters cholesterol | 100 μM | 1 mM |
| Methyl-β-cyclodextrin | Inhibits cholesterol translocation toward lipid rafts | 1 mM | 10 mM | |
| Macropinocytosis | Amiloride | Inhibits Na+/H+-ATPase exchangers, preventing membrane extension formation | 100 μM | >250 μM |
| Cytochalasin D | Inhibits actin polymerization for membrane extension | 50 μM | 75 μM | |
| Microtubules | Nocodazole | Inhibits tubulin subunit polymerization; inhibits endosome trafficking | 10 μM | 100 μM |
| Paclitaxel | Inhibits microtubule dynamics, preventing depolymerization | 0.5 μM | 1 μM | |
| Actin | Cytochalasin D | Inhibits actin polymerization (binds to filaments) | 50 μM | 75 μM |
| Latrunculin A | Inhibits actin polymerization (binds to monomers) | 0.1 μM | 0.5 μM | |
| Endosomal pathway | Bafilomycin | Inhibits endosomal acidification through vacuolar H+/ATPase pump block | 100 nM | 500 nM |
| Brefeldin A | Inhibits translocation to late endosomes | 500 nM | 1 μM | |
| Proteasome | MG-132 | Inhibits the 26S subunits of the proteasome, preventing proteolytic activity | 1 μM | 10 μM |
| Lactacystin | Inhibits the β subunits of the proteasome, preventing proteolytic activity | 5 μM | 10 μM |
Concentrations shown in Results displaying optimal inhibition of PPV replication without significant side effects (by the MTT assay).
Determined by the MTT assay.
Binding of PPV on cells.
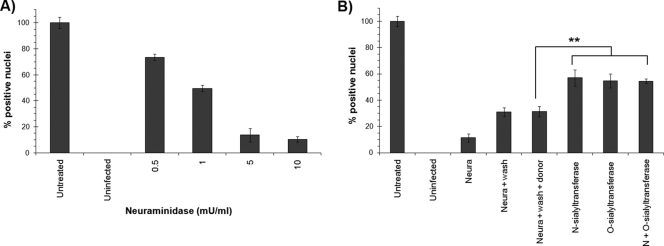
The efficiency of the infection was evaluated at 20 h p.i. by immunofluorescence. The number of capsid-positive cells was compared to the total number of cells on each slide. The effect of each inhibitor was expressed as the efficiency of infection relative to that for untreated cells. The first step of PPV infection was determined to be the binding of the capsid to sialic acid on cell surface glycoproteins. As shown in Fig. 2A, neuraminidase treatment of the cells, which cleaves sialic acids at the cell surface, greatly impeded viral infection in a dose-dependent manner. Moreover, the infection could be partially reconstituted after the rebuilding of sialic acid moieties using either α-(2,3)-O-sialyltransferase or α-(2,3)-N-sialyltransferase (10 mU). Therefore, both O- and N-linked sialic acids seem to be used by the virus to enter the cells (Fig. 2B). Cells were capable of rebuilding some receptors by themselves, since removal of the neuraminidase, without the addition of any enzyme, doubled the number of infected cells. The presence of the donor only (cytidine 5′-monophospho-N-acetylneuraminic acid) did not help receptor reconstruction or increase the level of infection. Finally, the addition of each sialyltransferase alone or in combination did not reconstitute the infection to the 100% level, even when more enzyme was added (up to 25 mU) (data not shown).
FIG. 2.
PPV binding on the cell surface. (A) PT cells were treated with increasing amounts of neuraminidase in order to remove sialic acid moieties on cell surface glycoproteins. After a wash, PPV was added to the cells for 2 h. Unbound virus was removed by a wash at 2 h p.i., and infection was continued for an additional 18 h. The percentage of infected cells was compared to that of untreated cells (arbitrarily set at 100%) by IF with the capsid-specific antibody 3C9 and DNA staining (Hoechst). (B) Efficiency of infection recovery after neuraminidase treatment followed by specific reconstruction of sialic acids, using O- or N-sialyltransferases, on the cell surface proteins prior to infection. Percentages of infected cells were determined by IF (**, P < 0.004 by the t test).
Viral entry into cells.
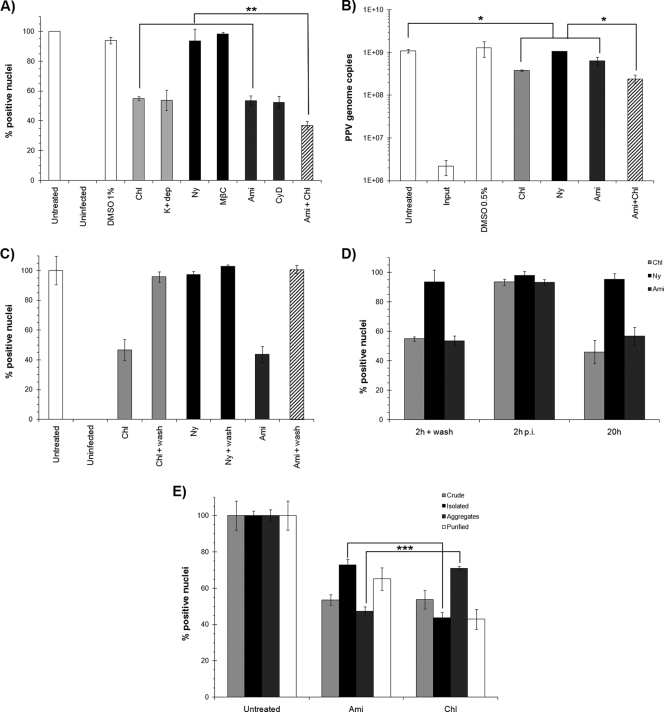
To investigate the pathways implicated in PPV entry, multiple inhibitors were used to target each pathway. A description of these inhibitors, and the concentrations used in this study, is shown in Table 1. After one infection cycle (20 h), the percentages of infected cells in the presence of the inhibitors were compared to those for untreated cells. The results shown in Fig. 3A (for optimal doses only) demonstrated that both clathrin-mediated endocytosis and macropinocytosis were important for PPV entry. Inhibition of caveolae had no effect on viral entry. These results were confirmed by qPCR of cell lysates at 20 h p.i. using PPV-specific primers (Fig. 3B). However, complete inhibition could not be achieved, even when clathrin and macropinocytosis inhibitors were combined, suggesting that one or more other entry pathways may be important for PPV. In both cases (percentage of infection and genome replication), the inhibition level was about 50%. As a control, the effects of the inhibitors on viral binding to the cell surface were also evaluated. The virus was incubated with inhibitor-treated cells at 4°C. The cells were then washed or not before incubation for 20 h at 37°C. The results in Fig. 3C show that inhibitors of clathrin, caveolae, or macropinocytosis had no effect on the binding of PPV to the cell surface. Furthermore, the inhibitors acted only during entry steps, as demonstrated in pulse experiments (Fig. 3D), and the addition of each inhibitor at 2 h p.i. or later had no detectable effect on PPV infection. Finally, the same results were observed whether the inhibitors were present throughout the infection or only during entry, suggesting that there were no significant secondary effects on the cells.
FIG. 3.
Entry pathways. (A) Inhibition of the major cellular entry pathways during PPV infection. Optimal concentrations of inhibitors of clathrin endocytosis (chlorpromazine and K+ depletion) (light shaded bars), caveolae (nystatin and methyl-β-cyclodextrin) (filled bars), and macropinocytosis (amiloride and cytochalasin D) (dark shaded bars) were used. The percentage of cells infected was determined at 20 h by indirect IF experiments as described for Fig. 2. The combination of clathrin and macropinocytosis inhibitors (striped bars) failed to completely abolish infection (**, P < 0.003 by the t test). (B) Inhibition of genome replication measured by qPCR with specific PPV primers after treatment with inhibitors as for panel A (*, P < 0.03 by the t test). (C) Binding assay. Inhibitors were present only for the first 2 h with PPV at 4°C and were removed at the time of washing to eliminate unbound virus. The infection was continued in normal cell culture medium for 20 h. (D) Pulse inhibition. Clathrin endocytosis and macropinocytosis inhibitors either were added prior to infection and were present only for the first 2 h of infection, after which cells were washed; were added 2 h p.i.; or were added prior to infection and left during the whole infection (20 h). (E) Inhibition of distinct particle types. Shown are results for crude preparations (infected cell culture supernatants), isolated particles and aggregates (both separated by low-speed centrifugation of the crude preparation), and purified particles. To determine the inhibition of the clathrin and macropinocytosis pathways for each particle type, infections were carried out with the same amount of PPV, as determined by qPCR (***, P < 0.0003 by the t test).
The importance of both specific and nonspecific pathways prompted us to further examine different arrangements of viral particles. When PPV was amplified in cell culture, a suspension containing the virus and cell fragments was obtained after virus-induced cell lysis. These preparations contained isolated viruses and clumps of PPV particles (aggregates), as observed by electron microscopy (data not shown), which could be separated through low-speed centrifugation. Four different viral stocks were used in order to compare the different inhibitors. The first was the crude preparation containing both types of particles. The aggregates were found in the centrifugation pellet, reconstituted with the same volume of cell culture medium, while single particles remained in the centrifugation supernatant. Finally, purified particles were isolated by ultracentrifugation on sucrose cushions. Approximately the same amounts of infectious particles and genome copies were obtained as isolated particles and as aggregate preparations (demonstrated by viral titration and quantitative PCR [data not shown]). The abilities of these different particle types to initiate infection in the presence of entry inhibitors were compared. The results shown in Fig. 3E indicated that single particles were more sensitive to inhibition by chlorpromazine, meaning that they entered preferably via clathrin-mediated endocytosis. Alternatively, infection with aggregates of virus was greatly inhibited by amiloride, indicating that macropinocytosis was more important for those particles. As expected, the level of inhibition of virus in the crude preparation was found to be intermediate between those for the two types of particles, and purified virus acted similarly to single particles. Nevertheless, the infection could not be inhibited by more than 60%, strengthening the hypothesis that an additional, unknown entry pathway would be important for PPV entry.
Cytoplasmic transport toward the nucleus.
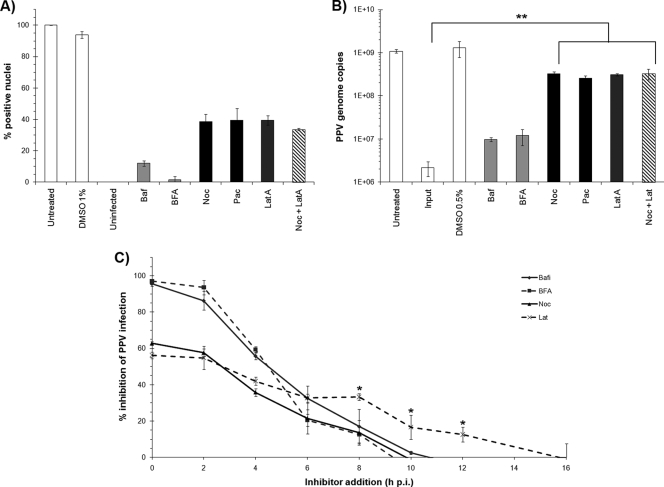
After endocytosis, the viral particles likely proceed in the endosomal pathway. Chemical inhibitors were used to evaluate the implication of these cellular components in PPV infection (Table 1). First, dose-response curves were obtained for each inhibitor (data not shown) as described in “Drug treatments and PPV infection” above, and when these curves were combined with MTT assay results, the optimal concentration of each inhibitor was determined. As described above, the relative percentages of infected cells were obtained after treatment with the optimal dose of each inhibitor. The results shown in Fig. 4A demonstrate that endosomal acidification and trafficking to the late endosomes were essential for proper PPV infection (inhibition with Baf and BFA). PPV then likely exits the endosomal pathway via its PLA2 activity to reach the cytoplasm, where the main transport pathways to the nucleus are the microtubules (MTs) and actin networks. Transport by the MTs would be more efficient, since they polymerize from the MTOC, located next to the nucleus. Both the MTs and the actin networks were found to be important, although the inhibitors could not completely abolish the infection (Noc and LatA). The dynamic instability of MTs due to polymerization-hydrolysis was also important, since fixation of the MT structure (Pac) also inhibited the infection. These results were confirmed by qPCR, as shown in Fig. 4B.
FIG. 4.
Cytoplasm trafficking toward the nucleus. (A) Inhibitors of the endosomal pathway and acidification (light shaded bars), microtubules (filled bars), and actin networks (dark shaded bars) were used to evaluate their necessity for PPV infection. Shown are the relative percentages of infection at 20 h p.i. in the presence of optimal concentrations of the inhibitors as measured by indirect IF, as described for Fig. 2. (B) Impacts of the inhibitors on genome replication as measured by qPCR and normalized to cell numbers, as described for Fig. 3 (**, P < 0.003 by the t test). (C) Pulse inhibition. Inhibitors were added at different times p.i. and were left until 20 h p.i. The percentages of infected cells were determined (*, P < 0.03 by the t test for the difference between the Noc and LatA curves).
Pulse inhibition experiments were performed by the addition of inhibitors at different times throughout the infection. This provided an estimation of the period during which the cellular components were important for the infection. As shown in Fig. 4C, the endosomal-pathway inhibitors were effective mostly in the first hours of infection. While the MT inhibitor was also most efficient during the first 10 h, like endosomal-pathway inhibitors, the actin inhibition curve was above the inhibition curve for the MTs, indicating that the actin network remained important later in the infection.
Proteasome.
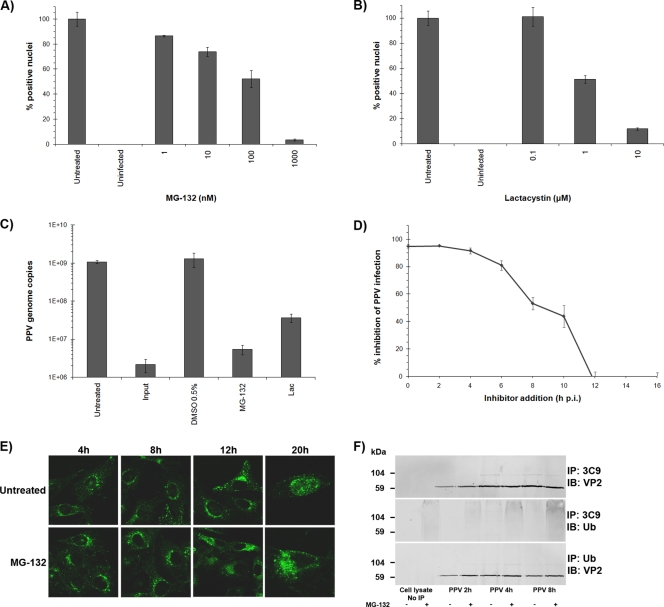
The proteasome machinery is critical for host cell defense and can lead to abortive infections via capsid degradation early in infection (13). On the other hand, partial degradation by the proteasome can facilitate some viral infections (47). Two proteasome inhibitors, MG-132 and lactacystin (Table 1), were used to evaluate the impact of the proteasome on PPV infection. As demonstrated in Fig. 5A and B, inhibition of the proteasome almost completely abolished PPV infection. These results were further confirmed by qPCR (Fig. 5C). Pulse inhibition was also performed as described for cytoplasmic trafficking and demonstrated that the proteasome was important up to 12 h of infection (Fig. 5D).
FIG. 5.
Proteasome involvement. (A and B) Inhibition of the proteolytic activity of the proteasome aborted PPV infection. The effects of the inhibitors were evaluated by indirect IF experiments as described for Fig. 2 and were expressed as the percentage of infected cells at 20 h p.i. relative to that for mock-treated cells (arbitrarily set at 100%). (C) Cells were treated with inhibitors as for panels A and B, and inhibition of genome replication was measured by qPCR for PPV, normalized to cell numbers, as described for Fig. 3. (D) Pulse inhibition. MG-132 was added at different times throughout the infection until cell fixation, and the percentage of infected cells was determined. (E) Capsid localization in the presence of MG-132. Capsids were visualized at different times throughout the infection by the capsid-specific antibody 3C9. In untreated cells, PPV reached perinuclear localization within 4 h p.i., and new virus was observed in the nucleus at 20 h p.i. The virus was also observed near the nucleus after MG-132-mediated inhibition but remained more diffuse in the cytoplasm (8 to 12 h p.i.), and very few positive nuclei were visible at 20 h p.i. (F) Coimmunoprecipitation of ubiquitin with anti-PPV capsid antibodies and of VP2 capsid proteins with anti-ubiquitin antibodies. Capsid proteins were observed in all ubiquitin precipitations. The ubiquitin signal in the capsid immunoprecipitation was more diffuse, as generally observed for ubiquitinated proteins.
The cellular localization of the virus in the presence of MG-132 was estimated by confocal microscopy. After treatment with the inhibitor, the cells were infected with PPV, fixed at different times during the infection, and processed for indirect immunofluorescence with a capsid-specific antibody (Fig. 5E). In the presence of MG-132, PPV was still able to reach the nuclear periphery, but the capsids remained more diffuse than those in untreated cells. At 20 h p.i., very few virus-positive nuclei were observed, supporting the finding that the infection was not productive.
The best-characterized mechanism by which the proteasome targets proteins is ubiquitination (43). After covalent linkage of several ubiquitin subunits to the targeted protein through lysine residues, the protein is unfolded and fragmented into small subunits that are further degraded by proteases to single amino acids. Coimmunoprecipitation of the viral capsid and ubiquitin showed that PPV capsids were ubiquitinated at 4 to 12 h p.i. (Fig. 5F), coinciding with the passage through the cytoplasm. Ubiquitin was present after the immunoprecipitation of the capsids, in increasing amounts up to 12 h p.i. Conversely, PPV protein was present after ubiquitin immunoprecipitation. When cells were treated with MG-132, a greater amount of ubiquitin was observed in capsid coprecipitation, suggesting that inhibition of proteasomal processing resulted in greater accumulation of ubiquitin-linked capsids early in infection.
DISCUSSION
Although porcine parvovirus was identified several decades ago, its interactions with host cells remain poorly understood. The early steps of PPV infection were investigated in the current study, since they are the keys to establishing a productive infection. Our data demonstrated both similarities and substantial differences between PPV and other closely or distantly related parvoviruses.
Direct probing of virus endocytosis is commonly achieved using pharmacological inhibitors, because (i) the effect can be easily evaluated and quantified; (ii) the short time of exposure delays side effects or compensatory mechanisms; and (iii) all cells are equally exposed. However, the potential for poor specificity remains a major concern, in particular if the actin cytoskeleton is affected. None of the common inhibitors of the endocytic pathways possesses absolute specificity for the targeted component. Therefore, two inhibitors with different actions were used for each cell pathway. The inhibitors were used at relatively low concentrations in order to minimize side effects. Nevertheless, the interpretation of the results required careful consideration of the degree of selectivity.
Prior treatment of cells with neuraminidase prevented infection, emphasizing the pivotal role of sialic acid receptors in infection. This first binding would occur regardless of the entry mode, since the infection could be almost completely inhibited by treatment of the cells with neuraminidase. Resialation experiments (6, 24) on sialidase-treated cells with the enzyme α-2,3-O-sialyltransferase or α-2,3-N-sialyltransferase, or with a combination of both enzymes, partially reconstituted infectivity. Thus, the sialylglycoprotein receptors available for virus attachment appear to contain both O- and N-linked carbohydrate moieties. However, to achieve optimal binding, sialation of the receptors may be more complex than what could be rebuilt in vitro (since the combination of the two enzymes could not fully reconstitute viral binding and infection) or may require other specific sialic acid derivatives.
Unexpected results were obtained in the process of deciphering the entry mechanisms of PPV. The entry of the virus into cells could not be completely prevented by any of the chemical inhibitors or by their combination. Inhibition of clathrin or macropinocytosis led to about 50% reduction of the infection. Moreover, even when these inhibitors were used in combination, inhibition was only slightly greater (60%). Accordingly, a third unusual or unknown entry mechanism is likely to be involved for PPV; this will require further investigation. It is also possible that the inhibitor concentrations used failed to completely block the entry pathways. However, dose-response curves and combinations of inhibitors showed that even beyond toxic doses, as many as 40% of cells could still be infected (data not shown). It is unlikely that the remaining levels of infection would be due solely to imperfect inhibition of the cells. It has been reported that chlorpromazine is able to block macropinocytosis in addition to affecting clathrin-coated pits, whereas amiloride and cytochalasin D may also affect endocytosis via clathrin-coated pits (14, 26). However, the contrasting effects of these drugs on isolated and aggregated PPV particles indicated that at the concentrations used, these drugs were sufficiently specific. With less cross-reactivity of these inhibitors, the preference for either coated-pit endocytosis or macropinocytosis by these forms of PPV should be even more significant than the P value of <0.003 that was observed.
This work is the first demonstration that macropinocytosis, a nonspecific entry mode, would be an important entry mechanism for a parvovirus. However, since binding to sialic acid moieties was necessary, prior binding with an unknown cell surface receptor may upregulate macropinocytosis (15). Most parvoviruses are known to use clathrin-mediated endocytosis extensively, if not exclusively. Nevertheless, it was recently shown that AAV5 uses both clathrin and caveolar endocytosis to enter cells (1). Inhibition of caveolae had no effect on PPV infection. Canine parvovirus, which uses the transferrin receptor (TfR) for receptor-mediated endocytosis, still infected cells efficiently when deletions or mutations within the cytoplasmic domain of TfR prevented interaction with adapter protein 2, thus abolishing clathrin-mediated uptake (22); this finding suggests the existence of a supplementary entry pathway. Therefore, the entry of these viruses appears to be more complicated than was previously assumed. Moreover, even among closely related parvoviruses, there are significant differences in the first steps of infection.
The observed preference for different entry mechanisms by isolated and aggregated PPV particles is particularly relevant, since most laboratory studies are based on purified virus (e.g., isolated particles) with cell culture, which may not reflect the natural context of infection in the host. Indeed, it is most likely that some aggregates form when cells are destroyed by the virus, and those aggregates would be in contact with neighboring cells.
As demonstrated in this study, endosomal acidification and trafficking to the late endosomes are both essential for PPV, since treatment of the cells with bafilomycin or brefeldin A, respectively, almost completely abolished the infection (Fig. 4). Transport through the endosomal pathway may trigger conformational changes that enable further steps of PPV infection, as shown for MVM (46). Indeed, the amino-terminal part of the VP1 protein must be externalized from the capsid. This capsid protein contains a phospholipase A2 motif, and its activity is essential for breaching vesicular membranes (16). Finally, since endosomes are transported on MTs, which are polarized toward the nucleus, vesicular traffic of PPV to the late endosomes or lysosomes would provide a means for the virus to reach the perinuclear area.
MTs and actin, the two major cell transport structures, were also shown to be involved in PPV infection. Indeed, inhibition of either structure reduced the infection up to 60%. However, since the inhibition was not complete, even when the two inhibitors were combined (Noc plus LatA), it is possible that destruction of the normal cell structure is toxic and hence that total inhibition cannot be achieved without killing the cells. Most interestingly, the virus seemed to utilize each transport structure at different times postinfection. MTs were important mostly in the first 8 to 10 h of infection, correlating with the time frame of endosomal acidification and traffic to the late endosomes. In contrast, the actin network was involved later in the infection, up to 12 to 16 h p.i. These results suggest that MTs are mostly important for the transport of the virus while it is in the endosome pathway, whereas actin could also be involved later in the infection cycle, such as during the transport of the newly synthesized proteins to the nucleus. The effect on the actin network may also indirectly affect the infection.
Proteasome activity was crucial for PPV infection (Fig. 5). In the presence of two commonly used proteasome inhibitors, MG-132 and lactacystin, the virus stayed more diffused in the perinuclear region, and very low replication was observed. These results strongly suggested that interaction with the proteasome occurred during the last transport steps before the delivery of the genome to the nucleus. Moreover, proteasomal processing would take place only in early infection, since addition of the inhibitor at 12 h p.i. had no effect on viral infection. Such a necessity for an interaction with the proteasome has already been demonstrated for MVM (46, 47), in contrast to the inhibitory effect of the proteasome on infection by AAVs (13). Protein degradation by the proteasome proceeds by ubiquitination of the target (28). Coimmunoprecipitation experiments demonstrated that PPV capsid proteins were indeed ubiquitinated early in infection (Fig. 5F), yet no significant degradation of the viral protein could be demonstrated in coimmunoprecipitation experiments (Fig. 5F, lower panel) or by Western blotting performed directly on cell lysates (data not shown). However, since the infectivity ratio of PPV is very low (1 infectious particle for at least 1,000 capsids), we cannot rule out the possibility that degradation remained undetected. Although degradation is a common result of ubiquitination, other purposes, such as differential interaction with cellular components, may be triggered by this modification (30, 34, 65). The exact role of the ubiquitination and proteasomal processing of PPV during entry thus remains to be determined.
In summary, the investigation of PPV entry and transport events within the cell has provided some unique results in comparison with known pathways of parvoviral infection. Indeed, this is the first description of the importance of macropinocytosis as an additional entry portal into cells. These results also highlighted the fact that particle types may play an underestimated role in the preferential uptake of viruses by cells. Taken together, these results may explain why it has been a substantial challenge to identify specific receptors for many viruses, including members of the family Parvoviridae.
Acknowledgments
This work was supported by a grant from the Natural Sciences and Engineering Research Council of Canada to P.T. and by scholarships from the Fondation Armand-Frappier to M.B. and S.F.
We also thank Marcel Desrosiers for technical expertise in confocal microscopy and Micheline Letarte for electronic microscopy.
Footnotes
Published ahead of print on 19 May 2010.
REFERENCES
- 1.Bantel-Schaal, U., I. Braspenning-Wesch, and J. Kartenbeck. 2009. Adeno-associated virus type 5 exploits two different entry pathways in human embryo fibroblasts. J. Gen. Virol. 90:317-322. [DOI] [PubMed] [Google Scholar]
- 2.Barton, E. S., J. L. Connolly, J. C. Forrest, J. D. Chappell, and T. S. Dermody. 2001. Utilization of sialic acid as a coreceptor enhances reovirus attachment by multistep adhesion strengthening. J. Biol. Chem. 276:2200-2211. [DOI] [PubMed] [Google Scholar]
- 3.Bergeron, J., B. Hebert, and P. Tijssen. 1996. Genome organization of the Kresse strain of porcine parvovirus: identification of the allotropic determinant and comparison with those of NADL-2 and field isolates. J. Virol. 70:2508-2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bergeron, J., J. Menezes, and P. Tijssen. 1993. Genomic organization and mapping of transcription and translation products of the NADL-2 strain of porcine parvovirus. Virology 197:86-98. [DOI] [PubMed] [Google Scholar]
- 5.Bishop, N. E. 2003. Dynamics of endosomal sorting. Int. Rev. Cytol. 232:1-57. [DOI] [PubMed] [Google Scholar]
- 6.Blackburn, S. D., S. E. Cline, J. P. Hemming, and F. B. Johnson. 2005. Attachment of bovine parvovirus to O-linked α2,3 neuraminic acid on glycophorin A. Arch. Virol. 150:1477-1484. [DOI] [PubMed] [Google Scholar]
- 7.Brown, K. E., S. M. Anderson, and N. S. Young. 1993. Erythrocyte P antigen: cellular receptor for B19 parvovirus. Science 262:114-117. [DOI] [PubMed] [Google Scholar]
- 8.Canaan, S., Z. Zadori, F. Ghomashchi, J. Bollinger, M. Sadilek, M. E. Moreau, P. Tijssen, and M. H. Gelb. 2004. Interfacial enzymology of parvovirus phospholipases A2. J. Biol. Chem. 279:14502-14508. [DOI] [PubMed] [Google Scholar]
- 9.Carroll, S. M., H. H. Higa, and J. C. Paulson. 1981. Different cell-surface receptor determinants of antigenically similar influenza virus hemagglutinins. J. Biol. Chem. 256:8357-8363. [PubMed] [Google Scholar]
- 10.Chiorini, J. A. 2006. Looking down the rabbit hole: understanding the binding, entry, and trafficking patterns of AAV particles in the cell, p. 165-170. In J. R. Kerr, S. F. Cotmore, M. E. Bloom, R. M. Linden, and C. R. Parrish (ed.), Parvoviruses. Hodder Arnold, London, United Kingdom.
- 11.Cotmore, S. F., and P. Tattersall. 2007. Parvoviral host range and cell entry mechanisms. Adv. Virus Res. 70:183-232. [DOI] [PubMed] [Google Scholar]
- 12.Delboy, M. G., D. G. Roller, and A. V. Nicola. 2008. Cellular proteasome activity facilitates herpes simplex virus entry at a postpenetration step. J. Virol. 82:3381-3390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Douar, A. M., K. Poulard, D. Stockholm, and O. Danos. 2001. Intracellular trafficking of adeno-associated virus vectors: routing to the late endosomal compartment and proteasome degradation. J. Virol. 75:1824-1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Elferink, J. G. 1979. Chlorpromazine inhibits phagocytosis and exocytosis in rabbit polymorphonuclear leukocytes. Biochem. Pharmacol. 28:965-968. [DOI] [PubMed] [Google Scholar]
- 15.Falcone, S., E. Cocucci, P. Podini, T. Kirchhausen, E. Clementi, and J. Meldolesi. 2006. Macropinocytosis: regulated coordination of endocytic and exocytic membrane traffic events. J. Cell Sci. 119:4758-4769. [DOI] [PubMed] [Google Scholar]
- 16.Farr, G. A., L. G. Zhang, and P. Tattersall. 2005. Parvoviral virions deploy a capsid-tethered lipolytic enzyme to breach the endosomal membrane during cell entry. Proc. Natl. Acad. Sci. U. S. A. 102:17148-17153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Govindasamy, L., K. Hueffer, C. R. Parrish, and M. Agbandje-McKenna. 2003. Structures of host range-controlling regions of the capsids of canine and feline parvoviruses and mutants. J. Virol. 77:12211-12221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Greber, U. F., and M. Way. 2006. A superhighway to virus infection. Cell 124:741-754. [DOI] [PubMed] [Google Scholar]
- 19.Harbison, C. E., S. M. Lyi, W. S. Weichert, and C. R. Parrish. 2009. Early steps in cell infection by parvoviruses: host-specific differences in cell receptor binding but similar endosomal trafficking. J. Virol. 83:10504-10514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Harding, M. J., and T. W. Molitor. 1992. A monoclonal antibody which recognizes cell surface antigen and inhibits porcine parvovirus replication. Arch. Virol. 123:323-333. [DOI] [PubMed] [Google Scholar]
- 21.Hueffer, K., L. Govindasamy, M. Agbandje-McKenna, and C. R. Parrish. 2003. Combinations of two capsid regions controlling canine host range determine canine transferrin receptor binding by canine and feline parvoviruses. J. Virol. 77:10099-10105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hueffer, K., L. M. Palermo, and C. R. Parrish. 2004. Parvovirus infection of cells by using variants of the feline transferrin receptor altering clathrin-mediated endocytosis, membrane domain localization, and capsid-binding domains. J. Virol. 78:5601-5611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hueffer, K., J. S. Parker, W. S. Weichert, R. E. Geisel, J. Y. Sgro, and C. R. Parrish. 2003. The natural host range shift and subsequent evolution of canine parvovirus resulted from virus-specific binding to the canine transferrin receptor. J. Virol. 77:1718-1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Johnson, F. B., L. B. Fenn, T. J. Owens, L. J. Faucheux, and S. D. Blackburn. 2004. Attachment of bovine parvovirus to sialic acids on bovine cell membranes. J. Gen. Virol. 85:2199-2207. [DOI] [PubMed] [Google Scholar]
- 25.Jones, A. T. 2007. Macropinocytosis: searching for an endocytic identity and role in the uptake of cell penetrating peptides. J. Cell. Mol. Med. 11:670-684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kaksonen, M., C. P. Toret, and D. G. Drubin. 2006. Harnessing actin dynamics for clathrin-mediated endocytosis. Nat. Rev. Mol. Cell Biol. 7:404-414. [DOI] [PubMed] [Google Scholar]
- 27.Kerr, J. R., S. F. Cotmore, M. E. Bloom, R. M. Linden, and C. R. Parrish (ed.). 2006. Parvoviruses. Hodder Arnold, London, United Kingdom.
- 28.Kornitzer, D., and A. Ciechanover. 2000. Modes of regulation of ubiquitin-mediated protein degradation. J. Cell. Physiol. 182:1-11. [DOI] [PubMed] [Google Scholar]
- 29.Lajoie, P., and I. R. Nabi. 2007. Regulation of raft-dependent endocytosis. J. Cell. Mol. Med. 11:644-653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lin, L., and S. Ghosh. 1996. A glycine-rich region in NF-κB p105 functions as a processing signal for the generation of the p50 subunit. Mol. Cell. Biol. 16:2248-2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.López-Bueno, A., M. P. Rubio, N. Bryant, R. McKenna, M. Agbandje-McKenna, and J. M. Almendral. 2006. Host-selected amino acid changes at the sialic acid binding pocket of the parvovirus capsid modulate cell binding affinity and determine virulence. J. Virol. 80:1563-1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mani, B., C. Baltzer, N. Valle, J. M. Almendral, C. Kempf, and C. Ros. 2006. Low pH-dependent endosomal processing of the incoming parvovirus minute virus of mice virion leads to externalization of the VP1 N-terminal sequence (N-VP1), N-VP2 cleavage, and uncoating of the full-length genome. J. Virol. 80:1015-1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mayor, S., and R. E. Pagano. 2007. Pathways of clathrin-independent endocytosis. Nat. Rev. Mol. Cell Biol. 8:603-612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mukhopadhyay, D., and H. Riezman. 2007. Proteasome-independent functions of ubiquitin in endocytosis and signaling. Science 315:201-205. [DOI] [PubMed] [Google Scholar]
- 35.Muro, S., R. Wiewrodt, A. Thomas, L. Koniaris, S. M. Albelda, V. R. Muzykantov, and M. Koval. 2003. A novel endocytic pathway induced by clustering endothelial ICAM-1 or PECAM-1. J. Cell Sci. 116:1599-1609. [DOI] [PubMed] [Google Scholar]
- 36.Nam, H. J., B. Gurda-Whitaker, W. Y. Gan, S. Ilaria, R. McKenna, P. Mehta, R. A. Alvarez, and M. Agbandje-McKenna. 2006. Identification of the sialic acid structures recognized by minute virus of mice and the role of binding affinity in virulence adaptation. J. Biol. Chem. 281:25670-25677. [DOI] [PubMed] [Google Scholar]
- 37.Norkin, L. C. 1999. Simian virus 40 infection via MHC class I molecules and caveolae. Immunol. Rev. 168:13-22. [DOI] [PubMed] [Google Scholar]
- 38.Nüesch, J. P., S. Lachmann, and J. Rommelaere. 2005. Selective alterations of the host cell architecture upon infection with parvovirus minute virus of mice. Virology 331:159-174. [DOI] [PubMed] [Google Scholar]
- 39.Pakkanen, K., E. Salonen, A. R. Makela, C. Oker-Blom, I. Vattulainen, and M. Vuento. 2009. Desipramine induces disorder in cholesterol-rich membranes: implications for viral trafficking. Phys. Biol. 6:046004. [DOI] [PubMed] [Google Scholar]
- 40.Parker, J. S., W. J. Murphy, D. Wang, S. J. O'Brien, and C. R. Parrish. 2001. Canine and feline parvoviruses can use human or feline transferrin receptors to bind, enter, and infect cells. J. Virol. 75:3896-3902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Parker, J. S., and C. R. Parrish. 2000. Cellular uptake and infection by canine parvovirus involves rapid dynamin-regulated clathrin-mediated endocytosis, followed by slower intracellular trafficking. J. Virol. 74:1919-1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Perrais, D., and C. J. Merrifield. 2005. Dynamics of endocytic vesicle creation. Dev. Cell 9:581-592. [DOI] [PubMed] [Google Scholar]
- 43.Pickart, C. M. 2001. Mechanisms underlying ubiquitination. Annu. Rev. Biochem. 70:503-533. [DOI] [PubMed] [Google Scholar]
- 44.Radtke, K., K. Dohner, and B. Sodeik. 2006. Viral interactions with the cytoskeleton: a hitchhiker's guide to the cell. Cell. Microbiol. 8:387-400. [DOI] [PubMed] [Google Scholar]
- 45.Rappoport, J. Z. 2008. Focusing on clathrin-mediated endocytosis. Biochem. J. 412:415-423. [DOI] [PubMed] [Google Scholar]
- 46.Ros, C., C. J. Burckhardt, and C. Kempf. 2002. Cytoplasmic trafficking of minute virus of mice: low-pH requirement, routing to late endosomes, and proteasome interaction. J. Virol. 76:12634-12645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ros, C., and C. Kempf. 2004. The ubiquitin-proteasome machinery is essential for nuclear translocation of incoming minute virus of mice. Virology 324:350-360. [DOI] [PubMed] [Google Scholar]
- 48.Sandvig, K., M. L. Torgersen, H. A. Raa, and B. van Deurs. 2008. Clathrin-independent endocytosis: from nonexisting to an extreme degree of complexity. Histochem. Cell Biol. 129:267-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Simpson, A. A., B. Hebert, G. M. Sullivan, C. R. Parrish, Z. Zadori, P. Tijssen, and M. G. Rossmann. 2002. The structure of porcine parvovirus: comparison with related viruses. J. Mol. Biol. 315:1189-1198. [DOI] [PubMed] [Google Scholar]
- 50.Suikkanen, S., T. Aaltonen, M. Nevalainen, O. Valilehto, L. Lindholm, M. Vuento, and M. Vihinen-Ranta. 2003. Exploitation of microtubule cytoskeleton and dynein during parvoviral traffic toward the nucleus. J. Virol. 77:10270-10279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Suikkanen, S., M. Antila, A. Jaatinen, M. Vihinen-Ranta, and M. Vuento. 2003. Release of canine parvovirus from endocytic vesicles. Virology 316:267-280. [DOI] [PubMed] [Google Scholar]
- 52.Suikkanen, S., K. Saajarvi, J. Hirsimaki, O. Valilehto, H. Reunanen, M. Vihinen-Ranta, and M. Vuento. 2002. Role of recycling endosomes and lysosomes in dynein-dependent entry of canine parvovirus. J. Virol. 76:4401-4411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Swanson, J. A., and C. Watts. 1995. Macropinocytosis. Trends Cell Biol. 5:424-428. [DOI] [PubMed] [Google Scholar]
- 54.Szelei, J., Z. Zadori, and P. Tijssen. 2006. Porcine parvovirus, p. 435-445. In J. R. Kerr, S. F. Cotmore, M. E. Bloom, R. M. Linden, and C. R. Parrish (ed.), Parvoviruses. Hodder Arnold, London, United Kingdom.
- 55.Tattersall, P. 2006. The evolution of parvovirus taxonomy, p. 5-14. In J. R. Kerr, S. F. Cotmore, M. E. Bloom, R. M. Linden, and C. R. Parrish (ed.), Parvoviruses. Hodder Arnold, London, United Kingdom.
- 56.Teale, A., S. Campbell, N. Van Buuren, W. C. Magee, K. Watmough, B. Couturier, R. Shipclark, and M. Barry. 2009. Orthopoxviruses require a functional ubiquitin-proteasome system for productive replication. J. Virol. 83:2099-2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tullis, G. E., L. R. Burger, and D. J. Pintel. 1993. The minor capsid protein VP1 of the autonomous parvovirus minute virus of mice is dispensable for encapsidation of progeny single-stranded DNA but is required for infectivity. J. Virol. 67:131-141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Varki, N. M., and A. Varki. 2007. Diversity in cell surface sialic acid presentations: implications for biology and disease. Lab. Invest. 87:851-857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Vendeville, A., M. Ravallec, F. X. Jousset, M. Devise, D. Mutuel, M. Lopez-Ferber, P. Fournier, T. Dupressoir, and M. Ogliastro. 2009. Densovirus infectious pathway requires clathrin-mediated endocytosis followed by trafficking to the nucleus. J. Virol. 83:4678-4689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Vihinen-Ranta, M., A. Kalela, P. Makinen, L. Kakkola, V. Marjomaki, and M. Vuento. 1998. Intracellular route of canine parvovirus entry. J. Virol. 72:802-806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Vihinen-Ranta, M., and C. R. Parrish. 2006. Cell infection processes of autonomous parvoviruses, p. 157-163. In J. R. Kerr, S. F. Cotmore, M. E. Bloom, R. M. Linden, and C. R. Parrish (ed.), Parvoviruses. Hodder Arnold, London, United Kingdom.
- 62.Vihinen-Ranta, M., W. Yuan, and C. R. Parrish. 2000. Cytoplasmic trafficking of the canine parvovirus capsid and its role in infection and nuclear transport. J. Virol. 74:4853-4859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Weigel-Kelley, K. A., M. C. Yoder, and A. Srivastava. 2003. α5β1 integrin as a cellular coreceptor for human parvovirus B19: requirement of functional activation of β1 integrin for viral entry. Blood 102:3927-3933. [DOI] [PubMed] [Google Scholar]
- 64.Weitzman, M. D. 2006. The parvovirus life cycle: an introduction to molecular interactions important for infections, p. 143-156. In J. R. Kerr, S. F. Cotmore, M. E. Bloom, R. M. Linden, and C. R. Parrish (ed.), Parvoviruses. Hodder Arnold, London, United Kingdom.
- 65.Welchman, R. L., C. Gordon, and R. J. Mayer. 2005. Ubiquitin and ubiquitin-like proteins as multifunctional signals. Nat. Rev. Mol. Cell Biol. 6:599-609. [DOI] [PubMed] [Google Scholar]
- 66.Wilhelm, S., P. Zimmermann, H. J. Selbitz, and U. Truyen. 2006. Real-time PCR protocol for the detection of porcine parvovirus in field samples. J. Virol. Methods 134:257-260. [DOI] [PubMed] [Google Scholar]
- 67.Yu, G. Y., and M. M. Lai. 2005. The ubiquitin-proteasome system facilitates the transfer of murine coronavirus from endosome to cytoplasm during virus entry. J. Virol. 79:644-648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zádori, Z., J. Szelei, M. C. Lacoste, Y. Li, S. Gariepy, P. Raymond, M. Allaire, I. R. Nabi, and P. Tijssen. 2001. A viral phospholipase A2 is required for parvovirus infectivity. Dev. Cell 1:291-302. [DOI] [PubMed] [Google Scholar]