Abstract
The capsid protein (CP) of Turnip crinkle virus (TCV) is a multifunctional protein needed for virus assembly, suppression of RNA silencing-based antiviral defense, and long-distance movement in infected plants. In this report, we have examined genetic requirements for the different functions of TCV CP and evaluated the interdependence of these functions. A series of TCV mutants containing alterations in the CP coding region were generated. These alterations range from single-amino-acid substitutions and domain truncations to knockouts of CP translation. The latter category also contained two constructs in which the CP coding region was replaced by either the cDNA of a silencing suppressor of a different virus or that of green fluorescent protein. These mutants were used to infect Arabidopsis plants with diminished antiviral silencing capability (dcl2 dcl3 dcl4 plants). There was a strong correlation between the ability of mutants to reach systemic leaves and the silencing suppressor activity of mutant CP. Virus particles were not essential for entry of the viral genome into vascular bundles in the inoculated leaves in the absence of antiviral silencing. However, virus particles were necessary for egress of the viral genome from the vasculature of systemic leaves. Our experiments demonstrate that TCV CP not only allows the viral genome to access the systemic movement channel through silencing suppression but also ensures its smooth egress by way of assembled virus particles. These results illustrate that efficient long-distance movement of TCV requires both functions afforded by the CP.
Plant viruses commonly encounter an array of host defense mechanisms in the plants that they infect and have evolved diverse strategies of counter-defense. Virus-encoded suppressors of RNA silencing (VSRs) are well-studied examples of such counter-defenses adopted by plant viruses (7, 20). VSRs function to suppress RNA silencing, a potent antiviral defense process that degrades viral RNAs in a sequence-specific manner (7). This defense process is initiated by the intracellular presence of double-stranded RNAs (dsRNAs), including dsRNA structures that are frequently present in viral RNAs, where some function as critical regulatory elements (32). Therefore, most RNA virus infections inevitably trigger RNA silencing.
Once detected, viral dsRNAs are processed by plant dicer-like (DCL) nucleases to small RNAs of discrete lengths (21, 22, and 24 nucleotides [nt]) called small interfering RNAs (siRNAs). The siRNAs are then recruited by Argonaute (AGO) proteins and serve as the sequence-specific guide to mediate the cleavage of cognate viral RNAs. The model plant Arabidopsis encodes four different DCLs, DCL1, DCL2, DCL3, and DCL4, each processing unique classes of dsRNAs and generating small RNAs of different sizes. Several studies have shown that RNA viruses are targeted by three different DCLs (DCL2, DCL3, and DCL4), while the RNAs produced by DNA viruses are processed by all four DCLs (5, 6, 15, 22).
Conversely, a number of VSRs, among them the capsid protein (CP) of Turnip crinkle virus (TCV), function by counteracting the DCL-mediated, dsRNA-processing step (5, 19). TCV is a small icosahedral virus with a positive-sense RNA genome of 4,054 nt. The 5′ proximal open reading frame (ORF) of TCV (P28), together with its read-through product (P88), is responsible for genome replication and transcription (21). Transcription of the TCV genome leads to the production of two subgenomic (sg) RNAs from the 3′ half of the genomic (g) RNA, which serve as mRNAs for the movement proteins (MPs) (P8 and P9) and the CP (P38), respectively. In addition to packaging the TCV genome, P38 functions as the VSR, as well as the effector of resistance (R) gene-mediated defense (9, 19, 23, 28).
TCV belongs to the Carmovirus genus of the Tombusviridae, a family of viruses that also includes the Tombusvirus genus (21). One main criterion for grouping these two genera into the same family is the significant similarity shared by the CPs of their member viruses. Interestingly, viruses in these two genera use different proteins as VSRs. All carmoviruses tested so far use their CPs as VSRs, while the CPs of tombusviruses such as Tomato bushy stunt virus (TBSV) do not suppress RNA silencing. Instead, tombusviruses encode a separate VSR termed P19 (27). The carmovirus CP and the tombusvirus P19 have been shown to act at different steps in the antiviral silencing pathways (20).
We are interested in determining the molecular requirements for the different functions of TCV CP. Previously, we demonstrated that certain TCV mutants containing single-amino-acid changes within the N terminus of CP failed to induce R gene-mediated resistance but were fully capable of suppressing RNA silencing. This study uncoupled the ability of the CP to elicit R gene-mediated resistance from its VSR function (3). However, we were unable to determine whether the VSR function of CP was dependent on its ability to form virus particles, because in-frame deletions within the CP compromised protein stability in vivo. Although several earlier studies have shown that TCV CP is necessary for long-distance but not cell-to-cell movement of the virus in Arabidopsis plants (4, 12), they did not clarify whether assembled virions or the VSR activity of CP alone allowed for viral long-distance movement, as the VSR activity of CP was not known then. A more recent report by Zhang and Simon (33) suggested that the VSR activity of CP, but not virion assembly, is sufficient for TCV long-distance movement. Nonetheless, they could not rule out the involvement of a small amount of assembled virions (30, 33).
In the current report, we have attempted to uncouple particle assembly and silencing suppressor functions of TCV CP and to evaluate their relative contributions to viral systemic spread in Arabidopsis. The VSR activity of TCV CP was disrupted by replacing amino acid residues conserved among carmoviruses but divergent from their counterparts in tombusvirus CPs. Arabidopsis plants in which three antiviral DCL genes are nonfunctional (dcl2 dcl3 dcl4 plants) were used to compensate for the loss of the VSR function of TCV CP. Our results show that, while VSR function of TCV CP is necessary for the viral RNA to reach the vascular tissue in systemic leaves, the particle assembly function of CP is required for efficient unloading of viral RNA into the leaf mesophyll. These results effectively uncouple the VSR function of CP from its particle assembly function and reveal the importance of virus assembly in vascular egress.
MATERIALS AND METHODS
Constructs used for Agrobacterium tumefaciens (strain C58C1)-mediated transient expression (agro-infiltration) in Nicotiana benthamiana leaves.
The PZP-green fluorescent protein (GFP), PZP-CP, and PZP-P19 constructs have been described before (19). Binary plasmids that contain full-length cDNAs of wild-type TCV, TCV-CPstop, and TCV-GFP (PZP-TCV, PZP-TCV-CPstop, and PZP-TCV-GFP) were also described elsewhere (17, 19). The PZP-CPstop (previously designated PZP-CPΔ) construct is a mutated form of PZP-CP that contains two consecutive stop codons after the fifth amino acid of TCV CP. The PZP-CPA, B, C, and F constructs contain single-amino-acid changes at amino acids (aa) 128 (K128Q), 130 (R130T), 137 (R137H), and 211 (K211Q) of TCV CP, respectively.
Constructs used for in vitro transcription to generate infectious RNAs.
The pTCV (previously known as T1d1) construct has been described (19). The pTCV-CPstop construct contains the same mutations as PZP-CPstop. pTCVV-P19 is a slightly modified version of pTCV-P19 (22), in which all AUGs prior to the authentic start codon of P19 were mutated, so that the P19 produced from the new construct initiates at the proper P19 initiation codon (Fig. 1A). TCV-GFP is the same as TCV-sGFP, described previously (17). The pTCV-CPA, -B, -C, and -F constructs are derivatives of pTCV that incorporated the mutations in PZP-CPA, -B, -C, and -F, respectively (Fig. 1A).
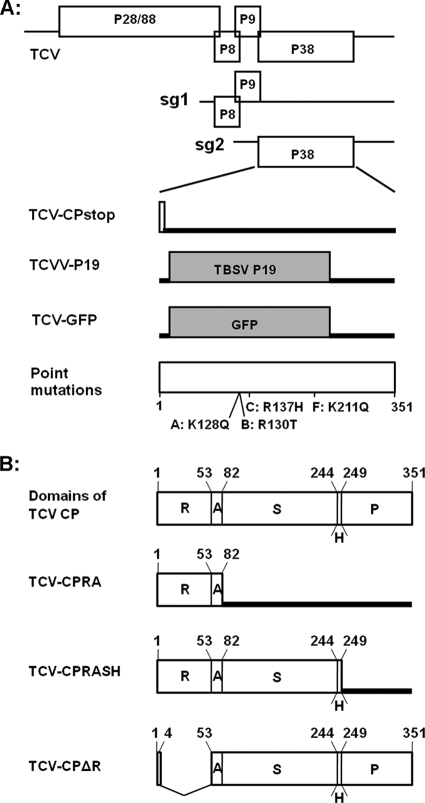
FIG. 1.
Schematic representation of the genome structure of TCV and its mutants. (A) Top three diagrams depict the genomic (g) and subgenomic (sg) RNAs of TCV, with all the open reading frames (ORFs) shown as open boxes. The bottom four diagrams show the CP region of mutants TCV-CPstop, TCVV-P19, and TCV-GFP and four single-amino-acid mutants (A, B, C, and F), with the open or shaded boxes denoting the expected translation products and the thick lines representing the RNA regions not expected to be translated. The exact amino acid changes in the single-amino-acid mutants are marked beneath the bottom diagram, together with their approximate positions. (B) Top diagram represents the full-length CP of TCV, with the sizes and the relative positions of the five structural domains shown. The numbers on the top are the positions of the first amino acids of the respective domains and the last amino acid of the whole CP. R, RNA-binding domain; A, arm; S, surface domain; H, hinge; P, protruding domain. The next three diagrams depict three mutants with truncated CPs. TCV-CPΔR contains an in-frame deletion, whereas both CPRA and CPRASH are mutants that prevent full CP translation with added stop codons.
The pTCV-CPRA construct was generated by introducing a stop codon at the end of the arm domain (aa 81) of TCV CP, so that only the N-terminal RNA-binding (R) domain and the arm would be translated in the infected plants. pTCV-CPRASH has a stop codon at the end of the 5-aa hinge connecting the surface (S) and the protruding (P) domains, so that a truncated CP without the P domain would be translated. pTCV-ΔR contains an in-frame deletion spanning from aa 5 to aa 52, resulting in a CP without the R domain (Fig. 1B). All mutants were sequenced to verify their identity. The sequences of oligonucleotides used to produce these mutants are available upon request.
Plant materials.
Both N. benthamiana and Arabidopsis plants were grown in a growth room set at 20°C, with 12 h of daylight. Agro-infiltration took place when the N. benthamiana plants were about 4 weeks old, with four to five true leaves. The Arabidopsis plants were inoculated with in vitro transcribed viral RNAs when they were 3 to 4 weeks old. Usually, six plants were inoculated with each transcript. At 10 to 11 days postinoculation (dpi), six uninoculated upper leaves, one per plant, were collected and pooled for RNA, virion, or protein extractions.
RNA blot analysis.
Total RNAs were extracted from infected or infiltrated plants and subjected to RNA blot analysis to detect TCV viral RNAs, green fluorescent protein (GFP) mRNA, or siRNAs using protocols outlined in previous reports (19, 22). Different amounts of Arabidopsis RNA were used to detect viral RNA (1 μg) and siRNAs (5 μg), whereas the same amount of N. benthamiana RNAs (5 μg) was used to detect GFP mRNA and siRNAs in the agro-infiltrated leaves.
Protein blots.
Protein extracts were prepared from systemically infected leaves of Arabidopsis (31). Protein samples (5 μg each) were separated on 12% polyacrylamide-0.1% SDS gels and subsequently transferred to polyvinylidene difluoride (PVDF) membranes for detection of TCV CP with the corresponding antiserum.
In vitro dsRNA binding.
The first 321 nt of Bean pod mottle virus (BPMV) RNA2 was used as the template for reverse transcription-PCR (RT-PCR) to generate a 381-bp-long DNA fragment containing the T7 promoter sequence at both ends. This PCR fragment was then transcribed in vitro into 32P-labeled, 321-bp-long dsRNA. The labeled dsRNA was then mixed with excess amounts of unlabeled transcripts, subjected to incubation at 90°C for 1 min, and allowed to cool down slowly to room temperature. These steps ensured that most labeled RNA molecules were in double-stranded form.
The protein extracts were prepared from N. benthamiana leaves agro-infiltrated with constructs that are designed to express proteins to be examined, at four days after infiltration. The binding experiments were carried out following the protocol of Merai et al. (14).
RESULTS
Alignment of CPs of multiple carmo- and tombusviruses identifies a region conserved in carmoviruses.
Structural studies have shown that the CPs of carmoviruses share a high level of similarity with their tombusviral counterparts, although tombusviral CPs do not suppress RNA silencing (1, 19). We reasoned that a comparison of the amino acid sequences of carmo- and tombusvirus CPs might identify a unique region in carmoviral CPs that could be responsible for silencing suppressor activity. We therefore compared the CPs of three carmoviruses and three tombusviruses. The three carmoviruses included were TCV, Hibiscus chlorotic ringspot virus (HCRSV), and Carnation mottle virus (CarMV), all of which have been shown to use CP as a VSR (13, 19; F. Qu, unpublished results). The three tombusviruses were TBSV, Cymbidium ringspot virus (CymRSV), and Cucumber necrosis virus (CNV), each of which uses a P19 homolog as the VSR (10, 29).
A partial amino acid sequence alignment is presented in Fig. 2. The overall sequence homology among the six CPs is evident, with invariable amino acids easily identifiable throughout the full length of CPs (Fig. 2 and data not shown). Importantly, most of the invariable amino acids are concentrated within the S domain of the CPs, spanning roughly from aa 100 to 280 of CymRSV CP. This region corresponds to aa 73 to 243 of TCV CP. Most of the conserved amino acids are shared by all six viruses, and very few of them are unique to viruses of either genus. Indeed, we were able to identify only 11 conserved carmoviral amino acids that are not shared by the tombusviruses. Notably, 4 of the 11 amino acids, corresponding to K128, R130, R137, and K211 in TCV CP (highlighted by asterisks immediately underneath the alignment in Fig. 2), are basic residues, suggesting that they might be involved in dsRNA binding (14). Since serving as VSRs is the function of carmovirus CPs not shared by tombusvirus CPs, we then tested the four conserved basic residues in the TCV CP for their role in silencing suppression.
FIG. 2.
Alignment of multiple CPs of viruses in the Tombusviridae family. The four basic amino acid residues conserved in carmoviruses but not in tombusviruses are highlighted by asterisks immediately underneath.
Site-directed mutagenesis identifies R130 and R137 as amino acids critical for the VSR function of TCV CP.
To examine the possible role of these amino acids in silencing suppression, we replaced the four basic residues (K128, R130, R137, and K211) of TCV CP with their counterparts in TBSV CP to create four different TCV CP mutants: CPA (K128Q), CPB (R130T), CPC (R137H), and CPF (K211Q) (Fig. 1A and 2). The VSR activity of these CP mutants was evaluated by agro-infiltration, using the fluorescence intensity of coexpressed green fluorescent protein (GFP) as a visual indicator. As shown in Fig. 3A, while CPA and CPF displayed virtually the same VSR activity as wild-type TCV CP (Fig. 3A, compare leaves 2, 5, and 8), CPB showed significantly weakened VSR activity (compare leaves 2 and 6). More dramatically, CPC appeared to have lost the VSR activity completely, as the level of GFP fluorescence in the leaves coinfiltrated with GFP+CPC diminished to the same extent as that of GFP+CPstop, which could not produce any CP (compare leaves 3 and 7).
FIG. 3.
Mutations within TCV CP differentially affect its functions in silencing suppression, virion assembly, and viral systemic spread. (A) Evaluation of the silencing suppression capability of different TCV CP mutants using the agro-infiltration assay. TBSV P19 (leaf no. 4) was used as an additional positive control. The leaves were photographed at 5 days after infiltration. (B) Detection of GFP mRNA and siRNAs in the agro-infiltrated leaves with RNA blot hybridizations. (C) dsRNA-binding properties of CPA, CPB, CPC, and CPF mutants. Top, 5% nondenaturing polyacrylamide gel containing protein extracts preinoculated with 32P-labeled dsRNA. Bottom, 12% SDS-PAGE gel serving showing the relative concentrations of mutant proteins used in the binding assay. (D) Systemic symptoms on Col-0 and dcl2 dcl3 dcl4 plants infected with various TCV mutants. Mock, buffer inoculated. Photographs were taken at 18 dpi. (E) Close-up images of dcl2 dcl3 dcl4 plants not inoculated (top left), inoculated with TCV-CPstop (bottom left), and inoculated with CPC (bottom right). Top right: systemically infected leaves taken from these plants. A relatively healthy class II vein on CPC-infected leaves is highlighted with an arrow. (F) Viral RNA, virion, and CP accumulations in plants infected with various TCV mutants as determined with RNA blot and Western blot hybridizations. Total RNA, protein, and virions were extracted from pooled systemic leaves (see Materials and Methods) at 11 to 12 dpi. Note that in the bottom two panels, lanes 5 and 6 were dilutions of the lane 4 sample. The panel at the very bottom is the stained gel serving as the control for protein load, with the TCV CP band easily visible.
The visual data were confirmed by RNA blot detection of GFP mRNA and siRNAs (Fig. 3B). Consistent with the intense GFP fluorescence shown in Fig. 3A, CPA and CPF suppressed the silencing of GFP mRNA, permitting high levels of GFP mRNA accumulation (Fig. 3B, top panel; compare lanes 5 and 8 with lane 2). This was accompanied by the absence of GFP-specific siRNAs (Fig. 3B, middle panel, lanes 2, 5, and 8). In contrast, CPC was unable to prevent the degradation of GFP mRNA, resulting in low levels of GFP mRNA similar to that in the GFP+CPstop combination (Fig. 3B, top panel, lanes 3 and 7). Accordingly, GFP siRNAs also accumulated to similar levels in both samples (Fig. 3B, middle panel, lanes 3 and 7). The CPB mutant was unique in that it permitted slightly higher accumulations of both GFP mRNA and siRNAs than CPC (Fig. 1B, lane 6 of top and middle panels). We speculate that partial suppression of RNA silencing by CPB might have stabilized a portion of GFP mRNA, providing the RNA silencing machinery with more template. Taken together, our agro-infiltration experiments implicate both R130 (CPB) and R137 (CPC) in the silencing suppression function of TCV CP.
It was shown previously that TCV CP bound dsRNAs, and this dsRNA-binding activity was thought to contribute to its silencing suppression activity (14). To evaluate whether the single-amino-acid mutations introduced by us compromised the dsRNA-binding activity of TCV CP, we agro-infiltrated N. benthamiana leaves with the mutant constructs. In addition, we sought to achieve similar expression levels for all CP mutants by including the P19 silencing suppressor in all infiltrations (Fig. 3C). Protein extracts were then prepared from infiltrated leaves and subjected to coincubation with 32P-labeled dsRNA, using a procedure established by Merai et al. (14). As shown in Fig. 3C (top panel), CPF retained the dsRNA-binding activity of wild-type CP despite lower protein levels, whereas the other three mutants displayed dramatically reduced affinity to dsRNA. The diminished affinity between CPA and dsRNA was particularly noteworthy because, unlike CPB and CPC, CPA protein accumulated to the level of wild-type CP (Fig. 3C, bottom panel). Considering the fact that both CPA and CPF remained strong silencing suppressors, our results suggest that dsRNA-binding might not be the sole mechanism of silencing suppression by TCV CP.
Infection of Arabidopsis plants with TCV mutants reveals a VSR-independent role of TCV CP in the systemic spread of the virus.
The previous experiments implicated R137, and R130 to a lesser extent, in the VSR function of CP. We next wanted to test if the compromised VSR phenotype would be evident upon virus infections and what impact these and the other mutations might have on the systemic spread of TCV. We hence introduced the CPA, CPB, CPC, and CPF mutations into pTCV, the full-length infectious clone of TCV. In vitro transcripts of these four mutants, together with those of TCV-CPstop and TCVV-P19, were then used to infect both wild-type Arabidopsis (Col-0) and dcl2 dcl3 dcl4 mutant plants that are defective in antiviral silencing targeting RNA viruses, including TCV (5, 6). For every transcript, six Col-0 and six dcl2 dcl3 dcl4 plants were infected. The whole set of experiments was repeated three times.
Based on the symptoms on systemically infected leaves, the six mutants were divided into three classes. The first class includes CPA and CPF, which infected both Col-0 and dcl2 dcl3 dcl4 plants systemically similarly to wild-type TCV. Symptoms were, however, more severe on CPA-infected plants and milder on CPF-infected plants than on those infected with wild-type TCV (Fig. 3D, compare the TCV, CPA, and CPF panels). These results confirmed that these three TCV CP variants effectively suppressed the DCL activity targeting TCV RNA in Col-0 plants. We speculate that the symptom differences among these three TCV variants were probably the consequence of some other VSR-independent functions of TCV CP.
Interestingly, further analyses revealed that the more severe symptoms caused by the CPA mutant was accompanied by a lower level of virus particle accumulation in systemically infected tissues. As illustrated in Fig. 3F (top panel, lanes 3, 4, 9, 10, 15, and 16), viral RNAs of CPA and CPF mutants accumulated to levels similar to those of wild-type TCV in both Col-0 and dcl2 dcl3 dcl4 plants. However, the CPA mutant produced a significantly reduced level of virions, suggesting that the K128Q mutation in CPA did compromise virus assembly to some extent (Fig. 3F, middle panel; compare lanes 9 and 10 with 3 and 4). In contrast, the CPF mutant appeared to accumulate slightly more viruses than wild-type TCV (lanes 15 and 16). Viruses purified from the same amount of systemically infected tissues of Col-0 as well as dcl2 dcl3 dcl4 plants were quantified by UV spectrometry, which showed that the relative ratio of these three viruses was approximately CPA:TCV: CPF = 0.4:1:1.3 (average of three virus preparations). No significant difference was seen between the two kinds of plants.
To determine whether the difference in virion levels was caused by altered accumulation of CP, we subjected the protein extracts of systemically infected leaves to Western blot analysis with CP antiserum. As shown on the bottom two panels of Fig. 3F (lanes 3, 4, 9, 10, 15, and 16), The CP protein levels were similar in all three infections (TCV, CPA, and CPF). These data, when considered together with the symptom severity of CPA- and CPF-infected plants (Fig. 3D), suggest that neither the level of CP nor the concentration of virions correlates with the severity of disease symptoms.
The second class of mutants includes CPB and CPC, which caused visible systemic symptoms in dcl2 dcl3 dcl4 plants but not in Col-0 plants (Fig. 3D, CPB and CPC panels). This is in agreement with the results presented in Fig. 3A and B showing that the CPB and CPC mutations compromised the silencing suppression function of TCV CP. Note here that even though the CPB-infected dcl2 dcl3 dcl4 plants appear to be more diseased that the CPC-infected plants, the CPC infections actually proceeded more rapidly, causing the systemic leaves to wilt before they had a chance to expand.
The results of RNA blot analysis (Fig. 3F) were consistent with prior results showing that the VSR function of TCV CP is partially compromised in CPB but completely lost in CPC. In infected Col-0 plants, CPB RNA was detected at much reduced levels (Fig. 3F, top panel, lane 11), whereas CPC RNA was completely absent (lane 13). In dcl2 dcl3 dcl4 plants, both CPB and CPC RNAs were detected at high levels, although CPC RNAs were slightly less abundant (Fig. 3F, top panel, compare lanes 4, 12, and 14). This was expected, because all three antiviral DCLs (DCL2, DCL3, and DCL4) are knocked out in dcl2 dcl3 dcl4 plants, compensating for the loss of the silencing suppressor function in the mutants.
CPC-infected dcl2 dcl3 dcl4 plants yielded very low levels of virions. As shown in Fig. 3F (middle panel, lanes 10, 12, and 14), the CPB virions accumulated to a level similar to that of CPA virions in dcl2 dcl3 dcl4 plants, while the CPC virions were far less abundant than in either CPA or CPB infections. In fact, the concentration of purified CPC virus is at least 10-fold less than that of CPB and 30-fold less than that of wild-type TCV. Strikingly, the CP protein levels were similar in CPB- and CPC-infected dcl2 dcl3 dcl4 plants (Fig. 3F, bottom two panels, lanes 12 and 14). These results suggest that the R137H mutation in CPC dramatically diminished the ability of CP to form stable virions. Nevertheless, as the photographs in Fig. 3D illustrate, the defect in virion assembly did not seem to affect the systemic spread of the CPC mutant in dcl2 dcl3 dcl4 plants. Together, our data with four single-amino-acid mutants suggest that virion assembly is not a prerequisite for efficient systemic virus movement. This confirms that in the absence of active antiviral silencing, only a very low threshold of virus assembly is required to facilitate successful systemic movement of these mutants.
The third class of mutants includes TCV-CPstop and TCVV-P19. TCV-CPstop contains two consecutive stop codons shortly after the start codon of CP, whereas TCVV-P19 harbors TBSV P19 in place of CP in the TCV infectious clone (see Materials and Methods and Fig. 1A for details). Therefore, unlike the single-amino-acid mutants discussed above, neither of them was capable of producing any CP in infected plants, thus precluding the production of virus particles. Consequently, these two mutants should help determine whether TCV CP facilitates viral systemic spread in ways that are independent of virion assembly or silencing suppression. As shown in Fig. 3D, TCV-CPstop caused systemic symptoms on dcl2 dcl3 dcl4 plants but not on Col-0 plants. This was expected, as no functional VSR is encoded in this mutant. TCVV-P19 infected 70 to 80% of both Col-0 and dcl2 dcl3 dcl4 plants (Fig. 3D, the TCVV-P19 panel), confirming previous reports showing that P19 functions downstream of DCL actions (10). As shown in Fig. 3F (top panel, lanes 5 to 8), the accumulation of viral RNA in plants infected with these two mutants was much lower than in wild-type TCV-infected plants.
Notably, TCV-CPstop and TCVV-P19 caused a distinctive set of systemic symptoms that differed from those of plants infected with wild-type TCV and the CP variant mutants (CPA, -B, -C, and -F). First, the symptoms caused by TCV-CPstop and TCVV-P19 were delayed for up to 1 week, as evidenced by the relatively larger size of infected plants (Fig. 3D). Second, the symptoms caused by TCV-CPstop and TCVV-P19 were almost exclusively concentrated on the main veins. This is in contrast to the symptoms of other infections, which started mostly from the leaf laminas and edges. Usually, the petioles and midribs, and sometimes the secondary veins directly connected to the midribs, became completely discolored (vein bleaching) before the rest of the leaf showed any indications of infection (Fig. 3D, the CPstop and TCVV-P19 panels; Fig. 3E). In some cases, the dehydrated petioles caused the leaves to fall off while the leaf mesophyll remained green (data not shown). The close-up images in Fig. 3E, showing dcl2 dcl3 dcl4 plants infected with TCV-CPstop or CPC as well as representative examples of systemically infected leaves, further highlight these symptom differences. These results suggest that the lack of virus particles in the TCV-CPstop and TCVV-P19 infections may seriously impair the exit of viral RNA from the vascular bundles, thus restricting their replication inside the main veins. The relatively low levels of viral RNA in the systemic leaves of plants with these infections are consistent with this observation (Fig. 3F, top panel, lanes 5 to 8).
In support of this view, the uppermost young leaves of plants infected with TCV-CPstop and TCVV-P19 took on a bulged appearance and became severely twisted as infection proceeded, suggestive of retarded development of veins relative to the mesophyll tissues in the same leaves (Fig. 3D and E). In contrast, CPC-infected young leaves stayed flat despite the yellowing or necrotic spots on the leaf laminas. Therefore, while disrupting the plant antiviral silencing pathways allowed the CP-less mutants to reach the vasculature of systemic leaves, the particle assembly function of the CP appeared necessary for TCV RNA to exit the vascular bundles into the mesophyll cells.
Exit from vascular tissues in systemic leaves likely involves assembled virions.
We next sought to more directly address the question of whether virus particles are required for TCV to exit the vascular bundles. Since none of our single-amino-acid mutants completely lost the capability of virion assembly, we decided to approach this question using mutants designed to express different domains of TCV CP. It is well established that TCV CP consists of five structural domains: the N-terminal RNA-binding domain (R domain) is followed by a short arm (A) that connects the R domain to the surface (S) domain. The S domain is in turn linked to the protruding (P) domain with a 5-aa hinge (H) domain (Fig. 1B, top diagram) (1). We reasoned that if exit of TCV RNA from vascular tissues did not require virus assembly, then it might be possible to identify a specific domain of CP that would promote vascular egress. To test this idea, we made three additional TCV mutants. As shown in Fig. 1B, the TCV-CPRA (RA) mutant contains a stop codon at the end of the A domain so that only an 82-aa-long N-terminal fragment of CP would be produced in the infected plants. Similarly, the TCV-CPRASH (RASH) mutant harbors a stop codon at the end of the H domain so that the resultant CP would lack the P domain. The third mutant contained an in-frame deletion (aa 5 to 52), leading to a CP without an R domain (Fig. 1B).
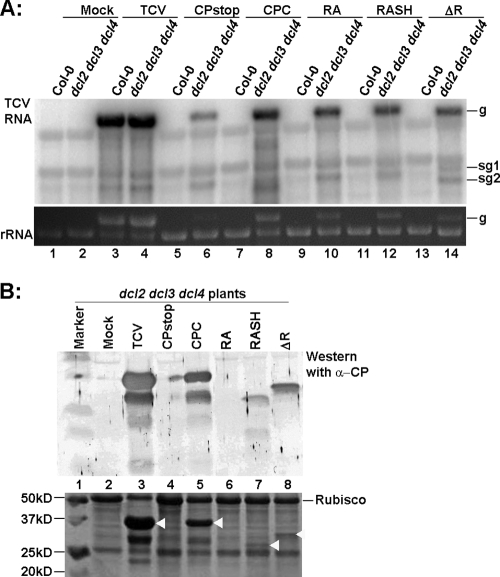
The infectious RNAs of the mutants were inoculated into Col-0 and dcl2 dcl3 dcl4 triple mutant plants to evaluate their ability to exit from vascular tissues in systemic leaves. As expected, none of the mutants were able to infect Col-0 plants systemically, reflecting the lack of a functional VSR (data not shown). However, all three mutant viruses reached the vascular tissues of systemic leaves in dcl2 dcl3 dcl4 plants, as evidenced by the fact that mutant viral RNAs as well as truncated CPs were detected in the young leaves of dcl2 dcl3 dcl4 plants (Fig. 4A, lanes 10, 12, and 14; Fig. 4B, lanes 7 and 8). The RA protein was not detected, possibly due to its smaller size or decreased stability. However, the systemic symptoms caused by these three mutants shared many of the same characteristics as those for TCV-CPstop: the older systemic leaves had a characteristic vein bleaching, with the leaf mesophyll staying relatively healthy, while younger leaves showed severe bulging and twisting (data not shown). These observations suggested that none of these mutants were able to exit the vascular bundle efficiently. Therefore, the entire CP appears to be required for vascular egress.
FIG. 4.
Relative accumulation levels of viral RNAs and CP (or its truncated forms) of different CP deletion mutants. (A) RNA blot hybridization showing the viral RNAs of TCV-RA, TCV-RASH, and TCV-ΔR mutants in both wild-type Col-0 and dcl2 dcl3 dcl4 plants at 12 dpi. (B) Western blot showing the accumulation of CP or its derivatives in the systemically infected leaves. The white arrows in the stained gel (bottom) mark the positions of CP variants.
The CP-less TCV-GFP mutant is restricted to the larger veins in systemic dcl2 dcl3 dcl4 leaves but spreads efficiently in CP-transgenic plant leaves.
We next wanted to determine the exact boundary at which the assembly-deficient TCV mutants were restricted. Previously, Cohen et al. (4) showed that transgenic plants expressing TCV CP complemented the long-distance movement of a TCV-GFP construct in which the 5′ half of the CP gene was replaced by GFP cDNA. We reasoned that the complementation mediated by transgenic CP might differ from that afforded by dcl2 dcl3 dcl4 plants, as the former would be expected to facilitate both silencing suppression and particle assembly. Experimental demonstration of this difference should help resolve the boundary that limits the movement of CP-less TCV mutants. We hence acquired P38CHS, a CP-transgenic line in the Col-0 background (8), and subjected both the dcl2 dcl3 dcl4 triple mutant and P38CHS to infections with TCV-GFP.
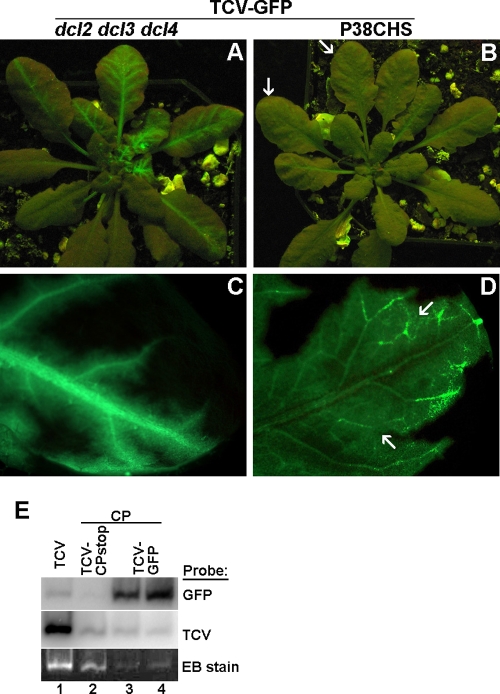
Starting from 14 dpi, GFP fluorescence was observed in the systemic leaves of most dcl2 dcl3 dcl4 and P38CHS plants inoculated with the TCV-GFP transcript. However, both the strength and distribution of GFP fluorescence were strikingly different in these two kinds of plants. As shown in Fig. 5 A and B, while leaves of dcl2 dcl3 dcl4 plants glowed brightly with primarily vein-centered fluorescence distribution, the GFP fluorescence on P38CHS leaves was barely visible (see arrows in Fig. 5B for two weakly fluorescent leaves; note here that the exposure time for Fig. 5B was twice as long as for Fig. 5A).
FIG. 5.
Different patterns of systemic movement of TCV-GFP in dcl2 dcl3 dcl4 and P38CHS plants. (A and B) TCV-GFP-infected dcl2 dcl3 dcl4 (A) and P38CHS (B) plants, photographed at 18 dpi. (C) A systemic leaf of a TCV-GFP-infected dcl2 dcl3 dcl4 plant viewed under a fluorescent microscope. (D) A systemic leaf of a TCV-GFP-infected P38CHS plant viewed under a fluorescent microscope. (E) A virion gel and its RNA blots showing virus particles assembled in N. benthamiana leaves infiltrated with CP+TCV-GFP constructs.
Nonetheless, the distribution of GFP fluorescence in P38CHS leaves was much more even than in dcl2 dcl3 dcl4 leaves. This is unambiguously illustrated by fluorescent microscopy of selected leaves. As shown in Fig. 5C, the bright green fluorescence in systemically infected dcl2 dcl3 dcl4 leaves was mostly restricted to primary and secondary veins, with limited bleeding into areas adjacent to these veins. In contrast, the fluorescence in systemically infected P38CHS leaves was distributed throughout the whole leaf, with tertiary veins and vein networks distinctly visible (Fig. 5D). The relatively intense fluorescence at the junctions of tertiary veins (arrows in Fig. 5D), accompanied by narrow fluorescence voids immediately adjacent to primary and secondary veins, suggests that unlike dcl2 dcl3 dcl4 plants, P38CHS plants permitted highly efficient movement of TCV-GFP into tertiary veins, followed by its egress via tertiary vein junctions. This movement pattern is similar to that of a fully competent Potato virus X derivative (PVX-GFP) in N. benthamiana (24) and a TCV construct that encodes a functional CP in Arabidopsis (34). Therefore, our results demonstrated that TCV-GFP became fully capable of vascular exit in the presence of transgenically expressed CP, most likely through virion assembly.
We then tried, but failed, to detect virions in TCV-GFP-infected P38CHS plants, using a routine virus isolation procedure. We reasoned that virions formed by TCV-GFP RNA and transgenically expressed CP might not accumulate to the levels detectable by our procedure. To increase their expression levels in plant cells, we sought to coexpress TCV-GFP and CP in N. benthamiana leaves through agro-infiltration-mediated delivery. Controls in this experiment included a wild-type TCV construct, as well as a mixture of TCV-CPstop and CP constructs. Coinfiltration of TCV-GFP and CP constructs led to high levels of TCV-GFP expression, as evidenced by strong GFP fluorescence in the infiltrated leaves (data not shown). The infiltrated leaves were subjected to virus isolation at 6 days after infiltration. As shown in the bottom panel of Fig. 5E, lane 1, large amounts of virions produced by wild-type TCV formed an intense band in the virion gel, which also served as the reference for the position of TCV virions. In lanes 2, 3, and 4, trans-assembled virions in CP+TCV-CPstop- and CP+TCV-GFP-infiltrated leaves were at much lower yet distinctly visible levels.
To further confirm the presence of TCV-GFP RNAs in these virus particles, we subjected the virus gel to RNA blot hybridizations with GFP-specific (Fig. 5E, top panel) and TCV-specific (middle panel) probes. The intensity of GFP-specific signals in CP+TCV-GFP lanes clearly demonstrated that TCV-GFP RNA was assembled into virus particles. These results strongly suggest that in transgenic P38CHS plants, the transgenically expressed CP subunits were capable of assembling TCV-GFP RNA in trans to form virus particles, which in turn facilitated the efficient exit of the viral genome from vascular bundles.
The PVX-GFP study (24) established that tertiary vein junctions, but not primary and secondary veins (also known as class I and II veins), constitute the main exit points of PVX-GFP from vascular bundles. Consistent with this observation, we found that TCV-GFP, when restricted to primary and secondary veins (as in leaves of dcl2 dcl3 dcl4 plants), was unable to spread into leaf lamina (Fig. 5C). We consider that the inability of naked TCV-GFP RNA to move into smaller tertiary veins in dcl2 dcl3 dcl4 leaves was the primary cause of its restriction in the vascular tissues.
Taken together, our results strongly suggest that TCV CP functions in two distinct capacities to facilitate viral systemic spread. First, suppression of RNA silencing by TCV CP facilitates entry of viral RNA into the vascular bundles from which it reaches systemic leaves. This function of TCV CP can be complemented by disrupting the silencing machinery of the plant host. Second, TCV CP is also required for efficient unloading of the viral RNA from the vascular tissues into the mesophyll cells of the systemic leaves. This particular function cannot be compensated by a VSR from a different virus or by shutting down the plant RNA-silencing machinery and hence is independent of its silencing suppression function.
DISCUSSION
Besides being the sole building block of the viral capsid, TCV CP plays critical roles in TCV long-distance movement and serves as a strong suppressor of RNA silencing-based host defense. TCV mutants with deletions within the CP coding region move cell to cell in Arabidopsis but are unable to spread into uninoculated leaves (4, 12). Recently, similar CP deletion mutants were shown to gain systemic movement in Arabidopsis plants carrying mutations in key silencing pathway genes, hence establishing that the silencing suppressor activity of the CP was a critical prerequisite for TCV RNA to move into systemic leaves (5, 22). However, no attempts were made to determine where the mutant RNA was located in the systemic leaves. Conversely, Simon and colleagues reported that TCV mutants with a 2-aa extension at the N terminus of CP produced little or no stable virus in the infected cells and yet were able to infect silencing-competent wild-type plants systemically (30, 34). These reports all appear to support the conclusion that the silencing suppression function of TCV CP is the primary requirement for TCV systemic spread. What is not yet clear in these studies is whether these mutants exited from the vascular bundles into mesophyll or epidermal cells.
In the current report, we have reevaluated the role of TCV CP in viral systemic movement using mutant Arabidopsis plants that lack antiviral RNA silencing activity. We compared infections by TCV mutants that produce no CP, partial CPs, or full-length CPs with various capacities of silencing suppression and/or particle assembly ability, as well as by one mutant in which the viral CP was replaced by a VSR from a different virus. Our results permit the following conclusions. (i) The ability of virus mutants to invade the vasculature of systemic leaves is strongly correlated with their silencing suppression activity. Mutants that were unable to suppress RNA silencing could move only into systemic leaves in dcl2 dcl3 dcl4 plants that lacked effective antiviral silencing. (ii) In the absence of effective antiviral silencing (as in dcl2 dcl3 dcl4 plants), all mutant viruses that could assemble into virus particles showed symptoms indicative of efficient exit from the vascular tissues of systemic leaves. The actual concentration of assembled virus did not seem to contribute to the efficiency of exit. (iii) In the absence of effective antiviral silencing, all viral mutants that were incapable of virion assembly shared similar symptoms that were primarily vein centered, suggesting that they were restricted to the vascular tissues in the systemic leaves.
Our first observation reiterates the existence of the first barrier to TCV systemic movement, which likely lies at the point of entry into the vascular bundles. This barrier can be overcome by silencing suppressors encoded by TCV or another virus (TBSV) or by disrupting the host antiviral silencing machinery (5, 22). Virion formation is not needed to overcome this barrier, as exemplified by the presence of TCV-CPstop RNA in the systemic leaves of dcl2 dcl3 dcl4 plants. This barrier may not pose a physical restriction of TCV RNA. Rather, it likely reflects the activity of antiviral silencing that is gradually enhanced by increasing concentrations of siRNAs in the surrounding cells (both mesophyll and vascular) as the infection progresses. This view is supported by the report of Deleris et al. (5), which showed that a TCV mutant carrying a GFP insert was able to enter the phloem of the inoculated leaves of wild-type Arabidopsis plants when a higher dose of mutant RNA was applied to the leaves.
Our second observation concerns the apparent lack of correlation between virus symptoms and the quantity of virus particles in the systemic leaves. We noted that the CPA mutant produced fewer virions than CPF but caused more severe symptoms. The same is true for CPC and CPB in dcl2 dcl3 dcl4 plants. Interestingly, the mutant CPs seem to accumulate to similar levels in infected cells (Fig. 3F, bottom two panels; compare CPA with CPF, CPB with CPC). While an inverse relationship between virion concentration and symptoms cannot be firmly concluded here due to the limited number of mutants analyzed, it is clear that severity of symptoms is not strictly dependent on the level of virus accumulation in cells. We speculate that the vascular bundles may serve as a bottleneck for TCV systemic movement, limiting the number of trafficking virions. Similar bottlenecks have been shown to exist for Cucumber mosaic virus (CMV) in CMV-infected tobacco plants (11).
The third observation is most striking because it showed very clearly that the absence of virus particles severely restricted the exit of viral RNA from vascular bundles of systemically infected leaves. This is best illustrated by the different degree of complementation of TCV-GFP movement by dcl2 dcl3 dcl4 and P38CHS plants (Fig. 5). Our observation is not inconsistent with the results of Deleris et al. (5) showing that in dcl4 plants TCV-GFP was able to exit vascular bundles of petioles of inoculated leaves. The petiole vasculature could be structurally different from the vascular tissues in the systemically infected leaves at the top of plants, the latter being the subject of our study. The indispensability of virus assembly for efficient vascular egress identifies the second barrier in the process of viral systemic movement. A similar barrier has been noted by Chen and Citovsky (2) in tobacco leaves that restrict the vascular unloading of Tobacco mosaic virus (2). Additionally, the vein-centered symptoms caused by TCV-CPstop and other assembly-defective mutants are reminiscent of those seen on N. benthamiana plants infected with TBSV mutants lacking a functional CP (16, 18, 26). This suggests that a requirement for assembled virions at the step of vascular unloading may be more common than previously recognized.
The mechanism of restricted unloading of viral RNAs from plant vasculature awaits further investigation. The study by Chen and Citovsky (2) seems to implicate a specific host factor(s) in this process. However, it is not clear whether direct interactions between viral gene product(s) and host proteins are needed. Our result with the TCV-GFP mutant suggests that this restriction might be caused by the failure of unassembled RNA to move effectively in the progressively smaller vascular bundles, thus preventing it from reaching the tertiary vein junctions where efficient exit takes place (24). We showed that TCV-GFP rarely entered tertiary veins of systemic dcl2 dcl3 dcl4 leaves but did so efficiently in P38CHS leaves (Fig. 5C and D), which provided CP for particle assembly in trans. By extension, the inability of other CP-less TCV mutants to reach tertiary vein junctions likely accounts for their low efficiency of vascular exit.
In summary, our current study has successfully uncoupled CP silencing suppression from virion assembly. We further showed that the silencing suppression by TCV CP enabled the long-distance movement of viral RNA to the vascular bundles of the systemic leaves yet was insufficient to support the exit of viral RNA from the vascular tissues. Our current data favor the idea that assembled virions are needed for this final step of TCV systemic movement.
Acknowledgments
We thank James C. Carrington for providing the seed of dcl2 dlc3 dcl4 mutant plants, and O. Voinnet, D. Garcia, and P. Dunoyer for the P38CHS seed. We are very grateful for the comments and suggestions provided by the anonymous reviewers who reviewed the first version of the manuscript.
This study is supported by seed grants from OARDC and the Ohio Plant Biotechnology Consortium.
Footnotes
Published ahead of print on 26 May 2010.
REFERENCES
- 1.Carrington, J. C., T. J. Morris, P. G. Stockley, and S. C. Harrison. 1987. Structure and assembly of turnip crinkle virus. IV. Analysis of the coat protein gene and implications of the subunit primary structure. J. Mol. Biol. 194:265-276. [DOI] [PubMed] [Google Scholar]
- 2.Chen, M.-H., and V. Citovsky. 2003. Systemic movement of a tobamovirus requires host cell pectin methylesterase. Plant J. 35:386-392. [DOI] [PubMed] [Google Scholar]
- 3.Choi, C. W., F. Qu, T. Ren, X. Ye, and T. J. Morris. 2004. The RNA silencing suppressor function of Turnip crinkle virus coat protein cannot be attributed to its interaction with the Arabidopsis protein TIP. J. Gen. Virol. 85:3415-3420. [DOI] [PubMed] [Google Scholar]
- 4.Cohen, Y., A. Gisel, and P. C. Zambryski. 2000. Cell-to-cell and systemic movement of recombinant green fluorescent protein-tagged turnip crinkle viruses. Virology 273:258-266. [DOI] [PubMed] [Google Scholar]
- 5.Deleris, A., J. Gallego-Bartolome, J. Bao, K. D. Kasschau, J. C. Carrington, and O. Voinnet. 2006. Hierarchical action and inhibition of plant Dicer-like proteins in antiviral defense. Science 313:68-71. [DOI] [PubMed] [Google Scholar]
- 6.Diaz-Pendon, J. A., F. Li, W.-X. Li, and S.-W. Ding. 2007. Suppression of antiviral silencing by cucumber mosaic virus 2b protein in Arabidopsis is associated with drastically reduced accumulation of three classes of viral small interfering RNAs. Plant Cell 19:2053-2063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ding, S. W., and O. Voinnet. 2007. Antiviral immunity directed by small RNAs. Cell 130:413-426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dunoyer, P., C.-H. Lecellier, E. A. Parizotto, C. Himber, and O. Voinnet. 2004. Probing the microRNA and small interfering RNA pathways with virus-encoded suppressors of RNA silencing. Plant Cell 16:1235-1250. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 9.Kachroo, P., K. Yoshioka, J. Shah, H. K. Dooner, and D. F. Klessig. 2000. Resistance to turnip crinkle virus in Arabidopsis is regulated by two host genes and is salicylic acid dependent but NPR1, ethylene, and jasmonate independent. Plant Cell 12:677-690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lakatos, L., G. Szittya, D. Silhavy, and J. Burgyán. 2004. Molecular mechanism of RNA silencing suppression mediated by p19 protein of tombusviruses. EMBO J. 23:876-884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li, H., and M. J. Roossinck. 2004. Genetic bottlenecks reduce population variation in an experimental RNA virus population. J. Virol. 78:10582-10587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li, W.-Z., F. Qu, and T. J. Morris. 1998. Cell-to-cell movement of turnip crinkle virus is controlled by two small open reading frames that function in trans. Virology 244:405-416. [DOI] [PubMed] [Google Scholar]
- 13.Meng, C., J. Chen, J. Peng, and S.-M. Wong. 2006. Host-induced avirulence of hibiscus chlorotic ringspot virus mutants correlates with reduced gene-silencing suppression activity. J. Gen. Virol. 87:451-459. [DOI] [PubMed] [Google Scholar]
- 14.Merai, Z., Z. Kerenyi, S. Kertesz, M. Magna, L. Lakatos, and D. Silhavy. 2006. Double-stranded RNA binding may be a general plant viral strategy to suppress RNA silencing. J. Virol. 80:5747-5756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moissiard, G., and O. Voinnet. 2006. RNA silencing of host transcripts by cauliflower mosaic virus requires coordinated action of the four Arabidopsis Dicer-like proteins. Proc. Natl. Acad. Sci. U. S. A. 103:19593-19598. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 16.Pignatta, D., P. Kumar, M. Turina, A. Dandekar, and B. W. Falk. 2007. Quantitative analysis of efficient endogenous gene silencing in Nicotiana benthamiana plants using tomato bushy stunt virus vectors that retain the capsid protein gene. Mol. Plant Microbe Interact. 20:609-618. [DOI] [PubMed] [Google Scholar]
- 17.Powers, J. G., T. L. Sit, F. Qu, T. J. Morris, K.-H. Kim, and S. A. Lommel. 2008. A versatile assay for the identification of RNA silencing suppressors based on complementation of viral movement. Mol. Plant Microbe Interact. 21:879-890. [DOI] [PubMed] [Google Scholar]
- 18.Qu, F., and T. J. Morris. 2002. Efficient infection of Nicotiana benthamiana by tomato bushy stunt virus is facilitated by the coat protein and maintained by p19 through suppression of gene silencing. Mol. Plant Microbe Interact. 15:193-202. [DOI] [PubMed] [Google Scholar]
- 19.Qu, F., T. Ren, and T. J. Morris. 2003. The coat protein of turnip crinkle virus suppresses posttranscriptional gene silencing at an early initiation step. J. Virol. 77:511-522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Qu, F., and T. J. Morris. 2005. Suppressors of RNA silencing encoded by plant viruses and their role in viral infections. FEBS Lett. 579:5958-5964. [DOI] [PubMed] [Google Scholar]
- 21.Qu, F., and T. J. Morris. 2008. Carmovirus, p. 453-457. In B. W. J. Mahy and M. H. V. van Regenmortel (ed.), Encyclopedia of virology, 3rd ed. Academic Press, Oxford, England.
- 22.Qu, F., X. Ye, and T. J. Morris. 2008. Arabidopsis DRB4, AGO1, and AGO7 participate in a DCL4-initiated antiviral RNA silencing pathway that is negatively regulated by DCL1. Proc. Natl. Acad. Sci. U. S. A. 105:14732-14737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ren, T., F. Qu, and T. J. Morris. 2000. HRT gene function requires interaction between a NAC protein and viral capsid protein to confer resistance to Turnip crinkle virus. Plant Cell 12:1917-1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Roberts, A. G., S. Santa Cruz, I. M. Roberts, D. A. M. Prior, R. Turgeon, and K. J. Oparka. 1997. Phloem unloading in sink leaves of Nicotiana benthamiana: comparison of a fluorescent solute with a fluorescent virus. Plant Cell 9:1381-1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reference deleted.
- 26.Scholthof, H. B., T. J. Morris, and A. O. Jackson. 1993. The capsid protein gene of tomato bushy stunt virus is dispensable for systemic movement and can be replaced for localized expression of foreign gene. Mol. Plant Microbe Interact. 6:309-322. [Google Scholar]
- 27.Scholthof, H. B. 2006. The Tombusvirus-encoded P19: from irrelevance to elegance. Nat. Rev. Microbiol. 4:405-411. [DOI] [PubMed] [Google Scholar]
- 28.Simon, A. E., X. H. Li, J. Lew, R. Stange, C. Zhang, M. Polacco, and C. D. Carpenter. 1992. Susceptibility and resistance of Arabidopsis thaliana to turnip crinkle virus. Mol. Plant Microbe Interact. 5:496-503. [Google Scholar]
- 29.Voinnet, O., Y. M. Pinto, and D. C. Baulcombe. 1999. Suppression of gene silencing: a general strategy used by diverse DNA and RNA viruses of plants. Proc. Natl. Acad. Sci. U. S. A. 96:14147-14152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang, J., and A. E. Simon. 1999. Symptom attenuation by a satellite RNA in vivo is dependent on reduced levels of virus coat protein. Virology 259:234-245. [DOI] [PubMed] [Google Scholar]
- 31.Weigel, D., and J. Glazebrook. 2002. Arabidopsis: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N. Y.
- 32.Wu, B., J. Pogany, H. Na, B. L. Nicholson, P. D. Nagy, and K. A. White. 2009. A discontinuous RNA platform mediates RNA virus replication: building an integrated model for RNA-based regulation of viral processes. PLoS Pathog. 5:e1000323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang, F., and A. E. Simon. 2003. Enhanced viral pathogenesis associated with a virulent mutant virus or a virulent satellite RNA correlates with reduced virion accumulation and abundance of free coat protein. Virology 312:8-13. [DOI] [PubMed] [Google Scholar]
- 34.Zhang, F., and A. E. Simon. 2003. A novel procedure for the localization of viral RNAs in protoplasts and whole plants. Plant J. 35:665-673. [DOI] [PubMed] [Google Scholar]