Abstract
High-frequency recombination is a hallmark of HIV-1 replication. Recombination can occur between two members of the same subtype or between viruses from two different subtypes, generating intra- or intersubtype recombinants, respectively. Many intersubtype recombinants have been shown to circulate in human populations. We hypothesize that sequence diversity affects the emergence of viable recombinants by decreasing recombination events and reducing the ability of the recombinants to replicate. To test our hypothesis, we compared recombination between two viruses containing subtype B pol genes (B/B) and between viruses with pol genes from subtype B or F (B/F). Recombination events generated during a single cycle of infection without selection pressure on pol gene function were analyzed by single-genome sequencing. We found that recombination occurred slightly (∼30%) less frequently in B/F than in B/B viruses, and the overall distribution of crossover junctions in pol was similar for the two classes of recombinants. We then examined the emergence of recombinants in a multiple cycle assay, so that functional pol gene products were selected. We found that the emerging B/B recombinants had complex patterns, and the crossover junctions were distributed throughout the pol gene. In contrast, selected B/F recombinants had limited recombination patterns and restricted crossover junction distribution. These results provide evidence for the evolved coadapted sites in variants from different subtypes; these sites may be segregated by recombination events, causing the newly generated intersubtype recombinants to undergo purifying selection. Therefore, the ability of the recombinants to replicate is the major barrier for many of these viruses.
There are three groups of HIV-1: M, O, and N; most HIV-1 infections are caused by viruses from group M (34). Our current understanding is that group M viruses originated from a single zoonotic transmission event that introduced simian immunodeficiency virus of chimpanzees (SIVcpz) into the human population (46). Through adaptation and diversification, the transmitted virus evolved into a complex group of viruses including at least nine subtypes (A to D, F to H, J, and K) and six sub-subtypes (A1 to A4, F1, and F2) (45, 47). Based on existing phylogenetic lineage analyses, these aforementioned forms are thought to be pure subtypes, from which the intersubtype recombinant forms were derived (1, 40). Cocirculation of different subtypes in many areas of the world has led to the emergence of an increasing number of recombinant forms since the early stages of the HIV-1 pandemic (44). Many intersubtype recombinants have been identified multiple times, some of which are established and cause local or widespread epidemics; these recombinants are referred to as circulating recombinant forms (CRFs) (40). Many other recombinants have been identified only once and are referred to as unique recombinant forms. With increasingly powerful analysis tools and extensive genomic sequencing of patient samples, the impact of recombination on the HIV-1 pandemic has become more evident (36, 38). A conservative estimate places the global prevalence of recombinant forms at ∼20% of the total infections worldwide (21).
Each HIV-1 virion contains two copies of full-length viral RNA (11). During infection, reverse transcriptase can use part of each RNA as the template for DNA synthesis, generating a recombinant DNA copy (10, 22). Recombination could generate a chimera that is genotypically different from the two parents when the two copies of RNA in a particle are derived from different proviruses (heterozygous virion) (22). A prerequisite for the generation of recombinants is that the host must be infected by more than one parent virus; only cells containing different proviruses can generate heterozygous virions (12, 31). During the infection of target cells, recombination can occur to produce a chimeric progeny (10).
Multiple factors can affect the generation and emergence of recombinants in a viral population. We previously showed that the sequence identity of the dimerization initiation signal (DIS) plays an important role in the copackaging of RNA from different subtypes (7, 8, 35). RNAs containing different DIS sequences are packaged into the same particle less efficiently than those with the same DIS. In addition, sequence diversity can affect recombination events during reverse transcription; it has been shown that template switching occurs less frequently between sequences with reduced sequence similarity (3). Studies using env regions from different HIV-1 subtypes have shown a similar correlation between sequence similarity and recombination efficiency (4, 42). Furthermore, recombinants need to be replication competent to allow the virus to propagate and emerge from the population. Although the replication capacities of the recombinants have not been directly tested, examination of chimeric env genes indicates that some recombinant proteins have reduced function compared with their parents (42). Therefore, it is possible that some recombinants are not able to replicate efficiently and are eliminated during the propagation of the viral population.
We hypothesized that it would be more difficult for intersubtype recombinants to emerge from the viral population than for intrasubtype recombinants. Coadapted sites may have evolved throughout the viral genome, and these sites can be diverse in variants from different subtypes. During recombination, these coadapted sites may be segregated, causing loss of replication fitness in the newly generated viruses. Therefore, the sequence diversity between HIV-1 from different subtypes can present barriers in multiple aspects of the process. In this report, we sought to define the role of sequence diversity in the generation and emergence of intra- and intersubtype recombinants in the viral population. We first examined the frequency and distribution of crossovers in the HIV-1 pol gene between two subtype B sequences (B/B) and between one subtype B and one subtype F sequence (B/F). We then tested the emergence of HIV-1 recombinants during multiple replication cycles. Our results revealed that the sequence diversity between the subtype B and F viruses caused only a slight decrease in the recombination events in the pol region compared with the subtype B viruses. However, a major bottleneck occurred when the function of the chimeric pol gene was required, which limited the diversity of the intersubtype recombinants. These results indicate that the newly generated intersubtype recombinants underwent purifying selection when the function of the pol gene was selected.
MATERIALS AND METHODS
Plasmid constructs.
HIV-1-based vectors used in the current study were derived from previously described plasmids pON-fHIG, pON-H0, and pON-T6 (39). Briefly, pON-fHIG was derived from HIV-1 subtype B molecular clone NL4-3 (2) and contains most of the viral genome, with inactivating deletions in vif, vpr, vpu, and env. Additionally, two markers were inserted in place of the nef gene: the mouse heat-stable antigen gene (hsa) followed by an internal ribosome entry site (IRES) from encephalomyocarditis virus and the green fluorescent protein gene (gfp). Plasmid pON-H0 is identical to pON-fHIG except it contains an inactivating mutation in gfp that is 6 bp downstream of the start codon. Plasmid pON-T6 has a structure similar to pON-fHIG except that hsa was replaced with the mouse Thy1.2/CD90.2 gene (thy) and gfp has an inactivating mutation 603 bp downstream of the start codon.
To facilitate cloning of the pol region from other HIV-1 strains into NL4-3, an ApaI site in the IRES element and an NdeI site in the plasmid backbone were eliminated from pON-fHIG by restriction enzyme digestion followed by fill-in and ligation reactions (41) to generate pHG(BNL). The ApaI-NdeI fragment containing the entire pol coding region from p93BR020 (subtype F1) (18) and pHXBnPLAP-IRES-N+ (subtype B) (6) were inserted into ApaI-NdeI-digested pHG(BNL) to generate pHG(F) and pHG(BHXB), respectively. Plasmids p93BR020 and pHXBnPLAP-IRES-N+ were obtained from the National Institutes of Health AIDS Research and Reference Reagent Program. Using PCR mutagenesis and cloning of the PCR fragment, a D67N mutation that inactivates the integrase coding region of pol was introduced into pHG(BNL) to generate pHG(BNL)pi*. Similarly, a D25N mutation that inactivates the pro gene was introduced into pHG(F) and pHG(BHXB) to generate pHG(F)p*i and pHG(BHXB)p*i, respectively.
To generate the vectors used in the single-cycle assays, the NdeI-XhoI fragment containing the marker region from pON-H0 was inserted into NdeI-XhoI-digested pHG(BNL)pi* to generate pH0(BNL)pi*. A similar NdeI-XhoI fragment from pON-T6 was inserted into NdeI-XhoI-digested pHG(F)p*i and pHG(BHXB)p*i to generate pT6(F)p*i and pT6(BHXB)p*i, respectively.
To facilitate cloning, a silent mutation was introduced into the env gene of pNL4-3 to remove an NdeI site. The NdeI-XhoI fragment containing vif, vpr, vpu, and env from this modified NL4-3 was isolated and used to replace the counterpart DNA fragments from pHG(BNL)pi*, pHG(F)p*i, and pHG(BHXB)p*i to generate vectors used in the multiple-round assays, namely, pRC(BNL)pi*, pRC(F)p*i, and pRC(BHXB)p*i, respectively.
The general structure of all plasmids was mapped by restriction enzyme digestion. In addition, to ensure the absence of inadvertent mutations introduced by PCR amplification, all plasmid DNA fragments derived from PCR were verified by sequencing.
Cells, transfections, and infections.
The modified human embryonic kidney cell lines 293T (14) and 293CC (33) were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum, penicillin (50 U/ml), and streptomycin (50 μg/ml). Cell line 293CC, a kind gift from Robert Doms, expresses human CD4 and CCR5. The human T-cell line Hut/CCR5, a Hut78 derivative that expresses CCR5 (49), was maintained in Roswell Park Memorial Institute 1640 medium supplemented with 10% fetal calf serum, penicillin (50 U/ml), streptomycin (50 μg/ml), puromycin (1 μg/ml), and G418 (500 μg/ml). All cultured cells were maintained at 37°C in humidified incubators with 5% CO2.
DNA transfections were performed using the TransIT-LT1 transfection reagent (Mirus) according to the manufacturer's recommendations. Briefly, 293T cells were plated at a density of 1 × 106 to 3 × 106 cells per 100-mm-diameter dish and transfected 24 h later with 10 to 15 μg of DNA. Viral supernatants were harvested 48 h posttransfection, clarified through a 0.45-μm filter to remove cellular debris, and used immediately or stored at −80°C prior to infection. For infection, 293T and Hut/CCR5 cells were plated at a density of 1 × 106 cells per 100-mm-diameter dish or 25-ml flask, respectively, and infected 24 h later. Serial dilutions were generated from each viral stock and used for infection. When required, spinoculation of Hut/CCR5 cells was performed in six-well plates at 1,200 × g at 24°C for 1.5 h in the presence of Polybrene. Cells were washed three times with phosphate-buffered saline 12 h postinfection and fresh medium was added; infected cells were processed and analyzed by flow cytometry 48 to 72 h postinfection.
Single-cycle recombination assay.
Dually infected producer cells were generated as previously described to examine recombination in a single round of HIV-1 replication (8, 39). Briefly, pseudotyped particles containing H0(BNL)pi* RNAs were generated by transfecting 293T cells with vector construct, pHCMV-G (51), and pSYNGP (28). Plasmid pHCMV-G expresses vesicular stomatitis virus envelope glycoprotein G (VSV-G), and pSYNGP expresses a codon-optimized HIV-1 gag-pol gene. Viral particles were used to infect 293T cells at a low multiplicity of infection (MOI), generally between 0.1 and 0.05. Infected cells expressing the HSA marker were enriched by cell sorting and subsequently infected at a low MOI (0.05 to 0.1) with pseudotyped viruses containing either the T6(F)p*i or T6(BHXB)p*i genome. Dually infected cells were enriched by cell sorting until >95% of the cells expressed both HSA and Thy markers.
To generate infectious HIV-1 particles, dually infected producer cells were transfected with pSYNGP and pIIINL(AD8) (12), which expresses CCR5-tropic HIV-1 Env. Viruses were harvested and used to infect Hut78/CCR5 cells at low MOIs (0.05 to 0.1); infected cells with GFP+ phenotypes were enriched by cell sorting. Viral particles produced by the sorted GFP+ cell pools were harvested, and cellular debris was removed by filtration; viral RNAs were isolated from these supernatants by using the QIAquick viral RNA minikit (Qiagen).
Multiple-cycle recombination assay.
Human 293T or 293CC cells were plated in six-well plates 24 h prior to transfection at 5 × 105 or 8 × 105 cells per well, respectively. An HIV-1 genome-containing plasmid and a plasmid expressing GFP or red fluorescent protein (RFP) were cotransfected into 293 cells, whereas 293CC cells were transfected with pSYNGP. Transfected cells were collected 24 h later, counted, and used for cell fusion experiments. To produce dually infected cells, 7.5 × 105 293T cells transfected with pRC(BNL)pi*, pRC(F)p*i, or pRC(BHXB)p*i were mixed with 5 × 105 293CC cells transfected with pSYNGP in the presence of roscovitine (50 mM), which prevents nuclear fusion (5). After incubating the cell mixtures for 16 h, the efficiency of cell fusion was assessed by fluorescence microscopy, and viral supernatants were collected, filtered, and used to infect Hut/CCR5 cells by spinoculation. Virus was harvested from infected Hut/CCR5 cells 4 days later and clarified by filtration; supernatants were used to infect fresh Hut/CCR5 to start a five-passage infection cycle. For each passage, viruses were collected and filtered 4 to 5 days postinfection, a portion of the viral supernatant was used to infect cells, and another portion was used for viral RNA isolation.
Flow cytometry analysis.
Infected cells were stained with phycoerythrin-conjugated anti-HSA antibody (BD Biosciences) and allophycocyanin-conjugated anti-Thy-1.2 antibody (eBioscience) and fixed with paraformaldehyde (2%, final concentration) prior to flow cytometry analysis. Flow cytometry analysis was performed using a FACSCalibur apparatus (BD Biosciences), whereas cell sorting was performed on a FACSVantage SE system with the FACSDiVa digital option (BD Biosciences). Generally, all the cell lines were cell pools containing at least 10,000 independent infection events. Results obtained from flow cytometry analysis were analyzed using FlowJo software (Tree Star).
SGS assay and DNA sequence analyses.
Viral RNA genomes were analyzed using a previously described limiting dilution assay termed single-genome sequencing (SGS) (26, 37). Briefly, viral RNA was reverse transcribed to cDNA and serially diluted, and a region encompassing the entire pol gene was amplified by PCR. To ensure that only the sequence from a single RNA genome was analyzed, end point dilutions in which less than 30% of the PCRs yielded positive signals were selected and these PCR products were sequenced directly. Alignments of the assembled viral sequences and the reference parental sequences were obtained using ClustalX 2.0 (29) and manually adjusted with Bioedit 7.0.9 (20). The crossover junctions, defined as regions of homology between two polymorphisms that correlate to different parental proviruses, were identified visually in the sequence alignment and recorded into a database.
Analyses for correlation between crossover distribution and sequence homology in a single HIV-1 replication cycle.
Each 50-nucleotide (nt) region was designated in a high or low sequence homology category based on comparison with the average sequence homology of the entire 2.5-kb target region. Similarly, each region was also classified as having a high or low recombination rate based on comparison with the average recombination frequency of the entire target sequence. We then compared the proportions of the high or low homology region groups that had a high or low recombination rate, and we did not observe statistically significant differences (P = 1.0 for both B/B and B/F; Fisher's exact test).
RESULTS
System used to examine recombination events occurring in one round of HIV-1 replication.
We sought to delineate the potential of and barriers for HIV-1 recombinants to emerge from a viral population; specifically, we wished to determine the effects of sequence divergence on the generation of intra- and intersubtype recombinants and on the replication fitness of these variants. To approach this experimental question, we first compared the effects of sequence variation on the generation of intra- and intersubtype recombinants during one round of HIV-1 replication. Although recombination can occur during reverse transcription in all HIV-1-infected cells, only heterozygous virions can generate genotypically different recombinants. A progeny that appears to be genetically identical to one of the parents could be generated from homozygous virions or from heterozygous virions that did not undergo recombination. To ensure that progeny from heterozygous virions were analyzed, we studied proviruses with known recombination events in a marker gene. We modified parts of the viral genome to contain sequences derived from different strains of HIV-1 and adapted a previously described recombination system that uses the reconstitution of a functional gfp to identify the progeny generated from heterozygous viruses (8, 39). HIV-1 constructs were derived from the subtype B molecular clone NL4-3, which contains the near-full-length viral genome with inactivating deletions in vif, vpr, vpu, and env and two marker genes inserted in the nef reading frame (Fig. 1A). The entire viral sequence in H0(BNL)pi* was derived from NL4-3, and an inactivating mutation (D67N) was introduced into the integrase coding region to abolish integrase function (15); additionally, H0(BNL)pi* encodes the mouse hsa gene and an inactivated gfp gene containing a frameshift mutation near the start codon of the gfp reading frame. The pol regions of T6(BHXB)p*i and T6(F)p*i were derived from subtype B molecular clone HXB-2 and subtype F molecular clone p93BR020, respectively. An inactivating mutation, D25N, was introduced into the pro gene of both vectors to render the protease inactive (50). Both T6(BHXB)p*i and T6(F)p*i vectors encode a mouse thy1.2 gene and an inactivated gfp gene with the frameshift mutation 0.6 kb downstream of the gfp start codon (10, 26).
FIG. 1.
General structures of the viruses used in single-cycle (A) and multiple-cycle (B) assays. The NL4-3-based structural backbones used to construct the viruses are shown at the top of each panel. The sequences of the pol gene derived from NL4-3, HXB-2, and F1 molecular clones are shown in black, gray, and white boxes, respectively. The pro gene in BNL viruses contains the inactivating mutation D25N, whereas the in gene in BHXB and F contains the inactivating mutation D64N. Viruses used in the single-cycle experiments contain inactivating deletions in vif, vpr, vpu, and env and encode two markers in the nef reading frame, a surface protein gene and a mutant gfp. Inactivating mutations are denoted by asterisks.
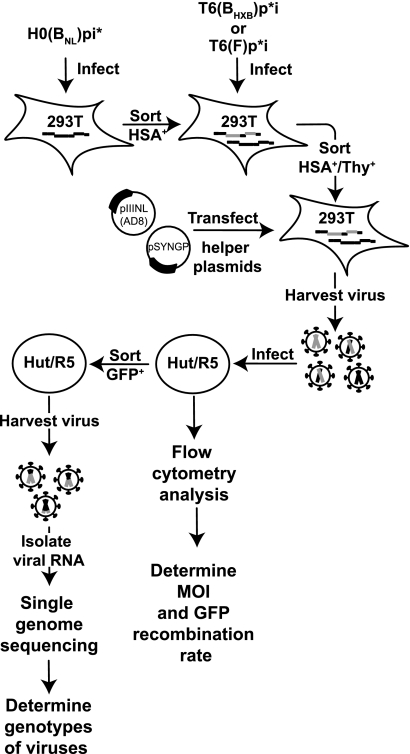
The general outline for studying recombination occurring in one round of HIV-1 replication is illustrated in Fig. 2. HIV-1 constructs described in Fig. 1A lack functional env gene products and one of the pol gene products (IN or PR); to generate infectious viruses, each HIV-1 construct was transfected into 293T cells along with two helper plasmids, pSYNGP and pHCMV-G, which express functional gag-pol and VSV- G protein, respectively. H0(BNL)pi* viruses were used to infect fresh 293T cells at a low MOI, and the resulting HSA+ cells were enriched by cell sorting; these cells were then infected at a low MOI with a second virus, either T6(BHXB)p*i or T6(F)p*i. Cells expressing both HSA and Thy markers were enriched by cell sorting until HSA+/Thy+ cells exceeded 95% of the population. A total of four cell lines was produced; two independent cell lines were generated for each parental virus pair. To study recombination in one round of replication, these producer cells were transfected with two helper constructs, pSYNGP and pIIINL(AD8), which expresses CCR5-tropic HIV-1 Env. Viruses were harvested and used to infect Hut/CCR5 target cells, and the infected target cells were processed and analyzed by flow cytometry. Infected cells were detected by HSA or Thy expression, and the proportion of HSA+ or Thy+ cells was used to calculate the MOI of the two parental viruses. None of the vectors encodes a functional gfp gene; however, recombination can occur during reverse transcription between the two copackaged RNAs to reconstitute a functional gfp. Therefore, the GFP+ phenotype was used to detect recombination. The GFP recombination rate, measured by the proportion of GFP+ cells in the infected cells (detected by HSA or Thy expression), is also an indicator of how frequently RNAs derived from two different proviruses are copackaged.
FIG. 2.
Protocol used to measure HIV-1 recombination in a single-cycle assay. Briefly, viruses containing the H0(BNL)pi* RNA were used to infect 293 cells at low MOI, and the resulting HSA+ cells were enriched by sorting. HSA+ cells were infected with viruses containing either T6(BHXB)p*i or T6(F)p*I at low MOI, and HSA+/Thy+ cells were enriched by cell sorting. Helper constructs pSYNGP and pIIINL(AD8), which expressed HIV-1 Gag/Gag-Pol and Env, respectively, were transfected into HSA+/Thy+ cell lines; viruses were harvested and used to infect fresh Hut/R5 cells. A portion of the infected Hut cells was analyzed by flow cytometry to determine the number of cells that expressed HSA, Thy, or GFP. These results were used to calculate the infection MOI and GFP recombination rate. Another portion of the infected cells was sorted to enrich GFP+ cells; viruses were harvested from GFP+ cells, and RNA was isolated and analyzed by single-genome sequencing to determine the genotypes of the viruses.
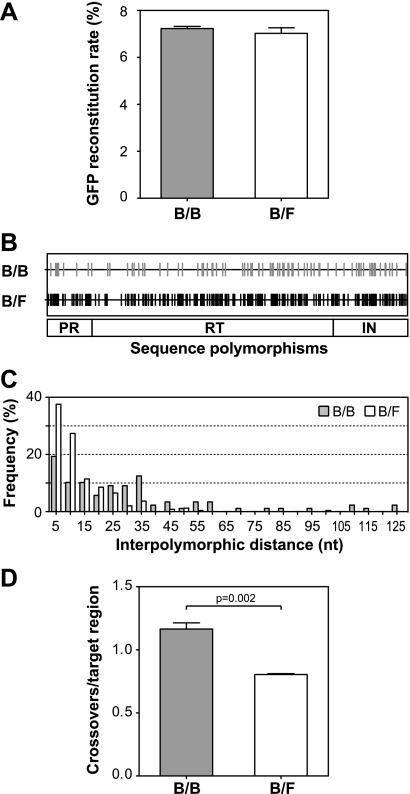
The efficiencies of GFP reconstitution from viruses generated from cell lines containing H0(BNL)pi* and T6(BHXB)p*i proviruses or H0(BNL)pi* and T6(F)p*i proviruses in four experiments are summarized in Fig. 3A. GFP was reconstituted at 7.2 ± 0.1% for H0(BNL)pi* and T6(BHXB)p*i, and 7.0 ± 0.2% for H0(BNL)pi* and T6(F)p*i (means ± standard errors); these two rates were not significantly different (P = 0.45; t test) and were similar to the frequency we had previously observed between the same two gfp markers when we used two viruses in which the viral sequences were derived from NL4-3 (6.9%) (39). These results indicate that the sequence differences in the pol gene of these constructs did not significantly interfere with copackaging efficiency; H0(BNL)pi* RNA was copackaged efficiently with either T6(BHXB)p*i RNA or T6(F)p*i RNA. Furthermore, the frequency of recombination in the gfp region was not affected by the sequence divergence in pol.
FIG. 3.
Recombination detected in a single HIV-1 replication cycle. (A) Rate of GFP reconstitution via HIV-1 recombination in infected cells. (B) Locations of the sequence polymorphisms in the target pol gene. Gray and black vertical lines indicate the positions of the polymorphisms in pol. (C) Distribution of distances between polymorphic sites in the target pol gene. The x axis shows the nucleotide distance between the polymorphisms; each bin represents distance increments of 5 nt. The y axis shows the number of interpolymorphic regions included in each bin. (D) Crossover events detected in the HIV-1 pol gene. B/B refers to recombination between H0(BNL)pi* and T6(BHXB)p*i, whereas B/F refers to that between H0(BNL)pi* and T(F)p*i. Recombination measurements are summarized from three independent experiments; error bars indicate standard errors.
Although the aforementioned three HIV-1 constructs contain almost identical gfp genes, their pol genes are diverse and contain multiple polymorphic sites. The distribution of the polymorphisms in pol between H0(BNL)pi* and T6(BHXB)p*i (gray line) or H0(BNL)pi* and T6(F)p*i (black line) is shown in Fig. 3B. In the 2.5-kb region, there are 88 polymorphic markers between the two subtype B sequences and 245 polymorphic markers between the NL4-3 and subtype F strains, generating 96.5% and 90.2% sequence identities, respectively, in these target areas. In both sets of sequences, the polymorphic sites are distributed throughout the target region. The distribution of distances between polymorphic sites is shown in Fig. 3C. The average and median of the distance distribution are 28 nt and 21 nt, respectively, for the two subtype B sequences and 10 nt and 7 nt, respectively, for the NL4-3 and subtype F sequences.
To delineate the effects of sequence divergence on the crossover events in the pol region, we analyzed viral genomes generated from heterozygous virions containing H0(BNL)pi* and T6(BHXB)p*i RNAs or H0(BNL)pi* and T6(F)p*i RNAs. For simplicity, events between H0(BNL)pi* and T6(BHXB)p*i are referred to as B/B recombination, as the pol regions of these constructs were from two different subtype B molecular clones, whereas events between H0(BNL)pi* and T6(F)p*i are referred to as B/F recombination. The experimental protocol we used is illustrated in the bottom portion of Fig. 2. To identify the recombination events in the pol region, we examined progeny derived from heterozygous viruses after one round of replication. GFP+ target cell pools were enriched by cell sorting (Fig. 2): two cell pools for B/B and two pools for B/F recombination. Because the parental vectors encode functional gag genes but not functional env genes, these cell pools generate noninfectious virus-like particles that contain viral RNA. We harvested virus-like particles from these cell pools and analyzed the genotypes of the viral RNA by SGS. Briefly, SGS is a limiting dilution amplification assay; viral RNA was reverse transcribed, serially diluted, and amplified by PCR. To ensure that the recombination events studied in these assays originated during HIV-1 replication, not from DNA recombination that occurred during PCR, we analyzed products generated from the end point dilution in which an average of 0.3 copies of cDNA template per amplification reaction mixture was used. It was previously shown that DNA recombination is very infrequent in SGS (0 events in 50 studied reactions) (37).
Using SGS, we amplified and sequenced the 2.5-kb region that encompasses most of the pol gene from multiple viruses. Sequences obtained by SGS were aligned with sequences of the parental strains, and recombinant regions were identified using polymorphic markers. Viral RNAs derived from two independent experiments were analyzed for B/B recombination, and a total of 73 genomes were analyzed; similarly, 121 genomes derived from two independent experiments were examined for B/F recombination. As summarized in Fig. 3D, on average, 1.2 crossovers were observed in the 2.5-kb target region in B/B recombinants, compared with 0.8 crossovers/target region in B/F recombinants. Therefore, B/F recombination occurs slightly (∼30%) less frequently than B/B recombination (P = 0.002; t test). As the copackaging of these two sets of parental viruses was equally efficient based on the similar GFP reconstitution rates (Fig. 3A), these results indicate that the increased sequence diversity is responsible for the reduction of crossover events in pol.
Distribution of crossover junctions in B/B and B/F recombinants generated in one round of HIV-1 replication.
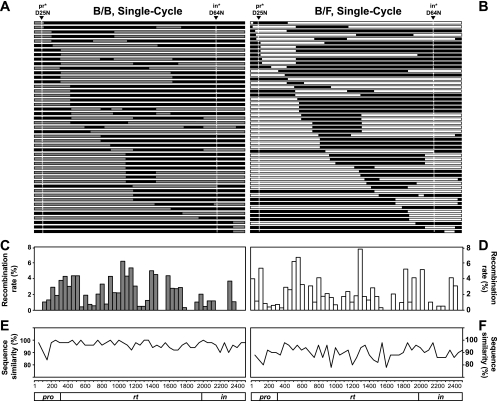
The recombination data obtained from SGS and sequence alignment were recorded in a relational database and used to build recombination maps (Fig. 4 A and B). These results revealed that both B/B and B/F recombinants have highly heterogeneous structures; very few recombinant patterns were identified more than once in these experiments. To further analyze the distribution of the crossover points, we calculated the frequencies of recombination occurring in each interpolymorphic region and normalized these values to 50-nt regions. Specifically, the number of crossover events for each interpolymorphic region was tabulated and divided by the total number of nucleotides in the region to generate a single-nucleotide crossover value. The number of crossover events in a given 50-nt region was calculated by combining the value of each single nucleotide encompassed in the region. The number of crossover events in each region was divided by the total crossover events detected in the 2.5-kb target region to generate the frequency of recombination in that region; the resulting distribution graphs are shown in Fig. 4C and D. Although there are fluctuations along different parts of the 2.5-kb target region, crossover junctions are distributed throughout the pol gene and are evident in most of the 50-nt regions (40 of the 50 regions in B/B and 42 of the 50 regions in B/F). These fluctuations are not directly linked to the variation of sequence similarity, at least in the range included in the target sequences (P = 1.0 for intra- and intersubtype recombination).
FIG. 4.
Recombination in the pol region detected in a single-cycle assay. (A and B) General structures of the recombinant target region in B/B (A) and B/F (B) recombination. Areas marked in black, gray, and white indicate sequences that originated from NL4-3, HXB-2, and F1, respectively. Arrowheads and vertical lines show the positions of the inactivating mutations: D25N in the pro gene and D64N in the in coding region. (C and D) Distribution of the crossover junctions in the pol region in B/B (C) and B/F (D) recombinants. The x axis shows the position in the target region; the y axis shows the percent crossover frequency in each 50-nt region. (E and F) Sequence similarities between the B/B (E) and B/F (F) target sequences. The y axis shows the percent sequence similarity.
System used to examine emergence of recombinants during multiple rounds of HIV-1 replication.
Both B/B and B/F recombinants generated from one round of HIV-1 replication revealed frequent crossovers in the pol region. We therefore examined B/B and B/F recombinants that emerged from multiple rounds of HIV-1 replication. Three constructs encoding full-length HIV-1 genomes were generated (Fig. 1B) so that they contained the same gag-pol sequences as those used in the single-round recombination assay. All three full-length constructs contain functional viral genes, including gag, env, vif, vpr, tat, rev, vpu, and nef. Each construct has an inactivating mutation in one pol gene product, generating an inactivated in gene region in RC(BNL)pi*and an inactivated pro gene in RC(BHXB)p*i and RC(F)p*i. As a result, none of the parental viruses can produce infectious particles. However, a crossover between the inactivating pr and in mutations can reconstitute a functional pol gene, allowing the resulting recombinants to generate infectious virions. As more than one round of replication occurs in this system, recombinants with functional pol genes are expected to be selected. To avoid potential DNA recombination occurring during cotransfection of HIV-1 constructs, we used a fusion assay to generate infectious, heterozygous virions.
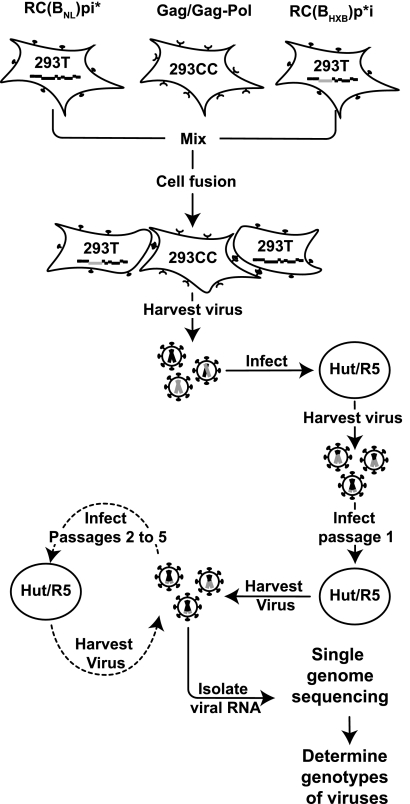
Our protocol for generating and selecting recombinants is summarized in Fig. 5; for simplicity, B/B recombination was used as an example. We first generated 293T cells expressing RC(BNL)pi* or RC(BHXB)p*i separately by transfection. We also generated 293CC cells expressing functional HIV-1 Gag/Gag-Pol by transfecting the cells with pSYNGP; 293CC is a modified 293T cell line expressing the HIV-1 receptor and coreceptor. None of these three types of cells can generate infectious HIV-1 particles on its own. However, when mixed together, HIV-1 Env receptor/coreceptor-mediated cell fusion can occur; these fused cells can express two different viral RNAs and functional viral proteins required to produce infectious HIV-1 particles, including heterozygous virions. Viruses were harvested from cell fusion reaction mixtures and used to infect Hut/CCR5 cells; this step is referred to as passage zero. During reverse transcription, recombination can occur to generate a DNA with a functional pol gene; proviruses derived from this DNA can generate infectious particles. However, it is expected that many proviruses in passage zero Hut/CCR5 cells would not contain reconstituted functional pol and would therefore generate only noninfectious particles. Viruses produced by passage zero cells were used to infect fresh target cells. After 48 h, an aliquot of the viruses was harvested from the infected cells, and these viruses were denoted as passage 1 viruses and were used for SGS analysis; the rest of the cells were cultivated for another 2 to 3 days to allow further virus replication. Viruses were harvested at the end of the cultivation and used to infect fresh Hut/CCR5 target cells to initiate passage 2 infection. A total of five passages were performed, and viral RNAs from passage 5 were also analyzed by SGS.
FIG. 5.
Protocol used to study the emergence of HIV-1 recombinants in a multiple-cycle assay. Briefly, 293 T cells were transfected with either the integrase mutant [RC(BNL)pi*] or protease mutant [RC(BHXB)p*i], whereas 293CC, 293T cells expressing CD4 and CCR5, were transfected with the Gag/Gag-Pol-expressing helper construct. Upon mixing these three types of cells, the HIV-1 Env-CD4-CCR5 interaction could mediate cell fusion and generate cells expressing an integrase mutant, protease mutant, and helper. Viruses were harvested from these fused cells and used to infect Hut/CCR5 (Hut/R5) cells, from which viruses were harvested and designated passage 0 virus. Passage 0 virus was used to infect fresh Hut/CCR5 cells (passage 1), from which passage 1 virus was harvested. A portion of the virus was used to infect fresh Hut/CCR5 cells (passage 2), and another portion of the virus was used to isolate viral RNA that was used for single-genome sequencing to determine the genotypes of the viruses. These steps were repeated to continue to propagate viruses to passage 5. The same protocol was used to examine recombinants between RC(BNL)pi* and RC(F)p*i.
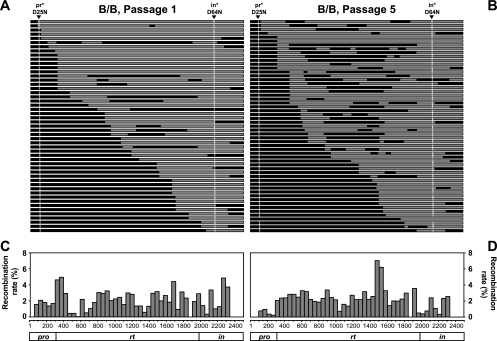
RNAs were isolated from two independent experiments to examine B/B recombination; 51 genomes from passage 1 viruses and 55 genomes from passage 5 viruses were analyzed by SGS. All of the 106 genomes analyzed were recombinants, and the general structures of these recombinants are summarized in Fig. 6 A and B. As expected, most of the recombinants contained the two wild-type markers that restored a functional pol gene, sequences encoding functional PR and IN from RC(BNL)pi*and RC(BHXB)p*i, respectively. A total of 91 crossover events were identified in the 51 analyzed passage 1 genomes; the crossover junctions are distributed throughout the pol gene and are evident in 46 of the 50 regions (Fig. 6C). The crossover patterns of recombinants at passage 5 are more complex than those at passage 1; 172 crossover junctions were identified in 55 genomes, indicating that recombination continued to reshuffle viral genomes between passage 1 and passage 5. Similar to those from passage 1, crossover junctions from passage 5 recombinants are distributed throughout the target sequences; 45 of the 50 regions have crossover events. These results indicate that a large number of recombinants with many different patterns can reconstitute functional Pol chimeras and many of these viruses can continue to replicate.
FIG. 6.
Intrasubtype recombinants identified in the multiple-cycle assay. (A and B) General structures of the target region in recombinants from passage 1 (A) and passage 5 (B). (C and D) Distribution of the crossover junctions in the viral genome in passage 1 (C) and passage 5 (D). Symbols and abbreviations are the same as in Fig. 4.
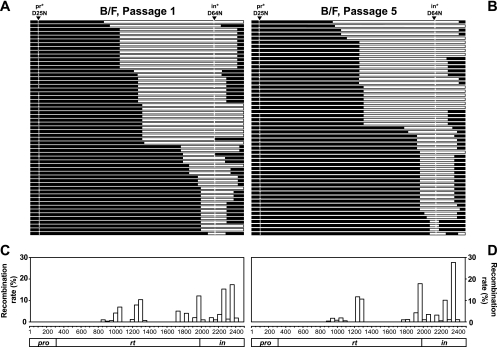
Using the same approach, we also analyzed passage 1 and passage 5 B/F genomes in two experiments. Of the 57 genomes examined from passage 1 viruses, all were recombinants and most had the wild-type active site sequences in PR and IN (Fig. 7A). Analyses of the locations of the crossover junctions revealed that the 99 crossover junctions were not distributed throughout the pol gene but were clustered in 21 of the 50 regions (Fig. 7C). This distribution is in sharp contrast with that of B/F recombination in the single-cycle assay, in which 42 of the 50 regions had crossover events (Fig. 4B). These results revealed that although recombination can occur throughout the pol gene in B/F recombination, the selection for Pol function creates a genetic bottleneck for the B/F recombinants. Viral genomes from passage 5 were also examined; in the 53 genomes, we observed 94 crossover events. These results show no apparent further shuffling of the viral genomes from passage 1 to passage 5 and are again different from those observed in B/B recombination.
FIG. 7.
Recombinants containing subtype B and F sequences identified in the multiple-cycle assay. (A and B) General structure of the target region in recombinants from passage 1 (A) and passage 5 (B). (C and D) Distribution of the crossover junctions in the viral genome in passage 1 (C) and passage 5 (D). Symbols and abbreviations are the same as in Fig. 4.
Taken together, our results indicate that recombination generates complex patterns in both B/B and B/F recombinants in a single-cycle assay that does not select for functional Pol. However, the requirement for Pol function creates a severe genetic bottleneck for B/F recombinants but not B/B recombinants, and the force of the purifying selection results in the observed decrease in diversity in the B/F recombinant population in the multiple-cycle assay.
DISCUSSION
Recombination allows rapid sorting of genetic information in the HIV-1 genome to facilitate the generation of diversity in the viral population within a short time span (32). It has been shown that recombination allows the assortment of different drug resistance mutations into the same genome to generate multidrug-resistant HIV-1 (19, 47). Recombination has also been shown to be able to change viral genomes to escape the cytotoxic T-lymphocyte response (38). Many aspects of HIV-1 recombination have been studied, such as the frequency and mechanisms of recombination (16, 17, 23, 24). However, for the most part, the fate of the newly generated recombinants has remained unexplored. In this report, we directly compared the recombinants generated in the absence of selection pressure with those that survived multiple rounds of HIV-1 replication. We found that a drastic genetic bottleneck occurs during viral replication to prevent the emergence of many newly generated intersubtype recombinants in the viral population. These results reveal that viral replication capacity or fitness is a strong selective force that can eliminate many of the new intersubtype recombinants.
Intersubtype recombinant forms are being increasingly isolated from patient samples worldwide (44). This increase is probably due to the expansion of geographic areas in which different HIV-1 subtypes cocirculate, whereas such cocirculation used to be limited mostly to central Africa. For example, the circulation of non-B subtypes is increasing in most Western countries that were previously limited to subtype B infections (34). Almost every possible combination of subtypes has been identified in mosaic forms, including recombinants involving other recombinants and CRFs (30). Our results imply that many newly generated recombinants may be eliminated during viral replication. These findings suggest that intersubtype recombinants can be generated more frequently and with greater diversity than the currently identified recombinant forms; however, most of these recombinants are short lived and are lost during the selection process. Although intersubtype recombinants are well documented in clinical isolates (9, 25, 43), the intrasubtype recombinants are reported less frequently. The high sequence similarity makes it very difficult to identify coinfection and the subsequent intrasubtype recombination events in circulating strains; therefore, most of these events remain undetected. In our experimental system, it is easy to identify intrasubtype recombinants, because we know the precise sequence of the two parental viruses. The results summarized in Fig. 4, 6, and 7 indicate that intrasubtype recombination occurs more frequently; furthermore, newly generated recombinants are more likely to be functional and survive the selection pressure. With the assumption that coinfection of variants from the same subtype occurs as frequently, if not more frequently, than coinfection of variants from different subtypes, the number of intrasubtype recombinants currently in circulation should be much higher than the reported number of intersubtype recombinants. Similarly, our results also suggest that intrasubtype recombination is more likely to contribute to the generation of variants that escape environmental selection pressure, such as drug treatment and the host immune response.
Previous work showed that many chimeric Env-containing sequences from different HIV-1 subtypes generated lower titers when used to pseudotype viruses (42). In our experiments, when Pol function was selected, we observed a drastic drop in B/F recombinant diversity (Fig. 7). These findings support the hypothesis that there is coadaptation of sequences in the viral genome; many of the intersubtype recombinants disrupted the linkages of the coadapted sequences, causing a decrease in viral replication fitness. The pol gene products of the two molecular clones used show 90% amino acid sequence identity. This result raises the possibility that the chimeric Pol proteins do not work well together because of the change in amino acids. These changes most likely result from genetic drift, leading to the accumulation of slightly deleterious mutations that are then corrected by epistatic mutations elsewhere in the protein. Intersubtype recombinants in which epistatic pairs of mutations are separated from one another will thus have reduced fitness relative to either parent. It has also been shown that the polymorphism in the reverse transcriptase of the CRF-01_AE causes lower RNase H activity than that of NL4-3 subtype B viruses (13). Therefore, there may be inherent differences among proteins encoded by different subtypes. Alternatively, sequences at the nucleotide level might affect virus replication capacity. A recent RNA folding model proposed that HIV-1 RNA is highly structured near the boundaries of different domains of the gag and pol genes (48), suggesting that the complex RNA structure in these areas can slow down translation to allow the proper folding of the newly translated protein domain. It is possible that the RNA structures of the recombinant sequences are disrupted, resulting in reduced viral replication. Replacing the pol gene sequence with that from a codon-optimized expression construct causes a delay in HIV-1 replication (27). These studies suggest that a change in the primary sequence can affect the replication capacity of a virus. Further experiments will be needed to distinguish whether the change in amino acid sequence or RNA sequence, or the combination of both, affects the replication of intersubtype recombinants.
Recombination is an important mechanism in the generation of HIV-1 diversity. The increasing genetic complexity of the circulating strains presents a significant challenge for the development of broadly protective vaccines and effective antiviral treatments. Understanding the potential of recombination in assorting genomes and the barriers to the emergence of recombinants will provide insight into the causes and limitations of this process. Our knowledge of the forces that control selection and fixation of recombinant forms will allow a better comprehension of the dynamics of HIV-1 diversity in the pandemic.
Acknowledgments
We thank Anne Arthur for her expert editorial help.
This work was supported in part by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research. A.G. was supported in part by a predoctoral fellowship from University of Milan. J.M.C. was a Research Professor of the American Cancer Society, with support from the F.M. Kirby Foundation.
Footnotes
Published ahead of print on 26 May 2010.
The authors have paid a fee to allow immediate free access to this article.
REFERENCES
- 1.Abecasis, A. B., P. Lemey, N. Vidal, T. de Oliveira, M. Peeters, R. Camacho, B. Shapiro, A. Rambaut, and A. M. Vandamme. 2007. Recombination confounds the early evolutionary history of human immunodeficiency virus type 1: subtype G is a circulating recombinant form. J. Virol. 81:8543-8551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adachi, A., H. E. Gendelman, S. Koenig, T. Folks, R. Willey, A. Rabson, and M. A. Martin. 1986. Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J. Virol. 59:284-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.An, W., and A. Telesnitsky. 2002. Effects of varying sequence similarity on the frequency of repeat deletion during reverse transcription of a human immunodeficiency virus type 1 vector. J. Virol. 76:7897-7902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baird, H. A., Y. Gao, R. Galetto, M. Lalonde, R. M. Anthony, V. Giacomoni, M. Abreha, J. J. Destefano, M. Negroni, and E. J. Arts. 2006. Influence of sequence identity and unique breakpoints on the frequency of intersubtype HIV-1 recombination. Retrovirology 3:91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Castedo, M., T. Roumier, J. Blanco, K. F. Ferri, J. Barretina, L. A. Tintignac, K. Andreau, J. L. Perfettini, A. Amendola, R. Nardacci, P. Leduc, D. E. Ingber, S. Druillennec, B. Roques, S. A. Leibovitch, M. Vilella-Bach, J. Chen, J. A. Este, N. Modjtahedi, M. Piacentini, and G. Kroemer. 2002. Sequential involvement of Cdk1, mTOR and p53 in apoptosis induced by the HIV-1 envelope. EMBO J. 21:4070-4080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen, B. K., R. T. Gandhi, and D. Baltimore. 1996. CD4 down-modulation during infection of human T cells with human immunodeficiency virus type 1 involves independent activities of vpu, env, and nef. J. Virol. 70:6044-6053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chin, M. P., J. Chen, O. A. Nikolaitchik, and W. S. Hu. 2007. Molecular determinants of HIV-1 intersubtype recombination potential. Virology 363:437-446. [DOI] [PubMed] [Google Scholar]
- 8.Chin, M. P., T. D. Rhodes, J. Chen, W. Fu, and W. S. Hu. 2005. Identification of a major restriction in HIV-1 intersubtype recombination. Proc. Natl. Acad. Sci. U. S. A. 102:9002-9007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chohan, B., L. Lavreys, S. M. Rainwater, and J. Overbaugh. 2005. Evidence for frequent reinfection with human immunodeficiency virus type 1 of a different subtype. J. Virol. 79:10701-10708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Coffin, J. M. 1979. Structure, replication, and recombination of retrovirus genomes: some unifying hypotheses. J. Gen. Virol. 42:1-26. [DOI] [PubMed] [Google Scholar]
- 11.Coffin, J. M., S. H. Hughes, and H. E. Varmus. 1997. Retroviruses. Cold Spring Harbor Laboratory, New York, NY. [PubMed]
- 12.Dang, Q., J. Chen, D. Unutmaz, J. M. Coffin, V. K. Pathak, D. Powell, V. N. KewalRamani, F. Maldarelli, and W. S. Hu. 2004. Nonrandom HIV-1 infection and double infection via direct and cell-mediated pathways. Proc. Natl. Acad. Sci. U. S. A. 101:632-637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Delviks-Frankenberry, K. A., G. N. Nikolenko, F. Maldarelli, S. Hase, Y. Takebe, and V. K. Pathak. 2009. Subtype-specific differences in the human immunodeficiency virus type 1 reverse transcriptase connection subdomain of CRF01_AE are associated with higher levels of resistance to 3′-azido-3′-deoxythymidine. J. Virol. 83:8502-8513.19553318 [Google Scholar]
- 14.DuBridge, R. B., P. Tang, H. C. Hsia, P. M. Leong, J. H. Miller, and M. P. Calos. 1987. Analysis of mutation in human cells by using an Epstein-Barr virus shuttle system. Mol. Cell. Biol. 7:379-387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Engelman, A., and R. Craigie. 1992. Identification of conserved amino acid residues critical for human immunodeficiency virus type 1 integrase function in vitro. J. Virol. 66:6361-6369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Galetto, R., V. Giacomoni, M. Veron, and M. Negroni. 2006. Dissection of a circumscribed recombination hot spot in HIV-1 after a single infectious cycle. J. Biol. Chem. 281:2711-2720. [DOI] [PubMed] [Google Scholar]
- 17.Galli, A., A. Lai, S. Corvasce, F. Saladini, C. Riva, L. Deho, I. Caramma, M. Franzetti, L. Romano, M. Galli, M. Zazzi, and C. Balotta. 2008. Recombination analysis and structure prediction show correlation between breakpoint clusters and RNA hairpins in the pol gene of human immunodeficiency virus type 1 unique recombinant forms. J. Gen. Virol. 89:3119-3125. [DOI] [PubMed] [Google Scholar]
- 18.Gao, F., D. L. Robertson, C. D. Carruthers, S. G. Morrison, B. Jian, Y. Chen, F. Barré-Sinoussi, M. Girard, A. Srinivasan, A. G. Abimiku, G. M. Shaw, P. M. Sharp, and B. H. Hahn. 1998. A comprehensive panel of near-full-length clones and reference sequences for non-subtype B isolates of human immunodeficiency virus type 1. J. Virol. 72:5680-5698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Geretti, A. M. 2006. HIV-1 subtypes: epidemiology and significance for HIV management. Curr. Opin. Infect. Dis. 19:1-7. [DOI] [PubMed] [Google Scholar]
- 20.Hall, T. A. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999:95-98. [Google Scholar]
- 21.Hemelaar, J., E. Gouws, P. D. Ghys, and S. Osmanov. 2006. Global and regional distribution of HIV-1 genetic subtypes and recombinants in 2004. AIDS 20:W13-W23. [DOI] [PubMed] [Google Scholar]
- 22.Hu, W. S., and H. M. Temin. 1990. Genetic consequences of packaging two RNA genomes in one retroviral particle: pseudodiploidy and high rate of genetic recombination. Proc. Natl. Acad. Sci. U. S. A. 87:1556-1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hu, W. S., T. Rhodes, Q. Dang, and V. Pathak. 2003. Retroviral recombination: review of genetic analyses. Front. Biosci. 8:d143-d155. [DOI] [PubMed] [Google Scholar]
- 24.Jetzt, A. E., H. Yu, G. J. Klarmann, Y. Ron, B. D. Preston, and J. P. Dougherty. 2000. High rate of recombination throughout the human immunodeficiency virus type 1 genome. J. Virol. 74:1234-1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jost, S., M. C. Bernard, L. Kaiser, S. Yerly, B. Hirschel, A. Samri, B. Autran, L. E. Goh, and L. Perrin. 2002. A patient with HIV-1 superinfection. N. Engl. J. Med. 347:731-736. [DOI] [PubMed] [Google Scholar]
- 26.Kearney, M., S. Palmer, F. Maldarelli, W. Shao, M. A. Polis, J. Mican, D. Rock-Kress, J. B. Margolick, J. M. Coffin, and J. W. Mellors. 2008. Frequent polymorphism at drug resistance sites in HIV-1 protease and reverse transcriptase. AIDS 22:497-501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Keating, C. P., M. K. Hill, D. J. Hawkes, R. P. Smyth, C. Isel, S. Y. Le, A. C. Palmenberg, J. A. Marshall, R. Marquet, G. J. Nabel, and J. Mak. 2009. The A-rich RNA sequences of HIV-1 pol are important for the synthesis of viral cDNA. Nucleic Acids Res. 37:945-956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kotsopoulou, E., V. N. Kim, A. J. Kingsman, S. M. Kingsman, and K. A. Mitrophanous. 2000. A Rev-independent human immunodeficiency virus type 1 (HIV-1)-based vector that exploits a codon-optimized HIV-1 gag-pol gene. J. Virol. 74:4839-4852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Larkin, M. A., G. Blackshields, N. P. Brown, R. Chenna, P. A. McGettigan, H. McWilliam, F. Valentin, I. M. Wallace, A. Wilm, R. Lopez, J. D. Thompson, T. J. Gibson, and D. G. Higgins. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23:2947-2948. [DOI] [PubMed] [Google Scholar]
- 30.Leitner, T., B. Korber, M. Daniels, C. Calef, and B. Foley. 2005. HIV-1 subtype and circulating recombinant form (CRF) reference sequences, p. 41-48. In T. Leitner, B. Foley, B. H. Hahn, P. A. Marx, F. E. McCutchan, J. W. Mellors, S. Wolinsky, and B. Korber (ed.), HIV Sequence Compendium. Los Alamos National Laboratory, Los Alamos, NM.
- 31.Levy, D. N., G. M. Aldrovandi, O. Kutsch, and G. M. Shaw. 2004. Dynamics of HIV-1 recombination in its natural target cells. Proc. Natl. Acad. Sci. U. S. A. 101:4204-4209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Malim, M. H., and M. Emerman. 2001. HIV-1 sequence variation: drift, shift, and attenuation. Cell 104:469-472. [DOI] [PubMed] [Google Scholar]
- 33.Martin, V., P. Ronde, D. Unett, A. Wong, T. L. Hoffman, A. L. Edinger, R. W. Doms, and C. D. Funk. 1999. Leukotriene binding, signaling, and analysis of HIV coreceptor function in mouse and human leukotriene B4 receptor-transfected cells. J. Biol. Chem. 274:8597-8603. [DOI] [PubMed] [Google Scholar]
- 34.McCutchan, F. E. 2006. Global epidemiology of HIV. J. Med. Virol. 78(Suppl. 1):S7-S12. [DOI] [PubMed] [Google Scholar]
- 35.Moore, M. D., W. Fu, O. Nikolaitchik, J. Chen, R. G. Ptak, and W. S. Hu. 2007. Dimer initiation signal of human immunodeficiency virus type 1: its role in partner selection during RNA copackaging and its effects on recombination. J. Virol. 81:4002-4011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nájera, R., E. Delgado, L. Pérez-Alvarez, and M. M. Thomson. 2002. Genetic recombination and its role in the development of the HIV-1 pandemic. AIDS 16(Suppl. 4):S3-S16. [DOI] [PubMed] [Google Scholar]
- 37.Palmer, S., M. Kearney, F. Maldarelli, E. K. Halvas, C. J. Bixby, H. Bazmi, D. Rock, J. Falloon, R. T. Davey, R. L. Dewar, J. A. Metcalf, S. Hammer, J. W. Mellors, and J. M. Coffin. 2005. Multiple, linked human immunodeficiency virus type 1 drug resistance mutations in treatment-experienced patients are missed by standard genotype analysis. J. Clin. Microbiol. 43:406-413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rambaut, A., D. Posada, K. A. Crandall, and E. C. Holmes. 2004. The causes and consequences of HIV evolution. Nat. Rev. Genet. 5:52-61. [DOI] [PubMed] [Google Scholar]
- 39.Rhodes, T. D., O. Nikolaitchik, J. Chen, D. Powell, and W. S. Hu. 2005. Genetic recombination of human immunodeficiency virus type 1 in one round of viral replication: effects of genetic distance, target cells, accessory genes, and lack of high negative interference in crossover events. J. Virol. 79:1666-1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Robertson, D. L., J. P. Anderson, J. A. Bradac, J. K. Carr, B. Foley, R. K. Funkhouser, F. Gao, B. H. Hahn, M. L. Kalish, C. Kuiken, G. H. Learn, T. Leitner, F. McCutchan, S. Osmanov, M. Peeters, D. Pieniazek, M. Salminen, P. M. Sharp, S. Wolinsky, and B. Korber. 2000. HIV-1 nomenclature proposal. Science 288:55-56. [DOI] [PubMed] [Google Scholar]
- 41.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 42.Simon-Loriere, E., R. Galetto, M. Hamoudi, J. Archer, P. Lefeuvre, D. P. Martin, D. L. Robertson, and M. Negroni. 2009. Molecular mechanisms of recombination restriction in the envelope gene of the human immunodeficiency virus. PLoS Pathog. 5:e1000418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Smith, D. M., D. D. Richman, and S. J. Little. 2005. HIV superinfection. J. Infect. Dis. 192:438-444. [DOI] [PubMed] [Google Scholar]
- 44.Thomson, M. M., and R. Najera. 2005. Molecular epidemiology of HIV-1 variants in the global AIDS pandemic: an update. AIDS Rev. 7:210-224. [PubMed] [Google Scholar]
- 45.Thomson, M. M., L. Perez-Alvarez, and R. Najera. 2002. Molecular epidemiology of HIV-1 genetic forms and its significance for vaccine development and therapy. Lancet Infect. Dis. 2:461-471. [DOI] [PubMed] [Google Scholar]
- 46.Van Heuverswyn, F., and M. Peeters. 2007. The origins of HIV and implications for the global epidemic. Curr. Infect. Dis. Rep. 9:338-346. [DOI] [PubMed] [Google Scholar]
- 47.Wainberg, M. A. 2004. HIV-1 subtype distribution and the problem of drug resistance. AIDS 18(Suppl. 3):S63-S68. [DOI] [PubMed] [Google Scholar]
- 48.Watts, J. M., K. K. Dang, R. J. Gorelick, C. W. Leonard, J. W. Bess, Jr., R. Swanstrom, C. L. Burch, and K. M. Weeks. 2009. Architecture and secondary structure of an entire HIV-1 RNA genome. Nature 460:711-716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wu, L., T. D. Martin, R. Vazeux, D. Unutmaz, and V. N. KewalRamani. 2002. Functional evaluation of DC-SIGN monoclonal antibodies reveals DC-SIGN interactions with ICAM-3 do not promote human immunodeficiency virus type 1 transmission. J. Virol. 76:5905-5914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xie, D., S. Gulnik, E. Gustchina, B. Yu, W. Shao, W. Qoronfleh, A. Nathan, and J. W. Erickson. 1999. Drug resistance mutations can effect dimer stability of HIV-1 protease at neutral pH. Protein Sci. 8:1702-1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yee, J. K., T. Friedmann, and J. C. Burns. 1994. Generation of high-titer pseudotyped retroviral vectors with very broad host range. Methods Cell. Biol. 43(Pt. A):99-112. [DOI] [PubMed] [Google Scholar]