Abstract
Human cytomegalovirus (HCMV) is a human pathogen that causes severe disease primarily in the immunocompromised or immunologically immature individual. To date, no vaccine is available. We describe use of a spread-deficient murine CMV (MCMV) as a novel approach for betaherpesvirus vaccination. To generate a spread-deficient MCMV, the conserved, essential gene M94 was deleted. Immunization with MCMV-ΔM94 is apathogenic and protective against wild-type challenge even in highly susceptible IFNαβR−/− mice. MCMV-ΔM94 was able to induce a robust CD4+ and CD8+ T-cell response as well as a neutralizing antibody response comparable to that induced by wild-type infection. Endothelial cells were identified as activators of CD8+ T cells in vivo. Thus, the vaccination with a spread-deficient betaherpesvirus is a safe and protective strategy and allows the linkage between cell tropism and immunogenicity. Furthermore, genomes of MCMV-ΔM94 were present in lungs 12 months after infection, revealing first-target cells as sites of genome maintenance.
The human cytomegalovirus (HCMV) is a ubiquitous betaherpesvirus that causes severe disease primarily in the immunocompromised or immunologically immature individual. Reactivation from latent infection can occur when the balance between viral maintenance and immune control is disturbed (56). HCMV is able to productively infect a large variety of different cell types and is thus associated with a broad spectrum of diseases. In adults, the main risk groups are immunocompromised patients such as transplant recipients and AIDS or tumor patients (73). Further, congenital HCMV infection represents the most common infectious cause of brain damage in children (20). Despite extensive efforts, no effective vaccine against HCMV is available to date, although its development is considered to be of greatest importance (70). Design of a protective HCMV vaccine must serve complex purposes of safety and immunogenicity due to the diversity of disease manifestations and the heterogeneous immune status of the patient groups (63).
The continuous passage of HCMV on fibroblasts generated the Towne strain, which was used in trials as a live attenuated vaccine. The attenuation was associated with randomly acquired mutations leading to the loss of antigens and receptors for tropism as well as genes with known or unknown functions (10). Vaccination with the Towne strain mitigated CMV disease in seronegative renal transplant recipients and protected against a low-dose virulent HCMV challenge in volunteers (51) but failed to confer protection against infection by natural contact (1, 22). Due to the undefined nature of the attenuation of the Towne strain, the focus for vaccine design shifted to a selective but well-investigated set of viral antigens and their linked effector mechanism.
Other strategies included adoptive transfer of T-cell clones that protected transplant recipients (58) and transferred anti-HCMV antibodies to prevent congenital HCMV transmission (47). Further, vaccination of mice with a recombinant adenovirus expressing gB of murine CMV (MCMV), the major target for neutralizing antibodies (8), protected against MCMV disease (66). Currently, a subunit vaccine targeting both B- and T-cell responses is under investigation but has yet to prove its potential as a suitable HCMV vaccine (current clinical trials are listed at http://clinicaltrials.gov).
In order to generate a CMV vaccine, further aspects deserve attention. Animal experiments proved that, although antibodies limit dissemination of recurrent virus, they were found to be not sufficient to control primary infection (32). Moreover, CD4+ as well as CD8+ T cells contain CMV infection by recognition of a multitude of T-cell epitopes (31, 57).
A large number of HCMV genes counteract innate and adaptive host immune responses (reviewed in reference 62). Efficient antigen processing is critical for protection against CMV disease in the presence of viral immune evasion proteins (28). Recently, we generated an attenuated MCMV by deletion of gene regions known to encompass such immune-evasive genes (11). The resulting vaccine, Δm01-17+m144-158-MCMV, lacked 32 genes dispensable for in vitro replication and induced a protective immune response against wild-type MCMV (MCMV-wt). Although this vaccine was strongly attenuated in vivo, the possibility that such a mutant might cause harm under severe immunosuppressive conditions cannot be excluded. Prolonged replication of attenuated vaccines in immunocompromised patients has indeed been observed (37). Therefore, virus spread needs to be completely excluded while maintaining full immunogenic properties. A similar approach has so far been applied only to alphaherpesviruses in animal models (reviewed in reference 16). In these studies, the deletion of genes essential for viral DNA replication (46) or packaging (35) gave rise to mutants in which the replication cascade was terminated at different checkpoints. The antibody response to such vaccines was stronger the later the block during the viral morphogenesis was introduced (3, 46). This concept led to protective vaccines against wild-type challenge in animal models (15, 18). Some of these are now entering clinical trials (http://clinicaltrials.gov). In this study a vaccine against betaherpesvirus was evaluated in its natural host. We constructed an MCMV that cannot spread from the first infected cell. For this purpose, we deleted the gene M94, which is conserved in all herpesvirus subfamilies. The gene M94 of MCMV represents the homolog of UL94 in HCMV, which is essential (17). The encoded protein, pM94, is part of the virion (34) and, like pUL94, expressed at late time points of infection (64, 77). Thus, the deletion of M94 should not affect the expression of other viral genes.
Here, we describe the properties of the MCMV-ΔM94 mutant. The MCMV-ΔM94 vaccine was well tolerated even by immune-deficient mice highly susceptible to MCMV. Vaccination induced robust humoral and cellular immune responses and conferred protection against wild-type virus challenge. MCMV-ΔM94 infection is restricted to the first-target cell, and the impact of infection of different cell types on induction of adaptive immune responses was examined. Using Cre-loxP-based conditional expression of a model antigen, we observed that mainly endothelial cells (EC) but not hepatocytes (Hc) contribute to the induction of virus-specific CD8+ T cells. The genome of MCMV-ΔM94 was found to be maintained in mice for at least 1 year, indicating that first-target cells of the vaccine are long-lived and could serve as an antigen reservoir for prolonged activation of the immune system.
MATERIALS AND METHODS
Cells and mice.
The fibroblast cell line NIH 3T3 and BALB/c-derived murine embryonic fibroblasts (MEF) were cultured as described previously (12). C57BL/6 (B6) mice, B6.SJL-Ptprc (Ptprc) mice, and 129.IFNαβR−/− mice were purchased from Elevage Janvier (Le Genest Saint Isle, France), Jackson Laboratories (Bar Harbor, ME), and B&K Universal Limited (Grimston, England), respectively. 129.IFNαβR−/− mice (45) were backcrossed on the B6 background (B6.IFNαβR−/−). T-cell receptor (TCR) transgenic OT-I (25) and OT-II (6) mice were backcrossed to Ptprc (CD45.1) and Thy1.1 (CD90.1) congenic mice, respectively. Alb-cre (52) and Tie2-cre (14) mice were maintained on the B6 background. Mice were kept under specific-pathogen-free conditions. Animal experiments were approved by the responsible office of the State of Bavaria (approval no. 55.2-1-54-2531-111-07) or by the Ethics Committee at the University of Rijeka.
Generation of the trans-complementing cell line NT/M94-7.
The conditional trans-complementing cell line NT/M94-7 was generated as described in reference (41). Briefly, the M94 open reading frame (ORF) was amplified from pSM3fr (61) using primers HAM94for (5′-GTGGGATCCACCATGTACCCCTACGACGTGCCCGACTACGCCACGTCCAGACTATCC-3′) and M94rev (5′-ACTCTAGAGTCGACTTCACATGTGCTCGAGAACA-3′), thereby introducing a hemagglutinin (HA) tag at the N terminus. The PCR product was digested with BamHI and XbaI and inserted into the BamHI- and NheI-cleaved pTRE2Hyg vector (BD Biosciences Clontech, Heidelberg, Germany), resulting in pTRE-HAM94, putting HAM94 expression under the control of the tetracycline (Tet)-inducible promoter. Stable NIH 3T3 transfectants harboring pTRE-HAM94 were selected with 50 μg/ml hygromycin B. The deletion virus MCMV-ΔM94 was reconstituted by transfecting different NT/M94 cell clones with the respective bacterial artificial chromosome (BAC). The most productively infected trans-complementing cell line, NT/M94-7, was subcloned by limiting dilution.
Generation of recombinant viruses.
Recombinant MCMV mutants were derived from the MCMV BAC clone pSM3fr, which originated from the Smith strain (44). Nucleotide positions are given according to Rawlinson et al. (55). The 1.4-kbp SmaI fragment of pCP15 carrying the FLP recombinase target (FRT)-flanked kanamycin resistance gene (kanR) was introduced into the BssHII site of pCR3 (Invitrogen, Basel, Switzerland), resulting in pCR3-FRT-kanR-FRT. A fragment containing an ATG start codon and a loxP site was generated by annealing the oligonucleotides ATGlox1 (5′-AATTCATGATAACTTCGTATAGCATACATTATACGAAGTTATCCGGAGATATCCACCGGTCTGGCGGCCGC-3′) and ATGlox2 (5′-TCGAGCGGCCGCCAGACCGGTGGATATCTCCGGATAACTTCGTATAATGTATGCTATACGAAGTTATCATG-3′). This fragment was inserted into the EcoRI and XhoI site positioned between the major immediate early promoter (IEP) of HCMV and the poly(A) signal of the bovine growth hormone of pCR3-FRT-kanR-FRT to obtain pCR3-FRT-kanR-FRT-ATG-loxP. The ovalbumin gene (ova) was synthesized, introducing GGAA after nucleotide position 9, resulting in a BspEI restriction site for further cloning. ova was inserted in frame using BspEI and NotI sites of pCR3-FRT-kanR-FRT-ATG-loxP, resulting in a full-length ova with inserted loxP site after the initial ATG under the control of IEP pCR3-FRT-kanR-FRT-ATG-loxP-ova. To obtain a construct with Cre-inducible ovalbumin (OVA) expression, a flox-stop cassette (61) was inserted into the EcoRI and BspEI sites of pCR3-ATG-loxP-ova, resulting in pCR3-ATG-flox-stop-ova. Using these constructs as templates and oligonucleotides 5′-Δm157-pCR3-FRT-kanR-FRT (5′-CGT GGT CAA GCC GGT CGT GTT GTA CCA GAA CTC GAC TTC GGT CGC GTT GCT TAC AAT TTA CGC GCG GG-3′; nucleotide position 216243 to 216290) and 3′-Δm157-flox-egfp (5′-CCC CGA TAT TTG AGA AAG TGT ACC CCG ATA TTC AGT ACC TCT TGA CTA AGA AGC CAT AGA GCC CAC CGC-3′; nucleotide position 216885 to 216930) as primers, a linear DNA fragment containing the IEP-ova cassette, the FRT-flanked kanR, and the viral homology sequences for the MCMV genome target site m157 was generated. In a similar procedure the firefly luciferase gene (luc) was cloned under the control of the IEP into pCP15 carrying the FRT-flanked kanR. These fragments were introduced into m157 of pSM3fr as described previously (61), resulting in pSM3fr-Δm157-ova, pSM3fr-Δm157-flox-ova, and pSM3fr-Δm157-luc. For excision of the FRT-flanked kanR, FLP recombinase was transiently expressed from plasmid pCP20.
Generation of spread-deficient virus mutants.
For generation of the recombinant MCMV lacking the M94 sequence, the parental MCMV BACs pSM3fr (MCMV-wt), pSM3fr-Δm157-ova (MCMV-ova), and pSM3fr-Δm157-rec-egfp (MCMV-Δm157-rec-egfp) (61) were applied to a second mutagenesis step. Therefore, plasmid pO6-tTA-mFRT-kanR-mFRT was obtained by insertion of kanR, on both sides flanked by mutant 34-bp FRT sites from pO6ie-F5, into pO6-tTA (41) to express the tTA transactivation gene under the control of the IEP necessary for trans complementation of pM94. A linear DNA fragment containing the tTA cassette, kanR, and viral homology sequences for the MCMV genome target site (MCMV upstream homology, nucleotide position 136189 to 136234; MCMV downstream homology, nucleotide position 137256 to 137309) was generated using primer 5′ΔM94-pO6-tTA (5′-TGC TTC CCG GCG GCT TCT GCG CGA CCT TCC AGC TGC AGG TAG ACC ACG GCG ACG TCC AGA CTA TCC GTG AAA AGT TTG AGA AGC ATC AGT AGC CGA TTT CGG CCT ATT GGT T-3′), primer 3′-ΔM94-pO6-tTA (5′-CAT GGA TGG GTT GGT TGA TTT GTA TGT CTG TTG GCT ACT CAC ATG TGC TCG AGA AGC CAG TGT GAT GGA TGA TCC TC-3′), and plasmid pO6-tTA-mFRT-kanR-mFRT as the template. This PCR fragment was inserted into the different parental pSM3fr clones, thereby deleting the M94 gene. Since ORFs of M94 and M93 overlap, 47 bp of homology had to be left at the 5′ end of M94 to keep the M93 ORF intact and 17 bp of homology is still present at the former 3′ end of M94. Again, FLP recombinase was expressed for excision of kanR. Construction of pSM3fr-ΔM94, pSM3fr-ova-ΔM94, pSM3fr-flox-ova-ΔM94, and pSM3fr-Δm157-rec-egfp-ΔM94 was confirmed by restriction digest analysis and sequencing.
Viruses were reconstituted from BAC DNA, propagated on NT/M94-7 complementing cells, and purified on a sucrose cushion as previously described (61). For analysis of virus replication, supernatants from infected cells were taken every 24 h. Quantification of infectious virus was done using the median tissue culture infectious dose (TCID50) method on NIH 3T3 or complementing NT/M94-7 cells. For the determination of virus replication in vivo, virus load was determined by standard plaque assay as PFU per gram organ as described previously (61). Spread deficiency of each virus stock of M94-deficient mutants (MCMV-ΔM94, MCMV-ova-ΔM94, MCMV-flox-ova-ΔM94, and MCMV-Δm157-rec-egfp-ΔM94) was confirmed by the absence of plaque formation after infection of noncomplementing MEF, although cytopathic effect (CPE) of individually infected cells was detectable.
UV inactivation of virus.
For in vivo application, a fraction of the MCMV-wt virus preparation used for immunization was inactivated by exposure to 1.5 kJ/cm2 UV light at a distance of 5 cm in a UV-cross-linker (Stratagene, Amsterdam, Netherlands) at 4°C. Viral infectivity was decreased by factor of 2.4 × 107. The same treatment was sufficient to abolish viral gene expression when MCMV-Δm157-rec-egfp was subjected to different doses (0.5, 1.0, and 1.5 kJ/cm2) of UV light and subsequently titrated on MEF (data not shown). At 4 days postinfection (p.i.) enhanced green fluorescent protein (EGFP) expression in single infected cells was detected after virus was irradiated with a low dose (0.5 kJ/cm2) of UV, whereas no EGFP expression was seen after strong irradiation (1.5 kJ/cm2). Untreated MCMV-Δm157-rec-egfp formed EGFP+ plaques.
Immunization and challenge of mice.
Eight- to 10-week-old female B6 mice were immunized by intraperitoneal (i.p.) or subcutaneous (s.c.) injection of either MCMV-wt or mutant MCMV. Each mouse received 100 μl of virus suspension s.c. or 300 μl i.p. C57BL/6 mice were immunized with 1 × 105 TCID50 MCMV-wt or MCMV-ΔM94, 129.IFNαβR−/− mice were immunized with 2.5 × 105 TCID50 of MCMV-ΔM94 or UV-irradiated MCMV-wt, and B6.IFNαβR−/− mice were immunized with 3 × 105 TCID50 of MCMV-ΔM94 or MCMV-wt. Mock-treated mice received the same volumes of phosphate-buffered saline (PBS). To boost mice, this procedure was repeated 14 days p.i. Sera collected from mice 12 weeks p.i. were used to determine amounts of virus-specific antibodies by virus neutralization assay, as described below.
Twenty-eight days or 20 weeks postpriming, mice were challenged by intravenous (i.v.) injection of 106 PFU of tissue culture-derived MCMV-wt. Five days postchallenge lungs, liver, and spleen were collected under sterile conditions and stored at −80°C. Organ homogenates were analyzed for infectious virus load by standard plaque assay of MEF. Salivary gland-derived MCMV (sgMCMV-wt) was generated as a homogenate of salivary glands from mice infected with tissue culture-derived MCMV-wt as already described (74). The isolated sgMCMV-wt is more virulent than tissue culture-derived MCMV-wt (50). Vaccinated B6.IFNαβR−/− mice were challenged with 2 × 105 PFU sgMCMV-wt, and 129.IFNαβR−/− mice were challenged with 2.5 × 105 TCID50 tissue culture-derived MCMV-wt.
Virus neutralization assay.
Heat-inactivated serum (56°C, 30 min) from 5 immunized mice 12 weeks p.i. was pooled and serially diluted 1:2 in Dulbecco's modified Eagle medium (DMEM) containing a final concentration of 10% guinea pig complement. Each dilution was mixed with 50 PFU of MCMV-luc, incubated for 90 min at 37°C, and subsequently added to NIH 3T3 cells in a 96-well format. After 1 h at 37°C the virus inoculum was removed and NIH 3T3 medium was added. The cultures were incubated for 24 h, and luciferase activity in cell extracts was determined using the luciferase assay (Promega, Mannheim, Germany) in a luminometer (Berthold, Bad Wildbad, Germany) according to the supplier's and manufacturer's instructions, respectively.
In vivo cytotoxicity assay.
To evaluate CD8+ T-cell effector function in vivo, splenocytes of congenic CD45.1+ Ptprc mice were incubated with 2 μM peptide and stained with 2 μM, 0.7 μM, or 0.1 μM carboxyfluorescein succinimidyl ester (CFSE) and the PKH26 red fluorescent cell linker minikit according to the manufacturer's instructions (Sigma-Aldrich). At day 6 p.i., labeled CD45.1+ cells were transferred into B6 (CD45.2+) recipients. After 16 h spleens of recipient mice were removed and flow cytometric analysis of the target cells was performed. Specific cytotoxicity of target cells was calculated using the equation % specific lysis = (1 − ratio for unprimed cells/ratio for primed cells)·100, where ratio = (% of cells with low-concentration CFSE labeling/% of cells with high-concentration CFSE labeling) (40). The OVA-derived class I peptide (SIINFEKL) and MCMV-specific peptides derived from m139 (TVYGFCLL), ie3 (RALEYKNL), M57 (SCLEFWQRV), and M45 (HGIRNASFI) (68) were purchased from Metabion, Germany, and were dissolved and stored according to the manufacturer's instructions.
Adoptive transfer and flow cytometric analysis.
OVA-specific CD8+ T cells were isolated from spleen and cervical, axillary, brachial, and inguinal lymph nodes of OT-I TCR transgenic mice backcrossed to congenic CD45.1+ mice. OT-I cells were purified by negative selection via the CD8α+ T-cell isolation kit (Miltenyi Biotec, Bergisch Gladbach, Germany). Transgenic T cells (3 × 105) were injected i.v. into recipient B6 mice 1 day prior to i.p. infection with 105 TCID50 MCMV. To follow expansion of the transferred OT-I T cells, 100 μl blood was taken 3, 6, and 8 days p.i., erythrocytes were lysed (PharmLyse, BD Biosciences, Heidelberg, Germany), and remaining cells were incubated with phycoerythrin (PE)-Texas Red-coupled anti-CD8α (5H10; Caltag, Sacramento, CA) and PE-coupled anti-CD45.1 antibodies (A20; BD Biosciences Pharmingen). Flow cytometric acquisition was performed using an Epics XL-MCL (Beckman-Coulter), and data were analyzed using FlowJo software (Tristar, Ashland, OR).
OVA-specific CD4+ T cells were isolated from spleen and cervical, axillary, brachial, and inguinal lymph nodes of OT-II TCR transgenic mice backcrossed to congenic CD90.1+ mice. After lysis of erythrocytes, 3 × 105 transgenic T cells were injected i.v. into recipient mice 1 day prior to infection with 105 TCID50 MCMV. Spleens were removed, and splenocytes were incubated with Fc block (2.4G2; BD Biosciences) and subsequently stained with PE-conjugated anti-CD90.1 (HIS51; eBioscience) and PE-Cy5.5-coupled anti-CD4 (RM 4-5; eBioscience). Flow cytometric acquisition was performed using a FACSCalibur (BD Biosciences), and data were analyzed using FlowJo software.
Quantification of viral genomes in organ homogenates.
Lungs were removed from mice 12 month after infection. Organs were homogenized, and DNA was extracted using the DNeasy blood and tissue kit from Qiagen (Hilden, Germany). Elution was done with 100 μl of the supplied elution buffer, and the genomic DNA concentration of each sample was quantified in duplicate using a NanoDrop ND-1000 UV-visual spectrophotometer. To quantify the viral DNA, a quantitative real-time PCR specific for the MCMV M54 gene (12) was performed using a specific TaqMan probe (5′-6-carboxyfluorescein [FAM]-AACGTACATCGCTCTCTGCTGGCCG-6-carboxytetramethylrhodamine [TAMRA]-3′) and the TaqMan 1000 RXN PCR core reagent kit on an ABI Prism 7700 sequence detector (Applied Biosystems, Carlsbad, CA). To calculate the viral genome copy number, a standard curve for the BAC plasmid pSM3fr containing the M54 gene was included.
Statistical analysis.
Statistical analyses were done using Prism 4 (GraphPad Software, La Jolla, CA). For comparison of in vitro growth rates of viruses, for the neutralizing antibody assay, for real-time PCR, and for T-cell proliferation assays, the means and standard deviations (SD) were calculated. In all figures depicting virus load in organs and in vivo cytotoxicity, the medians are given. Comparison of the neutralizing antibody responses in mice vaccinated with MCMV-wt or MCMV-ΔM94 was performed with the two-way analysis of variance (ANOVA) test. Comparison of percentages of T-cell proliferation and quantification of virus in organs and viral genomes were done with the two-tailed Wilcoxon rank sum test using procedures at http://elegans.swmed.edu/∼leon/stats/utest.cgi.
RESULTS
MCMV-ΔM94 is spread deficient.
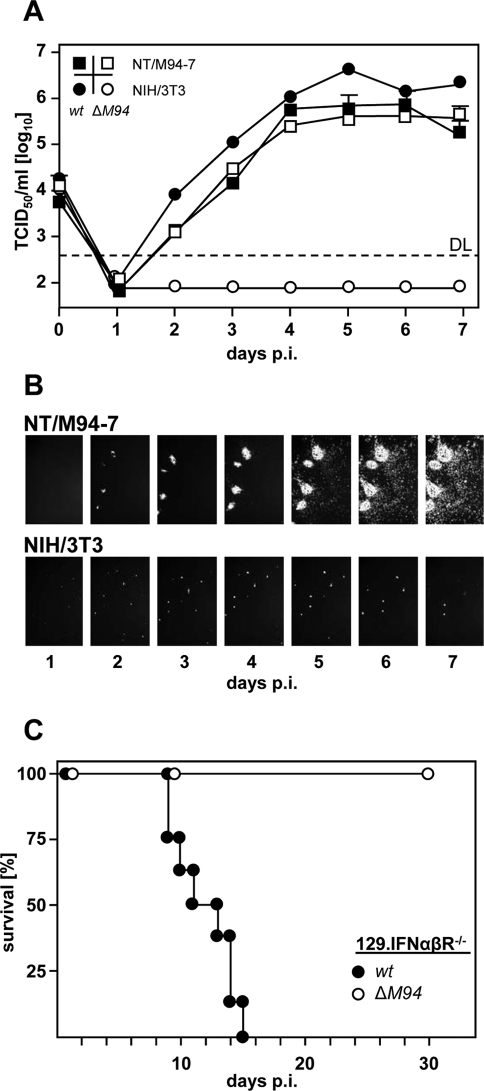
The HCMV virion protein pUL94 is essential for virus replication (17) and is expressed with late kinetics (77). Preliminary work in our group showed that pM94, the MCMV homologue, is also essential and plays a crucial role in a postnuclear step of virus maturation (S. Maninger, et al., unpublished data). In order to trans complement the essential M94 gene product and reconstitute an M94 deletion mutant, we generated the NIH 3T3-derived complementing cell line NT/M94-7, harboring the M94 gene under the control of the TRE promoter. The TRE promoter is active only in the presence of the Tet transactivator (tTA). To provide the tTA for trans complementation of pM94, the tTA expression cassette was introduced into pSM3fr (44), disrupting M94 and generating pSM3fr-ΔM94. MCMV-ΔM94 virus was reconstituted by transfecting NT/M94-7 cells (Fig. 1). Next, we performed multistep growth analysis, infecting NT/M94-7 cells as well as parental NIH 3T3 fibroblasts with MCMV-ΔM94 or MCMV-wt. While MCMV-ΔM94 replicated to MCMV-wt-like titers on NT/M94-7 cells, no infectious virus was detectable in the supernatant of NIH 3T3 cells (Fig. 2 A). As the defect of MCMV-ΔM94 to release infectious virus particles into the supernatant does not exclude cell-associated virus spread, we constructed a ΔM94 mutant expressing the enhanced green fluorescent protein (EGFP) (MCMV-Δm157-rec-egfp-ΔM94). While MCMV-Δm157-rec-egfp-ΔM94 spread with kinetics comparable to that of MCMV-wt on NT/M94-7 cells, MCMV-Δm157-rec-egfp-ΔM94 remained strictly confined to the first-infected NIH 3T3 cells (Fig. 2B). This result was confirmed also in endothelial cells (see Fig. S1 in the supplemental material). In summary, M94 is essential and deletion abrogates virus release and cell-to-cell spread. In addition, MCMV-ΔM94 can be efficiently produced by trans complementation.
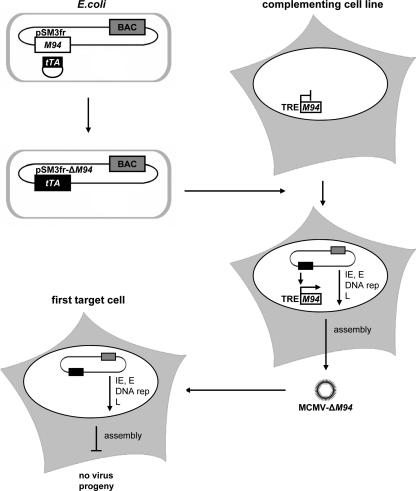
FIG. 1.
Concept of inducible trans complementation. In Escherichia coli the BAC pSM3fr-ΔM94 was generated by insertion of the tTA transactivator cassette into pSM3fr, thereby deleting M94. The trans-complementing cell line NT/M94-7 expresses pM94 under the control of the Tet-inducible promoter. Upon transfection with pSM3fr-ΔM94, expression of tTA by the viral genome induces expression of pM94 by the cell, leading to the production of trans-complemented MCMV-ΔM94. This virus is able to infect noncomplementing first-target cells. Due to the lack of the essential gene M94, the release of infectious virus particles is impossible although immediate early (IE), early (E), and late (L) viral gene expression as well as DNA replication (DNA rep) occurs.
FIG. 2.
MCMV-ΔM94 is spread deficient, and replication depends on trans complementation of pM94. (A) Parental NIH 3T3 and NT/M94-7 fibroblasts were infected at 0.1 TCID50/cell with MCMV-wt (w) or MCMV-ΔM94 (ΔM94). At the indicated days, infectious virus in the supernatant was quantified on NT/M94-7 cells by TCID50 endpoint titration. Shown are the means plus SD of titrated duplicates. At day 5 postinfection (p.i.) supernatants were additionally titrated on MEF. No PFU was found in 1 ml supernatant of MCMV-ΔM94-infected NT/M94-7 cells. DL, detection limit. (B) Parental NIH 3T3 and NT/M94-7 fibroblasts were infected with MCMV-Δm157-rec-egfp-ΔM94. At the indicated time points EGFP-expressing cells were monitored. hpi, hours p.i. (C) 129.IFNαβR−/− mice (n = 15 for MCMV-ΔM94, n = 8 for MCMV-wt) were infected with 2.5 × 105 TCID50 i.p., and survival was followed for 30 days p.i.
MCMV-ΔM94 does not revert to replication-competent virus.
A major safety concern is reversion of vaccine strains to replication-competent viruses during preparation (60) or in the vaccinee (30). To exclude acquisition of the M94 gene through recombination via homologous sequences between MCMV-ΔM94 and the complementing cell line, homologies were carefully avoided during virus construction. Replication-competent virus indicative of recombination between the deletion virus and the M94 gene expressed by NT/M94-7 was never observed. In order to investigate the safety of MCMV-ΔM94 for vaccination studies in a highly susceptible mouse strain, 129.IFNαβR−/− mice were infected with MCMV-wt or MCMV-ΔM94. While all IFNαβR−/− mice died within 14 days upon infection with MCMV-wt, after infection with MCMV-ΔM94 all mice survived with no or only minimal weight loss (Fig. 2C). In conclusion, MCMV-ΔM94 could be safely produced and even immune-deficient mice tolerated MCMV-ΔM94 infection.
MCMV-ΔM94 induces neutralizing antibody and T-cell responses.
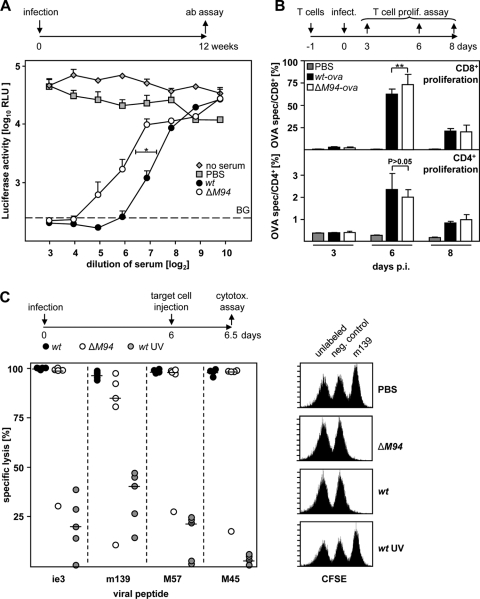
Poor induction of neutralizing antibodies that prevent viral entry is a problem in HCMV infection (39). Therefore, we compared the neutralizing antibody responses to MCMV-wt and MCMV-ΔM94 12 weeks postimmunization. Serial dilutions of sera were mixed with a luciferase-expressing MCMV (MCMV-luc) prior to infection of NIH 3T3 cells. The reduction of the luciferase signal reflected the neutralizing capacity of the antisera. Immunization with MCMV-ΔM94 induced a slightly smaller amount of neutralizing antibodies than immunization with MCMV-wt (Fig. 3A) (P < 0.05), whereas immunization with UV-irradiated MCMV-wt abolished the induction of neutralizing antibodies (data not shown), confirming published observations (21).
FIG. 3.
MCMV-ΔM94 induces neutralizing antibodies and T-cell responses. (A) B6 mice were immunized i.p. with 105 TCID50 MCMV-wt (wt) or MCMV-ΔM94 (ΔM94) or mock infected (PBS). Blood was collected 12 weeks p.i., and virus-neutralizing capacity of the serum was determined using MCMV-luc. Neutralizing antibody (ab) levels of MCMV-ΔM94-immunized mice were significantly lower than antibody levels of MCMV-wt-immunized mice by two-way ANOVA testing (P = 0.04). Values represent the means plus SD of measured serum pools. RLU, relative luciferase units; BG, background. (B) After adoptive transfer of 3 × 105 OT-I CD8+ T cells (top), B6 mice (n = 5) were infected i.p. with 105 TCID50 MCMV-ova (wt-ova) or MCMV-ova-ΔM94 (ΔM94-ova) or injected with PBS. At days 3, 6, and 8 p.i. flow cytometric analysis was performed on blood for the congenic marker CD45.1 and CD8. After adoptive transfer of 3 × 105 OT-II CD4+ T cells (bottom), B6 mice (n = 5) were infected i.p. as above. At days 3, 6, and 8 p.i. flow cytometric analysis was done on splenocytes for CD90.1 and CD4. Each bar represents the mean plus SD for the indicated group (**, P < 0.01). spec, specific. (C) B6 mice (n = 5) were infected i.p. with 105 TCID50 MCMV-wt (wt), MCMV-ΔM94 (ΔM94), or UV-irradiated MCMV-wt (wt UV). At day 6 p.i. an in vivo cytotoxicity assay was performed using splenocytes labeled with carboxyfluorescein succinimidyl ester (CFSE) and the indicated viral peptides. Symbols represent the specific lysis activity against the indicated peptide in individual animals. The cross bars indicate the medians of the analyzed groups. The right panel shows an exemplary set of flow cytometric data.
Both CD4+ and CD8+ T cells play important roles in host defense against CMV. Antiviral CD8+ T cells are effective in controlling MCMV during acute infection and mediate protection after immunization (57). In addition, CD4+ T helper cells are required for virus clearance in salivary glands (31). In order to compare the levels of CD4+ and CD8+ T-cell responses induced by MCMV-wt and MCMV-ΔM94, we chose OVA as a model antigen to be expressed by the vaccine. B6 mice were infected with MCMV-ova and MCMV-ova-ΔM94 1 day after adoptive transfer of OVA-specific CD4+ or CD8+ T cells. For MCMV-ova the expansion of OVA-specific CD4+ and CD8+ T cells peaked at day 6 p.i., concordant with published data (33). Remarkably, MCMV-ova-ΔM94 also stimulated the proliferative response of OVA-specific CD8+ and CD4+ (Fig. 3B) T cells to a degree comparable to that achieved by the spread-competent MCMV-ova. The amount of CD8+ T cells was even slightly larger than that obtained with MCMV-wt (P < 0.01).
We wished to confirm this observation for native MCMV antigens. B6 mice were infected with MCMV-ΔM94 or MCMV-wt. At six days p.i., target cells loaded with viral peptides derived from either m139, ie3, M57, or M45 (68) were injected and their cytolysis in vivo was analyzed (Fig. 3C). The cytolytic CD8+ T-cell response induced by MCMV-ΔM94 turned out to be comparable to that induced by MCMV-wt. In contrast, B6 mice injected with UV-irradiated MCMV generated no or only poor lysis of targets. UV inactivation of MCMV-ΔM94 or MCMV-wt also abolished OVA-specific T-cell expansion and the virus-neutralizing capacity of sera (data not shown). Thus, viral gene expression appeared to be crucial for the induction of the adaptive immune response. Altogether, spread-deficient MCMV induced an immune response comparable to that induced by MCMV-wt.
Role of viral target cell types in CD8+ T-cell activation.
The strong adaptive immune response against MCMV-ΔM94 was surprising, since MCMV-ΔM94 gene expression is limited to the first-target cells. Induction of a specific T-cell response is dependent on antigen presentation by infected cells and by professional antigen-presenting cells (APC) (76). In order to assess the contributions of infection of different cell types to the generation of an efficient CD8+ T-cell response, we combined the replication-deficient MCMV with conditional activation of a marker gene (61). We constructed MCMV-flox-ova-ΔM94, which expresses OVA only after Cre-mediated recombination.
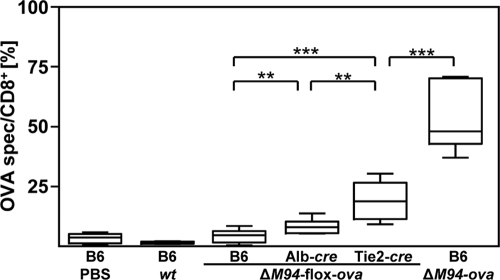
Endothelial cells (EC) and hepatocytes (Hc) are among the first-target cells infected by MCMV in vivo (61). Whether these cell types contribute to CD8+ T-cell activation was addressed by infecting mice that express Cre recombinase selectively in vascular EC (Tie2-cre) or Hc (Alb-cre). One day after adoptive transfer of OVA-specific CD8+ T cells, mice were infected with 105 TCID50 of spread-deficient MCMV-flox-ova-ΔM94. Hc are the main producers of infectious virus during the first few days of infection and are highly effective in activating a conditional marker gene via Cre recombinase (61). Yet selective induction of OVA expression in MCMV-infected Hc resulted in only weak proliferation of OVA-specific CD8+ T cells (Fig. 4). In contrast, a significantly (P < 0,001) higher proliferative response of OVA-specific CD8+ T cells was observed upon OVA expression in EC. Therefore, infection of EC makes a stronger contribution to the induction of an antiviral CD8+ T-cell response than infection of Hc. As infection of C57BL/6 mice with MCMV-ΔM94-ova, which leads to expression of OVA in all infected cells, induces a higher proportion of OVA-specific CD8+ T cells than expression selectively in EC (Tie2-cre mice infected with MCMV-ΔM94-flox-ova; P < 0.01), additional cell types seem to be involved in antigen expression and T-cell stimulation. In addition, the significantly different T-cell responses after cell-type-specific recombination in vivo prove that MCMV-ΔM94 is unable to spread from cell to cell.
FIG. 4.
EC contribute to antiviral CD8+ T-cell stimulation. One day prior to i.p. injection of 105 TCID50 of MCMV-flox-ova-ΔM94 (ΔM94-flox-ova), MCMV-ova-ΔM94 (ΔM94-ova), MCMV-wt (wt), or PBS, 3 × 105 congenic OT-I CD8+ T cells were transferred i.v. into B6, Alb-cre, and Tie2-cre mice. At day 6 p.i. a flow cytometric analysis was performed on peripheral blood lymphocytes (PBL) for the congenic marker CD45.1 and CD8. Boxes represent the ratios of OT-I cells per CD8+ cells as a pool of 3 independent experiments and extend from the 25th to the 75th percentile. The lines indicate the medians. Whiskers extend to show the extreme values. The P values were obtained by applying a two-tailed Wilcoxon rank sum test (**, P < 0.01; ***, P < 0.001).
MCMV-ΔM94 protects against challenge with MCMV-wt.
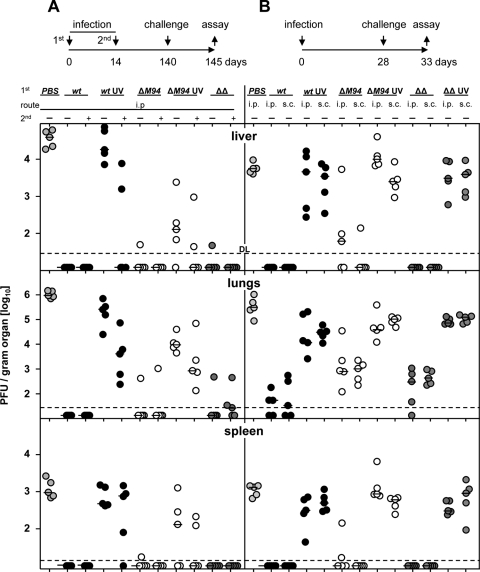
In order to test protection of MCMV-ΔM94 against lethal challenge, B6 mice were infected with either spread-deficient MCMV-ΔM94, the attenuated strain Δm01-17+m144-158-MCMV (11), or MCMV-wt. A boost infection with the same dose was applied 4 weeks later. Twenty weeks after priming mice were challenged i.v. with 106 TCID50 tissue culture-derived MCMV-wt. Most remarkably, a single immunization dose of MCMV-ΔM94 was already sufficient to strongly suppress MCMV-wt replication by 10,000-fold in lungs, 1,000-fold in liver, and at least 100-fold in spleen, whereas nonimmunized controls had high virus loads in all organs tested (all P < 0.01) (Fig. 5A). Overall, the protection mediated by MCMV-ΔM94 vaccination was comparable to that mediated by MCMV-wt or Δm01-17+m144-158-MCMV vaccination (all P > 0.05). Due to the strong protection achieved already after one administration, a boosting effect could not be detected. However, there was weak protective effect after a single dose when UV-inactivated MCMV-wt or UV inactivated MCMV-ΔM94 virus was administered. After a boost with UV-inactivated viruses, the effect was slightly improved but still remained lower than that of a single dose of MCMV-ΔM94 (P < 0.05).
FIG. 5.
MCMV-ΔM94 protects against challenge with MCMV-wt. B6 mice (n = 5) were immunized (1st) s.c. or i.p. with 105 TCID50 MCMV-wt (wt; solid symbols), MCMV-ΔM94 (ΔM94; open symbols), Δm01-17+m144-158-MCMV (ΔΔ; dark gray symbols), or PBS (light gray symbols). Virus preparations were UV irradiated before immunization (UV) as indicated. Optionally, mice were boosted (2nd) 2 weeks later with the same dose, route, and virus. Challenge infection was applied i.v. 20 (A) or 4 weeks (B) after priming with 106 PFU MCMV-wt. Five days postchallenge, a plaque assay was performed. Horizontal bars show the medians for the groups. Each symbol represents one individual mouse. DL, detection limit.
We asked whether the strong protection after a single administration of MCMV-ΔM94 could also be realized in a short-term vaccination protocol. In addition, we tested the influence of two different application routes. B6 mice were injected either i.p. or s.c., followed by challenge infection with MCMV-wt only 4 weeks later. Here, vaccination with MCMV-ΔM94 resulted in about a 100-fold reduction of challenge virus load in liver (P < 0.05), lungs (P < 0.01), and spleen (P < 0.01) (Fig. 5B), comparable to immunization with Δm01-17+m144-158-MCMV. MCMV-wt vaccination resulted in reduction of challenge virus load by 1,000-fold (P < 0.01). Generally, there was no significant difference between the i.p. and s.c. vaccination routes although s.c. injection appeared to induce slightly better protection in spleen (P > 0.05) (Fig. 5B) and hearts (data not shown).
In summary, the spread-deficient MCMV-ΔM94 was able to efficiently protect immunocompetent mice against challenge with MCMV-wt after vaccination with a single dose. Remarkably, vaccination with MCMV-ΔM94 was as efficient as vaccination with MCMV-wt for long-term protection, whereas the use of UV-inactivated virus could not compete even after a second application.
Protection of severely immune-compromised recipients.
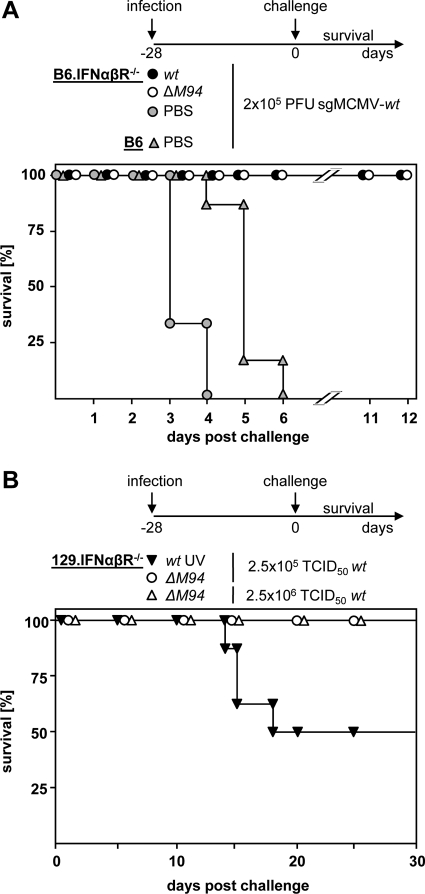
Type I interferons are key cytokines in the immune response against CMV, and deletion of their receptor results in a mouse (IFNαβR−/−) that is severely immunocompromised and at least 1.000-fold more susceptible to MCMV infection than the parental mouse strain (53). Since spread-deficient MCMV-ΔM94 was proven to be well tolerated by IFNαβR−/− mice (Fig. 1C), we tested whether MCMV-ΔM94 could protect IFNαβR−/− mice against lethal MCMV-wt challenge (Fig. 6 A). B6.IFNαβR−/− mice were immunized with MCMV-ΔM94 or a sublethal dose of MCMV-wt. Both groups survived, and mice immunized with MCMV-ΔM94 showed no significant weight loss, whereas MCMV-wt-infected mice lost approximately 15% of their body weight (data not shown). Four weeks later, mice were challenged by infection with a lethal dose of more-virulent salivary gland-derived MCMV (see Materials and Methods). Most strikingly, the vaccination with both MCMV-ΔM94 and MCMV-wt was protective and all animals survived (Fig. 6A).
FIG. 6.
MCMV-ΔM94 protects immunocompromised mice against MCMV-wt challenge. (A) B6.IFNαβR−/− (n = 6) mice were immunized i.p. with 3 × 105 TCID50 MCMV-wt (wt) or MCMV-ΔM94 (ΔM94). Control groups of B6.IFNαβR−/− (gray circles) or B6 (gray triangles) mice were treated with PBS. Four weeks later challenge infection was performed by i.p. injection of mice with 2 × 105 PFU salivary gland-derived MCMV (sgMCMV-wt), and survival was monitored. (B) 129.IFNαβR−/− mice immunized 4 weeks earlier with 2.5 × 105 TCID50 of MCMV-ΔM94 (ΔM94; open circles; n = 8), or UV-irradiated MCMV-wt (wt UV; n = 8) were challenged with a lethal dose of MCMV-wt (see Fig. 2C), and survival was monitored. A 10-fold-higher dose of MCMV-wt was applied to mice immunized with MCMV-ΔM94 (n = 7) (open triangles).
B6 mice profit from an Ly49H-dependent activation of natural killer cells, resulting in a strong innate immune response stimulated by the MCMV protein encoded by m157 (71). 129.IFNαβR−/− mice do not express Ly49H and are even more susceptible to MCMV infection than B6.IFNαβR−/− mice. 129.IFNαβR−/− mice were vaccinated with MCMV-ΔM94 and challenged 4 weeks later with a dose of 2.5 × 105 TCID50 tissue culture-derived MCMV-wt (Fig. 6B). In line with the earlier data (11), vaccination with UV-inactivated virus mediated only partial protection and could delay death for a short period. MCMV-ΔM94-vaccinated mice survived the lethal challenge even with a dose of 2.5 × 106 TCID50. In summary, vaccination with MCMV-ΔM94 is able to protect even highly susceptible immune-compromised mice against lethal MCMV challenge.
Maintenance of the MCMV-ΔM94 genome in vivo.
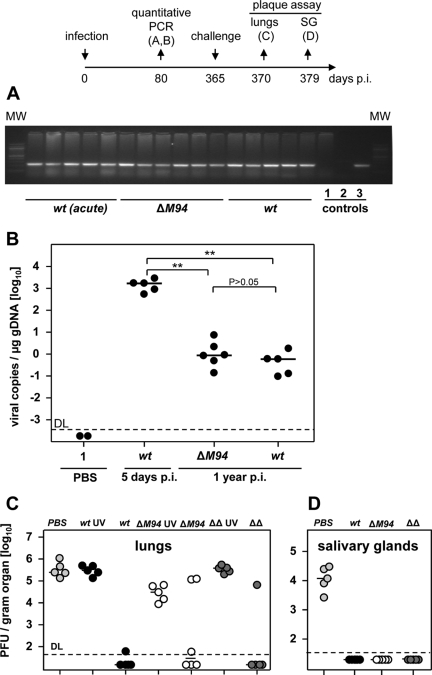
One argument against the application of live attenuated vaccines is their ability to establish a latent infection that bears the risk of reactivation (30). On the other hand nonproductive reactivation episodes might result in endogenous boosts of the antiviral immune response (68). Thus, it was intriguing to test whether the MCMV-ΔM94 genome is maintained in vaccinated hosts. Quantitative PCR analysis of total DNA extracted from lungs, a key manifestation site of CMV disease (5), was performed. Twelve months p.i. genomes of MCMV-ΔM94 could be detected in all mice tested (Fig. 7 A and B), proving that the genome of MCMV-ΔM94 is maintained. Interestingly, the genome numbers detected in lungs 1 year after infection with MCMV-ΔM94 and MCMV-wt were not significantly different (P > 0.05). This finding proved that at least some of the first-target cells are not lost after infection either due to virus-induced cell death or elimination by the immune response. In summary, these data also provide the first evidence that virus spread is not necessary for long-term genome maintenance and that first-target cells infected with MCMV-ΔM94 may be able to contribute to a more sustained immune response.
FIG. 7.
Long-term maintenance of MCMV-ΔM94 genome and protection, B6 mice were infected i.p. with 105 TCID50 MCMV-wt (wt) (n = 5) or MCMV-ΔM94 (ΔM94) (n = 6). Twelve months p.i. total DNA was extracted from lungs. (A) PCR analysis was performed, obtaining a specific 246-bp fragment of the polymerase gene M54. As controls DNA from lungs 5 days after infection with 105 TCID50 MCMV-wt [wt (acute)] (n = 5), PBS (1), no template (2), or the BAC plasmid pSM3fr (3) was used. MW, molecular weight marker. (B) Quantitative real-time PCR analysis was performed, and viral M54 gene copies per μg genomic DNA were calculated. Each symbol represents one individual mouse. Horizontal bars show the medians of the groups. Genome copy numbers of MCMV-wt (wt) and MCMV-ΔM94 (ΔM94) are not significantly different (P > 0.05). Both groups are significantly different from mice with acutely infected lungs [wt (acute)] (**, P < 0.01). DL, detection limit. (C and D) B6 mice (n = 5) were immunized i.p. with 105 TCID50 MCMV-wt (wt; solid symbols), MCMV-ΔM94 (ΔM94; open symbols), Δm01-17+m144-158-MCMV (ΔΔ; dark gray symbols), or PBS (light gray symbols). Virus preparations were UV irradiated before immunization (UV) as indicated. Challenge infection was applied i.v. 1 year after priming with 106 PFU MCMV-wt. The plaque assay was performed 5 days after challenge in lungs (C) and 14 days after challenge in salivary glands (D). Horizontal bars show the medians of the groups. Each symbol represents one individual mouse.
Vaccination with MCMV-ΔM94 prevents replication of virus in the respiratory tract.
From epidemiological studies it was suggested that saliva is an important route of transmission of HCMV (49). To test whether the vaccine MCMV-ΔM94 is able to block virus replication in salivary glands and lungs, C57BL/6 mice were immunized with MCMV-ΔM94 or control viruses and received 12 months later a challenge infection with 106 PFU MCMV-wt i.v. (Fig. 7C and D). A single application of MCMV-ΔM94 was sufficient to suppress challenge virus replication by more than a factor of 1,000 in lungs in 4 out of 6 animals (Fig. 7C). Further, no challenge virus could be isolated from salivary glands 14 days after challenge (Fig. 7D). This implies that shedding of virus from the respiratory tract via saliva and therefore horizontal transmission via this route are abrogated by vaccination with spread-deficient MCMV.
DISCUSSION
We report on the vaccination against a betaherpesvirus using a spread-deficient vaccine. The vaccine induced a strong adaptive immune response comparable to that induced by MCMV-wt, conferring protection even in highly immune-compromised mice. This implies that infection of the first-target cells is sufficient for successful vaccination.
An intact immune system usually protects against HCMV disease. Hence, the antigenic capacity of the wild-type virus is sufficient for the induction of a protective immune response. The inability of UV-inactivated virus to protect efficiently against challenge infection demonstrated the need for viral antigen expression, including nonstructural antigens (11, 21). As a consequence an ideal vaccine should exploit the full immunogenic potential but avoid the pathogenic potential of the wild-type virus.
The alphaherpesvirus field has pioneered the use of replication-defective viruses as vaccines (16). These vaccines were generated by the deletion of genes essential for virus replication and are thus apathogenic (46). Now, to construct a spread-deficient betaherpesvirus vaccine, we chose deletion of M94 for the following reasons. First, M94 is essential for spread of MCMV and, as inferred from studies of HCMV, it should be expressed with late kinetics during virus replication (64, 77). Second, pM94 does not belong to the group of glycoproteins, which comprise major targets for the neutralizing antibody response of HCMV. Third, M94 of MCMV is the homologue of UL94 in HCMV (77), which in principle allows translation to the human pathogen. Finally, the deletion of UL94 of HCMV might even be of advantage because pUL94 induces autoreactive antibodies that are associated with systemic sclerosis (42). Interestingly, genomes of the spread-deficient MCMV-ΔM94 were detected in lungs after i.p. infection, showing that virus can disseminate either as free particles (29) or as cell-associated virus. Monocytes and macrophages were shown to be attracted to the peritoneal cavity after infection and transport of the virus in blood (69, 75). These cells could also release virus at distant sites to infect EC or other cell types, a process called trans infection (24).
The spread-deficient betaherpesvirus vaccine presented here has a strong protective capacity, similar to MCMV-wt infection. The immune response of the vaccinee controls virus replication in all analyzed organs, preventing overt CMV disease. The absence of detectable amounts of infectious virus in salivary glands of long-term-vaccinated mice 2 weeks after challenge implies that horizontal transmission to other individuals via saliva is also abrogated. But a crucial difference between MCMV (38) and HCMV is that only in the latter is reinfection common (4, 7, 13, 54). Thus, it seems possible that a similar human vaccine will not be able to protect in all scenarios against reinfection. Nevertheless, it is possible that such an equivalent vaccine will protect against HCMV disease, similar to the protective effect of a preexisting infection. This is supported by the observation that women who were exposed to HCMV were at lower risk of giving birth to children with symptomatic disease than noninfected women (19). The seropositivity of the mother could not prevent infection of, but did prevent pathogenesis in, the children. In addition, frequent exposure to different CMV strains could further increase immunity against reinfection (1). It is therefore tempting to test a spread-deficient HCMV vaccine for the induction of an immune response equal to natural infection, which might still protect against symptomatic HCMV infection without the risk for reactivation and pathogenesis.
The immune response to MCMV-ΔM94 reached a level comparable to that to MCMV-wt. Protection was similar to that by the recently generated vaccine Δm01-17+m144-158-MCMV (11), which lacks 32 viral genes but which is not spread deficient in vitro. In Δm01-17+m144-158-MCMV, immune-evasive genes were deleted to increase the antiviral immune response and thereby to attenuate the virus. The combination of both approaches in the future, namely, the deletion of at least one essential gene combined with the deletion of selected immune evasins (62), provides room for improved immunogenicity and safety.
Infection of susceptible IFNαβR−/− mice with spread-deficient MCMV proved the safety of the vaccination concept. Furthermore, IFNαβR−/− mice were protected against an otherwise lethal challenge, similar to results from other infection models (9, 48). Although recent work revealed the capacity of MCMV to efficiently induce type I interferon (26), the efficacy of the spread-deficient MCMV vaccine in IFNαβR−/− mice implies that type I interferon-dependent immunity is not essential for the protection conferred by short-term vaccination.
Interestingly, the spread-deficient MCMV induced an adaptive immune response with efficiency similar to that of MCMV-wt. The CD4+ and CD8+ T-cell response was on the same level as that for MCMV-wt, and the neutralizing antibody response was only marginally reduced. This slightly lower neutralizing capacity might be caused by the smaller number of infected cells and therefore by the reduced amount of antigen that is released after infection with MCMV-ΔM94. A lower number of antigen-antibody complexes might lead to less-efficient affinity maturation, creating antibodies of lower neutralizing capacity. Nevertheless, the neutralization of virus appears sufficient to control virus replication.
Why did the adaptive immune response to the vaccine reach a level near to that for MCMV-wt infection despite the inability to spread? MCMV-ΔM94 was able to establish viral genome maintenance as efficiently as MCMV-wt. The classical definition of herpesviral latency includes the potential for reactivated gene expression, with subsequent release of infectious virus (59). Although the term “latency” is formally not applicable to the situation with MCMV-ΔM94 in the absence of productive infection, there is no evidence that pM94 affects reactivation of gene expression. Because the protective effect of MCMV-ΔM94 increased rather than faded over time, we believe that periodic restimulation of the immune response due to reactivation of gene expression contributed to the sustained protection induced by MCMV-ΔM94. Interestingly, virus-infected cells are not eliminated by the activated immune response. This means that the first-target cells that are infected by the spread-deficient vaccine are resistant to elimination. Similarly, cells infected with a spread-deficient mutant of the gammaherpesvirus MHV-68 were not attacked by the adaptive immune response (72). For MCMV-wt it was shown that T cells are activated against a highly antigenic virus epitope of M45 presented by professional APC but that the activated T cells did not eliminate infected target cells in organs of C57BL/6 mice (27). This protection was caused by m152, which is known to downmodulate major histocompatibility complex class I (MHC-I). The target cells that are protected from CD8+ T-cell elimination were not identified, and we could show that at least some of these protected cells are first-target cells of MCMV. Studies with MCMV-ΔM94 and new constructs should help to shed more light on latency and long-term immunity against MCMV.
Endothelial cells (EC), hepatocytes (Hc), and macrophages are first-target cells for HCMV and MCMV in vivo (29, 61). In addition, EC have recently been identified as sites of virus latency (65), and at least liver EC are able to directly stimulate a cytotoxic T-cell response (36). Using MCMV-ΔM94 constructs for conditional gene expression, we noticed substantial differences in the abilities of EC and Hc to activate a CD8+ T-cell response. In contrast to EC, Hc, one of the most important first targets for MCMV during acute infection (61), induced only a poor CD8+ T-cell response. This lack of stimulatory capacity is apparently not compensated by cross presentation through professional antigen-presenting cells. Cross presentation was shown to be important for the induction of a T-cell response against fibroblasts infected with a spread-deficient MCMV (67). On the other hand, bone marrow-derived APC, which are thought to be important cross-presenting cells, seem not to be necessary for the activation of a CD8+ T-cell response via cross presentation against MCMV infection (36). In addition to EC, other cell types seem to contribute to CD8+ T-cell stimulation, as antigen expression in most infected cells led to a stronger T-cell response than expression in infected EC only. Infected dendritic cells and macrophages were shown to activate a T-cell response against MCMV in vitro (43), and these cells can be infected in vivo (2). Therefore, infected professional APC may contribute to immune stimulation against MCMV in addition to EC. It appears noteworthy that the attenuated HCMV strains such as Towne and AD169, which are characterized by a 20-fold reduction of immunogenicity and the inability to confer immune protection (1), accumulated mutations resulting in their inability to infect EC, epithelial cells, smooth muscle cells, and macrophages (23). Thus, it appears likely that the restricted cell tropism may in fact represent the cause for their failure as HCMV vaccines. It will be tempting to determine the roles of different cell types in CD8+ T-cell stimulation and protection. In conclusion, we propose that spread-deficient HCMVs that are able to infect the proper primary cell types and establish long-term genome maintenance deserve further clinical studies as candidate vaccines.
Supplementary Material
Acknowledgments
This project was supported by the DFG SFB 455 (Z.R., U.K.) and DFG SFB 571 (D.V.), NGFN 01GS0405 (U.K.), Baur Stiftung (T.S.), and Croatian Ministry of Science grant 0621261-1268 (A.K.).
We thank S. Jonjic, S. Jordan, T. Brocker and A. Gruber for helpful discussions and support. We thank N. Röder and A. Käsewieter for expert technical assistance.
The authors have no conflicts of interest.
Footnotes
Published ahead of print on 12 May 2010.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Adler, S. P., S. E. Starr, S. A. Plotkin, S. H. Hempfling, J. Buis, M. L. Manning, and A. M. Best. 1995. Immunity induced by primary human cytomegalovirus infection protects against secondary infection among women of childbearing age. J. Infect. Dis. 171:26-32. [DOI] [PubMed] [Google Scholar]
- 2.Andrews, D. M., C. E. Andoniou, F. Granucci, P. Ricciardi-Castagnoli, and M. A. Degli-Esposti. 2001. Infection of dendritic cells by murine cytomegalovirus induces functional paralysis. Nat. Immunol. 2:1077-1084. [DOI] [PubMed] [Google Scholar]
- 3.Babu, J. S., J. Thomas, S. Kanangat, L. A. Morrison, D. M. Knipe, and B. T. Rouse. 1996. Viral replication is required for induction of ocular immunopathology by herpes simplex virus. J. Virol. 70:101-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bale, J. F., Jr., S. J. Petheram, I. E. Souza, and J. R. Murph. 1996. Cytomegalovirus reinfection in young children. J. Pediatr. 128:347-352. [DOI] [PubMed] [Google Scholar]
- 5.Balthesen, M., M. Messerle, and M. J. Reddehase. 1993. Lungs are a major organ site of cytomegalovirus latency and recurrence. J. Virol. 67:5360-5366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barnden, M. J., J. Allison, W. R. Heath, and F. R. Carbone. 1998. Defective TCR expression in transgenic mice constructed using cDNA-based alpha- and beta-chain genes under the control of heterologous regulatory elements. Immunol. Cell Biol. 76:34-40. [DOI] [PubMed] [Google Scholar]
- 7.Boppana, S. B., L. B. Rivera, K. B. Fowler, M. Mach, and W. J. Britt. 2001. Intrauterine transmission of cytomegalovirus to infants of women with preconceptional immunity. N. Engl. J. Med. 344:1366-1371. [DOI] [PubMed] [Google Scholar]
- 8.Britt, W. J., L. Vugler, E. J. Butfiloski, and E. B. Stephens. 1990. Cell surface expression of human cytomegalovirus (HCMV) gp55-116 (gB): use of HCMV-recombinant vaccinia virus-infected cells in analysis of the human neutralizing antibody response. J. Virol. 64:1079-1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Calvo-Pinilla, E., T. Rodriguez-Calvo, J. Anguita, N. Sevilla, and J. Ortego. 2009. Establishment of a bluetongue virus infection model in mice that are deficient in the alpha/beta interferon receptor. PLoS One 4:e5171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cha, T. A., E. Tom, G. W. Kemble, G. M. Duke, E. S. Mocarski, and R. R. Spaete. 1996. Human cytomegalovirus clinical isolates carry at least 19 genes not found in laboratory strains. J. Virol. 70:78-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cicin-Sain, L., I. Bubic, M. Schnee, Z. Ruzsics, C. Mohr, S. Jonjic, and U. H. Koszinowski. 2007. Targeted deletion of regions rich in immune-evasive genes from the cytomegalovirus genome as a novel vaccine strategy. J. Virol. 81:13825-13834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cicin-Sain, L., J. Podlech, M. Messerle, M. J. Reddehase, and U. H. Koszinowski. 2005. Frequent coinfection of cells explains functional in vivo complementation between cytomegalovirus variants in the multiply infected host. J. Virol. 79:9492-9502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Coaquette, A., A. Bourgeois, C. Dirand, A. Varin, W. Chen, and G. Herbein. 2004. Mixed cytomegalovirus glycoprotein B genotypes in immunocompromised patients. Clin. Infect. Dis. 39:155-161. [DOI] [PubMed] [Google Scholar]
- 14.Constien, R., A. Forde, B. Liliensiek, H. J. Grone, P. Nawroth, G. Hammerling, and B. Arnold. 2001. Characterization of a novel EGFP reporter mouse to monitor Cre recombination as demonstrated by a Tie2 Cre mouse line. Genesis 30:36-44. [DOI] [PubMed] [Google Scholar]
- 15.Da Costa, X. J., L. A. Morrison, and D. M. Knipe. 2001. Comparison of different forms of herpes simplex replication-defective mutant viruses as vaccines in a mouse model of HSV-2 genital infection. Virology 288:256-263. [DOI] [PubMed] [Google Scholar]
- 16.Dudek, T., and D. M. Knipe. 2006. Replication-defective viruses as vaccines and vaccine vectors. Virology 344:230-239. [DOI] [PubMed] [Google Scholar]
- 17.Dunn, W., C. Chou, H. Li, R. Hai, D. Patterson, V. Stolc, H. Zhu, and F. Liu. 2003. Functional profiling of a human cytomegalovirus genome. Proc. Natl. Acad. Sci. U. S. A. 100:14223-14228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Forrester, A., H. Farrell, G. Wilkinson, J. Kaye, N. Davis-Poynter, and T. Minson. 1992. Construction and properties of a mutant of herpes simplex virus type 1 with glycoprotein H coding sequences deleted. J. Virol. 66:341-348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fowler, K. B., S. Stagno, and R. F. Pass. 2003. Maternal immunity and prevention of congenital cytomegalovirus infection. JAMA 289:1008-1011. [DOI] [PubMed] [Google Scholar]
- 20.Fowler, K. B., S. Stagno, R. F. Pass, W. J. Britt, T. J. Boll, and C. A. Alford. 1992. The outcome of congenital cytomegalovirus infection in relation to maternal antibody status. N. Engl. J. Med. 326:663-667. [DOI] [PubMed] [Google Scholar]
- 21.Gill, T. A., P. J. Morley, and C. Sweet. 2000. Replication-defective mutants of mouse cytomegalovirus protect against wild-type virus challenge. J. Med. Virol. 62:127-139. [DOI] [PubMed] [Google Scholar]
- 22.Gonczol, E., and S. Plotkin. 2001. Development of a cytomegalovirus vaccine: lessons from recent clinical trials. Expert Opin. Biol. Ther. 1:401-412. [DOI] [PubMed] [Google Scholar]
- 23.Hahn, G., M. G. Revello, M. Patrone, E. Percivalle, G. Campanini, A. Sarasini, M. Wagner, A. Gallina, G. Milanesi, U. Koszinowski, F. Baldanti, and G. Gerna. 2004. Human cytomegalovirus UL131-128 genes are indispensable for virus growth in endothelial cells and virus transfer to leukocytes. J. Virol. 78:10023-10033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Halary, F., A. Amara, H. Lortat-Jacob, M. Messerle, T. Delaunay, C. Houles, F. Fieschi, F. Arenzana-Seisdedos, J. F. Moreau, and J. Dechanet-Merville. 2002. Human cytomegalovirus binding to DC-SIGN is required for dendritic cell infection and target cell trans-infection. Immunity 17:653-664. [DOI] [PubMed] [Google Scholar]
- 25.Hogquist, K. A., S. C. Jameson, W. R. Heath, J. L. Howard, M. J. Bevan, and F. R. Carbone. 1994. T cell receptor antagonist peptides induce positive selection. Cell 76:17-27. [DOI] [PubMed] [Google Scholar]
- 26.Hokeness-Antonelli, K. L., M. J. Crane, A. M. Dragoi, W. M. Chu, and T. P. Salazar-Mather. 2007. IFN-alphabeta-mediated inflammatory responses and antiviral defense in liver is TLR9-independent but MyD88-dependent during murine cytomegalovirus infection. J. Immunol. 179:6176-6183. [DOI] [PubMed] [Google Scholar]
- 27.Holtappels, R., J. Podlech, M. F. Pahl-Seibert, M. Julch, D. Thomas, C. O. Simon, M. Wagner, and M. J. Reddehase. 2004. Cytomegalovirus misleads its host by priming of CD8 T cells specific for an epitope not presented in infected tissues. J. Exp. Med. 199:131-136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Holtappels, R., D. Thomas, and M. J. Reddehase. 2009. The efficacy of antigen processing is critical for protection against cytomegalovirus disease in the presence of viral immune evasion proteins. J. Virol. 83:9611-9615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hsu, K. M., J. R. Pratt, W. J. Akers, S. I. Achilefu, and W. M. Yokoyama. 2009. Murine cytomegalovirus displays selective infection of cells within hours after systemic administration. J. Gen. Virol. 90:33-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Iyer, S., M. K. Mittal, and R. L. Hodinka. 2009. Herpes zoster and meningitis resulting from reactivation of varicella vaccine virus in an immunocompetent child. Ann. Emerg. Med. 53:792-795. [DOI] [PubMed] [Google Scholar]
- 31.Jonjic, S., W. Mutter, F. Weiland, M. J. Reddehase, and U. H. Koszinowski. 1989. Site-restricted persistent cytomegalovirus infection after selective long-term depletion of CD4+ T lymphocytes. J. Exp. Med. 169:1199-1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jonjic, S., I. Pavic, B. Polic, I. Crnkovic, P. Lucin, and U. H. Koszinowski. 1994. Antibodies are not essential for the resolution of primary cytomegalovirus infection but limit dissemination of recurrent virus. J. Exp. Med. 179:1713-1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Karrer, U., M. Wagner, S. Sierro, A. Oxenius, H. Hengel, T. Dumrese, S. Freigang, U. H. Koszinowski, R. E. Phillips, and P. Klenerman. 2004. Expansion of protective CD8+ T-cell responses driven by recombinant cytomegaloviruses. J. Virol. 78:2255-2264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kattenhorn, L. M., R. Mills, M. Wagner, A. Lomsadze, V. Makeev, M. Borodovsky, H. L. Ploegh, and B. M. Kessler. 2004. Identification of proteins associated with murine cytomegalovirus virions. J. Virol. 78:11187-11197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Keadle, T. L., L. A. Morrison, J. L. Morris, J. S. Pepose, and P. M. Stuart. 2002. Therapeutic immunization with a virion host shutoff-defective, replication-incompetent herpes simplex virus type 1 strain limits recurrent herpetic ocular infection. J. Virol. 76:3615-3625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kern, M., A. Popov, K. Scholz, B. Schumak, D. Djandji, A. Limmer, D. Eggle, T. Sacher, R. Zawatzky, R. Holtappels, M. J. Reddehase, G. Hartmann, S. Debey-Pascher, L. Diehl, U. Kalinke, U. Koszinowski, J. Schultze, and P. A. Knolle. 2010. Virally infected mouse liver endothelial cells trigger CD8+ T-cell immunity. Gastroenterology 138:336-346. [DOI] [PubMed] [Google Scholar]
- 37.Kew, O. M., R. W. Sutter, B. K. Nottay, M. J. McDonough, D. R. Prevots, L. Quick, and M. A. Pallansch. 1998. Prolonged replication of a type 1 vaccine-derived poliovirus in an immunodeficient patient. J. Clin. Microbiol. 36:2893-2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Klotman, M. E., D. Starnes, and J. D. Hamilton. 1985. The source of murine cytomegalovirus in mice receiving kidney allografts. J. Infect. Dis. 152:1192-1196. [DOI] [PubMed] [Google Scholar]
- 39.Landini, M. P., and M. La Placa. 1991. Humoral immune response to human cytomegalovirus proteins: a brief review. Comp. Immunol. Microbiol. Infect. Dis. 14:97-105. [DOI] [PubMed] [Google Scholar]
- 40.Lauterbach, H., C. Ried, A. L. Epstein, P. Marconi, and T. Brocker. 2005. Reduced immune responses after vaccination with a recombinant herpes simplex virus type 1 vector in the presence of antiviral immunity. J. Gen. Virol. 86:2401-2410. [DOI] [PubMed] [Google Scholar]
- 41.Lotzerich, M., Z. Ruzsics, and U. H. Koszinowski. 2006. Functional domains of murine cytomegalovirus nuclear egress protein M53/p38. J. Virol. 80:73-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lunardi, C., C. Bason, R. Navone, E. Millo, G. Damonte, R. Corrocher, and A. Puccetti. 2000. Systemic sclerosis immunoglobulin G autoantibodies bind the human cytomegalovirus late protein UL94 and induce apoptosis in human endothelial cells. Nat. Med. 6:1183-1186. [DOI] [PubMed] [Google Scholar]
- 43.Mathys, S., T. Schroeder, J. Ellwart, U. H. Koszinowski, M. Messerle, and U. Just. 2003. Dendritic cells under influence of mouse cytomegalovirus have a physiologic dual role: to initiate and to restrict T cell activation. J. Infect. Dis. 187:988-999. [DOI] [PubMed] [Google Scholar]
- 44.Messerle, M., I. Crnkovic, W. Hammerschmidt, H. Ziegler, and U. H. Koszinowski. 1997. Cloning and mutagenesis of a herpesvirus genome as an infectious bacterial artificial chromosome. Proc. Natl. Acad. Sci. U. S. A. 94:14759-14763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Muller, U., U. Steinhoff, L. F. Reis, S. Hemmi, J. Pavlovic, R. M. Zinkernagel, and M. Aguet. 1994. Functional role of type I and type II interferons in antiviral defense. Science 264:1918-1921. [DOI] [PubMed] [Google Scholar]
- 46.Nguyen, L. H., D. M. Knipe, and R. W. Finberg. 1992. Replication-defective mutants of herpes simplex virus (HSV) induce cellular immunity and protect against lethal HSV infection. J. Virol. 66:7067-7072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nigro, G., S. P. Adler, R. La Torre, and A. M. Best. 2005. Passive immunization during pregnancy for congenital cytomegalovirus infection. N. Engl. J. Med. 353:1350-1362. [DOI] [PubMed] [Google Scholar]
- 48.Paran, N., Y. Suezer, S. Lustig, T. Israely, A. Schwantes, S. Melamed, L. Katz, T. Preuss, K. M. Hanschmann, U. Kalinke, N. Erez, R. Levin, B. Velan, J. Lower, A. Shafferman, and G. Sutter. 2009. Postexposure immunization with modified vaccinia virus Ankara or conventional Lister vaccine provides solid protection in a murine model of human smallpox. J. Infect. Dis. 199:39-48. [DOI] [PubMed] [Google Scholar]
- 49.Pass, R. F., C. Hutto, R. Ricks, and G. A. Cloud. 1986. Increased rate of cytomegalovirus infection among parents of children attending day-care centers. N. Engl. J. Med. 314:1414-1418. [DOI] [PubMed] [Google Scholar]
- 50.Pilgrim, M. J., L. Kasman, J. Grewal, M. E. Bruorton, P. Werner, L. London, and S. D. London. 2007. A focused salivary gland infection with attenuated MCMV: an animal model with prevention of pathology associated with systemic MCMV infection. Exp. Mol. Pathol. 82:269-279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Plotkin, S. A., S. E. Starr, H. M. Friedman, E. Gonczol, and R. E. Weibel. 1989. Protective effects of Towne cytomegalovirus vaccine against low-passage cytomegalovirus administered as a challenge. J. Infect. Dis. 159:860-865. [DOI] [PubMed] [Google Scholar]
- 52.Postic, C., M. Shiota, K. D. Niswender, T. L. Jetton, Y. Chen, J. M. Moates, K. D. Shelton, J. Lindner, A. D. Cherrington, and M. A. Magnuson. 1999. Dual roles for glucokinase in glucose homeostasis as determined by liver and pancreatic beta cell-specific gene knock-outs using Cre recombinase. J. Biol. Chem. 274:305-315. [DOI] [PubMed] [Google Scholar]
- 53.Presti, R. M., J. L. Pollock, A. J. Dal Canto, A. K. O'Guin, and H. W. Virgin. 1998. Interferon gamma regulates acute and latent murine cytomegalovirus infection and chronic disease of the great vessels. J. Exp. Med. 188:577-588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rasmussen, L., C. Hong, D. Zipeto, S. Morris, D. Sherman, S. Chou, R. Miner, W. L. Drew, R. Wolitz, A. Dowling, A. Warford, and T. C. Merigan. 1997. Cytomegalovirus gB genotype distribution differs in human immunodeficiency virus-infected patients and immunocompromised allograft recipients. J. Infect. Dis. 175:179-184. [DOI] [PubMed] [Google Scholar]
- 55.Rawlinson, W. D., H. E. Farrell, and B. G. Barrell. 1996. Analysis of the complete DNA sequence of murine cytomegalovirus. J. Virol. 70:8833-8849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Reddehase, M. J. 2002. Antigens and immunoevasins: opponents in cytomegalovirus immune surveillance. Nat. Rev. Immunol. 2:831-844. [DOI] [PubMed] [Google Scholar]
- 57.Reddehase, M. J., W. Mutter, K. Munch, H. J. Buhring, and U. H. Koszinowski. 1987. CD8-positive T lymphocytes specific for murine cytomegalovirus immediate-early antigens mediate protective immunity. J. Virol. 61:3102-3108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Riddell, S. R., and P. D. Greenberg. 2000. T-cell therapy of cytomegalovirus and human immunodeficiency virus infection. J. Antimicrob. Chemother. 45(Suppl. T3):35-43. [DOI] [PubMed] [Google Scholar]
- 59.Roizman, B., and A. E. Sears. 1987. An inquiry into the mechanisms of herpes simplex virus latency. Annu. Rev. Microbiol. 41:543-571. [DOI] [PubMed] [Google Scholar]
- 60.Roizman, B., J. Warren, C. A. Thuning, M. S. Fanshaw, B. Norrild, and B. Meignier. 1982. Application of molecular genetics to the design of live herpes simplex virus vaccines. Dev. Biol. Stand. 52:287-304. [PubMed] [Google Scholar]
- 61.Sacher, T., J. Podlech, C. A. Mohr, S. Jordan, Z. Ruzsics, M. J. Reddehase, and U. H. Koszinowski. 2008. The major virus-producing cell type during murine cytomegalovirus infection, the hepatocyte, is not the source of virus dissemination in the host. Cell Host Microbe 3:263-272. [DOI] [PubMed] [Google Scholar]
- 62.Scalzo, A. A., A. J. Corbett, W. D. Rawlinson, G. M. Scott, and M. A. Degli-Esposti. 2007. The interplay between host and viral factors in shaping the outcome of cytomegalovirus infection. Immunol. Cell Biol. 85:46-54. [DOI] [PubMed] [Google Scholar]
- 63.Schleiss, M. 2008. Cytomegalovirus vaccine development, p. 361-382. In T. E. Shenk and M. F. Stinski (ed.), Human cytomegalovirus. Springer-Verlag, Berlin, Germany.
- 64.Scott, G. M., B. G. Barrell, J. Oram, and W. D. Rawlinson. 2002. Characterisation of transcripts from the human cytomegalovirus genes TRL7, UL20a, UL36, UL65, UL94, US3 and US34. Virus Genes 24:39-48. [DOI] [PubMed] [Google Scholar]
- 65.Seckert, C. K., A. Renzaho, H. M. Tervo, C. Krause, P. Deegen, B. Kuhnapfel, M. J. Reddehase, and N. K. Grzimek. 2009. Liver sinusoidal endothelial cells are a site of murine cytomegalovirus latency and reactivation. J. Virol. 83:8869-8884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Shanley, J. D., and C. A. Wu. 2003. Mucosal immunization with a replication-deficient adenovirus vector expressing murine cytomegalovirus glycoprotein B induces mucosal and systemic immunity. Vaccine 21:2632-2642. [DOI] [PubMed] [Google Scholar]
- 67.Snyder, C. M., J. E. Allan, E. L. Bonnett, C. M. Doom, and A. B. Hill. 2010. Cross-presentation of a spread-defective MCMV is sufficient to prime the majority of virus-specific CD8+ T cells. PLoS One 5:e9681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Snyder, C. M., K. S. Cho, E. L. Bonnett, S. van Dommelen, G. R. Shellam, and A. B. Hill. 2008. Memory inflation during chronic viral infection is maintained by continuous production of short-lived, functional T cells. Immunity 29:650-659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Stoddart, C. A., R. D. Cardin, J. M. Boname, W. C. Manning, G. B. Abenes, and E. S. Mocarski. 1994. Peripheral blood mononuclear phagocytes mediate dissemination of murine cytomegalovirus. J. Virol. 68:6243-6253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Stratton, K., J. Durch, and R. Lawrence. 1999. Vaccines for the 21st century: a tool for decisionmaking. National Academy Press, Washington, DC. [PubMed]
- 71.Sun, J. C., and L. L. Lanier. 2008. Tolerance of NK cells encountering their viral ligand during development. J. Exp. Med. 205:1819-1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tibbetts, S. A., F. Suarez, A. L. Steed, J. A. Simmons, and H. W. Virgin. 2006. A gamma-herpesvirus deficient in replication establishes chronic infection in vivo and is impervious to restriction by adaptive immune cells. Virology 353:210-219. [DOI] [PubMed] [Google Scholar]
- 73.Torres-Madriz, G., and H. W. Boucher. 2008. Immunocompromised hosts: perspectives in the treatment and prophylaxis of cytomegalovirus disease in solid-organ transplant recipients. Clin. Infect. Dis. 47:702-711. [DOI] [PubMed] [Google Scholar]
- 74.Trgovcich, J., D. Stimac, B. Polic, A. Krmpotic, E. Pernjak-Pugel, J. Tomac, M. Hasan, B. Wraber, and S. Jonjic. 2000. Immune responses and cytokine induction in the development of severe hepatitis during acute infections with murine cytomegalovirus. Arch. Virol. 145:2601-2618. [DOI] [PubMed] [Google Scholar]
- 75.van der Strate, B. W., J. L. Hillebrands, S. S. Nijeholt, L. Beljaars, C. A. Bruggeman, M. J. Van Luyn, J. Rozing, T. H. The, D. K. Meijer, G. Molema, and M. C. Harmsen. 2003. Dissemination of rat cytomegalovirus through infected granulocytes and monocytes in vitro and in vivo. J. Virol. 77:11274-11278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Villadangos, J. A., and L. Young. 2008. Antigen-presentation properties of plasmacytoid dendritic cells. Immunity 29:352-361. [DOI] [PubMed] [Google Scholar]
- 77.Wing, B. A., G. C. Lee, and E. S. Huang. 1996. The human cytomegalovirus UL94 open reading frame encodes a conserved herpesvirus capsid/tegument-associated virion protein that is expressed with true late kinetics. J. Virol. 70:3339-3345. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.