Abstract
The transcriptional mechanism utilized by turnip crinkle carmovirus to synthesize subgenomic (sg) mRNAs was investigated by analyzing viral RNAs and their associated regulatory RNA elements. In vivo analyses revealed the following: (i) that minus-strand sg RNAs are detectable in infections, (ii) that minus-strand sg RNA accumulation can be partially uncoupled from that of their plus-strand sg mRNA counterparts, and (iii) that an RNA secondary structure located upstream of the sg mRNA start site mediates transcription by functioning in the plus strand of the viral genome. Collectively, these observations are consistent with this carmovirus using a premature termination mechanism for sg mRNA transcription.
One of the gene expression strategies that positive-strand RNA viruses use for the production of their proteins is to synthesize subgenomic (sg) mRNAs. sg mRNAs are viral messages that are transcribed during infections to allow expression of proteins that are encoded 3′ proximally in viral RNA genomes (3). Several genera in the large plant virus family Tombusviridae transcribe their sg mRNAs by a premature termination (PT) mechanism (5, 10, 12). This process involves early termination of the viral RNA-dependent RNA polymerase (RdRp) while copying the viral genome to produce minus-strand complements of sg mRNAs that are then used as templates to transcribe sg mRNAs (10). The generation of these smaller minus strands is due in part to RNA structures in the viral genome located just 5′ of the termination site that cause the viral RdRps to disengage (10). These RNA structures are generally comprised of either local secondary structures or long-distance higher-order structures (5, 10, 12).
The carmoviruses represent a prominent genus in the family Tombusviridae, and most, such as Turnip crinkle virus (TCV), produce two sg mRNAs (7, 9); sg mRNA1 allows for expression of two small movement proteins, while sg mRNA2 mediates coat protein production (Fig. 1 A). An internal initiation mechanism, involving RdRp recognition of an internally located promoter in the full-length genomic minus strand (3), has been proposed to describe sg mRNA transcription in two carmoviruses, Hibiscus chlorotic ringspot virus (HCRSV) (2) and TCV (7, 9). Here we have further investigated sg mRNA production in TCV with the goal of more clearly defining the transcriptional mechanism used by this virus.
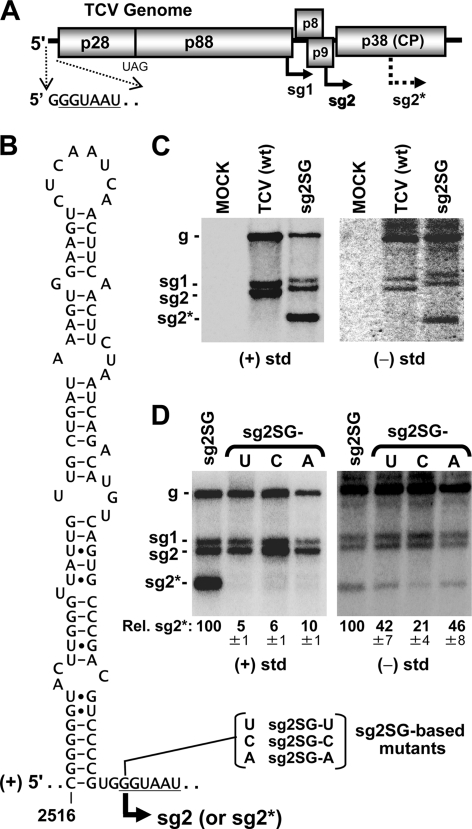
FIG. 1.
(A) Schematic representation of the TCV RNA genome. The initiation sites of sg mRNAs and the nucleotide sequence at the 5′ terminus of the viral genome are shown below. (B) mfold predicted plus-strand RNA structure for the sg mRNA2 regulatory segment. Substitutions of the initiation nucleotide in sg2SG mutants are shown. (C and D) Northern blot analysis of glyoxal-treated viral RNAs separated in 2% agarose gels after transfection of cucumber protoplasts that were incubated at 22°C for 22 h under constant light. Following transfer to nylon membranes, plus strands were detected with a 32P-end-labeled deoxyoligonucleotide complemenary to positions 3889 to 3909 of the TCV genome, while minus strands were detected with a uniformly 32P-labeled riboprobe transcript corresponding to positions 3801 to 4050. The identities of the viral genomic transcripts used for transfection are shown above each lane, and the positions of genomic (g) and sg RNA (sg) are indicated to the side. Relative sg2* levels of plus (+) and minus (−) strands were determined as means from three independent experiments and were normalized to cognate genomic RNA levels.
Our analysis of TCV transcription focused on sg mRNA2. To facilitate the analysis of the RNA elements regulating transcription of sg mRNA2, a previously defined 97-nucleotide (nt)-long segment of RNA encompassing the relevant regulatory sequences for sg mRNA2 synthesis (9) was inserted into the coat protein coding region at a BsmI site, position 3197 (Fig. 1A). This segment included an RNA secondary structure and adjacent linear sequence previously defined and analyzed in the minus-strand context (7, 9). The corresponding secondary structure in the plus strand, predicted by mfold analysis (13), is shown in Fig. 1B along with the adjacent linear sequence that shares a high degree of sequence identity with the 5′ terminus of the genome (Fig. 1A and B, underlined sequences). Ectopic expression of sg mRNA2 was used previously in TCV (9) and HCRSV (2) transcriptional studies and for consistency was also adopted here.
In vitro-generated transcripts of the TCV genome with the ectopic sg mRNA2 regulatory sequences, termed sg2SG, were generated by standard recombinant DNA techniques using an infectious TCV clone, as described previously (11). Insertion of the sg mRNA2 transcriptional segment in the coat protein coding region caused a frameshift that disrupted the coat protein reading frame; however, coat protein activities are dispensable for protoplast infections. Wild-type (wt) and mutant genomic transcripts were transfected into plant protoplasts, and viral RNAs were subsequently analyzed by Northern blotting with strand-specific probes (Fig. 1C), as outlined elsewhere (11). An anticipated extra sg mRNA, designated sg RNA2*, was detected in total nucleic acids prepared from transfections with sg2SG but not with wt TCV (Fig. 1C, left panel). Minus-strand counterparts of viral RNAs, including all sg RNAs, were also detectable (Fig. 1C, right panel), consistent with a PT model in which they would serve as intermediate templates for transcription (10). When the initiating nucleotide for transcription of sg RNA2* was replaced with different residues, plus-strand levels for sg RNA2* decreased to near-undetectable levels, while corresponding minus strands were less notably affected (Fig. 1D). This ability to partially uncouple sg minus- from plus-strand accumulation is consistent with a PT mechanism in which minus strands are synthesized before plus strands (10). Additionally, these results suggest that the identity of the nucleotide at the initiation site also contributes, but to a lesser degree, to the efficiency of minus-strand production.
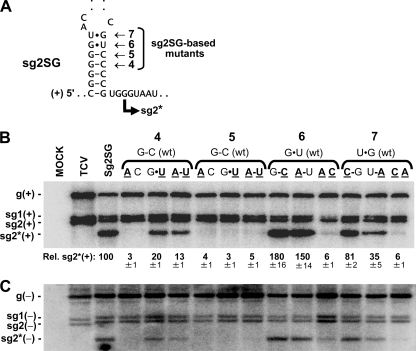
We next analyzed the mfold-predicted extended stem-loop RNA structure located just 5′ to the initiation site for sg RNA2* (Fig. 1B). Strand-preferential destabilization was used to assist in determining the strand in which this RNA structure functions. The principle of the method is that a GU wobble pair in one strand is usually stronger than the corresponding CA mismatch in the complementary strand. Thus, by analyzing the transcriptional activity of viral genomes containing either GU pairs (which are predicted to preferentially destabilize the complementary minus-strand structure) or CA pairs (which are predicted to preferentially destabilize the plus-strand structure) at specified positions, it should be possible to deduce the strand in which the structure functions. Four different base pairs near the bottom of the predicted structure were targeted for replacement with alternative pairs (Fig. 2 A). In vivo analysis of sg RNA2* transcription from viral genomes containing changes at position 4 indicated that a GU but not an AC was tolerated in the plus strand, and similar results were also obtained at positions 6 and 7 (Fig. 2A and B). Conversely, all changes at position 5 led to inactivity, indicating a key role for the wt pair (Fig. 2A and B). Collectively, AC or CA pairs in the plus strand at three different locations caused greater transcriptional inhibition than GU or UG pairs at the same plus-strand positions, supporting the concept that this RNA structure forms and functions in the viral genome, as required for a PT mechanism. Additionally, for all mutants, the relative accumulation profiles for sg RNA2* minus strands were similar to those of their plus-strand counterparts (Fig. 2C), which is in line with a proposed role for the RNA structure in mediating production of sg minus-strand templates via a PT process.
FIG. 2.
(A) The four positions at the bottom of the RNA structure that were targeted for mutational analysis in sg2SG are indicated. (B) Northern blot analysis of sg2SG mutants, as described in the legend to Fig. 1.
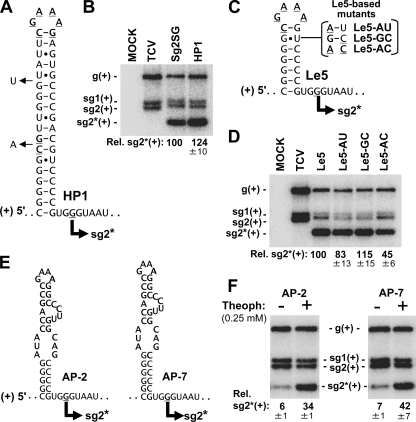
Additional deletion mutations were made based on the predicted plus-strand RNA secondary structure (Fig. 1A). Removing the upper half of the predicted extended stem-loop structure, as well as the U bulge and CA/C internal loop, generated a smaller stem-loop structure that was still able to efficiently direct sg RNA2* transcription (Fig. 3 A and B). Further truncations of the upper portion of this stem-loop structure defined a smaller hairpin that also functioned well (Fig. 3C and D). This result corresponded well with a minimal sequence previously defined as important for sg mRNA2 transcription from its wt location in the TCV genome (7). Substitutions in the stem of the hairpin in Le5 showed that the GU wobble pair was not important for activity of the structure and that this pair was favored over the corresponding AC, indicative of plus-strand activity (Fig. 3C and D).
FIG. 3.
(A) mfold-predicted structure for the truncated and modified stem-loop structure in mutant viral genome HP1. The added tetraloop and closing pair at the top, as well as substitutions within the stem, are underlined, while the U and A residues that were deleted from the stem are shown with arrows. (B) Northern blot analysis of genomic mutant HP1 as described in the legend to Fig. 1. (C) mfold-predicted structure for a small hairpin RNA structure present in mutant viral genome Le5. Substitutions in genomic mutants are shown to the right. (D) Northern blot analysis of genomic mutant Le5 and its derivatives. (E) Aptamer-based RNA structures in genomic mutants AP-2 and AP-7. The aptamer sequences are indicated in bold. (F) Northern blot analysis of AP-2 and AP-7. The incubation medium for transfected protoplasts either lacked (−) or contained (+) 0.25 mM theophylline.
Having identified the lower portion of the stem as an essential element for activity, we next sought to further confirm its proposed plus-strand activity using a different approach. To do this, upper portions of the small hairpin, including the ultrastable GNRA-type tetraloop (Fig. 3C), were replaced with an RNA aptamer domain (Fig. 3E) as described previously (8). The presence of a theophylline-binding RNA aptamer domain (6) resulted in low levels of sg mRNA2* transcription (Fig. 3F), likely by interfering with stable formation of the lower stem (8). However, the addition of the small molecule theophylline to the incubation medium, which binds to its cognate aptamer domain and stabilizes the structure (6), led to ∼5- to 6-fold levels of recovery of sg RNA2* transcription (Fig. 3F). Since aptamer binding is strand specific, this result further supports the concept that stability of the RNA structure in the plus strand is important for its activity.
In conclusion, several lines of evidence indicate that TCV sg mRNA2 transcription occurs via a PT mechanism (Fig. 4). (i) Minus strands were detected in infections, and their accumulation could be partially uncoupled from corresponding plus-strand accumulation. (ii) The extended RNA stem-loop positioned 5′ proximally to the initiation site of transcription functioned in the plus strand, consistent with its role as an attenuation structure. (iii) In similarity to other viruses that use the PT mechanism (10), there is a high degree of identity between the linear promoter for plus-strand genome synthesis and that for sg mRNA transcription. Collectively, these data support TCV using a PT mechanism for sg mRNA2 transcription, a concept that may also extend to other carmoviruses (1, 2, 4).
FIG. 4.
Schematic representation of TCV replication (top half) and proposed sg mRNA transcription mechanism (shaded region). The asterisk represents a functional attenuating RNA structure in the genome, while the small squares signify the high degree of sequence identity between the 3′ termini of the genomic and sg minus-strand templates.
Acknowledgments
We thank members of our laboratory for reading the manuscript.
We thank the NSERC and CRC for funding.
Footnotes
Published ahead of print on 26 May 2010.
REFERENCES
- 1.García-Castillo, S., M. A. Sánchez-Pina, and V. Pallás. 2003. Spatio-temporal analysis of the RNAs, coat and movement (p7) proteins of Carnation mottle virus in Chenopodium quinoa plants. J. Gen. Virol. 84:745-749. [DOI] [PubMed] [Google Scholar]
- 2.Li, W., and S. M. Wong. 2006. Analyses of subgenomic promoters of Hibiscus chlorotic ringspot virus and demonstration of 5′ untranslated region and 3′-terminal sequences functioning as subgenomic promoters. J. Virol. 80:3395-3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Miller, W. A., and G. Koev. 2000. Synthesis of subgenomic RNAs by positive-strand RNA viruses. Virology 273:1-8. [DOI] [PubMed] [Google Scholar]
- 4.Rico, P., and C. Hernández. 2009. Characterization of the subgenomic RNAs produced by Pelargonium flower break virus: identification of two novel RNAs species. Virus Res. 142:100-107. [DOI] [PubMed] [Google Scholar]
- 5.Sit, T. L., A. A. Vaewhongs, and S. A. Lommel. 1998. RNA-mediated trans-activation of transcription from a viral RNA. Science 281:829-832. [DOI] [PubMed] [Google Scholar]
- 6.Suess, B. 2005. Engineered riboswitches control gene expression by small molecules. Biochem. Soc. Trans. 33:474-476. [DOI] [PubMed] [Google Scholar]
- 7.Wang, J., C. D. Carpenter, and A. E. Simon. 1999. Minimal sequence and structural requirements of a subgenomic RNA promoter for turnip crinkle virus. Virology 253:327-336. [DOI] [PubMed] [Google Scholar]
- 8.Wang, S., L. Mortazavi, and K. A. White. 2008. Higher-order RNA structural requirements and small-molecule induction of tombusvirus subgenomic mRNA transcription. J. Virol. 82:3864-3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang, J., and A. E. Simon. 1997. Analysis of the two subgenomic RNA promoters for turnip crinkle virus in vivo and in vitro. Virology 232:174-186. [DOI] [PubMed] [Google Scholar]
- 10.White, K. A. 2002. The premature termination model: a possible third mechanism for subgenomic mRNA transcription in (+)-strand RNA viruses. Virology 304:147-154. [DOI] [PubMed] [Google Scholar]
- 11.White, K. A., J. M. Skuzeski, W. Li, N. Wei, and T. J. Morris. 1995. Immunodetection, expression strategy and complementation of turnip crinkle virus p28 and p88 replication components. Virology 211:525-534. [DOI] [PubMed] [Google Scholar]
- 12.Xu, W., and K. A. White. 2009. RNA-based regulation of transcription and translation of aureusvirus subgenomic mRNA1. J. Virol. 83:10096-10105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zuker, M. 2003. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 31:3406-3415. [DOI] [PMC free article] [PubMed] [Google Scholar]