Abstract
To identify cellular processes involved in retroviral infection, we employed a high-volume forward genetic screen of insertionally mutagenized somatic cells using a murine leukemia virus (MLV) vector. This approach identified a clonal cell line that exhibited approximately 10-fold reduced gene expression from MLV vectors following infection despite supporting normal levels of MLV reverse transcription and integration. The defect in this cell line was specific for the MLV long terminal repeat (LTR) promoter, as normal levels of reporter gene expression were obtained from both an internal cytomegalovirus (CMV) promoter contained within an LTR-defective MLV vector and LTR expression from an avian sarcoma and leukosis virus (ASLV) vector. Complementation and shRNA knockdown experiments demonstrated that the defective gene in these cells is ZASC1 (ZNF639), a transcription factor with strong links to cancer and inherited ataxias. We demonstrated that ZASC1 is a sequence-specific DNA binding protein with three closely related binding sites located within the MLV LTR promoter, but it does not bind to the ASLV promoter. Mutating these putative ZASC1 binding sites significantly reduced levels of MLV gene expression. While wild-type ZASC1 activated expression from the MLV promoter, a green fluorescent protein-ZASC1 fusion protein showed dominant-negative inhibition of MLV gene expression. These studies identify the cellular transcription factor ZASC1 as an activator of MLV gene expression and provide tools that should be useful in studying the links between ZASC1 and human diseases.
The Retroviridae family includes the human pathogens human immunodeficiency virus type 1 and 2 (HIV-1 and HIV-2), the causative agents of AIDS. The study of prototypical simple retroviruses such as murine leukemia virus (MLV) and avian sarcoma and leukosis virus (ASLV) has lead to significant advances in the understanding of retroviral infections and host cell processes (10, 22, 33). The early stages of the retroviral replication cycle consist of virus-receptor binding, virus-cell membrane fusion, reverse transcription, nuclear translocation, and viral DNA integration into a cellular chromosome, which generates the provirus. The late stages consist of proviral transcription by host RNA polymerase II, RNA processing and cytoplasmic export, the translation of viral proteins, viral assembly, egress, and maturation. These events are heavily dependent upon host cellular machinery.
Many cellular factors that regulate different steps of retroviral replication were identified previously through a combination of genetic and biochemical approaches (4, 7, 10, 18, 23, 34, 37). However, it is likely that other important cellular factors remain to be identified. To illustrate this point, we recently used a forward genetic approach, based upon retroviral insertional mutagenesis in CHO-K1 cells, to uncover an unprecedented role for the host cell sulfonation pathway in regulating HIV-1 and MLV gene expression (5). The role of this cellular pathway in provirus transcription was not detected by multiple genome-wide RNA interference (RNAi)-based screens (4, 18, 34, 37).
Here, we have used the same screening approach to identify a new transcriptional activator of the MLV provirus. The transcription of integrated proviral DNA is driven from the unique 3′ (U3) element in the viral genome. This is a strong RNA polymerase II promoter that contains binding sites for multiple host transcription factors. In MLV, the enhancer region of the promoter contains multiple overlapping binding sites for various cell-specific transcription factors, expanding the cell types that MLV can infect. The preferential binding of one transcription factor over another at specific overlapping sites can alter cell line tropism and disease outcome in mice (11, 19, 21, 25-29).
The MLV transcriptional regulator identified in this study is zinc finger associated with squamous cell carcinoma (ZASC1 or ZNF639), a protein with nine Kruppel-like zinc fingers that has a broad tissue distribution (14). ZASC1 copy number amplification has been linked to squamous cell carcinomas and an increased propensity for metastasis (3, 14). Furthermore, it is a major interacting protein of the histone acetyltransferase CBP (17), is involved in the nuclear transport of β-catenin (3), and has been associated with inherited ataxias (20). Although ZASC1 has been proposed to bind to DNA and function as a transcription factor, to date, no ZASC1 DNA binding sites have been identified, nor has any has any transcriptional modulation activity been described for the wild-type (WT) ZASC1 protein (3, 14). The data described in this report reveal that the wild-type ZASC1 protein binds to specific DNA elements in the MLV long terminal repeat (LTR) and activates proviral transcription.
MATERIALS AND METHODS
Plasmids and viral vectors.
The viral genome plasmids pMMP-nls-LacZ, pCMMP-EGFP, pCMMP-SEAP-IRES-GFP, pCMMP-IRES-GFP, pCMMP-CD4-EGFP, pHIV-TVA800-hcRED, pRET, RCASBP(A)-AP, pLEGFP, pQCLIN, and pLenti6/V5-GW/lacZ have been described previously (5). The viral genome plasmid pCMMP-luciferase was generated by replacing the enhanced green fluorescent protein (EGFP) gene in pCMMP-EGFP with the firefly luciferase gene (fLuc) from pGL3-Basic (Promega, Madison, WI). The viral genome plasmid encoding secreted Gaussia luciferase (gLuc) and puromycin resistance was generated by replacing the EGFP gene in pCMMP-EGFP with the gLuc gene from pGluc-Basic (NEB, Ipswich, MA) and inserting the internal ribosome entry site and the puromycin acetyltransferase gene from pQCXIP downstream of gLuc to make pCMMP-gLUC-IRES-Puro. The plasmid carrying the ZASC1 open reading frame under the control of the human cytolomegalovirus (HCMV) promoter was obtained commercially (Open Biosystems, Huntsville, AL). The ZASC1 open reading frame was cloned in-frame downstream of EGFP in pEGFP-N1 (Clontech, Palo Alto, CA) to make the GFP-ZASC1 expression vector. The coding sequence of the GFP-ZASC1 fusion protein then was inserted into the multiple-cloning site downstream of the TET operator of pLenti4/TO/V5-DEST (Invitrogen, Carlsbad, CA) to generate the tetracycline-inducible HIV-1 vector pLenti4/TO-GFP-ZASC1. HIV-1 vectors for the stable expression of WT ZASC1 or 3′-phosphoadenosine 5′-phosphosulfate synthase 1 (PAPSS1) were generated by the PCR amplification of the relevant coding sequence (ZASC1[ZNF639], image 4794621; PAPSS1: image 3869484) from commercially available cDNAs (Open Biosystems, Huntsville, AL) and cloned into MluI/EcoRV-digested pLenti6/V5-GW/lacZ, generating plasmids pLenti6-ZASC1 and pLenti6-PAPSS1, respectively.
To generate short hairpin RNA (shRNA) constructs targeting ZASCI, a linker (top, 5′-TGCTGGAGACCAACCTAGGACTAGTGGTCTCT-3′; bottom, 5′-CCTGAGAGACCACTAGTCCTAGGTTGGTCTCC-3′) was inserted into the vector pcDNA6.2-GW/EmGFP-miR (Invitrogen, Carlsbad, CA) in the hairpin insertion site of the vector. This linker adds two BsaI sites (underlined) that, when the resulting plasmid is digested with BsaI, result in incompatible sticky ends, which facilitates the insertion of hairpin-containing oligonucleotides. Double-stranded oligonucleotides (Table 1) then were inserted into this site to generate gene-specific shRNA constructs against ZASC1 or secreted alkaline phosphatase (SEAP).
TABLE 1.
Oligonucleotides used for cloning shRNA expression construct
| Target genea | Oligonucleotide name | Orientationb | Sequence (5′ to 3′)c | Binding positiond |
|---|---|---|---|---|
| SEAP | OJWB299 | Sense | TGCTGCATGTCTGCTCGAAGCGGCCGGTTTTGGCCACTGACTGACCGGCCGCTGAGCAGACATG | 1539 |
| OJWB300 | Antisense | CCTGCATGTCTGCTCAGCGGCCGGTCAGTCAGTGGCCAAAACCGGCCGCTTCGAGCAGACATGC | ||
| ZASC1 | OJWB309 | Sense | TGCTGTTTGCTGGTTGACTGATTCAGGTTTTGGCCACTGACTGACCTGAATCACAACCAGCAAA | 376 |
| OJWB310 | Antisense | CCTGTTTGCTGGTTGTGATTCAGGTCAGTCAGTGGCCAAAACCTGAATCAGTCAACCAGCAAAC | ||
| ZASC1 | OJWB546 | Sense | TGCTGTTGAGGAGAACTGTACATCACGTTTTGGCCACTGACTGACGTGATGTAGTTCTCCTCAA | 881 |
| OJWB547 | Antisense | CCTGTTGAGGAGAACTACATCACGTCAGTCAGTGGCCAAAACGTGATGTACAGTTCTCCTCAAC | ||
| ZASC1 | OJWB548 | Sense | TGCTGATAGCAACATGCCTGTTAACAGTTTTGGCCACTGACTGACTGTTAACACATGTTGCTAT | 1164 |
| OJWB549 | Antisense | CCTGATAGCAACATGTGTTAACAGTCAGTCAGTGGCCAAAACTGTTAACAGGCATGTTGCTATC | ||
| ZASC1 | OJWB550 | Sense | TGCTGTTATGAGGCAAGTCAGCTGAAGTTTTGGCCACTGACTGACTTCAGCTGTTGCCTCATAA | 1362 |
| OJWB551 | Antisense | CCTGTTATGAGGCAACAGCTGAAGTCAGTCAGTGGCCAAAACTTCAGCTGACTTGCCTCATAAC |
The gene that the shRNA targets.
Denotes whether the oligonucleotide binds on the coding strand (relative to the mRNA of the genes) or noncoding strand.
Sequence of the oligonucleotide written in the 5′ to 3′ orientation.
The region of the target gene the shRNA corresponds to, relative to the start codon.
To generate retroviral promoters for electrophoretic mobility shift assays (EMSAs) and reporter gene analysis, MLV U3 was amplified from the MLV vector pLEGFP-C1 (Clontech, Palo Alto, CA) using the primers 5′-GGACGCGTGGAATGAAAGACCCCACCTGTAGG-3′ and 5′-GGAAGCTTCCCGAGTGAGGGGTTGTGGGCTC-3′, and the ASLV U3 was amplified from RCASBP(A)-AP using the primers 5′-GGACGCGTGAAATGTAGTCTTATGCAATACTCTTG-3′ and 5′-GGAAGCTTTCGTCCAATCCATGTCAGACC-3′. PCR products were digested with MluI and HindIII (underlined) and cloned into pGluc-Basic (NEB, Ipswich, MA). The mutagenesis of potential binding sites was performed by overlap PCR or linker mutagenesis. MLV genomic clones containing mutations in the ZASC1 binding sites were made by overlap PCR or linker mutagenesis on these subcloned fragments and moved back into the 3′LTR of full-length pCMMP-SEAP-IRES-GFP by standard restriction enzyme cloning. All mutations were validated by sequencing. The plasmids used for mammalian two-hybrid analysis all were provided by, or cloned into, vectors in the Checkmate mammalian two-hybrid kit (Promega, Madison, WI).
Cell culture and virus production.
The source and growth conditions for Chinese hamster ovary cells (CHO-K1), CHO-TVA800, human embryonic kidney 293T cells, THP-1, and human Jurkat cells were described elsewhere (5, 30). Mouse NIH 3T3 and EL4 cells were obtained from the ATCC (Manassas, VA) and cultured as suggested by the distributor. Cells expressing either ZASC1 or PAPSS1 cDNA were generated by infecting CHO-K1 and IM1 cells with the vesicular stomatitis virus (VSV)-G-pseudotyped HIV-1 vector pLenti6-ZASC1 or pLenti6-PAPSS1 at an approximate multiplicity of infection (MOI) of 0.5 blasticidin-transducing units. Infected cells were selected for 2 weeks in 3 μg/ml blasticidin. To generate the cell line expressing tetracycline-inducible GFP-ZASC1 and a gLuc-expressing MLV provirus, CHO-TREX cells (Invitrogen, Carlsbad, CA) were transduced with pLenti4/TO-GFP-ZASC1 and the MLV reporter vector pCMMP-gLUC-IRES-Puro. Transduced cells were selected with 5 μg/ml zeomycin and 500 μg/ml puromycin. GFP-ZASC1 expression was induced with 1 μg/ml doxycycline. The procedures used to produce the retroviral vectors and titer of each viral stock are described in detail elsewhere (5).
Isolation of an MLV-resistant clone.
The procedure used to generate insertionally mutagenized CHO-K1 cells and to select for MLV-resistant clones was described in detail elsewhere (5). Briefly, a pool of 2 × 107 cells insertionally mutagenized with the poly(A)-trap vector pRET (15) was subjected to five rounds of challenge with a second, replication-defective, VSV-G-pseudotyped MLV vector that contains a human CD4 gene that is expressed from the viral promoter. Infected cells that expressed human CD4 on their surface were removed from the population at each round by magnetic cell sorting (MACS) using an iron-conjugated CD4-specific antibody. Each round of infection and sorting resulted in an approximately 3-fold enrichment of CD4-negative cells relative to the preceding round, with a total measured enrichment of 239-fold. The resultant cell population, which exhibited an overall 2-fold resistance to MLV infection, then was challenged a final time with another VSV-G-pseudotyped MLV vector encoding the far-red fluorescent protein HcRed. A total of 264 single-cell clones of HcRed-negative cells then were isolated by fluorescence-activated cell sorting (FACS) and tested for their susceptibility to infection by a VSV-G-pseudotyped MLV vector encoding β-galactosidase (MMP-nls-LacZ). The most resistant clone, IM1, was chosen for further validation.
Assays of viral infection.
Quantitative chemiluminescent infection assays were performed as previously described (5, 6). Briefly, 96-well plates were seeded at 1 × 104 cells/well for each cell line tested. The cells were incubated with an approximate MOI of 1 transducing unit in the presence of 4 μg/ml polybrene. At 48 h postinfection (hpi), four wells were assayed for β-galactosidase activity using the Galacto-star kit (Applied Biosystems, Foster City, CA), for alkaline phosphatase activity using the Phospha-Light kit (Applied Biosystems, Foster City, CA), or for luciferase activity using a Britelite (PerkinElmer, Boston, MA) according to the manufacturer's instructions. The other four wells were assayed for cell number and cell viability using CellTiter-Glo reagent (Promega, Madison, WI). The results obtained were normalized for relative cell number.
To determine the absolute fold resistance to viral infection, 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) staining was performed on cells that were infected with serial dilutions of viruses. For these experiments, cells were seeded at 1 × 104 cells/well in triplicate rows for each cell line tested. The cells then were infected for 2 h with 10-fold serial dilutions of MMP-nls-LacZ[VSV-G] in the presence of 4 μg/ml polybrene as described before, and the cells subsequently were stained with X-Gal as previously described (1). The blue cells contained in wells that had between 20 and 200 β-galactosidase-positive cells were counted to give an accurate measure of the viral titer.
Real-time quantitative PCR.
The procedures to measure MLV reverse transcription products have been described in detail elsewhere (5). Briefly, cells were seeded in triplicate wells at 5 × 105/well in a six-well plate and then infected at 4°C on a rocking platform at an MOI of 1 GFP-transducing unit (GTU) for 2 h with an MLV vector (pLEGFP; Clontech, Palo Alto, CA) pseudotyped with VSV-G that was treated with DNase I. DNA was harvested from infected cells 24 hpi using the DNeasy kit (Qiagen, Valencia, CA). To measure integrated proviral DNA copy numbers, cells were seeded and infected as described above and then passaged for 18 days. DNA then was harvested from 1 × 106 cells as described above. The DNA concentration was calculated by measuring the A260 on a SPECTRAmax plus 96-well UV spectrophotometer (Molecular Devices, Sunnyvale, CA). Quantitative, real-time PCR analysis was performed as previously described (5) using primers specific for the U3-U5 region of the LEGFP vector. These primers amplify the plus-strand strong stop (+SSS) reverse transcription product and are shown along with the viral LTR feature and the base pair position recognized in pLEGFP: OJWB39 (5′-CAGTTCGCTTCTCGCTTCTGTTC-3′; U3, bp 523 to 535), OJWB47 (5′-GTCGTGGGTAGTCAATCACTCAG-3′; R and U5, bp 697 to 719), and OJWB38 (5′-6-carboxyfluorescein-ATCCGAATCGTGGTCTCGCTGTTC-6-carboxytetramethylrhodamine-3′; R, bp 657 to 680). The number of molecules in each reaction mixture was determined by comparison to standard curves generated from the amplification of plasmid DNA containing the target sequence.
Immunoblot analysis.
Cells (1 × 106) were lysed in 300 μl SDS-PAGE loading buffer and transferred to a QIAshredder column (Qiagen, Valencia, CA), and the DNA was sheared by centrifugation at 10,000 × g for 2 min. The samples were boiled, separated by SDS-PAGE, transferred to a polyvinylidene difluoride (PVDF) membrane, and blotted with a rabbit polyclonal antibody raised against human ZASC1, a rabbit polyclonal antibody raised against human actin (sc-1616-R; Santa Cruz Biotechnology, Santa Cruz, CA), or a rabbit polyclonal antibody against GFP (Invitrogen, Carlsbad, CA). The blots were washed and treated with a secondary anti-rabbit horseradish peroxidase conjugate (Thermo Scientific, Rockford, IL), and the blots were imaged by chemiluminescence using the Supersignal West femto substrate (Thermo Scientific, Rockford, IL). The signal was detected on a ChemiDoc XRS system (Bio-Rad, Hercules, CA). The ZASC1 antibody was generated by immunizing New Zealand White rabbits with Escherichia coli-expressed ZASC1 at Harlan Laboratories (Madison, WI). Virus release into media was detected using the rat monoclonal antibody from the R187 hybridoma (ATCC, Manassas, VA) and anti-rat horseradish peroxidase conjugate (Thermo Scientific, Rockford, IL) following immunoblotting.
Electrophoretic mobility assays (EMSA).
The open reading frame of ZASC1 was PCR amplified and cloned between the NheI and XhoI sites in the pET28a vector (Novagen, La Jolla, CA). This pET-ZASC1 construct encodes an in-frame, N-terminal His6-T7-tag-ZASC1 fusion protein under the control of the T7 bacteriophage promoter. Protein for EMSA was generated by in vitro transcription/translation using the TnT T7 coupled reticulocyte lysate system (Promega, Madison, WI) by following the manufacturer's protocols. DNA probes were either restriction fragments of cloned U3 elements or annealed oligonucleotides end labeled with [γ-32P]ATP and T4 polynucleotide kinase (New England Biolabs, Ipswich, MA) by following the manufacturer's procedure. For each binding reaction mixture, 8 μl of the coupled in vitro transcription/translation reaction mixture was mixed on ice with 50 fm 32P end-labeled DNA probe and 5 μl 5× EMSA buffer {250 mM Tris-HCl 7.5, 50 mM MgCl2, 25 ng/μl sheared calf thymus [CT] DNA, 0.05% 3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate [CHAPS] 5% glycerol, 10 μM zinc acetate [Zn2(C2H3O2)]} in a final volume of 25 μl. Samples were incubated at room temperature for 10 min and on ice for 10 min. Bound DNA was separated from free probe on a 4% Tris-borate-EDTA (TBE) polyacrylamide gel run at 125 V at 4°C for 2 h. The gels were dried and exposed to phosphorimager plates for analysis.
Transient transfection assays.
CHO-K1, IM1, and HEK293 cells (2 × 104 cells/well) were reverse transfected in a 96-well format with a total of 100 ng DNA/well with 0.4 ml/well Transit LT-1 transfection reagent (Mirus, Madison, WI) by following the manufacturer's instructions. For transient promoter activation assays, 10 ng of the retrovirus promoter plasmids consisting of the MLV U3 promoter cloned in front of the secreted Gaussia luciferase reporter gene as described above was transfected. To monitor transfection efficiency, a plasmid (5 ng) encoding mRFP under the control of the HCMV promoter and a plasmid (5 ng) encoding firefly luciferase under the control of the HCMV promoter were included in each transfection. WT ZASC1 or GFP-ZASC1 expression plasmids were included at 25 ng/well. Vector plasmid DNA or calf thymus DNA was used to maintain a constant 100 ng/well in each transfection. Two days posttransfection, 10 μl of media was removed, diluted with 40 μl of phosphate-buffered saline (PBS), and assayed for secreted Gaussia luciferase (gLuc) by injecting 30 μl coelenterazine solution (Renilla luciferase assay system; Promega, Madison, WI), waiting 1.6 s, and then reading luminescence for 1 s. Firefly luciferase (fLuc) activity from the internal control plasmid was determined using a Britelite (PerkinElmer, Boston, MA) according to the manufacturer's instructions. The ratio of gLuc to fLuc was determined, and the activity of either the WT promoter or the relevant promoter in the absence of exogenous ZASC1 was set to 100%, and the reporter gene activity of the matched mutant promoter or ZASC1-expressing transfection sample was compared to this finding.
RESULTS
Isolation and characterization of MLV-resistant cell line IM1.
We previously described a procedure to isolate MLV-resistant cell clones from a library of Chinese hamster ovary (CHO-K1) cells that had been insertionally mutagenized with a retroviral vector (5). CHO-K1 cells were used because these cells are functionally hypodiploid at numerous loci (13), so the insertion of the viral vector into a single allele of a given cellular gene can be sufficient to produce a genetically null phenotype. The insertional mutagenesis was performed with the murine leukemia virus (MLV)-based vector pRET, which encodes green fluorescent protein (GFP), as well as a neomycin phosphotransferase (NPT) mRNA that contains an instability element downstream of a canonical splice donor site (15). The integration of pRET upstream of a cellular exon, and in the opposite transcriptional orientation relative to a cellular gene, gives rise to an NPT mRNA transcript in which the RNA instability element is removed by mRNA splicing, thereby conferring G418 resistance on the mutagenized cells (15).
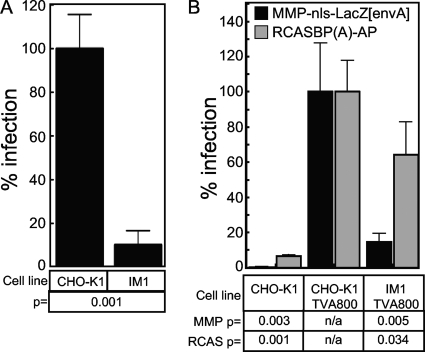
To identify cells that were resistant to retroviral infection, a pool of insertionally mutagenized cells was subjected to five rounds of challenge with a second, replication-defective, VSV-G-pseudotyped MLV vector that contains a human CD4 gene that is expressed from the viral promoter. Infected cells that expressed human CD4 on their surface were removed from the population at each round by magnetic cell sorting (MACS) using an iron-conjugated CD4-specific antibody. A final challenge with another VSV-G-pseudotyped MLV vector encoding the far-red fluorescent protein HcRed was followed by the single-cell cloning of HcRed-negative cells by FACS. In this report, we describe the characterization of a clonal cell line, designated IM1, that exhibited an approximately 10-fold resistance to infection by the VSV-G-pseudotyped MLV vector MMP-nls-LacZ based upon β-galactosidase reporter gene expression (Fig. 1A).
FIG. 1.
Resistance of the IM1 cell line maps to the MLV core. (A) CHO-K1 and IM1 cells were challenged with serial dilutions of the VSV-G-pseudotyped MLV vector (MMP-nls-LacZ[VSV-G]), encoding β-galactosidase. The cells then were stained 48 hpi with X-Gal, the number of blue cells were counted, and the data were reported as the percentage of LacZ-transducing units (LTU) obtained from WT CHO-K1 infections (5 × 105 LTU). The data shown are the averages from three experiments each performed with triplicate samples. (B) CHO-K1 cells and IM1 cells engineered to express TVA800 or wild-type CHO-K1 cells were challenged with either pMMp-nls-LacZ[envA], an EnvA-pseudotyped MLV vector encoding β-galactosidase, or with RCASBP(A)-AP, an ALSV-A vector encoding heat-stable alkaline phosphatase. Infection was monitored using chemiluminescent assays to detect reporter enzyme activities along with a chemiluminescent assay to measure relative viable cell numbers. The ratios of enzyme activities to relative viable cell number were calculated for each sample and compared to values from CHO-K1 TVA 800 cells (defined as 100% infection). The data shown are the average mean values obtained in an experiment performed with quadruplicate samples and are representative of three independent experiments. Error bars indicate the standard deviations from the data. P values were calculated using a standard Student's t test.
To determine if the defect associated with the IM1 cell line was specific for the MLV vector, wild-type CHO-K1 cells and mutant IM1 cells were engineered to express TVA800, the cellular receptor for an avian retrovirus, subgroup A avian sarcoma and leukosis viruses (ASLV-A) (2, 35). The TVA800-expressing cells were challenged with either an ASLV-A envelope protein (EnvA)-pseudotyped MLV vector encoding β-galactosidase or, instead, with an ASLV-A vector that encodes heat-stable alkaline phosphatase (8). Viral reporter gene expression following the infection of IM1-TVA800 cells by the EnvA-pseudotyped MLV vector was 7.8-fold reduced compared to that of CHO-K1-TVA800 cells (Fig. 1B). This effect mirrored that seen with VSV-G-pseudotyped MLV vectors (Fig. 1A), indicating that the defect seen with IM1 cells was independent of the viral glycoprotein type used to pseudotype the MLV vector. In contrast, the level of viral reporter gene expression following infection by the ASLV-A vector was only slightly reduced in IM1-TVA800 versus CHO-K1-TVA800 cells (Fig. 1B). Since both vectors employ EnvA-TVA receptor interactions to mediate entry, these observations indicate that the defect in the IM1 cell line is specific for protein or RNA components of the MLV core.
IM1 cells support viral reverse transcription and integration of viral DNA.
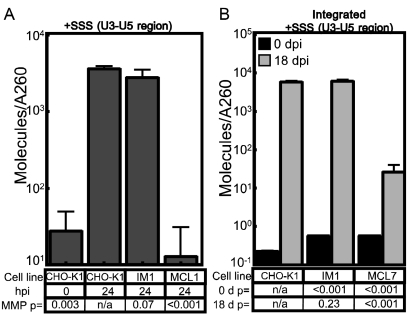
We utilized a previously established (5, 6) real-time PCR amplification procedure to monitor the levels of reverse transcription products and integrated viral DNA. Since IM1 cells potentially contain both the mutagenic pRET vector and the pCMMP-derived vector utilized in the screen, this assay used a primer/probe set that amplified the plus-strand strong stop replication intermediate (12, 32) specifically from the U3 and unique 5 (U5) LTR region of the pLEGFP, but not the pRET and pCMMP, MLV vector (5). Cells were challenged with the pLEGFP MLV vector, and total DNA subsequently was harvested at 0 and 24 hpi. The levels of viral DNA at 24 hpi were found to be similar in IM1 and CHO-K1 cells (Fig. 2A). For control purposes, a chemically mutagenized CHO-K1 cell line (MCL1) that exhibits a strong block to MLV reverse transcription (6) was challenged in parallel (Fig. 2A). To investigate viral DNA integration in IM1 cells, cells were infected with the same MLV vector and passaged for 18 days to eliminate episomal forms of viral DNA (31, 36), and the levels of total viral DNA then were measured. IM1 and CHO-K1 cells contained equivalent amounts of integrated viral DNA (Fig. 2B). By comparison, at 18 days postinfection, nearly 220-fold lower levels of viral vector DNA were detected in MCL7 cells, a chemically mutagenized CHO-K1 cell line that exhibits a strong, post-reverse transcription block to MLV DNA integration (6). These data demonstrate that the block to the infection of IM1 cells is subsequent to reverse transcription and viral DNA integration into the host cell genome.
FIG. 2.
IM1 cells support WT levels of MLV reverse transcription and integration. (A) CHO-K1, IM1, and MCL7 cell lines were challenged with the VSV-G-pseudotyped MLV vector LEGFP, and total DNA was isolated at 0 or 24 hpi, DNA concentration was quantitated by A260, and a real-time PCR amplification analysis was performed to measure the levels of viral DNA. (B) CHO-K1, IM1, and MCL7 cells were challenged with the same VSV-G-pseudotyped MLV vector as that used in panel A, and total DNA was harvested at 1 or 18 days postinfection for real-time PCR quantitation of viral DNA products. The chemically mutagenized cell line MCL1 displays a strong block to MLV reverse transcription, while the MCL7 cell line displays a strong block to MLV DNA integration (6). The data shown are the average mean values obtained in independent experiments performed with triplicate samples, and each is representative of three independent experiments. Error bars indicate the standard deviations of the data. P values were calculated using a standard Student's t test.
ZASC1 is responsible for the resistance phenotype of IM1 cells.
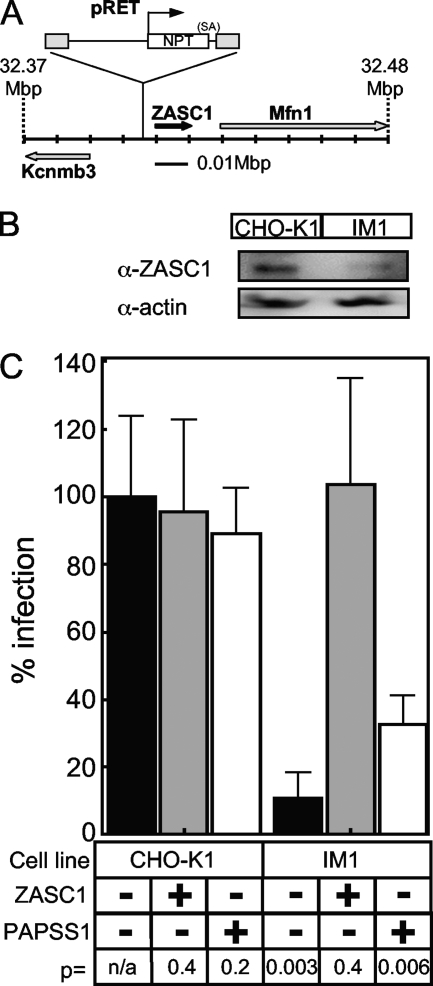
To identify the cellular gene disrupted by the mutagenic vector, total RNA was isolated from IM1 cells and subjected to reverse transcription-PCR amplification, with primers that recognize the NPT gene carried by the pRET vector and the captured poly(A) sequence, to identify the cellular gene flanking the mutagenic pRET vector insertion site as described previously (5, 15). DNA sequences of the PCR amplification products were compared to the sequenced mouse genome, since the hamster genome has not yet been sequenced. This analysis revealed that the pRET provirus was located 3,130 bp upstream of the transcriptional start site for the putative zinc finger transcription factor ZASC1 (ZNF639) (modeled on mouse chromosome 3 in Fig. 3A). It appears that a cryptic poly(A) site located within the ZASC1 promoter region was used to polyadenylate the pRET-encoded NPT mRNA transcript (data not shown). The next-nearest genes were those encoding the Kcnmb3 potassium channel and the mitochondrial fusion gene Mfn1, located 15,493 bp upstream and 22,024 bp downstream of the pRET proviral DNA, respectively (Fig. 3A). Based on the location of the pRET integration site, we reasoned that the expression of the ZASC1 gene most likely was impaired by the pRET insertion. This notion was supported by immunoblotting experiments that demonstrated that IM1 cells contain, on average, 35% ± 0.6% of the level of ZASC1 in wild-type CHO-K1 cells (Fig. 3B). Therefore, we focused on ZASC1 as a candidate gene that might influence infection by MLV vectors.
FIG. 3.
Reduced expression of ZASC1 is responsible for the MLV resistance phenotype of IM1 cells. (A) Since the hamster genome has not been sequenced, the integration of pRET was modeled on the mouse genome (chromosome 3, position 32.37 to 32.48 Mbp). The pRET provirus integration is expanded, and the orientation of the neomycin phosphotransferase (NPT) transcript and the splice acceptor site are shown in relation to the viral LTR (gray boxes). The relative locations and orientation of the pRET integration site ZASC1, potassium channel Kcnmb3, and the mitochondrial fusion gene Mfn1 are shown. (B) Immunoblot analysis of cell lysates from CHO-K1 and IM1 cells performed with rabbit polyclonal antibodies against human ZASC1 or human actin (α-ZASC1 and α-actin, respectively). The blots were washed, treated with a secondary anti-rabbit horseradish peroxidase conjugate, and imaged by chemiluminescence. The blots are representative of three independent experiments. (C) CHO-K1 and IM1 cells engineered to express either a human ZASC1 cDNA or a control human PAPSS1 cDNA were challenged with serial dilutions of the VSV-G-pseudotyped MLV vector (MMP-nls-LacZ[VSV-G]) encoding β-galactosidase. The cells then were stained 48 hpi with X-Gal, the numbers of blue cells were counted, and the data were reported as the percentage of LacZ-transducing units (LTU) obtained with WT CHO-K1 cells (5 × 105 LTU). The data shown are the averages from three experiments, each performed with triplicate samples. P values were calculated using a standard Student's t test.
To directly evaluate its role, we attempted to complement the defect in IM1 cells by expressing a wild-type human ZASC1 cDNA clone. For control purposes, these experiments also were conducted with wild-type CHO-K1 cells that also expressed human ZASC1. These cells were challenged with the VSV-G-pseudotyped MLV vector encoding β-galactosidase, and infected cells were enumerated by X-Gal staining. These experiments revealed that human ZASC1 fully complemented the MLV infection defect of the IM1 cell line but had little impact on the level of infection seen with wild-type CHO-K1 cells (Fig. 3C). In contrast, a control cDNA carrying human PAPSS1, a gene that we previously showed to be important for MLV infection in another mutant CHO-K1 cell line (5), only weakly affected virus infectivity in IM1 cells (Fig. 3C). Presumably the endogenous levels of PAPSS enzymes in IM1 cells are sufficient to support close-to-maximal levels of MLV infection. These data confirm that deficiency in ZASC1 is responsible for the virus infection-resistant phenotype of IM1 cells.
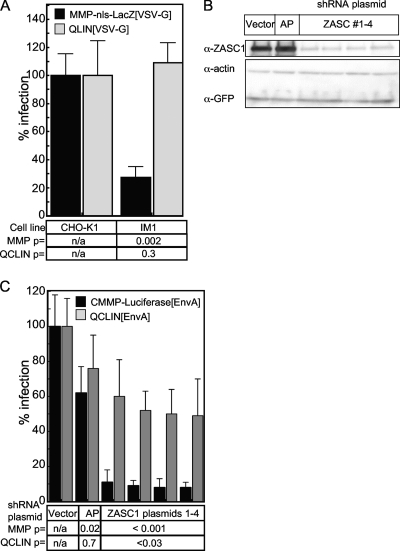
Since ZASC1 is a putative transcription factor, we next asked if its effect mapped to the MLV LTR promoter region. To address this question, the relative levels of LTR-driven transcription from the MMP-nls-LacZ MLV vector in WT CHO-K1 and IM1 cells were compared to those from the internal HCMV immediate-early promoter contained in QCLIN, a commercially available, self-inactivating (SIN) MLV vector with promoter-defective LTRs (16). Consistent with the previous results, the level of reporter gene expression from the MMP-nls-LacZ vector was impaired in IM1 cells compared to that of WT CHO-K1 cells. In contrast, the levels of β-galactosidase produced from the QCLIN vector were the same in both IM1 and CHO-K1 cells (Fig. 4A). This result mapped the activity of ZASC1 to the MLV LTR promoter, supporting a role for ZASC1 in promoting MLV gene expression.
FIG. 4.
ZASC1 regulates expression from the MLV LTR promoter. (A) CHO-K1 or IM1 cells were challenged with the VSV-G-pseudotyped MLV vector MMP-nls-LacZ, which drives LacZ expression from the MLV-LTR promoter, or pQCLIN, a self-inactivating vector with defective LTRs and an internal HCMV promoter that drives LacZ expression. (B) Western blot analysis of HEK293T transiently transfected with GFP and ZASC1 expression plasmid and plasmids expressing either one of four shRNAs targeting the open reading frame of ZASC1, a control shRNA targeting secreted alkaline phosphatase (AP), or the empty shRNA vector. Cell lysates were analyzed by Western blotting as described in Materials and Methods with rabbit antibodies against human ZASC1, human actin, and GFP. The blot is representative of three independent experiments. (C) HEK293T cells were transiently transfected with 80 ng of plasmids expressing either one of four shRNAs targeting the open reading frame of ZASC1, a control shRNA targeting secreted alkaline phosphatase (AP), or the empty shRNA vector along with 20 ng of an expression vector encoding the ASLV receptor TVA800 (2, 35). Three days posttransfection, cells were challenged with EnvA-pseudotyped MLV vectors, either CMMP-luciferase, which directs MLV LTR-driven firefly luciferase gene expression, or pQCLIN, which directs β-galactosidase expression from an internal HCMV promoter. Infection was monitored using chemiluminescent assays to detect reporter enzyme activities along with a chemiluminescent assay to measure relative viable cell numbers, and the data are reported as the ratio of reporter gene activity to the relative viable cell number observed, with CHO-K1 cells defined as 100% infection. The data shown are the average mean values obtained in an experiment performed with quadruplicate samples, and each is representative of three independent experiments. Error bars indicate the standard deviations of the data. P values were calculated using a standard Student's t test.
The role for ZASC1 in MLV gene expression was further validated by the transient RNAi-mediated knockdown of ZASC1 in human HEK293T cells. These cells were transiently transfected with plasmids carrying one of four ZASC1 shRNAs that reduce ZASC1 levels in transfected cells (Fig. 4B). To mark the transfected cells, the shRNA plasmids were cotransfected with an expression vector encoding the ASLV receptor TVA800. Three days posttransfection, the cells were challenged with two different EnvA-pseudotyped MLV vectors, either CMMP-luciferase, which directs firefly luciferase gene expression from the MLV LTR, or the self-inactivating QCLIN vector. Since human cells do not express the receptor for EnvA, only cells cotransfected with TVA-800 and the shRNA-encoding plasmids are infected under these conditions. The independent expression of four distinct ZASC1 shRNAs reduced reporter gene expression from the CMMP-luciferase vector between 9- and 12.5-fold (Fig. 4C). In stark contrast, these shRNAs only inhibited expression from the internal HCMV promoter of the QCLIN vector by 2-fold or less (Fig. 4C). These data, obtained with multiple ZASC1 shRNAs, cannot be explained by nonspecific, off-target effects, and they provide additional strong support for the model that ZASC1 promotes gene expression from the MLV LTR promoter.
ZASC1 binds to three sites in the U3 region of the MLV LTR.
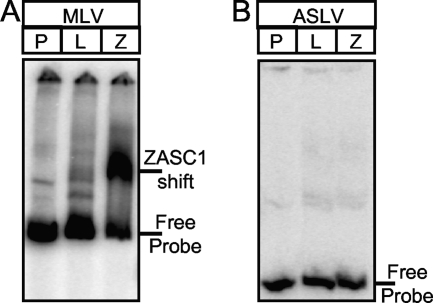
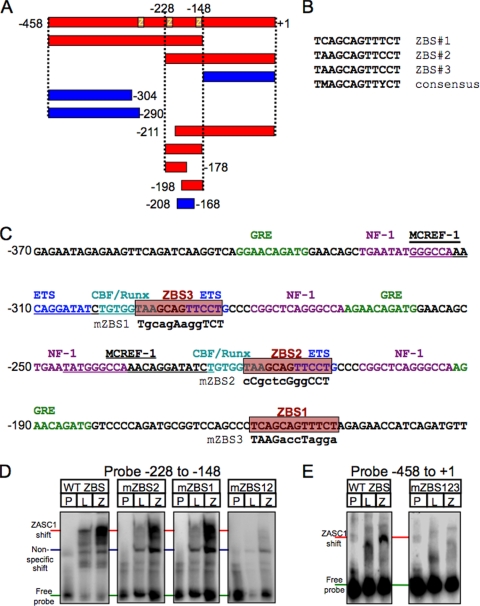
In retroviral LTR promoters, the majority of transcription control elements are contained in the unique 3′ (U3) DNA element. To determine if ZASC1 can bind directly to the U3 DNA element of the MLV LTR, electrophoretic mobility assays (EMSA) were performed. The MLV U3 DNA element was PCR amplified, end labeled with γ-32P, and mixed with in vitro transcription/translation reaction mixtures containing either ZASC1 or luciferase proteins. For control purposes, the ASLV U3 DNA element was studied in parallel, because that virus was not influenced to the same extent by ZASC1 deficiency (Fig. 1B). The samples then were resolved by 4% PAGE and exposed to phosphorimager plates. A specific high-molecular-weight mobility shift band was observed when ZASC1 was incubated with the MLV, but not the ASLV, U3 DNA element (Fig. 5A and B). To map the ZASC1 binding site in the MLV U3 DNA region, a series of overlapping restriction fragments and oligonucleotides were tested by EMSA for the ability to support ZASC1 binding (Fig. 6A). These studies indicated that at least two independent DNA sequences, one located between −228 and approximately −208 and another located between approximately −168 and −148, are important for ZASC1 binding. Consistent with these binding results, each of these U3 subregions contains a nearly identical 12-bp sequence (TAAGCAGTTCCT versus TCAGCAGTTTCT; between nucleotides −221 and −210 and between −160 and −149, respectively) (Fig. 6B and C). To test if these sequences are important for ZASC1 binding, each was mutated, and the impact of these mutations was tested individually or in combination upon ZASC1 binding. A DNA fragment corresponding to the −223 to −148 region of MLV U3 bound ZASC1 when either of these two sites was present but not when both were mutated (Fig. 6D). These data demonstrate that ZASC1 binds to these 12-bp elements in the MLV U3 region. Since this region partially overlaps the direct repeat in the MLV U3 enhancer region, there actually are three potential ZASC1 binding sites (site 3 is located between nucleotides −296 and −285) in the MLV U3 region, numbered ZBS1 to ZBS3 (Fig. 6B and C). ZBS2 and ZBS3 are located in the direct repeats of the MLV enhancer region and partially overlap CBF/Runx and ETS transcription factor binding sites, which have important functions in different myeloid cell lines (19, 28). ZBS1 is positioned downstream of the enhancer in a region of the promoter in which no other transcription factor binding sites have been identified so far. The simultaneous mutation of all three binding sites abolished ZASC1 binding to the entire MLV U3 DNA region (mZBS123 in Fig. 6E).
FIG. 5.
ZASC1 binds to the MLV LTR. EMSA of DNA fragments corresponding to the U3 promoter regions of MLV (−458 to +1) (A) and ASLV (−233 to −64) (B) that were end labeled with γ-32P and mixed with in vitro transcription translation reaction mixtures containing either luciferase (L) or ZASC1 (Z) proteins. Protein/DNA complexes were resolved on 4% TBE polyacrylamide gels, dried, and exposed to phosphorimager plates. Lanes containing DNA probe alone (P) are indicated, as are the positions of the free probe and the ZASC1-specific band.
FIG. 6.
ZASC1 binds to three highly related sites in the MLV promoter. (A) DNA restriction fragments or oligonucleotides corresponding to the indicated regions of the MLV promoter were tested for the ability to bind to ZASC1 as described for Fig. 4. Those that bound ZASC1 are indicated in red, and those that did not bind are in blue. (B) Alignment of three putative ZASC1 binding sites (ZBS) in the MLV promoter. (C) The MLV U3 region from nucleotides −370 to −131 is shown with the location of previously characterized transcription factor sites: glucocorticoid response element (GRE) (green), nuclear factor 1 (NF-1) (purple), ETS sites (dark blue), CBF/Runx sites (light blue), and MCREF-1 sites (underlined). Red boxes indicate the locations of the three ZASC1 binding sites. Mutations introduced into the ZASC1 binding sites to test the effect of mutation on ZASC1 binding and MLV infection are indicated underneath the WT sequence. (D) EMSA of oligonucleotides corresponding to the −228 to −148 MLV U3 fragment with either ZBS1 or ZBS2 mutated. The locations of free probe, nonspecific shifts, and the specific ZASC1 bandshift are indicated. The data shown are representative of at least three independent experiments. (E) EMSA of the complete WT MLV U3 DNA fragment or the corresponding version with all three ZBS sites mutated (mZBS123).
ZASC1 activates the MLV promoter primarily through binding ZBS1.
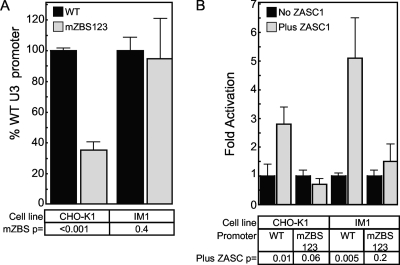
To test the effects of mutating ZASC1 binding sites on the activity of the MLV promoter, CHO-K1 and IM1 cells were transfected with a reporter plasmid that expressed Guassia luciferase under the control of either the WT MLV promoter or a promoter in which all three ZBS had been mutated (mZBS123) as in Fig. 6C. Notably, in WT CHO-K1 cells, the mZBS123 promoter exhibited only 36% of the activity of the WT promoter (Fig. 7A). However, in IM1 cells, the transcriptional activities of the WT and mZBS123 promoters were equivalent (Fig. 7A), albeit at a level 4-fold lower than the levels seen with the WT promoter in WT CHO-K1 cells: the relative gLuc activity of the WT promoter was 315,725 ± 21,892 in CHO-K1 cells versus 75,237 ± 6,408 in IM1 cells. We concluded from this experiment that ZASC1 affects the basal activity of the MLV promoter in a ZBS-dependent fashion. Consistently, in plasmid cotransfection experiments, exogenously expressed ZASC1 stimulated the activity of a WT MLV promoter by 2.8-fold in CHO-K1 cells and by 5.1-fold in the ZASC1-deficient IM1 cells (Fig. 7B). As expected, exogenous ZASC1 expression did not stimulate expression from the mZBS123 reporter construct in either cell line (Fig. 7B). These data support the EMSA-derived model that ZASC1 binds to three highly similar DNA binding sites in the MLV U3 DNA region, thereby stimulating transcription from the MLV promoter.
FIG. 7.
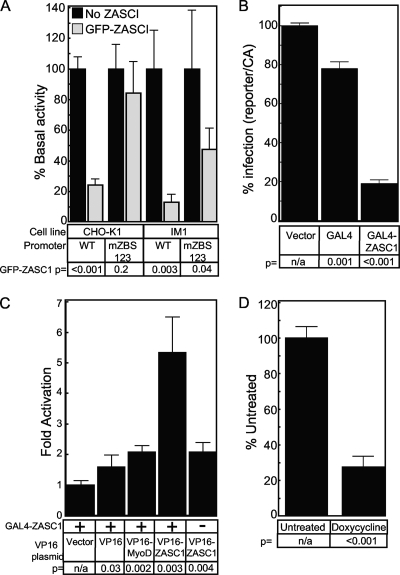
ZASC1 activates the expression of the MLV promoter. (A) Basal activity of a plasmid encoding Gaussia luciferase under the control of either the WT MLV LTR promoter or a promoter with all three ZBS mutated in transiently transfected CHO-K1 or IM1 cells. Promoter activity of the WT plasmid is set to 100% for each cell line. (B) CHO-K1 or IM1 cells transiently transfected with WT or mZBS reporter constructs with or without a ZASC1 expression plasmid. WT and mZBS promoter activity in the absence of ZASC1 in each cell line was set to 1, and fold-activation in the presence of ZASC1 was determined as described in Materials and Methods. The data shown are the average mean values obtained in an experiment performed with quadruplicate samples, and each is representative of three independent experiments. Error bars indicate the standard deviations of the data. P values were calculated using a standard Student's t test.
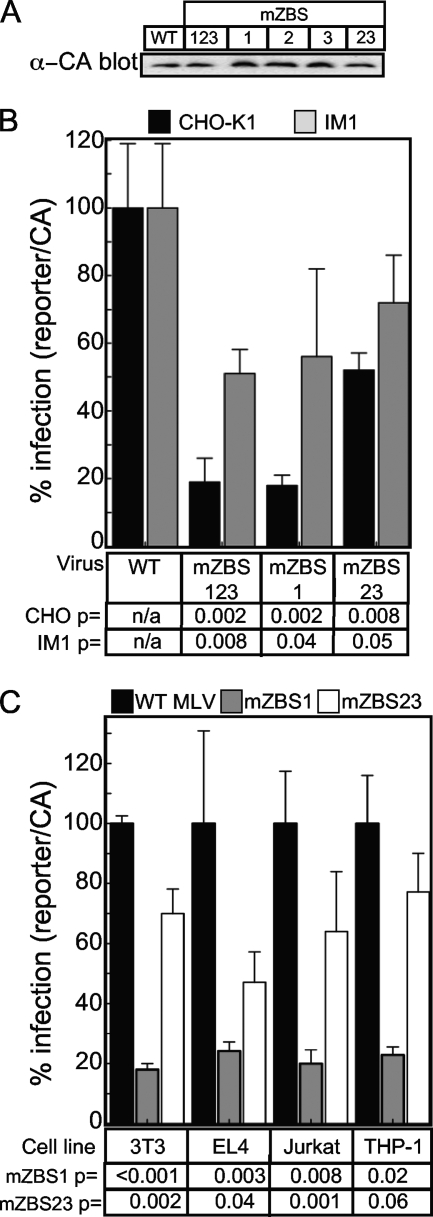
To determine if ZASC1 binding sites contribute to MLV provirus gene expression, the mutations that abolished transcription factor binding were introduced into the 3′ U3 element of the plasmid pCMMP-SEAP-IRES-GFP. Since the HCMV immediate-early promoter is used to express the MLV genome RNA from this plasmid in producer cell lines, virus production was unaffected by the introduced mutations (Fig. 8A). However, after reverse transcription and integration, the mutated U3 elements are present in both 5′ and 3′ LTRs, regulating provirus reporter gene expression. In wild-type CHO-K1 cells, the triple mutant (mZBS123) virus exhibited a 5.3-fold reduced level of reporter gene activity compared to that of the WT virus (Fig. 8B). A virus construct with only ZBS1 mutated exhibited a nearly identical 5.6-fold reduction in reporter gene expression. However, viral challenge with a virus that contained mutations in both ZBS2 and ZBS3 exhibited only a 1.9-fold reduction in reporter gene expression. Thus, ZBS1 is the most critical ZASC1 binding site for MLV provirus expression (Fig. 8B). In contrast, challenge of IM1 cells resulted in a less-than 2-fold reduction in reporter gene expression for all three ZBS mutant viruses. Since mutating ZASC1 binding sites should have little effect in a cell line that is already deficient in ZASC1, these data strongly indicate that the effect of the introduced ZBS mutations is mediated primarily through ZASC1 rather than through perturbing other transcription factor sites for the mZBS123 and mZBS1 viruses. In keeping with the much weaker effect of the mZBS23 mutations, the difference between the effects of these mutations in CHO-K1 and IM1 cells was not large enough (1.9-fold in CHO-K1 versus 1.4-fold in IM1 [Fig. 8B]) to conclude that the majority of the effect is through ZASC1 rather than the perturbation of other overlapping transcription factor sites (Fig. 6C). Individually mutating ZBS1, ZBS2, and ZBS3 confirmed that ZBS1 is the most critical site for MLV provirus expression (data not shown).
FIG. 8.
Mutation of ZASC1 binding sites inhibits MLV vector gene expression. (A) Capsid Western blot (α-CA) analysis of representative viral supernatants used in infection assays. (B) Reporter assays of CHO-K1 or IM1 cells challenged with MLV vectors expressing SEAP under the control of either the WT MLV U3 promoter, a U3 promoter with all 3 ZASC1 binding sites mutated (mZBS123), binding site 1 mutated (mZBS1), or both binding sites 2 and 3 mutated (mZBS23). (C) Reporter assays of mouse fibroblast (3T3), mouse T-cell (EL4), human T-cell (Jurkat), or human monocyte (THP-1) cell lines challenged with MLV vectors expressing firefly luciferase and containing either the mZBS1 or mZBS2 mutation. Infection was monitored using chemiluminescent assays to detect reporter enzyme activities. Infections were normalized to input virus as determined by anti-capsid Western blotting and are reported as the ratio of reporter gene activity to input capsid observed. The data obtained with the WT constructs is defined as 100% infection. The data shown are the average mean values obtained in an experiment performed with quadruplicate samples, and each is representative of three independent experiments. P values were calculated using a standard Student's t test.
Since ZASC1 exhibits a broad tissue distribution (14), we tested if the ZBS mutants exhibited similar phenotypes in more relevant cell lines for MLV infection, namely, mouse cells and cells of hematopoietic origin (Fig. 8C). Similar to the results observed in CHO-K1 cells, challenge of mouse fibroblasts (3T3 cells), mouse and human T-cell lines (EL4 and Jurkat, respectively), and a human monocyte cell line (THP-1) with the mZBS1 mutant resulted in 4.2- to 5.6-fold lower levels of reporter gene expression than those with WT MLV vectors. In contrast, the mZBS23 mutation exhibited at most a 2-fold reduction in reporter gene expression in the cell lines tested. These data are consistent with a model in which ZBS1 is the primary mediator of the ZASC1 regulation of MLV transcription in all cell lines tested.
A GFP-ZASC1 fusion protein exhibits dominant-negative activity on MLV gene expression.
During attempts to complement the IM1 cell line, we observed that the expression of an N-terminal GFP-ZASC1 fusion protein failed to complement MLV vector function in IM1 cells despite the ability of the fusion protein to bind to DNA (data not shown) and properly localize to the nucleus (data not shown). To assess if the GFP-ZASC1 fusion protein was defective for the activation of the MLV promoter, transient transfection assays were performed. Surprisingly, the exogenous expression of the GFP-ZASC1 fusion protein not only failed to activate the MLV promoter but also inhibited reporter gene expression 4.1-fold in CHO-K1 cells (Fig. 9A). The inhibition was even greater in IM1 cells (7.7-fold). These data suggest that the lower levels of WT ZASC1 in IM1 cells rendered the promoter more sensitive to the GFP-ZASC1 fusion protein and implies a dominant-negative mechanism of action for the recombinant protein. Indeed, similar results have been reported with a GAL4-ZASC1 fusion protein inhibiting expression from a promoter with GAL4 binding sites (3). We tested a similar GAL4-ZASC1 construct to confirm this earlier work (Fig. 9B) and utilized this fusion protein in a mammalian two-hybrid assay to show that ZASC1 self associates (Fig. 9C). These data support the idea that large N-terminal fusions of ZASC1 inhibit WT ZASC1 through the formation of inactive multimers. This notion is further supported by the observation that the fusion protein had a minimal effect on the mZBS123 MLV promoter in CHO-K1 cells (Fig. 9A). Similar to the more severe effect of GFP-ZASC1 on the WT promoter in IM1 cells compared to that on CHO-K1 cells, the dominant-negative effect on the mZBS123 promoter is more pronounced (2-fold) in IM1 cells, although it is still well below the 7.7-fold effect observed on the WT promoter. This minor inhibition might be due to GFP-ZASC1 titrating out cellular proteins common to multiple promoters, such as CBP, a phenomenon previously described as squelching (9, 24). Indeed, at high expression levels of GFP-ZASC1, we have observed similar moderate (2-fold) inhibition of other promoters with no recognizable ZASC1 binding sites (data not shown).
FIG. 9.
GFP-ZASC1 fusion protein inhibits expression from the MLV promoter. (A) CHO-K1 or IM1 cells transiently transfected with WT or mZBS reporter constructs with or without a GFP-ZASC1 expression plasmid. WT and mZBS promoter activity in the absence of GFP-ZASC1 in each cell line was set to 100%, and activity in the presence of GFP-ZASC1 was determined as described in Materials and Methods. (B) GAL4-ZASC1 fusion protein inhibits expression from an artificial promoter with five GAL4 binding sites upstream of a TATA box. HEK293 cells transiently transfected with a GAL4 reporter construct and either GAL4 or GAL4 expression plasmids. Promoter activity in transfections with empty vector were set to 100%, and activity levels in the presence of GAL4 and GAL4-ZASC1 expression plasmids were determined as described in Materials and Methods. (C) Mammalian two-hybrid analysis of GAL4-ZASC1 fusion protein. HEK293 cells transiently transfected with a GAL4-ZASC1 expression plasmid and an additional expression plasmid encoding a VP16 activation domain fusion protein. (D) CHO-TREX cells (CHO-K1 cells expressing the Tet repressor) were stably transduced with a lentivirus construct encoding a Tet-inducible GFP-ZASC1 fusion protein. GFP-ZASC1 expression was induced with 1 μg/ml doxycycline, and 24 h postinduction the cells were infected with the VSV-G-pseudotyped MLV vector (MMP-nls-LacZ[VSV-G]) encoding β-galactosidase. Reporter gene expression from the newly acquired virus was monitored 72 h postinduction using chemiluminescent assays, and the data are reported with no doxycycline treatment defined as 100% infection. The data shown are the average mean values obtained in an experiment performed with quadruplicate samples, and each is representative of three independent experiments. Error bars indicate the standard deviations of the data. P values were calculated using a standard Student's t test.
To determine if expressing the GFP-ZASC1 fusion protein could inhibit MLV vector expression, we employed a tetracycline-inducible system. CHO-Trex cells, a cell line that expresses the Tet repressor, were transduced with a lentiviral vector that encodes the GFP fusion protein under the control of a tetracycline-inducible promoter. The fusion protein was induced with 1 μg/ml of doxycycline, and 24 h postinduction the cells were challenged with MMP-nls-LacZ. Reporter gene expression from the newly acquired virus was reduced 3.6-fold (Fig. 9D). Thus, expressing the GFP-ZASC1 fusion protein exhibits dominant-negative effects on expression from incoming MLV vectors.
Taken together, these data demonstrate that ZASC1 is a novel, sequence-specific transcription factor that binds to the MLV U3 LTR promoter and activates viral gene expression.
DISCUSSION
Here, we have presented the results of an insertional mutagenesis screen and have provided multiple lines of evidence that ZASC1 is a DNA sequence-specific transcriptional activator of the MLV LTR promoter. First, we isolated a clonal cell line, IM1, that exhibited 10-fold lower levels of viral gene expression on MLV vector challenge (Fig. 1A) but supported WT levels of MLV reverse transcription and integration (Fig. 2A and B). The IM1 cell line contained the mutagenic pRET vector located adjacent to the ZASC1 gene (Fig. 3A) and was deficient in ZASC1 protein expression (Fig. 3B). Subsequent cDNA complementation of ZASC1 expression restored susceptibility to MLV vector challenge (Fig. 3C). The knockdown of endogenous ZASC1 by expressing ZASC1-specific shRNAs in human cells specifically reduced the levels of MLV vector LTR-driven gene expression and validated the role of ZASC1 in MLV infection (Fig. 4C). Consistent with the specific effect on MLV transcription, ZASC1 did not influence reporter gene expression from either an internal CMV promoter contained within an LTR-defective MLV vector (Fig. 4A and C) or LTR expression from an ASLV vector (Fig. 1B). Also, EMSA analyses demonstrated that ZASC1 binds to three highly related sites in the U3 region of the MLV LTR (Fig. 5 and 6), but it does not bind the corresponding region of the ASLV genome (Fig. 5). Mutagenesis analysis revealed that the ZASC1 binding site closest to the transcription start site was most critical for regulating MLV gene expression (Fig. 8). That site, ZBS1, does not overlap with other known transcription factor binding sites in the MLV LTR (Fig. 6C). Importantly, ZASC1 binding site mutations had little impact on MLV vector expression in IM1 cells (Fig. 8A). Since the defect in the IM1 cell line is due to ZASC1 deficiency, the lack of a further reduction in gene expression demonstrates that these ZBS mutations specifically affect ZASC1 binding and do not perturb the function of other transcription factor binding sites. Taken together, these data provide strong evidence supporting the identification of ZASC1 as a newly defined transcriptional activator of the MLV promoter.
While WT ZASC1 is a transcriptional activator, our data show that the expression of a GFP-ZASC1 fusion protein inhibits expression from the MLV promoter in transient transfection assays (Fig. 9A). Furthermore, the expression of GFP-ZASC1 inhibits MLV vector gene expression from a newly acquired infection (Fig. 9D). Since, as detailed above, WT ZASC1 acts as a transcriptional activator of the MLV promoter, it seems likely that the inhibitory effects of GFP-ZASC1 are due to the fusion protein acting in a dominant-negative manner. These results are consistent with a prior report that a GAL4 DNA binding domain (GAL4 DBD)-ZASC1 fusion protein repressed luciferase expression from a reporter construct consisting of GAL4 binding sites inserted upstream of the simian virus 40 (SV40) promoter (3). In this same study, WT ZASC1 expression had no effect on the GAL4/SV40 promoter, so clearly the ZASC1 fusion protein was targeted to this construct through the GAL4 DBD. Such dominant-negative activity may be due to the N-terminal ZASC1 fusion proteins: forming inactive multimers with WT ZASC1, sterically blocking the assembly of higher-order complexes on the DNA, failing to interact with a vital cellular coactivator when bound to DNA, or sequestering coactivators away from the target promoter due to a failure to bind DNA. Since the GFP-ZASC1 fusion protein can bind DNA (data not shown) and the dominant- negative activity of the GFP-ZASC1 fusion protein requires ZASC1 binding sites on the promoter (Fig. 9A), the latter possibility seems unlikely, at least for the GFP-ZASC1 fusion. Additionally, mammalian two-hybrid results (Fig. 9B and C) suggest that ZASC1 multimerizes. Thus, it seems likely that the N-terminal fusion proteins form inactive multimers on DNA. Elucidating the dominant-negative mechanism of action of the N-terminal ZASC1 fusion proteins could contribute to understanding the functions of the WT protein.
Our data, along with the coimmunoprecipitation of WT ZASC1 in a complex with the coactivator and histone acetyl-transferase CBP (17), show that WT ZASC1 functions as a classical transcriptional activator on the MLV promoter. In viral promoters and, by extension, cellular promoters, ZASC1 binds to sequences in the promoter and either recruits coactivators such as p300/CBP or stabilizes the association of other cellular activators or components of the preinitiation complex with the promoter.
It will be important for future studies to determine how ZASC1 interacts with other cellular transcription factors that bind the MLV LTR promoter. Previous studies have shown that the enhancer region of the MLV promoter contains multiple overlapping binding sites for various transcription factors (Fig. 6C), and the differential occupancy of these sites affects cell line tropism and disease outcome (11, 19, 21, 25-29). Indeed, it is interesting that ZBS2 and ZBS3 partially overlap the CBF/Runx and ETS sites in the MLV enhancer. However, our data suggest that these sites contribute minimally to ZASC1 function in the cell lines we have tested (Fig. 8). Instead, ZBS1, which does not overlap with known transcription factor binding sites, is responsible for the majority of ZASC1 function in these cell lines (Fig. 8). The analysis of the interaction of ZASC1 with other transcription factors that bind in the MLV LTR should not only contribute to the understanding of how ZASC1 regulates MLV gene expression but also serve as a well-established, tractable model for the regulation of cellular genes by ZASC1. Indeed, the identification here of the MLV ZASC1 binding sites should facilitate identifying cellular promoters regulated by ZASC1. Additionally, the dominant-negative form of ZASC1 provides a useful tool for characterizing ZASC1 activity on viral and cellular promoters. Thus, the results presented here and future work aimed at identifying the specific mechanism by which ZASC1 binding regulates MLV infection should help to uncover precisely how cells regulate provirus transcriptional competency and provide insights into how ZASC1 contributes to the development of ataxias and squamous cell carcinomas.
Acknowledgments
We thank Nolan Wessell for technical assistance.
This work was supported by NIH grant CA22443 (P.A.) and NIH grant AI72645 (J.A.T.Y.). P.A. is an investigator of the Howard Hughes Medical Institute.
Footnotes
Published ahead of print on 19 May 2010.
REFERENCES
- 1.Adkins, H. B., S. C. Blacklow, and J. A. Young. 2001. Two functionally distinct forms of a retroviral receptor explain the nonreciprocal receptor interference among subgroups B, D, and E avian leukosis viruses. J. Virol. 75:3520-3526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bates, P., J. A. Young, and H. E. Varmus. 1993. A receptor for subgroup A Rous sarcoma virus is related to the low density lipoprotein receptor. Cell 74:1043-1051. [DOI] [PubMed] [Google Scholar]
- 3.Bogaerts, S., A. Vanlandschoot, J. van Hengel, and F. van Roy. 2005. Nuclear translocation of alphaN-catenin by the novel zinc finger transcriptional repressor ZASC1. Exp. Cell Res. 311:1-13. [DOI] [PubMed] [Google Scholar]
- 4.Brass, A. L., D. M. Dykxhoorn, Y. Benita, N. Yan, A. Engelman, R. J. Xavier, J. Lieberman, and S. J. Elledge. 2008. Identification of host proteins required for HIV infection through a functional genomic screen. Science 319:921-926. [DOI] [PubMed] [Google Scholar]
- 5.Bruce, J. W., P. Ahlquist, and J. A. Young. 2008. The host cell sulfonation pathway contributes to retroviral infection at a step coincident with provirus establishment. PLoS Pathog. 4:e1000207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bruce, J. W., K. A. Bradley, P. Ahlquist, and J. A. Young. 2005. Isolation of cell lines that show novel, murine leukemia virus-specific blocks to early steps of retroviral replication. J. Virol. 79:12969-12978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bushman, F. D., N. Malani, J. Fernandes, I. D'Orso, G. Cagney, T. L. Diamond, H. Zhou, D. J. Hazuda, A. S. Espeseth, R. Konig, S. Bandyopadhyay, T. Ideker, S. P. Goff, N. J. Krogan, A. D. Frankel, J. A. Young, and S. K. Chanda. 2009. Host cell factors in HIV replication: meta-analysis of genome-wide studies. PLoS Pathog. 5:e1000437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Federspiel, M. J., P. Bates, J. A. Young, H. E. Varmus, and S. H. Hughes. 1994. A system for tissue-specific gene targeting: transgenic mice susceptible to subgroup A avian leukosis virus-based retroviral vectors. Proc. Natl. Acad. Sci. U. S. A. 91:11241-11245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gill, G., and M. Ptashne. 1988. Negative effect of the transcriptional activator GAL4. Nature 334:721-724. [DOI] [PubMed] [Google Scholar]
- 10.Goff, S. P. 2007. Host factors exploited by retroviruses. Nat. Rev. Microbiol. 5:253-263. [DOI] [PubMed] [Google Scholar]
- 11.Golemis, E. A., N. A. Speck, and N. Hopkins. 1990. Alignment of U3 region sequences of mammalian type C viruses: identification of highly conserved motifs and implications for enhancer design. J. Virol. 64:534-542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gotte, M., X. Li, and M. A. Wainberg. 1999. HIV-1 reverse transcription: a brief overview focused on structure-function relationships among molecules involved in initiation of the reaction. Arch. Biochem. Biophys. 365:199-210. [DOI] [PubMed] [Google Scholar]
- 13.Gupta, R. S., D. Y. Chan, and L. Siminovitch. 1978. Evidence for functional hemizygosity at the Emtr locus in CHO cells through segregation analysis. Cell 14:1007-1013. [DOI] [PubMed] [Google Scholar]
- 14.Imoto, I., Y. Yuki, I. Sonoda, T. Ito, Y. Shimada, M. Imamura, and J. Inazawa. 2003. Identification of ZASC1 encoding a Kruppel-like zinc finger protein as a novel target for 3q26 amplification in esophageal squamous cell carcinomas. Cancer Res. 63:5691-5696. [PubMed] [Google Scholar]
- 15.Ishida, Y., and P. Leder. 1999. RET: a poly A-trap retrovirus vector for reversible disruption and expression monitoring of genes in living cells. Nucleic Acids Res. 27:e35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Julius, M. A., Q. Yan, Z. Zheng, and J. Kitajewski. 2000. Q. vectors, bicistronic retroviral vectors for gene transfer. Biotechniques 28:702-708. [DOI] [PubMed] [Google Scholar]
- 17.Jung, S. Y., A. Malovannaya, J. Wei, B. W. O'Malley, and J. Qin. 2005. Proteomic analysis of steady-state nuclear hormone receptor coactivator complexes. Mol. Endocrinol. 19:2451-2465. [DOI] [PubMed] [Google Scholar]
- 18.Konig, R., Y. Zhou, D. Elleder, T. L. Diamond, G. M. Bonamy, J. T. Irelan, C. Y. Chiang, B. P. Tu, P. D. De Jesus, C. E. Lilley, S. Seidel, A. M. Opaluch, J. S. Caldwell, M. D. Weitzman, K. L. Kuhen, S. Bandyopadhyay, T. Ideker, A. P. Orth, L. J. Miraglia, F. D. Bushman, J. A. Young, and S. K. Chanda. 2008. Global analysis of host-pathogen interactions that regulate early-stage HIV-1 replication. Cell 135:49-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lewis, A. F., T. Stacy, W. R. Green, L. Taddesse-Heath, J. W. Hartley, and N. A. Speck. 1999. Core-binding factor influences the disease specificity of Moloney murine leukemia virus. J. Virol. 73:5535-5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lim, J., T. Hao, C. Shaw, A. J. Patel, G. Szabo, J. F. Rual, C. J. Fisk, N. Li, A. Smolyar, D. E. Hill, A. L. Barabasi, M. Vidal, and H. Y. Zoghbi. 2006. A protein-protein interaction network for human inherited ataxias and disorders of Purkinje cell degeneration. Cell 125:801-814. [DOI] [PubMed] [Google Scholar]
- 21.Manley, N. R., M. O'Connell, W. Sun, N. A. Speck, and N. Hopkins. 1993. Two factors that bind to highly conserved sequences in mammalian type C retroviral enhancers. J. Virol. 67:1967-1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Naghavi, M. H., and S. P. Goff. 2007. Retroviral proteins that interact with the host cell cytoskeleton. Curr. Opin. Immunol. 19:402-407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Naghavi, M. H., S. Valente, T. Hatziioannou, K. de Los Santos, Y. Wen, C. Mott, G. G. Gundersen, and S. P. Goff. 2007. Moesin regulates stable microtubule formation and limits retroviral infection in cultured cells. EMBO J. 26:41-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Natesan, S., V. M. Rivera, E. Molinari, and M. Gilman. 1997. Transcriptional squelching re-examined. Nature 390:349-350. [DOI] [PubMed] [Google Scholar]
- 25.Redondo, J. M., J. L. Pfohl, C. Hernandez-Munain, S. Wang, N. A. Speck, and M. S. Krangel. 1992. Indistinguishable nuclear factor binding to functional core sites of the T-cell receptor delta and murine leukemia virus enhancers. Mol. Cell. Biol. 12:4817-4823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Speck, N. A., B. Renjifo, E. Golemis, T. N. Fredrickson, J. W. Hartley, and N. Hopkins. 1990. Mutation of the core or adjacent LVb elements of the Moloney murine leukemia virus enhancer alters disease specificity. Genes Dev. 4:233-242. [DOI] [PubMed] [Google Scholar]
- 27.Speck, N. A., B. Renjifo, and N. Hopkins. 1990. Point mutations in the Moloney murine leukemia virus enhancer identify a lymphoid-specific viral core motif and 1,3-phorbol myristate acetate-inducible element. J. Virol. 64:543-550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sun, W., B. J. Graves, and N. A. Speck. 1995. Transactivation of the Moloney murine leukemia virus and T-cell receptor beta-chain enhancers by cbf and ets requires intact binding sites for both proteins. J. Virol. 69:4941-4949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sun, W., M. O'Connell, and N. A. Speck. 1993. Characterization of a protein that binds multiple sequences in mammalian type C retrovirus enhancers. J. Virol. 67:1976-1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tsuchiya, S., M. Yamabe, Y. Yamaguchi, Y. Kobayashi, T. Konno, and K. Tada. 1980. Establishment and characterization of a human acute monocytic leukemia cell line (THP-1). Int. J. Cancer 26:171-176. [DOI] [PubMed] [Google Scholar]
- 31.Weller, S. K., A. E. Joy, and H. M. Temin. 1980. Correlation between cell killing and massive second-round superinfection by members of some subgroups of avian leukosis virus. J. Virol. 33:494-506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Whitcomb, J. M., and S. H. Hughes. 1992. Retroviral reverse transcription and integration: progress and problems. Annu. Rev. Cell Biol. 8:275-306. [DOI] [PubMed] [Google Scholar]
- 33.Wolf, D., and S. P. Goff. 2008. Host restriction factors blocking retroviral replication. Annu. Rev. Genet. 42:143-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yeung, M. L., L. Houzet, V. S. Yedavalli, and K. T. Jeang. 2009. A genome-wide short hairpin RNA screening of jurkat T-cells for human proteins contributing to productive HIV-1 replication. J. Biol. Chem. 284:19463-19473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Young, J. A., P. Bates, and H. E. Varmus. 1993. Isolation of a chicken gene that confers susceptibility to infection by subgroup A avian leukosis and sarcoma viruses. J. Virol. 67:1811-1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zack, J. A., S. J. Arrigo, S. R. Weitsman, A. S. Go, A. Haislip, and I. S. Chen. 1990. HIV-1 entry into quiescent primary lymphocytes: molecular analysis reveals a labile, latent viral structure. Cell 61:213-222. [DOI] [PubMed] [Google Scholar]
- 37.Zhou, H., M. Xu, Q. Huang, A. T. Gates, X. D. Zhang, J. C. Castle, E. Stec, M. Ferrer, B. Strulovici, D. J. Hazuda, and A. S. Espeseth. 2008. Genome-scale RNAi screen for host factors required for HIV replication. Cell Host Microbe 4:495-504. [DOI] [PubMed] [Google Scholar]