Abstract
In this study, we characterized Autographa californica multiple nucleopolyhedrovirus (AcMNPV) orf76 (ac76), which is a highly conserved gene of unknown function in lepidopteran baculoviruses. Transcriptional analysis of ac76 revealed that transcription of multiple overlapping multicistronic transcripts initiates from a canonical TAAG late-transcription start motif but terminates at different 3′ ends at 24 h postinfection in AcMNPV-infected Sf9 cells. To investigate the role of ac76 in the baculovirus life cycle, an ac76-knockout virus was constructed using an AcMNPV bacmid system. Microscopy, titration assays, and Western blot analysis demonstrated that the resulting ac76-knockout virus was unable to produce budded viruses. Quantitative real-time PCR analysis demonstrated that ac76 deletion did not affect viral DNA synthesis. Electron microscopy showed that virus-induced intranuclear microvesicles as well as occlusion-derived virions were never observed in cells transfected with the ac76-knockout virus. Confocal microscopy analysis revealed that Ac76 was predominantly localized to the ring zone of nuclei during the late phase of infection. This suggests that ac76 plays a role in intranuclear microvesicle formation. To the best of our knowledge, this is the first baculovirus gene identified to be involved in intranuclear microvesicle formation.
Baculoviruses are arthropod-specific, rod-shaped, enveloped viruses with circular, supercoiled double-stranded DNA genomes that replicate in the nuclei of host cells (38). The Baculoviridae are divided into four genera: Alphabaculovirus (lepidopteran-specific nucleopolyhedrovirus [NPV]), Betabaculovirus (lepidopteran-specific granuloviruses), Gammabaculovirus (hymenopteran-specific NPV), and Deltabaculovirus (dipteran-specific NPV) (38). Autographa californica multiple nucleopolyhedrovirus (AcMNPV) is the archetype species of Alphabaculovirus (38). During the AcMNPV life cycle, two virion phenotypes, budded virus (BV) and occlusion-derived virus (ODV), are produced. Both types of virions have similar nucleocapsid structures and identical genetic information, but they differ in the origin and composition of their envelopes and in the roles they play in the baculovirus life cycle (32). During the early phase of infection, viral DNA replicates in the virogenic stroma, and newly synthesized viral genomes are condensed and packaged into rod-shaped capsids to form nucleocapsids. Nucleocapsids are transported into the cytoplasm and become enveloped by budding through the GP64-modified plasma membrane, thereby forming BVs. BVs are responsible for spreading infection between susceptible insect tissues and between cells in cell culture. During the late phase of infection, nucleocapsids remain localized within a peristromal ring zone, where they are enveloped in intranuclear microvesicles (whose formation is induced by viral infection) to form ODVs. The ODVs are subsequently embedded into a paracrystalline protein matrix to form occlusion bodies (OBs). ODVs play a role in the horizontal transmission of infection among insect hosts (42).
Baculovirus gene expression follows a temporal cascade, which is primarily regulated at the transcriptional level (32). Viral gene transcription can be subdivided into three major phases: early, late, and very late. Early genes are transcribed by host RNA polymerase II. A common regulatory motif of many baculovirus early genes includes a TATA promoter sequence and the transcriptional initiation consensus sequence CAGT (10). Transcription of late and very late genes is dependent on viral DNA replication, is mediated by a virus-encoded RNA polymerase, and is generally initiated from the baculovirus late promoter motif TAAG (22).
AcMNPV has a genome of approximately 134 kbp that contains 154 predicted open reading frames (ORFs) (2). AcMNPV orf76 (ac76) encodes a putative 9.4-kDa protein (2). Homologs of ac76 have been identified in all sequenced lepidopteran baculovirus genomes (14, 40). Analysis with the InterProScan program shows that the ac76 homologs constitute a DUF843 baculovirus protein family (IPR008561) of unknown function. In this study, we generated an ac76-knockout mutant to investigate the role of ac76 in the AcMNPV life cycle. We found that ac76 is essential for both BV and ODV development but is not required for viral DNA synthesis. Electron microscopy showed that ac76 is not required for nucleocapsid assembly but that it is required for intranuclear microvesicle formation, ODV envelopment, and the subsequent embedding of virions into OBs.
MATERIALS AND METHODS
Viruses and cell lines.
Bacmid bMON14272, containing an AcMNPV genome, was propagated in Escherichia coli DH10B as previously described (23). The Sf9 cell strain, the clonal isolate 9 of the parent cell line IPLB-Sf21-AE which is derived from the fall armyworm Spodoptera frugiperda (41), was cultured at 27°C in TNM-FH medium (Invitrogen) supplemented with 10% fetal bovine serum, penicillin (100 μg/ml), and streptomycin (30 μg/ml). BV titers were determined with a TCID50 endpoint dilution assay using Sf9 cells (29).
RNA preparation and Northern blot analysis.
Sf9 cells were infected with AcMNPV at a multiplicity of infection (MOI) of 10 50% tissue culture infective doses (TCID50) per cell. Cells were collected at various time points. Total cellular RNA was isolated using an RNeasy Mini-Kit (Qiagen) according to the manufacturer's instructions. The RNA samples were quantified by optical density measurements at 260 nm.
Total RNA (10 μg) was denatured with glyoxal for 1 h at 55°C and electrophoresed on 1.5% agarose gel in 1× BPTE buffer [10 mM piperazine-N,N′-bis(2-ethanesulfonic acid) (PIPES), 30 mM Bis-Tris, 10 mM EDTA, pH 8.0]. RNA was transferred to a positively charged nylon membrane (Hybond-N+; Amersham) and fixed to the membrane by UV cross-linking. The probe was a 59-nucleotide (nt) single-stranded DNA sequence (5′-TAAACACGAGAAACAATATGAGCAGATACAAAAAGATGCTGTTTTCCTTTTTGTCATAC-3′; synthesized by Invitrogen). Labeling of probe, hybridization, and signal detection were performed by using Amersham AlkPhos Direct Labeling and Detection Systems according to the manufacturer's recommendations.
Analysis by 3′ and 5′ RACE.
3′ Rapid amplification of cDNA ends (RACE) was performed using a 5′/3′ RACE Kit, 2nd Generation (Roche), with 1 μg of total RNA. Following the manufacturer's instructions, first-strand cDNA synthesis was performed with SuperScript III reverse transcriptase (Invitrogen) and an oligo(dT) anchor primer. The cDNA mixtures were amplified via PCR with a PCR oligo(dT) anchor primer and the ac76-specific forward primer (5′-ATATTTGTTGTTGGGCGCACTGG-3′). The PCR products were analyzed by agarose gel electrophoresis, and the bands of the product that was amplified from the RNA isolated at 24 h postinfection (p.i.) were purified and cloned into pMD18-T vector (TaKaRa) for sequencing.
The 5′ end of the ac76 transcript was determined with the 5′/3′ RACE Kit, 2nd Generation(Roche), using 1 μg of purified total RNA isolated at 24 h p.i. Briefly, first-strand cDNA synthesis was performed with the ac76-specific reverse primer GSP1 (5′-TTAGAATCGCATCAAGCGCTTG-3′). The cDNA was purified with a High Pure PCR Purification Kit (Promega), and a poly(A) tail was added to the 5′ end using terminal transferase and dATP. The tailed cDNAs were amplified by PCR using an oligo(dT) anchor primer and the nested GSP2 primer (5′-ACGGTAGACTCGGTGTTCTTAC-3′). The PCR products obtained were gel purified and cloned into the pMD18-T vector (TaKaRa) for sequencing.
Generation of the ac76-knockout AcMNPV bacmid.
An ac76-knockout AcMNPV bacmid was generated via homologous recombination in E. coli employing the bacmid bMON14272 as previously described (4, 43, 44). A transfer vector in which the ac76 locus region was replaced with a chloramphenicol resistance (Cm) gene for antibiotic selection in E. coli was constructed as follows. The 1,039-bp Cm gene cassette was excised by PstI/BamHI from the pUC18-US-Cm-DS plasmid (43), purified, and ligated into pUC18 to generate the recombinant plasmid pUC-Cm. Using the primers ac76-US1 (5′-TACTGCAGCTAGGCTAAATATGGCCAGTGC-3′; PstI site is underlined) and ac76-US2 (5′-ATAAGCTTCGCAGTCGTAATAACACACTCAAC-3′; HindIII site is underlined), a 478-bp fragment homologous to the 5′ region of ac76 was amplified from the AcMNPV genome via PCR. The resulting product was ligated into the PstI/HindIII-digested pUC-Cm plasmid to generate the recombinant plasmid pUC-US-Cm. Using the primers ac76-DS1 (5′-AAGGTACCATCAACCTGTCGCCTACTGAG-3′; KpnI site is underlined) and ac76-DS2 (5′-TAGGATCCGTCAGCCCGGCCATTATAAG-3′; BamHI site is underlined), a 409-bp fragment homologous to the 3′ region of ac76 was amplified from the AcMNPV genome via PCR. The PCR product was then cloned into the KpnI/BamHI-digested plasmid pUC-US-Cm to generate the final ac76-knockout transfer vector pUC-US-Cm-DS. This transfer vector was then digested with KpnI and HindIII, and the resulting linear 1,926-bp fragment containing the Cm gene cassette and the ac76 flanking region was gel purified and resuspended in distilled water at a final concentration of 200 ng/μl.
To facilitate homologous recombination between the Cm gene and the target sequence, DH10Bac cells (DH10B containing the bacmid bMON14272) were transformed with pBAD-gbaA, which supplied the λ Red recombination function (26). The resulting cells were then induced by the addition of l-arabinose to allow expression of the λ Red system, made competent, and electro-transformed with 1 μg of the purified linear 1,926-bp recombinant fragment, as previously described (30). The electroporated cells were incubated at 37°C for 1 h in 1 ml of SOC (35) medium and were subsequently spread onto agar medium containing 20 μg/ml chloramphenicol, 50 μg/ml kanamycin, and 7 μg/ml tetracycline. Plates were incubated at 37°C for 2 days, and colonies that were resistant to chloramphenicol and kanamycin were selected for further confirmatory testing by PCR and Southern blot analysis. The resulting ac76-knockout bacmid was named vAcac76-KO.
Verification of the recombinant bacmids.
The deletion of ac76 by the substitution of the Cm gene cassette was confirmed by using four pairs of specific PCR primers. CmU and CmD were used to detect the correct insertion of the Cm gene cassette. The primers ac76-US2 and ac76-DS1 were used to confirm the deletion region. The primer pairs CmU/ac76-US2 and ac76-DS1/CmD were used to examine the recombination junctions of the upstream and downstream flanking regions.
To further confirm the deletion of ac76 and the replacement of it by the Cm gene in vAcac76-KO, Southern blot hybridization analysis was performed as previously described (43). Briefly, a 59-nt single-strand DNA which is identical to the ac76 deletion region (mentioned above in Northern blot analysis) was used as a probe to detect the ac76. Meanwhile, the Cm gene fragment was amplified with primers CmU and CmD via PCR, and the resulting product was used as a probe to detect the Cm cassette. AcMNPV and vAcac76-KO bacmid DNA were digested with SacI, run overnight in an ethidium bromide-stained 0.8% agarose gel, and transferred on a positively charged nylon membrane (Hybond-N+; Amersham). The probes were labeled with alkaline phosphatase. Labeling of probe, hybridization, and signal detection were performed according to the manufacturer's instructions (Gene Images AlkPhos Direct Labeling and Detection System; Amersham).
Construction of knockout, repair and wt AcMNPV bacmids.
In order to facilitate the detection of recombinant virus infection and to examine whether the ac76 knockout has any effect on OB morphogenesis, the polyhedrin gene (polh; designated “PH” in constructs) of AcMNPV and the green fluorescent protein gene (gfp) were inserted into the polh locus of vAcac76-KO by site-specific transposition, as previously described (43). A 950-bp fragment containing ac76 with its own promoter and a poly(A) signal was amplified via PCR using the primers ac76-US2 and ac76-DS1. The PCR product was digested with EcoRI/PstI and cloned into pFB1-PH-GFP (43) to generate pFB1-ac76-PH-GFP. Electrocompetent DH10B cells containing the pMON7124 helper plasmid and vAcac76-KO were transformed with either the pFB1-PH-GFP or pFB1-ac76-PH-GFP donor plasmid to generate the ac76-knockout bacmid vAcac76-KO-PH-GFP or the ac76-repair bacmid vAcac76-REP-PH-GFP, respectively. Electrocompetent DH10B cells containing the helper plasmid pMON7124 and the bMON14272 bacmid were transformed with pFB1-PH-GFP to generate the wild-type (wt) control bacmid, which we named vAcPH-GFP. Successful transposition was confirmed by PCR. The correct recombinant bacmids were electroporated into E. coli DH10B cells and were screened for tetracycline sensitivity to ensure that the isolated bacmids were free of helper plasmids. Bacmid DNA was then extracted and purified using a Qiagen Large-Construct Kit and was quantified by optical density.
Analysis of viral growth.
Sf9 cells (1.0 × 106 cells/35-mm-diameter dish) were transfected in triplicate with 1.0 μg of each bacmid construct (vAcac76-KO-PH-GFP, vAcac76-REP-PH-GFP, or vAcPH-GFP) using Cellfectin liposome reagent (Invitrogen), or cells were infected in triplicate with BV at an MOI of 5. The cell debris was removed by centrifugation (3,000 × g for 10 min). The supernatants containing BVs were collected at different time points, and titers were determined with a TCID50 endpoint dilution assay using Sf9 cells (29).
Western blot analysis.
Sf9 cells (1 × 106) were transfected with 1.0 μg of each bacmid (vAcac76-KO-PH-GFP, vAcac76-REP-PH-GFP, or vAcPH-GFP). BVs were purified as previously described (24, 44). At 120 h posttransfection (p.t.), the cultures were harvested and centrifuged at 2,000 × g for 20 min at room temperature to pellet the cells. The cells were resuspended in double-distilled H2O (ddH2O) for Western blot analysis. The supernatants (3 ml) were loaded onto a 25% sucrose cushion and were centrifuged at 80,000 × g for 90 min at 4°C in a SW41 Ti rotor. The pellets of vAcac76-KO-PH-GFP-transfected and mock-transfected cells were resuspended in 9.9 μl of 250 mM Tris-Cl (pH 7.8) and 0.1 μl of protease inhibitor cocktail (Protease Inhibitor Cocktail Set III; Calbiochem), while the pellets of vAcac76-REP-PH-GFP-transfected and vAcPH-GFP-transfected cells were resuspended in four times the volume of the same solutions due to the abundance of BVs present. For Western blot analysis, one-quarter of the vAcac76-REP-PH-GFP and vAcPH-GFP samples and the totality of the vAcac76-KO-PH-GFP- and mock-transfected samples were electrophoresed on a 10% SDS-PAGE gel and were electrophoretically transferred to a nitrocellulose transfer membrane (Schleicher and Schuell) according to the manufacturer's instructions. Western blotting was performed using standard methods (35). One-sixteenth of the vAcac76-REP-PH-GFP- and vAcPH-GFP-transfected cell extracts and one-quarter of the vAcac76-KO-PH-GFP- and mock-transfected cell extracts were used for Western blot analysis. A polyclonal primary antibody against AcMNPV VP39 (18) was used at a dilution of 1:200. A goat anti-rabbit-horseradish peroxidase (HRP) secondary antibody (Amersham Biosciences) was used at a dilution of 1:5,000. Blots were detected using an enhanced chemiluminescence system (ECL; Amersham) according to the manufacturer's instruction.
DNA synthesis analysis by real-time PCR.
To assess viral DNA synthesis, quantitative real-time PCR was performed as previously described with several modifications (39). A recombinant virus, Ac-GP64-KO which contains a deletion of the gp64 envelope fusion protein gene, was used as a noninfectious control as the deletion of gp64 results in a virus unable to propagate infection from cell to cell and could provide a more accurate comparison (25, 39). Briefly, 1 × 106 Sf9 cells were transfected in triplicate with 1 μg of Ac-GP64-KO or vAcac76-KO-PH-GFP bacmid DNA, and cells were collected at selected time points. Total DNA from each sample was prepared with a Universal Genomic DNA Extraction Kit (TaKaRa) according to the manufacturer's protocol. The total DNA was resuspended in 150 μl of sterile water. Prior to PCR, 5 μl of total DNA from each time point was digested with 20 units of DpnI restriction enzyme (NEB) overnight in a 50-μl reaction volume to eliminate input bacmid DNA. Quantitative PCR (qPCR) was performed with 10 μl of the digested DNA and the Hot Start PCR Master Mix III (Chaoshi-Bio) according to the manufacturer's instructions using the primers targeting at a 100-bp region of the gp41 gene and conditions described previously (39).
Electron microscopy.
A total of 1 × 106 Sf9 cells (per 35-mm-diameter dish) were transfected with 1.0 μg of vAcPH-GFP, vAcac76-KO-PH-GFP, or vAcac76-REP-PH-GFP. At 72 h p.i., cells were dislodged with a rubber policeman and pelleted at 3,000 × g for 5 min. Then the cells were fixed, dehydrated, embedded, sectioned, and stained as described previously (19). Samples were examined with a JEM-100CX/II transmission electron microscope at an accelerating voltage of 80 kV.
Construction of GFP fusion recombinant bacmids and confocal microscopy.
To monitor the localization of Ac76 in AcMNPV-infected Sf9 cells, Ac76 was expressed in frame with GFP to create an Ac76-GFP fusion protein. A recombinant fusion bacmid vAcac76-KO-PH-Ac76GFP and a control bacmid vAcPH-p76GFP were constructed as previously described (43, 44). To generate a recombinant bacmid containing the polh gene cassette, a donor plasmid called pFB1-PhR was first constructed as follows. The polh cassette was excised from pFB1-PH-GFP (43) with EcoRI and SnaBI. The resulting fragment was cloned into pFastBac1 (Invitrogen), which was digested with EcoRI and SnaBI, yielding pFB1-PhR. The gfp ORF was excised from pUC19egfp (43) with XbaI and PstI, and the resulting fragment was cloned into pFB1-PhR to generate the pFB1-ph-gfp. The ac76 gene (without its stop codon TGA) with its native promoter was amplified from the AcMNPV bacmid using the primers ac76CF-D2 (5′-TCTAGAATCTATTGAGCTGGTATTTTTGTTTAG-3′; XbaI site is underlined) and ac76CF-U (5′-GAATTCACGCCCTTCAAAGATTTCAG-3′; EcoRI site is underlined). The PCR product was digested with EcoRI and XbaI and was cloned into pFB1-ph-gfp to create the donor plasmid pFB1-ph-ac76-gfp. Competent cells containing the vAcac76-KO bacmid and the helper plasmid pMON7124 were transformed with pFB1-ph-ac76-gfp, and the ac76-gfp chimera and polh were site-specifically transposed into the vAcac76-KO bacmid polh locus. The resulting virus was named vAcac76-KO-PH-Ac76GFP, and this virus expressed Ac76 with a GFP tag under the control of the native ac76 promoter. The ac76 promoter was amplified via PCR using the primers ac76CF-U and ac76CF-D1 (5′-TCTAGATTTTATTCCCTTACTCTATTCGTTGC-3′; the XbaI site is underlined). The EcoRI/XbaI-digested PCR product was then inserted into pFB1-ph-gfp to generate the donor plasmid pFB1-ph-p76-gfp. The control bacmid vAcPH-p76GFP, in which only GFP was expressed (under the control of the ac76 promoter), was generated using a procedure similar to that used for the creation of vAcac76-KO-PH-Ac76GFP.
Sf9 cells (1 × 106) were transfected with 1 μg of vAcPH-p76GFP or vAcac76-KO-PH-Ac76GFP DNA. At 96 h p.t., supernatants were collected, and BV titers were determined by a TCID50 endpoint dilution assay using Sf9 cells. For confocal microscopy analysis, Sf9 cells (1 × 105) were seeded onto glass coverslips. Cells were infected with vAcPH-p76GFP or vAcac76-KO-PH-Ac76GFP at an MOI of 10. At 12, 24, 48, and 72 h p.t., cells were visualized with a Leica TCS SP2 confocal laser scanning microscope to search for fluorescence using a wavelength of 488 nm for GFP. All images were digitally recorded and were merged using the Leica software.
RESULTS
Comparison of the amino acid sequences of Ac76 homologs.
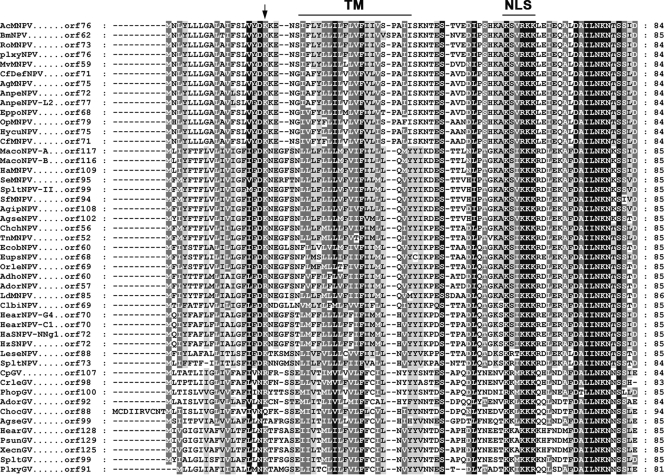
Homologs of Ac76 are found in all sequenced lepidopteran baculovirus genomes stored in the GenBank at the National Center for Biotechnology Information. A comparison of the predicted amino acid sequences showed that three regions are highly conserved: (i) a classical putative nuclear localization signal [(K/R)X2KKK] located near the C terminus (Fig. 1, above-lined NLS); (ii) a highly conserved motif of unknown function, DAILNKNTSSID, located at the C terminus; and (iii) a positively charged amino acid (K/R) (Fig. 1, arrow) close to a highly hydrophobic transmembrane domain (Fig. 1, above-lined TM) (predicted by TMHMM [http://genome.cbs.dtu.dk/services/TMHMM/]). This sequence pattern is similar to the inner nuclear membrane-sorting motif (INM-SM) of the viral envelope protein ODV-E66 of AcMNPV (5, 8) which has been demonstrated to function as an N-terminal signal anchor and targets INM-SM-containing proteins to ODV envelope (6, 9).
FIG. 1.
Amino acid sequence alignment of Ac76. The alignment was performed using ClustalX, version 1.83, and was adjusted using GeneDoc software. Black shading denotes identical amino acids, and gray shading denotes similar amino acids. Residue numbers are indicated on the left. The predicted transmembrane domain (TM) and putative nuclear localization signal (NLS) are indicated by the horizontal lines at the top of the sequences. The positively charged amino acids close to the end of the TM are noted by an arrow. TM helix predictions were made using the TMHMM program.
Transcriptional mapping analysis of ac76 transcripts.
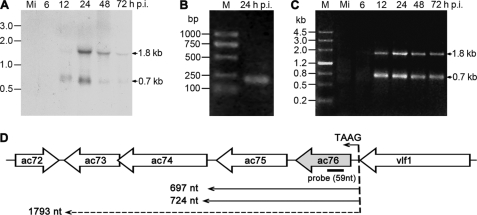
The temporal expression of ac76 was examined with total RNA extracted from AcMNPV-infected Sf9 cells at different time points by using Northern blot analysis as well as 5′ and 3′ RACE analysis. Northern blot analysis demonstrated that a 1.8-kb and a 0.7-kb transcript were present with increasing abundance between 12 h and 24 h p.i. and remained present until 72 h p.i. (Fig. 2A). The transcriptional initiation site of the ac76 transcripts was determined by 5′ RACE using total RNA collected at 24 h p.i. Only one 5′ RACE product was obtained (Fig. 2B). Five clones derived from the product were sequenced, and the results revealed that all of the transcripts were initiated at the first A of the canonical baculovirus late promoter motif TAAG, which is located 12 nt upstream from the ac76 translation initiation codon ATG (Fig. 2D). The termination sites of ac76 were examined using 3′ RACE. A transcriptional pattern similar to that observed using Northern blot analysis was revealed by 3′ RACE. Two 3′ RACE products that were 1.8 kb and 0.7 kb in length were first detected at 12 h p.i. and continued to be detected until 72 h p.i. (Fig. 2C). Five clones derived from the 1.8 kb PCR product that was present at 24 h p.i. were sequenced. The results showed that this 1.8-kb mRNA terminated at the T of the putative ac73 stop codon TAA and that the transcript could potentially encode ac76, ac75, ac74, and ac73 of the AcMNPV genome (Fig. 2D). Ten PCR clones from the 0.7-kb product isolated at 24 h p.i. were sequenced and revealed two different transcript stop points located 13 and 40 nt downstream from the ac75 stop codon (Fig. 2D). These two transcripts could potentially encode ac76 and ac75.
FIG. 2.
Temporal expression of the ac76 transcript in AcMNPV-infected Sf9 cells. Total RNA was extracted from mock-infected (lane Mi) and AcMNPV-infected cells at designated time points. (A) Northern blot analysis of the transcript from the ac76 region. A 59-nt single-stranded, alkaline phosphatase-labeled cRNA probe complementary to ac76 was used, and the sizes of specific hybridization bands are indicated on the right. (B) The 5′ RACE product of the ac76 transcripts. Total RNA derived from AcMNPV-infected cells at 24 h p.i. was used. The PCR product was purified and sequenced to determine the transcriptional start site. (C) Analysis by 3′ RACE of ac76 transcripts. The PCR products derived from RNA isolated at 24 h p.i. were purified and sequenced to determine the transcriptional stop site. M, DNA marker. The sizes of the PCR products are indicated on the right. (D) The transcriptional initiation and termination sites of ac76 transcripts.
Generation of the ac76-knockout, repair, and wt AcMNPV bacmids.
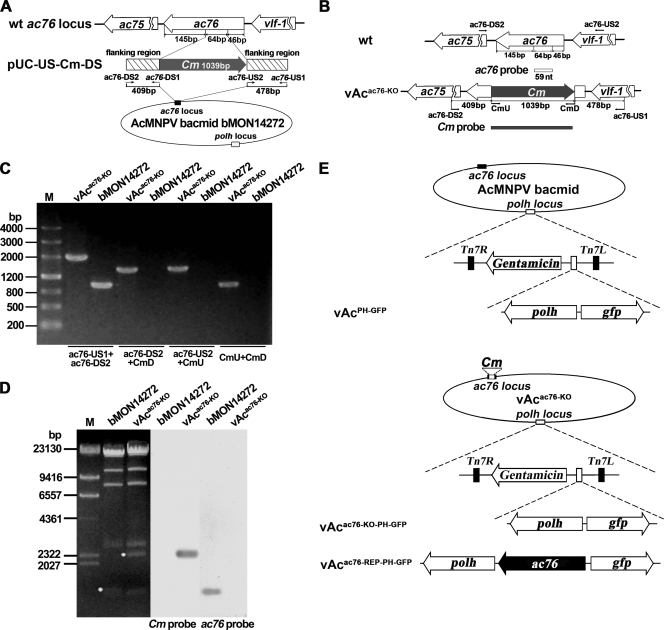
The ac76-knockout AcMNPV bacmid, vAcac76-KO, was constructed using λ Red homologous recombination as previously described (43). To avoid affecting the transcription of either ac75 or vlf1, the resulting deletion left 145 bp at the 3′ end and 46 bp at the 5′ end of the ac76 ORF, respectively (Fig. 3A). The remaining 64-bp coding sequence was replaced with the Cm cassette (Fig. 3A).
FIG. 3.
Construction of the ac76-knockout bacmid. (A) Strategy for construction of an ac76-knockout bacmid using the AcMNPV bacmid system. A 64-bp sequence of the ac76 ORF was deleted and replaced with the Cm gene sequence. (B) Positions of primer pairs and probes used to confirm the disruption of ac76 and the insertion of the Cm gene. (C) PCR analysis to determine the presence or absence of sequence modification in bMON14272 and vAcac76-KO. The primer pairs are shown below the panel, and the templates are noted above each lane. (D) Southern blot analysis of the bMON14272 bacmid and vAcac76-KO. The ac76 probe and the Cm gene probe were used to confirm the deletion of ac76 and its replacement by the Cm gene. (E) Schematic diagram of the recombinant viruses vAcPH-GFP, vAcac76-KO-PH-GFP, and vAcac76-REP-PH-GFP showing the polh and gfp genes inserted in the polh locus by Tn7-mediated transposition. The ac76 ORF that was inserted into vAcac76-KO was controlled by its own promoter.
PCR confirmed the correct insertion of the Cm gene into the ac76 locus of the bMON14272 bacmid (Fig. 3B and C). Southern blot analysis further indicated that the 64-bp fragment of the ac76 gene was successfully replaced by the Cm gene and confirmed the absence of an intact ac76 gene in the vAcac76-KO genome (Fig. 3D).
To examine if the ac76 deletion had any effect on OB morphogenesis and to facilitate observation of viral infection, the polh gene of AcMNPV and the gfp gene were inserted into the polh locus of vAcac76-KO via transposition. The resulting bacmid was named vAcac76-KO-PH-GFP (Fig. 3E). As a positive control, vAcPH-GFP was constructed by transposing polh and gfp into the polh locus of the bMON14272 bacmid (Fig. 3E). To ensure that the phenotype resulting from the ac76 knockout was due only to the deletion of ac76 and not to disruption of flanking genes, a repair bacmid was generated. A repair bacmid, vAcac76-REP-PH-GFP, was generated in which the ac76 gene with its own promoter and poly(A) signal, as well as polh and gfp, were inserted into the polh locus of vAcac76-KO by transposition (Fig. 3E). All constructs were confirmed by PCR analysis (data not shown).
Analysis of vAcac76-KO-PH-GFP, vAcac76-REP-PH-GFP, and vAcPH-GFP replication in transfected Sf9 cells.
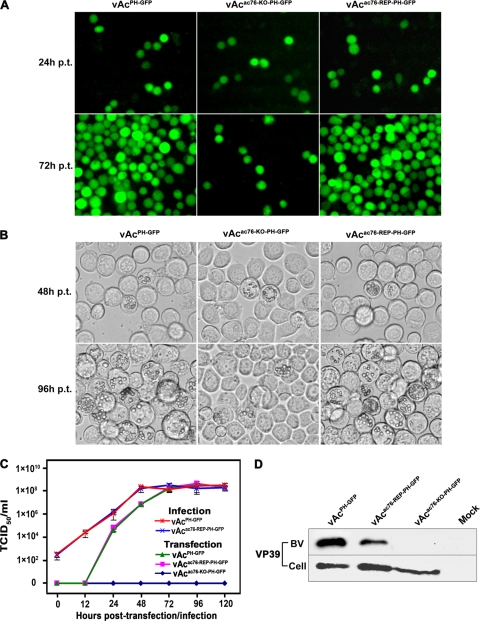
To examine the effect of ac76 deletion on viral propagation, Sf9 cells were transfected with vAcac76-KO-PH-GFP, vAcac76-REP-PH-GFP, or vAcPH-GFP and monitored by fluorescence microscopy. No difference was observed among the three viruses at 24 h posttransfection (p.t.), and the transfection efficiencies were comparable (approximately 10%) (Fig. 4A). There was almost no increase in the number of fluorescent cells in the vAcac76-KO-PH-GFP-transfected cells up to 72 h p.t., indicating that there was no spread of the virus from the initially transfected cells. In contrast, fluorescence was observed in almost all cells transfected with vAcac76-REP-PH-GFP or vAcPH-GFP by 72 h p.t., indicating that both viruses were able to generate infectious BVs from the initial transfection (Fig. 4A).
FIG. 4.
Analysis of viral replication in Sf9 cells. (A) Fluorescence microscopy of Sf9 cells transfected with vAcPH-GFP, vAcac76-KO-PH-GFP, or vAcac76-REP-PH-GFP at 24 h and 72 h p.t. (B) Light microscopy of Sf9 cells transfected with vAcPH-GFP, vAcac76-KO-PH-GFP, or vAcac76-REP-PH-GFP at 48 h and 96 h p.t. (C) Virus growth curves generated from Sf9 cells transfected or infected with virus. For the transfection growth curves, Sf9 cells were transfected with each bacmid DNA. For the infection growth curves, cells were infected with each virus at an MOI of 5. The supernatants were harvested at the designated time points, and virus titers were determined by TCID50 endpoint dilution assays. Each data point represents the average titer of three independent transfections or infections. Error bars indicate standard deviations. (D) Western blot analysis of purified BV particles and cell extracts. The cells and supernatants were harvested from Sf9 cells that had been transfected with each virus, and BVs were purified from the supernatants. Cell extracts (Cell) and purified BV particles were separated by SDS-PAGE and analyzed with anti-VP39 to detect the nucleocapsid protein VP39.
Light microscopy showed that OBs with a normal appearance formed in cells transfected individually with the three viruses and that the number of cells containing OBs did not differ among the three constructs at 48 h p.t (Fig. 4B). However, by 96 h p.t., most of the cells transfected with vAcac76-REP-PH-GFP or vAcPH-GFP contained OBs, but the number of vAcac76-KO-PH-GFP-transfected cells containing OBs did not increase (Fig. 4B).
These results suggested that the deletion of ac76 leads to a defect in infectious BV production and no secondary infection in Sf9 cells. To better assess the effect that ac76 deletion had on virus replication and to compare the replication kinetics of the virus constructs, virus growth was analyzed. Sf9 cells were transfected with vAcac76-KO-PH-GFP, vAcac76-REP-PH-GFP, or vAcPH-GFP individually. At selected time points, the BV titers were determined using a TCID50 endpoint dilution assay. The virus titer from the supernatant of vAcac76-KO-PH-GFP-transfected cells could not be determined at any time point up to 120 h p.t., indicating that no infectious progeny viruses were produced. In contrast, the Sf9 cells transfected with vAcac76-REP-PH-GFP and vAcPH-GFP showed a normal increase in BV titers (Fig. 4C). To further determine whether or not the ac76-repair virus was able to rescue the defect in infectious BV production observed in vAcac76-KO-PH-GFP-transfected cells, a second growth analysis was performed with the BVs produced from cells transfected with vAcac76-REP-PH-GFP or vAcPH-GFP at an MOI of 5. The ac76-repair virus showed similar replication kinetics to the wt virus, indicating that the defective phenotype could be rescued by inserting ac76 into the polh locus of the vAcac76-KO-PH-GFP and that the defect in BV production in cells transfected with the ac76-knockout virus was due only to the deletion of ac76 (Fig. 4C).
To further determine if any noninfectious BVs budded from the vAcac76-KO-PH-GFP-transfected cells and if the expression of the primary capsid protein, VP39, was inhibited in vAcac76-KO-PH-GFP-transfected cells, Western blot analysis was performed to compare the levels of VP39 in the supernatants and the cell extracts of bacmid-transfected cells (Fig. 4D). VP39 was detected in the extracts of the cells transfected with vAcac76-KO-PH-GFP, vAcac76-REP-PH-GFP, or vAcPH-GFP but not in the mock-transfected cell extracts, indicating that vp39 expression was not affected in the vAcac76-KO-PH-GFP-transfected cells. However, VP39 was detected only in the supernatants of vAcac76-REP-PH-GFP- and vAcPH-GFP-transfected cells. In contrast, no VP39 protein was detected, even when a longer exposure time was used, in the supernatants of cells transfected with vAcac76-KO-PH-GFP or the supernatants of mock-transfected cells, indicating that ac76 deletion results in a defect in BV production (Fig. 4D).
Quantitative analysis of viral DNA synthesis.
To determine whether or not ac76 deletion affected viral DNA synthesis, the level of viral DNA synthesis in vAcac76-KO-PH-GFP-transfected cells was assayed by quantitative real-time PCR (qPCR) analysis. A gp64-knockout bacmid was used as a noninfectious control virus (25, 39). The results of this analysis showed that the ac76 deletion virus was able to synthesize viral DNA at levels similar to those of the bacmid lacking gp64 during a 96-h period, indicating that the deletion of ac76 does not impact viral DNA synthesis (Fig. 5).
FIG. 5.
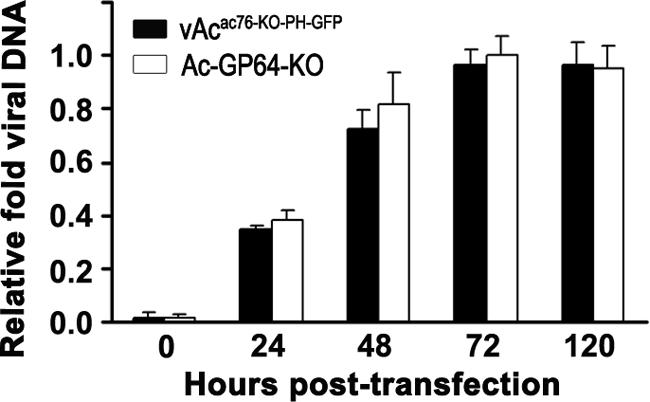
Real-time PCR analysis of viral DNA synthesis in Sf9 cells. Total DNA was isolated from Sf9 cells transfected with vAcac76-KO-PH-GFP or vAc-GP64-KO at selected time points, digested with the restriction enzyme DpnI to eliminate input bacmid DNA, and assayed by real-time PCR. The numerical data on the y axis were normalized by comparison to the lowest threshold cycle (CT) value (i.e., the maximum number of DNA copies) of sample measured by real-time PCR in each assay. Values represent the means from three independent transfections. Error bars indicate standard deviations.
Electron microscopy analysis of vAcac76-KO-PH-GFP-, vAcac76-REP-PH-GFP- and vAcPH-GFP-transfected cells.
The results described above showed that ac76 was required for BV production but that deletion of ac76 did not affect viral DNA synthesis or the progression of the viral infection to the very late phase, as evidenced by the presence of OBs in the nuclei of transfected cells. To further determine if the lack of ac76 had any effect on virion morphogenesis, electron microscopy analysis was performed with thin sections generated from vAcac76-KO-PH-GFP-, vAcac76-REP-PH-GFP-, and vAcPH-GFP-transfected cells.
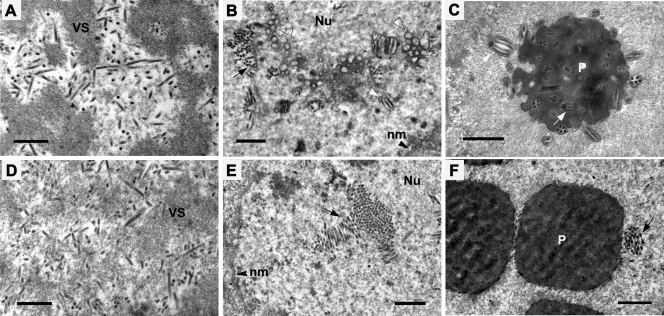
At 72 h p.t., cells transfected with vAcac76-REP-PH-GFP showed the typical characteristics of a baculovirus infection, such as the presence of a well-defined virogenic stroma inundated with electron-dense rod-shaped nucleocapsids (Fig. 6A), nucleocapsids accumulating and aligning with nuclear envelopes that had been synthesized de novo (Fig. 6B), and nucleocapsids acquiring their envelopes and embedding into the developing OBs (Fig. 6C). In cells transfected with vAcac76-KO-PH-GFP, the development of the virogenic stroma and abundant normal-appearing nucleocapsids could also be observed (Fig. 6D). Accumulation of nucleocapsids was observed in the ring zone; however, no virus-induced intranuclear microvesicles could be observed within the ring zone, and nucleocapsids were never enveloped to form ODVs (Fig. 6E). The OBs were purified from Sf9 cells transfected with vAcac76KO-PH-GFP or vAcPH-GFP to be subjected to Western blot analysis. Polyhedrin was detected in the OBs from cells transfected with either wt or knockout virus. However, VP39 was detected in the OBs only from cells transfected with wt but not the knockout virus (data not shown). This result showed that the ac76-knockout virus OBs did not contain any ODVs although the shape and the size of OBs were similar to those observed in wt virus-transfected (data not shown) or repair virus-transfected cells (Fig. 6F). The electron microscopy indicated that the deletion of ac76 had no effect on nucleocapsid assembly but that it did disrupt intranuclear microvesicle formation as well as ODV envelopment and OB formation.
FIG. 6.
Transmission electron microscopy analysis of Sf9 cells transfected with either vAcac76-REP-PH-GFP (A to C) or vAcac76-KO-PH-GFP (D to F) at 72 h p.t. (A and D) Normal nucleocapsids appeared at the electron-dense edges of the virogenic stroma (VS). (B) Bundles of nucleocapsids aligning with envelopes, microvesicles (triangle), and ODVs (white arrow) in the ring zone of vAcac76-REP-PH-GFP-transfected cells. (C) Normal virions embedded within polyhedra. (E) Nucleocapsids forming a bundle (black arrow) in the ring zone of vAcac76-KO-PH-GFP-transfected cells without evidence of intranuclear microvesicles or ODV presence. (F) Polyhedra devoid of normal virions. Nu, nucleus; nm, nuclear membrane; P, polyhedra. Scale bar, 500 nm.
Localization of Ac76 in AcMNPV-infected Sf9 cells.
To monitor the subcellular localization of Ac76, two recombinant viruses, vAcPH-p76GFP and vAcac76-KO-PH-Ac76GFP, were constructed (Fig. 7A). Ac76 was expressed in frame with GFP to produce an Ac76-GFP chimera that was under the control of the ac76 promoter in vAcac76-KO-PH-Ac76GFP. The C-terminal GFP tag of the chimera facilitated visualization of the trafficking of Ac76 in virus-infected cells. As a control, GFP alone was expressed under the control of ac76 promoter in vAcPH-p76GFP (Fig. 7A).
FIG. 7.
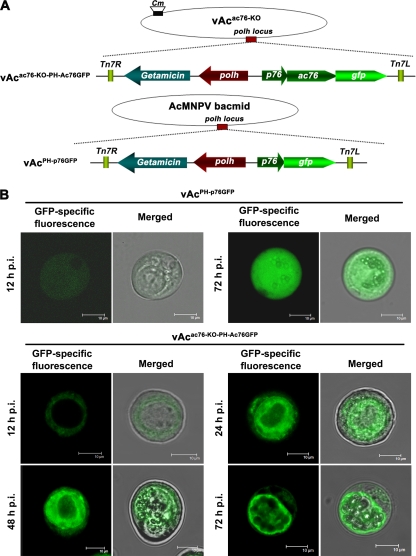
Subcellular localization of the Ac76-GFP fusion protein in Sf9 cells infected with GFP-tagged virus. (A) Schematic diagram of the generation of GFP-tagged recombinant viruses. An ac76-gfp chimera, under the control of the native ac76 promoter, and polh were inserted into the polh locus of the vAcac76-KO bacmid to generate vAcac76KO-PH-Ac76GFP. The control virus vAcPH-p76GFP was constructed by transposing gfp (under the control of the ac76 promoter) and polh into the AcMNPV bacmid. (B) Confocal images of Sf9 cells infected with recombinant virus. Cells were infected with vAcac76-KO-PH-Ac76GFP at an MOI of 10 and were observed for fluorescence by confocal laser scanning microscopy at 12, 24, 48, and 72 h p.i. As a control, cells were infected with vAcPH-p76GFP and examined with the confocal microscope. For each time point, GFP-specific fluorescence micrographs are shown to the left of the merged micrographs. Scale bar, 10 μm.
It was observed that fluorescence could spread from the cells initially transfected with vAcPH-p76GFP or vAcac76-KO-PH-Ac76GFP (data not shown), indicating that both recombinant viruses were able to produce infectious budded virions in Sf9 cells. Sf9 cells infected with vAcPH-p76GFP or vAcac76-KO-PH-Ac76GFP were examined for GFP-specific fluorescence with a confocal laser scanning microscope. Fluorescence was found to be distributed along the outer periphery of the nucleus at 12 h p.i. and subsequently entered the nucleus and localized in the intranuclear ring zone between 24 and 72 h p.i. in the vAcac76-KO-PH-Ac76GFP-infected cells (Fig. 7B). However, in the vAcPH-p76GFP-infected cells, fluorescence was observed throughout the whole cell at all time points selected between 12 h and 72 h p.i. (Fig. 7B).
DISCUSSION
AcMNPV ac76 is a highly conserved gene, homologs of which have been found in all sequenced lepidopteran baculoviruses, but the function of the gene is unknown. In this study, the transcription and function of ac76 were investigated.
Transcriptional analysis of ac76 in AcMNPV-infected cells revealed the presence of at least three overlapping RNA transcripts transcribed from the ac76 region. The three transcripts were first detected at 12 h p.i., and their transcription was initiated from a viral late gene promoter motif, TAAG. Analysis by 3′ RACE indicated that these transcripts possessed different transcription termination sites. Clusters of compactly arranged genes with the same transcriptional orientation are commonly found in baculovirus genomes. Each gene in such cluster is transcribed from its respective promoter, resulting in a series of overlapping coterminal transcripts. In most cases, the overlapping transcripts generally share either a common 5′ end (7, 31) or a coterminal 3′ end (1, 12, 13, 21, 27, 28). Transcription of the distal upstream gene may interfere with the transcriptional initiation of the downstream gene via a regulatory mechanism called promoter occlusion (1, 11, 12, 31).
We investigated the role of AcMNPV ac76 in viral replication using an ac76-knockout bacmid and found that ac76 plays an essential role in the viral life cycle. Infection can be initiated without ac76, as evidenced by the presence of OBs in the ac76-knockout bacmid-transfected Sf9 cells, but the infection was restricted to the initially transfected cells, and the virus was unable to spread. The viral growth curve and Western blot assays confirmed that the ac76-deletion virus was unable to produce BVs.
As mentioned above, three overlapping multicistronic transcripts were transcribed from ac76. All of the transcripts have the same 5′ end but different 3′ ends. So the defect in BV production induced by deletion of ac76 from the AcMNPV bacmid might have resulted from an interruption in the expression of the gene downstream of ac76. In our study, the deletion phenotype could be rescued by reinsertion of ac76 into the polh locus of the ac76-knockout virus confirmed that the observed phenotype was directly due to the deletion of ac76. This indicates that the deletion of the 64-bp ac76 sequence in the ac76-knockout virus might not interrupt downstream gene expression of the overlapping multicistronic transcripts, or the deletion could disrupt the expression of flanking genes but has no effect on the phenotype.
Electron microscopy showed that abundant nucleocapsids with a normal appearance were both present in the intrastromal space of the virogenic stroma of vAcac76-KO-PH-GFP-transfected cells and bundled in the ring zone, indicating that the deletion of ac76 had no effect on nucleocapsid assembly or on trafficking of the nucleocapsids out of the virogenic stroma. The normal, abundant nucleocapsids suggest that new viral DNAs have been embedded into capsids. This is consistent with the qPCR analysis which showed that ac76 is not involved in viral DNA synthesis. In vAcac76-REP-PH-GFP- and vAcPH-GFP-transfected cells, the nucleocapsids approached the intranuclear microvesicles and acquired envelopes to form ODVs. However, membrane vesicles were not observed at the ring zone of vAcac76-KO-PH-GFP-transfected cells, and nucleocapsids were never enveloped to form ODVs. Intranuclear microvesicle formation is induced by baculovirus infection, and these microvesicles ultimately become the envelope of the ODVs (8). There are two hypotheses about the origin of the nuclear microvesicles; some investigators believe that de novo synthesis occurs, while others believe that ODV envelopes are derived from the inner nuclear membrane (15, 42). Our results suggest that Ac76 plays a role in intranuclear microvesicle formation. Similar to viruses with knockouts of the 38K (43), ac142 (24), p48 (44), and ac53 (20) genes, no ODVs were present in the ac76-knockout virus occlusion bodies; interestingly, these genes have also been shown to affect the production of BV. Many ODV envelope proteins, including ODV-E56 (6), ODV-E66 (15), ODV-E25 (34), BV/ODV-E26 (3), and ODV-E18/ODV-E35 (7), are enriched in microvesicles, suggesting that virus-induced intranuclear microvesicles play an important role, either as a direct precursor or as an assembly focus for the ODV envelope (8). However, previous studies have indicated that disruption of ODV-E56 and ODV-E66 does not affect the formation of intranuclear microvesicles (6, 16), indicating that these proteins execute their functions after intranuclear microvesicle formation. It is possible that Ac76 interacts with other proteins and induces the formation of intranuclear microvesicles and that, after formation, the intranuclear microvesicles could be modified with specific ODV envelope proteins and could ultimately become the envelope of the ODV.
Using a recombinant virus expressing an Ac76-GFP chimera as a visual marker, we found that Ac76 predominantly localized to the ring zone during the late phase of infection. The intranuclear ring zone is the site of several morphogenic processes that are closely associated with ODV development (42). Some ODV-associated proteins, e.g., P74 (37), BV/ODV-E26 (3), and P91 (17, 33), also have similar localization patterns in virus-infected cells. In accordance with this localization pattern, Ac76 contains a highly hydrophobic transmembrane domain and a putative nuclear localization signal. The primary ODV envelope proteins have prominent hydrophobic transmembrane motifs that are involved in membrane insertion, anchoring, and localization in the ODV envelope (8, 36). The N-terminal hydrophobic transmembrane regions of ODV-E66 and ODV-E25 are sufficient to traffic fusion proteins to intranuclear membranes and microvesicles (16). In a study in which the hydrophobic C-terminal transmembrane region of p74 was fused to GFP, the resulting chimera localized to the intranuclear ring zone in a similar pattern to that of full-length p74-GFP (37). The Ac76 transmembrane region is similar to the INM-SM, which contains two features: a hydrophobic sequence of 18 to 20 amino acids that constitutes a transmembrane domain and a positively charged residue within 4 to 8 amino acids of the end of the hydrophobic sequence that is exposed to the cytoplasm/nucleoplasm (5, 8, 9). In this study, the hydrophobic transmembrane region of Ac76 was disrupted by the replacement of ac76 by the Cm gene, which resulted in the blockage of intranuclear microvesicle formation. Ac76 may therefore be an integral membrane protein that is associated with intranuclear microvesicle formation. Further studies need to be performed in order to determine whether Ac76 is a virion structural protein. Additionally, it is important to examine whether Ac76 localizes to the nuclear envelope and intranuclear microvesicles.
In conclusion, this study suggests that ac76 is essential for BV and ODV production and intranuclear microvesicle formation. Although the exact role of ac76 is unclear, we are hopeful that the results of our study will lead to a better understanding of the molecular mechanism of microvesicle formation by providing a foundation for further research on this topic.
Acknowledgments
This research was supported by the National Basic Research Program of China (973 Program; grant 2009CB118903), the National Nature Science Foundation of China (grant 30530540), the Hi-Tech Research and Development Program of China (863 Program; grant 2006AA10A210), and the Science Foundation of the State Key Laboratory of Biocontrol (SKLBC09A01).
Footnotes
Published ahead of print on 19 May 2010.
REFERENCES
- 1.Acharya, A., and K. Gopinathan. 2002. Transcriptional analysis and preliminary characterization of ORF Bm42 from Bombyx mori nucleopolyhedrovirus. Virology 299:213-224. [DOI] [PubMed] [Google Scholar]
- 2.Ayres, M. D., S. C. Howard, J. Kuzio, M. Lopez-Ferber, and R. D. Possee. 1994. The complete DNA sequence of Autographa californica nuclear polyhedrosis virus. Virology 202:586-605. [DOI] [PubMed] [Google Scholar]
- 3.Beniya, H., S. C. Braunagel, and M. D. Summers. 1998. Autographa californica nuclear polyhedrosis virus: subcellular localization and protein trafficking of BV/ODV-E26 to intranuclear membranes and viral envelopes. Virology 240:64-75. [DOI] [PubMed] [Google Scholar]
- 4.Bideshi, D. K., and B. A. Federici. 2000. The Trichoplusia ni granulovirus helicase is unable to support replication of Autographa californica multicapsid nucleopolyhedrovirus in cells and larvae of T. ni. J. Gen. Virol. 81:1593-1599. [DOI] [PubMed] [Google Scholar]
- 5.Braunagel, S. C., V. Cox, and M. D. Summers. 2009. Baculovirus data suggest a common but multifaceted pathway for sorting proteins to the inner nuclear membrane. J. Virol. 83:1280-1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Braunagel, S. C., D. M. Elton, H. Ma, and M. D. Summers. 1996. Identification and analysis of an Autographa californica nuclear polyhedrosis virus structural protein of the occlusion-derived virus envelope: ODV-E56. Virology 217:97-110. [DOI] [PubMed] [Google Scholar]
- 7.Braunagel, S. C., H. He, P. Ramamurthy, and M. D. Summers. 1996. Transcription, translation, and cellular localization of three Autographa californica nuclear polyhedrosis virus structural proteins: ODV-E18, ODV-E35, and ODV-EC27. Virology 222:100-114. [DOI] [PubMed] [Google Scholar]
- 8.Braunagel, S. C., and M. D. Summers. 2007. Molecular biology of the baculovirus occlusion-derived virus envelope. Curr. Drug Targets 8:1084-1095. [DOI] [PubMed] [Google Scholar]
- 9.Braunagel, S. C., S. T. Williamson, S. Saksena, Z. Zhong, W. K. Russell, D. H. Russell, and M. D. Summers. 2004. Trafficking of ODV-E66 is mediated via a sorting motif and other viral proteins: facilitated trafficking to the inner nuclear membrane. Proc. Natl. Acad. Sci. U. S. A. 101:8372-8377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Friesen, P. D. 1997. Regulation of baculovirus early gene expression., p. 141-166. In L. K. Miller (ed.), The baculovirus. Plenum Press, New York, NY.
- 11.Friesen, P. D., and L. K. Miller. 1985. Temporal regulation of baculovirus RNA: overlapping early and late transcripts. J. Virol. 54:392-400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gross, C. H., and G. F. Rohrmann. 1993. Analysis of the role of 5′ promoter elements and 3′ flanking sequences on the expression of a baculovirus polyhedron envelope protein gene. Virology 192:273-281. [DOI] [PubMed] [Google Scholar]
- 13.Gutierrez, S., I. Kikhno, and F. M. Lopez. 2004. Transcription and promoter analysis of pif, an essential but low-expressed baculovirus gene. J. Gen. Virol. 85:331-341. [DOI] [PubMed] [Google Scholar]
- 14.Herniou, E. A., J. A. Olszewski, J. S. Cory, and D. R. O'Reilly. 2003. The genome sequence and evolution of baculoviruses. Annu. Rev. Entomol. 48:211-234. [DOI] [PubMed] [Google Scholar]
- 15.Hong, T., S. C. Braunagel, and M. D. Summers. 1994. Transcription, translation, and cellular localization of PDV-E66: a structural protein of the PDV envelope of Autographa californica nuclear polyhedrosis virus. Virology 204:210-222. [DOI] [PubMed] [Google Scholar]
- 16.Hong, T., M. D. Summers, and S. C. Braunagel. 1997. N-terminal sequences from Autographa californica nuclear polyhedrosis virus envelope proteins ODV-E66 and ODV-E25 are sufficient to direct reporter proteins to the nuclear envelope, intranuclear microvesicles and the envelope of occlusion derived virus. Proc. Natl. Acad. Sci. U. S. A. 94:4050-4055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kawasaki, Y., S. Matsumoto, and T. Nagamine. 2004. Analysis of baculovirus IE1 in living cells: dynamics and spatial relationships to viral structural proteins. J. Gen. Virol. 85:3575-3583. [DOI] [PubMed] [Google Scholar]
- 18.Li, L. L., Z. F. Li, W. C. Chen, and Y. Pang. 2007. Cloning, expression of Autographa californica nucleopolyhedrovirus vp39 gene in Escherichia coli and preparation of its antibody. Biotechnology 17:5-7. [Google Scholar]
- 19.Li, Y., J. Wang, R. Deng, Q. Zhang, K. Yang, and X. Wang. 2005. vlf-1 deletion brought AcMNPV to defect in nucleocapsid formation. Virus Genes 31:275-284. [DOI] [PubMed] [Google Scholar]
- 20.Liu, C., Z. Li, W. Wu, L. Li, M. Yuan, L. Pan, K. Yang, and Y. Pang. 2008. Autographa californica multiple nucleopolyhedrovirus ac53 plays a role in nucleocapsid assembly. Virology 382:59-68. [DOI] [PubMed] [Google Scholar]
- 21.Lu, A., and E. B. Carstens. 1992. Transcription analysis of the EcoRI D region of the baculovirus Autographa californica nuclear polyhedrosis virus identifies an early 4-kilobase RNA encoding the essential p143 gene. J. Virol. 66:655-663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lu, A., and L. K. Miller. 1997. Regulation of baculovirus late and very late gene expression, p. 193-211. In L. K. Miller (ed.), The baculoviruses. Plenum Press, New York, NY.
- 23.Luckow, V. A., S. C. Lee, G. F. Barry, and P. O. Olins. 1993. Efficient generation of infectious recombinant baculoviruses by site-specific transposon-mediated insertion of foreign genes into a baculovirus genome propagated in Escherichia coli. J. Virol. 67:4566-4579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McCarthy, C. B., X. Dai, C. Donly, and D. A. Theilmann. 2008. Autographa californica multiple nucleopolyhedrovirus ac142, a core gene that is essential for BV production and ODV envelopment. Virology 372:325-339. [DOI] [PubMed] [Google Scholar]
- 25.Monsma, S. A., A. G. Oomens, and G. W. Blissard. 1996. The GP64 envelope fusion protein is an essential baculovirus protein required for cell-to-cell transmission of infection. J. Virol. 70:4607-4616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Muyrers, J. P., Y. Zhang, G. Testa, and A. F. Stewart. 1999. Rapid modification of bacterial artificial chromosomes by ET-recombination. Nucleic Acids Res. 27:1555-1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Oellig, C., B. Happ, T. Muller, and W. Doerfler. 1987. Overlapping sets of viral RNAs reflect the array of polypeptides in the EcoRI J and N fragments (map positions 81.2 to 85.0) of the Autographa californica nuclear polyhedrosis virus genome. J. Virol. 61:3048-3057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Olszewski, J., and L. K. Miller. 1997. Identification and characterization of a baculovirus structural protein, VP1054, required for nucleocapsid formation. J. Virol. 71:5040-5050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.O'Reilly, D. R., L. K. Miller, and V. A. Luckow. 1992. Baculovirus expression vector: a laboratory manual. W. H. Freeman, New York, NY.
- 30.Pijlman, G. P., J. C. Dortmans, A. M. Vermeesch, K. Yang, D. E. Martens, R. W. Goldbach, and J. M. Vlak. 2002. Pivotal role of the non-hr origin of DNA replication in the genesis of defective interfering baculoviruses. J. Virol. 76:5605-5611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rankin, C., B. F. Ladin, and R. F. Weaver. 1986. Physical mapping of temporally regulated, overlapping transcripts in the region of the 10K protein gene in Autographa californica nuclear polyhedrosis virus. J. Virol. 57:18-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rohrmann, G. F. 2008. Baculovirus molecular biology. National Library of Medicine, National Center for Biotechnology Information, Bethesda, MD. http://www.ncbi.nlm.nih.gov/bookshelf/br.fcgi?book=bacvir.
- 33.Russell, R. L., and G. F. Rohrmann. 1997. Characterization of P91, a protein associated with virions of an Orgyia pseudotsugata baculovirus. Virology 233:210-223. [DOI] [PubMed] [Google Scholar]
- 34.Russell, R. L., and G. F. Rohrmann. 1993. A 25-kDa protein is associated with the envelopes of occluded baculovirus virions. Virology 195:532-540. [DOI] [PubMed] [Google Scholar]
- 35.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 36.Slack, J., and B. M. Arif. 2007. The baculoviruses occlusion-derived virus: virion structure and function. Adv. Virus Res. 69:99-165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Slack, J. M., E. M. Dougherty, and S. D. Lawrence. 2001. A study of the Autographa californica multiple nucleopolyhedrovirus ODV envelope protein p74 using a GFP tag. J. Gen. Virol. 82:2279-2287. [DOI] [PubMed] [Google Scholar]
- 38.Theilmann, D. A., G. W. Blissard, B. Bonning, J. A. Jehle, D. R. O'Reilly, G. F. Rohrmann, S. Thiem, and J. M. Vlak. 2005. Baculoviridae, p. 1129-1185. In C. M. Fauquet, M. A. Mayo, J. Maniloff, U. Desselberger, and L. A. Ball (ed.), Virus taxonomy: classification and nomenclature of viruses. Eighth report of the International Committee on Taxonomy of Viruses. Elsevier/Academic Press, London, United Kingdom.
- 39.Vanarsdall, A. L., K. Okano, and G. F. Rohrmann. 2005. Characterization of the replication of a baculovirus mutant lacking the DNA polymerase gene. Virology 331:175-180. [DOI] [PubMed] [Google Scholar]
- 40.van Oers, M. M., and J. M. Vlak. 2007. Baculovirus genomics. Curr. Drug Targets 8:1051-1068. [DOI] [PubMed] [Google Scholar]
- 41.Vaughn, J. L., R. H. Goodwin, G. J. Tompkins, and P. McCawley. 1977. The establishment of two cell lines from the insect Spodoptera frugiperda (Lepidoptera; Noctuidae). In Vitro 13:213-217. [DOI] [PubMed] [Google Scholar]
- 42.Williams, G. V., and P. Faulkner. 1997. Cytological changes and viral morphogenesis during baculovirus infection, p. 61-107. In L. K. Miller (ed.), The baculoviruses. Plenum Press, New York, NY.
- 43.Wu, W., T. Lin, L. Pan, M. Yu, Z. Li, Y. Pang, and K. Yang. 2006. Autographa californica multiple nucleopolyhedrovirus nucleocapsid assembly is interrupted upon deletion of the 38K gene. J. Virol. 80:11475-11485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yuan, M., W. Wu, C. Liu, Y. Wang, Z. Hu, K. Yang, and Y. Pang. 2008. A highly conserved baculovirus gene p48 (ac103) is essential for BV production and ODV envelopment. Virology 379:87-96. [DOI] [PubMed] [Google Scholar]