Abstract
Phospholipase A2 (PLA2) has long been proposed to be involved in rickettsial entry into host cells, escape from the phagosome to evade destruction by lysosomal exposure, and lysis of the host cells. However, the corresponding rickettsial gene(s) encoding a protein with PLA2 activity has not been identified or functionally characterized. Here, we report that the Rickettsia typhi genome possesses two genes encoding patatin-like PLA2 proteins, RT0590 and RT0522. Sequence analysis of RT0522 and RT0590 reveals the presence of the conserved motifs essential for PLA2 activity. Transcriptional analysis indicates that RT0522, but not RT0590, is transcribed at all stages of intracellular growth of R. typhi in Vero cells. The differential gene expression pattern of RT0522 at various stages of growth suggests its potential role during R. typhi infection of host cells. In silico, RT0522 is predicted to be noncytoplasmic and its gene does not encode a recognizable signal peptide sequence. However, our data indicate that RT0522 is secreted into the host cytoplasm. In addition, we observe that RT0522 protein expression is cytotoxic to both yeast and Vero cells. Importantly, we demonstrate that recombinant RT0522 possesses phospholipase A activity that requires a eukaryotic host cofactor for activation. Both cytotoxicity and phospholipase A activity associated with RT0522 were reduced by PLA2 inhibitors. Site-directed mutagenesis of predicted catalytic Ser/Asp residues of RT0522 also eliminates cytotoxicity and phospholipase A activity. To our knowledge, RT0522 is the first protein identified from Rickettsia typhi with functional phospholipase A activity.
The genus Rickettsia includes the causative agents of some of the most severe bacterial diseases of humans, such as epidemic typhus and Rocky Mountain spotted fever. Rickettsiae are small, Gram-negative obligate intracellular alphaproteobacteria. In nature, rickettsiae are maintained in a complex life cycle involving transmission between arthropod and vertebrate hosts. Humans can become infected when they come into contact with infected arthropod vectors such as fleas, ticks, or lice or their excrement (2, 11).
In both mammals and arthropods, rickettsial invasion of host cells, including nonphagocytic cells, is rapidly followed by escape from the phagocytic vacuole. The rickettsiae thus evade exposure to lysosomal contents and are free to grow and replicate within the nutrient-rich host cell cytoplasm (11, 14). Rickettsial phospholipase A2 (PLA2) has been hypothesized to mediate entry into host cells and escape from the phagosome and to inflict injury on host cells; however, the corresponding gene encoding rickettsial PLA2 enzyme and its biological activity remain unknown (21, 40, 46, 48, 49).
Phospholipases comprise a diverse collection of secreted and membrane-bound enzymes that are classified into four major groups (A to D) based on their site of cleavage within the phospholipids. PLA2 enzymes hydrolyze the sn-2 ester bond of phospholipids, releasing free fatty acids and lysophospholipids (15, 43). Several bacterial pathogens have been shown to utilize phospholipases for invasion of host cells, virulence, and initiation of a proinflammatory response (15, 35, 43). For example, Listeria monocytogenes secretes two phospholipase proteins that are involved in phagosomal escape (14). Importantly, PLA2 enzymes from Pseudomonas aeruginosa (ExoU) and group A Streptococcus (SlaA) have been shown to play a clear role in the pathogenicity of these organisms (36, 42, 43). The intracellular lung pathogen Legionella pneumophila secretes VipD, a patatin-like protein (PLP) with sequence similarity to ExoU, into host cells via the type IVB secretion system, interferes with the late secretory pathway when expressed in Saccharomyces cerevisiae, and is suggested to be involved in the intracellular infection process (38, 45).
The PLA2 enzymes are themselves classified into three categories: secreted (sPLA2), calcium dependent cytosolic (cPLA2), and calcium independent (iPLA2). This classification is based on the catalytic mechanism (Ser/Asp or Ser/His/Asp) as well as functional and structural features (35, 37, 43). P. aeruginosa ExoU contains conserved iPLA2 and cPLA2 Ser/Asp catalytic residues; this enzyme also shares homology with the plant storage phospholipase patatin (35, 43).
Several rickettsial genomes possess genes with potential membranolytic activities: the genes for patatin-like protein (pat1), hemolysin A (tlyA), hemolysin C (tlyC), and phospholipase D (pld) (47). Empirical evidence suggests that some of these genes play a role during rickettsial intracellular growth and virulence. For example, hemolytic activity was conferred on a nonhemolytic Proteus mirabilis strain upon transformation of tlyC from Rickettsia typhi (28). Introduction of Rickettsia prowazekii tlyC or pld into Salmonella enterica serovar Typhimurium conferred an ability on this bacterium to escape from vacuoles, indicating a role of tlyC and/or pld in the process of phagosomal escape (47). Recently, Driskell et al. reported the generation of a Δpld mutant of R. prowazekii (8). This mutant strain showed no noticeable impairment of growth and phagosomal escape in tissue culture but did exhibit attenuated virulence in a guinea pig infection model, suggesting that additional factors are likely to be involved in rickettsial intracellular growth and virulence (8).
Our interest is in elucidating the mechanism of rickettsial infection, intracellular growth, and virulence. As mentioned above, phospholipase A2 has long been proposed to be involved in rickettsial entry into host cells, escape from the phagosome, and damage to host cells; however, this enzymatic activity has not been demonstrated within the known rickettsial proteome. Therefore, rather than relying on genome annotation to identify genes predicted to encode phospholipase enzymes, we searched all rickettsial genome sequences available in GenBank for genes encoding proteins with phospholipase A2 motifs. Our search resulted in the identification of two open reading frames with the putative patatin-like phospholipase A2 motifs from each of the following rickettsial species: R. typhi, R. prowazekii, R. massiliae MTU5, R. bellii OSU 85-389, and R. bellii RML369-C. Here, we report the identification and sequence analysis of these rickettsial proteins with putative phospholipase A2 motifs and demonstrate the functional characterization of the corresponding R. typhi proteins: RT0522, annotated as a hypothetical protein, and RT0590, annotated as a putative patatin-like protein (19).
MATERIALS AND METHODS
Bacterial strain, host cells, and growth condition.
R. typhi strain Wilmington (ATCC VR-144) was propagated in Vero76 (ATCC CRL-1587) cells. Host Vero76 cells were grown in DMEM (Dulbecco's modified Eagle's medium with 4.5 g glucose/liter with glutamine; Biofluids Inc., Rockville, MD) supplemented with 5% fetal bovine serum (Gemini, Calabasas, CA) at 37°C and 5% CO2 in an air atmosphere.
Genomic DNA extraction.
Propagation and partial purification of R. typhi from Vero76 cells were performed as previously described (31). Genomic DNA of R. typhi was extracted using the Wizard genomic DNA purification kit (Promega, Madison, WI).
Isolation of RNA from rickettsiae grown at various time points.
The titers of partially purified R. typhi from Vero76 cells were determined using the Live/Dead BacLight bacterial viability assay (Molecular Probes, Eugene, OR) as previously described (29). A monolayer of Vero76 cells (approximately 1 × 106 cells) was infected with R. typhi at a multiplicity of infection of approximately 100 rickettsiae. Rickettsia-infected Vero76 cells were incubated at 37°C and 5% CO2 for 0, 5, 15, and 30 min and 1, 4, 8, 24, 48, and 120 h. At each time point, culture medium was removed and rickettsia-infected cells were washed with fresh growth medium. Total RNA was isolated from rickettsia-infected Vero76 cells using Trizol reagent (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions. The RNA was DNase treated to remove contaminating DNA by using RNase-free DNase (Qiagen, Valencia, CA). DNase-treated total RNA was purified with the RNeasy MinElute cleanup kit (Qiagen). The purified total RNA was checked for the presence of DNA contamination by PCR using standard conditions.
Real-time qRT-PCR.
Rickettsial gene expression was monitored by one-step real-time quantitative reverse transcription-PCR (qRT-PCR). qRT-PCR was performed on 200 ng of total RNA isolated (as mentioned above) from R. typhi-infected Vero76 cells using the Brilliant II SYBR green qRT-PCR master mix kit (Stratagene, La Jolla, CA) following the manufacturer's instructions. The specific primers (primers used in this study are listed in Table 1) for R. typhi genes used in qRT-PCR were AZ3758 and AZ3759 for RT0590, AZ3760 and AZ3761 for RT0522, and AZ5632 and AZ5633 for RT0119 (rpsL). For host Vero76 cells, a primer pair specific for the glyceraldehyde-3-phosphate dehydrogenase (GAPDH) gene, AZ6808 and AZ6809, were used for qRT-PCR. Cycling conditions were as follows: 1 cycle at 50°C for 1 h; 1 cycle at 95°C for 10 min; 40 cycles at 95°C for 15 s, 56°C for 30 s, and 60°C for 30 s; and 1 cycle to generate the dissociation curve. The qRT-PCR amplification and detection were performed on an MX3005P thermal cycler (Stratagene). The specificities of these primer pairs used in qRT-PCR were verified by PCR on DNA isolated from R. typhi.
TABLE 1.
Primers used in this studyb
| Primer | Sequencea (5′ to 3′) |
|---|---|
| AZ3758 | GGT ATT TGA TTT TAC TGG AGG AAC |
| AZ3759 | CAT TAT CTT GAC TTT TTC TCG C |
| AZ3760 | GCA ACA CAA CGA GAG ACA AGT GC |
| AZ3761 | ACC AAA AGC CAT AGC CAG AGT TC |
| AZ5632 | CTC CTG CCT TAG AAT CCA ACC C |
| AZ5633 | TGA ACG ACC TTG CTT ACG CC |
| AZ6808 | GGT ACA GAA GGT TTG CTG GG |
| AZ6809 | CAC AGT CAA GGC TGA GAA TG |
| AZ5359 | ggt acc ATG AAG CAA GAG TTC AAA ACT TCT |
| AZ5360 | ctc gag CCA CCT ATT CCT GTG ATT CCT AAC |
| AZ7240 | GCT GTA GCT GGG AGC GCT GTG GGA GCA ATC |
| AZ7241 | GGA AAA AAA TAT GTT GCT GGA GGG TAT CGC |
| AZ5801 | GGT ACC ATG AAG CAG GAG TTC AAA ACA |
| AZ5802 | CTC GAG TTA CCA CCT GTT GCG GTG GTT TCT |
| AZ5803 | CTC GAG CCA CCT GTT GCG GTG GTT TCT |
| AZ5971 | AAG CTT ATG CAT ATC CAA TCG TTG GGG |
| AZ5972 | GGA TCC TCA TGT GAA CTC CTT ATT CCG |
| AZ5973 | GGA TCC TGT GAA CTC CTT ATT CCG CCA |
The nucleotides incorporated to generate restriction sites are indicated in lowercase.
Primers were designed based on the genome sequences available in the GenBank database (www.ncbi.nlm.nih.gov) with accession number NC_006142.
The gene expression data generated by real-time qRT-PCR were analyzed by using the Q-Gene software tool with amplification efficiency correction (20). The qRT-PCR data were exported for estimation of the amplification efficiency for each primer pair using the LinReg PCR software tool (32). The amplification efficiencies and cycle threshold (CT) values from the experiments were imported into Q-Gene for calculation of normalized expression (20). To calculate the normalized expression of the target gene, efficiency-corrected CT values for target genes were divided by that for rickettsial housekeeping gene RT0119 (rpsL). To calculate the rickettsial burden, the efficiency-corrected CT values for rickettsial housekeeping gene RT0119 (rpsL) were divided by that of a host housekeeping gene (GAPDH).
Yeast cytotoxicity assay.
The R. typhi RT0522 gene was cloned by PCR into the KpnI and XhoI sites of yeast expression vector pYES2/CT with C-terminal epitope (V5/6×His) tags (Invitrogen-Life Technologies, Carlsbad, CA) according to the manufacturer's instructions, using primers AZ5359 and AZ5360. The constructed plasmid pYES-522 was confirmed by sequencing. The sequence of the RT0522 gene and the deduced amino acid sequence from R. typhi were analyzed using MacVector 7.1.1 software (Genetics Computer Group, Inc. Madison, WI). Sequence comparisons were performed using BLAST analysis (www.ncbi.nlm.nih.gov).
The coding sequence of the RT0522 gene was codon optimized (RT0522HS) to mammalian cells by Blue Heron Biotechnology, Bothell, WA. The RT0522HS sequence was cloned into the KpnI and XhoI sites of pYES2/CT vector with C-terminal V5 epitope and six-His tags. The constructed plasmid pYES-522HS was confirmed by sequencing.
Mutations at catalytic sites (Ser-86 and Asp-250) of RT0522 were introduced in plasmid pYES-522HS using the QuikChange Lightning multisite-directed mutagenesis kit (Stratagene) according to the manufacturer's instructions. The primer used for site-directed mutagenesis of Ser-86 to Ala is AZ7240, and that for mutagenesis of Asp-250 to Ala is AZ7241. The constructed plasmids pYES-S86A and pYES-S86A-D250A were confirmed by sequencing.
The constructed plasmids were transformed into Saccharomyces cerevisiae strain INVSc1 (Invitrogen) by using the Frozen-EZ yeast transformation kit (Zymo Research, Orange, CA) according to the manufacturer's protocol. Yeast transformants were grown in synthetic complete (SC) medium agar containing 2% glucose and lacking uracil (SC-U+Glu) to select plasmids at 30°C for 3 days. Yeast cells containing plasmid were grown in SC-U+Glu medium at 30°C overnight. The overnight culture was pelleted, washed with an equal volume of SC-U without a carbon source, and resuspended in SC-U without a carbon source. The resuspended yeast cells containing plasmid were induced in medium agar containing 2% galactose and lacking uracil (SC-U+Gal) or repressed on SC-U+Glu agar and incubated at 30°C for 3 days (25). For the CFU assay, the resuspended yeast transformants were serially diluted in SC-U without a carbon source and plated on inducing (SC-U+Gal) and repressing (SC-U+Glu) agar. After incubation at 30°C for 3 days, the colonies were counted to determine the percentage of CFU on inducing agar with respect to that on repressing agar. For the PLA2 inhibitor assay, the resuspended yeast transformants were plated on inducing (SC-U+Gal) agar with the inhibitors. The PLA2 inhibitors used, methyl arachidonyl fluorophosphonate (MAPF) and bromoenol lactone (BEL), were obtained from Sigma-Aldrich.
Production of recombinant proteins for PLA2 assay.
The coding sequence of RT0522 (codon optimized to mammalian cells) was subcloned by PCR from the pYES-522HS plasmid into the pTrcHis2TOPO-TA vector (Invitrogen) for expression in Escherichia coli using primer pair AZ5801 and AZ5803 to generate the plasmid pTrc-522HS. The mutation at catalytic sites (Ser-86 and Asp-250) of RT0522 was introduced into plasmid pTrc-522HS by the procedure mentioned above to generate the plasmids pTrc-S86A and pTrc-S86A-D250A. For a positive control, the coding sequence of ExoU was subcloned by PCR from pYES-ExoUGFP (pYES-ExoU) (36) into the pTrcHis2TOPO-TA vector using primer pair AZ5971 and AZ5973 to generate the plasmid pTrc-ExoU. The constructed plasmids pTrc-522HS, pTrc-S86A, pTrc-S86A-D250A, and pTrc-ExoU were confirmed by sequencing.
Escherichia coli TOP10 chemically competent cells (Invitrogen) were transformed with pTrc-522HS, pTrc-S86A, pTrc-S86A-D250A, or pTrc-ExoU according to the manufacturer's instructions. The transformed TOP10 cells were grown overnight at 37°C with shaking in LB broth supplemented with 100 μg ml−1 ampicillin. Two milliliters of the overnight culture was used to inoculate 200 ml of LB broth supplemented with 100 μg ml−1 ampicillin. The culture was incubated at 30°C with shaking until the optical density at 600 nm (OD600) reached 0.5 to 0.7. Protein expression was induced by the addition of IPTG (isopropyl-β-d-thiogalactopyranoside; 1 mM final concentration), and the mixture was incubated for 4 h at 30°C with shaking. The culture was pelleted at 10,000 × g for 10 min at 4°C. The recombinant proteins with a C-terminal six-His tag were isolated and purified with the nickel-nitrilotriacetic acid (Ni-NTA) spin kit (Qiagen) according to the manufacturer's protocol. The elution buffer used for protein purification was exchanged with PLA2 assay buffer (50 mM Tris-HCl, 100 mM NaCl, 1 mM CaCl2, pH 8.9) by using an Amicon Ultra-4 30,000-molecular-weight-cutoff (MWCO) centrifugal filter unit (Millipore, Billerica, MA). The concentration of purified proteins was determined with the bicinchoninic acid (BCA) protein assay kit (Pierce, Rockford, IL), and purity was assessed by using an Imperial protein-stained gel (see Fig. S1 in the supplemental material).
Phospholipase activity of recombinant proteins.
Vero76 cells were incubated in culture medium at 37°C and 5% CO2 for 72 h. The culture was washed twice with 5 ml of ice-cold PLA2 assay buffer. The cells were harvested by using a scraper in 0.5 ml of ice-cold PLA2 assay buffer and lysed by sonication at setting 6.5 for 1 min using a sonic dismembranator (Fisher Scientific). The disrupted host cells were cleared from debris by centrifugation at 1,000 × g for 15 min at 4°C. The protein concentration of Vero76 cell lysate was determined by using the BCA protein assay kit.
Phospholipase activity of each recombinant protein was measured using the following fluorogenic phospholipid substrate (specific to PLA2 enzyme activity) according to the manufacturer's protocol (Invitrogen): 1-O-(6-BODIPY [dipyrromethene boron difluoride] 558/568-aminohexyl)-2-BODIPY FL C5-sn-glycero-3-phosphocholine (red/green BODIPY PC-A2). Briefly, the cleavage of the sn-2 bond of red/green BODIPY PC-A2 by the PLA2 enzyme results in an increase in fluorescence emission. The red/green BODIPY PC-A2 substrate was suspended to a final concentration of 1 mM in dimethyl sulfoxide. The substrate-liposome mix was prepared by mixing 25 μl of 1 mM red/green BODIPY PC-A2 substrate, 25 μl of 10 mM 1,2-dioleoyl-sn-glycero-3-phosphocholine (DOPC), and 25 μl of 1,2-dioleoyl-sn-glycero-3-phospho-(1′-rac-glycerol) (sodium salt) (DOPG) in 5 ml of PLA2 assay buffer. Fifty microliters of substrate-liposome mix was added to 50 μl of phospholipase A (PLA) samples containing (approximately) 10 μg of recombinant proteins and 25 μg of Vero76 cell lysate. The reaction mixture was incubated at room temperature for 4 h. For PLA2 inhibitor assay, MAPF was added to the reaction mixture at a final concentration of 0.1 μM or 0.5 μM. The fluorescence emission was measured by using a FLUOstar Omega plate reader (BMG Labtech, Germany) using an excitation at 485 nm and emission at 520 nm. The fluorescence intensity/μg of recombinant protein was calculated using the following equation:
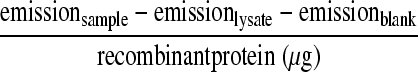 |
(1) |
Cytotoxicity assay with Vero76 cells.
The coding sequence of RT0522 (codon optimized) was subcloned by PCR from pYES-522HS plasmid into pcDNA6.2/N-EmGFP-GW/TOPO vector (Invitrogen) for mammalian expression using primer pair AZ5801 and AZ5802, resulting in the plasmid pcDNA-522HS. As a positive control, the coding sequence of ExoU was subcloned by PCR from pYES-ExoU (36) into the same vector by using primer pair AZ5971 and AZ5972 to generate the plasmid pcDNA-ExoU. The constructed plasmids pcDNA-522HS and pcDNA-ExoU were confirmed by sequencing.
One day prior to transfection, 1 × 105 Vero76 cells were seeded onto a 12-well tissue culture plate using DMEM supplemented with 5% fetal bovine serum and incubated at 37°C and 5% CO2 in an air atmosphere. Vero76 cells were transfected with pcDNA-522HS, pcDNA-ExoU, or pcDNA6.2/N-EmGFP/GW/CAT (control) plasmid DNA using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. Transfected Vero76 cells were grown for 24 h in DMEM supplemented with 5% fetal bovine serum at 37°C and 5% CO2 in an air atmosphere.
To determine the transfection efficiency, the transfected Vero76 cells were fixed in freshly prepared 3.5% paraformaldehyde (in 1× phosphate-buffered saline [PBS]) for 20 min at room temperature followed by a wash in 1× PBS at room temperature. The transfected Vero76 cells were then mounted under VectaShield with DAPI (4′,6-diamidino-2-phenylindole; Vector Laboratories, Burlingame, CA). Expression of EmGFP (N-terminally tagged emerald green fluorescent protein of the plasmid) and DAPI (total cells) was visualized using a fluorescence microscope (Nikon Eclipse E600). Transfection efficiency was determined as the percentage of EmGFP-expressing cells (transfected cells) of total cells (DAPI) counted.
The cytotoxicity of transfected Vero76 cells was measured by a lactate dehydrogenase (LDH) release assay using the CytoTox 96 nonradioactive cytotoxicity kit (Promega, Madison, WI) according to the manufacturer's protocol. Briefly, transfected Vero76 cells grown for 24 h in DMEM with 5% fetal bovine serum in a 12-well plate were centrifuged at 250 × g for 4 min. The overlying medium (50 μl) was added to 50 μl of substrate (from the LDH assay kit) and incubated at room temperature for 30 min in the dark. The reaction was terminated by adding 50 μl of stop solution (from the LDH assay kit). For 100% release of LDH, Triton X-100 (0.8%, vol/vol) was added to control wells for complete lysis of Vero76 cells. The absorbance at 490 nm (A490) was recorded within 1 h after the addition of stop solution. The percent cytotoxicity of transfected Vero76 cells was calculated using the following equation:
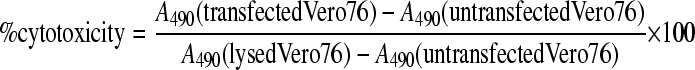 |
Antibodies.
Rabbit anti-RT0522 antibody was raised against the first (N-terminal) 174 amino acids of RT0522. Rabbit anti-EF-Ts antibody was raised against the first (N-terminal) 156 amino acids of EF-Ts (elongation factor Ts, encoded by RT0049). Both anti-RT0522 and anti-EF-Ts antibodies were generated and affinity purified by Primm Biotech, Inc., Cambridge, MA.
RT0522 translocation assay.
R. typhi-infected or uninfected Vero76 cells were incubated in culture medium at 37°C and 5% CO2 for 48 h. The cells were harvested by trypsinization, washed twice in cold 1× PBS, and resuspended in cold 1× PBS (supplemented with complete mini-EDTA-free protease inhibitor). The host cells were disrupted by mild sonication at setting 6.5 for 15 s by using a sonic dismembranator (Fisher Scientific) (7). The disrupted host cells were centrifuged at 16,000 × g for 10 min at 4°C to separate the supernatant containing rickettsial secreted proteins from the pellet containing intact rickettsiae. The supernatant was filtered through an 0.45-μm-pore-size filter (Millipore) and concentrated using an Amicon Ultra-4 (30,000-MWCO) centrifugal filter unit. Samples from the pellet and supernatant were separated on a 4 to 20% Tris-glycine precast gel (Invitrogen) with 1× Tris-glycine-SDS running buffer (Bio-Rad) and transferred to a polyvinylidene difluoride (PVDF) membrane using the iBlot transfer system (Invitrogen). The membrane was probed with rabbit anti-RT0522 antibody or with rabbit anti-EF-Ts antibody (as a control) and developed using the SuperSignal West Pico chemiluminescent substrate kit (Pierce). As a host cytoplasmic protein control, membranes were probed with mouse monoclonal antibody against glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (Abcam Inc., Cambridge, MA).
RESULTS
Identification of putative phospholipase A2 homologs in Rickettsia.
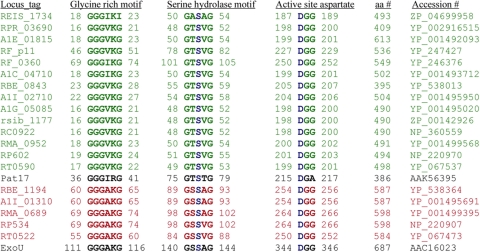
Annotations of 13 Rickettsia genome sequences available in GenBank (http://www.ncbi.nlm.nih.gov) indicate the presence of putative patatin-like proteins (PLPs). We aligned the rickettsial patatin-like proteins with potato patatin isozyme Pat17 (5, 33, 35). The alignment shows that rickettsial patatin-like proteins share 15% to 19% identity with Pat17 and all of the conserved motifs, the glycine-rich, serine hydrolase, and active-site aspartate motifs, required for PLA2 activity (Fig. 1, colored green).
FIG. 1.
Alignment of putative PLA2 motifs of various rickettsial proteins with Pat17 (patatin) and ExoU. The rickettsial genome-annotated patatin-like proteins (colored green) are RT0590 (R. typhi), RP602 (R. prowazekii), RMA_0952 (R. massiliae MTU5), RC0922 (R. conorii), rsib_1177 (R. sibirica), A1G_05085 (R. rickettsii), A1I_02710 (R. bellii OSU 85-389), RBE_0843 (R. bellii RML369-C), A1C_04710 (R. akari), RF_0360 (Rf-pat1 of R. felis URRWXCal2), RF_p11 (Rf-pat2 of R. felis URRWXCal2), A1E_01815 (R. canadensis), RPR_03690 (R. peacockii), and REIS_1734 (Rickettsia endosymbiont of Ixodes scapularis). The Rickettsia spp. with ExoU homologs (colored red) are R. typhi (RT0522), R. prowazekii (RP534), R. massiliae MTU5 (RMA_0689), R. bellii OSU 85-389 (A1I_01310), and R. bellii RML369-C (RBE_1194). The catalytic dyad serine-aspartate is colored blue. Accession numbers and amino acid (aa) numbers indicate the GenBank accession number and total number of amino acids of each rickettsial protein carrying putative PLA2 motifs, respectively.
Although the RP602 protein (shown in Fig. 1) is annotated as a patatin-like PLA2 protein for R. prowazekii, there are some reports that another gene of R. prowazekii, RP534, encodes a patatin-like PLA2 protein that is homologous to P. aeruginosa ExoU, a potent cytotoxin and virulence factor with a patatin-like PLA2 protein (5, 24, 35). Banerji et al. also reported that several Rickettsia spp. contain two genes encoding patatin-like proteins based on the number of protein coding sequences present in the whole genome (4). Based on these reports, we searched the nonredundant protein sequence database of Rickettsia using the BLASTP program (http://www.ncbi.nlm.nih.gov/blast/Blast.cgi) for ExoU homologs. Our homology search supports the report by others that RP534 of R. prowazekii is homologous to ExoU. We also identified a homolog of ExoU in four other rickettsial species: RT0522 from R. typhi, RMA_0689 from R. massiliae MTU5, A1I_01310 from R. bellii OSU 85-389, and RBE_1194 from R. bellii RML369-C (Fig. 1, colored red). However, other rickettsial species (e.g., R. felis, R. conorii, and the Rickettsia endosymbiont of Ixodes scapularis) contained only a fragmented segment of this second ExoU homolog (data not shown). Alignment of the putative PLA2 motifs of Rickettsia ExoU homologs, shown in Fig. 1, revealed the presence of all conserved motifs required for iPLA2, cPLA2, and patatin lipase family members whose catalytic centers consist of a Ser-Asp dyad (24, 35, 36). The ExoU homologs of Rickettsia spp. show 17% to 19% identity with P. aeruginosa ExoU. The two patatin-like proteins (RT0590 and RT0522) identified in R. typhi showed 15% identity with each other. This bioinformatics analysis indicates that the two patatin-like proteins (RT0590 and RT0522) identified in R. typhi contain all the conserved motifs essential for PLA2 activity and could serve as functional phospholipase A enzymes. To address this hypothesis, we examined the expression and function of the R. typhi proteins that possess patatin-like protein motifs.
Transcriptional profiles of putative phospholipase A2 homologs RT0522 and RT0590 of R. typhi during different stages of growth in host cells.
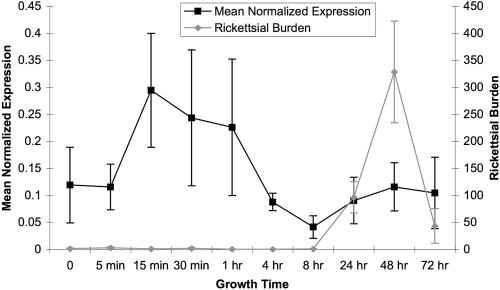
The expression pattern of RT0522 and RT0590 in Vero76 cells was determined by one-step real-time qRT-PCR. All the primer pairs used in qRT-PCR were tested on genomic DNA isolated from R. typhi-infected Vero76 cells, and the expected PCR products were observed (see Fig. S2A in the supplemental material). Gene expression data generated by real-time qRT-PCR showed no detectable cycle threshold (CT) values for the RT0590 gene at all time points tested. Agarose gel analysis of the qRT-PCR products showed no amplicon for the RT0590 gene; however, RT0522, rpsL (RT0119), and GAPDH genes produced the expected products (Fig. S2B). These data suggest that RT0590 is not transcribed under the experimental conditions followed in this assay. It is observed from the gene expression pattern shown in Fig. 2 that RT0522 is differentially expressed at various stages of growth of R. typhi in Vero76 cells. Although the level of transcription of RT0522 is highest at 15 min postinfection, it remains elevated up to 1 h postinfection. The level of expression of RT0522 is lowest at 8 h postinfection. After the doubling time (8 to 10 h) of rickettsiae (11, 13, 23), the data suggest an increase in the expression level of RT0522 (Fig. 2). However, the R. typhi burden remains low until 8 h postinfection and peaks at 48 h postinfection (Fig. 2).
FIG. 2.
Transcriptional profile and R. typhi burden during various stages of growth in Vero76 cells. The mean normalized expression (±standard errors) of RT0522 was calculated relative to the expression of the rickettsial reference (rpsL) gene. To account for rickettsial growth in host cells at different stages of the rickettsial life cycle, the mean rickettsial burden (±standard errors) was calculated by dividing the efficiency-corrected CT values for the RT0119 rickettsial housekeeping gene (rpsL) by that of the host housekeeping gene (GAPDH). The expression levels of RT0522 at all time points were found to be significantly different (P < 0.05 [two-tailed t test]) from that of the 8-h-postinfection time point.
Cytotoxicity assay by yeast viability loss.
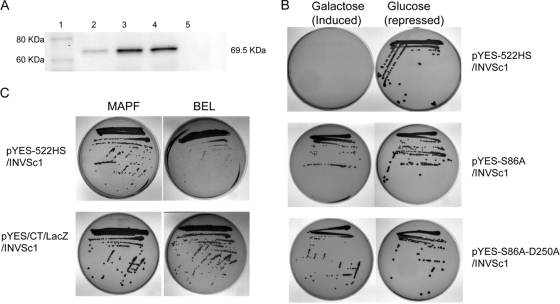
Heterologous model systems have been successfully utilized for many genetically intractable organisms to study gene function and to elucidate virulence factors (26, 27, 29-31, 36, 41, 44). S. cerevisiae is a genetically tractable system and has been utilized to demonstrate the cytotoxic effect of P. aeruginosa ExoU (25, 26, 35, 36, 39, 44). We used the S. cerevisiae model system to determine the potential cytotoxic effect of RT0522. Yeast transformants carrying pYES2/CT/LacZ (control plasmid) or pYES-522 (carrying RT0522) grew on both inducing (SC-U+Gal) and repressing (SC-U+Glu) agar (see Fig. S3 in the supplemental material). However, yeast cells transformed with pYES-ExoU (carrying P. aeruginosa ExoU; positive control) grew on repressing agar and showed no growth on inducing agar, indicating the cytotoxic effect of ExoU on yeast cells (Fig. S3) (36). We checked the expression of RT0522 from pYES-522 transformed into INVSc1 cells under inducing conditions and found that there was no detectable expression of RT0522 in yeast cells (data not shown). This result prompted us to consider codon optimization of RT0522 (as Rickettsia genomes are AT rich) for expression in mammalian cells (10, 19, 29). The yeast strain INVSc1 transformed with plasmid pYES-522HS carrying the codon-optimized RT0522 gene expresses the expected protein (Fig. 3A, lane 2) and shows no growth on inducing agar (Fig. 3B). These data indicate that expression of RT0522 is cytotoxic to yeast cells.
FIG. 3.
Cytotoxicity assay in yeast strain INVSc1 transformed with the plasmid pYES-522HS (codon-optimized RT0522), pYES-S86A (site-directed mutagenesis of codon-optimized RT0522 at Ser 86 to Ala), or pYES-S86A-D250A (site-directed mutagenesis of codon-optimized RT0522 at Ser 86 to Ala and Asp 250 to Ala). (A) Western blot analysis of the expression of codon-optimized RT0522 and its mutant derivatives in yeast strain INVSc1 under inducing conditions (SC-U+Gal medium). The total proteins from yeast cells carrying the appropriate plasmid were probed with anti-V5 antibody (Invitrogen) using the Western Breeze chemiluminescent immunodetection kit (Invitrogen). Lane 1, MagicMark XP Western protein standard (Invitrogen); lane 2, pYES-522HS/INVSc1; lane 3, pYES-S86A/INVSc1; lane 4, pYES-S86A-D250A/INVSc1; lane 5, pYES2/CT/INVSc1 (vector control). The sizes of the expected protein (including C-terminal V5 epitope and 6×His tag) RT0522 (lane 2) and its mutant derivatives (lanes 3 and 4) are shown on the right (69.5 kDa). (B) Transformed yeast cells were streaked onto inducing (SC-U+Gal) or repressing (SC-U+Glu) agar and incubated at 30°C for 3 days. (C) Transformed yeast cells with the plasmids pYES-522HS or pYES/CT/LacZ (control plasmid) were streaked onto inducing (SC-U+Gal) agar containing PLA2 inhibitor MAPF at 15 μM or BEL at 20 μM and incubated at 30°C for 3 days.
Inhibitors of RT0522-mediated cytotoxicity.
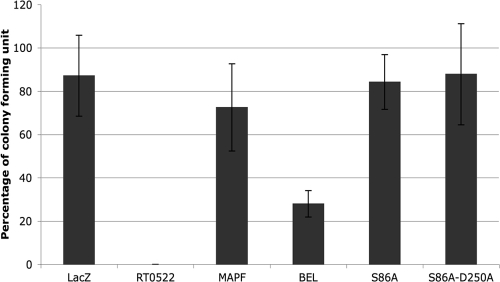
Here we sought to determine whether inhibitors of calcium-independent (iPLA2) and calcium-dependent cytosolic (cPLA2) phospholipase A2 block the cytotoxicity mediated by RT0522 within S. cerevisiae. Methyl arachidonyl fluorophosphonate (MAPF), an irreversible inhibitor of both iPLA2 and cPLA2 (18, 24, 36), blocks cytotoxicity caused by the expression of RT0522 protein in pYES-522HS/INVSc1 cells on inducing agar (Fig. 3C) and restores the growth to 72.5% ± 20.1% of the original value as determined by CFU assay (Fig. 4). However, bromoenol lactone (BEL), an irreversible inhibitor of iPLA2 (3, 24), shows partial protection of pYES-522HS/INVSc1 cells on inducing agar (Fig. 3C) and restores the growth to 27.9% ± 6.1% of the original value as determined by CFU assay (Fig. 4). The inhibitor assay provides support for the idea that RT0522 is a functional phospholipase A enzyme.
FIG. 4.
Cytotoxicity in yeast transformants determined by CFU assay. The percentages of CFU in yeast cells transformed with LacZ (pYES/CT/LacZ/INVSc1, for control plasmid), RT0522 (pYES-522HS/INVSc1, which showed no growth on inducing agar), S86A (pYES-S86A/INVSc1), and S86A-D250A (pYES-S86A-D250A/ INVSc1) were determined as described in Materials and Methods. MAPF and BEL represent yeast cells transformed with RT0522 (pYES-522HS/INVSc1) that were plated onto inducing (SC-U+Gal) agar containing PLA2 inhibitors MAPF at 15 μM and BEL at 20 μM, respectively.
We also cloned codon-optimized RT0522 in the pYES2/CT vector without the C-terminal V5/6×His tag and performed the cytotoxicity and inhibitor assays. They showed the same result as that observed for pYES-522HS (with the C-terminal V5/6×His tag), indicating that there was no effect of the tagged sequence on the cytotoxicity mediated by RT0522 expression within S. cerevisiae (data not shown).
Correlation of RT0522-mediated cytotoxicity with patatin-like PLA2 domain.
Our bioinformatic analysis of RT0522 shows (Fig. 1) the presence of patatin-like motifs with the Ser/Asp catalytic dyad characteristic of PLA2 enzymes (12, 35). The Ser/Asp catalytic dyad is required for cytotoxicity caused by patatin-like PLA2 enzymes (25, 35, 36). To examine the effect of the Ser/Asp catalytic dyad on RT0522-mediated cytotoxicity, we constructed two plasmids, pYES-S86A and pYES-S86A-D250A, by site-directed mutagenesis of plasmid pYES-522HS. In plasmid pYES-S86A, the serine residue of the serine hydrolase motif (amino acids 84 to 88 of RT0522 [Fig. 1]) was replaced by alanine. In plasmid pYES-S86A-D250A, the serine residue as well as an aspartate residue of the active-site aspartate motif (amino acids 250 to 252 of RT0522 [Fig. 1]) was replaced by alanine. The yeast strain INVSc1 transformed with plasmid pYES-S86A or pYES-S86A-D250A shows the expression of the expected proteins (Fig. 3A, lanes 3 and 4). The yeast transformants with plasmid pYES-S86A or pYES-S86A-D250A grow on inducing agar (Fig. 3B) and restore the growth to 84.2% ± 12.6% or 87.8% ± 23.3% of the original value, respectively, as determined by CFU assay (Fig. 4). Together these data indicate that the cytotoxicity caused by the expression of the RT0522 gene in yeast is abolished by mutation at the predicted Ser/Asp catalytic sites of the RT0522 protein.
Lactate dehydrogenase release assays to demonstrate cytotoxicity in mammalian cells.
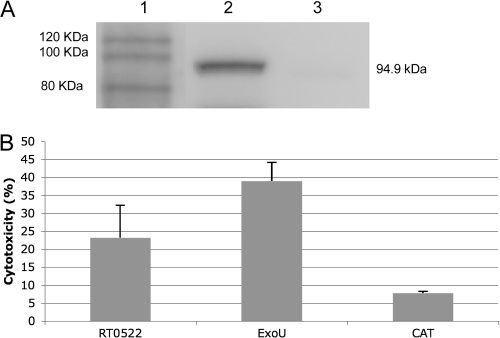
To confirm that the cytotoxicity observed in yeast was biologically relevant in the context of the R. typhi life cycle, we also examined the potential cytotoxic effects of RT0522 in mammalian cells by using the lactate dehydrogenase (LDH) release assay. LDH is a cytosolic enzyme of mammalian cells that is found in culture media only upon cell lysis. For the LDH release assay, we transfected Vero76 cells with the plasmid pcDNA-522HS, pcDNA-ExoU, or pcDNA6.2/N-EmGFP/GW/CAT (control plasmid). We observed 44% transfection efficiency with the control plasmid pcDNA6.2/N-EmGFP/GW/CAT. Vero76 cells transfected with the plasmid pcDNA-522HS carrying codon-optimized RT0522 show the expression of the expected protein (Fig. 5A, lane 2). The assay of LDH release from the transfected Vero cells reveals that the expression of RT0522 (pcDNA-522HS) and ExoU (pcDNA-ExoU; positive control) results in 23.23% ± 8.9% and 38.98% ± 5.22% cytotoxicity, respectively (Fig. 5B). The cytotoxicity associated with the expression of RT0522 and ExoU in Vero76 cells is significantly different (P < 0.05, two-tailed t test) from that for the CAT control plasmid (7.76% ± 0.48%) (Fig. 5B). However, the expression of RT0522 in Vero76 cells is relatively less cytotoxic than that of ExoU.
FIG. 5.
Cytotoxicity assay of RT0522 expression in Vero76 cells. (A) Western blot analysis of the expression of RT0522 in Vero76 cells. Total proteins from Vero76 cells carrying the appropriate plasmid were probed with anti-RT0522 antibody. Lane 1, MagicMark XP Western protein standard (Invitrogen); lane 2, pcDNA-522HS/Vero76; lane 3, pcDNA6.2/N-EmGFP/GW/CAT/Vero76 (for control plasmid). The size of the expected protein RT0522 (including N-terminally tagged EmGFP protein) in lane 2 is shown on the right (94.9 kDa). (B) The cytotoxicity of RT0522 expression in Vero76 cells was measured by LDH release assay and is presented as the percentage of lysed Vero76 cells. RT0522, pcDNA-522HS/Vero76; ExoU, pcDNA-ExoU/Vero76; CAT, pcDNA6.2/N-EmGFP/GW/CAT/Vero76. Error bars represent the standard errors of the means. The cytotoxicities of RT0522 and ExoU were found to be significantly different (P < 0.05 [two-tailed t test]) from that of the CAT control plasmid. The cytotoxicity of ExoU was significantly different (P < 0.05 [two-tailed t test]) from that of RT0522.
Phospholipase A activity of recombinant proteins.
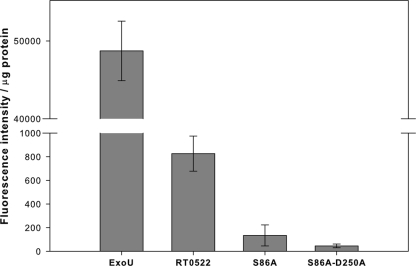
To assess the phospholipase A (PLA) activity of RT0522, we produced recombinant protein RT0522 (from pTrc-522HS) in an E. coli protein expression system. We also produced recombinant ExoU (from pTrc-ExoU) as a positive control. The PLA activity of each recombinant protein was measured by using the fluorogenic phospholipid substrate red/green BODIPY PC-A2 (specific for PLA2 enzyme activity). Neither of the purified recombinant proteins (RT0522 or ExoU) exhibited PLA activity on their own (in the absence of Vero76 cell lysate) as measured by fluorescence intensity. For ExoU, this was expected because it is known that its enzymatic activity is dependent on a eukaryotic cofactor (35, 36), and by further fractionation of eukaryotic cell extracts, it has been reported that Cu2+, Zn2+ superoxide dismutase (SOD1) acts as a cofactor to activate phospholipase activity of ExoU (34). In this study, we observe that both RT0522 and ExoU produce PLA activity in the presence of Vero76 cell lysate, as measured by fluorescence intensity/μg of recombinant proteins (Fig. 6). The phospholipase activity of ExoU is approximately 60-fold higher than that of RT0522, indicating that ExoU is a stronger PLA enzyme than RT0522.
FIG. 6.
Phospholipase A assay of recombinant proteins RT0522 (from pTrc-522HS), S86A (from pTrc-S86A), S86A-D250A (from pTrc-S86A-D250A), and ExoU (from pTrc-ExoU). The recombinant proteins were added to the fluorogenic phospholipid substrate red/green BODIPY PC-A2 in the presence of Vero76 cell lysate and incubated at room temperature for 4 h as described in Materials and Methods. The PLA activity was measured as fluorescence intensity/μg of recombinant proteins. Error bars represent standard errors of the means. The PLA activity of RT0522 was found to be significantly different (P < 0.05 [two-tailed t test]) from that of S86A or S86A-D250A.
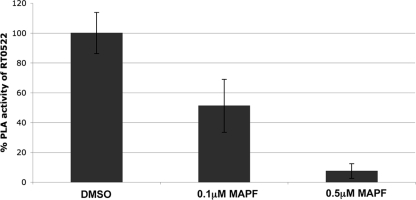
To determine whether the inhibitors of calcium-independent (iPLA2) and calcium-dependent cytosolic (cPLA2) phospholipase A2 block PLA activity of RT0522, we used methyl arachidonyl fluorophosphonate (MAPF), an irreversible inhibitor of both iPLA2 and cPLA2 (18, 24, 36). Methyl arachidonyl fluorophosphonate at 0.1 μM and 0.5 μM concentrations significantly (P < 0.05 [two-tailed t test]) reduced the PLA activity of RT0522 to 51.2% ± 17.8% and 7.4% ± 4.9% of the original values, respectively (Fig. 7). These data further indicate that RT0522 possesses phospholipase A activity, as this function can be blocked by iPLA2 and cPLA2 inhibitors.
FIG. 7.
Inhibition of phospholipase A (PLA) activity of RT0522 by methyl arachidonyl fluorophosphonate (MAPF). The inhibitor MAPF at an 0.1 μM or 0.5 μM final concentration (dissolved in dimethyl sulfoxide [DMSO]) was added to a reaction mixture containing recombinant protein RT0522 (from pTrc-522HS), fluorogenic phospholipid substrate, and Vero76 cell lysate as described in Materials and Methods. The PLA activity measured as fluorescence intensity/μg of recombinant proteins is presented as a percentage of RT0522 PLA activity. Error bars represent standard errors of the means. The % PLA activity of RT0522 (dimethyl sulfoxide) was found to be significantly different (P < 0.05 [two-tailed t test]) from that in the presence of 0.1 μM or 0.5 μM MAPF.
To examine the effect of the Ser/Asp catalytic residues in RT0522-mediated phospholipase A activity, we produced its mutant derivative proteins S86A (from pTrc-S86A) and S86A-D250A (pTrc-S86A-D250A) in an E. coli protein expression system. The S86A and S86A-D250A mutant derivatives of RT0522 recombinant proteins exhibited significant decreases in phospholipase A activity (Fig. 6). The phospholipase A activity of mutant protein S86A-D250A is 18-fold lower than that of RT0522. These data showing the effect of mutagenesis of catalytic Ser/Asp sites on PLA activity clearly suggest that RT0522 is a phospholipase A enzyme.
RT0522 translocation assay.
The subcellular localization of R. typhi protein RT0522 was analyzed by using the SignalP (6), LipoP (16), and Phobius (17) web-based tools. No signal peptide sequence was identified for RT0522 by all three programs. However, RT0522 was predicted to be noncytoplasmic (in the rickettsial cell) by the Phobius program. This analysis suggests that the in silico predicted noncytoplasmic protein RT0522 may be exported from the rickettsial cytoplasm by a Sec-independent pathway.
Here we sought to demonstrate whether the RT0522 protein remains associated with R. typhi or is secreted into the host cytoplasm during an infection. We prepared two cellular fractions, supernatant and pellet, from uninfected or R. typhi-infected Vero76 cells after incubation for 48 h. The reason that we chose the 48-h time point (when the rickettsial burden was maximal [Fig. 2]) was to have enough rickettsiae to detect the rickettsial proteins by Western blotting. The supernatant contained the rickettsial secreted proteins as well as the host soluble proteins. The pellet contained intact rickettsiae along with host debris and unbroken host cells. Both fractions, supernatant and pellet, from uninfected or R. typhi-infected Vero76 cells were probed with antibodies raised against RT0522, EF-Ts, or GAPDH. We observed that GAPDH, which is a host cytoplasmic protein, appears in the supernatant of both the uninfected and the infected cells (Fig. 8, lanes 3 and 4). The faint bands of GAPDH in the pellets (Fig. 8, lanes 1 and 2) may have appeared due to the unbroken host cells or the residual supernatant left with the pellet. For control (soluble rickettsial cytoplasmic protein), we used antibodies against EF-Ts (elongation factor Ts, encoded by RT0049, which functions during the elongation stage of protein translation) (22). We observe that EF-Ts is present only in the pellet (Fig. 8, lane 2) of infected cells, supporting the idea that EF-Ts remains associated with R. typhi and that R. typhi remains intact during our fraction preparation. However, RT0522 is present in both the pellet (Fig. 8, lane 2) and the supernatant (Fig. 8, lane 4) of infected cells. These data suggest that RT0522 protein is expressed and secreted into the host cell cytoplasm during R. typhi growth in host cells.
FIG. 8.
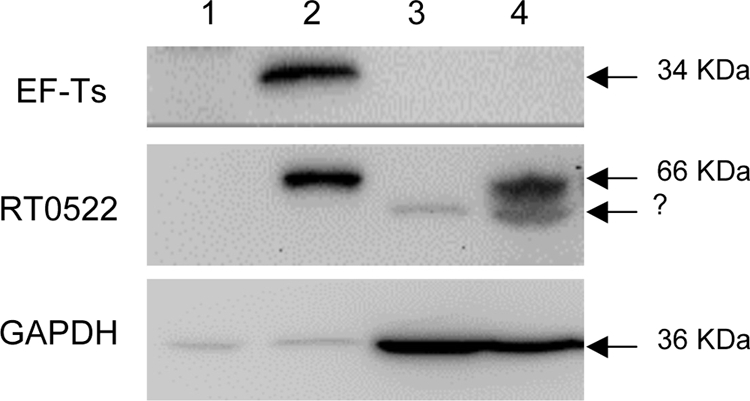
Translocation assay of RT0522 and EF-Ts by Western blotting. The membrane was probed with rabbit anti-EF-Ts antibody, rabbit anti-RT0522 antibody, or mouse anti-GAPDH monoclonal antibody by using the SuperSignal West Pico chemiluminescent substrate kit. Lane 1, pellet of uninfected Vero76 cells; lane 2, pellet of R. typhi-infected Vero76 cells; lane 3, supernatant of uninfected Vero76 cells; lane 4, supernatant of R. typhi-infected Vero76 cells. The sizes of the expected proteins EF-Ts (34 kDa), RT0522 (66 kDa), and GAPDH (36 kDa) are shown on the right. The bands marked with a question mark below the 66-kDa band in lanes 3 and 4 may have resulted from the nonspecific binding to host soluble proteins.
DISCUSSION
In this communication, we report the bioinformatic-based identification of an R. typhi gene, RT0522, encoding a homolog of putative patatin-like PLA2 enzyme. We further demonstrate that RT0522 undergoes both gene and protein expression during Rickettsia typhi infection in Vero76 cells and that the protein product is secreted in the host cell cytoplasm. The recombinant protein RT0522 possesses phospholipase A enzymatic activity and is cytotoxic to both yeast and mammalian cells.
In 2005, Whitworth et al. reported that tlyC (hemolysin C) and pld (phospholipase D) were transcribed by R. prowazekii and might play a part in phagosomal escape (47). However, the same publication reported that the genome-annotated patatin-like protein (pat1, RP602) of R. prowazekii was not transcribed in Vero cells during the time period (30 min postinfection) corresponding to phagosomal escape (47). The transcriptional data presented in this article revealed that the genome-annotated patatin-like protein RT0590 (ortholog of RP602) of R. typhi is also not transcribed during various stages of rickettsial growth in Vero76 cells. These data suggest that the genome-annotated patatin-like proteins for R. prowazekii and R. typhi may not be required for their growth in Vero cell culture. However, these data do not rule out the role of this protein during rickettsial growth in other eukaryotic host cells or during in vivo infection. The transcriptional data for RT0522 suggest a role of this patatin-like protein during various stages of growth of R. typhi in Vero76 cells. Although the rickettsial burden is maximal at 48 h postinfection, the elevated transcription of RT0522 from 15 min to 1 h postinfection suggests the potential involvement of RT0522 protein during the early stage of R. typhi infection in Vero cells. The increase in transcription of RT0522 after doubling time suggests a possible role in R. typhi survival, including lysis of host cells so that the bacteria can escape for further infection. The other Rickettsia species that lack a complete ortholog of RT0522 (identified in only five species of Rickettsia [Fig. 1]) might utilize other factors or the RT0590 ortholog (Fig. 1) for their infection and intracellular growth in host cells.
P. aeruginosa ExoU, a member of the patatin-like PLA2 family of enzymes, has been shown to be cytotoxic to a variety of cultured mammalian and yeast cells (35, 43). The PLA2 activity of ExoU is essential for cytotoxicity (24, 36). In this study, the codon-optimized RT0522 gene product has been demonstrated to be cytotoxic to yeast and Vero76 cells. The expression of codon-optimized RT0522 was detected by Western blotting in both yeast and Vero cells. However, the expression of ExoU has never been reported by Western blotting because the amount of ExoU required for toxicity is below the protein detection limit (35, 36). The fact that we could detect RT0522 protein expression, therefore, supports the observation that this protein is relatively less cytotoxic than ExoU.
Phospholipase A2 activity has been implicated in both rickettsial entry into the host cell and escape from the phagosome (40, 50). For example, phospholipase A activity was demonstrated by a fluorometric assay for whole R. prowazekii and R. typhi bacterial cells (21). In addition, the inhibition of phospholipase A2-mediated cytotoxicity in Vero cells was demonstrated when R. prowazekii and R. conorii were pretreated with a PLA2 inhibitor (bromophenacyl bromide) or with antibody to the PLA2 of king cobra venom (46). Despite this evidence for rickettsial PLA2 activity, a rickettsial gene encoding phospholipase A enzyme has not previously been identified. Our results clearly demonstrate the phospholipase A activity of RT0522 and show that this enzymatic activity required a eukaryotic host cofactor for its activation. Addition of inhibitors of iPLA2 and cPLA2 blocks the cytotoxic effect and phospholipase A activity of RT0522, similar to an iPLA2/cPLA2-like enzyme (24, 35, 36). The Ser/Asp catalytic dyad is required for patatin-like PLA2 enzyme activity, and mutation at these catalytic sites abolishes the cytotoxic effect caused by the enzyme (24, 25, 36). Our site-directed mutagenesis data indicate further that RT0522 is a patatin-like phospholipase A protein and that the cytotoxicity associated with RT0522 is due to the catalytic Ser/Asp sites. Collectively, these data clearly show that RT0522 is a phospholipase A enzyme.
Our data suggest that the relatively low cytotoxicity and phospholipase A activity of RT0522, compared to those of ExoU, may be necessary for rickettsiae to survive inside the host cell without causing rapid damage until they are ready to lyse the host cell and escape to cause further infection. The low level of phospholipase A activity of RT0522 may also be involved in cell signaling activity without affecting host cell integrity during the initial stage of infection and subsequent stages of growth in host cells.
Subcellular localization of P. aeruginosa ExoU to the plasma membrane of host cells is considered to be essential for its cytotoxicity (27). The data presented in this study demonstrate the translocation of R. typhi RT0522 protein into the host cytoplasm. However, the secretion pathway of RT0522 protein into the host cytoplasm, its posttranslocational modification (if any), and the mechanism of interaction with host organelles remain unknown. In silico analysis revealed that the protein RT0522 does not contain a signal peptide sequence, suggesting that this protein is not secreted to the host cell cytoplasm through the Sec-dependent protein secretion pathway of rickettsiae (1, 29-31). However, other secretory pathways, e.g., the type IV protein secretion pathway, have been identified in rickettsiae (9). Our ongoing research will address the mechanism of RT0522 secretion; its role in rickettsial intracellular growth, including phagosomal escape and exit via host cell lysis; and the molecular basis of its interaction with the host cellular structures.
Supplementary Material
Acknowledgments
The research presented in this article was supported by funds from the National Institutes of Health/National Institute of Allergy and Infectious Diseases (AI017828 and AI59118).
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of Allergy and Infectious Diseases or the National Institutes of Health.
We gratefully acknowledge Dara W. Frank, Department of Microbiology and Molecular Genetics, Medical College of Wisconsin, Milwaukee, WI, for generously providing plasmid pYES-ExoU-GFP as a positive control for this work. We are also thankful to Magda S. Beier for her critical comments and helpful suggestions on the manuscript.
Footnotes
Published ahead of print on 30 April 2010.
Supplemental material for this article may be found at http://jb.asm.org/.
The authors have paid a fee to allow immediate free access to this article.
REFERENCES
- 1.Ammerman, N. C., M. S. Rahman, and A. F. Azad. 2008. Characterization of Sec-translocon-dependent extracytoplasmic proteins of Rickettsia typhi. J. Bacteriol. 190:6234-6242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Azad, A. F., and C. B. Beard. 1998. Rickettsial pathogens and their arthropod vectors. Emerg. Infect. Dis. 4:179-186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Balsinde, J., and E. A. Dennis. 1996. Bromoenol lactone inhibits magnesium-dependent phosphatidate phosphohydrolase and blocks triacylglycerol biosynthesis in mouse P388D1 macrophages. J. Biol. Chem. 271:31937-31941. [DOI] [PubMed] [Google Scholar]
- 4.Banerji, S., P. Aurass, and A. Flieger. 2008. The manifold phospholipases A of Legionella pneumophila—identification, export, regulation, and their link to bacterial virulence. Int. J. Med. Microbiol. 298:169-181. [DOI] [PubMed] [Google Scholar]
- 5.Banerji, S., and A. Flieger. 2004. Patatin-like proteins: a new family of lipolytic enzymes present in bacteria? Microbiology 150:522-525. [DOI] [PubMed] [Google Scholar]
- 6.Bendtsen, J. D., H. Nielsen, G. von Heijne, and S. Brunak. 2004. Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 340:783-795. [DOI] [PubMed] [Google Scholar]
- 7.Dreher-Lesnick, S. M., S. M. Ceraul, M. S. Rahman, and A. F. Azad. 2008. Genome-wide screen for temperature-regulated genes of the obligate intracellular bacterium, Rickettsia typhi. BMC Microbiol. 8:61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Driskell, L. O., X. J. Yu, L. Zhang, Y. Liu, V. L. Popov, D. H. Walker, A. M. Tucker, and D. O. Wood. 2009. Directed mutagenesis of the Rickettsia prowazekii pld gene encoding phospholipase D. Infect. Immun. 77:3244-3248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gillespie, J. J., N. C. Ammerman, S. M. Dreher-Lesnick, M. S. Rahman, M. J. Worley, J. C. Setubal, B. S. Sobral, and A. F. Azad. 2009. An anomalous type IV secretion system in Rickettsia is evolutionarily conserved. PLoS One 4:e4833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gustafsson, C., S. Govindarajan, and J. Minshull. 2004. Codon bias and heterologous protein expression. Trends Biotechnol. 22:346-353. [DOI] [PubMed] [Google Scholar]
- 11.Hackstadt, T. 1996. The biology of rickettsiae. Infect. Agents Dis. 5:127-143. [PubMed] [Google Scholar]
- 12.Hauser, A. R. 2009. The type III secretion system of Pseudomonas aeruginosa: infection by injection. Nat. Rev. Microbiol. 7:654-665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Higgins, J. A., S. Radulovic, B. H. Noden, J. M. Troyer, and A. F. Azad. 1998. Reverse transcriptase PCR amplification of Rickettsia typhi from infected mammalian cells and insect vectors. J. Clin. Microbiol. 36:1793-1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hybiske, K., and R. S. Stephens. 2008. Exit strategies of intracellular pathogens. Nat. Rev. Microbiol. 6:99-110. [DOI] [PubMed] [Google Scholar]
- 15.Istivan, T. S., and P. J. Coloe. 2006. Phospholipase A in Gram-negative bacteria and its role in pathogenesis. Microbiology 152:1263-1274. [DOI] [PubMed] [Google Scholar]
- 16.Juncker, A. S., H. Willenbrock, G. Von Heijne, S. Brunak, H. Nielsen, and A. Krogh. 2003. Prediction of lipoprotein signal peptides in Gram-negative bacteria. Protein Sci. 12:1652-1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kall, L., A. Krogh, and E. L. Sonnhammer. 2004. A combined transmembrane topology and signal peptide prediction method. J. Mol. Biol. 338:1027-1036. [DOI] [PubMed] [Google Scholar]
- 18.Lio, Y. C., L. J. Reynolds, J. Balsinde, and E. A. Dennis. 1996. Irreversible inhibition of Ca(2+)-independent phospholipase A2 by methyl arachidonyl fluorophosphonate. Biochim. Biophys. Acta 1302:55-60. [DOI] [PubMed] [Google Scholar]
- 19.McLeod, M. P., X. Qin, S. E. Karpathy, J. Gioia, S. K. Highlander, G. E. Fox, T. Z. McNeill, H. Jiang, et al. 2004. Complete genome sequence of Rickettsia typhi and comparison with sequences of other rickettsiae. J. Bacteriol. 186:5842-5855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Muller, P. Y., H. Janovjak, A. R. Miserez, and Z. Dobbie. 2002. Processing of gene expression data generated by quantitative real-time RT-PCR. Biotechniques 32:1372-1379. [PubMed] [Google Scholar]
- 21.Ojcius, D. M., M. Thibon, C. Mounier, and A. Dautry-Varsat. 1995. pH and calcium dependence of hemolysis due to Rickettsia prowazekii: comparison with phospholipase activity. Infect. Immun. 63:3069-3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pan, X., A. Luhrmann, A. Satoh, M. A. Laskowski-Arce, and C. R. Roy. 2008. Ankyrin repeat proteins comprise a diverse family of bacterial type IV effectors. Science 320:1651-1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pang, H., and H. H. Winkler. 1996. Transcriptional analysis of the 16S rRNA gene in Rickettsia prowazekii. J. Bacteriol. 178:1750-1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Phillips, R. M., D. A. Six, E. A. Dennis, and P. Ghosh. 2003. In vivo phospholipase activity of the Pseudomonas aeruginosa cytotoxin ExoU and protection of mammalian cells with phospholipase A2 inhibitors. J. Biol. Chem. 278:41326-41332. [DOI] [PubMed] [Google Scholar]
- 25.Rabin, S. D., and A. R. Hauser. 2005. Functional regions of the Pseudomonas aeruginosa cytotoxin ExoU. Infect. Immun. 73:573-582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rabin, S. D., and A. R. Hauser. 2003. Pseudomonas aeruginosa ExoU, a toxin transported by the type III secretion system, kills Saccharomyces cerevisiae. Infect. Immun. 71:4144-4150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rabin, S. D., J. L. Veesenmeyer, K. T. Bieging, and A. R. Hauser. 2006. A C-terminal domain targets the Pseudomonas aeruginosa cytotoxin ExoU to the plasma membrane of host cells. Infect. Immun. 74:2552-2561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Radulovic, S., J. M. Troyer, M. S. Beier, A. O. Lau, and A. F. Azad. 1999. Identification and molecular analysis of the gene encoding Rickettsia typhi hemolysin. Infect. Immun. 67:6104-6108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rahman, M. S., S. M. Ceraul, S. M. Dreher-Lesnick, M. S. Beier, and A. F. Azad. 2007. The lspA gene, encoding the type II signal peptidase of Rickettsia typhi: transcriptional and functional analysis. J. Bacteriol. 189:336-341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rahman, M. S., J. A. Simser, K. R. Macaluso, and A. F. Azad. 2005. Functional analysis of secA homologues from rickettsiae. Microbiology 151:589-596. [DOI] [PubMed] [Google Scholar]
- 31.Rahman, M. S., J. A. Simser, K. R. Macaluso, and A. F. Azad. 2003. Molecular and functional analysis of the lepB gene, encoding a type I signal peptidase from Rickettsia rickettsii and Rickettsia typhi. J. Bacteriol. 185:4578-4584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ramakers, C., J. M. Ruijter, R. H. Deprez, and A. F. Moorman. 2003. Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neurosci. Lett. 339:62-66. [DOI] [PubMed] [Google Scholar]
- 33.Rydel, T. J., J. M. Williams, E. Krieger, F. Moshiri, W. C. Stallings, S. M. Brown, J. C. Pershing, J. P. Purcell, et al. 2003. The crystal structure, mutagenesis, and activity studies reveal that patatin is a lipid acyl hydrolase with a Ser-Asp catalytic dyad. Biochemistry 42:6696-6708. [DOI] [PubMed] [Google Scholar]
- 34.Sato, H., J. B. Feix, and D. W. Frank. 2006. Identification of superoxide dismutase as a cofactor for the pseudomonas type III toxin, ExoU. Biochemistry 45:10368-10375. [DOI] [PubMed] [Google Scholar]
- 35.Sato, H., and D. W. Frank. 2004. ExoU is a potent intracellular phospholipase. Mol. Microbiol. 53:1279-1290. [DOI] [PubMed] [Google Scholar]
- 36.Sato, H., D. W. Frank, C. J. Hillard, J. B. Feix, R. R. Pankhaniya, K. Moriyama, V. Finck-Barbancon, A. Buchaklian, et al. 2003. The mechanism of action of the Pseudomonas aeruginosa-encoded type III cytotoxin, ExoU. EMBO J. 22:2959-2969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schaloske, R. H., and E. A. Dennis. 2006. The phospholipase A2 superfamily and its group numbering system. Biochim. Biophys. Acta 1761:1246-1259. [DOI] [PubMed] [Google Scholar]
- 38.Shohdy, N., J. A. Efe, S. D. Emr, and H. A. Shuman. 2005. Pathogen effector protein screening in yeast identifies Legionella factors that interfere with membrane trafficking. Proc. Natl. Acad. Sci. U. S. A. 102:4866-4871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Siggers, K. A., and C. F. Lesser. 2008. The yeast Saccharomyces cerevisiae: a versatile model system for the identification and characterization of bacterial virulence proteins. Cell Host Microbe 4:8-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Silverman, D. J., L. A. Santucci, N. Meyers, and Z. Sekeyova. 1992. Penetration of host cells by Rickettsia rickettsii appears to be mediated by a phospholipase of rickettsial origin. Infect. Immun. 60:2733-2740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sisko, J. L., K. Spaeth, Y. Kumar, and R. H. Valdivia. 2006. Multifunctional analysis of Chlamydia-specific genes in a yeast expression system. Mol. Microbiol. 60:51-66. [DOI] [PubMed] [Google Scholar]
- 42.Sitkiewicz, I., M. J. Nagiec, P. Sumby, S. D. Butler, C. Cywes-Bentley, and J. M. Musser. 2006. Emergence of a bacterial clone with enhanced virulence by acquisition of a phage encoding a secreted phospholipase A2. Proc. Natl. Acad. Sci. U. S. A. 103:16009-16014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sitkiewicz, I., K. E. Stockbauer, and J. M. Musser. 2007. Secreted bacterial phospholipase A2 enzymes: better living through phospholipolysis. Trends Microbiol. 15:63-69. [DOI] [PubMed] [Google Scholar]
- 44.Valdivia, R. H. 2004. Modeling the function of bacterial virulence factors in Saccharomyces cerevisiae. Eukaryot. Cell 3:827-834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.VanRheenen, S. M., Z. Q. Luo, T. O'Connor, and R. R. Isberg. 2006. Members of a Legionella pneumophila family of proteins with ExoU (phospholipase A) active sites are translocated to target cells. Infect. Immun. 74:3597-3606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Walker, D. H., H. M. Feng, and V. L. Popov. 2001. Rickettsial phospholipase A2 as a pathogenic mechanism in a model of cell injury by typhus and spotted fever group rickettsiae. Am. J. Trop. Med. Hyg. 65:936-942. [DOI] [PubMed] [Google Scholar]
- 47.Whitworth, T., V. L. Popov, X. J. Yu, D. H. Walker, and D. H. Bouyer. 2005. Expression of the Rickettsia prowazekii pld or tlyC gene in Salmonella enterica serovar Typhimurium mediates phagosomal escape. Infect. Immun. 73:6668-6673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Winkler, H. H., and R. M. Daugherty. 1989. Phospholipase A activity associated with the growth of Rickettsia prowazekii in L929 cells. Infect. Immun. 57:36-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Winkler, H. H., and E. T. Miller. 1982. Phospholipase A and the interaction of Rickettsia prowazekii and mouse fibroblasts (L-929 cells). Infect. Immun. 38:109-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Winkler, H. H., and J. Turco. 1988. Rickettsia prowazekii and the host cell: entry, growth and control of the parasite. Curr. Top. Microbiol. Immunol. 138:81-107. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.