Abstract
GerR is a sporulation-specific transcriptional factor of Bacillus subtilis that has been identified as a negative regulator of genes transcribed by σE-containing RNA polymerase and as a positive effector of the expression of three late sporulation genes. Here we confirmed that gerR transcription is dependent on σE-containing RNA polymerase but also observed that it requires the transcriptional regulator SpoIIID. The study of the role of GerR in regulating the expression of several late sporulation genes allowed us to observe that its effect is strongly positive on spoVIF, cotC, and cotG, weakly positive on cotB, and negative on cotU. The results of chromatin immunoprecipitation (ChIP) experiments indicated that GerR binds to the promoter regions of some, but not all, of the GerR-controlled genes, leading us to propose that GerR controls late sporulation genes in two ways: (i) directly, by acting on the transcription of cotB, cotU and spoVIF; and (ii) indirectly, through the activation of SpoVIF, which stabilizes the transcriptional activator GerE and consequently induces the expression of the GerE-dependent genes cotC and cotG.
Spore formation in Bacillus subtilis involves the formation of an asymmetrical cell division septum that divides the sporulating cell (sporangium) into a larger cell (mother cell) and a smaller cell (forespore). In a process that takes 7 to 8 h to complete, the forespore differentiates into a mature spore, while the mother cell contributes to spore development but eventually undergoes lysis to liberate the mature spore when morphogenesis is completed (20). Much is known about the programs of differential gene expression that drive the process of spore formation in the two cell types. In the forespore, the RNA polymerase sigma factors σF and σG and the DNA-binding proteins RsfA and SpoVT are responsible for the coordinate expression of many genes that contribute to spore morphogenesis and resistance and germination properties (21). In the mother cell, the RNA polymerase sigma factors σE and σK and three positively and/or negatively acting transcription factors GerE, GerR, and SpoIIID are active (5). The appearance of these regulatory proteins in the mother cell is governed by a hierarchical regulatory cascade of transcription factors σE, SpoIIID, GerR, σK, and GerE (5). SpoIIID and GerE are both negatively and positively acting proteins that switch off the transcription of certain genes activated by σE or σK and, in conjunction with σE- or σK-containing RNA polymerase, switch on additional genes (5). GerR exhibits significant similarity to members of the basic leucine zipper family of transcription factors; in particular, it is 52% similar to RsfA (5), a forespore-specific regulator. Transcriptional profiling experiments performed with RNA collected at hour 3.5 of sporulation from B. subtilis strains not expressing σK and carrying either the wild-type gerR gene or a null mutant allele of gerR allowed the identification of 14 genes belonging to the σE regulon overexpressed in the gerR mutant (5). Five of the 14 genes negatively regulated by GerR (phoB, spoIIIAA, spoIIIAB, spoIVCA, and ydhF) are also under the control of another transcriptional regulator, SpoIIID (5). An example of a transcription unit under the dual control of GerR and SpoIIID is the eight-cistron spoIIIA operon. In this case, GerR has only a small effect on spoIIIA transcription, and maximum repression of the operon is achieved by the combined action of GerR and SpoIIID (5). Since the transcriptional profiling experiments were performed in strains that did not express σK (5), members of the σK regulon were not identified as controlled by GerR. An independent study showed, however, that gerR (ylbO) is needed for normal expression of three genes of the σK regulon, namely, cgeA, cotG, and cotY (12). Western and dot blot experiments showed that in a strain carrying a gerR null mutation, the expression of the three σK-dependent genes is strongly reduced (12).
We were intrigued by the observations that GerR has a negative role on the expression of genes transcribed by σE between 2 and 4 h after the onset of sporulation (5) and a positive role on the expression of genes transcribed by σK and GerE several hours later (7 to 8 h after the start of sporulation) (12). A possible explanation, suggested by Kuwana et al. (12), is that the action of GerR on late genes is indirect. Here we report on a more detailed analysis of the mechanism of expression of gerR and the mode of regulatory action of its product on late sporulation genes.
MATERIALS AND METHODS
Bacterial strains and transformation.
B. subtilis strains utilized in this study are listed in Table 1. Plasmid amplification for DNA sequencing, subcloning experiments, and transformation of Escherichia coli competent cells were performed with E. coli strain DH5α (19). Bacterial strains were transformed by previously described procedures: CaCl2-mediated transformation of E. coli competent cells (19) and two-step transformation of B. subtilis (4).
TABLE 1.
B. subtilis strains used in this study
| Strain | Relevant genotype | Reference |
|---|---|---|
| PY79 | Wild type | 22 |
| KS450 | gerE36 | 2 |
| GC260 | gerR::neo | This study |
| GC261 | gerR::neo gerE36 | This study |
| GC284 | gerR::neo amyE::gerR | This study |
| GC272 | gerR::lacZ | This study |
| GC275 | gerR::lacZ spoIIID::erm | This study |
| GC301 | cotB::lacZ | This study |
| GC302 | cotB::lacZ gerR::neo | This study |
| GC267 | cotB::lacZ gerE36 | This study |
| GC328 | cotB::lacZ gerE36 gerR::neo | This study |
| GC266 | cotG::lacZ | This study |
| GC322 | cotG::lacZ gerR::neo | This study |
| GC323 | cotG::lacZ gerE36 | This study |
| GC325 | cotG::lacZ gerE36 gerR::neo | This study |
| GC285 | cotA::lacZ | This study |
| GC305 | cotA::lacZ gerR::neo | This study |
| GC303 | cotA::lacZ gerE36 | This study |
| GC327 | cotA::lacZ gerE36 gerR::neo | This study |
| GC280 | cotD::lacZ | This study |
| GC330 | cotD::lacZ gerR::neo | This study |
| RH259 | cotC::lacZ | This study |
| GC317 | cotC::lacZ gerR::neo | This study |
| GC318 | cotC::lacZ gerE36 | This study |
| GC319 | cotC::lacZ gerE36 gerR::neo | This study |
| GC290 | spoVIF::lacZ | This study |
| GC291 | spoVIF::lacZ gerR::neo | This study |
| RH248 | cotU::lacZ | 8 |
| RH249 | cotU::lacZ gerE36 | 8 |
| GC310 | cotU::lacZ gerR::neo | This study |
| GC312 | cotU::lacZ gerE36 gerR::neo | This study |
| BK107 | SPβ::spoIVCB-lacZ | 9 |
| BK541 | spoIIIDΔerm | 9 |
| GC281 | SPb::spoIVCB-lacZ gerR::neo | This study |
| BK595 | spoIIID::lacZ | 9 |
| GC263 | spoIIID::lacZ gerR::neo | This study |
| SPβgerE-lacZ | SPβ::gerE-lacZ | 3 |
| GC262 | SPb::gerE-lacZ gerR::neo | This study |
Growth and sporulation conditions.
Sporulation for β-galactosidase assays and chromatin immunoprecipitation (ChIP) experiments was induced by the resuspension method as previously described (4). The onset of sporulation (T0) is defined as the time of resuspension; samples were collected at the indicated time after resuspension.
Spore purification, sporulation efficiency, lysozyme resistance, and germination assay.
Sporulation was induced in DSM (Difco sporulation medium) by the exhaustion method as described elsewhere (4). After a 30-h incubation at 37°C, the spores were collected, washed four times, and purified as described by Nicholson and Setlow (16) using overnight incubation in H2O at 4°C to lyse residual sporangial cells. Assays for resistance to lysozyme and sporulation efficiency were performed 30 min after the onset of sporulation without spore purification, as previously described (4). Measurement of spore germination was performed by monitoring the loss of optical density as described by Nicholson and Setlow (16).
Extraction of spore coat proteins.
Spore coat proteins from B. subtilis PY79, the gerR null mutant (GC260), and the complemented strain (GC284) were extracted from a suspension of spores by using an SDS-dithiothreitol (DTT) extraction buffer as previously described (16). The concentration of extracted proteins was determined by using Bio-Rad DC protein assay kit (Bio-Rad), and 15-μg samples of total spore coat proteins were fractionated on 12.5% SDS-polyacrylamide gels and stained with Coomassie blue.
Genetic and molecular procedures.
Plasmid extraction, restriction digestion, DNA ligation, PCR amplification, and primer extension were carried out by standard methods (19). Synthetic oligonucleotides utilized in PCR amplification and primer extension are listed in Table 2. The PCR products were visualized on ethidium bromide-stained agarose gels and gel purified by using the QIAquick gel extraction kit (Qiagen) as specified by the manufacturer.
TABLE 2.
Synthetic oligonucleotides used in PCR amplification and primer extension
| Oligonucleotide | Sequence (5′-3′)a | Restriction site | Annealing positionsb |
|---|---|---|---|
| gerR2s | ggatccGAAGAAAAGATTGCGCCG | BamHI | −293 to −275 |
| ylBOaBamHI | ggatcc TGAAAGCTGTGTTCCTCC | BamHI | +99 to +82 |
| ylBOsenso | AGCCTTTGAGGAGGTTGG | +99 to +116 | |
| ylBOanti | CATGATATCGATCAGTGCC | +513 to +495 | |
| ylBOH3 | aagcttCCGGTTCATTTCAGGCG | HindIII | +604 to +588 |
| cotBps | gaattcTATGAACGGATTAGGCCG | EcoRI | −268 to −250 |
| cotBpa | ggatccAATACTTTTACATGCTCC | BamHI | +206 to +187 |
| cotAps | gaattcGTCTATCTTGTCATCGCC | EcoRI | −451 to −433 |
| cotApa | ggatccTGTAGCCCCACAGGCG | BamHI | +154 to +139 |
| cotDps | gaattcCAGGACAGTGCACGGC | EcoRI | −594 to −578 |
| cotDpa | ggatccGCCATCATATGCGGTCTG | BamHI | +29 to +12 |
| cotCps | gaattcGAATTCCGGCAGACGTAC | EcoRI | −564 to −531 |
| cotCpa | ggatccGTATTTTTTGTAATAACC | BamHI | +11 to +4 |
| cotGps | gaattcTGGATGAACAAACACCTG | EcoRI | −187 to −169 |
| cotGpa | ggatccTGCGACTTTTTATGACTGC | BamHI | +118 to +99 |
| spoVIFs | gaattcATGAGATTGAAGAGCTGG | EcoRI | −570 to −552 |
| spoVIFa | ggatccGGCAAGATTCATTACATCC | BamHI | +71 to +52 |
| gerR PE | CTGAGTCCAAGCATCTTG | +33 to +15 | |
| gerRcoding | tctagaATGAGATAAATAAGCTGAC | XbaI | −35 to −17 |
| gerRanti | ctcgagCGTTTCTTGAGCCGC | XhoI | +578 to +563 |
| chip cotUs | TATGATCTGTTTCTAGCCG | −350 to −331 | |
| chip cotU1 | GAAAAACAAAACGGATAGC | −107 to −126 | |
| chip cotB | ACATAGATGATAAACATACTG | −23 to −43 | |
| yjczs | GGTCGTATTATTCATTCTCC | −154 to −134 | |
| chip spoVIF1 | CGGTATTGTATGAATGCAC | −26 to −45 | |
| chip gerEs | GTATCCGTAAGATTTTCCG | −490 to −471 | |
| chip gerE1 | GATATCATATGTAAGAAGGG | −60 to −80 | |
| ftsHs | CAAAGCAGACTTTGTCGG | −223 to −205 | |
| ftsHa | GTACCATAGAAAATCATTCC | −39 to −59 |
The restriction sites inserted with synthetic oligonucleotides are shown in lowercase type.
Considering the first base of the first codon as +1.
Construction of a gerR null mutation.
An internal fragment of the gerR gene was PCR amplified using B. subtilis chromosomal DNA as a template and oligonucleotides ylBOsenso and ylBOanti as primers. The PCR product of the expected size (414 bp) was cloned into the pGEM-T Easy vector (Promega) and then excised by NotI. The NotI restriction fragment was cloned into vector pBEST501 (8a) obtaining plasmids pGC95. This plasmid was then used to transform competent cells of B. subtilis strain PY79 by single reciprocal recombination (Campbell-like) at the gerR locus to obtain strain GC260 (gerR::neo). The gerR mutation was moved by chromosomal DNA-mediated transformation to the isogenic strain carrying a null mutation in gerE (gerE36), generating the double mutant strain GC261 (gerE gerR).
To insert a wild-type copy of gerR at the amyE locus of strain GC260, a PCR amplification product was obtained using chromosomal DNA as the template and oligonucleotides gerR2s and ylBOH3 as primers. The PCR product was cloned into the pGEM-T Easy vector (Promega) and then digested with BamHI and HindIII restriction enzymes, and the 897-bp fragment containing the gerR promoter and coding region was transferred into plasmid pDG364 that had previously been linearized with the same enzymes. The plasmid obtained, pGC101, was linearized with ScaI and used to transform GC260 competent cells. The resulting recombinant strain GC284 was obtained by double-crossover recombination at the nonessential amyE gene on the B. subtilis chromosome.
Construction of lacZ transcriptional fusion and β-galactosidase assays.
The integrative vector pJM783 (1) was used to obtain transcriptional fusions to the lacZ gene. Genomic fragments containing the promoter regions of gerR (391 bp), cotB (474 bp), cotA (605 bp), cotD (623 bp), cotC (575 bp), cotG (308 bp), and spoVIF (642 bp) were PCR amplified using the B. subtilis chromosome as a template and oligonucleotides listed in Table 2 as primers. The purified fragments were cloned into pGEM-T Easy vector (Promega), excised by EcoRI/BamHI digestion, gel purified, and cloned upstream of the lacZ gene into the pJM783 vector (1) linearized with the same enzymes. The resulting plasmids, pGC102 (gerR::lacZ), pGC103 (cotB::lacZ), pGC104 (cotA::lacZ), pGC105 (cotD::lacZ), pGC106 (cotC::lacZ), pGC107 (cotG::lacZ), and pGC108 (spoVIF::lacZ), were used to transform competent cells of B. subtilis strain PY79. Recombinant strains were obtained by single reciprocal recombination (Campbell-like) at the corresponding loci: gerR, cotB, cotA, cotD, cotC, cotG, and spoVIF (Table 1). Chromosomal DNA of B. subtilis GC272 (gerR::lacZ) was used to transform competent cells of the spoIIID mutant, generating strain GC275. The various transcriptional lacZ fusions were moved by chromosome-mediated transformation into an isogenic strain carrying a null mutation in gerE or gerR (Table 1).
The specific β-galactosidase activity was determined using o-nitrophenol-β-d-galactosidase (ONPG) as the substrate as previously reported (17). Samples (1 ml each) of cells bearing the fusion were collected, during sporulation, at the indicated times and assayed as described by Cutting and Vander Horn (4).
Primer extension analysis.
Total RNA was extracted from a wild-type strain 2 and 3 h after the onset of sporulation using the Qiagen minikit (Qiagen, Milan, Italy) according to the manufacturer's instructions. Total RNAs were dissolved in 50 μl of RNase-free water and stored at −80°C. The final concentration and quality of the RNA samples were estimated either spectrophotometrically or by agarose gel electrophoresis with ethidium bromide staining. Total RNAs were treated with RNase-free DNase (1 U/μg of total RNA; Turbo DNA-free; Ambion) for 30 min at 37°C, and the reaction was stopped with DNase inactivation reagent. For primer extension experiments, 10 μg of total RNA was used with [γ32-P]dATP (GE Healthcare)-labeled oligonucleotide gerR PE (Table 2), deoxynucleoside triphosphate, and avian myeloblastosis virus reverse transcriptase (BRL) to prime cDNA synthesis as previously described (15). The reaction products were fractionated on 6 M urea-6% polyacrylamide gels, along with DNA sequencing reactions using pGC102 as the template primed with the same oligonucleotide.
GerR expression in E. coli.
The gerR coding region was amplified by PCR from B. subtilis chromosome with primers gerRcoding and gerRanti containing XbaI and XhoI sites, respectively. The 614-bp PCR product was cleaved with XbaI and XhoI restriction enzymes and ligated into pET28a(+) (Novagen) linearized with the same enzymes. The recombinant plasmid named pGC109 carried an in-frame fusion of the histidine tag to the 3′ end of the gerR coding region expressed under the transcriptional control of the T7 promoter. This plasmid was used to transform competent cells of E. coli C43(DE3) derived from E. coli BL21(DE3) [E. coli F− ompT hsdSB(rB− mB−) gal dcm (DE3)] as described by Miroux and Walker (13). This strain has at least one uncharacterized mutation, which prevents cell death, which is associated with the expression of toxic recombinant proteins. The recombinant strain was grown in TY medium supplemented with kanamycin (20 μg/ml) to an optical density of 0.4 at 600 nm. The T7 promoter was induced by adding isopropyl-β-d-thiogalactopyranoside (IPTG) (final concentration of 0.2 mM) to the culture incubated for 2 h at 37°C. The six-His-tagged GerR protein was purified under denaturing conditions via Ni-nitrilotriacetic acid affinity chromatography as recommended by the manufacturer (Qiagen Inc.). Purified GerR protein was utilized to obtain polyclonal antibodies in rabbits (Primm, Treviso, Italy).
Western blot.
Sporulation of wild-type and mutant strains (GC260 gerR::neo and BK541 spoIIID::erm) was induced by the exhaustion method (4). Sporulating cell samples were harvested at various times during sporulation and sonicated in 25 mM Tris (pH 7.5), 0.1 M NaCl, 1 mM EDTA, 15% (vol/vol) glycerol, and 0.1 mg/ml of phenylmethysulfonyl fluoride (PMSF). Sonicated cells were centrifuged at 10,000 rpm for 20 min, the supernatants were recovered, and the total protein concentration was determined as described above. Samples (25 μg) of total proteins were fractionated on 15% denaturing polyacrylamide gels, electrotransferred to nitrocellulose filters (Bio-Rad), and used for Western blot analysis following standard procedures. GerR-specific antibody was used at a working dilution of 1:5,000, and an horseradish peroxidase (HRP)-conjugated anti-rabbit secondary antibody was used (Santa Cruz). Western blot filters were visualized by the SuperSignal West Pico chemiluminescence (Pierce) method as specified by the manufacturer.
Chromatin immunoprecipitation (ChIP).
Five hours after resuspension of PY79 cells in Sterlini-Mandelstam medium at 37°C, cross-links were generated by treatment with formaldehyde (1% final concentration) for 30 min. The rest of the procedure was performed by the method of Molle et al. (14), modified as described below. After the samples were precleared, they were centrifuged, and GerR-DNA complexes were immunoprecipitated for 90 min at room temperature with rabbit antibody to GerR (working dilution of 1:50). The formaldehyde-induced cross-links were reversed by incubation at 65°C overnight, and DNA purification was performed by NucleoSpin extract II kit (Macherey-Nagel) for PCR cleanup. PCR was performed with Taq DNA polymerase (Invitrogen) using serial dilutions of the immunoprecipitate and the total DNA control as the template. Oligonucleotide primers were typically 18 to 20 bases long and amplified a 200- to 400-bp product in the promoter region of cotU (chip cotUs/chip cotU1 amplify a 243-bp product), spoVIF (yjczs/chip spoVIF1 amplify a 154-bp product), gerE (chip gerEs/chip gerE1 amplify a 420-bp product), cotB (cotBps/chip cotB amplify a 245-bp product), and ftsH (ftsHs/ftsHa amplify a 184-bp product) (negative control of the experiment). All primers are listed in Table 2.
RESULTS
The gerR gene of B. subtilis is under σE-SpoIIID control.
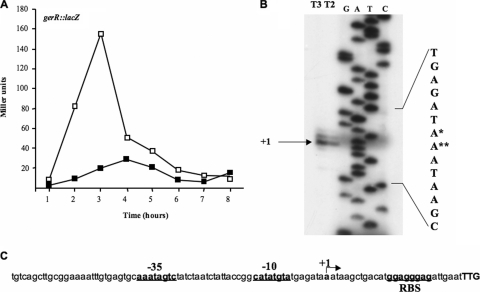
A previous DNA microarray analysis performed with a σK mutant strain of B. subtilis has indicated that gerR transcription is driven by σE-containing RNA polymerase (5). To analyze gerR expression in more detail, we constructed a transcriptional gene fusion between the 5′ region of gerR and the coding sequence of the Escherichia coli lacZ gene. We measured β-galactosidase activity at various times after the onset of sporulation in an otherwise wild-type strain and a collection of congenic sporulation mutants. In all cases, the fusion was integrated at the gerR locus as the result of a single, reciprocal crossover event (see Materials and Methods). In agreement with the expectation that gerR is under the control of σE, we observed that β-galactosidase activity commenced 1 h after the onset of sporulation (T1), reached its maximum after 3 h (T3), and then rapidly decreased (Fig. 1 A, open squares). Moreover, expression of gerR::lacZ was not detected in a σE mutant (spoIIG; data not shown) but was normal in mutants known to be impaired in σK or GerE production (spoIVB::erm mutant and gerE36 mutant, respectively) (data not shown). gerR::lacZ-driven synthesis of β-galactosidase was also impaired in a spoIIID mutant (Fig. 1A, closed squares), indicating that efficient transcription of gerR requires the presence of the spoIIID-encoded DNA-binding protein. On the basis of these data, we infer that gerR is transcribed by σE-containing RNA polymerase acting in conjunction with SpoIIID.
FIG. 1.
(A) gerR-directed β-galactosidase synthesis in an otherwise wild-type strain (open squares) and in a spoIIID null background (closed squares). Samples were collected at various times after the onset of sporulation. Enzyme activity is expressed in Miller units. Data are the means of three independent experiments. (B) Primer extension analysis of the gerR promoter region performed with total RNA extracted from sporulating cells 2 and 3 h after the onset of sporulation (T2 and T3). Primer extension and sequencing reactions were primed with the synthetic oligonucleotide gerR PE (Table 2). Although this oligonucleotide primed the sequence of the template strand, the sequence of the coding strand is shown for consistency with panel C. The single and double asterisks indicate the more and less frequently used transcriptional start point, respectively. (C) gerR promoter region. The translational start site (TTG) is shown in boldface capital letters, the transcriptional start site is indicated as +1, and the putative promoter and ribosomal binding site (RBS) sequences are underlined boldface lowercase letters.
A primer extension experiment was performed to map the gerR transcriptional start site. The extension product obtained (Fig. 1B) allowed us to localize the 5′ terminus of gerR mRNA 27 bp upstream of the beginning of the open reading frame (ORF) (Fig. 1C). Sequences upstream of the 5′ terminus (+1) resembled the conserved features of a σE promoter, matching the −10 consensus sequence at five positions [the −10 consensus sequence is CATA-(a/g)-T, and the gerR sequence is CATATGTA (a hyphen indicates any base, and lowercase letters and slashes indicate base options)] and matching the −35 consensus sequence at five positions [the −35 consensus sequence is (g/t)(c/a)ATA(t/a)(t/a)(c/t), and the gerR sequence is AAATAGCT] (18). We also observed that the sequence 5′-GCAAATAGTC-3′, which partially overlaps the −35 region of the gerR promoter, matches the proposed binding site for the SpoIIID protein (6). A partial overlap between the −35 region and SpoIIID binding site has been previously reported for the sigK and spoIVCA promoters (6).
Phenotype of a gerR null mutant.
To analyze in more detail the role of GerR on late sporulation gene expression, we constructed a gerR null mutation as described in Materials and Methods to yield strain GC260. A copy of the gerR gene, complete with the promoter region, was inserted ectopically at the amyE locus of strain GC260, yielding strain GC284. While these strains did not show differences in their sporulation efficiency with respect to their isogenic wild-type strain (PY79) (data not shown), a 10-fold reduction in the number of viable spores after lysozyme treatment was observed with strain GC260 compared with the wild type and the complemented strain (data not shown). A similar reduced resistance to lysozyme of spores of a gerR mutant was previously reported (12). Also in agreement with a previous report (5), our data show that a strain with a gerR null mutation had a strongly reduced germination efficiency compared with the wild-type and the complemented strain when l-alanine was used to induce germination (not shown).
As previously reported (12), in a strain carrying a gerR null mutation, the synthesis or incorporation of several coat proteins is affected. When a copy of the gerR gene was expressed ectopically in the gerR null mutant, a wild-type pattern of spore coat proteins was observed, indicating an apparently complete complementation of the mutant phenotype (not shown).
gerR is needed for normal expression of cotB, cotC, cotG, and cotU.
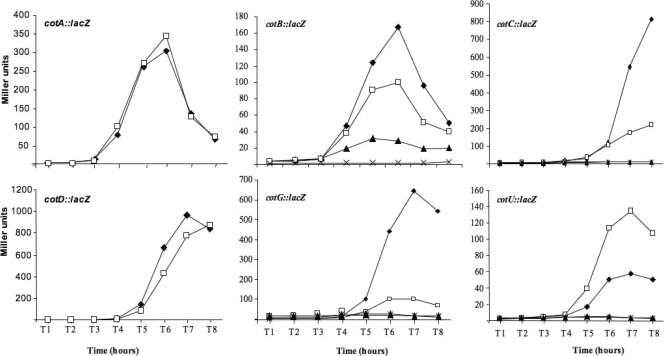
To analyze the effect of GerR on late sporulation genes, we used a collection of strains carrying transcriptional fusions of cotA, cotB, cotC, cotD, cotG, or cotU genes to the lacZ gene of E. coli. Chromosomal DNA was extracted from strains carrying the gene fusions and used to transform strain GC260 (gerR::neo) or GC261 (gerE36 gerR::neo). As shown in Fig. 2, while the expression of cotA and cotD was not affected by the null mutation in gerR, the expression of cotB, cotC, and cotG appeared higher in the wild type than in the gerR null mutant. An opposite regulatory effect was observed for cotU, whose expression was higher in the gerR null mutant than in the wild-type strain (Fig. 2). From those results, we can conclude that gerR expression has a positive role on cotB, cotC, and cotG and a negative role on cotU. The effect of gerR on the three positively controlled genes seems, however, not always the same. Expression of cotC and cotG is totally dependent on the presence of gerE and only partially dependent on gerR, while the expression of cotB is partially dependent on both gerE and gerR. In the latter case, the role of the two transcriptional factors, GerE and GerR, is somehow synergistic, with a total absence of expression in a strain carrying null mutations in gerE and gerR (Fig. 2).
FIG. 2.
β-Galactosidase activity in strains carrying the indicated gene fusions. β-Galactosidase activity in wild-type background (closed diamonds), gerR null background (open squares), gerE null background (closed triangles), and gerR gerE null background (multiplication symbols) was examined. Samples were collected at various times after the onset of sporulation. Enzyme activity is expressed in Miller units. Data are the means of three independent experiments. Fusions cotA::lacZ and cotD::lacZ were analyzed only in the wild-type and gerR null backgrounds, since the expression of both genes is known to be GerE independent.
GerR effect on GerE.
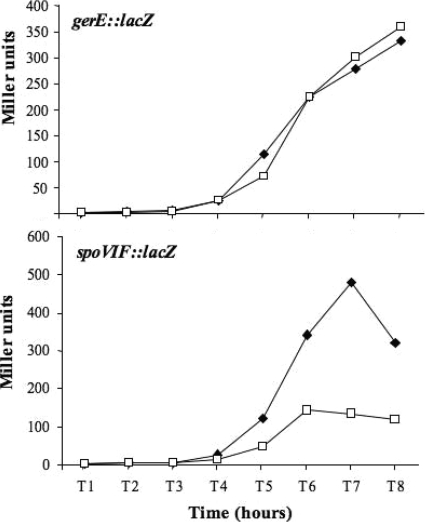
The expression of the late sporulation genes previously identified as controlled by GerR, cgeA, cotG, and cotY (12), as well as the expression of cotB, cotC, and cotU (Fig. 2), is strongly dependent on GerE (7, 8). This raises the possibility that GerR acts on those genes indirectly through GerE. To evaluate this possibility, we first analyzed a gerE::lacZ translational gene fusion in an otherwise wild-type strain and in a isogenic strain carrying a gerR null mutation. As shown in the top panel of Fig. 3, the gerE-driven production of β-galactosidase was identical in the wild type and in the gerR-defective background, clearly indicating that GerR does not affect the expression of the gerE gene.
FIG. 3.
β-Galactosidase activity in strains carrying the indicated gene fusions in otherwise wild-type strain (closed squares) and in a gerR null background (open squares). Samples were collected at various times after the onset of sporulation. Enzyme activity is expressed in Miller units. Data are the means of three independent experiments.
It has been previously reported that GerE fails to accumulate in the sporulating cells of a strain carrying a null mutation in the spoVIF gene (10, 11). Since GerE is normally synthesized in the spoVIF mutant, a positive posttranslational effect of SpoVIF on GerE has been proposed (10, 11). Since the SpoVIF structural gene is σK dependent and GerE independent (10, 11), we hypothesized that GerR indirectly controls GerE activity by affecting the expression of the spoVIF gene. Therefore, we constructed a transcriptional spoVIF::lacZ gene fusion (Materials and Methods) and monitored the spoVIF-driven production of β-galactosidase in time course experiments in the wild type and an isogenic strain carrying a gerR null mutation. As shown in the bottom panel of Fig. 3, spoVIF expression was significantly lower in the gerR mutant than in the congenic wild-type strain.
These results indicate that while GerR does not control the expression of gerE, it positively controls the expression of spoVIF whose product regulates the level of GerE accumulation in the mother cell (10, 11). On the basis of these results, we conclude that GerR acts indirectly on GerE and that its role on at least some of the GerR-controlled cot genes could be indirect and mediated by GerE.
GerR directly regulates spoVIF, cotB, and cotU.
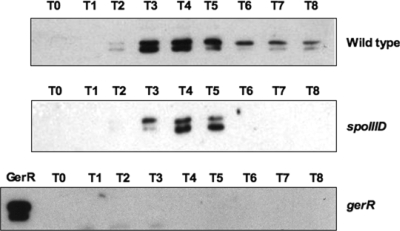
While the action of GerR on cotB, cotC, and cotG is likely to be indirect and mediated by GerE, its action on cotU has to be independent from GerE, since cotU is positively regulated by GerE (8) and negatively regulated by GerR (Fig. 2). This raises the possibility that the effect of GerR on cotU could be direct or mediated by a transcriptional factor other than GerE. A direct effect of GerR on late sporulation genes would imply that this protein, whose gene is expressed at maximal levels 3 h after the onset of sporulation and is then drastically reduced (Fig. 1A), is still present and active several hours later (Fig. 2). We decided to verify the presence of GerR during sporulation, and to this aim, we raised an anti-GerR antibody. A His6-tagged version of the gerR gene was overexpressed in E. coli, and the protein was partially purified by Ni2+-affinity chromatography used to immunize rabbits as described in Materials and Methods. Western blot analysis was performed on proteins extracted from wild-type and isogenic strains carrying null mutations in gerR or spoIIID. Anti-GerR antibody recognized two protein bands with apparent molecular masses of 25 and 28 kDa (Fig. 4). Both bands were not present in a strain carrying a null mutation in gerR and were partially dependent on spoIIID. They appeared 2 h after the onset of sporulation, peaked between 3 and 4 h after the onset of sporulation, and then decreased, consistent with the gene expression profile shown in Fig. 1A. The two forms of GerR are probably due to an additional start codon; we noticed that codons 15 and 21 are both potential start codons with putative ribosome binding sites (not shown). In a wild-type strain, although GerR was present at a lower level than the levels 3 and 4 h after the onset of sporulation, it was present in sporulating cells at least up to 8 h after the beginning of sporulation (Fig. 4), which is consistent with a possible direct regulatory role of GerR on the expression of late sporulation genes.
FIG. 4.
Western blot analysis performed with an anti-GerR antibody on extracts of cells collected at various times after the onset of sporulation. The GerR lane of the bottom panel was loaded with proteins extracted 4 h after the onset of sporulation (T4) from a wild-type strain. In all experiments, 25-μg samples of total proteins were fractionated on 15% polyacrylamide gels, electrotransferred to nitrocellulose membranes, reacted first with primary antibodies and then with peroxidase-conjugated secondary antibodies, and visualized by the enhanced chemiluminescence method.
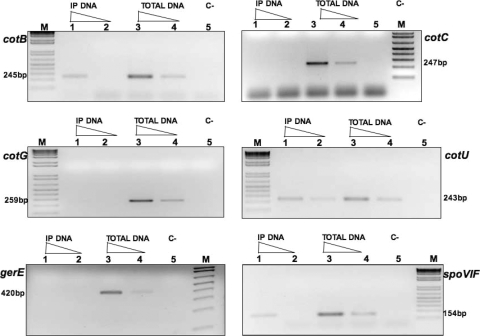
To verify whether GerR was able to bind to the promoter region of GerR-controlled genes, we decided to perform ChIP experiments. In vivo cross-linking of proteins to DNA was induced by adding formaldehyde to sporulating cells (Materials and Methods), and the anti-GerR antibody was then used to immunoprecipitate GerR and DNA fragments bound to it. Tenfold serial dilutions of immunoprecipitated and total input DNA were used as the template in PCR amplification reactions primed by oligonucleotides annealing to the promoter region of cotB, cotC, cotG, cotU, gerE, or spoVIF. For a negative control, we also used an oligonucleotide annealing to the promoter region of the ftsH gene, a vegetative gene of B. subtilis unrelated to GerR. As shown in Fig. 5, we observed an amplification product using immunoprecipitated (IP) DNA as a template, when the reaction was primed with oligonucleotides specific for cotB, cotU, or spoVIF. In contrast, we did not detect any amplification product when the reaction was primed with oligonucleotides specific for cotC, cotG, gerE (Fig. 5), or ftsH (not shown).
FIG. 5.
PCR amplification products separated on agarose gels. Templates used for PCRs were chromosomal DNAs extracted from sporulating cells 5 h after the onset of sporulation and treated with formaldehyde to cross-link protein to DNA. The DNA obtained was either immunoprecipitated with anti-GerR antibody (IP DNA) or directly used in PCR (Total DNA). The amount of DNA in lanes 1 to 4 is indicated by the height of the triangle over the lane. In all panels, lanes 2 and 4 show the results of PCR with DNA diluted 10-fold with respect to lanes 1 and 3. Lanes 5 show the results obtained by PCR without chromosomal DNA (C−) as a template. The M lanes were loaded with 1 kb plus ladder (Invitrogen) as molecular size marker. The size of the expected amplification product is indicated for each panel.
These results indicate that GerR binds to the promoter regions of the cotB, cotU, and spoVIF genes, presumably directly regulating their expression. The promoter regions of the other two GerR-controlled genes, cotC and cotG, are not bound by GerR, at least under our experimental conditions. We hypothesize that GerR acts on their expression in an indirect way, through SpoVIF. spoVIF expression is directly controlled by GerR (Fig. 5) which, in turn, positively acts on GerE at a posttranscriptional level (10, 11).
DISCUSSION
An initial result of this work was the transcriptional characterization of the gerR gene. We confirmed that the gene belongs to the σE regulon and demonstrated the occurrence of a previously undetected requirement of the transcriptional factor SpoIIID for full gerR expression.
We also analyzed the effects of GerR on several late sporulation genes. The analysis of a collection of strains carrying transcriptional fusions of late sporulation genes to lacZ showed that GerR is needed for normal expression of cotB, cotC, cotG, cotU, and spoVIF, but not for normal expression of cotA, cotD, and gerE. The effect of GerR on these genes was not always the same. While GerR is critical for the expression of cotC, cotG, and spoVIF, its positive effect on cotB expression was limited. A completely different situation was observed with cotU, whose expression is negatively regulated by GerR.
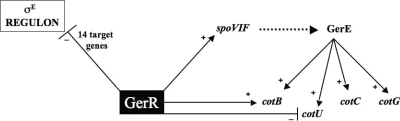
The role of GerR on late sporulation genes has been hypothesized to be indirect and mediated by other transcriptional factors (12). To address this issue, first of all, we confirmed, by Western blotting with an anti-GerR antibody, that GerR was still present in the mother cell cytoplasm 8 h after the onset of sporulation, when the late sporulation gene are expressed. Then we used a ChIP approach to check whether GerR was able to bind in vivo to the promoter regions of the controlled genes. Our results indicated that GerR binds to the promoter regions of cotB, cotU, and spoVIF, but not to the promoter regions of cotC and cotG. On the basis of these results, we propose that GerR acts on late sporulation genes in two ways: (i) directly by binding to the promoter regions of cotB, cotU, and spoVIF; and (ii) indirectly, on cotC and cotG, through SpoVIF and GerE (Fig. 6). The spoVIF gene codes for a protein needed for the accumulation of GerE in the mother cell cytoplasm (11). In a strain carrying a spoVIF null mutation, the gerE gene is normally expressed, but its product fails to accumulate in the cell cytoplasm (11). On the basis of this observation, we propose that GerR directly promotes spoVIF transcription, therefore positively affecting GerE accumulation and its transcriptional role on late sporulation genes, such as cotC and cotG (Fig. 6).
FIG. 6.
Schematic representation of the action of GerR on the analyzed genes. Continuous arrows and lines indicate a positive or a negative transcriptional effect, respectively. The dotted arrow indicates a positive effect at the posttranslational level (10, 11).
In the cases of cotC and cotG, expression of the two genes seems to be strictly dependent on the presence of GerE. In a gerR mutant, the minimal amount of GerE present in the cell because of the reduced expression of spoVIF (11), is most likely responsible for the strong transcriptional effect on cotC and cotG. Expression of cotB needs both GerE and GerR. As shown in Fig. 2, cotB expression is totally abolished only when both transcriptional activators are missing. In the case of cotU, the two transcriptional factors have opposite effects with GerE acting as an activator and GerR acting as a repressor. However, in a gerR mutant, where the repressor is absent and only a reduced amount of activator is present, cotU expression is significantly higher than in a wild-type strain.
In conclusion, our data reinforce the view that in the mother cell, the programs of gene expression driven by σE and σK are tightly connected to each other by the action of various transcriptional factors. GerR is one such factor, which controls genes of both σE and σK regulons, acting alone on some genes (5) and in conjunction with SpoIIID (5) or GerE on others, and modulates GerE function through a direct control on the expression of spoVIF, whose product positively affects GerE action.
Acknowledgments
We thank L. Di Iorio for technical assistance.
This work was supported by a grant (KBBE-2007-207948) from the EU 7th Framework to E.R.
Footnotes
Published ahead of print on 30 April 2010.
REFERENCES
- 1.Cosmina, P., F. Rodriguez, F. de Ferra, G. Grandi, M. Perego, G. Venema, and D. van Sinderen. 1993. Sequence and analysis of the genetic locus responsible for surfactin synthesis in Bacillus subtilis. Mol. Microbiol. 8:821-831. [DOI] [PubMed] [Google Scholar]
- 2.Cutting, S., and J. Mandelstam. 1986. The nucleotide sequence and the transcription during sporulation of the gerE gene of Bacillus subtilis. J. Gen. Microbiol. 132:3013-3024. [DOI] [PubMed] [Google Scholar]
- 3.Cutting, S., S. Panzer, and R. Losick. 1989. Regulatory studies on the promoter for a gene governing synthesis and assembly of the spore coat in Bacillus subtilis. J. Mol. Biol. 207:393-404. [DOI] [PubMed] [Google Scholar]
- 4.Cutting, S., and P. B. Vander Horn. 1990. Genetic analysis, p. 27-74. In C. Harwood and S. Cutting (ed.), Molecular biological methods for Bacillus. John Wiley and Sons, Chichester, United Kingdom.
- 5.Eichenberger, P., M. Fujita, S. T. Jensen, E. M. Conlon, D. Z. Rudner, S. T. Wang, C. Ferguson, K. Haga, T. Sato, J. S. Liu, and R. Losick. 2004. The program of gene transcription for a single differentiating cell type during sporulation in Bacillus subtilis. PLoS Biol. 2:e328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Halberg, R., and L. Kroos. 1994. Sporulation regulatory protein SpoIIID from Bacillus subtilis activates and represses transcription by both mother-cell-specific forms of RNA polymerase. J. Mol. Biol. 243:425-436. [DOI] [PubMed] [Google Scholar]
- 7.Henriques, A. O., and C. P. Moran, Jr. 2007. Structure, assembly and function of the spore surface layers. Annu. Rev. Microbiol. 61:555-588. [DOI] [PubMed] [Google Scholar]
- 8.Isticato, R., A. Pelosi, R. Zilhão, L. Baccigalupi, A. O. Henriques, M. De Felice, and E. Ricca. 2008. CotC-CotU heterodimerization during assembly of the Bacillus subtilis spore coat. J. Bacteriol. 190:1267-1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8a.Itaya, M., K. Kondo, and T. Tanaka. 1989. A neomycin resistance gene cassette selectable in a single copy state in the Bacillus subtilis chromosome. Nucleic Acids Res. 12:4410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kunkel, B., L. Kroos, H. Poth, P. Youngman, and R. Losick. 1989. Temporal and spatial control of the mother-cell regulatory gene spoIIID of Bacillus subtilis. Genes Dev. 3:1735-1744. [DOI] [PubMed] [Google Scholar]
- 10.Kuwana, R., S. Yamamura, H. Ikejiri, K. Kobayashi, N. Ogasawara, K. Asai, Y. Sadaie, H. Takamatsu, and K. Watabe. 2003. Bacillus subtilis spoVIF (yjcC) gene, involved in coat assembly and spore resistance. Microbiology 149:3011-3021. [DOI] [PubMed] [Google Scholar]
- 11.Kuwana, R., H. Ikejiri, S. Yamamura, H. Takamatsu, and K. Watabe. 2004. Functional relationship between SpoVIF and GerE in gene regulation during sporulation of Bacillus subtilis. Microbiology 150:163-170. [DOI] [PubMed] [Google Scholar]
- 12.Kuwana, R., T. Okumura, H. Takamatsu, and K. Watabe. 2005. The ylbO gene product of Bacillus subtilis is involved in the coat development and lysozyme resistance of spore. FEMS Microbiol. Lett. 242:51-57. [DOI] [PubMed] [Google Scholar]
- 13.Miroux, B., and J. E. Walker. 1996. Over-production of proteins in Escherichia coli: mutant hosts that allow synthesis of some membrane proteins and globular proteins at high levels. J. Mol. Biol. 260:289-298. [DOI] [PubMed] [Google Scholar]
- 14.Molle, V., Y. Nakaura, R. P. Shivers, H. Yamaguchi, R. Losick, Y. Fujita, and A. L. Sonenshein. 2003. Additional targets of the Bacillus subtilis global regulator CodY identified by chromatin immunoprecipitation and genome-wide transcript analysis. J. Bacteriol. 185:1911-1922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Naclerio, G., L. Baccigalupi, R. Zilhao, M. De Felice, and E. Ricca. 1996. Bacillus subtilis spore coat assembly requires cotH gene expression. J. Bacteriol. 178:4375-4380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nicholson, W. L., and P. Setlow. 1990. Sporulation, germination and outgrowth, p. 391-450. In C. Harwood and S. Cutting (ed.), Molecular biological methods for Bacillus. John Wiley and Sons, Chichester, United Kingdom.
- 17.Ricca, E., S. Cutting, and R. Losick. 1992. Characterization of bofA, a gene involved in intercompartmental regulation of pro-sigma K processing during sporulation in Bacillus subtilis. J. Bacteriol. 174:3177-3184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Roels, S., A. Driks, and R. Losick. 1992. Characterization of spoIVA, a sporulation gene involved in coat morphogenesis in Bacillus subtilis. J. Bacteriol. 174:575-585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 20.Stragier, P., and R. Losick. 1996. Molecular genetics of sporulation in Bacillus subtilis. Annu. Rev. Genet. 30:297-341. [DOI] [PubMed] [Google Scholar]
- 21.Wang, S. T., B. Setlow, E. M. Conlonc, J. L. Lyond, D. Imamura, T. Sato, P. Setlow, R. Losick, and P. Eichenberger. 2006. The forespore line of gene expression in Bacillus subtilis. J. Mol. Biol. 358:16-37. [DOI] [PubMed] [Google Scholar]
- 22.Youngman, P., J. B. Perkins, and R. Losick. 1984. A novel method for the rapid cloning in Escherichia coli of Bacillus subtilis chromosomal DNA adjacent to Tn917 insertion. Mol. Gen. Genet. 195:424-433. [DOI] [PubMed] [Google Scholar]