Abstract
A genetically homogenous bacterial population may contain physiologically distinct subpopulations. In one such case, a minor part of an otherwise antibiotic-sensitive bacterial population maintains a nondividing state even in a growth-supporting environment and is therefore not killed by bactericidal antibiotics. This phenomenon, called persistence, can lead to failure of antibiotic treatment. We followed the development of sensitivity to killing by ampicillin and norfloxacin when Escherichia coli cells were transferred from a stationary-phase culture into fresh growth medium. In parallel, we monitored growth resumption by individual bacteria. We found that bacteria in a population resumed growth and became sensitive to antibiotics at different times after transfer to fresh medium. Moreover, both growing and dormant bacteria coexisted in the same culture for many hours. The kinetics of awakening was strongly influenced by growth conditions: inocula taken from the same stationary-phase culture led to very different persister frequencies when they were transferred into different fresh media. Bactericidal antibiotics kill cells that have woken up, but the later-awakening subpopulation is tolerant to them and can be identified as persisters when the antibiotic is removed. Our observations demonstrate that persister count is a dynamic measure and that the persister frequency of a particular culture is not a fixed value.
A clonal population of microbial cells, although genetically uniform, can have several different phenotypes coexisting in the same culture. Examples of phenotypic heterogeneity include genetic competence (32) and sporulation (30) in Bacillus subtilis, induction of galactose utilization in Saccharomyces cerevisiae (2), and lactose operon activation by a nondegradable inducer in Escherichia coli (27). Bacterial persistence also belongs to this category; persisters are antibiotic-tolerant cells in an otherwise susceptible population (20). They can survive antibiotic treatment and give rise to a population that is again mostly sensitive to such treatment.
Persisters were first described over 65 years ago (3), but their identity has remained obscure. In cultures of wild-type E. coli, they are reported to be present at low frequencies (10−4 to 10−6) (20), but estimates vary. A single-cell study revealed that persisters do not grow before antibiotic treatment, even in a growth-supporting environment (1), as was conjectured in the first description of persisters (3). This may explain their tolerance to bactericidal antibiotics, which do not usually kill nongrowing bacteria. Persisters are mainly formed during stationary phase, as prolonged cultivation in logarithmic phase eliminates them from the population (16).
The molecular mechanism of persister formation is not well understood. Several studies have suggested the importance of toxin-antitoxin (TA) modules in this process. Indeed, elevated levels of mRNAs for several TA modules have been observed in persisters, and TA overexpression can induce persister-like phenotypes (17). In addition, a hipA7 mutant has been described that forms persisters with a higher frequency (10−2 to 10−3) than that of the wild type (25). It was subsequently determined that hipA encodes the toxin component of a TA module (10). However, no single TA module is solely responsible for persister formation, as no single knockout abolishes it (15, 17). Persister formation can also be affected by other genes—deletion of phoU (21) or relA (18) decreases the frequency of persisters, while overexpression of several toxic proteins has been shown to increase it (34). Nevertheless, recent genome-wide screens for persistence genes have failed to point to a single specific pathway (5, 14).
According to the current consensus in the field, persisters are formed at low frequencies during the late-logarithmic and stationary phases and constitute only a minor fraction of stationary-phase cells (20). While normal cells are believed to resume growth and become susceptible to antibiotics immediately after transfer into fresh medium, persisters remain dormant longer (28). Here we demonstrate that heterogeneous growth resumption is an intrinsic property of the whole E. coli population. We propose that persisters do not form a distinct subpopulation in stationary phase but that their number reflects wake-up kinetics.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth media.
E. coli strain MG1655 (4) was used in all experiments. Plasmid pETgfp mut2 AGGAGG (3) carries a GFP mut2 gene under the control of an IPTG (isopropyl-β-d-thiogalactopyranoside)-inducible tac promoter and a kanamycin resistance gene (35). Cells were grown in LB (Difco Laboratories) or morpholinepropanesulfonic acid (MOPS) medium (26), with the latter supplemented with either 0.1% glucose (MOPS Glc) or 0.2% glycerol (MOPS Gly). The media also contained kanamycin (25 μg/ml) for plasmid retention.
Persistence assay.
Cells were grown to stationary phase in MOPS Glc (3 to 5 days), and 1:100 dilutions (1:1,000 dilutions in the case of the experiments for Fig. 1B) were made in fresh medium (LB, MOPS Glc, or MOPS Gly) containing ampicillin (Amp) (200 μg/ml; Actavis) or norfloxacin (5 μg/ml; Sigma). Samples were withdrawn at the indicated times, diluted in sterile phosphate-buffered saline (PBS) (11), and plated on LB-kanamycin plates (25 μg/ml; Amresco). Colonies were counted on the following day, after overnight incubation at 37°C.
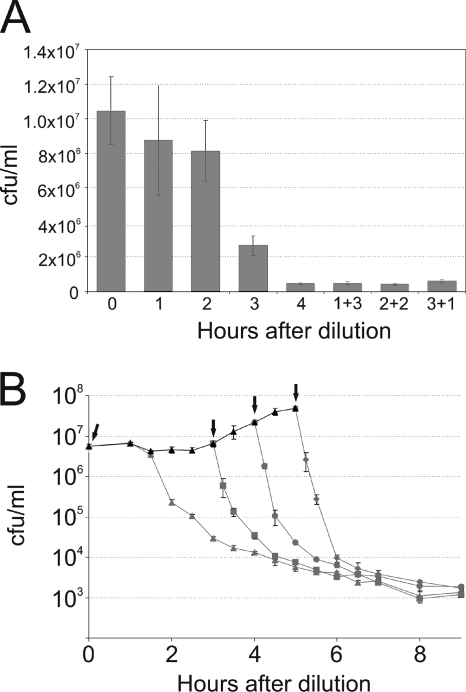
FIG. 1.
The extent of cell death depends on the time spent in fresh medium. (A) Stationary-phase cells were diluted in fresh MOPS Glc medium containing Amp, and the number of live cells was determined by plate counts after each hour for 4 h (bars 1, 2, 3, and 4). Dilutions were also made in Amp-free medium, with Amp added after 1 (1 + 3), 2 (2 + 2), or 3 (3 + 1) h. The cell counts of these cultures were determined by plate counts 4 h after dilution. Standard errors were calculated from the results of at least three independent experiments and are indicated by error bars. (B) Stationary-phase cells were diluted in fresh MOPS Glc medium, and their growth was monitored (black triangles). Aliquots were taken 0 (gray triangles), 3 (gray squares), 4 (gray circles), and 5 (gray diamonds) hours after dilution (indicated by arrows), and Amp was added to the aliquots. The numbers of surviving cells were determined by plate counts. Standard errors were calculated from the results of at least three independent experiments and are indicated by error bars.
Growth resumption.
Cells were grown in the presence of inducer (1 mM IPTG) so that in stationary phase all cells expressed high levels of green fluorescent protein (GFP). Stationary-phase cultures were diluted 1:100 in fresh medium without inducer, and 30-μl samples were withdrawn at the indicated times. The samples were mixed with an equal volume of 30% glycerol in PBS and stored at −70°C pending analysis. After dilution with sterile PBS, the samples were analyzed on a flow cytometer (LSR II; BD Biosystems). The software packages FACSDiva and FloJo were used to analyze the data.
RESULTS
Time spent in fresh medium is critical for ampicillin-induced cell death.
The existence of persisters becomes evident when cells are diluted from stationary-phase culture into fresh medium; this is required for the bulk of the population to resume growth and become susceptible to antibiotics. Under such conditions, the rate of cell death depends on both the action of the antibiotic (the time it takes to kill a susceptible cell) and the time necessary for a stationary-phase cell to resume growth and become susceptible (to wake up). To determine the exact contributions of these two factors to the apparent time-dependent killing, we used MOPS-based minimal medium to achieve a better time resolution than that obtained with rich LB medium, in which the decrease in CFU is rapid.
Previously, antibiotics have usually been applied to a culture some time after transfer to fresh medium and incubated for several hours to determine the frequency of persisters. In such cultures, the time course of awakening is obscured by the initial proliferation, as the fraction of sensitive bacteria contains both cells that became vulnerable just before or even during antibiotic treatment and the progeny of those bacteria that resumed growth immediately after dilution. In our experiments, we subjected bacteria to antibiotics immediately upon transfer to the growth-supporting medium.
Stationary-phase E. coli cells grown in MOPS Glc were diluted in fresh MOPS Glc medium containing ampicillin (Amp), and samples were withdrawn every hour for CFU measurement. Dilutions were also made in MOPS Glc without Amp, with the antibiotic added later, so the cultures had been subjected to different Amp treatment times (1, 2, and 3 h) when they were sampled 4 h after dilution.
Cells were progressively killed by Amp during the 4-h period, and only ∼5% remained alive at the end of the experiment (Fig. 1A). The extent of killing was the same for all cultures at this time point, regardless of the amount of time for which the Amp was present: 4 h with Amp led to the same number of colonies as 3 h without plus 1 h with Amp. This nontrivial result shows that the time spent in fresh medium determines the number of Amp-susceptible cells and suggests that wake-up speed is the limiting factor in the kinetics of cell death.
This experiment also demonstrates that 1 h is enough for Amp to kill all susceptible cells and that the presence of Amp does not intervene in the wake-up process. This result conflicts with the one described by Balaban and coworkers (12). In their hands, the immediate addition of Amp resulted in fewer persisters than those in cultures to which Amp was added after some time. Since they used a different E. coli strain (HM22, the hipA7 mutant), different medium (LB), and different persister detection method (most probable number [MPN] in liquid culture), the results are not directly comparable.
The same conclusion can be drawn from Fig. 1B, for which we determined the killing curves after adding Amp to the culture at different times. A stationary-phase culture in MOPS Glc was diluted in fresh MOPS Glc, and its growth was monitored by CFU measurements. At 0, 3, 4, and 5 h, an aliquot was removed from the culture and Amp was added to it (indicated by arrows).
The addition of Amp immediately after dilution caused relatively slow killing. In contrast, the addition of Amp after some time led to two-phase killing curves, with a rapid initial decrease in CFU followed by a slower decline. During the quick phase, the CFU counts dropped until reaching the level of the culture with Amp added immediately after dilution. The slow killing phase then followed, and a further decline occurred in similar fashion, regardless of the time of Amp addition. This suggests that after quick elimination of already growing cells, Amp-mediated cell death reflects growth resumption.
The small quantitative discrepancies between Fig. 1A and B are due to the high sensitivity of growth resumption rate to the length of stationary phase.
The frequency of persisters reflects wake-up kinetics.
If sensitivity to antibiotic-induced killing depends on bacterial awakening from dormancy, then the number of persisters should depend on the growth resumption potency of the medium.
To test this experimentally, we grew E. coli cells to stationary phase in MOPS Glc and diluted the culture in three different Amp-containing media: LB, MOPS Glc, and MOPS Gly. If persisters are predetermined in stationary phase as a distinct subpopulation, then all three cultures should have displayed equal numbers of persisters. Alternatively, if the frequency of persisters reflects the kinetics of awakening, then each medium should have had an individual persister count.
The killing curves were different in the different media (Fig. 2A). While a 1-h Amp treatment in LB was enough to kill more than half of the cells, it took 6 h to reach the same level of killing in MOPS Gly. Incubation with Amp in MOPS Glc produced an intermediate level of killing during the first hours but caught up with that in LB later. Because dilutions were made from the same stationary-phase culture, the differences in persister frequency must have arisen from the properties of the different fresh media.
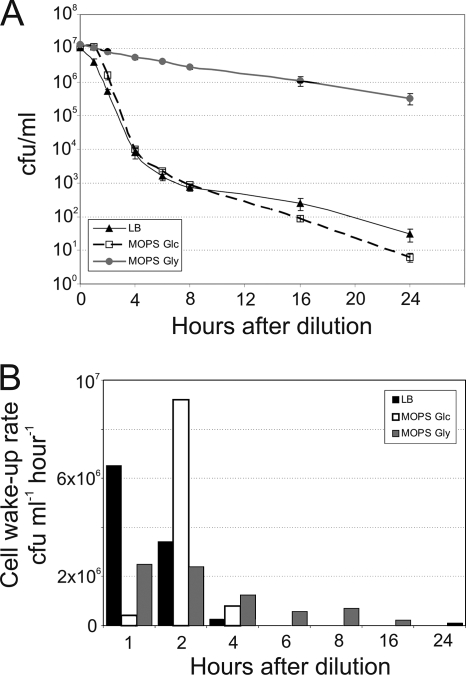
FIG. 2.
The number of persisters in E. coli depends on the growth medium. (A) Cells were grown to stationary phase in MOPS Glc and then diluted in LB, MOPS Glc, and MOPS Gly, all containing Amp. The numbers of surviving cells were determined by plate counts. Standard errors were calculated from the results of at least three independent experiments and are indicated by error bars. (B) The CFU/ml data from panel A were used to calculate bacterial wake-up rates (CFU ml−1 h−1).
The data in Fig. 2A allowed us to calculate the rates of appearance of Amp-sensitive cells, which we interpreted as wake-up rates, in different media (Fig. 2B). Exit from dormancy was the fastest in LB: the majority of cells woke up during the first hour in fresh medium. In MOPS Glc, the growth resumption was delayed and peaked during the second hour. Cells diluted in MOPS Gly woke up during a prolonged period. Delayed exit from dormancy is behind the ability to withstand antibiotic treatment better in poorer media.
To rule out the possibility that the slower killing in MOPS Gly was due to the slower growth in this medium, we measured the killing rates of logarithmic-phase cultures in all 3 media. The CFU half-life in Amp-treated LB was 4 min, that in MOPS Glc was 6 min, and that in MOPS Gly was 9 min. This shows that the Amp-mediated killing of logarithmic-phase cultures is significantly faster than the events shown in Fig. 2. This again, in addition to the data in Fig. 1, supports our notion that Amp-mediated killing during growth resumption is not limited by slow action of Amp but rather dominated by a process of making the cells vulnerable to Amp (exit from dormancy).
The dependency of wake-up speed on medium composition can be visualized directly by following cell divisions by use of a GFP dilution method (29). Cells containing a plasmid encoding GFP were grown to stationary phase in the presence of inducer, leading to the formation of bright green cells. Inducer was omitted from the fresh medium, and therefore no more GFP was synthesized after dilution. Every cell division led to a 2-fold reduction of cell GFP content, which could be observed at single-cell resolution by flow cytometry.
We grew E. coli strain MG1655 to stationary phase in MOPS Glc in the presence of inducer (IPTG) and followed its behavior after dilution in fresh LB, MOPS Glc, and MOPS Gly (all without Amp). Flow cytometry revealed different wake-up kinetics in the different media (Fig. 3A). The growing population with diminished GFP content was first visible in LB, followed by MOPS Glc, and it appeared last in MOPS Gly.
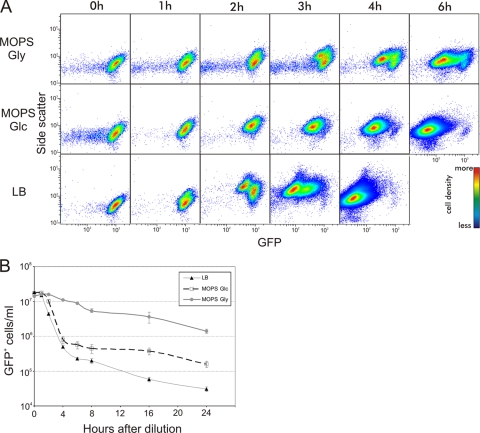
FIG. 3.
E. coli growth resumption after stationary phase is heterogeneous over time. (A) Stationary-phase cells carrying pETgfp mut2 AGGAGG (3) and filled with GFP were diluted in LB, MOPS Glc, or MOPS Gly, and samples for flow cytometry were taken at the times indicated. Vertical axis, side scatter; horizontal axis, GFP content. Dividing cells have elevated side scatter signals and reduced GFP content. (B) Stationary-phase cells filled with GFP were diluted in Amp-containing fresh media, and samples were taken at the indicated times for flow cytometry. Cells resuming growth were lysed and disappeared from the GFP-positive population. Standard errors were calculated from the results of at least three independent experiments and are indicated by error bars.
For prolonged surveillance of growth resumption, we performed the GFP dilution experiment in the presence of Amp. In this way, the growing cells were lysed and disappeared from the culture. A decrease in the number of GFP-positive cells could thus be monitored and growth resumption described over time without interference from the growing GFP-negative population. The growth resumption dynamics in different media are shown in Fig. 3B. Overall, they are very similar to the CFU measurements shown in Fig. 2A: cells woke up most quickly in LB and most slowly in MOPS Gly. Quantitative differences between the GFP dilution and CFU data can be explained by the existence of GFP-positive cells that did not resume growth during the experiment (29). These cells were invisible in CFU measurements but certainly contributed to the GFP-positive population. It is also possible that cells which had lost their viability (did not form colonies after plating) were still detectable as GFP-positive particles and were scored among the intact GFP-positive cells by flow cytometry. The results from GFP measurements accord well with the CFU dynamics and support the notion that differences in cell killing are determined by the wake-up speed.
The time-dispersed awakening observed led to considerable heterogeneity among the isogenic cells. This is shown in Fig. 3A—heterogeneity was manifested by the appearance of multiple subpopulations with different GFP contents (different numbers of cell divisions completed). The wide distribution of wake-up times led to the coexistence of dormant and growing cells over several hours.
Awakening-dependent killing by norfloxacin.
Amp and other β-lactams target growing cells by inhibiting peptidoglycan synthesis (36). There are other classes of bactericidal antibiotics with different modes of action. If growth resumption is critical for cells to become sensitive to antibiotics, then different antibiotics should give us similar results, regardless of their specific targets in the cells.
We tested this assumption by using norfloxacin, a bactericidal antibiotic that targets DNA gyrase (36). Stationary-phase bacteria were diluted in fresh medium containing norfloxacin, and the cultures were sampled for surviving bacteria for 8 h (Fig. 4). There were profound differences in cell death kinetics among LB, MOPS Glc, and MOPS Gly. As with Amp, the cells diluted in LB were killed most rapidly and those in MOPS Gly were the most tolerant to norfloxacin. There were small differences in death kinetics between ampicillin and norfloxacin, which can be explained by their different modes of action: Amp lyses cells rapidly (29), whereas the damage caused by norfloxacin can be reversed after removal of the drug (7).
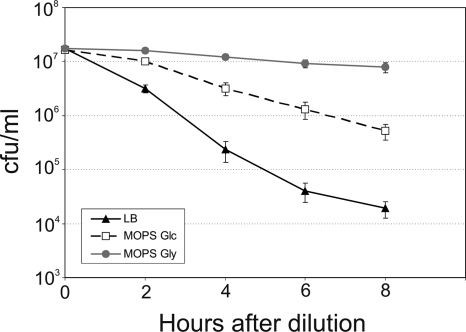
FIG. 4.
Killing by norfloxacin is dependent on growth resumption kinetics. Cells were grown to stationary phase in MOPS Glc and diluted in LB, MOPS Glc, and MOPS Gly, all containing norfloxacin. The numbers of surviving cells were determined by plating. Standard errors were calculated from the results of at least three independent experiments and are indicated by error bars.
DISCUSSION
Persisters are believed to be a survival form of nonsporulating bacteria formed in the stationary phase of growth (20). Indeed, stationary-phase cells are tolerant to most bactericidal antibiotics. Cells shifted back to growth conditions start to grow and become susceptible to antibiotics again. Here we show that the kinetics of the awakening process is the key to the persistence phenomenon. The number of cells identified as persisters depends on the time the cells have spent under growth-permitting conditions. The properties of fresh media have a strong influence on wake-up kinetics, and therefore the same stationary-phase culture can produce very different numbers of persisters if diluted in different media.
Differences in growth resumption might reflect a heterogeneity already existing during stationary phase. Indeed, stationary-phase E. coli cells can be separated by gradient centrifugation into several subpopulations (23), each of which could have its own growth resumption profile. The idea of the persister population consisting of various subpopulations with different physiologies in continuum has been proposed (37), and the existence of “deep” and “shallow” persisters with different sensitivities to antibiotics has been suggested (22). An alternative explanation, that cells are identical and wake up randomly, seems less likely, since the killing curves (Fig. 2A, LB and MOPS Glc) suggest the existence of at least two subpopulations with different wake-up rates.
The variance of the wake-up times might reflect the extent of damage accumulated by the cells during stationary phase (6). Cells with more accumulated damage may take longer for repair and then resume growth later. They might escape the antibiotic treatment just by chance, and therefore the heterogeneity in growth resumption might not be under positive selection. Alternatively, the phenotype could have been under selection because of its ecological significance. Exit from dormancy enables cell multiplication, but it also makes cells vulnerable to a dangerous environment. It is possible that E. coli has adapted to uncertain environments by diversification of its wake-up times, with some cells resuming growth rapidly while others stay in dormancy longer. This “bet-hedging” strategy ensures survival of the population in an uncertain environment (19, 33).
Exit from dormancy is a well-studied process in sporulating bacteria such as Bacillus subtilis. Various environmental signals can induce spore germination, and this is mediated by specific germination receptors (24, 31). Strains deficient in a particular receptor cannot germinate in response to the specific signal. Interestingly, germination of Bacillus spores also seems to be heterogeneous, as it is possible to isolate “superdormant” spores that do not germinate under normal conditions but require a high concentration of nutrients (13). They resemble late-awakening E. coli cells in their ability to stay dormant in a potentially growth-supporting environment.
Large heterogeneity in growth resumption times is also evident in microbes from natural environments. Epstein and colleagues incubated thousands of environmental samples under growth-supporting conditions and noticed that the same bacterial species continued to emerge over many months (8). Despite large differences in growth resumption times, the actual growth speeds were similar for different isolates. This led to the development of the “scout” hypothesis—scouts are cells in a generally dormant population that can wake up and test the environment for suitability for growth (9).
Our results suggest that classifying bacteria into “normal” and “persister” cells is an oversimplification. The frequency of cells identified as persisters changes over time, and this change is heavily dependent on growth conditions. “Two-hour persisters in LB” have a very different frequency from that for “4-h persisters in MOPS Gly,” making the very definition of persisters an operational one that may not have a uniform physical equivalent. The idea of persister frequency being dynamic has also been proposed in the context of a phoU mutant which has the same persister frequency as the wild type after short Amp exposure but a greatly decreased one (compared to the wild type) after long treatment (21). The complex nature of the persister phenomenon is also illustrated by our recent observation that the length of the stationary phase has a major influence on the growth resumption kinetics observed after transfer to fresh medium (H. Luidalepp, A. Jõers, and T. Tenson, unpublished data).
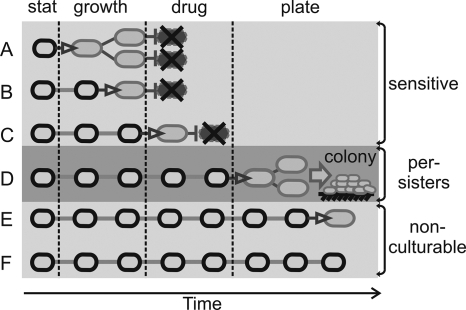
Persisters are reported to occur at low frequencies in wild-type E. coli (104 to 106) (20), but a single value has limited usefulness in the light of our results. Cells continue to resume growth and become susceptible to antibiotics, so the time-dependent killing curve is more appropriate for describing a particular culture. According to our model (Fig. 5), cells resume growth continuously, and classification into normal, persister, and noncultivable populations depends on the experimental setup.
FIG. 5.
Model for bacterial growth resumption. Stationary-phase bacteria resume growth at different times after dilution in fresh medium. (A to C) In a typical persister detection assay, cells waking up before or during antibiotic treatment are killed and deemed antibiotic sensitive. (D) Cells are considered tolerant to antibiotic (persister cells) if they resume growth after drug treatment but early enough to form a colony at the time of inspection (usually after overnight incubation). Resuming growth very late (E) or not at all (F) during the experiment would classify cells as nonculturable. Stat, stationary phase.
Acknowledgments
We thank Hannes Luidalepp, Slava Epstein, Ülo Maiväli, and Johanna Roostalu for valuable discussions and for reviewing the manuscript.
This work was supported by the European Regional Development Fund through the Center of Excellence in Chemical Biology.
Footnotes
Published ahead of print on 30 April 2010.
REFERENCES
- 1.Balaban, N. Q., J. Merrin, R. Chait, L. Kowalik, and S. Leibler. 2004. Bacterial persistence as a phenotypic switch. Science 305:1622-1625. [DOI] [PubMed] [Google Scholar]
- 2.Biggar, S. R., and G. R. Crabtree. 2001. Cell signaling can direct either binary or graded transcriptional responses. EMBO J. 20:3167-3176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bigger, J. 1944. Treatment of staphylococcal infections with penicillin by intermittent sterilisation. Lancet 244:497-500. [Google Scholar]
- 4.Blattner, F. R., G. Plunkett III, C. A. Bloch, N. T. Perna, V. Burland, M. Riley, J. Collado-Vides, J. D. Glasner, C. K. Rode, G. F. Mayhew, J. Gregor, N. W. Davis, H. A. Kirkpatrick, M. A. Goeden, D. J. Rose, B. Mau, and Y. Shao. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453-1462. [DOI] [PubMed] [Google Scholar]
- 5.De Groote, V. N., N. Verstraeten, M. Fauvart, C. I. Kint, A. M. Verbeeck, S. Beullens, P. Cornelis, and J. Michiels. 2009. Novel persistence genes in Pseudomonas aeruginosa identified by high-throughput screening. FEMS Microbiol. Lett. 297:73-79. [DOI] [PubMed] [Google Scholar]
- 6.Desnues, B., C. Cuny, G. Gregori, S. Dukan, H. Aguilaniu, and T. Nystrom. 2003. Differential oxidative damage and expression of stress defence regulons in culturable and non-culturable Escherichia coli cells. EMBO Rep. 4:400-404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Drlica, K., M. Malik, R. J. Kerns, and X. Zhao. 2008. Quinolone-mediated bacterial death. Antimicrob. Agents Chemother. 52:385-392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Epstein, S. S. 2009. General model of microbial uncultivability. Uncultivated microorganisms. Springer, Berlin, Germany.
- 9.Epstein, S. S. 2009. Microbial awakenings. Nature 457:1083. [DOI] [PubMed] [Google Scholar]
- 10.Falla, T. J., and I. Chopra. 1999. Stabilization of Rhizobium symbiosis plasmids. Microbiology 145:515-516. [DOI] [PubMed] [Google Scholar]
- 11.Fritsch, E. F., T. Maniatis, and J. Sambrook. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 12.Gefen, O., C. Gabay, M. Mumcuoglu, G. Engel, and N. Q. Balaban. 2008. Single-cell protein induction dynamics reveals a period of vulnerability to antibiotics in persister bacteria. Proc. Natl. Acad. Sci. U. S. A. 105:6145-6149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ghosh, S., and P. Setlow. 2009. Isolation and characterization of superdormant spores of Bacillus species. J. Bacteriol. 191:1787-1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hansen, S., K. Lewis, and M. Vulic. 2008. Role of global regulators and nucleotide metabolism in antibiotic tolerance in Escherichia coli. Antimicrob. Agents Chemother. 52:2718-2726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harrison, J. J., W. D. Wade, S. Akierman, C. Vacchi-Suzzi, C. A. Stremick, R. J. Turner, and H. Ceri. 2009. The chromosomal toxin gene yafQ is a determinant of multidrug tolerance for Escherichia coli growing in a biofilm. Antimicrob. Agents Chemother. 53:2253-2258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Keren, I., N. Kaldalu, A. Spoering, Y. Wang, and K. Lewis. 2004. Persister cells and tolerance to antimicrobials. FEMS Microbiol. Lett. 230:13-18. [DOI] [PubMed] [Google Scholar]
- 17.Keren, I., D. Shah, A. Spoering, N. Kaldalu, and K. Lewis. 2004. Specialized persister cells and the mechanism of multidrug tolerance in Escherichia coli. J. Bacteriol. 186:8172-8180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Korch, S. B., T. A. Henderson, and T. M. Hill. 2003. Characterization of the hipA7 allele of Escherichia coli and evidence that high persistence is governed by (p)ppGpp synthesis. Mol. Microbiol. 50:1199-1213. [DOI] [PubMed] [Google Scholar]
- 19.Kussell, E., and S. Leibler. 2005. Phenotypic diversity, population growth, and information in fluctuating environments. Science 309:2075-2078. [DOI] [PubMed] [Google Scholar]
- 20.Lewis, K. 2007. Persister cells, dormancy and infectious disease. Nat. Rev. Microbiol. 5:48-56. [DOI] [PubMed] [Google Scholar]
- 21.Li, Y., and Y. Zhang. 2007. PhoU is a persistence switch involved in persister formation and tolerance to multiple antibiotics and stresses in Escherichia coli. Antimicrob. Agents Chemother. 51:2092-2099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ma, C., S. Sim, W. Shi, L. Du, D. Xing, and Y. Zhang. 2010. Energy production genes sucB and ubiF are involved in persister survival and tolerance to multiple antibiotics and stresses in Escherichia coli. FEMS Microbiol. Lett. 303:33-40. [DOI] [PubMed] [Google Scholar]
- 23.Makinoshima, H., A. Nishimura, and A. Ishihama. 2002. Fractionation of Escherichia coli cell populations at different stages during growth transition to stationary phase. Mol. Microbiol. 43:269-279. [DOI] [PubMed] [Google Scholar]
- 24.Moir, A. 2006. How do spores germinate? J. Appl. Microbiol. 101:526-530. [DOI] [PubMed] [Google Scholar]
- 25.Moyed, H. S., and K. P. Bertrand. 1983. hipA, a newly recognized gene of Escherichia coli K-12 that affects frequency of persistence after inhibition of murein synthesis. J. Bacteriol. 155:768-775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Neidhardt, F. C., P. L. Bloch, and D. F. Smith. 1974. Culture medium for enterobacteria. J. Bacteriol. 119:736-747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Novick, A., and M. Weiner. 1957. Enzyme induction as an all-or-none phenomenon. Proc. Natl. Acad. Sci. U. S. A. 43:553-566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pearl, S., C. Gabay, R. Kishony, A. Oppenheim, and N. Q. Balaban. 2008. Nongenetic individuality in the host-phage interaction. PLoS Biol. 6:e120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Roostalu, J., A. Joers, H. Luidalepp, N. Kaldalu, and T. Tenson. 2008. Cell division in Escherichia coli cultures monitored at single cell resolution. BMC Microbiol. 8:68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schaeffer, P., J. Millet, and J. P. Aubert. 1965. Catabolic repression of bacterial sporulation. Proc. Natl. Acad. Sci. U. S. A. 54:704-711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shah, I. M., M. H. Laaberki, D. L. Popham, and J. Dworkin. 2008. A eukaryotic-like Ser/Thr kinase signals bacteria to exit dormancy in response to peptidoglycan fragments. Cell 135:486-496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Smits, W. K., C. C. Eschevins, K. A. Susanna, S. Bron, O. P. Kuipers, and L. W. Hamoen. 2005. Stripping Bacillus: ComK auto-stimulation is responsible for the bistable response in competence development. Mol. Microbiol. 56:604-614. [DOI] [PubMed] [Google Scholar]
- 33.Thattai, M., and A. van Oudenaarden. 2004. Stochastic gene expression in fluctuating environments. Genetics 167:523-530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vazquez-Laslop, N., H. Lee, and A. A. Neyfakh. 2006. Increased persistence in Escherichia coli caused by controlled expression of toxins or other unrelated proteins. J. Bacteriol. 188:3494-3497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vimberg, V., A. Tats, M. Remm, and T. Tenson. 2007. Translation initiation region sequence preferences in Escherichia coli. BMC Mol. Biol. 8:100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Walsh, C. 2003. Antibiotics: actions, origins, resistance. ASM Press, Washington, DC.
- 37.Zhang, Y. 2007. Advances in the treatment of tuberculosis. Clin. Pharmacol. Ther. 82:595-600. [DOI] [PubMed] [Google Scholar]