Abstract
The mobile element IS30 has 26-bp imperfect terminal inverted repeats (IRs) that are indispensable for transposition. We have analyzed the effects of IR mutations on both major transposition steps, the circle formation and integration of the abutted ends, characteristic for IS30. Several mutants show strikingly different phenotypes if the mutations are present at one or both ends and differentially influence the transposition steps. The two IRs are equivalent in the recombination reactions and contain several functional regions. We have determined that positions 20 to 26 are responsible for binding of the N-terminal domain of the transposase and the formation of a correct 2-bp spacer between the abutted ends. However, integration is efficient without this region, suggesting that a second binding site for the transposase may exist, possibly within the region from 4 to 11 bp. Several mutations at this part of the IRs, which are highly conserved in the IS30 family, considerably affected both major transposition steps. In addition, positions 16 and 17 seem to be responsible for distinguishing the IRs of related insertion sequences by providing specificity for the transposase to recognize its cognate ends. Finally, we show both in vivo and in vitro that position 3 has a determining role in the donor function of the ends, especially in DNA cleavage adjacent to the IRs. Taken together, the present work provides evidence for a more complex organization of the IS30 IRs than was previously suggested.
Mobile DNA elements have been described in most organisms and represent a considerable proportion of their genetic material. These elements play an important role in the evolution of the host genome due to their capacities to generate DNA rearrangements and influence the expression of neighboring genes. Their ability to form compound transposons contributes to the sequestering and dispersion of accessory genes, such as those specifying resistance to antibiotics, virulence, and various catabolic activities. The simplest mobile elements are the bacterial insertion sequences (ISs), which typically harbor one or two open reading frames (ORF) coding for the transposase (Tpase). More than 2,400 ISs have been described and classified into families (IS Finder, http://www-is.biotoul.fr/) on the basis of similarities in their genetic organization and Tpases (30). The terminal inverted repeats (IRs) are essential for the transposition of most ISs. The IRs, together with the Tpase, form a complex where the cleavage and strand transfer reactions occur. The IRs generally contain two functional modules: the internal region serves as the binding site of Tpase, while the terminal part is required for DNA cleavage and the strand transfer process (2). Besides these principal cis-acting elements, some ISs carry additional regulatory DNA sequences in the IRs or in the subterminal regions (18).
The IS30 family currently comprises more than 80 elements distributed throughout the Gram-positive and Gram-negative bacteria and the Archaea (IS Finder, http://www-is.biotoul.fr). IS30 (1, 5), the founding element of the family, is 1,221 bp long and has 26-bp imperfect IRs (the left end of the IR [IRL] and the right end of the IR [IRR]; Fig. 1A) and one ORF with a coding capacity for a 44.3-kDa Tpase. The element has a preference for two distinct types of target sequences: the natural hot spots (HSs), characterized by a 24-bp symmetric consensus (23), and the IRs of the element itself (21, 22). Potential helix-turn-helix motifs (HTH) responsible for HS and IR targeting are located in the N-terminal region of the Tpase (19). While the first motif, HTH1, is required only for transposition into the HS sequences, the conserved H-HTH2 motif is essential for both IR and HS targeting (15, 19).
FIG. 1.
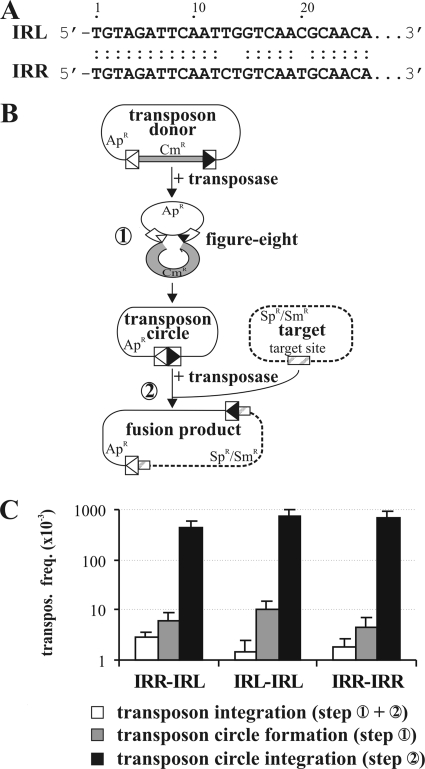
Transposition assays for comparing the IS30-based transposons composed of simple IRs. (A) Comparison of the IS30 IR sequences. Dots indicate matching bases. (B) Schematic representation of the intermolecular transposition pathway. The graph shows the two major steps characteristic for IS30 transposition (steps 1 and 2). The transposon donor plasmid and its derivative, the circular transposon (thin line), carry the 26-bp IRs of IS30 (boxes with open and filled triangles representing IRL and IRR, respectively). The Cmr gene flanking the transposon in the donor plasmid is shown as a gray box. The target plasmid (dotted line) carries the GOHS hot spot sequence (cross-hatched box). (C) Transposition frequencies of IS30-based transposons with different combinations of the IRs. The graph shows the overall frequency of transposition into the hot spot (steps 1 and 2) and the frequency of the major steps assayed separately. Data were obtained from at least three parallel experiments.
IS30 transposition occurs through two major steps (14, 24) (Fig. 1B). The first is the formation of an active intermediate by joining of the IRs. This process involves the Tpase-catalyzed cleavage of one strand at the 3′ IS end, which then attacks the same strand 2 bp outside the other IR. This strand transfer generates a single-strand bridge between the ends and leads to a figure-eight structure (33). This active transposition intermediate carrying the joined IRs probably proceeds via replicative resolution, as described for IS911 (11, 25) and IS2 (16). The resolution can lead to the circularization of a single IS or to the formation of a head-to-tail repeat of two IS30 copies. In the second step of transposition, the active forms interact with the target DNA, resulting in the known transposition products: simple insertion, deletion, inversion, or replicon fusion (14, 24).
In this work, we describe the modularity of the IR ends of IS30 by analyzing several mutants. According to our results, the IS30 IRs can be divided into functional regions that are differently involved in the main transposition steps. We show that positions 2 and 3 play a pivotal role in cleavage of the ends and, consequently, in their donor function. While the terminal part (1 to 17 bp) of the IRs is indispensable for both major steps, the internal region, i.e., the binding site for the N-terminal part of Tpase (20 to 26 bp), appears to be required only for the junction formation. Although the exact role of the terminal part of IRs is less clear, several mutations in this region considerably affected both the junction formation and integration. The fact that the internal IR region is not involved in the integration suggests that the Tpase binds to other sequences during this reaction.
MATERIALS AND METHODS
Microbial and DNA techniques.
Standard DNA procedures and the 32P labeling of oligonucleotides were carried out as described by Sambrook et al. (27). Enzymes were purchased from Fermentas, New England Biolabs, and Amersham; chemicals were purchased from Sigma, Roth, and Roche; and [γ-32P]ATP was purchased from Amersham. Sequencing was performed on an ABI Prism 3100 genetic analyzer (Perkin-Elmer). For the sequence analysis, the GCG software package (10) was applied. Sequence alignments of the IRs of the IS30 family members were made by the MultAlin server (http://bioinfo.genotoul.fr/multalin/multalin.html [3]); the sequence logo was generated by the WebLogo server (http://weblogo.berkeley.edu [4]).
Bacteria were grown at 37°C (the other culture temperatures used are indicated in the text) in LB medium supplemented with the following appropriate antibiotics used at the indicated final concentrations: ampicillin (Ap), 150 μg/ml; chloramphenicol (Cm), 20 μg/ml; kanamycin (Km), 30 μg/ml; spectinomycin (Sp), 50 μg/ml; and streptomycin (Sm), 50 μg/ml.
Plasmids. (i) Transposase producer and target plasmids.
The transposase producer was p15A-based Kmr plasmid pJKI324 carrying IS30 Orf-A under the control of the tac promoter (33). For the in vitro assays, the full-length Tpase was purified from strain ER2566 (New England Biolabs) harboring pJKI380 (33). For the electrophoretic mobility shift assay (EMSA), the N-terminal fragment of Tpase (containing amino acids [aa] 1 to 134) was expressed from pMSZ516, which is a derivative of pAW387 (32) containing the lacIq gene and a stop codon introduced after the HindIII site of IS30. In the integration assays, temperature-sensitive target plasmid pZNA133 (19) carrying the GOHS sequence (Escherichia coli genomic oligonucleotide hot spot [23]) was utilized.
(ii) Transposon donor plasmids.
The transposon donors were derivatives of the Apr ColE1-based vector pBluescript SK (Stratagene) or pEMBL19 (7). Their basic structures were the same, as all harbored IS30 ends bracketing the Cmr gene from pAW302 (31). The transposon donor pMSZ88 contained the simple IRL and IRR (33). Its analogues, which contained two identical IRs, were pMSZ105 (IRL-IRL) and pMSZ506 (IRR-IRR). For the single-end-mutation series, where only the IRR was mutated, the wild-type (wt) prototype was pMSZ358 (66 bp of the right end [RE] and 464 bp of the left end [LE]) (33). The prototype of the double-end-mutation series was pMSZ537, constructed from the 66-bp RE and the 122-bp LE. In both series of donors, the IRs are flanked by identical sequences. The wt form of the transposon donors used for determining mutations that influence the donor activity of the ends was pJKI499, which contains the 464 bp of the LE and the 66 bp of the RE flanked by different sequences.
Each mutant donor plasmid was structurally analogous to its wild-type counterpart. The point mutations were introduced into the IRs by the megaprimer PCR technique (28) using three oligonucleotide primers for each mutant. The outer primers containing the restriction sites (EcoRI and BamHI, respectively) for cloning were the same for each mutant, and these were combined with one of the inner mutagenesis primers. The IR ends were mutated from the 1st to the 27th positions by a single transversion (at the first position, M1) or pairwise transversions (M1-2, M2-3, M4-5, etc., to M26-27; Fig. 2A).
FIG. 2.
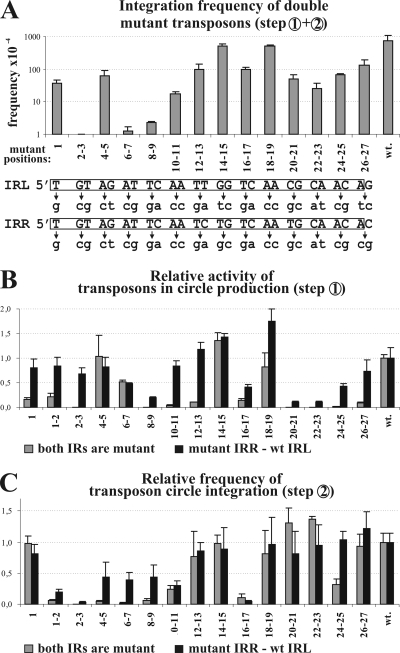
Activities of transposons harboring the same mutations in one or both ends. (A) Integration frequency of double mutant transposons. The graph represents the mean value of at least five parallel experiments measuring the overall frequency of transposon integration (steps 1 and 2; Fig. 1B) into the target. Base changes in the transposon ends are shown below the graph; the numbering refers to the mutant positions. The wt IRs are boxed. (B) Effects of IR mutations on the transposon circularization (step 1; Fig. 1B). The activity was characterized by the relative intensity of the junction fragments (see supplemental material). The graph shows the mean values of three parallel experiments for each mutant transposon donor normalized to that of the wild type (wt = 1). Note that less than 4% of the wt activity results in undetectable fragment intensity on the gel. (C) Effects of IR mutations on the integration of transposon circles (step 2; Fig. 1B). The assay measures the efficiency of integration into the target site. Frequency data are normalized to that of the wt transposon circle (wt = 1) and represent the mean values of five parallel measurements.
The in vivo assay for transposon circularization and integration was described previously (33) and is further described in the Results section.
Gel retardation experiments.
The truncated IS30 Tpase (aa 1 to 134) was purified as described by Stalder et al. (32). The protein concentration was about 0.4 mg/ml in buffer A (20 mM Tris, pH 7.5, 1 mM EDTA, 10% glycerol, 0.05% β-mercaptoethanol, 900 mM KCl). 32P-labeled fragments with the wt or mutant IRR were PCR amplified from pMSZ358 and its mutant derivatives. PCRs were carried out with primer 5′-GTATCAACAGGGACACCAGGATTTA-3′ and 32P-labeled primer 5′-AATAGCCTCTCCACCCAAGCGG-3′, which amplify a 238-bp fragment, including the 65 bp of the right IS30 end with the wt or mutant IRR. The IR-IR junctions were amplified from pMSZ88c (33) and its variants pMSZ569 and pMSZ567, which harbor the 26-26, 26-19, and 19-19 bp IR junctions, respectively. PCRs were carried out with primer 5′-CGCCAGGGTTTTCCCAGTCACGAC-3′ and 32P-labeled primer 5′-ATTTCACACAGGAAACAGCTATGAC-3′. Labeled PCR fragments (∼20 ng) were incubated in a 16-μl final volume with 3 μl of the purified N-terminal Tpase fragment, 1 μl of poly(dI-dC), and 8 μl of reaction buffer (25 mM Tris, pH 7.5, 2 mM EDTA, 2 mM dithiothreitol, 15 mM KCl) at 30°C for 30 min and then loaded on a 5% native TBE (Tris-borate-EDTA)-acrylamide gel after addition of 2 μl loading buffer (50% glycerol, 1 mg/ml bovine serum albumin) and run for 3 h at 4°C and 7.5 V/cm.
Primer extension experiment.
The figure-eight molecule was produced in the standard in vitro assay (33) and digested with BstYI. This template DNA was ethanol precipitated and dissolved in 5 μl distilled water. The extension reaction was carried out with 5 U of Pwo polymerase (Roche) and 0.1 pM 32P-labeled c5 primer (5′-GTATCAACAGGGACACCAGGATTTA-3′) in 25 μl (1× reaction buffer, 2 mM MgSO4, 0.5 mM deoxynucleoside triphosphates) at 72°C for 10 min. The sequence ladder of the M2-3 IRR mutant donor was generated using a Sequenase (version 2.0) DNA sequencing kit (USB), as indicated by the supplier. The reaction mixtures were run on a 6% denaturing polyacrylamide gel at 1,800 V. The gel was exposed to a storage phosphorscreen, which was scanned on a Storm 840 phosphorimaging system.
RESULTS
Functional equivalence of IS30 inverted repeats.
The 26-bp IRs of IS30 differing at three positions (Fig. 1A) were shown to be the only cis elements required for the whole transposition cycle (Fig. 1B, steps 1 and 2) of the element (33). To analyze the function of the 3-bp sequence divergence, we constructed two artificial IS30-based transposons delimited by identical IRs on both ends (i.e., two IRLs or IRRs). Due to the orientation of the IRs, the entire ColE1-based Apr plasmid backbone was included in the transposons (Fig. 1B). The overall activity of the transposons (circularization and integration, steps 1 and 2) and the integration activity of the circular transposons (step 2) were assayed using a three-plasmid system (33). The temperature-sensitive Spr/Smr target plasmid (pZNA133) harboring the GOHS hot spot (the most attractive known hot spot for IS30), the Kmr p15A-based transposase producer plasmid (pJKI324), and one of the transposon donor plasmids or circular transposons was introduced into Salmonella enterica serovar Typhimurium strain MA1703 (an LT2 derivative [recA1 srl]; L. Bossi, unpublished data) devoid of resident IS30 copies. After 3 h of induction of Tpase expression, transposition events were selected at 42°C for the Spr/Smr and Apr markers of the target plasmid and the transposon. Since pZNA133 cannot replicate at 42°C, only those cells in which the transposon carrying the ColE1 origin of replication is inserted into the target plasmid are viable on Sp/Sm plates. The transposon circle formation results in the loss of the Cmr gene separating the ends (Fig. 1B, step 1). This could be assayed for independently of the integration step. The transposon donor plasmids were introduced into Escherichia coli strain TG2 (27) harboring the Tpase producer pJKI324. After the overnight induction of Tpase expression starting from at least four colonies, plasmid DNA was isolated and transformed into TG2 cells and the ratio of the number of Cms transformants to the number of Cmr transformants (indicating the frequency of transposon circle formation) was determined by replica plating.
Both assays showed that the activities of the transposons assembled from identical IRs do not significantly differ from the wild-type situation, where the left and right IRs delimited the transposon (Fig. 1C). These findings suggest that the IRs of IS30 are equivalent in the transposition reactions.
Effects of IR mutations on transposition activity.
To identify the functional regions in the IRs, point mutations were introduced into both ends. Since single point mutations do not appear to cause a detectable phenotype for other elements (e.g., γδ [17] and IS911 [20]), we applied two consecutive transversions along the whole IRs, except for the first position, where a single mutation was also tested (Fig. 2A). Mutations were generated in the region from 1 to 27 bp (M1 to M26-27) in the 122-bp LE and in the 66-bp RE; otherwise, the donor plasmids and the experimental setup were analogous to those applied in the previous experiments. The resulting double mutant transposons carried the same mutations in both ends except at position 14 and positions 26 and 27, where the base changes differed in the two IRs (Fig. 2A). The transposon integration assay showed that only the mutants with the M14-15 and M18-19 mutations appeared to be as active as the wt element (7.2 × 10−2 ± 3.6 × 10−2) (Fig. 2A). Mutations, including M1, M4-5, M12-13, M16-17, and M20-27, decreased the integration frequency to ca. 1/10 of that of the wild type, while M6-7, M8-9, and M10-11 had more serious effects, leading to 40- to 600-fold decreases. Mutant M2-3 had the strongest phenotype, as the integration activity of this mutant was not detectable at all in this assay (<3 × 10−7 ± 0.5 × 10−7).
IR mutations influence the efficiency of transposon circle formation.
The previous assay measured the combined activities of the two major steps of IS30 transposition (Fig. 1B, steps 1 and 2). To determine which step is influenced by the mutations, each mutant transposon was assayed separately for circle formation (step 1) and for the integration of the transposon circle (step 2). Two series of transposon donors and circular transposons were applied. In the series of single mutants, the wt IRL was combined with a mutant IRR harboring one of the mutations from M1 to M26-27, while in the set of double mutant transposons, both IRs contained these mutations.
To describe the efficiency of the mutant transposons in IR-IR junction formation (Fig. 1B, step 1), each transposon donor was introduced into TG2 cells harboring the Tpase producer plasmid. After the overnight induction of Tpase expression, plasmid DNA was isolated. Digestion of the transposon circle with NcoI and XbaI results in a characteristic junction fragment, the intensity of which is indicative of the efficiency of circle formation (see the supplemental material). The relative intensity of this fragment was determined for each IR mutant. The results showed that almost all mutations were more deleterious when they were present in both ends. Those mutants having no significant effect in the previous integration assay (M14-15 and M18-19) showed nearly wt activity in junction formation as well (Fig. 2B). The wild-type level of circle formation was also detected with M4-5 (both in the single and in the double mutants), which previously showed reduced activity (1/10 of wt) in the overall transposition (Fig. 2A). This suggests that nucleotides at positions 4 and 5 influence the integration but not the circle formation of IS30. Interestingly, mutations at the tips of the IRs (M1, M1-2, and especially M2-3) were quite harmful when they were present in both ends, but they appeared to be almost ineffective when only the IRR was mutated. A similar phenomenon was observed with the M10-11, M12-13, and M26-27 mutants. This effect can be interpreted as a complementation or suppression characterized by the S2/D parameter (Table 1), where S and D represent the relative activities of the single and the double IR mutant transposons, respectively, compared to the activity of the isogenic wt transposon. If the mutation affects the functional domains of the IRs acting independently, the defect in the double mutant will be the square of the defect of the corresponding single mutant, so the parameter S2/D will be close to 1. In contrast, high values of S2/D indicate those positions fulfilling cooperative functions in the IRs (12).
TABLE 1.
Relative activity and S2/D factor of mutant transposons and transposon circles
| Mutant | Transposon circle production |
Transposon circle integration |
||||
|---|---|---|---|---|---|---|
| S | D | S2/D | S | D | S2/D | |
| M1 | 0.84 | 0.17 | 4.15 | 0.94 | 0.98 | 0.90 |
| M1-2 | 0.84 | 0.13 | 5.43 | 0.19 | 0.06 | 0.60 |
| M2-3 | 0.69 | <0.04a | >11.78 | 0.04 | <0.0000a | >122.65 |
| M4-5 | 0.83 | 1.04 | 0.66 | 0.43 | 0.04 | 4.45 |
| M6-7 | 0.48 | 0.51 | 0.45 | 0.40 | 0.02 | 7.55 |
| M8-9 | 0.20 | <0.04a | >0.98 | 0.44 | 0.14 | 1.44 |
| M10-11 | 0.85 | 0.04 | 17.62 | 0.46 | 0.40 | 0.53 |
| M12-13 | 1.18 | 0.11 | 12.81 | 0.80 | 0.77 | 0.83 |
| M14-15 | 1.43 | 1.36 | 1.51 | 0.77 | 0.98 | 0.61 |
| M16-17 | 0.42 | 0.14 | 1.22 | 0.10 | 0.05 | 0.24 |
| M18-19 | 1.75 | 0.82 | 3.73 | 0.90 | 0.81 | 0.99 |
| M20-21 | 0.11 | <0.04a | >0.31 | 0.92 | 1.04 | 0.81 |
| M22-23 | 0.11 | <0.04a | >0.29 | 0.78 | 1.37 | 0.45 |
| M24-25 | 0.42 | <0.04a | >4.50 | 0.75 | 0.31 | 1.82 |
| M26-27 | 0.73 | 0.10 | 5.54 | 0.88 | 0.93 | 0.84 |
Detection limit of the assay.
Some mutations, located mainly in the internal part of the IRs (positions 8 and 9, 16 and 17, and 20 to 25), caused significant reductions in circle formation both in single and in double mutants, although the effect was more robust in the double mutants (Fig. 2B). The M20-21 to M24-25 double mutants resulted in ca. 20- to 40-fold reductions both in circle formation and in overall transposition, suggesting that positions 20 to 25 have a significant effect on circle formation but not on integration. In contrast, M6-7 caused a 50% reduction in junction formation (both in the single and in the double mutants) but a 600-fold reduction in the overall activity (Fig. 2A), which suggests that this mutation interferes with both the circle formation and the integration steps.
Effect of IR mutations on transposon circle integration.
The previous analyses showed that the reduced activity of some mutants in the circle formation (step 1) could not explain the observed decrease in the overall transposition (steps 1 and 2). Therefore, we assayed for the effects of mutations on the integration of transposon circles (step 2) alone. The circular transposons, carrying the preformed IRL-IRR junctions, were isolated as Cms derivatives of single and double mutant transposon donors (see above). The assay was performed as described for the overall transposition assay, except that the circular transposons were applied as donors.
In the integration step, the majority of the mutations causing strong defects localized to the terminal part of the IRs (Fig. 2C, positions 2 to 11) and overlapped with the positions possessing high values of the S2/D parameter (Table 1, positions 2 to 7). In the internal region, only the M16-17 mutation had a similarly deleterious effect, which caused a 10-fold drop in the activities of both the single and the double mutants (low S2/D values). Strikingly, the M20-21 to M26-27 mutations, which significantly reduced circle formation, did not appear to affect the integration. On the other hand, mutations M1-2 and M2-3 (but not M1), which were defective in transposon circularization only in the case of the double mutants, caused significant reductions in the integration activity even in single mutants. The fact that M1-2 and M2-3 had a serious effect on circle integration while M1 had no detectable phenotype indicates that positions 2 and 3 possess important and specific functions. The neighboring mutations (positions 4 to 7), especially in double mutants, also caused a considerable decrease in integration efficiency, while in the case of circle formation, they had no striking effect. This implies that positions 2 to 3 and 4 to 7 are responsible for somewhat different functions. All the results presented above suggest that IS30 IRs probably possess functionally distinct domains having diverse roles in the two major transposition steps.
IR mutations interfering with binding to the N-terminal part of the transposase.
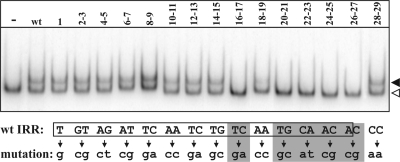
Binding of the Tpase to the IRs is known to be indispensable in transposition. Since the DNase I footprint accomplished with the N-terminal part of IS30 Tpase could not precisely identify the nucleotides having specific roles in IR recognition (32), a gel retardation assay was carried out for each IR mutant. As for several other transposases (8, 26, 34), full-length IS30 Tpase does not bind efficiently to the IRs in vitro (19); thus, the N-terminal part (the first 134 amino acids) that was previously shown to bind strongly to IRs (19, 32) was applied for the EMSA. The wt and the mutant IRs were PCR amplified (see Materials and Methods), and 32P-labeled fragments were incubated with the purified N-terminal Tpase fragment (Fig. 3). The assay indicated that only mutations located at the internal part of the IR (16 to 27 bp) decreased the IR binding significantly. At the same time, mutations located at the first 15 bp or 28 and 29 bp did not cause a detectable change in binding of the N-terminal Tpase fragment. These results suggest that the lower efficiency of circle formation observed in the cases of the M16-17 and M20-21 to M26-27 mutants (Fig. 2B) might be the result of imperfect IR recognition. On the other hand, our results also emphasize that the wt integration rate of the M20 to M27 mutant transposon circles (Fig. 2C) was achieved without efficient Tpase binding in the region of the IRs from 20 to 27 bp.
FIG. 3.
IR mutations affecting Tpase binding. The N-terminal part of IS30 transposase (134 aa) and a 238-bp PCR fragment, including the 65-bp IRR, were applied in EMSA. Lane −, Tpase-less control; lane wt, wt IRR; numbered lanes, the numbers refer to the mutant positions of IRR. Open and filled arrowheads, the free oligonucleotides and the shifted complexes, respectively. The IRR sequence is boxed; base changes are shown below. Those positions where the mutation reduced IR binding are highlighted by gray.
Several IR mutations influence spacer formation in transposon circles.
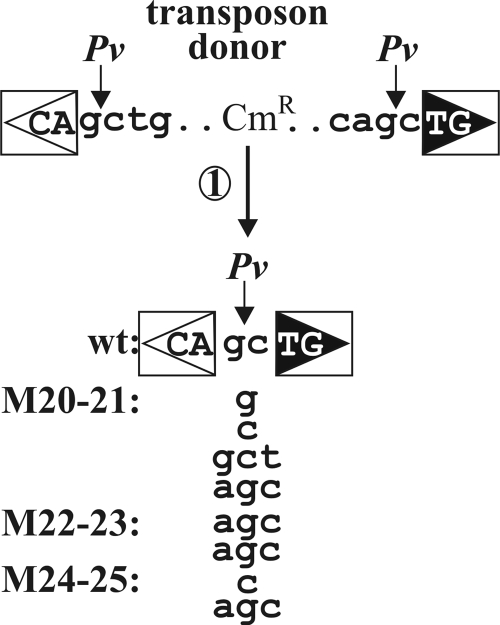
For the integration assay described above, we have isolated the circular form of the wt and mutant transposons as the Cms derivatives of the donor plasmids. The IRs in the donor plasmids are surrounded by the same sequences, forming a PvuII restriction site in the transposon donors and the circular derivatives as well (Fig. 4) (21). Therefore, a PvuII digestion allowed us to select transposon circles that harbored the regular IRL-IRR junction. Four Cms derivatives from each transposon donor were generally analyzed. Several mutants, particularly M20-21 to M24-25, yielded transposon circles at a very low rate (Fig. 2B). In these cases, all the Cms clones obtained were further analyzed.
FIG. 4.
Irregular transposon circle junctions caused by mutations M20-21 to M24-25. Four parallel experiments were carried out with each mutant to isolate transposon circles containing the IR-IR junction. The isolated DNA samples were transformed into TG2 cells, and plasmid DNA was purified from all the Cms colonies (if any) from each parallel experiment and analyzed by PvuII (Pv) digestion. One circle junction not cleavable by PvuII was sequenced from each experiment. Transposons harboring M20-21, M22-23, or M24-25 in both IRs consistently resulted in aberrant spacers of 1 or 3 bp. For M20-21, the four parallel experiments yielded four different irregular junctions and M22-23 bore three independent Cms derivatives, of which two contained identical junctions and one carried a small deletion (data not shown), while M24-25 produced only two independent Cms derivatives with different spacers. Uppercase letters, the first and last two nucleotides of the IS30 ends; lowercase letters, the flanking sequence in the donor plasmid or the spacer sequence detected by sequencing. Other symbols are as defined in the legend to Fig. 1.
This study showed that no PvuII-cleavable junction could be found for the M20-21 to M24-25 double mutants, while the single mutants at this region and all the other mutants (either single or double) yielded exclusively regular IR-IR junctions. Restriction and sequence analyses revealed two classes of non-PvuII-cleavable junctions: (i) incomplete junctions with a small deletion affecting one of the IRs (M20-21, 6/20; M22-23, 13/15; M24-25, 10/20) or (ii) complete junctions with irregular spacer lengths (M20-21, 14/20; M22-23, 2/15; M24-25, 10/20) (Fig. 4). Thus, mutations drastically reducing the IR-IR junction formation in vivo (Fig. 2B) and abolishing the IR binding of the N-terminal Tpase fragment in vitro (Fig. 3) caused aberrant spacer formation. In this respect, these mutations differed from those at positions 1 to 3 and 8 to 13, which also reduced junction formation but did not affect IR binding or spacer formation. On the other hand, M16-17 formed a third category of mutants, as it significantly decreased the junction formation and the IR binding in vitro but did not yield aberrant spacers.
The binding site of the N-terminal Tpase fragment is not required for transposon circle integration.
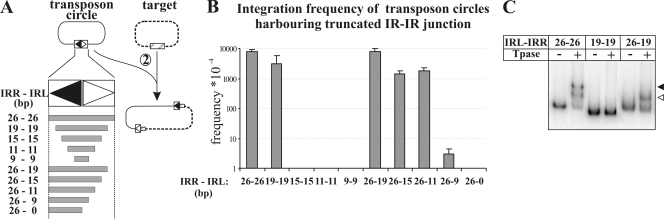
The assays for the integration of the single and double IR mutant transposon circles and the EMSA indicated that mutations negatively affecting the binding of the N-terminal Tpase fragment are not deleterious for the integration (compare Fig. 2C and Fig. 3). This raised the possibility that the integration process does not require the whole IR. Thus, we assessed the minimal sequence requirements for the integration. Transposon circles harboring the IR-IR junctions were constructed from internally truncated IRs. One series of the constructs contained equally shortened IRL and IRR, while in the other series only the IRL was truncated and joined to the intact 26-bp IRR (Fig. 5A). Removing the regions from 20 to 26 bp from both IRs caused no significant reduction in the integration of transposon circles, while the absence of the region from 16 to 26 bp completely blocked the integration. In contrast, one complete IR in the junction could rescue the lack of the region from 10 to 26 bp in the other IR (Fig. 5B). This may suggest the cooperative binding of Tpase molecules to the two IRs during the assembly of the integration complex.
FIG. 5.
Minimal requirements for transposon circle integration. (A) Structure of transposon circles assembled from IRs truncated in their internal region. The total length of the IRs forming the junctions is indicated. The integration capacity of these constructs (step 2) was assayed as described in the legend to Fig. 2 and is shown in panel B. Data were obtained from at least four parallel measurements. (C) EMSA for IR-IR junctions assembled from intact and/or truncated IRs. The first 134 aa of the IS30 Tpase and the 32P-labeled IR-IR junction fragments (26-26, 19-19, 26-19 bp) were applied in the shift assay. Open and filled arrowheads indicate the shifted complexes possibly containing one and two Tpase molecules, respectively.
A binding assay for the fragments with the 26-26, 26-19, and 19-19 bp IRL-IRR junctions was carried out using the 134-aa N-terminal Tpase fragment (see Materials and Methods). The results (Fig. 5C) showed that only those fragments where at least one complete IR was present in the junction gave shifted bands. The fragment with the 26-26 bp junction resulted in two shifted bands (probably representing DNA fragments bound by one or two N-terminal Tpase proteins), the fragment with the 26-19 bp junction gave one shifted band, while the junction truncated on both sides (19-19 bp) gave no shift. These results support the idea that the N-terminal Tpase fragment might have no binding site in the region of the IRs from 19 to 26 bp. Knowing that the deletion of this IR region gives the wt level of circle integration (Fig. 5), it seems plausible that the integration step requires different Tpase-DNA contacts than the junction formation does.
IR bases involved in donor activity of the end.
We have previously shown that the first detectable intermediate structure in IS30 transposition is the figure-eight form, in which a 2-bp single-strand bridge holds the IRs together (33). This is probably formed via a single-strand transfer reaction, where the Tpase cuts one DNA strand adjacent to one IR (donor end) and the liberated 3′ OH group attacks the same strand 2 bp away from the tip of the other IR (target end). Since both the IRL and the IRR proved to be equivalent as donor or target ends (Fig. 1), the figure eight can be formed in alternative ways, depending on which IR is cleaved by the Tpase first. Consequently, the wt element can yield two equivalent figure eights and transposon circles which differ only in the origin of the spacer bases (Fig. 6A). Since these bases always derive from the flanking sequence of the targeted IR, they clearly indicate the role (donor or target) of the ends during the strand transfer reaction.
FIG. 6.
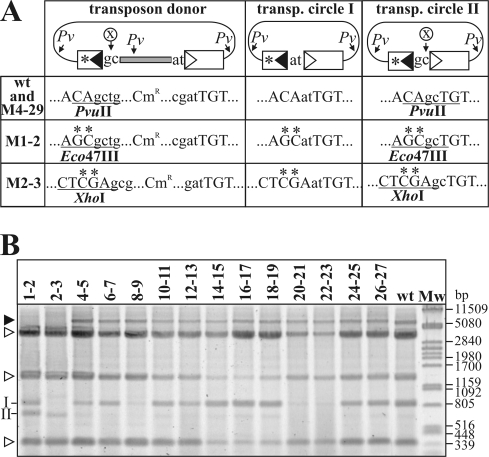
IR mutations affecting donor activity. (A) Schematic structure of the transposon donors and the expected transposon circle derivatives. The mutations were introduced into the RE (asterisk). The RE-flanking sequence and the IR-IR junctions derived from targeting the IRR (transposon circle II) are cleavable by a restriction enzyme, labeled X. In the case of the wt and the M4-5 to M26-27 mutant transposons, X is PvuII, which has a further three or two cleavage sites (indicated by Pv) in the donor plasmids and transposon (transp.) circles, respectively. In the donors harboring mutation M1-2 or M2-3, the base changes affect the positions which would contribute to the PvuII site; thus, the RE-flanking sequence and the IR-IR junction can be analyzed by X = Eco47III (for M1-2) or X = XhoI (for M2-3) digestions. Uppercase letters, the last bases of IRs; lowercase letters, the flanking or spacer bases. The restriction sites used to identify the transposon circle type (type I or II) are underlined. (B) Restriction analysis of the transposon circles deriving from the IRR mutant transposon donors. Plasmid DNA was extracted from IPTG-induced cells harboring the Tpase producer plasmid and one of the donor plasmids and digested with PvuII (for the M4-5 to M28-29 mutants), PvuII-Eco47III (for the M1-2 mutant), or PvuII-XhoI (for the M2-3 mutant). II and I, characteristic fragments deriving from transposon circle type II and I, respectively; open arrowheads, fragments of unreacted donor plasmid; filled arrowhead, Tpase producer plasmid.
To test which mutations influence the donor activity of the IRs, a new series of transposon donors was constructed where the flanking sequences adjacent to the IRs were different. This allowed us to follow the origins of the spacer bases. Each transposon donor was introduced into TG2 cells harboring the Tpase producer. Plasmid DNA was extracted from IPTG-induced overnight cultures and was digested with PvuII (for the wt and the M4-5 to M26-27 mutants), PvuII-Eco47III (for the M1-2 mutant), or PvuII-XhoI (for the M2-3 mutant). These cleavages can distinguish the two types of transposon circles, as only type II is cleavable (Fig. 6A).
The results presented in Fig. 6B showed that the wt donor produced predominantly type I circles; however, a small amount of the type II circle was also visible. Restriction analysis of individual transposon circles obtained after the transformation of the DNA samples showed that the type II/type I ratio was less than 1/60. Since the hot spot sequence flanking the IRL is the most attractive target for IS30 identified so far (21), the circle formation, as expected, occurred predominantly through the targeting of IRL, resulting in mostly type I circles. Most of the mutant transposons showed similar biases; however, some mutations clearly changed this distribution. M1-2 yielded ca. equal quantities of the two circle types, while M2-3 produced exclusively type II circles (the type II/type I ratio was 60/0). As expected, M8-9, M20-21, and M22-23 produced small quantities of circles (Fig. 2B), among which only type I was detectable, indicating a bias similar to that observed with the wt transposon. Since the spacer bases of the M1 mutant are not detectable by restriction analysis, we isolated and sequenced three independent M1 transposon circles. This showed that M1 did not significantly change the donor activity of IRR, as all samples contained the type I circle. Thus, we concluded that only the 2nd and 3rd positions have a determining role in the donor activity of the IS30 IRs.
Mutation M2-3 prevents IR cleavage.
The results obtained in the previous assays suggest that the mutations at positions 2 and 3 might interfere with the strand cleavage adjacent to the IR. Analysis of the figure-eight forms produced by the single and double mutant transposon donors can uncover the deleterious effect of the M2-3 mutation (Fig. 7A). Such an in vitro analysis (33) showed that the M2-3 mutation did not significantly change the figure-eight production in the single mutant compared to that in the wt transposon. On the other hand, it had a deleterious effect in the double mutant (Fig. 7B), which agrees with the results obtained in the circle formation assay (see M2-3 in Fig. 2B).
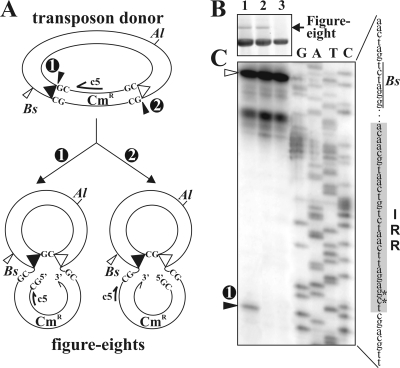
FIG. 7.
Detection of the cleavage site adjacent to the IRR in the figure-eight molecules. (A) Alternative ways to form a figure-eight structure from the wt transposon donor. The potential cleavage sites and the consequently differing figure-eight forms are numbered (1 and 2). The primer (c5) and the restriction sites (Al, AlwNI; Bs, BstYI) used in the experiment are shown. The bases flanking the IRs and the free DNA ends are indicated; the other symbols are as defined in the legend to Fig. 1. (B) Detection of figure-eight production in vitro from the wt transposon donor pMSZ358 and its single or double IR mutant (M2-3 = GT → CG) analogues. The in vitro reaction mixtures were digested with AlwNI, which converts the figure eight to an α form migrating high in the gel. Lane 1, wt transposon donor; lane 2, transposon donor with the wt IRL and the M2-3 mutation in IRR; lane 3, transposon donor with the M2-3 mutation in both IRs. (C) Detection of the free 5′ vector end adjacent to IRR by primer extension using the c5 primer. Template DNAs from the standard in vitro reaction are shown in panel B. DNAs were predigested with BstYI. The adjacent lanes labeled G, A, T, and C contain the conventional sequencing reaction mixtures of the M2-3 mutant donor plasmid using the c5 primer. The 91-bp single band in lane 1 indicated by a circled 1 comigrates with the position corresponding to the final base of the flanking DNA of IRR. The 157-bp fragment generated from unreacted templates cut by BstYI is indicated by an open arrowhead. The IRR sequence is highlighted by gray; asterisks indicate the M2-3 mutation.
If mutation M2-3 prevents the cleavage next to the mutant IR, the population of figure eights originating from the single IR mutant donor should be homogeneous, since all of them are formed by targeting the mutant end (Fig. 7A, type 2). In contrast, the wt donor should produce a mix of figure eights, since both IRs are cleavable (Fig. 7A, types 1 and 2). Assuming that the first cleavage by the Tpase liberates a 3′ OH end of the donor IR, which is then transferred to the other (target) IR (25, 33), the free 5′ P end of the vector sequence next to the donor IR is detectable by primer extension assay. The assay was carried out for the wt and the single and double M2-3 mutant donors using 32P-labeled primer annealing to the flanking sequence of the IRR (c5 in Fig. 7A). After deproteinization of the in vitro reaction mixtures, DNA samples were digested with BstYI, which cleaves the RE 66 bp away from the tip of IRR. The extension should yield a 157-bp fragment from unreacted templates and a shorter (91-bp) fragment from those cleaved by the Tpase next to the RE. Our data show that only the wt reaction mixture contained both runoff products, and the presence of the 91-bp fragment proved that the cleavage occurred precisely next to the 3′ end of IRR (Fig. 7C). As expected, the lack of the 91-bp fragment in the reaction mixtures containing the single or double IR mutant donors shows that the M2-3 mutation blocks the cleavage next to the mutant IR. The absence of the 91-bp fragment from the extension reaction using the single IR mutant (which otherwise produced a similar quantity of figure eights as the wt donor; compare lanes 1 and 2 in Fig. 7B) clearly indicates that the figure-eight molecules were uniform and originated from targeting of the mutant IRR. This also precludes the hypothetical reverse chemistry, where the initial Tpase cleavage would produce a free 5′ OH end for the donor IR, which then might attack the other end. In this case, the figure eights originating from the single mutant donor would be similar to the type 1 form (Fig. 7A), which could not be detected. As the double IR mutant donor failed to produce a detectable quantity of figure eights and no cleavage was observed at the mutant IRR, we concluded that IR positions 2 and 3 play a central role in the first cleavage.
DISCUSSION
Our analysis uncovered several functionally distinct regions in the inverted repeats of IS30. This element has adopted a two-step transposition pathway, where the first step is the joining of IRs taking place via the figure-eight formation catalyzed by the Tpase and its consecutive, host-mediated resolution (33). The second step is the insertion of the circular transposon by opening the IR-IR junction and integrating the free 3′ OH ends into the target DNA. Since the major steps are separated from each other in time and space, we could follow the role of the functional regions of IRs through these discrete steps. The majority of mutations having the strongest effect on IS30 transposition tend to overlap with the highly conserved regions of IRs in the IS30 family (Fig. 8); however, some nonconserved IR positions also play essential roles.
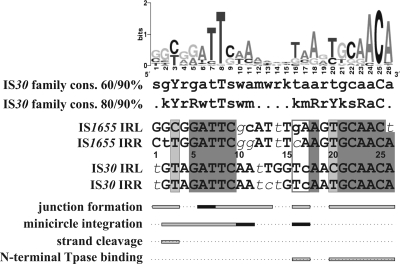
FIG. 8.
Schematic representation of the functional regions of IS30 IRs and their comparison to the family consensus and the sequence of the closely related family member IS1655. The sequence logo and the two consensus (cons.) sequences were generated for 53 IS30 family members collected from public databases (IS Finder and GenBank; the whole collection will be published elsewhere). Uppercase letters, >90% conservation; lowercase letters, >60% and >80% conservation in the two consensus sequences, respectively (r = A or G, y = C or T, m = A or C, k = G or T, s = C or G, w = A or T). Within the IR sequences, uppercase letters show bases matching both the consensus and the sequence logo, lowercase letters show bases differing from the same bases in the logo but matching the 80/90% consensus sequences, and lowercase italic letters indicate bases differing from the corresponding bases of both the consensus and the sequence logo. Dark gray shading, perfect matches in the four IRs (note that the IRs of IS1655 are only 25 bp long); light gray, divergent positions still matching the 90% consensus; open box in IR sequences, bases where the base change in the IS1655 IRs to the corresponding bases of IS30 IRs led to the correct integration of the IS1655 circle by the IS30 Tpase; horizontal bars below IR sequences, those positions where the IS30 IR mutations significantly affected the given functions; dark bars, positions where the single and double mutants had similar activities (Fig. 2 and Table 1).
Almost all IR positions have important functions in junction formation. The M16-17, M20-21, and M20-23 mutations had the most deleterious effects both in the single and in the double mutants, while M24-25 and M26-27 had a similar effect only in the double mutants. M18-19 had no detectable phenotype in vivo. Moreover, mutations at the region from 20 to 25 bp but not at the region from 16 to 19 bp caused irregular spacers between the abutted IRs (Fig. 4). The qualitative EMSA analysis indicated that mutations at positions 16 to 17 and 20 to 27 drastically reduced the sequence-specific binding of the N-terminal Tpase fragment, while the M18-19 mutation had a less significant effect. These facts suggest that the internal part of IRs can be divided into two subregions. The YGCAACA motif (20 to 26 bp), which is conserved throughout the IS30 family (Fig. 8), has an important role both in the binding and the positioning of the Tpase in the synaptic complex and in correct spacer formation. The other subregion (positions 16 to 19) is also important in binding but does not affect the spacer formation.
Most of the mutations in the terminal 15 bp of the IRs cause severe defects in junction formation; however, they do not influence the binding of the N-terminal Tpase fragment in vitro. The deleterious mutations are clustered into two regions: at the very tip of IRs (1 to 3 bp) and subterminally (6 to 13 bp), partially overlapping with the conserved GTAGATTCAA motif (Fig. 8, 2 to 11 bp). The most deleterious mutation was M8-9, which hit the highly (>90%) conserved 8T position. Mutations of the region from 10 to 13 bp also caused 10- to 25-fold reductions in circle formation in double mutants but were rescued by the presence of a wt IR in single mutants (S2/D > 12.8; Fig. 2B and Table 1).
The single mutations at positions 1 to 3 had no detectable effect, while the double mutants showed significantly decreased activity in junction formation (S2/D = 4.2, 5.4, and >11.8, respectively; Table 1), similarly to IS5 (13), IS10 (12), and IS903 (9). The most detrimental mutation was M2-3, which included the highly conserved T at position 3 (>90% Y). These results suggest that the first positions are necessary for the initial cleavage required for the donor activity of the IR (Fig. 6 and 7). This is clearly supported by the fact that the M2-3 double mutant, lacking cleavable IRs, was almost completely inactive (Fig. 2B). In contrast, the single mutant transposon donor can produce a type II transposon circle (Fig. 6) and type 2 figure-eight intermediate (Fig. 7), which explains how the wt IR (functioning as the donor end) efficiently rescued the mutant IR in the transposon. In contrast to the IS3 family, where the conservative 5′-TG… CA-3′ terminal dinucleotides are absolutely required for cleavage of the donor end (16, 25, 29), mutations of the same bases at the tips of IS30 (M1 and M1-2) do not impede this function. Conversely, the conserved 3rd position appears to be the most important in the cleavage of IS30 IRs, suggesting its essential role probably in the whole IS30 family.
In the integration assays, the same set of mutants showed a quite different phenotype pattern. The most striking difference was that mutations at the region from 20 to 26 bp, which decreased or prevented the binding of the N-terminal Tpase fragment to the IR, had no significant phenotype, even in the double mutant (Fig. 2C). This indicates that the binding site of the N-terminal Tpase fragment required for the IR-IR junction formation is dispensable for the integration step. The nearly wt activity of the transposon circle assembled from the first 19 bp of the IRs (Fig. 5) also underpins this notion. Thus, we believe that IRs possess a second Tpase binding site, which is sufficient for the circle integration. If we accept that the very end of the IRs (1 to 3 bp) is involved in the cleavage, the putative secondary binding site can overlap with the conservative block of the region from 4 to 11 bp. Double mutations in this region (particularly at 4 to 9 bp) caused a significant drop in the integration activity. The fact that one wt IR can efficiently complement the mutations at the region from 4 to 9 bp of the other end (S2/D = 1.4 to 7.5; Table 1) highlights cooperative Tpase binding during formation of the integrative complex. Comparing those regions of IRs which are important for junction formation or integration, the longest overlap, ATTCAA (positions 6 to 11; Fig. 8), is embedded in the conserved GTAGATTCAA motif. Since the footprint assay (32) and EMSA (Fig. 5C) did not show any protection or binding by the N-terminal Tpase fragment in this region, a domain missing from the truncated Tpase could possibly be involved in the recognition of this second binding site.
Besides the binding of Tpase to the IRs, the other indispensable function for the integration is the cleavage of the ends. In this step, both strands of the IR-IR junction have to be cut prior to the strand transfer into the target DNA (35). Thus, mutations preventing the cleavage should be deleterious even in the single mutant. The M2-3 mutants show the expected phenotype, as the integration activity of the single mutant is only 4% of that of the wt and the double mutant is essentially inactive (Fig. 2C). All these data support the hypothesis that positions 2 to 3 have a central role in the cleavage of donor ends in both major transposition steps.
Mutations at 16 and 17 bp also affected the integration, as both single and double mutants showed reduced activity (5 to 10% of the wt values). The particular importance of TC bases at positions 16 and 17 is obvious if we compare the IR sequences of IS30 and the related element IS1655. The IRs of the two elements are mostly identical or contain bases that correspond to the family consensus in the conserved regions. The only exceptions are positions 16 and 17, where the IS30 IRs carry TC, while IS1655 carries GA or CA bases (Fig. 8). In the M16-17 mutant, the TC bases were converted to GA (Fig. 2A), which caused significant decreases both in circle formation and in integration activities (Fig. 2). In spite of the extensive homology of IRs, IS30 Tpase can neither produce the IS1655 transposon circle nor integrate it into a preferred target site of IS30 (M. Szabó, unpublished data). However, changing the GA and CA bases to TC in the two IS1655 IRs yields detectable integration of the transposon circle by the IS30 Tpase (Szabó, unpublished). Thus, we can conclude that the TC bases at positions 16 and 17 have an important function in both major transposition steps and contribute to the specificity of the IS30 Tpase to its particular IR ends.
On the basis of our data, we suggest that the Tpase would make intimate contacts with the IRs along their whole length, as is seen in the Tn5 preintegration complex (6). In the synaptic complex, the IRs are brought together to allow a single-strand nick at one IR and the strand transfer. One Tpase active site needs to be catalytically engaged and correctly aligned for cleaving 2 bp outside the second IR. We believe that the N-terminal region of the Tpase (presumably, the H-HTH2 motif [19]) binds to the internal conserved region of IRs (YGCAACA), while an unknown domain of the protein makes contact with the terminal conserved region (ATTCAA), so that the DDE catalytic domain can interact with the very tip of the IRs. Less is known about the integration complex, where the IRs are immediately juxtaposed with a 2-bp spacer separating them. Thus, the step 2 substrate differs considerably from the step 1 substrate, and consequently, the integration complex must involve different Tpase contacts and/or Tpase conformations. The most striking difference is that the N-terminal Tpase binding in the internal region of the IRs is dispensable for the integration. The slightly different distributions of the deleterious mutations in the terminal region during circle formation or integration (i.e., the regions from 6 to 13 bp and 4 to 11 bp, respectively; Fig. 8) also underline this difference.
The complexity of the functional regions of the IS30 IRs suggests that the general view about the organization of IRs, i.e., that the internal region includes the Tpase binding site and the terminal region is involved in catalytic steps, is oversimplified and does not fully apply to IS30. Further analyses will shed light on the interaction of the IS30 Tpase and its cognate IRs in the different transposome complexes, which may then help to establish a comprehensive molecular model for the copy-and-paste mechanism adopted by IS30.
Supplementary Material
Acknowledgments
We are grateful to Z. Nagy and K. Schlett for critical reading of the manuscript. We thank I. Könczöl, E. Sztána-Keresztúri and M. Turai for their excellent technical assistance.
Footnotes
Published ahead of print on 23 April 2010.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Caspers, P., B. Dalrymple, S. Iida, and W. Arber. 1984. IS30, a new insertion sequence of Escherichia coli K12. Mol. Gen. Genet. 196:68-73. [DOI] [PubMed] [Google Scholar]
- 2.Chandler, M., and J. Mahillon. 2002. Insertion sequences revisited, p. 305-366. In N. L. Craig, R. Craigie, M. Gellert, and A. M. Lambowitz (ed.), Mobile DNA II. American Society for Microbiology, Washington, DC.
- 3.Corpet, F. 1988. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 16:10881-10890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Crooks, G. E., G. Hon, J. M. Chandonia, and S. E. Brenner. 2004. WebLogo: a sequence logo generator. Genome Res. 14:1188-1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dalrymple, B., P. Caspers, and W. Arber. 1984. Nucleotide sequence of the prokaryotic mobile genetic element IS30. EMBO J. 3:2145-2149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Davies, D. R., I. Y. Goryshin, W. S. Reznikoff, and I. Rayment. 2000. Three-dimensional structure of the Tn5 synaptic complex transposition intermediate. Science 289:77-85. [DOI] [PubMed] [Google Scholar]
- 7.Dente, L., G. Cesareni, and R. Cortese. 1983. pEMBL: a new family of single stranded plasmids. Nucleic Acids Res. 11:1645-1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Derbyshire, K. M., and N. D. Grindley. 1992. Binding of the IS903 transposase to its inverted repeat in vitro. EMBO J. 11:3449-3455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Derbyshire, K. M., L. Hwang, and N. D. Grindley. 1987. Genetic analysis of the interaction of the insertion sequence IS903 transposase with its terminal inverted repeats. Proc. Natl. Acad. Sci. U. S. A. 84:8049-8053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Devereux, J., P. Haeberli, and O. Smithies. 1984. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 12:387-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Duval-Valentin, G., B. Marty-Cointin, and M. Chandler. 2004. Requirement of IS911 replication before integration defines a new bacterial transposition pathway. EMBO J. 23:3897-3906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huisman, O., P. R. Errada, L. Signon, and N. Kleckner. 1989. Mutational analysis of IS10's outside end. EMBO J. 8:2101-2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jilk, R. A., D. York, and W. S. Reznikoff. 1996. The organization of the outside end of transposon Tn5. J. Bacteriol. 178:1671-1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kiss, J., and F. Olasz. 1999. Formation and transposition of the covalently closed IS30 circle: the relation between tandem dimers and monomeric circles. Mol. Microbiol. 34:37-52. [DOI] [PubMed] [Google Scholar]
- 15.Kiss, J., M. Szabó, and F. Olasz. 2003. Site-specific recombination by the DDE family member mobile element IS30 transposase. Proc. Natl. Acad. Sci. U. S. A. 100:15000-15005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lewis, L. A., and N. D. Grindley. 1997. Two abundant intramolecular transposition products, resulting from reactions initiated at a single end, suggest that IS2 transposes by an unconventional pathway. Mol. Microbiol. 25:517-529. [DOI] [PubMed] [Google Scholar]
- 17.May, E. W., and N. D. Grindley. 1995. A functional analysis of the inverted repeat of the gamma delta transposable element. J. Mol. Biol. 247:578-587. [DOI] [PubMed] [Google Scholar]
- 18.Nagy, Z., and M. Chandler. 2004. Regulation of transposition in bacteria. Res. Microbiol. 155:387-398. [DOI] [PubMed] [Google Scholar]
- 19.Nagy, Z., M. Szabó, M. Chandler, and F. Olasz. 2004. Analysis of the N-terminal DNA binding domain of the IS30 transposase. Mol. Microbiol. 54:478-488. [DOI] [PubMed] [Google Scholar]
- 20.Normand, C., G. Duval-Valentin, L. Haren, and M. Chandler. 2001. The terminal inverted repeats of IS911: requirements for synaptic complex assembly and activity. J. Mol. Biol. 308:853-871. [DOI] [PubMed] [Google Scholar]
- 21.Olasz, F., T. Farkas, J. Kiss, A. Arini, and W. Arber. 1997. Terminal inverted repeats of insertion sequence IS30 serve as targets for transposition. J. Bacteriol. 179:7551-7558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Olasz, F., T. Fischer, M. Szabó, Z. Nagy, and J. Kiss. 2003. Gene conversion in transposition of Escherichia coli element IS30. J. Mol. Biol. 334:967-978. [DOI] [PubMed] [Google Scholar]
- 23.Olasz, F., J. Kiss, P. König, Z. Buzás, R. Stalder, and W. Arber. 1998. Target specificity of insertion element IS30. Mol. Microbiol. 28:691-704. [DOI] [PubMed] [Google Scholar]
- 24.Olasz, F., R. Stalder, and W. Arber. 1993. Formation of the tandem repeat (IS30)2 and its role in IS30-mediated transpositional DNA rearrangements. Mol. Gen. Genet. 239:177-187. [DOI] [PubMed] [Google Scholar]
- 25.Polard, P., and M. Chandler. 1995. An in vivo transposase-catalyzed single-stranded DNA circularization reaction. Genes Dev. 9:2846-2858. [DOI] [PubMed] [Google Scholar]
- 26.Rousseau, P., E. Gueguen, G. Duval-Valentin, and M. Chandler. 2004. The helix-turn-helix motif of bacterial insertion sequence IS911 transposase is required for DNA binding. Nucleic Acids Res. 32:1335-1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 28.Sarkar, G., and S. S. Sommer. 1990. The “megaprimer” method of site-directed mutagenesis. Biotechniques 4:404-407. [PubMed] [Google Scholar]
- 29.Sekine, Y., K. Aihara, and E. Ohtsubo. 1999. Linearization and transposition of circular molecules of insertion sequence IS3. J. Mol. Biol. 294:21-34. [DOI] [PubMed] [Google Scholar]
- 30.Siguier, P., J. Perochon, L. Lestrade, J. Mahillon, and M. Chandler. 2006. IS-finder: the reference centre for bacterial insertion sequences. Nucleic Acids Res. (Database Issue) 34:D32-D36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stalder, R., and W. Arber. 1989. Characterization of in vitro constructed IS30-flanked transposons. Gene 76:187-193. [DOI] [PubMed] [Google Scholar]
- 32.Stalder, R., P. Caspers, F. Olasz, and W. Arber. 1990. The N-terminal domain of the insertion sequence 30 transposase interacts specifically with the terminal inverted repeats of the element. J. Biol. Chem. 265:3757-3762. [PubMed] [Google Scholar]
- 33.Szabó, M., J. Kiss, Z. Nagy, M. Chandler, and F. Olasz. 2008. Sub-terminal sequences modulating IS30 transposition in vivo and in vitro. J. Mol. Biol. 375:337-352. [DOI] [PubMed] [Google Scholar]
- 34.Ton-Hoang, B., C. Turlan, and M. Chandler. 2004. Functional domains of the IS1 transposase: analysis in vivo and in vitro. Mol. Microbiol. 53:1529-1543. [DOI] [PubMed] [Google Scholar]
- 35.Ton-Hoang, B., P. Polard, and M. Chandler. 1998. Efficient transposition of IS911 circles in vitro. EMBO J. 17:1169-1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.