Abstract
Sphingobium sp. strain SYK-6 converts various lignin-derived biaryls with guaiacyl (4-hydroxy-3-methoxyphenyl) and syringyl (4-hydroxy-3,5-dimethoxyphenyl) moieties to vanillate and syringate. These compounds are further catabolized through the protocatechuate (PCA) 4,5-cleavage (PCA45) pathway. In this article, the regulatory system of the PCA45 pathway is described. A LysR-type transcriptional regulator (LTTR), LigR, activated the transcription of the ligK-orf1-ligI-lsdA and ligJABC operons in the presence of PCA or gallate (GA), which is an intermediate metabolite of vanillate or syringate, respectively, and repressed transcription of its own gene. LigR bound to the positions −77 to −51 and −80 to −48 of the ligK and ligJ promoters, respectively, and induced DNA bending. In the presence of PCA or GA, DNA bending on both promoters was enhanced. The LigR-binding regions of the ligK and ligJ promoters in the presence of inducer molecules were extended and shortened, respectively. The LTTR consensus sequences (Box-K and Box-J) in the ligK and ligJ promoters were essential for the binding of LigR and transcriptional activation of both operons. In addition, the regions between the LigR binding boxes and the −35 regions were required for the enhancement of DNA bending, although the binding of LigR to the −35 region of the ligJ promoter was not observed in DNase I footprinting experiments. This study shows the binding features of LigR on the ligK and ligJ promoters and explains how the PCA45 pathway genes are expressed during degradation of lignin-derived biaryls by this bacterium.
Lignin is the most abundant aromatic compound in nature, and its biodegradation represents a key step in carbon cycling. It is thought that white rot fungi initiate the biodegradation of native lignin, and the resultant low-molecular-weight products are further catabolized by soil bacteria (19, 21). Sphingobium sp. strain SYK-6 (formerly Sphingomonas paucimobilis SYK-6) is one of the best-characterized degraders of lignin-derived aromatic compounds, and this strain is able to utilize various lignin-derived biaryls, including β-aryl ether (44), biphenyl (37), and diarylpropane, as sole sources of carbon and energy (28). In SYK-6, lignin-derived biaryls with guaiacyl (4-hydroxy-3-methoxyphenyl) and syringyl (4-hydroxy-3,5-dimethoxyphenyl) moieties are converted to vanillate and syringate, respectively (see Fig. 1A) (28). After O demethylation of vanillate and syringate, protocatechuate (PCA) and 3-O-methylgallate (3MGA) are generated, respectively (1, 30). PCA is further degraded to pyruvate and oxaloacetate via the PCA 4,5-cleavage (PCA45) pathway (11, 12, 29, 31, 33). On the other hand, 3MGA is degraded through multiple ring cleavage pathways; however, eventually the 3MGA catabolic pathway conjoins with the PCA45 pathway (13-15). Therefore, the PCA45 pathway is essential for the catabolism of lignin-derived compounds in SYK-6.
FIG. 1.
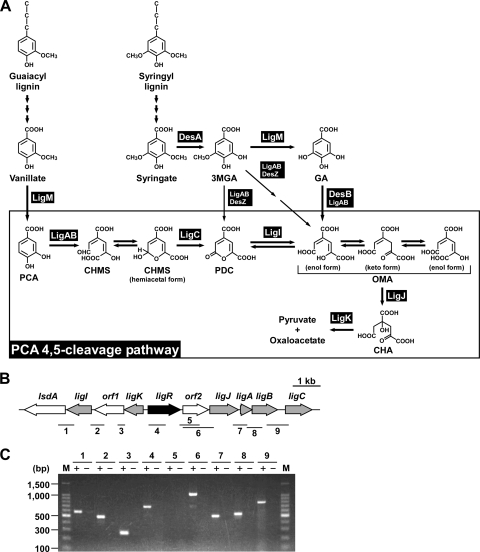
Catabolic pathway for vanillate and syringate and gene organization of the PCA45 pathway genes in Sphingobium sp. strain SYK-6. (A) Catabolism of vanillate and syringate. Enzymes: LigM, vanillate/3MGA O-demethylase; LigA and LigB, small and large subunits of PCA 4,5-dioxygenase; LigC, CHMS dehydrogenase; LigI, PDC hydrolase; LigJ, OMA hydratase; LigK, CHA aldolase/oxaloacetate decarboxylase; DesA, syringate O-demethylase; DesZ, 3MGA 3,4-dioxygenase; DesB, GA dioxygenase. Abbreviations: PCA, protocatechuate; CHMS, 4-carboxy-2-hydroxymuconate-6-semialdehyde; 3MGA, 3-O-methylgallate; GA, gallate; PDC, 2-pyrone-4,6-dicarboxylate; OMA, 4-oxalomesaconate; CHA, 4-carboxy-4-hydroxy-2-oxoadipate. (B) Organization of the PCA45 pathway genes. Regulatory genes, enzyme genes, and unidentified genes are indicated by black, gray, and white arrows, respectively. The bars and numbers below the genes indicate the locations of the amplified RT-PCR products shown in panel C. (C) RT-PCR analysis of the PCA45 pathway genes from SYK-6. Total RNA isolated from SYK-6 cells grown in the presence of PCA was used as a template. The sizes of molecular weight markers in lane M are indicated on the left. Lane numbers correspond to the numbers of the amplified regions indicated in panel B. “+” and “−“, with or without reverse transcriptase, respectively.
The PCA45 pathway was enzymatically characterized by Kersten et al. (17) and Maruyama and colleagues (22-26). To date, the PCA45 pathway genes have been isolated from SYK-6 (33), Sphingomonas sp. strain LB126 (55), Comamonas testosteroni BR6020 (41), Pseudomonas straminea NGJ1 (27), and Arthrobacter keyseri 12B (7). Our research group characterized all of the enzyme genes of the PCA45 pathway in SYK-6 and reported that the PCA45 pathway genes consist of four transcriptional units, which are the ligK-orf1-ligI-lsdA operon, ligJ-ligA-ligB operon, and monocistronic ligC and ligR (12). LigR shows similarity to proteins belonging to the family of the LysR-type transcriptional regulator (LTTR) (45). Disruption of ligR led to significant growth retardation of SYK-6 on vanillate and syringate (12), suggesting that LigR plays a crucial role in the regulation of the PCA45 pathway genes. However, regulatory system of the pathway genes remains unknown for all the strains mentioned above.
LTTR is one of the most common types among prokaryotic regulators. Proteins belonging to this family typically activate the expression of a target gene(s) in response to a small inducer molecule and repress their own gene expression. In solution, LTTRs are homodimers or homotetramers, and their DNA binding forms are suggested to be tetramers (20). LTTRs associate with two distinct binding sites at the target promoter (45). The recognition binding site (RBS) contains the LTTR consensus binding sequence (T-N11-A) within an interrupted inverted repeat. The RBS is often centered at position −65 relative to the transcription start site and is commonly essential for the binding of LTTR. The activation binding site (ABS) is generally located between the RBS and the target promoter. This site is important for transcriptional activation and is thought to be involved in assisting DNA binding (40). A large number of LTTRs have been shown to induce DNA bending upon binding of the protein. Binding of inducer provokes a conformational change and typically alters the binding region and DNA bending angle (49).
In this study, we characterized for the first time the transcriptional regulation of the PCA45 pathway genes. This study provides an insight into how the downstream pathway of bacterial lignin degradation is controlled. In addition, results of the binding analysis demonstrate that the behavior of LigR with respect to DNA protection and DNA bending is distinct from that of well-characterized LTTRs, and they also suggest the importance of LigR binding to the −35 regions in transcriptional activation.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The strains and plasmids used in this study are listed in Table 1. Sphingobium sp. strain SYK-6 was grown in Luria-Bertani (LB) medium or in W minimal salt medium (36) containing 10 mM vanillate or SEM (10 mM sucrose, 10 mM glutamate, and 50 mg of methionine/liter) at 30°C. The SYK-6 mutants were grown in LB medium. When required, 50 mg of kanamycin/liter and 30 mg of chloramphenicol/liter were added to the media. Escherichia coli JM109 was used for cloning experiments. E. coli HB101 was employed to transfer the plasmids to SYK-6 and its derivatives. E. coli BL21(DE3) was used for protein overproduction. E. coli strains were grown in LB medium at 37°C. For cultures of E. coli cells carrying bla, the media were supplemented with 100 mg of ampicillin/liter.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristic(s)a | Reference or source |
|---|---|---|
| Sphingobium sp. | ||
| SYK-6 | Wild type; Nalr Smr | 16 |
| DLR | SYK-6 derivative; ligR::kan; Nalr Smr Kmr | 12 |
| AKB | SYK-6 derivative; ligB::kan; Nalr Smr Kmr | 38 |
| RBB | SYK-6 derivative; ΔdesB ligB::bla; Nalr Smr Cbr | 15 |
| E. coli | ||
| JM109 | recA1 supE44 endA1 hsdR17(rK− mK+) gyrA96 relA1 thi-1 Δ(lac-proAB) F′[traD36 proAB+lacIqlacZΔM15) | 57 |
| HB101 | recA13 supE44 hsd20(rB− mB−) ara-14 proA2 lacY1 galK2 rpsL20 xyl-5 mtl-1 | 3 |
| BL21(DE3) | F−ompT hsdSB(rB− mB−) gal dcm (DE3) | Novagen |
| Plasmids | ||
| pUC18 | Cloning vector, Apr | 57 |
| pT7Blue | Cloning vector, T7 promoter, Apr | Novagen |
| pET21a(+) | Expression vector, T7 promoter, Apr | Novagen |
| pKT230 | Broad-host-range vector, Kmr | 2 |
| pRK2013 | Tra+ Mob+ Kmr | 9 |
| pPR9TT | Translational fusion LacZ reporter vector, Apr Cmr | 43 |
| pQF50 | Broad-host-range transcriptional fusion vector containing a promoterless lacZ, Apr | 8 |
| pPR9TZ | pPR9TT with a 3.6-kb SmaI-ScaI fragment containing lacZ from pQF50 replacing the 3.2-kb BamHI fragment | This study |
| pHN139R | pUC18 with a 10.5-kb EcoRI fragment carrying ligK-orf1-ligI-lsdA, ligJAB, ligR, and a part of ligC | 31 |
| pVA01 | pKT230 with a 12.0-kb EcoRI fragment carrying ligK-orf1-ligI-lsdA, ligJABC, and ligR | 16 |
| pT7K | pT7Blue with a 259-bp PCR fragment carrying the ligK-ligR intergenic region | This study |
| pT7R | pT7Blue carrying the same fragment as pT7K in the opposite direction | This study |
| pT7JTN12 | pT7Blue with a 164-bp PCR fragment carrying the ligJ promoter region | This study |
| pT716 | pT7Blue with a 1.4-kb PCR fragment carrying 16S rRNA gene of SYK-6 | This study |
| pET21R | pET21a(+) with a 1.3-kb NdeI-BamHI fragment carrying ligR | This study |
| pT7KSD | pT7K with site-directed mutations in Box-K | This study |
| pT7JSD | pT7JTN12 with site-directed mutations in Box-J | This study |
| pZK | pPR9TZ with a 0.3-kb HindIII-KpnI fragment carrying the ligK promoter region | This study |
| pZKSD | pZK with site-directed mutations in Box-K | This study |
| pZR | pPR9TZ with a 0.3-kb HindIII-KpnI fragment carrying the ligR promoter region | This study |
| pZJTN12 | pPR9TZ with a 0.2-kb HindIII-KpnI fragment carrying the ligJ promoter region | This study |
| pZJTN12SD | pZJTN12 with site-directed mutations in Box-J | This study |
Nalr, Smr, Kmr, Cbr, Apr, and Cmr, resistance to nalidixic acid, streptomycin, kanamycin, carbenicillin, ampicillin, and chloramphenicol, respectively.
DNA manipulations and nucleotide sequencing.
Plasmid preparation, restriction enzyme digestion, DNA ligation, and transformation were all performed as described by Sambrook et al. (42). The introduction of plasmids into SYK-6 was carried out by the triparental mating procedure with a helper plasmid, pRK2013. Nucleotide sequences were determined by the dideoxy termination method using a CEQ2000XL genetic analysis system (Beckman Coulter, Fullerton, CA).
Induction.
Cells of SYK-6 and its derivatives grown in W-SEM were washed twice in 0.9% NaCl and resuspended in fresh medium to an absorbance at 600 nm (A600) of 0.2. The cells were incubated at 30°C until the A600 of the culture reached 1.0. After the addition of 10 mM PCA or GA as an inducer, the cells were further incubated for 3 h.
RNA preparation.
Total RNA was isolated from cells of SYK-6 and its derivatives using Isogen (Nippon Gene Co., Ltd., Tokyo, Japan) according to the manufacturer's instructions. To remove any contaminating genomic DNA, the RNA samples were treated with RNase-free DNase I (Takara Bio Inc., Otsu, Japan).
Reverse transcription (RT)-PCR and real-time quantitative RT-PCR (qRT-PCR).
After 3 h of induction, total RNA was isolated from the cells of SYK-6. A total RNA (5 μg) was reverse transcribed by ReverTra Ace reverse transcriptase (Toyobo, Osaka, Japan) with random 9-mer primers (Takara Bio Inc.). For each sample, a reverse transcriptase negative reaction was also prepared. The synthesized cDNA was purified by phenol-chloroform extraction and ethanol precipitation, and it was dissolved in 20 μl of DNase-free water. PCR was performed with 1 μl of the cDNA mixture, specific primers listed in Table S1 in the supplemental material, and ExTaq DNA polymerase (Takara Bio Inc.). A control PCR was performed with reverse transcriptase negative samples to verify no genomic DNA contamination. The resulting DNA was applied to 2.0% agarose gel electrophoresis and visualized with ethidium bromide.
A qRT-PCR was performed using a SYBR green PCR master mix (Applied Biosystems, Foster City, CA) with an ABI Prism 7000 sequence detection system (Applied Biosystems). The specific primer pairs used for qRT-PCR analyses were designed with the Primer Express version 2.0 software program (Applied Biosystems) (see Table S1 in the supplemental material). Real-time PCR was performed using 2 μl of cDNA sample, gene-specific primers (18 pmol), and SYBR green PCR master mix (10 μl) in a total reaction volume of 20 μl. Thermal cycling conditions were as follows: 15 min at 95°C, followed by 40 repeats of 15 s at 95°C and 1 min at 60°C. A change in the fluorescence emission was detected and analyzed with an ABI Prism 7000 sequence detection system. A melting-curve analysis was performed at the end of real-time PCR to verify the specific amplification. To normalize the amount of RNA in each sample, 16S rRNA was used as an internal standard. Each measurement was carried out in triplicate, and the means and standard deviations were calculated.
Construction of promoter probe vector.
The plasmid for promoter analysis was designed on the basis of the broad-host-range lacZ translational fusion vector pPR9TT. For the construction of a lacZ transcriptional fusion vector, pPR9TZ, pPR9TT was digested at the BamHI sites and blunt ended to replace a 3.6-kb SmaI-ScaI fragment containing full-length lacZ from pQF50.
LacZ reporter assay.
After 3 h of induction, SYK-6 cells harboring reporter plasmids were harvested by centrifugation (4,600 × g, 10 min, 4°C) and broken by ultrasonication in 20 mM Tris-HCl (pH 8.0). The supernatant was collected by centrifugation (19,000 × g, 30 min, 4°C). In order to determine the lacZ expression, β-galactosidase activities were measured using 4-methylumbelliferyl-β-d-galactopyranoside. The product, 4-methylumbelliferone (4MU), was detected with an RF-1500 spectrofluorophotometer (Shimadzu, Kyoto, Japan). One unit of β-galactosidase activity was defined as the amount of enzyme that catalyzed the production of 1 μmol of 4MU per min at 30°C. Specific activity was expressed in units per milligram of protein. The protein concentration was determined by the Bradford method (4). Each measurement was carried out in triplicate, and the means and standard deviations were calculated.
Primer extension.
After 3 h of induction, total RNAs were isolated from the cells of SYK-6 and SYK-6 harboring pVA01. The primer extension reactions were carried out with the Beckman dye D4 (D4)-labeled oligonucleotides PEligK43 and PEligK70 (positions +19 to +43 and +51 to +70 relative to the start codon of ligK, respectively) and PEligJ108 and PEligJ173 (positions +84 to +108 and +153 to +173 relative to the start codon of orf2, respectively). The primers (2 pmol) were annealed to total RNA (5 μg) after 5 min of denaturation at 65°C and were extended with PrimeScript reverse transcriptase (Takara Bio Inc.) at 42°C for 45 min. The enzyme was inactivated by being exposed to a temperature of 95°C for 5 min. The extended products were purified by phenol-chloroform extraction and ethanol precipitation and dissolved in 25 μl of CEQ sample loading solution (Beckman Coulter) combined with 0.5 μl of DNA size standard kit 400 (Beckman Coulter). Samples were analyzed utilizing a CEQ2000XL fragment analysis system.
Overexpression and purification of LigR.
The ligR coding sequence was amplified from pHN139F as a template with the ligR-NdeI-F and ligR-NdeI-R primers (see Table S1 in the supplemental material). The 1.3-kb PCR product was cloned into pT7Blue and confirmed by sequencing. The 1.3-kb NdeI-BamHI fragment from the resulting plasmid was inserted into the corresponding sites of pET21a(+) to yield pET21R. E. coli BL21(DE3) cells harboring pET21R were grown overnight in LB medium, and then 1 liter of the same medium was inoculated with 10 ml of overnight culture. After incubation at 30°C, isopropyl-β-d-thiogalactopyranoside was added to the culture to a final concentration of 1 mM when the A600 of the culture reached 0.5. Gene expression was induced at 30°C for 3 h. The cells were harvested by centrifugation (4,600 × g, 5 min, 4°C), resuspended in 50 mM Tris-HCl (pH 7.5) (buffer A), and broken by two passages through a French pressure cell (Aminco, Urbana, IL). The supernatant was then collected by centrifugation (27,200 × g, 30 min, 4°C) to remove cell debris, and streptomycin was added to the supernatant to a final concentration of 1% (wt/vol). The lysate was kept on ice for 10 min and centrifuged (27,200 × g, 30 min, 4°C). The supernatant was fractionated with ammonium sulfate (20 to 40%), and the precipitated proteins were resuspended in buffer A. The purification procedures described below were carried out using a Bioassist eZ system (Tosoh, Tokyo, Japan). A fraction containing LigR was applied to a TSKgel SP-5PW cation exchange column (7.5 by 75 mm; Tosoh) previously equilibrated with buffer A. The protein was eluted with 90 ml of a linear gradient of 0 to 1.0 M NaCl. The fractions containing LigR that eluted at approximately 0.4 M were pooled and concentrated by ultrafiltration (Amicon Ultra-4; Millipore, Bedford, MA). The purified protein sample was analyzed by SDS-PAGE.
Molecular weight determination of LigR.
After the remaining precipitant of the purified LigR sample was removed with an Ultrafree-MC filter (Millipore), the supernatant was used for static light-scattering experiments. The 20 μl of LigR protein (6.8 mg/ml) and each standard protein (10 mg/ml) were applied to a gel filtration column (Superdex200 10/300 GL; GE Healthcare, Buckinghamshire, United Kingdom) equilibrated with buffer A containing 0.4 M NaCl. The separated proteins were introduced into a MiniDAWN Tristar detector (Wyatt technology, Santa Barbara, CA) in order to measure the static light scattering at 690 nm. The absorbance at 280 nm and the refractive index (RI) were also measured with an SPD-20A UV detector (Shimadzu) and an RID-10A RI detector (Shimadzu), respectively. The ratio of the static light-scattering intensity (LS) to the protein concentration, which was determined with the RI, was proportional to the molecular weight of the protein (56). Each measurement was carried out in triplicate, and the means and standard deviations were calculated.
EMSAs.
Electrophoretic mobility shift assays (EMSAs) for LigR were performed with a DIG gel shift kit (2nd generation) (Roche, Mannheim, Germany) according to the manufacturer's instructions. DNA fragments were generated by PCR. Purified fragments were labeled at the 3′ end with digoxigenin (DIG)-11-ddUTP. The 29-bp DNA fragments, KRTN129, KRTN129SD, JTN129, and JTN129SD, were obtained by annealing of complementary oligonucleotides. The oligonucleotides used for EMSAs are listed in Table S1 in the supplemental material. The DNA-protein binding reactions were carried out at 20°C in a final volume of 10 μl containing 1 fmol of DIG-labeled probe, 1 μg of poly(dI-dC), 0.1 μg of poly-(l-lysine), and the LigR dimer (100 nM) in binding buffer (20 mM HEPES, 1 mM EDTA, 10 mM (NH4)2SO4, 1 mM dithiothreitol [DTT], 0.2% [wt/vol] Tween 20, and 30 mM KCl, pH 7.6) for 20 min. After incubation, 2.5 μl of loading buffer (60% [vol/vol] 0.25× TBE buffer [89 mM Tris, 89 mM boric acid, and 20 mM EDTA, pH 8.0], 40% [vol/vol] glycerol, and 0.2% [vol/vol] bromophenol blue) was added, and samples were separated on a 10% native Long Ranger polyacrylamide gel (FMC Bioproducts, Rockland, ME) in 0.5× TBE buffer at 80 V at 4°C for 2 to 4 h. After electrophoresis, the labeled DNAs were electroblotted onto Hybond-N+ membranes and detected using a CSPD detection system (Roche) with a LumiVision PRO image analyzer (Aisin Seiki Co., Ltd, Kariya, Japan). The intensity of single bands was measured using the LumiVision Analyzer software program (Aisin Seiki Co., Ltd). To determine the apparent dissociation constants (Kd), EMSAs were performed using 8 concentrations of purified LigR (range, 0 to 500 nM) and the KRTN12 or JTN12 probe in triplicate. The Kd values were estimated as the concentration of the LigR dimer at which 50% of the free probe was shifted (18).
DNase I footprinting assays.
DNA fragments used for DNase I footprinting were generated by PCR. 5′-DIG-labeled and unlabeled primers FPKRR and FPKRF were used to amplify the 238-bp DIG-labeled fragments containing the ligK promoter region, and the primers FPJF and FPJR were used to amplify the 251-bp DIG-labeled fragments containing the ligJ promoter region (see Table S1 in the supplemental material). The binding reactions were carried out at 20°C in a final volume of 100 μl containing 0.5 pmol of DIG-labeled probe, 100 μM DTT, 2 μg of salmon sperm DNA, 5 μg of bovine serum albumin (BSA), and the LigR dimer (100 nM) in binding buffer (20 mM Tris-HCl, 100 mM NaCl, 5 mM CaCl2, 3 mM MgCl2, 100 μM EDTA, pH 7.9) for 20 min. After the reaction, samples were preincubated at 25°C for 5 min, and partial digestion of the DNA was initiated by the addition of 50 mU of DNase I (Takara Bio Inc.). The incubation was continued for 1 min, and reactions were stopped by the addition of 25 μl of stop solution (1.5 M sodium acetate, 20 mM EDTA, 100 μg/ml yeast tRNA). The DNA was subsequently purified by phenol-chloroform extraction, ethanol precipitated, resuspended in 4.5 μl of loading buffer (8 M urea, 50 mM Tris-borate, 1 mM EDTA, 0.025% [vol/vol] bromophenol blue, 0.025% [vol/vol] xylene cyanol, pH 8.3), and separated by gel electrophoresis on a 6% polyacrylamide-8 M urea sequencing gel with a G/A sequencing ladder. After electrophoresis, the labeled DNAs were contact blotted onto Hybond-N+ membranes for 60 min and detected as described above.
Circular permutation analysis.
PCR-generated 200-bp fragments containing Box-K or Box-J at different positions relative to the ends of the fragments were labeled at the 3′ end with DIG-11-ddUTP using a DIG gel shift kit (2nd generation) (Roche). After three independent EMSAs performed with these fragments, the mobilities of the LigR-DNA complexes were determined by measuring the distance traveled from the well during electrophoresis. The relative mobilities were plotted and fitted to a second-order polynomial curve. The bending angles (α) were calculated using the formula μm/μe = cos(α/2), where μm is the mobility of the LigR-DNA complexes with a bend at the center of the fragment and μe is the mobility of the LigR-DNA complexes with a bend at the end of the fragment (47). The bending center was calculated as a coordinate corresponding to the minimal relative mobility in the fitted curve.
RESULTS
Transcriptional regulation of PCA45 pathway genes.
In a previous study, we estimated that the PCA45 pathway genes in SYK-6 consist of four transcriptional units, which are the ligK-orf1-ligI-lsdA operon, the ligJAB operon, and monocistronic ligC and ligR (12). We reevaluated the operon structure of the PCA45 pathway genes by reverse transcription (RT)-PCR analysis using total RNA isolated from SYK-6 cells grown in the presence of PCA. The amplification products were observed from all the regions tested except the region between ligR and orf2 (Fig. 1 C). No amplification was detected without RT. These results suggest that ligC is transcribed as part of the orf2-ligJABC operon and the PCA45 pathway genes consist of three transcriptional units, including the ligK-orf1-ligI-lsdA operon, the orf2-ligJABC operon, and monocistronic ligR.
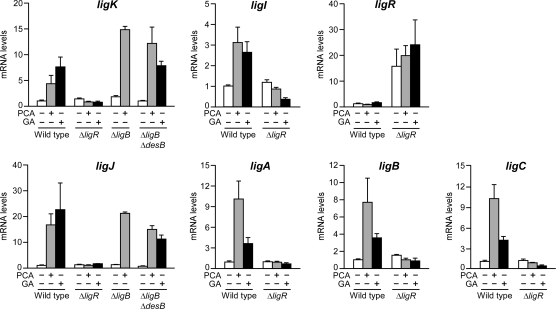
A quantitative RT-PCR (qRT-PCR) using total RNA isolated from the cells of SYK-6 and the ligR insertion mutant (DLR) was performed to measure mRNA levels of ligK, ligI, ligR, ligJ, ligA, ligB, and ligC. Transcription of all the genes except for ligR was induced 3- to 20-fold in the presence of PCA in SYK-6; however, these inductions were not seen in the ligR mutant strain DLR (Fig. 2). On the other hand, ligR was transcribed at a very low and constant level (1 × 10−2- to 1 × 10−3-fold the basal transcript amount of each enzyme gene), but DLR exhibited a higher transcriptional level of ligR than the wild type. These results demonstrate that LigR activates the transcription of the ligK and ligJ operons in the presence of PCA while the transcription of ligR is repressed by its own product. The mRNA levels of the PCA45 pathway genes in DLR cells under the induced condition were almost the same as those in noninduced SYK-6 cells, suggesting that LigR is required for the transcriptional activation of the ligK and ligJ operons in the presence of PCA.
FIG. 2.
qRT-PCR analysis of the expression of the PCA45 pathway genes. Total RNA was isolated from SYK-6 and its derivatives (ΔligR, DLR; ΔligB, AKB; and ΔligB ΔdesB, RBB) grown in the presence or absence of PCA or GA. Expression of the PCA45 pathway genes was measured using a qRT-PCR. Values for each mRNA level were normalized to 16S rRNA and shown as a fold increase over that for uninduced SYK-6 cells (level of 1.0). The data are mean values ± standard deviations for three independent experiments.
To determine whether PCA itself acts as an inducer, a qRT-PCR analysis was performed using total RNA from the ligB mutant (AKB), which lacks the ability to transform PCA (38). The mRNA levels of ligK and ligJ in AKB were increased 8- and 17-fold, respectively, in the presence of PCA (Fig. 2). These results suggest that PCA itself is an inducer of the ligK and ligJ operons.
In SYK-6, syringate catabolism conjoins with the PCA45 pathway at 2-pyrone 4,6-dicarboxylate (PDC) or 4-oxalomesaconate (Fig. 1A); therefore, expression of the ligK and ligJ operons is essential for syringate catabolism. However, PCA does not appear in the syringate catabolic pathway. Based on these facts, an alternative compound seemed to act as an inducer of the ligK and ligJ operons in syringate catabolism. SYK-6 degrades syringate to 3MGA (30), which is further degraded via gallate (GA) (1, 15, 30). Due to the structural similarity of GA to PCA (Fig. 1A), the ability of GA to induce the ligK and ligJ operons was examined (Fig. 2). The mRNA levels of the PCA45 pathway genes in SYK-6 were increased 3- to 22-fold in the presence of GA. To examine whether or not GA itself acts as an inducer, the desB ligB double mutant (RBB), which lacks GA transformation activity (15), was subjected to a qRT-PCR analysis. RBB grown in the presence of GA exhibited the transcriptional induction of ligK and ligJ, demonstrating that GA is also an inducer of the ligK and ligJ operons.
Determination of the ligK and ligJ promoters.
In order to determine the promoter regions of the ligK and ligJ operons and ligR, we constructed the lacZ reporter plasmids pZK, pZJTN12, and pZR, which carry a 204-bp region upstream of the start codon of ligK, a 160-bp region upstream of the start codon of orf2, and a 212-bp region upstream of the start codon of ligR, respectively. The expression of the ligK, ligJ, and ligR promoters in SYK-6 cells harboring the reporter plasmids was determined by means of β-galactosidase activity assays (Table 2). The ligK and ligJ promoter activities were increased 3.6- and 2.8-fold, respectively, in the presence of PCA. These results suggested that pZK and pZJTN12 contain the regions sufficient for the induction of the respective promoters in response to PCA. On the other hand, the ligR promoter activities were similar in the presence or absence of PCA, but the values were significantly higher than those of the ligK and ligJ promoters.
TABLE 2.
Expression of the ligK, ligJ, and ligR promoters in SYK-6
| Promoter | Plasmid | β-Galactosidase activity (mU/mg of protein) |
|
|---|---|---|---|
| SEMa | SEM + PCA | ||
| Vector control | pPR9TZ | 0.2 ± 0.0 | 0.2 ± 0.0 |
| ligK | pZK | 1.2 ± 0.3 | 4.3 ± 0.2 |
| pZKSD | 1.1 ± 0.1 | 1.1 ± 0.1 | |
| ligJ | pZJTN12 | 1.6 ± 0.2 | 4.5 ± 0.3 |
| pZJTN12SD | 1.9 ± 0.3 | 1.8 ± 0.2 | |
| ligR | pZR | 78 ± 4 | 85 ± 3 |
See Materials and Methods.
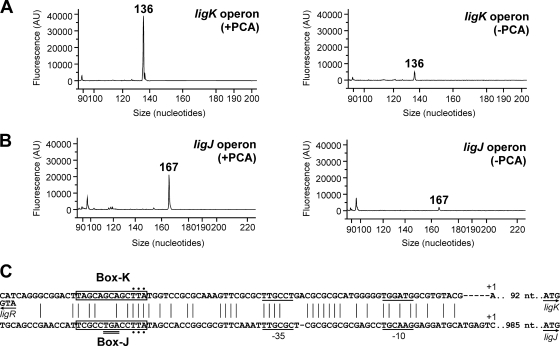
The transcription start sites of the ligK and ligJ operons were determined by primer extension analyses using fluorescently labeled oligonucleotides (see Table S1 in the supplemental material) and total RNA isolated from SYK-6 cells. Due to the fact that no significant extension product was obtained, we isolated total RNA from SYK-6 cells harboring pVA01, which carries the PCA45 pathway gene cluster, to increase transcription of the ligK and ligJ operons. When the primer PEligK43 was used, a 136-nucleotide (nt) fragment corresponding to a transcript initiating at position −93 from the ligK start codon was observed (Fig. 3 A). Because it was suggested that the transcription start site of the ligJ operon was located upstream of orf2 by utilization of RT-PCR and promoter analyses, we used a primer, PEligJ173, which annealed to positions +153 to +173 relative to the start codon of orf2. A 167-nt fragment corresponding to a transcript initiating at position +6 from the orf2 start codon was observed (Fig. 3B). These transcription start sites were confirmed by using different primers, PEligK70 and PEligJ108 (data not shown). The results obtained by primer extension analysis indicated that the region containing the orf2 translation start site is not transcribed from the ligJ promoter. Due to the lack of other potential start codons, orf2 appeared not to be translated. The amount of extension products clearly increased in the PCA-induced cells, corresponding to the results of the qRT-PCR analysis. Putative −35 and −10 sequences of the ligK and ligJ promoters were found upstream of the transcription start sites (Fig. 3C). The LTTR consensus binding sequences (45) of the ligK promoter (Box-K) and ligJ promoter (Box-J) are centered at positions −65 and −69 relative to the transcription start site of each operon, respectively. The locations of these motifs are consistent with those reported for other LTTR-activated promoters (20, 45, 49). Furthermore, Box-K is located immediately upstream of ligR (centered at position −18 relative to the start codon), suggesting that the binding of LigR to the motif results in autorepression. Alignment of the sequences upstream of the transcription start sites of the ligK and ligJ operons with the corresponding regions of the putative PCA45 pathway gene clusters in Sphingomonas sp. strain LB126 and Novosphingobium aromaticivorans DSM 12444 indicates that the −35, −10, and LTTR consensus binding sequences are well conserved among all these strains. We tried to determine the transcription start site of ligR; however, no significant extension product was obtained.
FIG. 3.
Determination of the transcription start sites of the ligK and ligJ operons. (A and B) Primer extension analyses of the ligK operon transcript using PEligK43 (A) or the ligJ operon transcript using PEligJ173 (B). Fluorescent primer extensions were performed with the D4-labeled primer, and total RNA was isolated from SYK-6 harboring pVA01, which carries ligK-orf1-ligI-lsdA, ligJABC, and ligR, grown in the presence (left) or absence (right) of PCA. Primer extension products were combined with DNA size standards and analyzed with the CEQ 2000XL fragment analysis system. (C) Alignment of sequences upstream from the transcription start sites of the ligK and ligJ operons. Vertical lines indicate conserved residues. Transcription start sites are indicated (+1). Putative −35 and −10 regions are underlined. Putative LigR binding sequences are boxed. The start codons of ligK, ligJ, and ligR are shown by arrows. The stop codon of ligR is indicated with a double underline. The residues indicated by dots were substituted nucleotides.
Purification of native LigR.
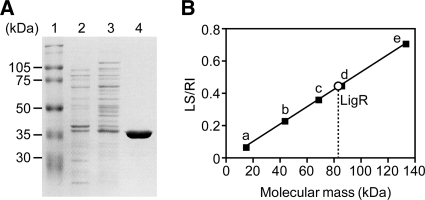
The ligR gene was cloned into the expression vector pET21a(+) to construct pET21R and expressed in E. coli BL21(DE3) under the control of the T7 promoter. Production of a 39-kDa protein was confirmed by SDS-PAGE (Fig. 4 A). The molecular mass of this product was close to that predicted from the amino acid sequence of LigR (41,907 Da). Recombinant LigR was purified in two steps: ammonium sulfate precipitation and cation exchange chromatography. To determine the molecular mass of LigR, we carried out a static light-scattering experiment. LigR was eluted as a single peak with a molecular mass of 83 ± 0.3 kDa (Fig. 4B), indicating that LigR is a dimer in solution. This feature is similar to those reported for other LTTRs (45, 49). A static light-scattering analysis was also performed in the presence of PCA (see Fig. S1 in the supplemental material), but the oligomeric state of LigR was not affected.
FIG. 4.
Native molecular mass of LigR. (A) Purification of LigR. Lanes: 1, molecular mass markers; 2, crude extract of E. coli BL21(DE3) harboring pET21R; 3, 20 to 40% ammonium sulfate fraction; 4, SP-5PW fraction. (B) Determination of the molecular mass of LigR by the static light-scattering method. LS and RI represent the intensity of static light scattering and the refractive index, respectively. Molecular masses of standard proteins are as follows: a, RNase A (14 kDa); b, ovalbumin (43 kDa); c, bovine serum albumin (66 kDa); d, dimer of ovalbumin (86 kDa); e, dimer of bovine serum albumin (132 kDa).
Binding of LigR to the ligK and ligJ promoters.
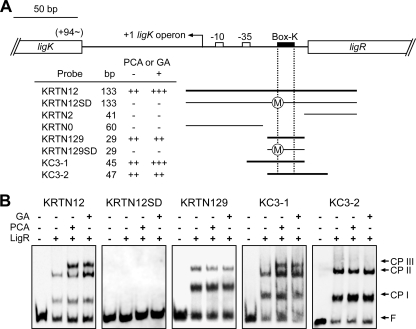
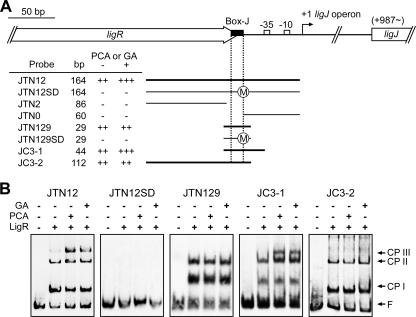
LigR was employed in electrophoretic mobility shift assays (EMSAs) with DNA fragments containing the promoter region of ligK (KRTN12; positions −120 to +13 relative to the transcription start site of the ligK operon) and ligJ (JTN12; positions −166 to −3 relative to the transcriptional start site of the ligJ operon) (Fig. 5 and 6). Both fragments contained a putative LigR binding site, Box-K or Box-J. Binding of LigR gave two shifted complexes (complexes I and II) for both the promoters in the absence of effectors. No further shifted complex was detected even when the concentration of LigR was increased (data not shown). The apparent dissociation constants (Kd) for LigR were estimated to be 3.7 × 10−8 M for the ligK promoter and 7.9 × 10−8 M for the ligJ promoter. These values are similar to those reported for other LTTRs (40, 46).
FIG. 5.
EMSA of the binding of LigR to the ligK promoter region. (A) DNA fragments used for the EMSA. Binding (++, +++; thicker bars) and nonbinding (−; thinner bars) of the respective fragments are indicated. “++” or “+++” represents the observation of two or three LigR-DNA complexes, respectively. The circled M in fragments KRTN12SD and KRTN129SD indicates that the T-N11-A motifs were changed by mutation. (B) EMSA with purified LigR and DIG-labeled fragments containing the ligK promoter region. The presence (+) or absence (−) of purified LigR (100 nM), PCA, and GA in the reaction mixtures is indicated. Inducers were added at a concentration of 1 mM in each reaction. The positions of complex I (CP I), complex II (CP II), complex III (CP III), and free probe (F) are indicated.
FIG. 6.
EMSA of the binding of LigR to the ligJ promoter region. (A) DNA fragments used for the EMSA. Binding (++, +++; thicker bars) and nonbinding (−; thinner bars) of the respective fragments are indicated. “++” and “+++” represent the observation of two or three LigR-DNA complexes, respectively. The circled M in fragments JTN12SD and JTN129SD indicates that the T-N11-A motifs were changed by mutation. (B) EMSA with purified LigR and DIG-labeled fragments containing the ligJ promoter region. The presence (+) or absence (−) of purified LigR (100 nM), PCA, and GA in the reaction mixtures is indicated. Inducers were added at a concentration of 1 mM in each reaction. The positions of complex I (CP I), complex II (CP II), complex III (CP III), and free probe (F) are indicated.
EMSAs were also performed in the presence of PCA or GA. Interestingly, an additional retarded band (complex III) appeared for both promoters (Fig. 5 and 6). We tested the effect of vanillate, syringate, 4-carboxy-2-hydroxymuconate-6-semialdehyde, PDC, and 3MGA (Fig. 1) on LigR binding, but the binding patterns of LigR for the ligK and ligJ promoters were not changed (data not shown). These results demonstrated that LigR directly and specifically recognizes PCA and GA. Furthermore, it is also suggested that interaction of LigR with PCA or GA altered the mode of protein-DNA interaction.
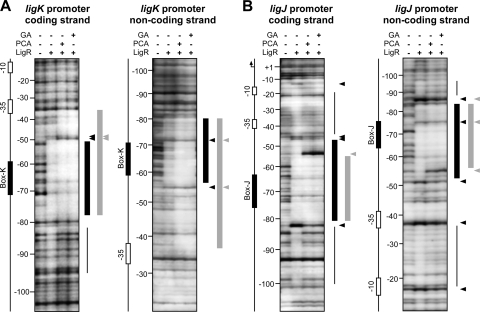
The localization of the LigR binding sites within the ligK and ligJ promoter regions was determined by DNase I footprinting on both strands of the 238-bp fragment (positions −182 to +42 relative to the transcription start site of the ligK operon) and the 251-bp fragment (positions −177 to +74 relative to the transcription start site of the ligJ operon). In the case of the ligK promoter, LigR strongly protected a region on the coding strand between −77 and −51 and also a region between −80 and −56 on the noncoding strand (Fig. 7 A). A similar footprint pattern showing protections of a region between −80 and −48 on the coding strand and a region between −83 and −52 on the noncoding strand was observed with the ligJ promoter fragment (Fig. 7B). The protected regions in both strands of the ligK and ligJ promoter fragments overlapped Box-K and Box-J, respectively, suggesting that LigR binds to these motifs. LigR generated several DNase I-hypersensitive sites in the ligK and ligJ promoters, which suggests that binding of LigR induces DNA bending. Weaker protections were observed just upstream and both upstream and downstream from the strongly protected area in the ligK and ligJ promoters, respectively. In the presence of PCA or GA, the 3′ end of the protected region of the ligK promoter was significantly extended from −51 to −36. Unlike the ligK promoter, the 3′ end of the protected region of the ligJ promoter was shortened from −47 to −55.
FIG. 7.
DNase I footprinting of LigR binding to the ligK and ligJ promoter regions. The 238-bp fragment or the 251-bp fragment containing the ligK (A) or ligJ (B) promoter region, respectively, was labeled with DIG on the coding (left) or noncoding (right) strands. The presence (+) or absence (−) of purified LigR (100 nM), PCA, and GA in the reaction mixture is indicated. Inducers were added at a concentration of 1 mM in each reaction. Thicker bars indicate strong protection by LigR, and thinner bars show weakly protected regions. The DNase I-hypersensitive sites are shown with arrowheads. The colors of bars and arrowheads indicate the reactions performed in the absence (black) or presence (gray) of PCA or GA, respectively.
Involvement of Box-K and Box-J in LigR binding and promoter activities.
In order to confirm the role of Box-K and Box-J in binding of LigR to the ligK and ligJ promoter regions, we performed EMSA using deletion probes (Fig. 5 and 6). When the 29-bp fragments (KRTN129 and JTN129) which contain Box-K and Box-J, respectively, were used as probes, two shifted complexes were observed. However, these complexes were no longer observed in the EMSA using fragments which lack Box-K (KRTN2 and KRTN0) or Box-J (JTN2 and JTN0) (data not shown). Furthermore, mutant probes bearing three substitutions at Box-K (KRTN12SD and KRTN129SD) or Box-J (JTN12SD and JTN129SD) gave no shifted complex. These results demonstrated that Box-K and Box-J are essential for binding of LigR to the ligK and ligJ promoters, respectively.
The reporter plasmids pZKSD and pZJTN12SD, harboring mutations at the same positions as the mutant probes used in the EMSA, were constructed to examine the effect of the mutations on the promoter activities. LigR-mediated activation in response to PCA was abolished in the cells harboring these mutated promoters (Table 2), suggesting that the binding of LigR to Box-K and Box-J is critical for activation of the ligK and ligJ promoters.
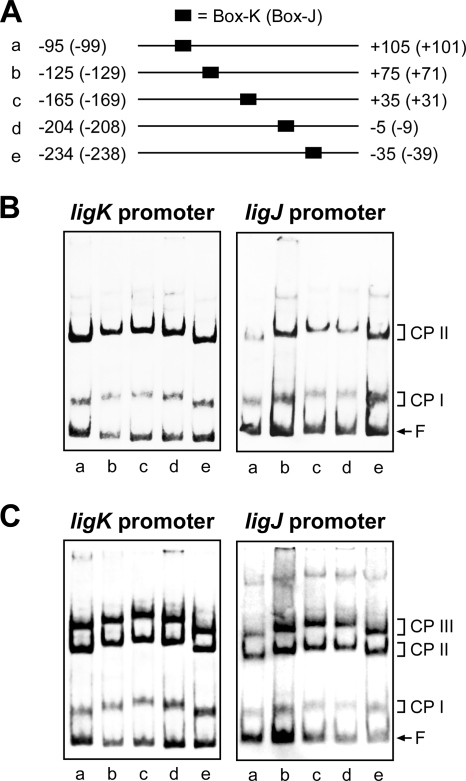
Bending of the ligK and ligJ promoter regions.
A large number of LTTRs have been shown to bend DNA, and sometimes inducers change the bending angle (20, 32, 45, 49). The ability of LigR to bend DNA at its binding sites was examined by circular permutation assays. As shown in Fig. 8 A, the DNA fragments used in the assays were prepared using PCR amplification in such a way that the position of Box-K and Box-J in the probe could be changed while maintaining the same overall length of the fragment. If binding of the protein induces a bend in the DNA, a complex formed on a probe with the binding site near the end of the probe will migrate faster than that in the center of the probe. The mobility of complexes I and II was minimal when the LigR-binding sites (Box-K and Box-J) were centrally located and maximal when the site was closest to the end of the fragment (Fig. 8B). The estimated bending angles for complexes I and II were 41° and 51° on the ligK promoter fragment and 31° and 46° on the ligJ promoter fragment, respectively. The possible locations of the bend centers were estimated to be in the middle of Box-K and the 5′-proximal part of Box-J. To examine the effect of the inducers on DNA bending by LigR, circular permutation assays were also performed in the presence of PCA (Fig. 8C). The DNA bending angles in complexes I and II were not affected by the presence of PCA. On the other hand, the bending angles in complex III (68° and 61° on the ligK and ligJ promoter fragments, respectively) were significantly larger than those in complex II, although the bending center was not changed. Almost the same bending angles were estimated when GA was added to the assay mixture (data not shown).
FIG. 8.
Circular permutation analysis of LigR binding to the ligK and ligJ promoter regions. (A) DNA probes used in circular permutation analysis. The 200-bp DNA fragments (a to e) contain Box-K or Box-J in different positions relative to the end of the fragment. The numbers noted next to the probes indicate positions of the ends of the probes relative to the transcription start sites of the ligK and ligJ (in parentheses) promoters. (B and C) EMSA demonstrating DNA bending by LigR. Binding reactions were carried out in the absence (B) or presence (C) of 1 mM PCA. The positions of complex I (CP I), complex II (CP II), complex III (CP III), and free probe (F) are indicated.
DNA regions required for formation of complex III.
Complex III was not observed in the EMSA when the short fragments KRTN129 (positions −51 to −79 relative to the transcription start site of the ligK operon) and JTN129 (positions −55 to −83 relative to the transcription start site of the ligJ operon) were used as probes (Fig. 5 and 6). To determine the regions required for the formation of complex III, we performed an EMSA using KC3-1 (positions −35 to −79) and KC3-2 (positions −51 to −96) as the Box-K probes (Fig. 5) and JC3-1 (positions −40 to −83) and JC3-2 (positions −55 to −166) as the Box-J probes (Fig. 6). In the presence of PCA or GA, complex III was observed only with the KC3-1 and JC3-1 probes. These results strongly suggest that the regions between the LigR binding boxes and the −35 regions of the respective promoters are essential for the formation of complex III.
DISCUSSION
qRT-PCR analyses demonstrated that LigR activates the transcription of the ligK and ligJ operons in the presence of PCA or GA and represses its own transcription. In SYK-6, vanillate and syringate, which are the intermediate metabolites of guaiacyl and syringyl lignins, are converted to PCA and 3MGA, respectively (Fig. 1). PCA is further degraded via the PCA45 pathway, while 3MGA is catabolized through multiple ring cleavage pathways (15); however, these pathways eventually conjoin with the PCA45 pathway. GA is only generated by O demethylation of 3MGA catalyzed by LigM in the multiple 3MGA degradation pathways, indicating that the degradation pathway for 3MGA via GA is crucial for the catabolism of syringyl lignin.
EMSAs showed that binding of LigR produces two shifted complexes (complexes I and II). Since LigR is a dimer in solution and most LTTRs bind to the promoter DNA as a dimer or a tetramer (32, 34, 35, 49, 50), it appears that complexes I and II resulted from the binding of LigR as a dimer and a dimer of dimers, respectively. The LTTR consensus binding sequences (45), Box-K and Box-J, are centered at positions −65 and −69 relative to the transcriptional start sites of the ligK and ligJ operons, respectively. These motifs do not contain apparent inverted repeats but are similar in locations to those of other LTTRs (49). Substitution mutant analyses demonstrated that Box-K and Box-J are essential for the binding of LigR and activation of the ligK and ligJ promoters. These results suggest that Box-K and Box-J function as RBS of the ligK and ligJ promoters. Because complex III was observed only in the presence of effector, this complex seems to be the transcriptionally active form. Complex III was observed in the EMSA with the KRTN12, KC3-1, JTN12, and JC3-1 probes; however, it was not detected with the KRTN129, KC3-2, JTN129, and JC3-2 probes (Fig. 5 and 6). Therefore, the regions between the LigR binding boxes and the −35 regions of the respective promoters appear to contain the activation binding sites (ABS). DNase I footprinting and circular permutation assays demonstrated that LigR protected the region encompassing the RBS and induced DNA bending on the ligK and ligJ promoters. Interaction of LigR with effector extended the protected region to a position proximal to −35 and enhanced DNA bending on the ligK promoter. On the other hand, the shortening of the protected region was observed for the ligJ promoter, while the bending angle was increased. The alterations of hypersensitivity patterns observed in the presence of the effector also differed between these promoters. These results may suggest that binding of effectors results in different conformational changes in the LigR-DNA complex on the ligK and ligJ promoters. However, the region required for the formation of complex III was not protected in the footprint of the ligJ promoter in the presence of effector. Further analysis will be necessary to clarify the manner of binding of LigR with the −35 region of the ligJ promoter.
The behavior of LigR toward both the ligK and ligJ promoters is distinct from the model of well-characterized LTTRs, such as AtzR, CbnR, OccR, and OxyR, known as the sliding dimer model. The LTTRs cause the effector-dependent shortening of the protected region from positions −80 to −20 to positions −80 to −30 and a relaxation of the DNA bending, which are all thought to be important in the release of the recognition site of RNA polymerase and the conformational change of regulator-DNA complex suitable for transcriptional activation (6, 40, 48, 51-54). TrpI is one exception to this pattern (10). In the absence of an effector molecule, indoleglycerol phosphate (InGP), TrpI protected a region between −77 and −52 of the trpBA promoter. When InGP was present, TrpI enhanced DNA bending with extended protection to −32 (5, 39). It was speculated that the extension of the binding region proximal to −35 allowed the interaction of TrpI with RNA polymerase (10). In the case of the ligK promoter, the PCA- and GA-dependent structural alteration of the LigR-DNA complex seems to cause the same effect as TrpI on transcriptional activation. Even though the behavior of LigR (binding to the ligK promoter) and TrpI with respect to DNA protection and DNA bending are different from those of LTTRs mentioned above, the eventual binding of a regulator to the −35 region in response to an inducer appears to be important for transcriptional activation by LTTRs.
The apparent Kds for LigR were determined to be 3.7 × 10−8 M for the ligK promoter and 7.9 × 10−8 M for the ligJ promoter. Because LigR had an approximately 2-fold-higher affinity for the ligK promoter than the ligJ promoter, LigR may bind preferentially to the ligK promoter region in SYK-6 cells. Considering the fact that the binding of LigR to the ligK promoter region would be directly involved in the autorepression of ligR, the higher affinity to the ligK promoter region may represent a mechanism for keeping LigR low in abundance. This hypothesis is consistent with the low level of ligR transcription demonstrated by the qRT-PCR analysis. On the other hand, the promoter activity of ligR was significantly higher than those of the ligK and ligJ promoters despite a ligR promoter fragment containing Box-K (Table 2). High-level ligR promoter activity was also seen in SYK-6 cells harboring a ligR expression plasmid (data not shown). These observations suggest that the binding of LigR to Box-K is not sufficient for maximal autorepression. It might be supposed that cooperative binding of LigR to Box-K and Box-J facilitates the repression of ligR transcription through protein-protein interaction. Furthermore, Box-J overlaps with the stop codon of ligR. Binding of LigR to Box-J may directly influence the transcription of ligR. These hypotheses need to be addressed in a future study.
Supplementary Material
Acknowledgments
We are grateful to S. Valla for the gift of pPR9TT.
This work was supported in part by a Grant-in-Aid for Scientific Research (16580055) from the Ministry of Education, Culture, Sports and Technology, Japan.
Footnotes
Published ahead of print on 30 April 2010.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Abe, T., E. Masai, K. Miyauchi, Y. Katayama, and M. Fukuda. 2005. A tetrahydrofolate-dependent O-demethylase, LigM, is crucial for catabolism of vanillate and syringate in Sphingomonas paucimobilis SYK-6. J. Bacteriol. 187:2030-2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bagdasarian, M., R. Lurz, B. Rückert, F. C. Franklin, M. M. Bagdasarian, J. Frey, and K. N. Timmis. 1981. Specific-purpose plasmid cloning vectors. II. Broad host range, high copy number, RSF1010-derived vectors, and a host-vector system for gene cloning in Pseudomonas. Gene 16:237-247. [DOI] [PubMed] [Google Scholar]
- 3.Bolivar, F., and K. Backman. 1979. Plasmids of Escherichia coli as cloning vectors. Methods Enzymol. 68:245-267. [DOI] [PubMed] [Google Scholar]
- 4.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 5.Chang, M., and I. P. Crawford. 1990. The roles of indoleglycerol phosphate and the TrpI protein in the expression of trpBA from Pseudomonas aeruginosa. Nucleic Acids Res. 18:979-988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dubbs, P., J. M. Dubbs, and F. R. Tabita. 2004. Effector-mediated interaction of CbbRI and CbbRII regulators with target sequences in Rhodobacter capsulatus. J. Bacteriol. 186:8026-8035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eaton, R. W. 2001. Plasmid-encoded phthalate catabolic pathway in Arthrobacter keyseri 12B. J. Bacteriol. 183:3689-3703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Farinha, M. A., and A. M. Kropinski. 1990. Construction of broad-host-range plasmid vectors for easy visible selection and analysis of promoters. J. Bacteriol. 172:3496-3499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Figurski, D. H., and D. R. Helinski. 1979. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc. Natl. Acad. Sci. U. S. A. 76:1648-1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gao, J., and G. N. Gussin. 1991. Mutations in TrpI binding site II that differentially affect activation of the trpBA promoter of Pseudomonas aeruginosa. EMBO J. 10:4137-4144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hara, H., E. Masai, Y. Katayama, and M. Fukuda. 2000. The 4-oxalomesaconate hydratase gene, involved in the protocatechuate 4,5-cleavage pathway, is essential to vanillate and syringate degradation in Sphingomonas paucimobilis SYK-6. J. Bacteriol. 182:6950-6957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hara, H., E. Masai, K. Miyauchi, Y. Katayama, and M. Fukuda. 2003. Characterization of the 4-carboxy-4-hydroxy-2-oxoadipate aldolase gene and operon structure of the protocatechuate 4,5-cleavage pathway genes in Sphingomonas paucimobilis SYK-6. J. Bacteriol. 185:41-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kasai, D., E. Masai, Y. Katayama, and M. Fukuda. 2007. Degradation of 3-O-methylgallate in Sphingomonas paucimobilis SYK-6 by pathways involving protocatechuate 4,5-dioxygenase. FEMS Microbiol. Lett. 274:323-328. [DOI] [PubMed] [Google Scholar]
- 14.Kasai, D., E. Masai, K. Miyauchi, Y. Katayama, and M. Fukuda. 2004. Characterization of the 3-O-methylgallate dioxygenase gene and evidence of multiple 3-O-methylgallate catabolic pathways in Sphingomonas paucimobilis SYK-6. J. Bacteriol. 186:4951-4959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kasai, D., E. Masai, K. Miyauchi, Y. Katayama, and M. Fukuda. 2005. Characterization of the gallate dioxygenase gene: three distinct ring cleavage dioxygenases are involved in syringate degradation by Sphingomonas paucimobilis SYK-6. J. Bacteriol. 187:5067-5074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Katayama, Y., S. Nishikawa, M. Nakamura, K. Yano, M. Yamasaki, N. Morohoshi, and T. Haraguchi. 1987. Cloning and expression of Pseudomonas paucimobilis SYK-6 genes involved in the degradation of vanillate and protocatechuate in P. putida. Mokuzai Gakkaishi 33:77-79. [Google Scholar]
- 17.Kersten, P. J., S. Dagley, J. W. Whittaker, D. M. Arciero, and J. D. Lipscomb. 1982. 2-Pyrone-4,6-dicarboxylic acid, a catabolite of gallic acids in Pseudomonas species. J. Bacteriol. 152:1154-1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Larsen, R., J. Kok, and O. P. Kuipers. 2005. Interaction between ArgR and AhrC controls regulation of arginine metabolism in Lactococcus lactis. J. Biol. Chem. 280:19319-19330. [DOI] [PubMed] [Google Scholar]
- 19.Leonowicz, A., N. S. Cho, J. Luterek, A. Wilkolazka, M. Wojtas-Wasilewska, A. Matuszewska, M. Hofrichter, D. Wesenberg, and J. Rogalski. 2001. Fungal laccase: properties and activity on lignin. J. Basic Microbiol. 41:185-227. [DOI] [PubMed] [Google Scholar]
- 20.Maddocks, S. E., and P. C. Oyston. 2008. Structure and function of the LysR-type transcriptional regulator (LTTR) family proteins. Microbiology 154:3609-3623. [DOI] [PubMed] [Google Scholar]
- 21.Martínez, Á. T., M. Speranza, F. J. Ruiz-Dueñas, P. Ferreira, S. Camarero, F. Guillén, M. J. Martínez, A. Gutiérrez, and J. C. del Río. 2005. Biodegradation of lignocellulosics: microbial, chemical, and enzymatic aspects of the fungal attack of lignin. Int. Microbiol. 8:195-204. [PubMed] [Google Scholar]
- 22.Maruyama, K. 1979. Isolation and identification of the reaction product of α-hydroxy-γ-carboxymuconic ɛ-semialdehyde dehydrogenase. J. Biochem. 86:1671-1677. [DOI] [PubMed] [Google Scholar]
- 23.Maruyama, K. 1983. Purification and properties of 2-pyrone-4,6-dicarboxylate hydrolase. J. Biochem. 93:557-565. [DOI] [PubMed] [Google Scholar]
- 24.Maruyama, K. 1990. Purification and properties of 4-hydroxy-4-methyl-2-oxoglutarate aldolase from Pseudomonas ochraceae grown on phthalate. J. Biochem. 108:327-333. [DOI] [PubMed] [Google Scholar]
- 25.Maruyama, K. 1985. Purification and properties of γ-oxalomesaconate hydratase from Pseudomonas ochraceae grown with phthalate. Biochem. Biophys. Res. Commun. 128:271-277. [DOI] [PubMed] [Google Scholar]
- 26.Maruyama, K., N. Ariga, M. Tsuda, and K. Deguchi. 1978. Purification and properties of α-hydroxy-γ-carboxymuconic ɛ-semialdehyde dehydrogenase. J. Biochem. 83:1125-1134. [DOI] [PubMed] [Google Scholar]
- 27.Maruyama, K., M. Miwa, N. Tsujii, T. Nagai, N. Tomita, T. Harada, H. Sobajima, and H. Sugisaki. 2001. Cloning, sequencing, and expression of the gene encoding 4-hydroxy-4-methyl-2-oxoglutarate aldolase from Pseudomonas ochraceae NGJ1. Biosci. Biotechnol. Biochem. 65:2701-2709. [DOI] [PubMed] [Google Scholar]
- 28.Masai, E., Y. Katayama, and M. Fukuda. 2007. Genetic and biochemical investigations on bacterial catabolic pathways for lignin-derived aromatic compounds. Biosci. Biotechnol. Biochem. 71:1-15. [DOI] [PubMed] [Google Scholar]
- 29.Masai, E., K. Momose, H. Hara, S. Nishikawa, Y. Katayama, and M. Fukuda. 2000. Genetic and biochemical characterization of 4-carboxy-2-hydroxymuconate-6-semialdehyde dehydrogenase and its role in the protocatechuate 4,5-cleavage pathway in Sphingomonas paucimobilis SYK-6. J. Bacteriol. 182:6651-6658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Masai, E., M. Sasaki, Y. Minakawa, T. Abe, T. Sonoki, K. Miyauchi, Y. Katayama, and M. Fukuda. 2004. A novel tetrahydrofolate-dependent O-demethylase gene is essential for growth of Sphingomonas paucimobilis SYK-6 with syringate. J. Bacteriol. 186:2757-2765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Masai, E., S. Shinohara, H. Hara, S. Nishikawa, Y. Katayama, and M. Fukuda. 1999. Genetic and biochemical characterization of a 2-pyrone-4,6-dicarboxylic acid hydrolase involved in the protocatechuate 4,5-cleavage pathway of Sphingomonas paucimobilis SYK-6. J. Bacteriol. 181:55-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Muraoka, S., R. Okumura, N. Ogawa, T. Nonaka, K. Miyashita, and T. Senda. 2003. Crystal structure of a full-length LysR-type transcriptional regulator, CbnR: unusual combination of two subunit forms and molecular bases for causing and changing DNA bend. J. Mol. Biol. 328:555-566. [DOI] [PubMed] [Google Scholar]
- 33.Noda, Y., S. Nishikawa, K. Shiozuka, H. Kadokura, H. Nakajima, K. Yoda, Y. Katayama, N. Morohoshi, T. Haraguchi, and M. Yamasaki. 1990. Molecular cloning of the protocatechuate 4,5-dioxygenase genes of Pseudomonas paucimobilis. J. Bacteriol. 172:2704-2709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Parsek, M. R., S. M. McFall, D. L. Shinabarger, and A. M. Chakrabarty. 1994. Interaction of two LysR-type regulatory proteins CatR and ClcR with heterologous promoters: functional and evolutionary implications. Proc. Natl. Acad. Sci. U. S. A. 91:12393-12397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Parsek, M. R., D. L. Shinabarger, R. K. Rothmel, and A. M. Chakrabarty. 1992. Roles of CatR and cis,cis-muconate in activation of the catBC operon, which is involved in benzoate degradation in Pseudomonas putida. J. Bacteriol. 174:7798-7806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Peng, X., T. Egashira, K. Hanashiro, E. Masai, S. Nishikawa, Y. Katayama, K. Kimbara, and M. Fukuda. 1998. Cloning of a Sphingomonas paucimobilis SYK-6 gene encoding a novel oxygenase that cleaves lignin-related biphenyl and characterization of the enzyme. Appl. Environ. Microbiol. 64:2520-2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Peng, X., E. Masai, D. Kasai, K. Miyauchi, Y. Katayama, and M. Fukuda. 2005. A second 5-carboxyvanillate decarboxylase gene, ligW2, is important for lignin-related biphenyl catabolism in Sphingomonas paucimobilis SYK-6. Appl. Environ. Microbiol. 71:5014-5021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Peng, X., E. Masai, H. Kitayama, K. Harada, Y. Katayama, and M. Fukuda. 2002. Characterization of the 5-carboxyvanillate decarboxylase gene and its role in lignin-related biphenyl catabolism in Sphingomonas paucimobilis SYK-6. Appl. Environ. Microbiol. 68:4407-4415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Piñeiro, S., I. Olekhnovich, and G. N. Gussin. 1997. DNA bending by the TrpI protein of Pseudomonas aeruginosa. J. Bacteriol. 179:5407-5413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Porrúa, O., M. García-Jaramillo, E. Santero, and F. Govantes. 2007. The LysR-type regulator AtzR binding site: DNA sequences involved in activation, repression and cyanuric acid-dependent repositioning. Mol. Microbiol. 66:410-427. [DOI] [PubMed] [Google Scholar]
- 41.Providenti, M. A., J. Mampel, S. MacSween, A. M. Cook, and R. C. Wyndham. 2001. Comamonas testosteroni BR6020 possesses a single genetic locus for extradiol cleavage of protocatechuate. Microbiology 147:2157-2167. [DOI] [PubMed] [Google Scholar]
- 42.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 43.Santos, P. M., I. Di Bartolo, J. M. Blatny, E. Zennaro, and S. Valla. 2001. New broad-host-range promoter probe vectors based on the plasmid RK2 replicon. FEMS Microbiol. Lett. 195:91-96. [DOI] [PubMed] [Google Scholar]
- 44.Sato, Y., H. Moriuchi, S. Hishiyama, Y. Otsuka, K. Oshima, D. Kasai, M. Nakamura, S. Ohara, Y. Katayama, M. Fukuda, and E. Masai. 2009. Identification of three alcohol dehydrogenase genes involved in the stereospecific catabolism of arylglycerol-β-aryl ether by Sphingobium sp. strain SYK-6. Appl. Environ. Microbiol. 75:5195-5201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schell, M. A. 1993. Molecular biology of the LysR family of transcriptional regulators. Annu. Rev. Microbiol. 47:597-626. [DOI] [PubMed] [Google Scholar]
- 46.Sperandio, B., C. Gautier, S. McGovern, D. S. Ehrlich, P. Renault, I. Martin-Verstraete, and E. Guédon. 2007. Control of methionine synthesis and uptake by MetR and homocysteine in Streptococcus mutans. J. Bacteriol. 189:7032-7044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Thompson, J. F., and A. Landy. 1988. Empirical estimation of protein-induced DNA bending angles: applications to λ site-specific recombination complexes. Nucleic Acids Res. 16:9687-9705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Toledano, M. B., I. Kullik, F. Trinh, P. T. Baird, T. D. Schneider, and G. Storz. 1994. Redox-dependent shift of OxyR-DNA contacts along an extended DNA-binding site: a mechanism for differential promoter selection. Cell 78:897-909. [DOI] [PubMed] [Google Scholar]
- 49.Tropel, D., and J. R. van der Meer. 2004. Bacterial transcriptional regulators for degradation pathways of aromatic compounds. Microbiol. Mol. Biol. Rev. 68:474-500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.van Keulen, G., L. Girbal, E. R. van den Bergh, L. Dijkhuizen, and W. G. Meijer. 1998. The LysR-type transcriptional regulator CbbR controlling autotrophic CO2 fixation by Xanthobacter flavus is an NADPH sensor. J. Bacteriol. 180:1411-1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.van Keulen, G., A. N. Ridder, L. Dijkhuizen, and W. G. Meijer. 2003. Analysis of DNA binding and transcriptional activation by the LysR-type transcriptional regulator CbbR of Xanthobacter flavus. J. Bacteriol. 185:1245-1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang, L., J. D. Helmann, and S. C. Winans. 1992. The A. tumefaciens transcriptional activator OccR causes a bend at a target promoter, which is partially relaxed by a plant tumor metabolite. Cell 69:659-667. [DOI] [PubMed] [Google Scholar]
- 53.Wang, L., and S. C. Winans. 1995. High angle and ligand-induced low angle DNA bends incited by OccR lie in the same plane with OccR bound to the interior angle. J. Mol. Biol. 253:32-38. [DOI] [PubMed] [Google Scholar]
- 54.Wang, L., and S. C. Winans. 1995. The sixty nucleotide OccR operator contains a subsite essential and sufficient for OccR binding and a second subsite required for ligand-responsive DNA bending. J. Mol. Biol. 253:691-702. [DOI] [PubMed] [Google Scholar]
- 55.Wattiau, P., L. Bastiaens, R. van Herwijnen, L. Daal, J. R. Parsons, M. E. Renard, D. Springael, and G. R. Cornelis. 2001. Fluorene degradation by Sphingomonas sp. LB126 proceeds through protocatechuic acid: a genetic analysis. Res. Microbiol. 152:861-872. [DOI] [PubMed] [Google Scholar]
- 56.Wen, J., T. Arakawa, and J. S. Philo. 1996. Size-exclusion chromatography with on-line light-scattering, absorbance, and refractive index detectors for studying proteins and their interactions. Anal. Biochem. 240:155-166. [DOI] [PubMed] [Google Scholar]
- 57.Yanisch-Perron, C., J. Vieira, and J. Messing. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33:103-119. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.