Abstract
In Shigella flexneri, the polysaccharide copolymerase (PCP) protein WzzSF confers a modal length of 10 to 17 repeat units (RUs) to the O-antigen (Oag) component of lipopolysaccharide (LPS). PCPs form oligomeric structures believed to be related to their function. To identify functionally important regions within WzzSF, random in-frame linker mutagenesis was used to create mutants with 5-amino-acid insertions (termed Wzzi proteins), and DNA sequencing was used to locate the insertions. Analysis of the resulting LPS conferred by Wzzi proteins identified five mutant classes. The class I mutants were inactive, resulting in nonregulated LPS Oag chains, while classes II and III conferred shorter LPS Oag chains of 2 to 10 and 8 to 14 RUs, respectively. Class IV mutants retained near-wild-type function, and class V mutants increased the LPS Oag chain length to 16 to 25 RUs. In vivo formaldehyde cross-linking indicated class V mutants readily formed high-molecular-mass oligomers; however, class II and III Wzzi mutants were not effectively cross-linked. Wzz dimer stability was also investigated by heating cross-linked oligomers at 100°C in the presence of SDS. Unlike the WzzSF wild type and class IV and V Wzzi mutants, the class II and III mutant dimers were not detectable. The location of each insertion was mapped onto available PCP three-dimensional (3D) structures, revealing that class V mutations were most likely located within the inner cavity of the PCP oligomer. These data suggest that the ability to produce stable dimers may be important in determining Oag modal chain length.
Lipopolysaccharide (LPS) of Shigella flexneri is an important virulence factor, providing protection against host defenses and affecting interaction with host cells. LPS is composed of three regions: the hydrophobic lipid A membrane anchor, the core sugar region, and the O-antigen (Oag) polysaccharide chain (18). The basic Oag repeat unit of S. flexneri is a tetrasaccharide consisting of three rhamnose sugars and one N-acetylglucosamine sugar (19). The contribution of Shigella Oag to establishing virulence has been extensively investigated, and results indicate that regulated Oag modal length is required for virulence (8, 23, 25). Loss of Oag modal chain length regulation affects virulence due to the masking of the outer membrane (OM) protein IcsA (8, 23), and the type III secretion system is also affected by Oag chain length (24).
The current model for Oag biogenesis in S. flexneri involves the initiation of Oag repeat unit synthesis on the cytoplasmic face of the inner membrane (IM) and continues with a series of successive sugar transferase reactions. The repeat units are assembled on the lipid carrier undecaprenol phosphate (Und-P), and transported across the IM by the Wzx flippase to the periplasmic face of the IM. Polymerization of Oag repeat units is catalyzed by the Wzy polymerase, linking the individual oligosaccharide repeat units into a chain; the nascent chain is transferred from its lipid carrier to the nonreducing end of the newly flipped oligosaccharide repeat unit. The resulting chain is then ligated to the lipid A core by WaaL ligase (18, 26) to form LPS.
The regulation of the chain length of the Oag polysaccharide is controlled by the Wzz protein, a member of the polysaccharide copolymerase 1a (PCP1a) family (13, 21). S. flexneri Wzz (WzzSF) confers an average chain modal length of 10 to 17 Oag repeat units. In addition to determining the Oag chain modal length, PCP proteins are involved in enterobacterial common antigen (ECA) modal chain length regulation and biosynthesis and in capsule polysaccharide (CPS) and exopolysaccharide (EPS) biosynthesis (13). The PCP1a proteins are located in the IM and have two transmembrane (TM) regions, TM1 and TM2 (14). TM1 is located close to the N-terminal end, and TM2 is located near the C-terminal end, while the hydrophilic region between TM regions is located in the periplasm (14). PCPs exhibit a conserved motif, proximal to and partly overlapping the TM2 region, rich in proline and glycine residues (2, 3, 13). Site-directed mutagenesis studies targeting a number of these conserved residues, singularly or in combination, indicate that changes to this region have a significant effect on the resulting Oag modal chain length (4). Many mutagenesis studies on residues throughout Wzz indicate that function may be an overall property of the protein and may not be limited to one particular region (4, 6, 21). Despite studies conducted to probe the Wzz structure function relationship, little is known about the mode of action in determining Oag modal chain length. Recently, the periplasmic domain structures of a collection of PCP proteins including Salmonella enterica serovar Typhimurium WzzB (WzzST) and Escherichia coli O157 FepE and WzzE have been solved, and it has been deduced that these structures show marked similarities at the protomer and oligomer levels (21). These protomers are elongated and consist of two structural components: a trapezoidal α/β base domain close to the membrane and an extended α-helical hairpin containing an ∼100-Å-long helix forming anti-parallel coiled-coil interactions with two helices that fold back toward the membrane (21). The protomers self-assemble into bell-shaped oligomers displaying comparable structural features, with WzzST forming pentameric oligomers, WzzE assembling into octameric oligomers, and FepE assembling into nonameric structures (21). In contrast, a recent study from Larue et al. reports that WzzST, FepE, and WzzK40 favor hexameric structures (9). A previous study on the oligomeric status of S. flexneri WzzB (WzzSF) via in vivo cross-linking with formaldehyde indicated that WzzSF has the ability to form hexamers and high-order oligomers, suggesting that oligomerization is important in function (4). Related to this, Marolda et al. have shown that the ECA-associated Wzx can fully complement an LPS Oag-associated Wzx-deficient mutant if the remaining ECA gene cluster is deleted, providing genetic evidence that proteins involved in Oag/ECA biosynthesis and processing may function as a complex (11).
Several models of the likely mechanisms of Oag chain regulation have been proposed. Bastin et al. initially suggested that Wzz acts as a molecular timer, allowing polymerization to occur to a particular point, hence increasing the number of repeat units added to the chain (1). An alternative model proposed by Morona et al. suggested that Wzz acts as a molecular chaperone, facilitating the interaction between Wzy and WaaL, and modal length is the result of the ratio of Wzy and WaaL (14). Published data indicated that the ratio of Wzy and Wzz was important in determining Oag modal chain length, which is supportive of the latter model (5). With recent developments in solving the PCP three-dimensional (3D) structure and oligomeric arrangement, a new model has been proposed by Tocilj et al. in which the Wzz oligomers act as molecular scaffolds for multiple Wzy polymerase molecules and the growing Oag chain is transferred from one Wzy to another Wzy molecule (21).
In a previous study, site-directed mutagenesis analysis was conducted on WzzSF (4). Although mutational alterations targeting the TM regions caused dramatic changes in the resulting LPS Oag chain length, mutations targeting the periplasmic domain generally did not have an obvious effect on the resulting LPS Oag chain length. This was also shown for mutations in FepE (17). Hence, we decided that a more severe approach to WzzSF mutagenesis was needed to investigate the relationship between Wzz structure and function by increasing the likelihood of acquiring Wzz mutants displaying phenotypic changes. In this study, we have investigated the structure and function of WzzSF by constructing a library of in-frame linker mutants with 5-amino-acid (aa) insertions throughout the WzzSF protein. We have identified regions in WzzSF which alter the modal length in different ways and present biochemical evidence acquired by in vivo chemical cross-linking that indicates oligomeric differences exist between Wzz mutants and the wild type (WT). We also present evidence that suggests the dimeric form of WzzSF is important in establishing modal length.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The strains used in this study are S. flexneri RMA2741 serotype Y SFL1 wzz::Kmr, cured of virulence plasmid and pHS-2, carrying F′ (lacIq Tetr), from our laboratory collection, and Escherichia coli strain Top10 F′[(lacIq Tetr) mcrA Δ(mrr-hsd RMS-mcrBC) φ80 lacZ ΔM15 ΔlacX74 deoR recA1 araD139 Δ(ara-leu) 7697 galU galK rpsL endA1 nupG] from Stratagene. Strains were grown in Luria-Bertani broth (10 g/liter Tryptone, 5 g/liter yeast extract, 5 g/liter NaCl) with aeration for 16 to 18 h. Eighteen-hour cultures were diluted 1/50 into fresh broth and grown to log phase (optical density at 600 nm [OD600] of 0.7). During induction conditions, cultures had 0.5 mM isopropyl-β-d-thiogalactopyranoside (IPTG) added and were grown for a further 1 to 1.5 h.
Mutagenesis.
An in vitro-based, random mutagenesis kit (mutagenesis generation system; Finnzymes) was employed to produce a library of in-frame wzzSF mutant constructs as per the manufacturer's instructions. Plasmid pRMCD30, a pQE-30 (Qiagen)-based construct with the wzzSF open reading frame (ORF) expressed as a His6-tagged protein His6-WzzSF (4) and containing SacI and SmaI sites flanking the coding region, was incubated with the kanamycin resistance conferring the Mu entranceposon DNA sequence element (harboring NotI sites very close to its ends) to allow its random formation of transposition complexes. These were electroporated into Top10 F′, and transformants which were resistant to kanamycin were screened via PCR using primers #2197 (5′-AGGGTAGAGCTCAGAGTAGAAAAT-3′) and #2198 (5′-GTTACCCGGGGAGCAGGTGTGA-3′), corresponding to the 5′ and 3′ region of wzzSF, respectively. Selected plasmids were restricted with NotI to remove the entrancesposon, leaving a 15-bp insertion which contained a NotI restriction site. The NotI miniprimer (5′-TGCGGCCGCA-3′) was used to identify the approximate site of insertion via PCR. The precise position of the 15-bp insertion within the coding region was determined by cycle sequencing (AGRF, Queensland, Australia). The mutational alterations were mapped on 3D images created using Pymol software (DeLano Scientific LLC 2008).
LPS PAGE and silver staining.
LPS was prepared as described previously (15, 22). Briefly, 1 × 109 cells were harvested by centrifugation, resuspended in lysing buffer (10% [wt/vol] glycerol, 2% [wt/vol] SDS, 4% [wt/vol] β-mercaptoethanol, 0.1% [wt/vol] bromophenol blue, 1 M Tris-HCl, pH 7.6) and incubated with 2 μg/ml of proteinase K for approximately 16 h. The isolated LPS samples were electrophoresed on a 15% polyacrylamide gel for 16 to 18 h at 12 mA. The gel was stained with silver nitrate and developed with formaldehyde (15).
SDS-PAGE and Western immunoblotting.
Log-phase bacteria were induced as described above, harvested by centrifugation, and resuspended in 1× sample buffer (10). SDS-PAGE and Western immunoblotting were performed as previously described (4, 17), and samples were run alongside a molecular mass marker (Invitrogen). The nitrocellulose was blocked with 5% skim milk and incubated overnight with affinity-purified anti-Wzz polysera, prepared by Daniels et al. (4), at a concentration of 1:1,000 in 2.5% (wt/vol) skim milk. The membrane was incubated with a goat anti-rabbit secondary antibody conjugated to horseradish peroxidase (HRP) (Biomediq DPC). The CPS3 chemiluminescence substrate (Sigma) was applied to the membrane for 5 min, exposed to film, and developed.
Chemical cross-linking analyses.
Bacteria grown as described above were induced, and 5 × 108 cells were harvested and washed with chilled buffer (10 mM KPO4, 10 mM Tris, pH 6.8) and then incubated with 0.5% (vol/vol) formaldehyde (Sigma) in buffer at 25°C for 1 h (4, 16). Samples were also incubated at 25°C without formaldehyde as a control. Both cross-linked and control samples were washed once more with chilled buffer and heated to 60°C for 5 min and electrophoresed on an SDS-12% polyacrylamide gel and either subjected Western immunoblotting as described above or stored at −20°C until needed. Cross-linked samples were also heated to 100°C for 5 min, electrophoresed, and subjected to Western immunoblotting as described above.
RESULTS
Location of 5-aa insertions within WzzSF.
As random mutagenesis of WzzSF has not previously been published, a random in-frame linker mutagenesis method was employed to create a library of WzzSF proteins with 5-aa insertions in different positions. An in vitro system (see Materials and Methods) was employed that resulted in random insertions of the Mu entranceposon sequence within the wzzSF ORF in pRMCD30. Following excision of the entranceposon with NotI, transformants were screened by PCR and restriction enzyme analysis to identify approximate sites of insertion, as each insertion possessed a NotI restriction site. DNA sequencing identified 18 unique mutant constructs encoding proteins termed Wzzi, which were then assessed for protein production and impact on LPS O-antigen modal chain length distribution. Due to the nature of this in-frame mutagenesis, each Wzzi mutant has a unique 5-aa sequence at the site of insertion. These insertion sequences are listed in Table 1.
TABLE 1.
Different classes of Wzz mutants based on the length of Oag they confer when complementing S. flexneri strain RMA2741a
| Wzzi mutant | Insertion site with 5-aa insertion | Classb | Protein detectionc |
|---|---|---|---|
| i32 | M32 TAAAM T33 | I | + |
| i66 | D66 CGRTD V67 | I | + |
| i80 | I80 CGRII Y81 | IV | + |
| i81 | Y81 CGRIY G82 | IV | + |
| i92 | Q92 VRPQQ E93 | III | + |
| i128 | Q128 LRPHQ Q129 | V | + |
| i131 | P131 LRPQP L132 | V | + |
| i138 | G138 CGRIG Q139 | III | +/− |
| i161 | Q161 DAAAQ E162 | I | +/− |
| i163 | L163 VRPQL E164 | I | − |
| i191 | E191 LRPQE Q192 | II | − |
| i199 | Q199 IAAAQ I200 | I | + |
| i219 | Q219 MRPQQ T220 | II | + |
| i231 | L231 VRPHL G232 | II | +/− |
| i247 | P247 CGRSP L248 | II | +/− |
| i255 | Y255 CGRNY Q256 | II | − |
| i279 | Y279 CGRSY V280 | I | − |
| i290 | D290 CGRSD S291 | I | + |
The 5-amino-acid insertion sequences of each Wzzi mutant are listed, along with the classes and confirmation of Wzzi protein detection.
Class I, random Oag modal chain length; class II, very short (2 to 10 repeats); class III, shorter (8 to 14 repeats); class IV, wild type (11 to 19 repeats); class V, longer (16 to 25 repeats).
+, wild type; +/−, less than wild type; −, not detected.
LPS Oag modal chain length conferred by Wzzi mutants.
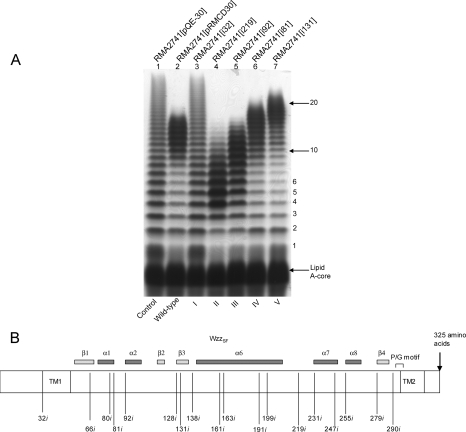
The Wzzi-encoding plasmids were transformed into RMA2741, an S. flexneri Y strain with a wzzSF::Kmr mutation and lacking both pHS-2 (encoding WzzpHS-2) and the large virulence plasmid. Following analysis of the resulting LPS Oag modal chain length distribution by SDS-PAGE and silver staining, the Wzzi mutants were grouped into five different phenotypic classes (Table 1 and Fig. 1). Seven of the 18 mutants, categorized into class I, had lost the ability to regulate Oag modal chain length under the conditions used in these experiments and hence displayed LPS with random-length Oag chains (represented by i32; Fig. 1A, lane 3). Five mutants conferred significantly reduced average Oag modal chain lengths from wild type to between 2 to 6 and 2 to 10 Oag repeat units (represented by i219, class II; lane 4). Two mutants conferred a reduced Oag modal chain length of 8 to 14 repeat units (represented by i92, class III; lane 5), and two mutants conferred a near-wild-type Oag modal chain length between 11 and 19 repeat units (represented by i81, class IV; lane 6). Interestingly, the two mutants categorized into class V conferred an increased Oag modal chain length from the wild-type modal length to 16 to 25 repeat units (represented by i131, class V; lane 7).
FIG. 1.
Oag modal chain length conferred by the different classes of Wzzi mutants expressed in S. flexneri RMA2741. (A) LPS samples were prepared, electrophoresed on a 15% SDS-polyacrylamide gel, and silver stained as described in Materials and Methods. The strains in each lane are as follows: 1, RMA2741(pQE-30); 2, RMA2741(pRMCD30); 3, RMA2741 (i32, class I); 4, RMA2741 (i219, class II); 5, RMA2741 (i92, class III); 6, RMA2741 (i81, class IV); and 7, RMA2741 (i131, class V). (B) Schematic representation of the locations of each insertion within WzzSF, illustrating the insertions within WzzSF that were designated titles according to the last uninterrupted amino acid preceding the 5-aa insertion. The proline rich motif is indicated (P/G motif). Secondary structural features are based on the 3D structure of WzzST.
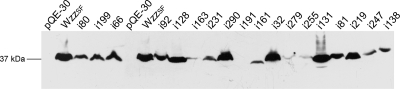
Whole-cell lysates from S. flexneri strains carrying plasmids encoding the Wzzi mutants were subjected to Western immunoblotting using anti-Wzz antibodies. The majority of plasmids encoding Wzzi proteins resulted in detectable Wzz protein under induced conditions: however, a number of Wzzi proteins could not be detected (class I i163, class II i191, class II i255, and class I i279 mutants; Table 1 and Fig. 2). The class I i161, class II i231, class II i247, and class III i138 mutant proteins were detected but at a lower intensity than wild type (summarized in Table 1 and Fig. 2).
FIG. 2.
Western immunoblotting performed on whole-cell lysates from S. flexneri strain RMA2741 carrying Wzzi plasmids. Strains were grown and induced, and whole-cell lysates were detected with affinity-purified Wzz polyclonal antibodies. Each lane contained approximately 1 × 108 bacterial cells.
In vivo chemical cross-linking of Wzzi mutants.
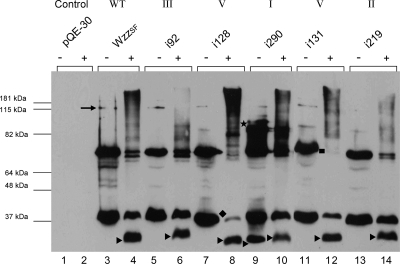
Wild-type WzzSF and representative Wzzi mutants which were expressed at a level comparable to wild-type WzzSF (class I i290, class II i219, class III i92, class V i128, and class V i131 mutants) were analyzed for the ability to form higher-order oligomers by cross-linking with 0.5% formaldehyde (4). Wild-type WzzSF monomeric (36 kDa) and dimeric (72 kDa) forms were readily detected in the non-cross-linked sample and also what appears to be a tetrameric form (144 kDa) (Fig. 3, lane 3). The cross-linked sample revealed bands at ∼30 kDa and ∼36 kDa, a doublet band at ∼72 kDa, and a smeared high-molecular-mass band of >180 kDa, which indicated the presence of higher-order oligomerization (Fig. 3, lane 4). The class I mutant i290 also exhibited the presence of both monomeric (∼36 kDa) and dimeric (∼72 kDa) bands in both non-cross-linked and cross-linked samples and higher-molecular-mass bands around the >180-kDa region (Fig. 3, lanes 9 and 10). The presence of a 30-kDa protein was detected in the non-cross-linked sample of i290 (Fig. 3, lane 9), although this band was only identified in the presence of formaldehyde for WzzSF and other Wzzi proteins (Fig. 3, lanes 4, 6, 8, 10, 12, and 14). There is also an additional i290 band of approximately 89 kDa which is not detected in other Wzzi cross-linked profiles (Fig. 3, lane 9).
FIG. 3.
Analysis of the Wzzi mutant by cross-linking with formaldehyde. S. flexneri RMA2742 strains carrying plasmid-encoded Wzzi proteins were harvested, washed in 10 mM KPO4, and exposed to 0.5% formaldehyde at 25°C (+); controls were incubated without formaldehyde (−) as described in Materials and Methods. Samples were heated at 60°C and electrophoresed on a 12% polyacrylamide gel. Western immunoblotting was performed with affinity-purified WzzSF polyclonal antisera. ▸, ∼30-kDa WzzSF conformation; ★, extra band in i290; ♦, depleted monomeric form of i128; ▪, lack of the dimeric form of i131. Each lane contained approximately 4 × 108 bacterial cells.
The class II mutant i219 (Fig. 3, lanes 13 and 14) had both monomeric (36 kDa) and dimeric (72 kDa) protein forms in cross-linked and non-cross-linked samples, and the 30-kDa protein was also detected in the cross-linked sample (Fig. 3, lane 14). Higher-molecular-mass bands were decreased in the cross-linked sample (Fig. 3, lane 14) compared to the wild type. The class III mutant i92 displayed a cross-linking profile comparable to WzzSF, having bands of ∼36 kDa, ∼72 kDa, and ∼144 kDa in the non-cross-linked sample (Fig. 3, lane 5) and ∼30 kDa, ∼36 kDa, and ∼72 kDa in the cross-linked sample (Fig. 3, lane 11). However, the high-molecular-mass band around the 180-kDa region was not detected, similar to i219.
The two class V mutants, i128 and i131, exhibited bands of sizes 36 kDa and 72 kDa and weak bands at ∼144 kDa in the non-cross-linked sample (Fig. 3, lanes 7 and 11). However, the cross-linked profile was very different from those of the other Wzzi proteins: monomeric protein (36 kDa) and dimeric protein (72-kDa) forms were significantly reduced in the cross-linked sample (Fig. 3, lanes 8 and 12), very little monomeric protein and dimeric protein was detected in the i128 cross-linked sample, and no dimeric form of i131 was detected under cross-linking conditions. Larger bands greater than 180 kDa were detected in i131 cross-linked samples (Fig. 3, lane 12), and in the case of class V mutant i128, Wzz-related bands greater than 180 kDa were very readily detected and were comparable to the higher-molecular-mass oligomers detected in the wild-type cross-linked sample (Fig. 3, lane 4). In summary, the data indicate that following cross-linking, class V mutants i128 and i131, which confer a longer modal length, exhibit high-molecular-mass oligomers and reduced monomeric and dimeric forms, whereas high-molecular-mass oligomers are not detected in the class II and III mutants (i92 and i219), which confer modal lengths shorter than wild type.
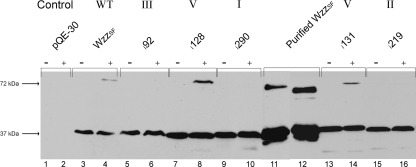
Stability of Wzz dimers.
Previous studies have shown that WzzSF dimers can be detected even after SDS-PAGE and also that formaldehyde cross-linked WzzSF dimers are still able to be detected after being heated to 100°C (4). Representative Wzzi mutants were chemically cross-linked, heated to 100°C, and along with non-cross-linked samples subjected to Western immunoblotting to ascertain whether the mutants exhibited different dimeric properties compared to wild-type WzzSF. Wild-type WzzSF dimers were detected in the cross-linked samples (Fig. 4, lane 4), as were dimers of class V mutants i128 (Fig. 4, lane 8) and i131 (Fig. 4, lane 14). Mutants i290, i219, and i92 from classes I, II, and III, respectively, did not form stable dimers under these conditions (Fig. 4, lanes 6, 10, and 16).
FIG. 4.
Comparison of stabilities of WzzSF and Wzzi dimers. S. flexneri RMA2742 strains carrying plasmid-encoded Wzzi proteins were harvested, washed in 10 mM KPO4, and exposed to 0.5% formaldehyde at 25°C (+); controls were incubated without formaldehyde (−) as described in Materials and Methods. Both cross-linked and non-cross-linked samples of WzzSF and Wzzi mutants were heated to 100°C for 5 min, electrophoresed on a 12% SDS-polyacrylamide gel, and subjected to Western immunoblotting. The strains in each lane are as follows: 1 and 2, RMA2741(pQE-30); 3 and 4, RMA2741(pRMCD30); 5 and 6, RMA2741 (i92, class III); 7 and 8, RMA2741 (i128, class V); 9 and 10, RMA2741 (i290, class I); 11 and 12, purified WzzSF; 13 and 14, RMA2741 (i131, class V); and 15 and 16, RMA2741 (i219, class II). Each lane contained approximately 1 × 108 bacterial cells.
Location of insertions mapped to PCP 3D structures.
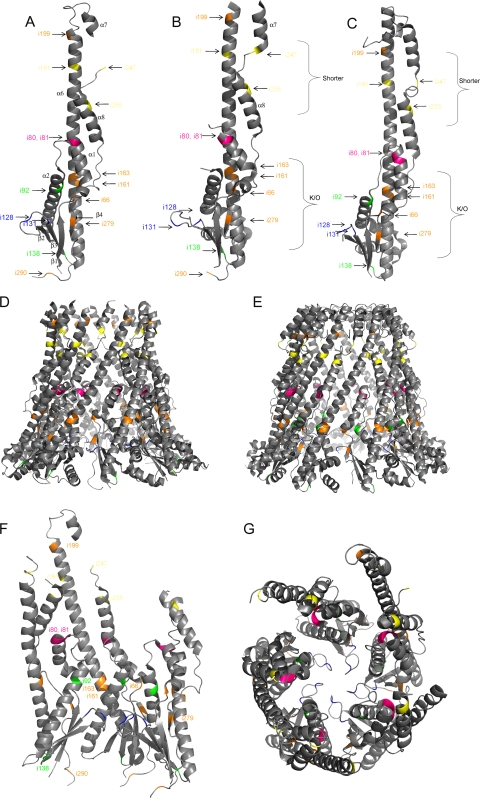
The location of the Wzzi 5-aa insertions could be mapped onto 3D structures of several PCP proteins (21). The last uninterrupted amino acid of each Wzzi mutant was mapped onto the monomeric and oligomeric structures of WzzST, WzzE, and FepE (Fig. 5). The three structures exhibit comparable secondary structure characteristics (21). Analysis was primarily conducted on WzzST, as it exhibits the greatest sequence identity to WzzSF (13). The six class I (no activity) mutant insertions that were mapped onto the structures were i66, i161, i163, i199, i279, and i290. Mutation i66 is predicted to be located on the very first turn of the first α helix, α1 (Fig. 5A, B, and C). Mutations i161 and i166 were predicted to be located about one-third of the way into α6, the long α helix extending from the α/β base domain, while i199 was predicted to be located toward the uppermost region of α6 (Fig. 5A, B, and C). Mutations i279 and i290 were predicted to be located within a β sheet of the α/β base domain (β4) and the loop closest to TM2 of the determined 3D structure, respectively (Fig. 5A, B, and C).
FIG. 5.
Location of the Wzzi insertions mapped on the 3D structures of WzzST (Protein Data Bank [PDB] no. 3b8p), WzzE (PDB no. 3b8o), and FepE (PDB no. 3b8n). The last uninterrupted amino acid of WzzSF was mapped on each monomeric and oligomeric structure. (A) WzzST monomer; (B) WzzE monomer; (C) FepE monomer; (D) WzzE octomer; (E) FepE nonamer; (F) three WzzST monomers; (G) WzzST pentamer (top view). The mutant classes are illustrated in different colors: class I mutants are orange, class II mutants are yellow, class III mutants are green, class IV mutants are magenta, and class V mutants are blue.
In the oligomeric structures, i66 is predicted to be on the periphery of the monomer, with close proximity to α2 and the long extended α6 helix of neighboring monomers (Fig. 5D to G). Mutants i161 and i163, with insertions predicted to be located on the bottom half of α6, appear to be embedded within the monomer, close to α2. Mutant i199, was predicted to have the insertion sequence located at the very summit of α6, isolated from other structural features (Fig. 5F). Mutant i279 was predicted to contain the insertion in the central region of the β4 sheet, on the fringe of the monomeric structure, very close to the neighboring α2 (Fig. 5F). Mutation i290 is predicted to be located on the lowest points in the oligomer and would most likely have close proximity to the transmembrane regions. The insertions from this class were mapped to both internal and external locations on the oligomeric structures (Fig. 5D to G).
Only three of the five class II (very short Oag chain modal length) mutants (i191, i247, and i255) were able to be mapped onto the 3D structure, as i219 and i231 are located in regions where structural data are unavailable for all three PCP proteins. Mutation i191 was predicted to be located on the uppermost quarter of α6. Mutation i247 was predicted to be located on the loop between α7 and α8, and the insertion of mutant i255 was mapped onto the second turn on α8 (Fig. 5A, B, and C).
Class II mutations were also mapped onto the oligomeric structures, which indicated that i191, i247, and i255 are all on the upper half of the oligomer (Fig. 5B to F). Mutation i191, predicted to be mapped to the uppermost region of α6, appeared to be quite remote from other helices or structural components of Wzz, and i247 was predicted to be positioned on the cusp of α7 and α8, close to α6 of neighboring monomers (Fig. 5A to C and F). Mutation i255, located on the second turn of α8, appeared to be situated closer to the inner surface of the cavity (Fig. 5F). These insertions are all mapped to be located on external regions of the oligomeric structures (Fig. 5D to G).
The two class III mutants i92 and i138 (conferring an Oag modal length slightly shorter than wild type) contain insertions predicted to be located in the second α helix (α2; Fig. 5A and C) and at the base of the monomer, respectively. The insertion of mutant i92 was predicted to be mapped onto the uppermost region of α2: on the second turn for WzzST and on the first turn in FepE (Fig. 5A, B, and C). This region of the structure is absent in WzzE. The insertion of mutant i138 was predicted to be mapped either on the very top of β3 (WzzST and WzzE) or on the loop between β3 and α6 (FepE), at the α/β base domain (Fig. 5A, B, and C). When these insertions were mapped onto the oligomeric structures, i92, positioned within α2, appears to be situated close to the lining of the inner cavity (Fig. 5F). Mutation i138 was predicted to be situated at the base of the oligomer, embedded between α6 and β3, close to the membrane surface. These insertions appear to be primarily located to internal regions of the oligomeric structures (Fig. 5D to G).
The class IV mutants i80 and i81, conferring wild-type LPS Oag modal length, have insertions which are predicted to be located within the last turn of α1 (Fig. 5A, B, and C). In the oligomeric prediction, this region is securely embedded internally between α2 and α8 (Fig. 5F and G).
Class V mutations i128 and i131, conferring longer Oag modal lengths, were mapped on the loop between two β strands, β2 and β3, toward the base of the structure (Fig. 5A, B, and C). This loop is located within the inner cavity in the oligomeric structures (Fig. 5G).
DISCUSSION
In this study, we constructed and characterized a collection of mutant Wzzi proteins. Previous mutagenesis studies (4, 6) suggested that there is not one particular region within Wzz which controls function: rather, Wzz Oag chain length regulation is a function of the entire protein structure. In our study, it is interesting to note that only 7 of the 18 characterized mutants appeared to be phenotypic knockout mutants (class I) and did not have Oag chain length regulation capability (Fig. 1). Considering that Wzz has such a key regulatory function and functionality is perceived to be a result of the 3D protein structure as a whole, it might be predicted that the insertion of 5 aa would abolish regulation of Oag chain length in most mutants. However, not only did approximately half of the mutants retain function, a range of Oag chain length phenotypes were observed. Class II mutants conferred Oag modal length of 2 to 10 repeat units, class III mutants resulted in shorter-than-wild-type Oag length (8-14), and the two class IV mutants conferred near-wild-type Oag chain length, while class V mutants increased the resulting Oag modal length to 16 to 25 repeat units.
Although approximately half of the Wzzi mutants exhibited control over Oag chain length regulation, in many cases, protein band intensities are not comparable to wild-type levels of detection. Strains with mutants i163, i191, i255, and i279 produced protein below the limit of detection, and mutants i138, i161, i231, and i247 were detected at a level apparently lower than that of wild-type WzzSF (Fig. 2). All other mutants (i32, i66, i80, i81, i92, i128, i131, i199, i219, and i290) were detected at a level comparable to wild type (Fig. 2). However, less-than-wild-type protein detection does not appear to correlate with a particular resulting Oag modal length, as these mutants are from a range of different phenotypic classes. It is possible that many of these mutant proteins exhibit instability and are targets for proteases such as DegP, as this protease acts on WzzpHS-2 mutant proteins (17). We cannot rule out that these 5-aa insertions have disrupted, concealed, or altered particular epitopes that the antibody binds to, hence altering protein detection in some mutants. It is also possible that several class I mutants lack control of Oag modal chain length due to low protein production and/or protein misfolding, although it is interesting that a collection of the seven class I mutants (i32, i66, i199, and i290) have detectable protein and are phenotypically inactive, whereas other mutants from which protein could not be detected (e.g., i191 of class II) still exhibit control over Oag modal chain length. It would appear that a very small amount of Wzz protein is required to establish a regulated Oag modal chain length, consistent with other studies (21).
The insertions in each Wzzi mutant were mapped onto the 3D PCP oligomeric structures to determine if the interrupted regions appeared to be structurally critical or important in intra- or intermonomer interactions. Despite the fact that each mutant exhibits a unique 5-aa insertion and the different amino acids may be affecting the structure and resulting phenotype, the classes generally consist of insertions that are predicted to be located in a similar region, with the exception of class I. The expected insertion positions relative to the Wzz 3D structure for class I mutants span many regions of the structure (Fig. 5A). Protein function appears to be sensitive to 5-aa insertions at these positions. The α/β base domain, comprised of β sheets β1 to β4, α helices α1 and α2, and the lowest region of α6 (in WzzST), and α1 to α4 and the lowest region of α6 (in FepE and WzzE), appears to play an important role in intermonomeric interactions and oligomerization (21). The mutations that are predicted to be located within this region are i66, i80, i81, i92, i128, i131, and i290 (Fig. 5). There are obvious differences in the LPS phenotypes of these mutations: i66 and i290 do not impart any Oag modal chain length control, whereas the others do. Mutation i66 is also predicted to exist on the periphery of the monomer and hence may be in a position to influence hydrophobic interactions between monomers, thereby affecting stability and oligomerization (Fig. 5A). The β strands comprising the central feature of the α/β base domain are key structural elements in bringing the N and C termini closer together, which may also bring the TM segments closer (21). Mutation i279, predicted to be located on the final β strand (β4), may be interfering with this process, perhaps by steric hindrance. In the case of the class IV i80 and i81 mutations, it is unclear as to why this region allows a 5-aa insertion with little alteration in resulting Oag chain length; however, the insertions appear to be located in a region embedded within α2 and α8 and may not be destabilizing intramonomeric interactions (Fig. 5A to C and F). Mutation i92 (class III) is predicted to be located close to the top of an α helix; however, i92 differs from both i80 and i81 in the way the α2 helix is more exposed to the inner cavity than α1, hence possibly being slightly less tolerant to a 5-aa insertion (Fig. 5F). The other class III i138 insertion is predicted to be located close to the base of the oligomeric structure, toward the end of β3 (Fig. 5A to C). Considering its predicted proximity to the α/β base domain (Fig. 5A to C and F), it is surprising that the resulting phenotype is merely a slight decrease in the LPS Oag modal chain length. This mutant insertion, however, is predicted to be located toward the end of β3 (WzzST) or on the loop between β3 and α6 (FepE), and this position may be able to sustain extra amino acids without drastically altering nearby structural features, as it appears to be on the cusp of the oligomer (Fig. 5D and F). In the case of i290, as it is close to TM2, the insertion may be influencing a key region needed for function (Fig. 1B). In previous WzzSF mutagenesis studies, P292, one of the highly conserved prolines present in the proline-glycine-rich motif, was found to knock out Wzz function when converted to an alanine (4). This proline is theorized to be in cis conformation, hence providing rigidity and stability and influencing the orientation of the base domain and TM2 (12, 21). Mutation i290, being located so closely to this conserved residue, may undermine this critical arrangement. It is also possible that this mutation destabilizes any anchoring of the oligomer at the membrane surface, as residues close to the inner membrane are considered to interact with lipid head groups (21).
There are a number of mutations predicted to be located on the long extended hairpin α6, such as i161, i191, and i199 (Fig. 5A). Mutant i191 is in class II, and i161 and i199 are class I mutants (Fig. 1 and Table 1). From the 3D crystal structure analyses, it appears as though the α6 helix is involved in maintaining intramonomeric stability, by interacting with α2 via conserved hydrophobic residues (21). Indeed, all mutations predicted to exist within α6 are in classes I and II and have similar phenotypes. The upper region of the oligomer is speculated to be involved in interacting with outer membrane proteins such as those involved in LPS export (21). It is possible that perturbation in the central α helix either results in disruption of local protein conformation or affects interaction with outer membrane proteins. Various conserved residues, including I237 and L240 in FepE, are present which form a leucine zipper motif (21). If this leucine zipper is critical for monomeric or oligomeric stability and/or interactions in WzzSF, it is possible that the insertion in i199, predicted to be located within two turns of this region, may severely disrupt these interactions.
Mutations i247 and i255 are predicted to be mapped to the upper region of the oligomer, within or close to α7 and α8 (Fig. 5). These α helices play a role in intermonomeric stability, as they interact with the long extended α6 helix on neighboring monomers (21). The mutations i247 and i255, both class II, are located on the outermost region of the oligomer, facing outwards and high up on the structure (Fig. 5). It is interesting to note that mutations that are located on the exterior region of the oligomer can play such a role in Wzz function; however, previous mutagenesis conducted on WzzSF indicates that a K267N mutation, predicted to be located on the outer side of the oligomer on the lower region of α8, results in an increase in Oag modal chain length (21). Hence, it appears that residues on the exterior face of Wzz oligomers have the ability to influence Wzz function, as previously proposed (21).
The class V mutants i128 and i131, conferring longer Oag chain length modality, were predicted to be located in the loop between β2 and β3 in the α/β base domain, directly in the central cavity of the oligomer (Fig. 5G). The phenotype resulting from the 5-aa insertions at this location is not observed for any other Wzzi mutant. Previous WzzSF mutagenesis studies have not yielded mutants which increase the Oag modal chain length to this degree. We can speculate that the cause for such a dramatic modal length change may be attributed to the change this 5-aa insertion exerts on the cavity width and that the increase in the number of amino acid residues within this cavity is widening it by increasing the size of the α/β base domain. In general, it appears as though class V, class IV, and class III mutants are mapped to internal regions on the oligomeric structure, whereas class II mutants have their insertions mapped exclusively to external regions, and class I mutant insertions are mapped to both internal and external regions (Fig. 5D to F).
Previous cross-linking studies show Wzz can oligomerize (4, 7, 9), and recent studies have suggested that E. coli O86:H2 Wzz can form tetrameric oligomers (20), while others show Wzz forms hexameric oligomers (9). In our study, higher-order oligomers were easily detected in wild-type WzzSF and a number of select mutants (Fig. 3). It is interesting that the mutants conferring longer Oag chain length have comparable cross-linking profiles to wild type, whereas mutants resulting in random or shorter chain lengths do not appear to oligomerize as well, as judged by cross-linking. The lack of detectable oligomers in the resulting cross-linking profiles of i92, i219, and i290 may also be attributed to weak stability of the mutant proteins. It is possible that various Wzzi mutant proteins may be able to form oligomers (e.g., i290); however, they either cannot be stably maintained or are perhaps incapable of successful interactions with other Oag processing proteins (or putative OM binding partners) to confer wild-type Oag modal chain length. An unusual feature of mutant i290 is the presence of the extra 30-kDa band in the non-cross-linked sample (Fig. 3, lane 9). This band appears to be Wzz related as it is readily detected by the Wzz antibody and is present in all other mutants (including wild-type Wzz), although never detected in the absence of cross-linking. The 30-kDa variation might have an altered conformation and may be nonfunctional, and it is possible that the presence of this variant in the non-cross-linked sample of i290 is linked to the fact that i290 appears to be nonfunctional. In contrast, monomers and dimers of class V Wzzi mutants were detected at a much lower intensity when subjected to cross-linking than other mutants, and higher-order oligomers were easily detected. Previous findings have shown that WzzSF dimers appear to be very stable, as being heated at 100°C in the presence of SDS does not cause complete disassociation (4). In this study, WzzSF and Wzzi cross-linked and non-cross-linked samples were heated to 100°C for 5 min and subjected to Western immunoblotting to ascertain whether or not the Wzzi mutant dimers exhibit the similar stable trait of the wild-type dimers by being able to withstand the presence of SDS at 100°C. Only the wild-type WzzSF and class V i128 and i131 mutant dimers were detected after this treatment (Fig. 4). From these data, it appears as though there is a positive correlation between dimeric stability and wild-type-or-longer Oag modal chain length determination. These experiments show that the mutants which exhibit higher-order oligomers, as judged by in vivo formaldehyde cross-linking (Fig. 3), possess stable dimers, and it may be possible that this feature is a key factor in the ability to form oligomers, perhaps by providing a scaffolding element. Our results support previous findings that Wzz function is not restricted to one particular region on the structure and that oligomerization is related to modal length chain determination.
Acknowledgments
This work was supported by a program grant from the National Health and Medical Research Council of Australia awarded to R.M. M. Papadopoulos was the recipient of a Faculty of Science Postgraduate Scholarship from the University of Adelaide.
Footnotes
Published ahead of print on 7 May 2010.
REFERENCES
- 1.Bastin, D. A., L. K. Romana, and P. R. Reeves. 1991. Molecular cloning and expression in Escherichia coli K-12 of the rfb gene cluster determining the O antigen of an E. coli O111 strain. Mol. Microbiol. 5:2223-2231. [DOI] [PubMed] [Google Scholar]
- 2.Becker, A., H. Kuster, K. Niehaus, and A. Puhler. 1995. Extension of the Rhizobium meliloti succinoglycan biosynthesis gene cluster: identification of the exsA gene encoding an ABC transporter protein, and the exsB gene which probably codes for a regulator of succinoglycan biosynthesis. Mol. Gen. Genet. 249:487-497. [DOI] [PubMed] [Google Scholar]
- 3.Becker, A., and A. Puhler. 1998. Specific amino acid substitutions in the proline-rich motif of the Rhizobium meliloti ExoP protein result in enhanced production of low-molecular-weight succinoglycan at the expense of high-molecular-weight succinoglycan. J. Bacteriol. 180:395-399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Daniels, C., and R. Morona. 1999. Analysis of Shigella flexneri Wzz (Rol) function by mutagenesis and cross-linking: Wzz is able to oligomerize. Mol. Microbiol. 34:181-194. [DOI] [PubMed] [Google Scholar]
- 5.Daniels, C., C. Vindurampulle, and R. Morona. 1998. Overexpression and topology of the Shigella flexneri O-antigen polymerase (Rfc/Wzy). Mol. Microbiol. 28:1211-1222. [DOI] [PubMed] [Google Scholar]
- 6.Franco, A. V., D. Liu, and P. R. Reeves. 1998. The Wzz (Cld) protein in Escherichia coli: amino acid sequence variation determines O-antigen chain length specificity. J. Bacteriol. 180:2670-2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guo, H., K. Lokko, Y. Zhang, W. Yi, Z. Wu, and P. G. Wang. 2006. Overexpression and characterization of Wzz of Escherichia coli O86:H2. Protein Expr. Purif 48:49-55. [DOI] [PubMed] [Google Scholar]
- 8.Hong, M., and S. M. Payne. 1997. Effect of mutations in Shigella flexneri chromosomal and plasmid-encoded lipopolysaccharide genes on invasion and serum resistance. Mol. Microbiol. 24:779-791. [DOI] [PubMed] [Google Scholar]
- 9.Larue, K., M. S. Kimber, R. Ford, and C. Whitfield. 2009. Biochemical and structural analysis of bacterial O-antigen chain length regulator proteins reveals a conserved quaternary structure. J. Biol. Chem. 284:7395-7403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lugtenberg, B., J. Meijers, R. Peters, P. van der Hoek, and L. van Alphen. 1975. Electrophoretic resolution of the “major outer membrane protein” of Escherichia coli K12 into four bands. FEBS Lett. 58:254-258. [DOI] [PubMed] [Google Scholar]
- 11.Marolda, C. L., L. D. Tatar, C. Alaimo, M. Aebi, and M. A. Valvano. 2006. Interplay of the Wzx translocase and the corresponding polymerase and chain length regulator proteins in the translocation and periplasmic assembly of lipopolysaccharide O antigen. J. Bacteriol. 188:5124-5135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Morona, R., L. Purins, A. Tocilj, A. Matte, and M. Cygler. 2009. Sequence-structure relationships in polysaccharide co-polymerase (PCP) proteins. Trends Biochem. Sci. 34:78-84. [DOI] [PubMed] [Google Scholar]
- 13.Morona, R., L. Van Den Bosch, and C. Daniels. 2000. Evaluation of Wzz/MPA1/MPA2 proteins based on the presence of coiled-coil regions. Microbiology 146:1-4. [DOI] [PubMed] [Google Scholar]
- 14.Morona, R., L. Van Den Bosch, and P. A. Manning. 1995. Molecular, genetic, and topological characterization of O-antigen chain length regulation in Shigella flexneri. J. Bacteriol. 177:1059-1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Murray, G. L., S. R. Attridge, and R. Morona. 2003. Regulation of Salmonella typhimurium lipopolysaccharide O antigen chain length is required for virulence; identification of FepE as a second Wzz. Mol. Microbiol. 47:1395-1406. [DOI] [PubMed] [Google Scholar]
- 16.Prossnitz, E., K. Nikaido, S. J. Ulbrich, and G. F. Ames. 1988. Formaldehyde and photoactivatable cross-linking of the periplasmic binding protein to a membrane component of the histidine transport system of Salmonella typhimurium. J. Biol. Chem. 263:17917-17920. [PubMed] [Google Scholar]
- 17.Purins, L., L. Van Den Bosch, V. Richardson, and R. Morona. 2008. Coiled-coil regions play a role in the function of the Shigella flexneri O-antigen chain length regulator WzzpHS2. Microbiology 154:1104-1116. [DOI] [PubMed] [Google Scholar]
- 18.Raetz, C. R., and C. Whitfield. 2002. Lipopolysaccharide endotoxins. Annu. Rev. Biochem. 71:635-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Simmons, D. A. 1993. Genetic and biosynthetic aspects of Shigella flexneri O-specific lipopolysaccharides. Biochem. Soc. Trans. 21:58S. [DOI] [PubMed] [Google Scholar]
- 20.Tang, K. H., H. Guo, W. Yi, M. D. Tsai, and P. G. Wang. 2007. Investigation of the conformational states of Wzz and the Wzz O-antigen complex under near-physiological conditions. Biochemistry 46:11744-11752. [DOI] [PubMed] [Google Scholar]
- 21.Tocilj, A., C. Munger, A. Proteau, R. Morona, L. Purins, E. Ajamian, J. Wagner, M. Papadopoulos, L. Van Den Bosch, J. L. Rubinstein, J. Fethiere, A. Matte, and M. Cygler. 2008. Bacterial polysaccharide co-polymerases share a common framework for control of polymer length. Nat. Struct. Mol. Biol. 15:130-138. [DOI] [PubMed] [Google Scholar]
- 22.Tsai, C. M., and C. E. Frasch. 1982. A sensitive silver stain for detecting lipopolysaccharides in polyacrylamide gels. Anal. Biochem. 119:115-119. [DOI] [PubMed] [Google Scholar]
- 23.Van Den Bosch, L., P. A. Manning, and R. Morona. 1997. Regulation of O-antigen chain length is required for Shigella flexneri virulence. Mol. Microbiol. 23:765-775. [DOI] [PubMed] [Google Scholar]
- 24.Van Den Bosch, L., and R. Morona. 2003. The actin-based motility defect of a Shigella flexneri rmlD rough LPS mutant is not due to loss of IcsA polarity. Microb. Pathog. 35:11-18. [DOI] [PubMed] [Google Scholar]
- 25.West, N. P., P. Sansonetti, J. Mounier, R. M. Exley, C. Parsot, S. Guadagnini, M. C. Prevost, A. Prochnicka-Chalufour, M. Delepierre, M. Tanguy, and C. M. Tang. 2005. Optimization of virulence functions through glucosylation of Shigella LPS. Science 307:1313-1317. [DOI] [PubMed] [Google Scholar]
- 26.Whitfield, C., and J. H. Naismith. 2008. Periplasmic export machines for outer membrane assembly. Curr. Opin. Struct. Biol. 18:466-474. [DOI] [PMC free article] [PubMed] [Google Scholar]