Abstract
The pathway of lysine biosynthesis in the methanococci has not been identified previously. A variant of the diaminopimelic acid (DAP) pathway uses diaminopimelate aminotransferase (DapL) to catalyze the direct conversion of tetrahydrodipicolinate (THDPA) to ll-DAP. Recently, the enzyme DapL (MTH52) was identified in Methanothermobacter thermautotrophicus and shown to belong to the DapL1 group. Although the Methanococcus maripaludis genome lacks a gene that can be unambiguously assigned a DapL function based on sequence similarity, the open reading frame MMP1527 product shares 30% amino acid sequence identity with MTH52. A Δmmp1527 deletion mutant was constructed and found to be a lysine auxotroph, suggesting that this DapL homolog in methanococci is required for lysine biosynthesis. In cell extracts of the M. maripaludis wild-type strain, the specific activity of DapL using ll-DAP and α-ketoglutarate as substrates was 24.3 ± 2.0 nmol min−1 mg of protein−1. The gene encoding the DapL homolog in Methanocaldococcus jannaschii (MJ1391) was cloned and expressed in Escherichia coli, and the protein was purified. The maximum activity of MJ1391 was observed at 70°C and pH 8.0 to 9.0. The apparent Kms of MJ1391 for ll-DAP and α-ketoglutarate were 82.8 ± 10 μM and 0.42 ± 0.02 mM, respectively. MJ1391 was not able to use succinyl-DAP or acetyl-DAP as a substrate. Phylogenetic analyses suggested that two lateral gene transfers occurred in the DapL genes, one from the archaea to the bacteria in the DapL2 group and one from the bacteria to the archaea in the DapL1 group. These results demonstrated that the DapL pathway is present in marine methanogens belonging to the Methanococcales.
Two lysine biosynthesis pathways evolved separately in organisms, the diaminopimelic acid (DAP) and aminoadipic acid (AAA) pathways. The DAP pathway synthesizes l-lysine from aspartate and pyruvate, and diaminopimelic acid is an intermediate. This pathway is utilized by most bacteria, some archaea, some fungi, some algae, and plants (28, 29). The AAA pathway synthesizes l-lysine from α-ketoglutarate and acetyl coenzyme A (acetyl-CoA), and α-aminoadipic acid is an intermediate. This pathway is utilized by most fungi, some algae, the bacterium Thermus thermophilus, and probably some archaea, such as Sulfolobus, Thermoproteus, and Pyrococcus (27, 36). No organism is known to possess both pathways.
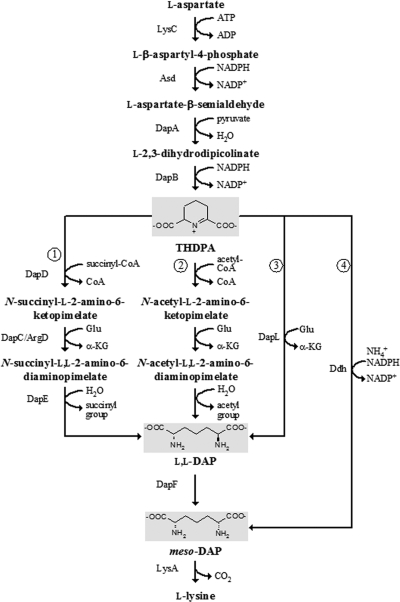
There are four known variations of the DAP pathway in bacteria: the succinylase, acetylase, aminotransferase, and dehydrogenase pathways (Fig. 1). These pathways share the steps converting l-aspartate to l-2,3,4,5-tetrahydrodipicolinate (THDPA), but the subsequent steps leading to the production of meso-diaminopimelate, the immediate precursor of l-lysine, are different. The succinylase pathway acylates THDPA with succinyl-CoA to generate N-succinyl-ll-2-amino-6-ketopimelate and forms meso-DAP by subsequent transamination, desuccinylation, and epimerization. This pathway is utilized by proteobacteria and many firmicutes and actinobacteria (12, 14, 20, 29). The acetylase pathway is analogous to the succinylase pathway but uses N-acetyl intermediates. This pathway is limited to certain Bacillus species, in which the corresponding genes have not been identified (33, 39). The aminotransferase pathway converts THDPA directly to ll-DAP by diaminopimelate aminotransferase (DapL) without acylation. This pathway is shared by cyanobacteria (19), chlamydia (24), the archaeon Methanothermobacter thermautotrophicus (15, 18), and the plant Arabidopsis thaliana (19). The dehydrogenase pathway forms meso-DAP directly from THDPA, NADPH, and NH4+ by using diaminopimelate dehydrogenase (Ddh). This pathway is utilized by some Bacillus and Brevibacterium species and Corynebacterium glutamicum (25, 26, 40). Most bacteria use only one of the four variants, although certain bacteria, such as C. glutamicum and Bacillus macerans, possess both the succinylase and dehydrogenase pathways (3, 30).
FIG. 1.
Variations in the DAP pathway for lysine biosynthesis. 1, succinylase pathway; 2, acetylase pathway; 3, aminotransferase pathway; 4, dehydrogenase pathway. Abbreviations and designations: THDPA, l-2,3,4,5-tetrahydrodipicolinate; l,l-DAP, ll-2,6-diaminopimelate; meso-DAP, meso-2,6-diaminopimelate; LysC, aspartate kinase; Asd, aspartate semialdehyde dehydrogenase; DapA, dihydrodipicolinate synthase; DapB, dihydrodipicolinate reductase; DapD, THDPA succinylase; DapC, succinyl-DAP aminotransferase; DapE, succinyl-DAP desuccinylase; DapF, DAP epimerase; LysA, DAP decarboxylase; DapL, ll-DAP aminotransferase; Ddh, DAP dehydrogenase.
The diaminopimelate aminotransferase (DapL) catalyzes the transfer of an amino group from l-glutamate to THDPA, forming ll-DAP (19, 24). It uses pyridoxal 5′-phosphate (PLP) as a coenzyme and has constrained substrate specificity. DapL is not closely related to the DapC/ArgD aminotransferase, which functions in the succinylase pathway. Comparative genomic analysis identified dapL homologs in both bacterial and archaeal genomes. Homologs of dapD and dapE have not been found in genomes with dapL homologs, suggesting that transamination of THDPA does not require succinylation in these organisms (18). Phylogenetic analysis also suggested classification of DapL into two groups, DapL1 and DapL2, which share ∼30% amino acid sequence identity (18). These two groups both exhibit DapL activity, and they cannot be differentiated by kinetic properties (18, 37). The distribution of the two groups is not obviously associated with specific prokaryotic lineages (18).
Methanogens are strictly anaerobic archaea that obtain all or most of their energy for growth from the production of large quantities of methane. All methanogens belong to the Euryarchaeota and are currently classified in six orders: Methanobacteriales, Methanococcales, Methanomicrobiales, Methanosarcinales, Methanopyrales, and Methanocellales (23, 41, 42). Biochemical studies of Methanocaldococcus jannaschii and Methanococcus voltae belonging to Methanococcales, Methanospirillum hungatei belonging to Methanomicrobiales, and Methanothermobacter thermautotrophicus belonging to Methanobacteriales suggested that these organisms derive their l-lysine from a DAP pathway, but the studies did not discriminate among the four DAP pathway variations (2, 9, 10, 32). Genome sequence analysis also suggested a DAP pathway in Methanosarcina mazei belonging to Methanosarcinales (8). Recent studies identified a dapL homolog belonging to the DapL1 group in M. thermautotrophicus. The gene product complemented an Escherichia coli dapD dapE double mutant and catalyzed the transamination of DAP to THDPA, suggesting that Methanobacteriales use the DapL pathway for l-lysine biosynthesis (15, 18). Homologs of asd, dapA, dapB, dapF, and lysA have been identified in the genomes of M. maripaludis and M. jannaschii belonging to the Methanococcales, but homologs responsible for the conversion of THDPA to ll-DAP have not been annotated (4, 17). Here we identified methanococcal DapL homologs and demonstrated that the DapL pathway is present in Methanococcales.
MATERIALS AND METHODS
Chemicals.
A mixture of ll, dd, and meso isomers of 2,6-diaminopimelic acid (dl-DAP) was purchased from Sigma-Aldrich. ll-DAP was a gift from André O. Hudson and Thomas Leustek.
A mixture of the different N-acetyl-diaminopimelic acid isomers was prepared by acetylation of dl-DAP with acetic anhydride. To do this, 150 μl of acetic anhydride was added to 3 ml of dl-DAP (190 mg [1 mmol] dissolved in 1 M NaOH) with stirring. After incubation for 1 h at room temperature, thin-layer chromatography (TLC) analysis followed by detection of the amines with ninhydrin revealed the presence of only two positive bands. Isolation of each band from the TLC plate and conversion to the dimethyl trifluoroacetyl derivatives showed that the bottom band was DAP with M+ = 410 m/z with fragment ions at M+-32 = 378 m/z and M+-COOCH3 at 351 m/z and the upper band was the N-acetyl-DAP derivative with M+ = 356 m/z with fragment ions at M+-31 = 325 m/z and M+-COOCH3 at 297 m/z. The sample was diluted with water, applied to a Dowex 50 H+ column (1.5 by 16 cm), and eluted with a 400-ml 0 to 2 M HCl gradient. This resulted in complete separation of DAP and N-acetyl-DAP. The N-acetyl-DAP was concentrated by lyophilization. The yield of a soft, white, noncrystalline solid was 15%. Despite the presence of multiple stereoisomers in each sample, only a single TLC spot was observed. The 1H nuclear magnetic resonance (NMR) spectrum of the final product was also consistent with the spectrum of the authentic compound.
A mixture of the different N-succinyl-DAP isomers was prepared by succinylation of dl-DAP as previously described for preparation of N-succinyl-l-DAP (22). Analysis of the reaction mixture by TLC revealed the presence of three compounds: the unreacted DAP, N-succinyl-DAP, and N,N-disuccinyl-DAP. The first two compounds formed ninhydrin-positive spots after TLC. Despite the presence of multiple steroisomers, only a single spot was observed by TLC.
The TLC plates used for analysis of amino acids were Silica Gel 60 F254 glass plates (5 by 20 cm; EMD Chemicals, Inc., Darmstadt, Germany), and the solvent consisted of acetonitrile, water, and formic acid (88%) (40:10:5, vol/vol/vol). With this solvent system, compounds had the following Rfs: diaminopimelate, 0.107; aspartate, 0.30; N-acetyl-diaminopimelate, 0.35; N-succinyl-diaminopimelate, 0.35; α-glutamate, 0.40; N-disuccinyl-diaminopimelate, 0.43; and β-alanine, 0.45.
Strains, media, and culture conditions.
M. maripaludis was grown in 28-ml aluminum seal tubes with 275 kPa of H2-CO2 (80:20, vol/vol) at 37°C in 5 ml of minimal medium containing 10 mM sodium acetate (McNA), McNA containing 1 mM l-lysine (McNALys), or McNA containing 0.2% (wt/vol) Casamino Acids and 0.2% (wt/vol) yeast extract (McC) as described previously (43). Antibiotics were not included when the growth of the wild type and the growth of mutants were compared. The inocula used were 0.1-ml portions of cultures (∼107 cells) grown in McC or McNALys. The inocula for cultures of the S600 mutant and strain S601 were started with frozen stocks for all experiments to ensure that a revertant at second loci had not been selected. Puromycin (2.5 μg/ml) or neomycin (500 μg/ml in plates and 1 mg/ml in broth) was added when necessary. Growth was determined by measuring the increase in the absorbance at 600 nm.
Construction of the Δmmp1527::pac mutant.
The Δmmp1527::pac mutant was constructed by transforming M. maripaludis wild-type strain S2 with pIJA03-MMP1527, which was constructed from the integration vector pIJA03. Plasmid pIJA03 lacks an origin of replication for methanococci and contains a pac cassette, which encodes puromycin resistance (13, 21). To construct pIJA03-MMP1527, a 764-bp region upstream of the MMP1527 gene and a 766-bp region downstream of the MMP1527 gene were PCR amplified and cloned into the multiple-cloning sites MCS1 and MCS2 of pIJA03, respectively. Primer sequences are available upon request. The orientation of each insert was confirmed by restriction mapping.
pIJA03-MMP1527 was transformed into M. maripaludis strain S2 by the polyethylene glycol method (35). After transformation, cultures were plated on McC containing puromycin. Puromycin-resistant isolates were restreaked on the same medium, and isolated colonies were then transferred to broth cultures in 5 ml McC containing puromycin. After growth, 3-ml portions of cultures were used for determination of the genotype and phenotype. The remaining culture was used for preparation of frozen stocks (34). The genotype of the Δmmp1527::pac mutant (S600) was confirmed by Southern hybridization (data not shown).
For complementation, the MMP1527 gene was PCR amplified. The primers introduced an NsiI site and an XbaI site at the 5′ and 3′ ends, respectively. The PCR products and the shuttle vector pMEV2 (21) were digested with NsiI and XbaI and gel purified. Cloning of the PCR products into pMEV2 placed MMP1527 downstream of the strong promoter PhmvA. The resulting plasmid, pMEV2-MMP1527, was transformed into S600 and screened on McC plates containing neomycin as previously described (21). The complementated strain was designated S601.
Preparation of M. maripaludis cell extract.
M. maripaludis was grown in 100 ml of McC to an absorbance at 600 nm of ∼0.4. The cells were collected by centrifugation at 10,000 × g for 30 min at 4°C and resuspended in 2 ml of buffer containing 100 mM Tris (pH 8.5). The cells were lysed by freezing them at −20°C. After thawing, the cell lysate was incubated with 20 U of RQ1 DNase (Promega) at 37°C for 15 min to digest the DNA. Unbroken cells were removed by centrifugation at 8,000 × g for 30 min at 4°C. The protein concentrations were determined by the bicinchoninic acid (BCA) protein assay (Pierce) (31).
Cloning, expression, and purification of recombinant MJ1391 in E. coli.
The M. jannaschii MJ1391 gene was amplified by PCR from genomic DNA. PCR was performed as described previously (16) using an annealing temperature of 45°C. The primers introduced an NdeI site and a BamHI site at the 5′ and 3′ ends, respectively. The amplified PCR product was purified with a QIAquick spin column (Invitrogen), digested with restriction enzymes NdeI and BamHI, and then ligated into compatible sites in plasmid pT7-7 (Novagen) by using bacteriophage T4 DNA ligase (Invitrogen) to obtain plasmid pMJ1391. DNA sequences were verified by dye terminator sequencing at the University of Iowa DNA facility. Plasmid pMJ1391 was transformed into E. coli Origami 2(DE3)/pLysS Singles (Novagen) cells. The transformed cells were grown in 1 liter of Luria-Bertani medium supplemented with 100 μg/ml ampicillin at 37°C with shaking until the absorbance at 600 nm was 0.6 to 0.8. Isopropyl-β-d-1-thiogalactopyranoside (IPTG) was added to a final concentration of 1 mM to induce recombinant protein production. After overnight incubation at room temperature, the cells were harvested by centrifugation at 8,000 × g for 20 min at 4°C.
Cell extracts were prepared by sonication of the E. coli cell pellets (∼2 g, wet weight) suspended in 10 ml of buffer (20 mM Tris-HCl, pH 7.6), followed by centrifugation at 10,000 × g for 20 min at 4°C. The E. coli proteins were denatured by heating the cell extracts at 70°C for 10 min, and the insoluble material was removed by centrifugation at 14,000 × g for 20 min at 4°C. About 5 ml of each extract was applied to a Mono Q 5/50 GL anion-exchange column (GE Healthcare) equilibrated with buffer A (20 mM Tris-HCl, pH 7.5). Bound protein was eluted with a 20-ml linear 0 to 0.5 M NaCl gradient in buffer A at a flow rate of 2.0 ml/min, and 1 ml of each fraction was collected. Elution of the protein was monitored using the UV absorbance at 280 nm. One major UV-absorbing peak eluted at 0.37 M NaCl and contained the desired protein, based on measurement of DapL activity and SDS-PAGE analysis. Purified proteins were dialyzed against 10 mM potassium phosphate buffer (pH 7.0) and 10% (wt/vol) glycerol and stored at −20°C until they were used. Protein concentrations were determined by the BCA protein assay (Pierce) (31).
DapL activity assays.
DapL activity was measured quantitatively in the physiologically reverse direction (ll-DAP → THDPA) based on the DAP-dependent formation of glutamate from α-ketoglutarate (α-KG). The assay was coupled to conversion of glutamate back to α-KG by glutamate dehydrogenase in the presence of NADP+, and the production of NADPH was monitored with a Beckman DU640B spectrophotometer at 340 nm.
For enzyme assays of the M. maripaludis cell extract, 1-ml reaction mixtures contained 100 mM Tris (pH 8.5), 0.25 mg of protein, 2 mM α-KG, 2 mM dl-DAP or 0.5 mM ll-DAP, 10 mM NADP+, 0.25 mM PLP, and 20 U l- glutamate dehydrogenase from bovine liver (Sigma-Aldrich). The reaction mixtures were incubated at 37°C, and the increase in absorbance at 340 nm was monitored continuously.
For assays of the purified recombinant MJ1391-derived protein, the DapL and glutamate dehydrogenase reactions were carried out separately due to the different optimum temperatures. The 0.9-ml DapL reaction mixture contained 100 mM Tris-HCl (pH 8.5), 5 μg MJ1391 protein, 2 mM α-KG, 2 mM dl-DAP or 0.5 mM ll-DAP, and 0.25 mM PLP. The reaction mixtures were incubated at 70°C for 30 min. Since the recombinant MJ1391 protein showed no detectable activity at temperatures below 45°C, the DapL reaction was terminated by rapid cooling of the mixture in an ice-water bath. Then 10 mM NADP+ and 20 U l-glutamate dehydrogenase were added to obtain a final volume of 1 ml. The reaction mixtures were incubated for an additional 20 min at 37°C, and the absorbance at 340 nm was measured. The amount of NADPH produced was calculated using the extinction coefficient (6.22 mM−1 cm−1) (7). The DapL reaction at room temperature was used as a negative control.
To determine the optimum pH, the following buffers (at a concentration of 200 mM) were used: bis(2-hydroxyethyl)-amino-tris(hydroxymethyl)-methane (bis-Tris) hydrochloride for pH 5.5, 6.0, and 6.5; Tris-HCl for pH 7.0, 7.5, 8.0, and 8.5; and sodium 2-(N-cyclohexylamino)ethanesulfonate (CHES) for pH 9.0, 9.5, and 10.0. The coupling reaction with l-glutamate dehydrogenase was not limiting in the pH range from pH 5.5 to 10.0.
The effects of salts on MJ1391 activity were determined by addition of each of the following salts at a concentration of 100 mM: KCl, K2SO4, KH2PO4, NaCl, Na2SO4, NaH2PO4, MgCl2, and MgSO4. The reaction buffers contained 100 mM Tris (pH 8.5), and the pH of each buffer was adjusted with HCl, H2SO4, or H3PO4 using the acid with the same anion as the salt added. The l-glutamate dehydrogenase coupling reaction was not limited by these salts.
Kinetic constants of MJ1391 were determined as described above, except that the ll-DAP concentrations were 0.2, 0.4, 0.6, 0.8, and 1.0 mM and the α-KG concentrations were 0.25, 0.5, 1.0, 1.5, and 2.0 mM. The data were analyzed using SigmaPlot 10.0 with the Enzyme Kinetics module (Systat Software Inc.) fitted with a Ping-Pong Bi-Bi mechanism.
Fluorescence assay of PLP.
PLP in purified recombinant protein MJ1391 was identified and quantified by using the fluorescence of the PLP-cyanide product as described by Adams (1). Fluorescence spectra were recorded with a Shimadzu RF-1501 spectrofluorometer.
Phylogenetic analysis.
DapL homologs were identified using BLASTp searches against selected genomes. A phylogenetic tree was constructed by the MEGA4 program using the minimum evolution (ME) method with the default settings. For comparisons of ratios of evolutionary distances (RED) (11), alignments were generated using the ClustalX program. Evolutionary distances (Ed) were calculated by the MEGA4 program using the PAM-t matrix with the default settings. The mean Ed of the genes encoding large ribosomal subunit protein 2 (L2P), leucyl-tRNA synthetase, and SecY was used as the control Ed (11).
RESULTS AND DISCUSSION
The Δmmp1527 mutant of M. maripaludis is a lysine auxotroph.
Sequence similarity searches identified DapL homologs in Methanococcales that exhibit 27 to 33% amino acid sequence identity with the DapL protein in M. thermautotrophicus (MTH52). Since DapL is an aminotransferase that is closely related to many enzymes with other catalytic activities (18), this level of relatedness was not sufficient to predict the function of the Methanococcales homologs. Therefore, the gene encoding the DapL homolog in M. maripaludis (MMP1527) was deleted by gene replacement with the pac cassette, which encodes puromycin resistance in methanococci. Deletion of MMP1527 was confirmed by Southern hybridization (data not shown). Since the downstream genes were transcribed on the strand opposite the MMP1527 strand, this mutation was not expected to be polar and affect transcription of downstream genes.
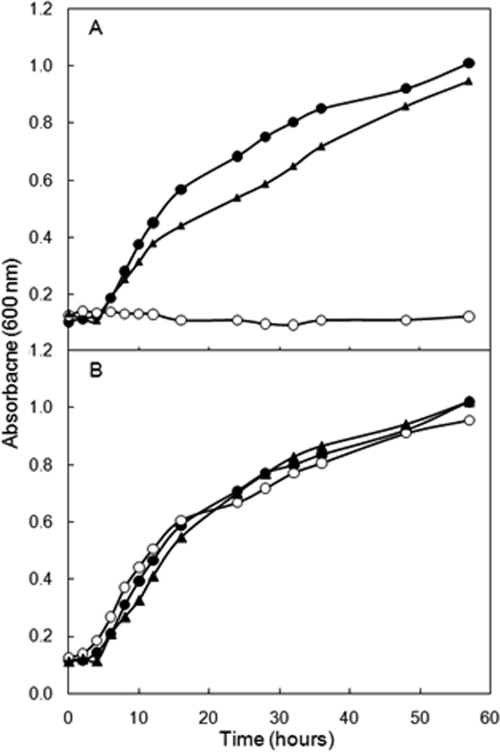
The Δmmp1527 mutant, strain S600, was unable to grow in the absence of exogenous l-lysine (Fig. 2). Unlike some amino acids, lysine can be readily taken up from the medium by methanococci (44). When 1 mM l-lysine was added to McNA, S600 grew at a rate comparable to that of wild-type strain S2. Addition of the other 19 standard amino acids to McNA did not result in growth. To confirm that these differences were due to deletion of MMP1527, the S601 complementation strain was constructed by transforming plasmid pMEV2-MMP1527 into S600. In this strain, MMP1527 was expressed from a strong methanococcal promoter. S601 grew in the absence of exogenous l-lysine (Fig. 2). These results indicated that MMP1527 was required for l-lysine biosynthesis and that MMP1527 was not essential for the biosynthesis of other amino acids.
FIG. 2.
Growth requirement of the Δmmp1527::pac mutant S600 for lysine. (A) Growth in McNA. (B) Growth in McNA containing lysine (1 mM). •, wild-type strain S2; ○, Δmmp1527 mutant S600; ▴, strain S601 (complemented strain S600 with mmp1527 expressed from pMEV2-Mmp1527). The growth curves are representative curves for experiments that were independently replicated three times. All values are the averages of triplicate cultures.
DapL activity in M. maripaludis cell extract.
The specific activities of DapL in cell extracts of M. maripaludis strains S2, S600, and S601 using 0.5 mM ll-DAP as the substrate were 24 ± 2, <1, and 36 ± 2 nmol min−1 mg of protein−1, respectively. These results confirmed the presence of DapL activity in M. maripaludis and its attribution to MMP1527.
The M. maripaludis wild-type S2 cell extract was examined for substrate specificity. When DAP was replaced in assays with acetyl-DAP (2 mM) and succinyl-DAP (2 mM), the specific activities were <4 nmol min−1 mg of protein−1. The absence of detectable activity suggested that M. maripaludis DapL did not use acetyl-DAP or succinyl-DAP as a substrate.
The MJ1391 gene product catalyzed the diaminopimelate aminotransferase reaction.
The DapL homolog from M. jannaschii (MJ1391) was expressed in E. coli and was partially purified by heat denaturation of E. coli proteins, followed by anion-exchange chromatography to obtain a purity of ∼70%. The presence of PLP in the recombinant MJ1391 protein was confirmed by the fluorescent excitation and emission spectra of the PLP-cyanide product. Analysis of the spectra indicated that there was 18.7 μmol of PLP/mg protein (data not shown). Using a molecular mass of 47.9 kDa for the protomer, this corresponded to 0.89 PLP molecule per protomer.
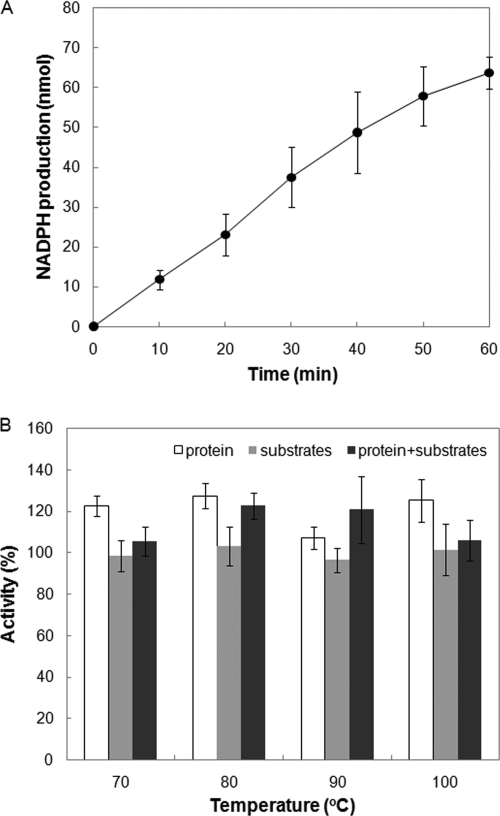
The recombinant MJ1391 protein catalyzed the transamination of ll-DAP, as determined by the DapL activity assay (Fig. 3 A). Moreover, the MJ1391 protein and the substrates α-KG and ll-DAP were stable at 100°C for at least 30 min (Fig. 3B). The level of activity with acetyl-DAP (2 mM) or succinyl-DAP (2 mM) as the substrate was below the detection limit (∼0.05 μmol min−1 mg of protein−1) (data not shown).
FIG. 3.
DapL activity (A) and heat stability (B) of MJ1391. (A) The DapL assay was performed at 70°C for 0 to 60 min before termination and measurement of the product glutamate. (B) The temperature stabilities of MJ1391 and the substrates α-KG and ll-DAP were examined by preincubation of the MJ1391 protein (open bars), the substrates (gray bars), or the protein and the substrates (incubated separately) (black bars) at 70 to 100°C in Tris-HCl buffer (pH 8.5) for 30 min before the DapL activity assay was performed at 70°C. One hundred percent activity was defined as the activity that was observed without preincubation (0.30 μmol min−1 mg of protein−1). The error bars indicate the standard deviations of three assays.
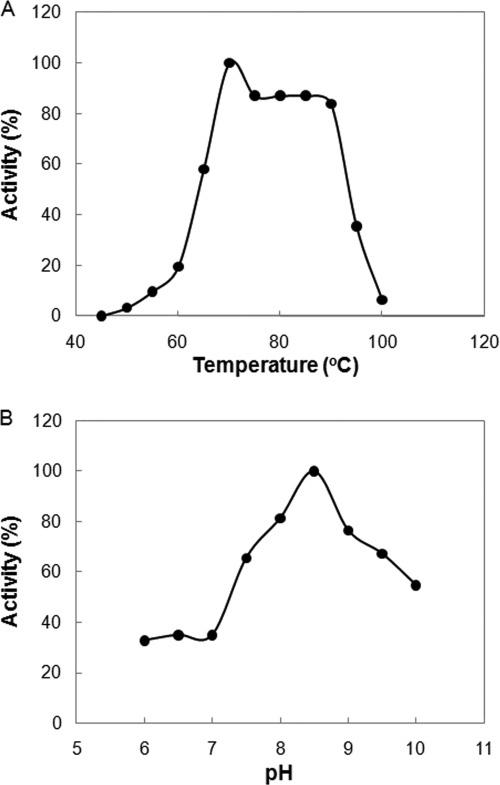
The DapL activity of the MJ1391-derived enzyme was examined under different reaction conditions. The optimum temperature for the DapL activity of MJ1391 was ∼70°C, and no activity was detected at temperatures below 45°C (Fig. 4 A). At 85°C, the optimum growth temperature of M. jannaschii, the DapL activity was 85% of the maximum activity. The optimum pH was ∼8.5. About 40% of the maximum activity and 50% of the maximum activity were observed at pH 7.0 and 10.0, respectively (Fig. 4B). All subsequent assays were performed at 70°C and pH 8.5. Monovalent anions frequently stimulate enzymes from methanococci (45). However, addition of 100 mM KCl, K2SO4, KH2PO4, NaCl, Na2SO4, NaH2PO4, MgCl2, or MgSO4 had no effect on the enzyme activity (data not shown).
FIG. 4.
Optimum temperature (A) and optimum pH (B) for MJ1391. Assays were conducted for 30 min. One hundred percent activity was defined as the activity that was observed at 70°C and pH 8.5 (0.33 μmol min−1 mg of protein−1). The assays were conducted at pH 8.5 (A) and at 70°C (B).
The kinetic constants of MJ1391 were within the ranges for DapL homologs from other sources (18, 24). The Kms for ll-DAP and α-KG were 82.8 ± 10 μM (6 to 116 μM for other organisms) and 0.42 ± 0.02 mM (0.3 to 2.6 mM for other organisms), respectively. The Vmax was 0.39 ± 0.01 μmol min−1 mg of protein−1 (0.25 to 10.6 μmol min−1 mg of protein−1 for other organisms). The kcat for MJ1391 was 18.3 min−1 after correction for a purity of 70%.
Phylogenetic analysis of dapL homologs.
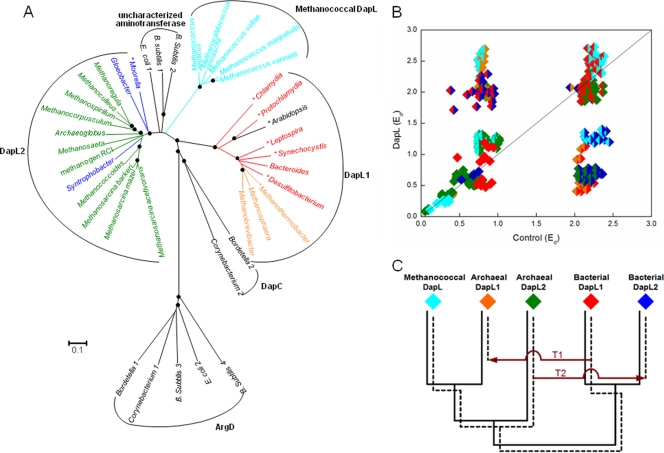
Phylogenetic analysis of DapL identified homologs in all sequenced genomes of members of the Methanococcales, including M. jannaschii, Methanocaldococcus infernus, Methanocaldococcus fervens, Methanocaldococcus vulcanius, M. maripaludis, M. voltae, Methanococcus aeolicus, and Methanococcus vannielii. The sequences exhibited 74 to 89% amino acid sequence identity. According to the phylogenetic tree topology, these methanococcal DapL homologs did not cluster with the DapL1 and DapL2 groups, with which they exhibited 29 to 33% and 30 to 42% amino acid sequence identity, respectively (Fig. 5 A).
FIG. 5.
Phylogenetic analyses of the DapL homologs. Red, bacterial DapL1; blue, bacterial DapL2; orange, archaeal DapL1; green, archaeal DapL2; cyan, methanococcal DapL. (A) Phylogenetic tree constructed by the minimum evolution method using the MEGA4 program. Scale bar = 0.1 amino acid substitution per site. Organisms whose enzymes have biochemically confirmed DapL activity are indicated by an asterisk (18, 24). Filled circles at branch points indicate ≥70% replication for 1,000 bootstraps. The grouping of DapL1 and DapL2 is based on the findings of Hudson et al. (18). Several aminotransferases from E. coli, B. subtilis, Bordetella parapertussis, and C. glutamicum, which are organisms that are known to lack DapL activity, were included to provide an overall context. The locus tags of the sequences used in the phylogenetic analysis are as follows: Protochlamydia, pc0685; Chlamydia, CT390; E. coli 1, b2379; E. coli 2, b3359; Gloeobacter, glr4108; B. subtilis 1, BSU37690; B. subtilis 2, BSU13580; B. subtilis 3, BSU11220; B. subtilis 4, BSU03900; Bacteroides, BF2666; Bordetella 1, BPP2543; Bordetella 2, BPP1996; Synechocystis, sll0480; Leptospira, LIC12841; Methanococcoides, Mbur1013; Methanosarcina barkeri, Mbar_A2605; Methanosphaera, Msp0924; Methanospirillum, Mhun2943; Moorella, Moth0889; Archaeoglobus, AF0409; Methanosarcina acetivorans, MA1712; Methanosarcina mazei, MM2649; Methanosaeta, Mthe0801; Methanothermobacter, MTH52; Corynebacterium 1, NCgl1343; Corynebacterium 2, NCgl1058; Desulfitobacterium, Dhaf1761; Arabidopsis, AT4G33680; Syntrophobacter, Sfum0054; Methanocorpusculum, Mlab0633; Methanoculleus, Memar1915; Methanobrevibacter, Msm1455; methanogen RCI, RCIX1962; Methanoregula, Mboo2096; Methanococcus aeolicus, Maeo1494; Methanococcus vannielii, Mevan0840; Methanococcus maripaludis, MMP1527; Methanocaldococcus jannaschii, MJ1391; and Methanococcus voltae, Mvol0315. (B) RED plot of DapL homologs. Intradomain comparisons within each DapL subgroup are indicated by diamonds that are a single color. Comparisons of different subgroups are indicated by diamonds that are two colors. (C) Hypothetical gene tree illustrating the proposed evolutionary history of DapL homologs. The solid lines show the organismal phylogeny in the absence of LGT. The dashed lines show the gene tree for DapL. T1 indicates the acquisition of bacterial DapL1 by archaea, and T2 indicates the acquisition of archaeal DapL2 by bacteria.
The methanococcal enzymes were also distinguished from the DapL1 and DapL2 groups by comparison of conserved amino acid residues. Based on the crystal structure of DapL from A. thaliana, three conserved residues in DapL1, Glu97, Asn309, and Lys129, are important for ll-DAP and glutamate binding (37). Lys129, which is found at similar positions in aspartate aminotransferases, is conserved in all DapL homologs. DapL2 lacks the other two residues, Glu97 and Asn309 (18). Methanococcal DapL contains only Glu97. Further study is needed to understand how the substrates bind to DapL2 and methanococcal DapL.
The phylogeny of the bacterial and archaeal DapL proteins in the ME tree was not correlated with the 16S rRNA-based phylogeny, indicating that lateral gene transfers (LTGs) may have made substantial contributions to the evolution of DapL in some lineages. The evolutionary history of DapL homologs was evaluated by comparison of the ratios of their evolutionary distances (RED) with those of control genes which were believed to have undergone primarily vertical evolution (11). If the evolutionary rate of DapL was constant and LTGs did not occur, then a plot of the DapL evolutionary distances (Eds) against the control Eds should be linear. As shown in Fig. 5B, the intradomain comparisons for each DapL group were close to a trend line indicative of a constant rate of DapL evolution relative to that of the control genes. Moreover, comparisons among the bacterial DapL1, the archaeal DapL2, and the methanococcal DapL lay on the same trend line, indicating that these groups did not obtain the gene from an LGT. However, the interdomain comparisons between bacteria and archaea within each of the DapL1 and DapL2 groups resulted in Eds that were lower than the values expected, and bacterial and archaeal intradomain comparisons for each of the groups resulted in Eds that were higher than the values expected for normal vertical evolution. These results suggested that interdomain LTGs occurred within both groups, that there was one transfer from the archaeal DapL2 group to form the bacterial DapL2 group, and that there was one transfer from the bacterial DapL1 group to form the archaeal DapL1 group (Fig. 5C). Lastly, comparisons between the bacterial DapL2 group and the archaeal DapL1 group are on the central trend line because each group resulted from LGTs but the LGTs were in opposite directions. In toto, these results suggested that in DapL evolution, while there were two LGT events, there were not discontinuities in the rate of evolution or the formation of paralogous gene families.
RED analyses that included the uncharacterized DapL paralogs of E. coli (b2379) and Bacillus subtilis (BSU37690) showed that these paralogs had lower Eds with archaeal DapL2 and methanococcal DapL groups and higher Eds with bacteria DapL than expected, suggesting that these paralogs may have had an archaeal origin (data not shown).
The selective pressure for some organisms to obtain and maintain the DapL pathway instead of the acylation pathways is unclear. THDPA is unstable at neutral pH and at equilibrium forms a mixture of the cyclic compound THDPA and the acyclic compound l-α-keto-ɛ-aminopimelate (5, 6). The equilibrium favors cyclic THDPA, while the acyclic form containing the keto group is the substrate for transamination. Therefore, acylation may facilitate the transamination by exposing the keto group at the expense of one succinyl-CoA or acetyl-CoA (15, 18, 19). However, whether cyclic THDPA is the actual substrate of DapL or DapL catalyzes the ring-opening reaction awaits further investigation (37, 38).
In conclusion, methanococci use the DapL pathway for l-lysine biosynthesis, as a deletion mutation of the dapL homolog in M. maripaludis resulted in lysine auxotrophy and the purified homologous protein in M. jannaschii catalyzed the DAP aminotransferase reaction. Moreover, activities with succinylated and acetylated DAP were not observed for M. maripaludis cell extract and the recombinant M. jannaschii protein, suggesting that the succinylase and acetylase pathways are not used by methanococci.
Acknowledgments
We thank André O. Hudson and Thomas Leustek for the kind gift of ll-DAP. We thank Kim Harich for assistance with gas chromatography-mass spectrometry analyses.
This work was supported by grants from the U.S. Department of Energy to W.B.W, by grant MCB0722787 from the National Science Foundation to R.H.W., and by a dissertation completion grant from the University of Georgia to Y.L.
Footnotes
Published ahead of print on 23 April 2010.
REFERENCES
- 1.Adams, E. 1979. Fluorometric determination of pyridoxal phosphate in enzymes. Methods Enzymol. 62:407-410. [DOI] [PubMed] [Google Scholar]
- 2.Bakhiet, N., F. Forney, D. Stahly, and L. Daniels. 1984. Lysine biosynthesis in Methanobacterium thermoautotrophicum is by the diaminopimelic acid pathway. Curr. Microbiol. 10:195-198. [Google Scholar]
- 3.Bartlett, A. T. M., and P. J. White. 1985. Species of Bacillus that make a vegetative peptidoglycan containing lysine lack diaminopimelate epimerase but have diaminopimelate dehydrogenase. J. Gen. Microbiol. 131:2145-2152. [Google Scholar]
- 4.Bult, C. J., O. White, G. J. Olsen, L. Zhou, R. D. Fleischmann, G. G. Sutton, J. A. Blake, L. M. FitzGerald, R. A. Clayton, J. D. Gocayne, A. R. Kerlavage, B. A. Dougherty, J.-F. Tomb, M. D. Adams, C. I. Reich, R. Overbeek, E. F. Kirkness, K. G. Weinstock, J. M. Merrick, A. Glodek, J. L. Scott, N. S. M. Geoghagen, J. F. Weidman, J. L. Fuhrmann, D. Nguyen, T. R. Utterback, J. M. Kelley, J. D. Peterson, P. W. Sadow, M. C. Hanna, M. D. Cotton, K. M. Roberts, M. A. Hurst, B. P. Kaine, M. Borodovsky, H.-P. Klenk, C. M. Fraser, H. O. Smith, C. R. Woese, and J. C. Venter. 1996. Complete genome sequence of the methanogenic archaeon, Methanococcus jannaschii. Science 273:1058-1073. [DOI] [PubMed] [Google Scholar]
- 5.Caplan, J., A. Sutherland, and J. C. Vederas. 2001. The first stereospecific synthesis of l-tetrahydrodipicolinic acid; a key intermediate of diaminopimelate metabolism. J. Chem. Soc. Perkin Trans. 1:2217-2220. [Google Scholar]
- 6.Chrystal, E. J. T., L. Couper, and D. J. Robins. 1995. Synthesis of a key intermediate in the diaminopimelate pathway to l-lysine: 2,3,4,5-tetrahydrodipicolinic acid. Tetrahedron 51:10241-10252. [Google Scholar]
- 7.Dawson, R. M. C., D. C. Elliott, W. H. Elliott, and K. M. Jones. 1986. Spectral data and pKa values for purines, pyrmidine, nucleosides and nucleotides p. 103-114. In R. M. C. Dawson, D. C. Elliott, W. H. Elliott, and K. M. Jones (ed.), Data for biochemical research, 3rd ed. Oxford University Press, New York NY.
- 8.Deppenmeier, U., A. Johann, T. Hartsch, R. Merkl, R. A. Schmitz, R. Martinez-Arias, A. Henne, A. Wiezer, S. Baumer, C. Jacobi, H. Bruggemann, T. Lienard, A. Christmann, M. Bomeke, S. Steckel, A. Bhattacharyya, A. Lykidis, R. Overbeek, H. P. Klenk, R. P. Gunsalus, H. J. Fritz, and G. Gottschalk. 2002. The genome of Methanosarcina mazei: evidence for lateral gene transfer between bacteria and archaea. J. Mol. Microbiol. Biotechnol. 4:453-461. [PubMed] [Google Scholar]
- 9.Ekiel, I., K. F. Jarrell, and G. D. Sprott. 1985. Amino acid biosynthesis and sodium-dependent transport in Methanococcus voltae, as revealed by 13C NMR. Eur. J. Biochem. 149:437-444. [DOI] [PubMed] [Google Scholar]
- 10.Ekiel, I., I. C. Smith, and G. D. Sprott. 1983. Biosynthetic pathways in Methanospirillum hungatei as determined by 13C nuclear magnetic resonance. J. Bacteriol. 156:316-326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Farahi, K., G. D. Pusch, R. Overbeek, and W. B. Whitman. 2004. Detection of lateral gene transfer events in the prokaryotic tRNA synthetases by the ratios of evolutionary distances method. J. Mol. Evol. 58:615-631. [DOI] [PubMed] [Google Scholar]
- 12.Fuchs, T. M., B. Schneider, K. Krumbach, L. Eggeling, and R. Gross. 2000. Characterization of a Bordetella pertussis diaminopimelate (DAP) biosynthesis locus identifies dapC, a novel gene coding for an N-succinyl-ll-DAP aminotransferase. J. Bacteriol. 182:3626-3631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gernhardt, P., O. Possot, M. Foglino, L. Sibold, and A. Klein. 1990. Construction of an integration vector for use in the archaebacterium Methanococcus voltae and expression of a eubacterial resistance gene. Mol. Gen. Genet. 221:273-279. [DOI] [PubMed] [Google Scholar]
- 14.Gilvarg, C. 1961. N-Succinyl-α-amino-6-ketopimelic acid. J. Biol. Chem. 236:1429-1431. [PubMed] [Google Scholar]
- 15.Graham, D. E., and H. K. Huse. 2008. Methanogens with pseudomurein use diaminopimelate aminotransferase in lysine biosynthesis. FEBS Lett. 582:1369-1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Graupner, M., H. Xu, and R. H. White. 2000. Identification of an archaeal 2-hydroxy acid dehydrogenase catalyzing reactions involved in coenzyme biosynthesis in methanoarchaea. J. Bacteriol. 182:3688-3692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hendrickson, E. L., R. Kaul, Y. Zhou, D. Bovee, P. Chapman, J. Chung, E. Conway de Macario, J. A. Dodsworth, W. Gillett, D. E. Graham, M. Hackett, A. K. Haydock, A. Kang, M. L. Land, R. Levy, T. J. Lie, T. A. Major, B. C. Moore, I. Porat, A. Palmeiri, G. Rouse, C. Saenphimmachak, D. Soll, S. Van Dien, T. Wang, W. B. Whitman, Q. Xia, Y. Zhang, F. W. Larimer, M. V. Olson, and J. A. Leigh. 2004. Complete genome sequence of the genetically tractable hydrogenotrophic methanogen Methanococcus maripaludis. J. Bacteriol. 186:6956-6969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hudson, A. O., C. Gilvarg, and T. Leustek. 2008. Biochemical and phylogenetic characterization of a novel diaminopimelate biosynthesis pathway in prokaryotes identifies a diverged form of ll-diaminopimelate aminotransferase. J. Bacteriol. 190:3256-3263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hudson, A. O., B. K. Singh, T. Leustek, and C. Gilvarg. 2006. An ll-diaminopimelate aminotransferase defines a novel variant of the lysine biosynthesis pathway in plants. Plant Physiol. 140:292-301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ledwidge, R., and J. S. Blanchard. 1999. The dual biosynthetic capability of N-acetylornithine aminotransferase in arginine and lysine biosynthesis. Biochemistry 38:3019-3024. [DOI] [PubMed] [Google Scholar]
- 21.Lin, W., and W. Whitman. 2004. The importance of porE and porF in the anabolic pyruvate oxidoreductase of Methanococcus maripaludis. Arch. Microbiol. 181:68-73. [DOI] [PubMed] [Google Scholar]
- 22.Lin, Y. K., R. Myhrman, M. L. Schrag, and M. H. Gelb. 1988. Bacterial N-succinyl-l-diaminopimelic acid desuccinylase. Purification, partial characterization, and substrate specificity. J. Biol. Chem. 263:1622-1627. [PubMed] [Google Scholar]
- 23.Liu, Y. 2010. Taxonomy of methanogens, p. 550-558. In K. N. Timmis, T. McGenity, J. R. van den Meer, and V. de Lorenzo (ed.), Handbook of hydrocarbon and lipid microbiology, vol. 1. Springer-Verlag, Berlin, Germany.
- 24.McCoy, A. J., N. E. Adams, A. O. Hudson, C. Gilvarg, T. Leustek, and A. T. Maurelli. 2006. l,l-Diaminopimelate aminotransferase, a trans-kingdom enzyme shared by Chlamydia and plants for synthesis of diaminopimelate/lysine. Proc. Natl. Acad. Sci. U. S. A. 103:17909-17914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Misono, H., H. Togawa, T. Yamamoto, and K. Soda. 1979. Meso-α,ɛ-diaminopimelate d-dehydrogenase: distribution and the reaction product. J. Bacteriol. 137:22-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Misono, H., H. Togawa, T. Yamamoto, and K. Soda. 1976. Occurrence of meso-α,ɛ-diaminopimelate dehydrogenase in Bacillus sphaericus. Biochem. Biophys. Res. Commun. 72:89-93. [DOI] [PubMed] [Google Scholar]
- 27.Nishida, H., M. Nishiyama, N. Kobashi, T. Kosuge, T. Hoshino, and H. Yamane. 1999. A prokaryotic gene cluster involved in synthesis of lysine through the amino adipate pathway: a key to the evolution of amino acid biosynthesis. Genome Res. 9:1175-1183. [DOI] [PubMed] [Google Scholar]
- 28.Patte, J.-C. 1996. Biosynthesis of threonine and lysine, p. 528-541. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. ASM Press, Washington, DC.
- 29.Scapin, G., and J. S. Blanchard. 1998. Enzymology of bacterial lysine biosynthesis. Adv. Enzymol. Relat. Areas Mol. Biol. 72:279-324. [DOI] [PubMed] [Google Scholar]
- 30.Schrumpf, B., A. Schwarzer, J. Kalinowski, A. Puhler, L. Eggeling, and H. Sahm. 1991. A functionally split pathway for lysine synthesis in Corynebacterium glutamicium. J. Bacteriol. 173:4510-4516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Smith, P. K., R. I. Krohn, G. T. Hermanson, A. K. Mallia, F. H. Gartner, M. D. Provenzano, E. K. Fujimoto, N. M. Goeke, B. J. Olson, and D. C. Klenk. 1985. Measurement of protein using bicinchoninic acid. Anal. Biochem. 150:76-85. [DOI] [PubMed] [Google Scholar]
- 32.Sprott, G. D., I. Ekiel, and G. B. Patel. 1993. Metabolic pathways in Methanococcus jannaschii and other methanogenic bacteria. Appl. Environ. Microbiol. 59:1092-1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sundharadas, G., and C. Gilvarg. 1967. Biosynthesis of α,ɛ-diaminopimelic acid in Bacillus megaterium. J. Biol. Chem. 242:3983-3984. [PubMed] [Google Scholar]
- 34.Tumbula, D. L., J. Keswani, J. S. Shieh, and W. B. Whitman. 1995. Maintenance of methanogen stock cultures in glycerol at −70°C, p. 85-87. In S. Robb F. T., K. R., DasSarma, S., Place, A. R., Schreier, H. J., Fleischmann, and E. M. (ed.), Archaea—a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 35.Tumbula, D. L., R. A. Makula, and W. B. Whitman. 1994. Transformation of Methanococcus maripaludis and identification of a PstI-like restriction system. FEMS Microbiol. Lett. 121:309-314. [Google Scholar]
- 36.Velasco, A. M., J. I. Leguina, and A. Lazcano. 2002. Molecular evolution of the lysine biosynthetic pathways. J. Mol. Evol. 55:445-449. [DOI] [PubMed] [Google Scholar]
- 37.Watanabe, N., M. M. Cherney, M. J. van Belkum, S. L. Marcus, M. D. Flegel, M. D. Clay, M. K. Deyholos, J. C. Vederas, and M. N. James. 2007. Crystal structure of ll-diaminopimelate aminotransferase from Arabidopsis thaliana: a recently discovered enzyme in the biosynthesis of l-lysine by plants and Chlamydia. J. Mol. Biol. 371:685-702. [DOI] [PubMed] [Google Scholar]
- 38.Watanabe, N., M. D. Clay, M. J. van Belkum, M. M. Cherney, J. C. Vederas, and M. N. G. James. 2008. Mechanism of substrate recognition and PLP-induced conformational changes in ll-diaminopimelate aminotransferase from Arabidopsis thaliana. J. Mol. Biol. 384:1314-1329. [DOI] [PubMed] [Google Scholar]
- 39.Weinberger, S., and C. Gilvarg. 1970. Bacterial distribution of the use of succinyl and acetyl blocking groups in diaminopimelic acid biosynthesis. J. Bacteriol. 101:323-324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.White, P. J. 1983. The essential role of diaminopimelate dehydrogenase in the biosynthesis of lysine by Bacillus sphaericus. J. Gen. Microbiol. 129:739-749. [Google Scholar]
- 41.Whitman, W., T. Bowen, and D. Boone. 2006. The methanogenic bacteria, p. 165-207. In M. Dworkin, S. Falkow, E. Rosenberg, K.-H. Schleifer, and E. Stackebrandt (ed.), The prokaryotes, 3rd ed. Springer-Verlag, New York, NY.
- 42.Whitman, W. B., D. R. Boone, Y. Koga, and J. Keswani. 2001. Taxonomy of methanogenic archaea, p. 211-213. In D. R. Boone, R. W. Castenholtz, and G. M. Garrity (ed.), Bergey's manual of systematic bacteriology, 2nd ed., vol. 1. Springer-Verlag, New York, NY. [Google Scholar]
- 43.Whitman, W. B., J. Shieh, S. Sohn, D. S. Caras, and U. Premachandran. 1986. Isolation and characterization of 22 mesophilic methanococci. Syst. Appl. Microbiol. 7:235-240. [Google Scholar]
- 44.Whitman, W. B., S. Sohn, S. Kuk, and R. Xing. 1987. Role of amino acids and vitamins in nutrition of mesophilic Methanococcus spp. Appl. Environ. Microbiol. 53:2373-2378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xing, R. Y., and W. B. Whitman. 1991. Characterization of enzymes of the branched-chain amino acid biosynthetic pathway in Methanococcus spp. J. Bacteriol. 173:2086-2092. [DOI] [PMC free article] [PubMed] [Google Scholar]