Abstract
Faithful segregation of chromosomes and plasmids is a vital prerequisite to produce viable and genetically identical progeny. Bacteria use a specialized segregation system composed of the partitioning proteins ParA and ParB to segregate certain plasmids. Strikingly, homologues of ParA and ParB are found to be encoded in many chromosomes. Although mutations in the chromosomal Par system have effects on segregation efficiency, the exact mechanism by which the chromosomes are segregated into the daughter cells is not fully understood. We describe the polar localization of the ParB origin nucleoprotein complex in the actinomycete Corynebacterium glutamicum. ParB and the origin of replication were found to be stably localized to the cell poles. After replication, the origins move toward the opposite pole. Purified ParB was able to bind to the parS consensus sequence in vitro. C. glutamicum possesses two ParA-like partitioning ATPase proteins. Both proteins interact with ParB but show a slightly different subcellular localization and phenotype. While ParA might be part of a conventional partitioning system, PldP seems to play a role in division site selection.
Bacterial cell division is a temporally and spatially tightly regulated process (1, 13, 16, 36, 37). Spatial regulation is achieved by division site selection and prevents fatal division across the nucleoids and aberrant division close to the cell poles (3, 40). Temporal control ensures that division does not precede chromosome replication and segregation. Replicated chromosomes are rapidly segregated into the daughter cells. However, the machinery that performs this active segregation is not fully elucidated. In contrast, plasmid segregation is somewhat better understood. Plasmids such as pB171 (8) encode a machinery composed of a tripartite system. Centromere-like DNA sequences, named parS sites, are composed of short inverted repeats. Centromere-binding proteins (ParB) are recruited to the parS sites, forming nucleoprotein complexes. Finally, a partitioning ATPase is recruited to the ParB-parS complex. The hydrolytic activity of ParA oligomers is believed to drive the active segregation process. Strikingly, many bacterial chromosomes encode orthologs of the plasmid partitioning genes parA and parB. A comparatively well-examined chromosomal partitioning system is that of Bacillus subtilis. B. subtilis encodes a ParA ATPase (called Soj) and a ParB protein (called Spo0J). B. subtilis contains eight parS sites that cluster around the oriC region and bind Spo0J. Subsequently Spo0J spreads across the DNA, thereby forming a huge nucleoprotein complex that could serve as a platform for anchoring the segregation machinery. The ParA protein Soj is a DNA-binding protein that dissociates from DNA upon ATP hydrolysis. A direct interaction of Soj and Spo0J has been described (35). Interestingly, analysis of knockout mutations revealed that only the loss of the ParB protein Spo0J increases the amount of anucleate cells slightly, while the loss of Soj has no significant effect on chromosome segregation (17, 18). However, knockout mutations in either parA or parB result only in subtle effects on chromosome segregation. Thus, although the two proteins might act together they have certainly multiple roles during chromosome segregation and cell division. Recently, it was shown that Spo0J (ParB) helps to recruit SMC proteins (for structural maintenance of chromosomes) to the oriC region, thereby ensuring correct chromosome organization, which seems essential for proper segregation (15, 39). The B. subtilis ParA homologue Soj was shown to play an role in the initiation of DNA replication by interacting with DnaA (32). Hence, the ParAB system is a central component connecting replication and segregation. Interestingly, Par proteins have been implicated with different developmental processes in other bacteria. In Caulobacter crescentus ParAB are involved in cell cycle progression and cell division. A ParA-like protein, MipZ, was shown to interact with ParB and directly inhibit FtsZ polymerization (42). Thus, chromosome segregation and cell division are directly coupled. Consequently, null mutations in ParA and ParB are lethal in C. crescentus. In Vibrio cholerae it was shown that ParA and ParB encoded on the large chromosome contribute to active chromosome segregation and anchor the oriC region of the chromosomes to the cell poles (10).
Although these diverse properties of the Par system have been studied in some detail in the classical model organisms, the situation in other bacteria remains unknown. Corynebacteria are high GC Gram-positive bacteria and, depending on the growth medium, rod-shaped or club-shaped. A remarkable feature of corynebacteria and their close relatives is a special cell wall that has, in addition to the common peptidoglycan, an arabino-galactan and a mycolic acid layer. Notorious pathogens such as Mycobacterium tuberculosis, Mycobacterium leprae, and Corynebacterium diphtheriae are members of this family, and hence an understanding of fundamental cell biological mechanisms might reveal insights how to combat these organisms. We now report the subcellular localization of the chromosome partitioning system and the oriC in the actinomycete Corynebacterium glutamicum. We show localization and phenotypic consequences of the canonical ParAB proteins. Furthermore, we identified a ParA-like division protein (PldP) that plays a role in division site selection.
MATERIALS AND METHODS
Growth medium and conditions.
Escherichia coli cells were grown at 37°C in Luria-Bertani (LB) medium containing, where appropriate, kanamycin at 25 μg ml−1 or carbenicillin at 100 μg ml−1. All C. glutamicum cells were grown at 30°C in brain heart infusion (BHI) medium (Becton Dickinson) for maintenance, and in MM1 (34), supplemented with 4% glucose, or LB medium, as indicated in the text, for all microscopic and growth analyses. For growth analysis, all strains were cultivated during the day in 5 ml of LB medium and then rediluted in 10 ml of LB medium and grown overnight. The following day the cultures were rediluted to an optical density (OD) of 1.0 in MM1 or LB medium as described in the text.
Strains, plasmids, and oligonucleotides.
All strains and plasmids used in the present study are listed in Table 1 and Table 2, respectively. Oligonucleotides are summarized in Table 3.
TABLE 1.
Bacterial strains
| Strain | Genotype | Source or reference |
|---|---|---|
| Escherichia coli | ||
| DH5α | F− φ80lacZΔM15(lacZYA-argF)U169 recA1 endA1 hsdR17(rK− mK+) phoA supE44 thi-1 gyrA96 relA1 λ− | Invitrogen |
| SU101 | Tn9 Cmr for selection of F′ plasmid, JL1434 lambda lysogen with lexA operator (wt/wt) sulA promoter-lacZ fusion, JL1434 lexA71::Tn5(Def)sulA211 ΔlacU169 F′ [lacIqlacZΔM15::Tn9] | 6 |
| SU202 | Tn9 Cmr for selection of F′ plasmid, JL1434 lambda lysogen with lexA operator (408/wt) sulA promoter-lacZ fusion, JL1434 lexA71::Tn5(Def)sulA211 ΔlacU169 F′[lacIqlacZΔM15::Tn9] | 6 |
| Corynebacterium glutamicum | ||
| RES 167 | Restriction-deficient C. glutamicum mutant, otherwise considered wild type | 41 |
| CDC001 | RES 167 derivative with in-frame deletion of parA | This study |
| CDC002 | RES 167 derivative with in-frame deletion of pldP | This study |
| CDC003 | RES 167 derivative with in-frame deletion of parB | This study |
| CDC004 | RES 167 derivative with parA-CFP | This study |
| CDC005 | RES 167 derivative with pldP-CFP | This study |
| CDC006 | CDC002, parA::parA-cfp | This study |
| CDC007 | RES 167 derivative with IPTG-inducible extrachromosomal copy of ParB-CFP | This study |
| CDC008 | CDC001 with IPTG-inducible extrachromosomal copy of ParB-CFP | This study |
| CDC009 | CDC002 with IPTG-inducible extrachromosomal copy of ParB-CFP | This study |
| CAS006 | RES 167 derivative containing native parAB operon with chromosomal integration pK18mob-parB-gfp; Kmr | This study |
| CAS009 | tetO-Spcr array integrated between cg0002 and cg0004 with pLAU44-integ ori, extrachromosomal YFP-TetR expression using pEKEx2-yfp-tetR | This study |
| CAS010 | C. glutamicum strain with IPTG-inducible extrachromosomal expression of ParA from plasmid pEKEx2-parA | This study |
| CAS011 | C. glutamicum strain with IPTG-inducible extrachromosomal expression of PldP from plasmid pEKEx2-pldP | This study |
TABLE 2.
Plasmids
| Plasmid | Characteristicsa | Source or reference |
|---|---|---|
| pK19mobsacB | Integration vector, ori pUC, Kmr, mob sacB | 38 |
| pK19mobsacBΔparA | Integration vector, ori pUC, Kmr, mob sacB ΔparA | This study |
| pK19mobsacBΔpldP | Integration vector, ori pUC, Kmr, mob sacB ΔpldP | This study |
| pK19mobsacBΔparB | Integration vector, ori pUC, Kmr, mob sacB ΔparB | This study |
| pSG1186 | bla cat cfp | 9 |
| pSG1186-parA | bla cat parA′ cfp | This study |
| pSG1186-pldP | bla cat pldP′ cfp | This study |
| pK19mobsacB-CFP-parA | Integration vector, ori pUC, Kmr, mob sacB parA′ cfp | This study |
| pK19mobsacB-CFP-pldP | Integration vector, ori pUC, Kmr, mob sacB pldP′ cfp | This study |
| pK18mob | ori pUC, Kmr, mob; C. glutamicum insertion vector | This study |
| pK18mob-parA | ori pUC, Kmr, insertion vector, with coding region of parA | This study |
| pK18mob-parA+100 | ori pUC, Kmr, insertion vector, with parA + native promoter | This study |
| pK18mob-parA+100-gfp | ori pUC, Kmr, insertion vector, with parA-gfp + native promoter | This study |
| pK18mob-pldP+100-gfp | ori pUC, Kmr, insertion vector, with pldP-gfp + native promoter | This study |
| pK18mob-parB-gfp | ori pUC, Kmr, insertion vector, with parB-gfp | This study |
| pMS604 | Tcr, lexA(1-87)-wt-Fos zipper | 6 |
| pMS604-parA | Tcr, expression of parA-lexA(1-87)-wt-Fos zipper | This study |
| pMS604-pldP | Tcr, expression of pldP-lexA(1-87)-wt-Fos zipper | This study |
| pMS604-parB | Tcr, expression of parB-lexA(1-87)-wt-Fos zipper | This study |
| pMS604-ftsZ | Tcr, expression of ftsZ-lexA(1-87)-wt-Fos zipper | This study |
| pDP804 | Apr, lexA(1-87)-408-Jun zipper | 6 |
| pDP804-parA | Apr, expression of parA-lexA(1-87)-408-Jun zipper | This study |
| pDP804-pldP | Apr, expression of pldP-lexA(1-87)-408-Jun zipper | This study |
| pDP804-parB | Apr, expression of parB-lexA(1-87)-408-Jun zipper | This study |
| pDP804-ftsZ | Apr, expression of ftsZ-lexA(1-87)-408-Jun zipper | This study |
| pLAU44 | tetO; Gmr | 20 |
| pLAU44-CGP3-Spec | tetO; Spr, cg1905-cg1906 intergenic region | 11 |
| pEKEx2 | KmrE. coli-C. glutamicum shuttle vector for regulated gene expression (Ptac lacIq pBL1 oriVC.g. pUC18 oriVE.c.) | B. Eikmanns |
| pEKEx2-CFP | Ptac lacIq pBL1 oriVC.g. pUC18 oriVE.c., CFP | This study |
| pEKEx2-CFP-ParB | Ptac lacIq pBL1 oriVC.g. pUC18 oriVE.c., ParB+-CFP | This study |
Tcr, tetracycline resistance; Apr, ampicillin resistance; Spr, spectinomycin resistance; Kmr, kanamycin resistance.
TABLE 3.
Oligonucleotides
| Oligonucleotide | Sequence (5′-3′)a | Restriction enzyme |
|---|---|---|
| ΔParA-1- F | CAGAAGCTTCTATCGCACGCCCAGATC | HindIII |
| ΔParA-1-R | GGAATGGAGTATGGAAGTTGGCGACGTCAACCATCCCTA | |
| ΔParA-2- F | CCAACTTCCATACTCCATTCCAGTAAACTTCTTTGAATA | |
| ΔParA-2-R | CAGTCTAGACACCAACTCGTCAAGTGC | XbaI |
| ParA-800- F | CAGTCTAGATTAAGTTGAGTCGTTATA | |
| ParA-800-R | CAGGCTAGCTACCGGACGGGAACGGCC | |
| ΔPldP-1-F | CAGAAGCTTAGGTGTATGACAGGGAAA | HindIII |
| ΔPldP-1-R | GGAATGGAGTATGGAAGTTGGAGTCAAACCTTCTTCCTT | |
| ΔPldP-2-F | CCAACTTCCATACTCCATTCCCCACGCCTCCTTGTGCGG | |
| ΔPldP-2-R | CAGTCTAGACGCGGGAGCAGGCGAGCT | XbaI |
| PldP-600-F | CAGGAATTCGCTCGCAGAAGTGTGGTTTTA | |
| PldP-800-R | CAGTCTAGAACTGACACCGCAACTTGG | |
| ΔParB-1-F | CAGAAGCTTGACTGCCCACCGTCTTTG | HindIII |
| ΔParB-1-R | GGAATGGAGTATGGAAGTTGGCGCTCTTAGACGCACCTT | |
| ΔParB-2-F | CCAACTTCCATACTCCATTCCTTTTAAGTTTGGCGCCAT | |
| ΔParB-2-R | CAGTCTAGACCTCCACATCAATCAGGC | XbaI |
| ParB-800-F | CAGGAATTCATTCATGGGCTTAAAGTTCTC | |
| ParB-800-R | CAGCTGCAGCCAACCCCGATGACCTGG | |
| ParA-1-F | CAGAAGCTTGTTTGGCGGATGCGTTGG | HindIII |
| ParA-1-R | CAGGAATTCTTTCGCAGGTTTTAGGCC | EcoRI |
| ParA-2-F | CAGACTAGTTAGCAGTAAACTTCTTTG | SpeI |
| ParA-2-R | CAGTCTAGAACCAACTCGTCAAGTGCC | XbaI |
| PldP-1-F | CAGAAGCTTTATTGACTTGTCCGCTGC | HindIII |
| PldP-1-R | CAGGAATTCGTCGTTGACGCGGCTGAT | EcoRI |
| PldP-2-F | CAGACTAGTTAGGTTGTTTTTCTAAAA | SpeI |
| PldP-2-R | CAGTCTAGACGCAACTTGGTGCACGTG | XbaI |
| ftsZ Eco91I F | CAGGGTCACCGGATGACCTCACCGAACAACTAC | Eco91I |
| ftsZ ET XhoI R | CAGCTCGAGTTACTGGAGGAAGCTGGG | XhoI |
| parA ET XhoI F | CAGCTCGAGATGGAAGACACTACTTGGGAA | XhoI |
| parA BglII R mS | CAGAGATCTCTATTTCGCAGGTTTTAG | BglII |
| pldP ET XhoI F | CAGCTCGAGGTGAGTGATGCAGGGAAGAAG | XhoI |
| pldP BglII R mS | CAGAGATCTCTAGTCGTTGACGCGGCTGAT | BglII |
| parB XhoI F | CAGCTCGAGATGGCTCAGAACAAGGGTTCC | XhoI |
| parB BglII R mS | CAGAGATCTTTATTGGCCCTGGATCAAGGA | BglII |
| ftsZ XhoI F | CAGCTCGAGATGACCTCACCGAACAACTACCTC | XhoI |
| ftsZ BglII R mS | CAGAGATCTTTACTGGAGGAAGCTGGGTACATC | BglII |
| Integ ori XbaI F | CGCTCTAGATTGGGAAATATAGATCAA | XbaI |
| Integ ori XbaI R | CGCTCTAGAGCCAGAATTCCGCGACTA | XbaI |
| gfp_reverse | CTGGGATCCCTATTTGTATAGTTCATCC | BamHI |
| gfp_reverse | CTGAAGCTTCTATTTGTATAGTTCATCC | HindIII |
| gfp _reverse db | GGGCCCTAATACGACTCACTATAGGGCTGCTATTTGTATAGTTCATCC | |
| parA_forward | CAGGAATTCGCGGCAGCGATGGAAGACACTACTTGG | EcoRI |
| parA100_forward | CAGGAATTCCACTTCGGTGGAATGGGT | EcoRI |
| parA_reverse_oS | CTGGGTACCTTTCGCAGGTTTTAGGCC | KpnI |
| parA_reverse_mS | CAGGGTACCCTATTTCGCAGGTTTTAG | KpnI |
| parA_forward | CAGCATATGGAAGACACTACTTGGGAA | NdeI |
| parA_reverse | CAGCTCGAGCTATTTCGCAGGTTTTAG | XhoI |
| parA-gfp_forward | CAGCTCGAGATGGAAGACACTACTTGGGAA | XhoI |
| parA_reverse | CAGCTGCAGATGGAAGACACTACTTGG | PstI |
| parA reverse db | GGGCCCTAATACGACTCACTATAGGGCTATTTCGCAGGTTTTAG | |
| pldP100_forward | CAGGAATTCGTCGAGAGCTGTAAAGTC | EcoRI |
| pldP_reverse | CAGGGTACCGCCGCCGTCGTTGACGCGGCT | KpnI |
| pldP-gfp_forward | CAGCTCGAGGTGAGTGATGCAGGGAAGAAG | XhoI |
| pldP_forward | CCCTGCAGGATGAGTGATGCAGGGAAG | SbfI |
| pldP_reverse db | GGGCCCTAATACGACTCACTATAGGGCTAGTCGTTGACGCGGCT | |
| parB_reverse de | GGGCCCTAATACGACTCACTATAGGGCAGTTATTGGCCCTGGATCAA | |
| parB_forward | CAGCATATGGCTCAGAACAAGGGTTCC | NdeI |
| parB_reverse | CAGCTCGAGTTATTGGCCCTGGATCAA | XhoI |
| Cfp-SacI-F | CGCGAGCTCATGGTGAGCAAGGGCGAG | SacI |
| Cfp-EcorI-R | GCGGAATTCTTACTTGTACAGCTCGTC | EcoRI |
| BeX-SalI-F | CAGGTCGACATGGCTCAGAACAAGGGT | SalI |
| BeX-BamHI-R | CAGGGATCCTTGGCCCTGGATCAAGGA | BamHI |
Homologous sequences for crossover PCR are indicated by boldfacing; restriction sites are indicated by underscoring.
Strain construction.
All plasmid cloning was carried out in E. coli DH5α. For the construction of in-frame deletion mutants, the nonreplicative pK19mobsacB vector was used, as described previously (38). Basically, a pK19mobsacB plasmid was constructed which contained homologous flanking sequences 500 bp upstream and downstream of the gene to be deleted. In order to construct a parA, pldP, or parB deletion plasmid, the upstream homologous flanking sequences were PCR amplified by using the primer pairs ΔParA-1-F/ΔParA-1-R, ΔPldP-1-F/ΔPldP-1-R, and ΔParB-1-F/ΔParB-1-R, respectively. The downstream homologous flanking fragments were PCR amplified by using the primer pairs ΔParA-2-F/ΔParA-2-R, ΔPldP-2-F/ΔPldP-2-R, and ΔParB-2-F/ΔParB-2-R, respectively. The fragments were purified and subjected to crossover PCR and amplified by using the primer pairs ΔParA-1-F/ΔParA-2-R, ΔPldP-1-F/ΔPldP-2-R, and ΔParB-1-F/ΔParB-2-R, respectively. The resulting fragments of 1,000 bp were HindIII/XbaI digested and ligated into the pK19mobsacB plasmid. These plasmids (pK19mobsacBΔparA, pK19mobsacBΔpldP, and pK19mobsacBΔparB) were transformed in C. glutamicum via electrotransformation. According to a method described before (38), deletion of the desired chromosomal genes occurs via a double-crossover event. Integration of the plasmid was selected for by kanamycin resistance. The second round of recombination was selected for by growth on 10% sucrose. The chromosomal deletions of parA (CDC001), pldP (CDC002), and parB (CDC003) in C. glutamicum were confirmed via PCR by using the primer pairs ParA-800-F/ParA-800-R, PldP-600-F/PldP-800-R, and ParB-800-F/ParB-800-R, respectively.
Strains expressing fusion proteins with cyan fluorescent protein (CFP) were constructed. To this end, strains CDC004 (ParA-CFP) and CDC005 (PldP-CFP) containing a single allele present in the chromosome with a 3′-cfp fusion were made. The B. subtilis pSG1186 vector, which contains two multiple cloning sites, one on either side of cfp, was used as a subcloning vector. Flanking sequences of at least 500 bp, complementary to the desired cfp integration site, were cloned into the appropriated multiple cloning site. The 3′ 500 bp of parA/pldP, excluding the stop codon, were PCR amplified by using the primer pair ParA-1-F/ParA-1 or PldP-1-F/PldP-1-R, respectively. Also, the first 500 bp downstream of parA/pldP was amplified by using the primer pair ParA-2-F/ParA-2-R or PldP-2-F/PldP-2-R, respectively. The resulting fragments were digested with HindIII/EcoRI and SpeI/XbaI, respectively, and subsequently ligated into the pSG1186 vector. The recombinant plasmid was transformed into DH5α. The entire fragment, consisting of the complementary flanking sequences and CFP, was then excised by restriction digest with HindIII/XbaI and subsequently ligated into the pK19mobsacB vector. Similar to the procedure described above, a two-step homologous recombination procedure was applied resulting in cfp integration into the chromosome at the 3′ end of the desired gene. Chromosomal integration of cfp fusion genes was confirmed by PCR.
A strain for expression of ParB with a C-terminal green fluorescent protein (GFP) fusion was constructed by integration of a second parB-gfp allele in the genome of C. glutamicum. To this end, the pk18mob-parB-gfp plasmid was constructed and transformed into C. glutamicum, resulting in strain CAS006. This strain contains a translational ParB-GFP copy under the control of the native promoter.
The vector pLAU44 was used for integrating a tetO cassette in the genome (19). A noncoding locus between the genes cg0002 and cg0004 was used for integration. In order to visualize the tetO arrays pEKEx2-yfp-tetR (11) was transformed into the mutant strain, giving strain CAS009.
Extrachromosomal, IPTG (isopropyl-β-d-thiogalactopyranoside)-inducible expression of ParB with a C-terminal CFP fusion was constructed by PCR amplification of parB, without the stop codon, and CFP using the primer pairs BeX-SalI-F/BeX-BamHI-R and Cfp-SacI-F/Cfp-EcorI-R, respectively. These two fragments were SalI/BamHI and SacI/EcoRI restriction digested and ligated in the pEKEx2 vector. The resulting plasmid was transformed into C. glutamicum wild type, strain CDC001, and CDC002, resulting in strains CDC007, CDC008, and CDC009, respectively.
ParB purification.
ParB was synthesized in E. coli BL21(DE3)/pLysS cells by using pET16B vector-based systems. Cultures were grown in 500 ml of LB medium at 37°C. At midexponential growth, protein expression was induced by adding 1 mM IPTG. After 3 h of induction, the cells were harvested and resuspended in buffer containing 50 mM Tris-HCl (pH 7.5), 100 mM NaCl, 5 mM MgCl2, 1 mM dithiothreitol (DTT), 10% (vol/vol) glycerol, and protease inhibitor. Subsequently, the cells were lysed by using a Fast-Prep homogenizer. The cell lysate was cleared by centrifugation for 30 min at 4°C and 20,000 × g. His-tagged proteins were purified in a batch procedure using Ni-NTA-agarose (Qiagen). Briefly, 0.5 ml of the Ni-NTA-agarose was mixed with 10 ml of the cleared lysate. After 30 min of incubation at 4°C, the batch was centrifuged for 4 min at 4°C and 800 × g. The agarose was then washed twice with washing buffer containing 50 mM Tris-HCl (pH 7.5), 100 mM NaCl, 5 mM MgCl2, and 1 mM DTT supplemented with 15 mM imidazole for the first washing step and 30 mM imidazole for the second washing step. After every step, the samples were centrifuged for 4 min at 4°C and 800 × g, and the supernatant was discarded. The imidazole concentration was stepwise increased up to 0.5 M to elute the proteins. The purified proteins were verified by mass spectrometry (Center of Molecular Medicine, University of Cologne).
Electrophoretic mobility shift assay.
DNA-binding experiments were performed with affinity-purified ParB. A double-stranded oligonucleotide containing the consensus parS site was generated by hybridization of two complementary oligonucleotides with the sequences: 5′-AGA ATG TTC CAC GTG AAA CAA AGA-3′ and 5′-TCT TTG TTT CAC GTG GAA CAT TCT-3′. Portions (30 to 50 μg) of each protein were incubated for 10 min at 30°C with 1 μg of plasmid or DNA fragment. These samples were separated in a 1% agarose gel. A denatured sample of ParB (denatured by boiling) was used as a negative control.
Fluorescence microscopy.
Constructed mutants that express xFP fusion proteins were analyzed by fluorescence microscopy. To this end, 3 μl of cell culture was mounted on agarose-coated slides (1.5% agarose). DNA was stained with Hoechst (1 μg ml−1; Sigma-Aldrich), and membranes were stained with Nile Red (12.5 μg ml−1; Molecular Probes).
Pictures were taken on an Axio-Imager M1 fluorescence microscope (Carl Zeiss), and the images were analyzed with AxioVision version 4.6 software. Final image preparation was done in Adobe Photoshop 6.0 (Adobe Systems). Cell length measurements were carried out using AxioVision software (Carl Zeiss).
Bacterial two-hybrid and β-galactosidase-assay.
Protein interactions were analyzed by a LexA-based bacterial two-hybrid analysis (6). Therefore, the genes parA, pldP, parB, and ftsZ were cloned into the plasmids pMS604 and pDP804, which both contain a LexA binding domain. The resulting plasmids were transformed, in different combinations, in the E. coli strains SU101 and SU202, and expression of the fusions was induced with IPTG. For this purpose, 5 ml of LB culture supplemented with appropriate antibiotics and 1 mM IPTG were inoculated (OD of 0.01) with an overnight culture also containing 1 mM IPTG. After reaching an OD of 0.5 to 0.6, the OD was measured, and 20 μl of culture was mixed with 80 μl of permeabilization buffer (100 mM Na2HPO4, 20 mM KCl, 2 mM MgSO4, 0.8 mg of CTAB [cetyltrimethylammonium bromide] ml−1, 0.4 mg of deoxycholic acid [sodium salt] ml−1, 5.4 μl of β-mercaptoethanol ml−1), followed by incubation for 30 min at 30°C. Afterward, 600 μl of prewarmed substrate solution (30°C) (60 mM Na2HPO4, 40 mM NaH2PO4, 1 mg of o-nitrophenyl-β-d-galactopyranoside [ONPG] ml−1, 2 μl of β-mercaptoethanol ml−1) were added, while the time of adding was noted. When the samples turned yellow, the reaction was stopped by adding 700 μl of stop solution (1 M Na2CO3), and the time was noted as well. Subsequently, the samples were centrifuged (10 min, maximum speed), and the supernatant was measured photometrically at 420 nm. Water was used as a blank. Miller units were calculated according to the following equation: 1,000 × [OD420/(OD600 × volume [0.02 ml] × reaction time in min)].
RESULTS
Genetic modifications in the parAB operon influence cell division and growth.
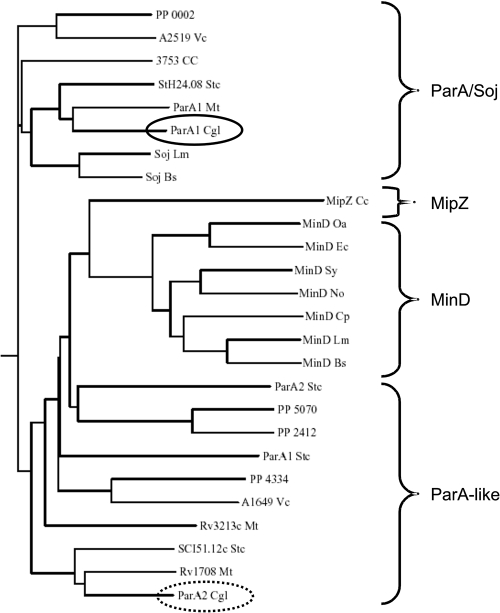
Although C. glutamicum is of major significance in biotechnology and a model organism for mycolic acid containing pathogens, the cell biology aspects of this bacterium have not been studied in sufficient detail. Corynebacteria are often mentioned in the literature for their lack of actinlike cytoskeletal structures and the lack of a Min system (21). In fact electron microscopy images of corynebacteria suggest that placement of septa is not always precisely at midcell (31) in contrast to other rod-shaped organisms, such as B. subtilis or E. coli. Therefore, we wanted to find out by which mechanism C. glutamicum regulates division site selection. In order to understand this fundamental cell biological process in Corynebacterium, we started an investigation of the chromosome partitioning system. We noticed that C. glutamicum possesses a parAB operon (cg3427 and cg3426) and an orphan parA-like gene (cg1610), which we now rename pldP (PldP) for ParA-like division protein for reasons that are made clear below. We have focused on this system because of the obvious evolutionary connection between ParA and MinD proteins on the one hand and ParB and the nucleoid occlusion protein Noc on the other (Fig. 1).
FIG. 1.
Phylogenetic relationship of MinD and ParA proteins. ParA and MinD proteins share an evolutionary relationship. Many organisms code for more than one ParA protein or have MinD and ParA proteins encoded. However, MinD proteins clearly cluster together, whereas bona fide ParA/Soj proteins cluster in one group as well. Many parA-like genes are closely related to the MinD proteins. At least for the MipZ protein from Caulobacter crescentus (MipZ Cc) a role in division site selection has been shown. The dendrogram was calculated by using a CLUSTAL W alignment based on the amino acid sequences of various ParA and MinD proteins. Sequences were derived from the genome information broker (http://gib.genes.nig.ac.jp). Proteins that were not annotated are shown with their gene codes. Note that the two ParA proteins encoded by C. glutamicum cluster differently. ParA1 (solid ellipse) clusters with the ParA/Soj proteins, whereas ParA2 (broken ellipse; based on the results obtained here, we renamed the protein PldP [see the text]) clusters closer to the MinD-like proteins. Strain abbreviations: PP, Pseudomonas putida KT2440; VC, Vibrio cholerae O395; Stc, Streptomyces coelicolor A3(2); Mt, Mycobacterium tuberculosis H37Rv; Cgl, Corynebacterium glutamicum ATCC 13032; Lm, Listeria monocytogenes 4b F2365; Bs, Bacillus subtilis 168; Cc, Caulobacter crescentus CB15; Oa, Ochrobactrum anthropi ATCC 49188; Ec, Escherichia coli K-12; Sy, Synechocystis sp. strain PCC 6803; Cp, Clostridium perfringens 13; No, Nostoc sp. strain PCC 7120.
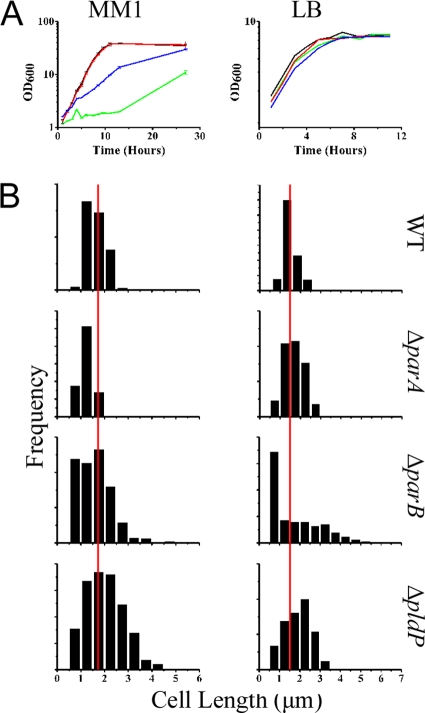
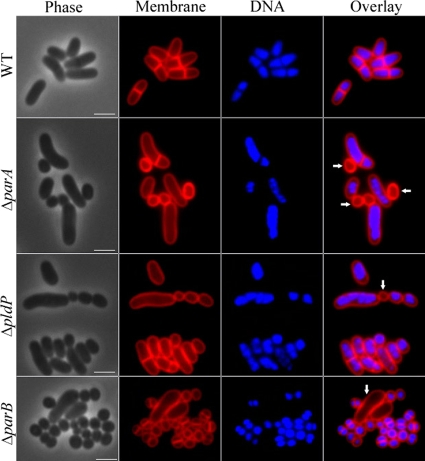
The parAB operon is located in the vicinity of the origin of replication (ori) between 99.85 and 99.88 min on the C. glutamicum chromosome. The pldP gene (cg1610) is located almost on the opposite side of the chromosomal map (45.7 min), without a parB-like gene in its vicinity. In order to study the molecular role of the Par system in Corynebacterium, we first tried to knock out its different components. We obtained markerless null mutations of parB, parA, and pldP. The clean deletions of the different genes were checked by PCR (data not shown). We started analyzing the phenotypic consequences of the different mutations in growth experiments. Growth in minimal medium supplemented with glucose revealed that loss of ParB and ParA has a drastic influence on the growth rates (Fig. 2A). In contrast, the loss of PldP does not influence growth in comparison to the wild-type strain. Strikingly, the differences in growth were almost absent when cells were cultured in medium that only allows slow growth rates for C. glutamicum, which is the case for LB medium (Fig. 2A). A plausible explanation for this effect comes from a comparison of the doubling times that were measured in the different media. Wild-type cells have a doubling time of around 97 min in the MM1 medium, whereas they had a doubling time of 151.8 min in LB medium. Hence, the reduced growth rate partially compensates for the loss of ParB and ParA. This indicates a defective chromosome replication initiation or chromosome segregation. Both effects can be tolerated when growth rates are reduced (4). Next, we analyzed the cell length distribution of the different strains in both media. Therefore, we stained the cell membranes with the lipophilic dye Nile Red before microscopic analysis. The histograms with the cell length distributions are shown in Fig. 2B. We observed striking differences in the cell length distribution. Table 4 summarizes the mean cell lengths and the numbers of anucleate cells. From these data it becomes immediately obvious that there is a clear difference between the parA and the pldP mutant. Although the pldP mutant strain has only a moderate increase in the amount of anucleate cells, cells of the parA mutant strain have a high degree of DNA-free cells. Strikingly, the amount of anucleate cells is even higher in a parB deletion strain. In particular, when cells are grown in MM1 medium (the medium that supports faster growth compared to LB medium) a large amount of the parB deletion cells were found to be DNA-free. Microscopic images of the different strains are shown in Fig. 3. Loss of ParA leads to a large variation in cell length (DNA-free cells were not counted). The cells range from very small to slightly elongated. An overall similar cell length distribution can be seen in cells lacking PldP; however, here only a few minicells (or anucleate cells) were observed (Fig. 3 and Table 4). The most drastic phenotype was observed in strains lacking ParB. Usually, the cells were almost coccoid; however, often elongated, DNA-free cells were observed.
FIG. 2.
Growth phenotypes of parAB and pldP mutants. (A) Growth curves of wild-type (WT) C. glutamicum (black line) and ΔparA (CDC001; green line), ΔparB (CDC003; blue line), and ΔpldP (CDC002; red line) mutants in MM1 with 4% glucose (left side) and LB medium (right side). Values are the means of three growth experiments. The standard deviations are shown. Growth was at 30°C under constant shaking. (B) Histograms showing the distribution of cell lengths of wild-type, ΔparA, ΔparB, and ΔpldP strains in MM1 medium (left panel) and LB medium (right panel). The red lines indicate the mean cell length of wild-type cells. Cell length was measured using Nile Red as a membrane stain.
TABLE 4.
Cell length and percent anucleate cells in C. glutamicum par mutantsa
| Strain | MM1 |
LB |
||
|---|---|---|---|---|
| Avg cell length (μm) | % Anucleate cells | Avg cell length (μm) | % Anucleate cells | |
| Wild type | 1.63 ± 0.38 | 0 | 1.5 ± 0.36 | 0 |
| ΔparA (CDC001) | 1.24 ± 0.28 | 17.75 | 1.69 ± 0.5 | 16.28 |
| ΔparB (CDC002) | 1.6 ± 0.72 | 43.8 | 1.7 ± 1.19 | 11.6 |
| ΔpldP (CDC003) | 2.02 ± 0.78 | 1.54 | 1.87 ± 0.62 | 0.85 |
Cells were grown in MM1 or LB medium at 30°C. Cell length was measured by using Nile Red as a membrane stain, and values are expressed as mean average lengths ± the standard deviation. DNA was stained with Hoechst dye. For each strain, more than 200 cells were counted (n > 200).
FIG. 3.
Phenotypes of C. glutamicum parAB and pldP mutants. Shown are phase-contrast images (Phase), membrane stain with Nile Red (Membrane), DNA stain with Hoechst dye (DNA), and a merge between membrane and DNA stain (Overlay). From top to bottom, wild-type (WT), ΔparA (CDC001), ΔpldP (CDC002), and ΔparB (CDC003) C. glutamicum cells are shown. Anucleate cells are indicated with arrows. Cells were grown in MM1 medium with glucose at 30°C under constant shaking. Scale bars, 2 μm.
Overexpression of ParA or PldP from inducible plasmids, however, resulted in an increase in cell length. We constructed two strains that express ParA and PldP from plasmids, under the control of inducible promoters, respectively. Strain CAS010(pEKEX2-ParA) had an increased cell length of 3.4 μm ± 1.05 upon induction with IPTG in comparison to 2.1 μm ± 0.32 for the uninduced strain (data not shown). Similarly, overexpression of PldP (strain CAS011(pEKEX2-PldP) led to an increase in cell length (3.41 μm ± 0.79) when the strain was induced with IPTG (data not shown).
Based on the different phenotype with respect to anucleate cells and the different growth phenotype, we can clearly differentiate between ParA and PldP, arguing that these two proteins, although they share sequence similarity (37.5% identity, 56.1% similarity), have different cellular roles.
In vivo localization of ParA and PldP.
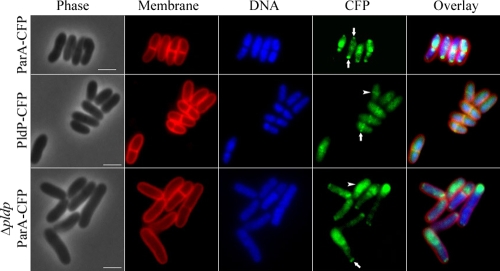
In order to further differentiate between ParA and PldP, we constructed strains where the native alleles were replaced by cfp fusion genes, which place the expression of the CFP fusion proteins under the control of the native promoters (see Materials and Methods). Thus, the ParA and PldP fusion constructs are expressed as single copies under the control of their natural promoters, respectively. Subcellular localization of ParA-CFP revealed that the protein often accumulates in foci close to the cell poles (Fig. 4, upper panel, arrows). Between these polar foci, we often observed larger patches of ParA-CFP that colocalizes with the nucleoid (although it has to be taken into account that the DNA occupies most of the cytoplasm). In striking contrast, PldP-CFP was localized to the site of septation. Although, we saw a relatively high background, a clear localization pattern of PldP was observed. In 45% of all cells counted (n > 200) we observed a midcell localization of PldP. In roughly 50% of those cells where PldP localized in a bandlike structure at midcell, we did not observe membrane invagination, suggesting that PldP localization precedes septation. The fact that we did not observe PldP localization in all cells is probably hampered by the background and the weak CFP fluorescence intensity. Interestingly, PldP seems to localize relatively early to the site of septation, because bands of PldP-CFP fluorescence were not only observed on closed septa but also before visible membrane stains marked inward growing septa and chromosomes are visibly segregated (Fig. 4, middle panel, arrowhead). Interestingly, the PldP distribution is reminiscent of that of division site selection proteins such as MinC or MinD in rod-shaped bacteria such as B. subtilis (14, 27). We also visualized ParA-CFP distribution in a pldP deletion background. In these slightly elongated cells (compare with Table 4) ParA-CFP still forms foci at the cell poles; however, the patches across the nucleoid were larger and longer.
FIG. 4.
Subcellular localization of ParA and PldP in C. glutamicum. Subcellular localization of ParA and PldP was analyzed using strains where the native alleles have been replaced by cfp fusion genes (see Materials and Methods). Shown are phase-contrast images (Phase), membrane stain (Membrane), DNA stain with Hoechst dye (DNA), CFP fluorescence (CFP, false colored in green), and a merge image of membrane stain, DNA stain, and CFP fluorescence (Overlay). ParA-CFP localization is shown in the upper panel. Characteristic polar foci of ParA-CF are indicated by arrows in the CFP channel. PldP localization in is shown in the middle panel. The arrow points to midcell localization of PldP. The lower panel shows the localization of ParA-CFP in the absence of PldP. These cells are often elongated. Still, ParA-CFP is localized in to polar foci and larger patches which often concentrate at the opposite pole. Scale bars, 2 μm.
Dynamic in vivo localization of parB.
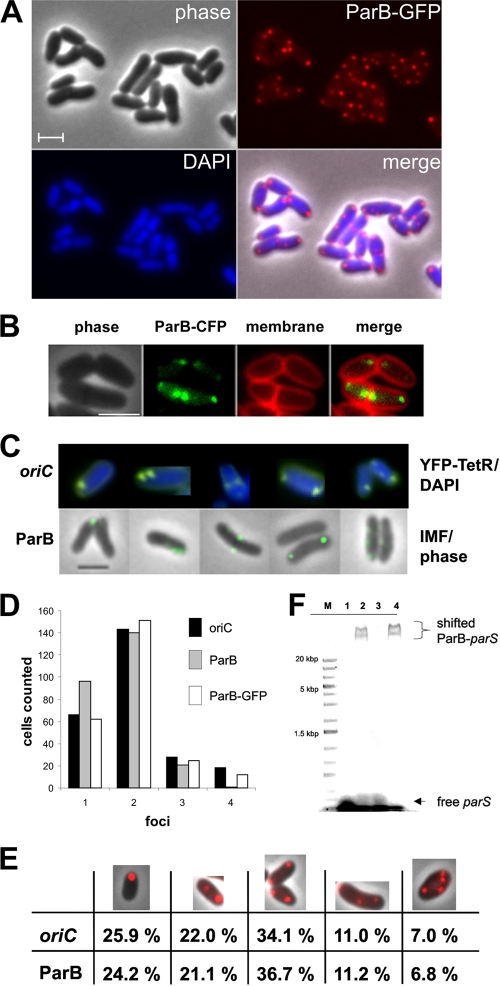
The observed foci of polar ParA prompted us to investigate the localization of ParB. ParB proteins usually form nucleoprotein complexes with parS sites (23; see also below) that are clustered around the origin region. First, we constructed a strain, CAS006, that encodes a parB-gfp allele which is under the control of the native promoter (see Materials and Methods). In strain CAS006, the ParB-GFP fusion is the only expressed version of parB since the integration of the plasmid leaves a promoterless parB allele as a second copy. Since strain CAS006 had no obvious growth or cell length phenotype (data not shown), we conclude that the ParB-GFP fusion may have retained at least partial functionality in vivo. Subcellular localization of ParB-GFP was analyzed in strain CAS006. ParB-GFP foci were found to be located mainly close to the cell poles (Fig. 5A and B). Careful observation of the foci revealed that cells contained up to four foci. Statistical analysis (Fig. 5D and E) showed that most cells contain either one or two foci. However, these foci were always localized close to the cell poles. If a third or fourth ParB focus was present, these were located more toward the midcell. This observation might reflect ParB foci that travel from the cell poles toward a site of cell division.
FIG. 5.
Polar localization of the origin and ParB in C. glutamicum. (A) Localization of ParB-GFP. Typical cells of strain CAS006 (ParB-GFP) are shown in gallery view with phase-contrast, GFP fluorescence (red), DAPI stain (blue), and merge images. (B) Localization of ParB-CFP (false colored in green) in strain CDC007. Cells were induced with 1 mM IPTG. Membrane stain was performed with Nile Red. (C) Polar localization of the oriC and ParB. Strain CAS009 expressing YFP-TetR grown in MM1 medium was analyzed microscopically. A compilation of cells showing YFP-TetR foci is shown in the upper panel. DAPI staining is depicted in blue, and YFP fluorescence is shown in yellow (ori). The lower panel (ParB) shows wild-type cells after immunofluorescence staining with polyclonal antibodies against ParB. A fluorescein-conjugated secondary antibody was used to visualize ParB localization (see Materials and Methods). Cells showing a similar arrangement of foci compared to the YFP-TetR foci in the upper panel were aligned with these. Scale bars, 2 μm. (D) Relative distribution of YFP-TetR foci (ori, n = 255), ParB (counted from IMF, n = 258), and ParB-GFP (n = 241) reveals that similar numbers of foci were detected. Notably, no more than four foci per cell were observed. (E) Percentage comparison between the localization of the oriC and ParB (counted from ParB-GFP). The categories are symbolized by the microscopic images of ParB-GFP expressing cells in the top row. Categories were (from left to right) one polar focus, two foci, with one polar focus and one focus somewhere in the cytoplasm, foci at both poles, three foci, and four foci. Note the similar percentages for statistics done with oriC and ParB-GFP. (F) Recombinant ParB protein binds to parS consensus sequences. A 24-bp oligonucleotide containing the consensus parS sequence (see the text) was mixed with ParB. Lanes were loaded as follows: M, molecular mass standard; 1, parS DNA; 2, parS plus 20 μg of ParB; 3, parS plus 50 μg of denatured ParB; 4, parS plus 50 μg of ParB.
In order to provide an independent proof that the localization of ParB-GFP faithfully reflects the in vivo situation, immunofluorescence microscopy (IMF) using polyclonal antibodies against ParB was performed. Fluorescent foci of ParB visualized with fluorescein-conjugated secondary antibodies were identical to the localization pattern of the origin region, a finding consistent with ParB binding to origin proximal parS sites (Fig. 5C, lower panel). Quantitative comparison between the numbers of ParB-GFP foci and ParB foci revealed by IMF were almost identical (Fig. 5C). Hence, the results obtained with the ParB-GFP constructs and IMF all point to a polar recruitment of the origin in C. glutamicum cells. The ParB foci that were found to be at the cell pole remained stable there during time lapse experiments (only new foci were migrating), suggesting that ParB might be stably attached to a structure that is localized to the cell poles.
We also constructed a ParB-CFP expressing strain (CDC007) that carries an IPTG-inducible copy of parB-cfp on a pEKEX plasmid. Similar to the genomic integration of parB-gfp, cell length or growth was not altered significantly upon induction of strain CDC007. Microscopic analysis revealed that the localization of ParB-CFP was identical to the ParB-GFP localization in strain CAS006 (Fig. 5B).
In vivo localization of the replication origin.
ParB is known to bind to a cis-acting parS sequence on the chromosome (23). Since parS sites cluster around the origin, we aimed to analyze the subcellular localization of the oriC. To this end a tetO array was integrated close to the ori region within an intergenic space between genes cg0002 and cg0004. An array of approximately 240 tetO operator regions of transposon Tn10 (tetO array) (19) was inserted using plasmid pLAU44-integ ori (see Materials and Methods). The resulting strain was transformed with plasmid pEKEx2-yfp-tetR (11), which encodes a yfp-tetR gene under the control of a Ptac promoter, leading to strain CAS009. After mild induction of the yfp-tetR construct, fluorescent foci were readily observed in cells of strain CAS009. Cells with up to four foci were identified, with most of the cells having either one or two foci close to the cell poles (Fig. 5C). Quantitative analysis of the subcellular distribution (Fig. 5D and E) revealed that the YFP-TetR foci were usually close to the cell poles with either one focus at a cell pole (25.9%) or with two foci at the opposite poles (34.1%). If three or four YFP-TetR foci were observed, two foci were usually close to the poles while the remaining ones were localized between pole and midcell. The localization of oriC was found to be identical with that of ParB. Thus, although we did not succeed in performing direct colocalization studies, it is highly probable that ParB and the origin colocalize to the cell poles in C. glutamicum (Fig. 5E).
The ParB protein binds to parS consensus sequences in vitro.
Since we used the localization of ParB as a marker for origin placement in vivo, we wanted to confirm that C. glutamicum ParB binds to parS sites. The parS sites are short, 16-bp direct repeats that are clustered around the origin of replication and are found in a large variety of bacteria (12). For Gram-positive bacteria a consensus sequence of a parS site has been formulated (5′-TGTTNCACGTGAAACA-3′). C. glutamicum harbors three putative parS sites around the origin region (a perfect consensus sequence is found in the trpCF locus at 97.8 min). To test whether ParB binds to the consensus parS site in vitro, we purified an N-terminal His10-tagged ParB. The specific binding activity of recombinant ParB was assayed with an electrophoretic mobility shift assay. ParB was added to a previously described 24-bp DNA fragment that contains the consensus parS sequence (33). This resulted in a shifted nucleoprotein complex that appeared to be a double band of high molecular weight (Fig. 5F). The specificity of the ParB binding was checked by shift assays with different DNA fragments. None of the fragments that lacked a consensus parS site was shifted upon addition of ParB (data not shown) supporting the notion that ParB binds specifically to a consensus parS site. In order to exclude unspecific binding of protein on the parS site we also denatured the ParB protein before incubation with parS DNA. Denatured ParB was not able to shift the parS DNA fragment (Fig. 5F). Thus, C. glutamicum ParB is able to bind specifically to a consensus parS site in vitro. This result is again in good agreement with the notion that ParB binds to the origin region in vivo.
ParA but not PldP are necessary for polar ParB localization.
Next, we wanted to know whether ParA or PldP might influence localization of ParB foci. The rational behind this is the idea that, similar to the situation described for chromosome I in V. cholerae (10), ParA might help to segregate the nucleoids by binding to the ParB-oriC nucleoprotein complex. ParA might segregate either through pulling or pushing the ParB-oriC complex to the opposite pole. Therefore, we used the ParB-CFP expressing strain (CDC007) and counted the foci. We discriminated between polar, midcell or cell quarter positions (compare with Table 5). The average number of foci was 2.2 with more than 80% of the foci being polar. In striking contrast, in a parA deletion background, the number of polar ParB-CFP foci decreased to 28% (Table 5). Simultaneously, the number of ParB foci found at the midcell or cell quarter positions increased. Also, the total number of ParB foci was elevated. The loss of PldP, however, had only a mild effect on ParB-CFP positioning (Table 5). Again, we observed a clear difference between ParA and the ParA-like protein PldP. These data suggest that ParA helps to segregate the ParB-oriC complex to the opposite poles.
TABLE 5.
Influence of ParA or PldP deletion on ParB localization in vivoa
| Strain | Mean cell length (μm) ± SD | % ParB-CFP localization (n ≤ 170) |
Avg no. of ParB foci/cell | ||
|---|---|---|---|---|---|
| Polar | Midcell | Cell quarters | |||
| ParB-CFP (CDC007) | 2.15 ± 0.5 | 81.90 | 9 | 9.1 | 2.21 |
| ParB-CFP ΔparA (CDC008) | 3.2 ± 0.9 | 27.6 | 23.6 | 48.8 | 3.22 |
| ParB-CFP ΔpldP (CDC009) | 3.19 ± 0.9 | 67.6 | 11.6 | 20.8 | 2.68 |
Cells were grown in LB medium at 30°C. Cell length was measured using Nile Red as a membrane stain. DNA was stained with Hoechst dye. Polar localization of at least one focus was counted as “polar.”
Protein-protein interaction map of the partitioning proteins from C. glutamicum.
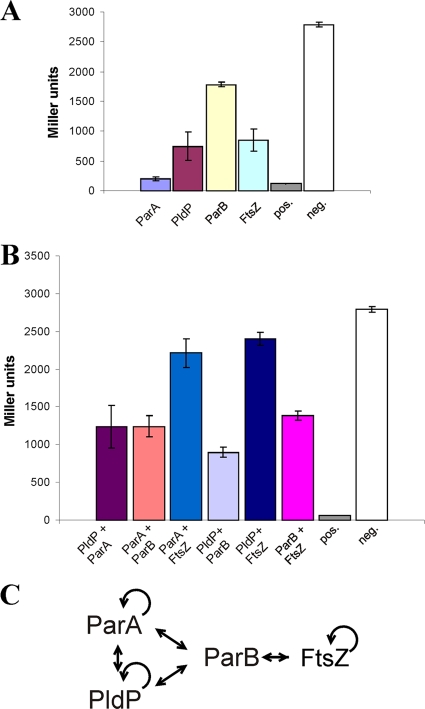
We aimed to analyze the interaction of ParA and PldP with ParB by applying a bacterial two hybrid (B2H) assay. Furthermore, we wanted to test the interaction of the Par proteins with FtsZ, because PldP, and sometimes ParB were found to localize at midcell positions (PldP, Fig. 4; ParB, Fig. 5B and C). In view of the fact that all Par proteins and FtsZ are soluble proteins we decided to use the LexA based B2H assay (5, 6). First, homo-oligomerization of ParA ATPases, ParB, and FtsZ was determined (Fig. 6A). Control experiments revealed that very tight interaction (Fos/Jun) results in almost complete repression of the lacZ gene. Consequently, only 116 Miller units were measured in the positive control, while a negative control resulted in 2,700 Miller units. ParA proteins exhibited the strongest self-interaction (200 Miller units). PldP and FtsZ showed significant self interaction (750 and 850 Miller units, respectively), as judged by their β-galactosidase activities. ParB, however, did not show drastically reduced activity (1,782 Miller units), suggesting that the self-interaction is, if at all, weak. Given the fact that ParB needs a specific parS site for self assembly, this might not be surprising. Next, we analyzed pairwise interactions. The results of the different protein pairs that were tested are summarized in Fig. 6B. Again, a Fos/Jun positive control indicated the strongest protein-interaction that can be achieved in vivo (57 Miller units), whereas the negative control shows the maximum level of β-galactosidase activity that could be reached (2,791 Miller units, indicating full expression). Most noteworthy are the direct interactions between ParA and PldP with ParB. Interestingly, we also observed interactions between ParA and PldP.
FIG. 6.
Protein-protein interactions between ParAB, PldP, and FtsZ. A bacterial two-hybrid approach (see Materials and Methods) was performed to study protein-protein interaction of the Par proteins. Homodimerization of was tested in panel A. Heterologous interaction between different protein pairs was studied in panel B. The positive control was interaction of Fos/Jun and negative control was expression of the plasmids pDB804 and pMS604-ftsZ. Standard deviations are derived from three independent experiments. (C) Based on the two-hybrid data, an interaction map was build that shows the interaction between the Par proteins and FtsZ.
No significant interaction of ParA or PldP with FtsZ was observed (2,212 and 2,403 Miller units, respectively). Based on the localization data, in particular, PldP could have been a direct recruitment of FtsZ, which, based on our results, seems not the case. A summary of the observed interactions is shown in Fig. 6C.
DISCUSSION
The rod-shaped actinomycete C. glutamicum has a characteristic cell morphology and cell division. A striking feature is that fast-growing cells often divide into unequal-sized daughter cells, which is in strong contrast to other model organisms such as E. coli or B. subtilis. We set out to investigate how these bacteria ensure chromosome segregation and cell division. In a first attempt, we analyzed subcellular localization and function of the partitioning (par) proteins in C. glutamicum.
Polar localization of origin regions in C. glutamicum.
ParB proteins bind to inverted repeats on the DNA, called parS sites (23). Usually, parS sites cluster around the origin region in the chromosomes. Phylogenetic analysis has shown that parS sites occur in a large number of bacteria and share a conserved consensus sequence (24). We identified three parS sites in the C. glutamicum chromosome, which is in agreement with the proposed two to four parS sites described by Livny et al. (24). The three sites are located around the ori region. In vitro assays have shown that C. glutamicum ParB is able to bind selectively to the parS sites that contain the consensus sequence for parS sites in Gram-positive bacteria (Fig. 5F). The selectivity of this binding supports the idea that subcellular localization of ParB faithfully reflects the position of the origin within the cell (compare Fig. 5A to C). ParB foci were found to be polar in C. glutamicum (Fig. 5). We corroborated the polar positioning of the origin in C. glutamicum by inserting a tetO array close to the origin (Fig. 5C). Similar to other bacteria that show polar origin localization (e.g., C. crescentus, Vibrio cholerae chromosome I), the ParB foci close to the cell poles are static. This is most likely to be achieved by anchoring of the ParB-parS complex to the pole by protein-protein interaction. An interesting candidate for such a polar anchoring could be the DivIVA protein in C. glutamicum. DivIVA is essential in C. glutamicum and is involved in recruitment of the polar peptidoglycan synthesis machinery (22; A. Schwaiger and M. Bramkamp, unpublished results). In B. subtilis DivIVA anchors the origin via RacA during sporulation (43), showing that DivIVA could function as a chromosome-orienting protein. In Caulobacter crescentus a coiled-coil protein, PopZ, was shown to be essential for polar localization of the origins (2, 7). Interestingly, the DivIVA sequence of the Corynebacterium protein has a large central insertion that is absent in other DivIVA proteins (e.g., in B. subtilis DivIVA). However, a direct interaction between DivIVA and Par proteins has not yet been observed.
Interaction pattern of the Par proteins.
A striking observation with the ParB-xFP-expressing strains was that ParB foci were found at midcell in dividing C. glutamicum cells until division was completed. It thus seemed likely that ParB might bind to the cytokinetic ring. We tested interaction of ParB and FtsZ in a LexA-based bacterial two hybrid system and found that the two proteins interact. In growing cells, it seems logical that the ParB-parS nucleoprotein complex is directed to the active cytokinetic ring, because at this site the new cell pole will shape. Our two-hybrid data also reveal that both ParA-like proteins, ParA and PldP, are interacting with ParB. Although interaction between ParA and ParB, which are encoded in an operon, is not unexpected, the strong interaction between PldP and ParB resembles the situation in C. crescentus, where the ParA-like protein MipZ is also an interaction partner of ParB (42). However, the different subcellular localization of PldP and MipZ suggests that division site selection in C. glutamicum differs from that in C. crescentus. Future experiments need to address the question whether PldP itself acts as an inhibitory factor for division (which is indicated by the increase in cell length after PldP overexpression and the minicell phenotype) or whether functional homologues of MinC might exist in corynebacteria.
Function of ParA and PldP in C. glutamicum.
A remarkable feature of Corynebacterium is its fast growth to unusual high cell densities (OD values of up to 30 to 40 are normal). This fast growth must be mirrored by a fast and efficient chromosome segregation and cell division system. Careful inspection of dividing C. glutamicum cells revealed that the daughter cells are not necessarily of equal size, a fact that is highly unusual for a rod-shaped organism. Most rod-shaped bacteria use a dual system to ensure precise division at midcell. The MinCD system prevents aberrant division at the cell poles, and the nucleoid occlusion system prevents fatal division across the nucleoids (3, 25, 26). Corynebacteria lack MinCD proteins, and a nucleoid occlusion system has not yet been described. However, the B. subtilis nucleoid occlusion protein Noc is homologous to ParB proteins (43), and MinD is evolutionarily related to ParA proteins (28) (Fig. 1). Since C. glutamicum possesses a classical parAB operon and an orphan parA-like gene (termed pldP), we reasoned that ParA and PldP might have distinct functions in C. glutamicum.
Our results with C. glutamicum strains lacking parA, parB, or pldP indicate that each null mutation is connected with a distinct phenotype. Similar phenotypes have been described for the unrelated alphaproteobacterium C. crescentus. Depletion of ParB or overexpression of ParA in C. crescentus leads to formation of filamentous cells (29). Strikingly, the localization of the ori region is also polar in C. crescentus (30). The coupling of chromosome segregation and cell division in Caulobacter has recently been shown to depend on a ParA-like protein, designated MipZ (42). Like canonical ParA proteins, MipZ binds to the ParB-parS complex and spreads from there across the nucleoid. This generates a concentration gradient toward midcell. MipZ was shown to directly inhibit FtsZ polymerization in vitro (42). However, C. glutamicum PldP reveals a subcellular localization that is more reminiscent of the MinD localization in other rod-shaped cells rather than that of MipZ. The most convincing indication for our hypothesis is that PldP acts as a division site selection protein is the observation of minicells in the null mutation and the slightly elongated cell length. Both features are also found in MinD (or MinC) deletions in E. coli and B. subtilis. It should be noted that this report is the first to describe a minicell phenotype in a Corynebacterium strain. Mutations that affect chromosome segregation but are not involved in division site selection give rise to a severe cell growth defect (compare growth curves in Fig. 2) and produce anucleate cells. Minicells, however, are produced by aberrant division close to a cell pole and hence are rather distinct structures. Mutations in parA differ from pldP mutants in growth rate and the amount of anucleate cells. Furthermore, subcellular localization of ParA is similar to other chromosome partitioning ATPases, such as ParA from V. cholerae, whereas PldP localizes similarly to MinD (or MinC) in E. coli or B. subtilis. However, a more detailed biochemical and genetic analysis is needed to understand the exact molecular role of ParA and PldP. If PldP is part of the division site selection system, it needs to be tested whether the effect on septum placement is performed directly by PldP (for example, by having a direct effect on FtsZ) or whether a partner protein (similar to the MinCD system) remains to be discovered.
Acknowledgments
We thank Anja Wittmann for excellent technical assistance and Suey van Baarle for critical reading of the manuscript. All lab members are acknowledged for constructive criticism.
Work in our lab is supported by grants from the Deutsche Forschungsgemeinschaft.
Footnotes
Published ahead of print on 30 April 2010.
REFERENCES
- 1.Adams, D. W., and J. Errington. 2009. Bacterial cell division: assembly, maintenance, and disassembly of the Z ring. Nat. Rev. Microbiol. 7:642-653. [DOI] [PubMed] [Google Scholar]
- 2.Bowman, G. R., L. R. Comolli, J. Zhu, M. Eckart, M. Koenig, K. H. Downing, W. E. Moerner, T. Earnest, and L. Shapiro. 2008. A polymeric protein anchors the chromosomal origin/ParB complex at a bacterial cell pole. Cell 134:945-955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bramkamp, M., and S. van Baarle. 2009. Division site selection in rod-shaped bacteria. Curr. Opin. Microbiol. 12:683-688. [DOI] [PubMed] [Google Scholar]
- 4.Britton, R. A., D. C. Lin, and A. D. Grossman. 1998. Characterization of a prokaryotic SMC protein involved in chromosome partitioning. Genes Dev. 12:1254-1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Daines, D. A., M. Granger-Schnarr, M. Dimitrova, and R. P. Silver. 2002. Use of LexA-based system to identify protein-protein interactions in vivo. Methods Enzymol. 358:153-161. [DOI] [PubMed] [Google Scholar]
- 6.Dmitrova, M., G. Younes-Cauet, P. Oertel-Buchheit, D. Porte, M. Schnarr, and M. Granger-Schnarr. 1998. A new LexA-based genetic system for monitoring and analyzing protein heterodimerization in Escherichia coli. Mol. Gen. Genet. 257:205-212. [DOI] [PubMed] [Google Scholar]
- 7.Ebersbach, G., A. Briegel, G. J. Jensen, and C. Jacobs-Wagner. 2008. A self-associating protein critical for chromosome attachment, division, and polar organization in Caulobacter. Cell 134:956-968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ebersbach, G., S. Ringgaard, J. Moller-Jensen, Q. Wang, D. J. Sherratt, and K. Gerdes. 2006. Regular cellular distribution of plasmids by oscillating and filament-forming ParA ATPase of plasmid pB171. Mol. Microbiol. 61:1428-1442. [DOI] [PubMed] [Google Scholar]
- 9.Feucht, A., and P. J. Lewis. 2001. Improved plasmid vectors for the production of multiple fluorescent protein fusions in Bacillus subtilis. Gene 264:289-297. [DOI] [PubMed] [Google Scholar]
- 10.Fogel, M. A., and M. K. Waldor. 2006. A dynamic, mitotic-like mechanism for bacterial chromosome segregation. Genes Dev. 20:3269-3282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Frunzke, J., M. Bramkamp, J. E. Schweitzer, and M. Bott. 2008. Population Heterogeneity in Corynebacterium glutamicum ATCC 13032 caused by prophage CGP3. J. Bacteriol. 190:5111-5119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gerdes, K., J. Moller-Jensen, and R. Bugge Jensen. 2000. Plasmid and chromosome partitioning: surprises from phylogeny. Mol. Microbiol. 37:455-466. [DOI] [PubMed] [Google Scholar]
- 13.Goehring, N. W., and J. Beckwith. 2005. Diverse paths to midcell: assembly of the bacterial cell division machinery. Curr. Biol. 15:R514-R526. [DOI] [PubMed] [Google Scholar]
- 14.Gregory, J. A., E. C. Becker, and K. Pogliano. 2008. Bacillus subtilis MinC destabilizes FtsZ-rings at new cell poles and contributes to the timing of cell division. Genes Dev. 22:3475-3488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gruber, S., and J. Errington. 2009. Recruitment of condensin to replication origin regions by ParB/SpoOJ promotes chromosome segregation in Bacillus subtilis. Cell 137:685-696. [DOI] [PubMed] [Google Scholar]
- 16.Harry, E., L. Monahan, and L. Thompson. 2006. Bacterial cell division: the mechanism and its precision. Int. Rev. Cytol. 253:27-94. [DOI] [PubMed] [Google Scholar]
- 17.Ireton, K., and A. D. Grossman. 1994. DNA-related conditions controlling the initiation of sporulation in Bacillus subtilis. Cell Mol. Biol. Res. 40:193-198. [PubMed] [Google Scholar]
- 18.Ireton, K., N. W. t. Gunther, and A. D. Grossman. 1994. Spo0J is required for normal chromosome segregation as well as the initiation of sporulation in Bacillus subtilis. J. Bacteriol. 176:5320-5329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lau, I. F., S. R. Filipe, B. Soballe, O. A. Okstad, F. X. Barre, and D. J. Sherratt. 2003. Spatial and temporal organization of replicating Escherichia coli chromosomes. Mol. Microbiol. 49:731-743. [DOI] [PubMed] [Google Scholar]
- 20.Lau, S. Y., and H. I. Zgurskaya. 2005. Cell division defects in Escherichia coli deficient in the multidrug efflux transporter AcrEF-TolC. J. Bacteriol. 187:7815-7825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Letek, M., M. Fiuza, E. Ordonez, A. F. Villadangos, A. Ramos, L. M. Mateos, and J. A. Gil. 2008. Cell growth and cell division in the rod-shaped actinomycete Corynebacterium glutamicum. Antonie Van Leeuwenhoek 94:99-109. [DOI] [PubMed] [Google Scholar]
- 22.Letek, M., E. Ordonez, J. Vaquera, W. Margolin, K. Flardh, L. M. Mateos, and J. A. Gil. 2008. DivIVA is required for polar growth in the MreB-lacking rod-shaped actinomycete Corynebacterium glutamicum. J. Bacteriol. 190:3283-3292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lin, D. C., and A. D. Grossman. 1998. Identification and characterization of a bacterial chromosome partitioning site. Cell 92:675-685. [DOI] [PubMed] [Google Scholar]
- 24.Livny, J., Y. Yamaichi, and M. K. Waldor. 2007. Distribution of centromere-like parS sites in bacteria: insights from comparative genomics. J. Bacteriol. 189:8693-8703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lutkenhaus, J. 2007. Assembly dynamics of the bacterial MinCDE system and spatial regulation of the Z ring. Annu. Rev. Biochem. 76:539-562. [DOI] [PubMed] [Google Scholar]
- 26.Margolin, W. 2001. Spatial regulation of cytokinesis in bacteria. Curr. Opin. Microbiol. 4:647-652. [DOI] [PubMed] [Google Scholar]
- 27.Marston, A. L., H. B. Thomaides, D. H. Edwards, M. E. Sharpe, and J. Errington. 1998. Polar localization of the MinD protein of Bacillus subtilis and its role in selection of the mid-cell division site. Genes Dev. 12:3419-3430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Michie, K. A., and J. Lowe. 2006. Dynamic filaments of the bacterial cytoskeleton. Annu. Rev. Biochem. 75:467-492. [DOI] [PubMed] [Google Scholar]
- 29.Mohl, D. A., J. Easter, Jr., and J. W. Gober. 2001. The chromosome partitioning protein, ParB, is required for cytokinesis in Caulobacter crescentus. Mol. Microbiol. 42:741-755. [DOI] [PubMed] [Google Scholar]
- 30.Mohl, D. A., and J. W. Gober. 1997. Cell cycle-dependent polar localization of chromosome partitioning proteins in Caulobacter crescentus. Cell 88:675-684. [DOI] [PubMed] [Google Scholar]
- 31.Moker, N., J. Kramer, G. Unden, R. Kramer, and S. Morbach. 2007. In vitro analysis of the two-component system MtrB-MtrA from Corynebacterium glutamicum. J. Bacteriol. 189:3645-3649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Murray, H., and J. Errington. 2008. Dynamic control of the DNA replication initiation protein DnaA by Soj/ParA. Cell 135:74-84. [DOI] [PubMed] [Google Scholar]
- 33.Murray, H., H. Ferreira, and J. Errington. 2006. The bacterial chromosome segregation protein Spo0J spreads along DNA from parS nucleation sites. Mol. Microbiol. 61:1352-1361. [DOI] [PubMed] [Google Scholar]
- 34.Nottebrock, D., U. Meyer, R. Kramer, and S. Morbach. 2003. Molecular and biochemical characterization of mechanosensitive channels in Corynebacterium glutamicum. FEMS Microbiol. Lett. 218:305-309. [DOI] [PubMed] [Google Scholar]
- 35.Ogura, Y., N. Ogasawara, E. J. Harry, and S. Moriya. 2003. Increasing the ratio of Soj to Spo0J promotes replication initiation in Bacillus subtilis. J. Bacteriol. 185:6316-6324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rothfield, L., A. Taghbalout, and Y. L. Shih. 2005. Spatial control of bacterial division-site placement. Nat. Rev. Microbiol. 3:959-968. [DOI] [PubMed] [Google Scholar]
- 37.Ryan, K. R., and L. Shapiro. 2003. Temporal and spatial regulation in prokaryotic cell cycle progression and development. Annu. Rev. Biochem. 72:367-394. [DOI] [PubMed] [Google Scholar]
- 38.Schafer, A., A. Tauch, W. Jager, J. Kalinowski, G. Thierbach, and A. Pühler. 1994. Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene 145:69-73. [DOI] [PubMed] [Google Scholar]
- 39.Sullivan, N. L., K. A. Marquis, and D. Z. Rudner. 2009. Recruitment of SMC by ParB-parS organizes the origin region and promotes efficient chromosome segregation. Cell 137:697-707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sullivan, S. M., and J. R. Maddock. 2000. Bacterial division: finding the dividing line. Curr. Biol. 10:R249-R252. [DOI] [PubMed] [Google Scholar]
- 41.Tauch, A., O. Kirchner, B. Loffler, S. Gotker, A. Puhler, and J. Kalinowski. 2002. Efficient electrotransformation of Corynebacterium diphtheriae with a mini-replicon derived from the Corynebacterium glutamicum plasmid pGA1. Curr. Microbiol. 45:362-367. [DOI] [PubMed] [Google Scholar]
- 42.Thanbichler, M., and L. Shapiro. 2006. MipZ, a spatial regulator coordinating chromosome segregation with cell division in Caulobacter. Cell 126:147-162. [DOI] [PubMed] [Google Scholar]
- 43.Wu, L. J., and J. Errington. 2004. Coordination of cell division and chromosome segregation by a nucleoid occlusion protein in Bacillus subtilis. Cell 117:915-925. [DOI] [PubMed] [Google Scholar]