Abstract
The genomes of Vibrio cholerae O1 Matlab variant MJ-1236, Mozambique O1 El Tor variant B33, and altered O1 El Tor CIRS101 were sequenced. All three strains were found to belong to the phylocore group 1 clade of V. cholerae, which includes the 7th-pandemic O1 El Tor and serogroup O139 isolates, despite displaying certain characteristics of the classical biotype. All three strains were found to harbor a hybrid variant of CTXΦ and an integrative conjugative element (ICE), leading to their establishment as successful clinical clones and the displacement of prototypical O1 El Tor. The absence of strain- and group-specific genomic islands, some of which appear to be prophages and phage-like elements, seems to be the most likely factor in the recent establishment of dominance of V. cholerae CIRS101 over the other two hybrid strains.
Vibrio cholerae, a bacterium autochthonous to the aquatic environment, is the causative agent of cholera, a life-threatening disease that causes severe, watery diarrhea. Cholera bacteria are serogrouped based on their somatic O antigens, with more than 200 serogroups identified to date (6). Only toxigenic strains of serogroups O1 and O139 have been identified as agents of cholera epidemics and pandemics; serogroups other than O1 and O139 have the potential to cause mild gastroenteritis or, rarely, local outbreaks. Genes coding for cholera toxin (CTX), ctxAB, and other virulence factors have been shown to reside in bacteriophages and various mobile genetic elements. In addition, V. cholerae serogroup O1 is differentiated into two biotypes, classical and El Tor, by a combination of biochemical traits, by sensitivity to biotype-specific bacteriophages, and more recently by nucleotide sequencing of specific genes and by molecular typing (5, 17, 19).
There have been seven pandemics of cholera recorded throughout human history. The seventh and current pandemic began in 1961 in the Indonesian island of Sulawesi and subsequently spread to Asia, Africa, and Latin America; the six previous pandemics are believed to have originated in the Indian subcontinent. Isolates of the sixth pandemic were almost exclusively of the O1 classical biotype, whereas the current (seventh) pandemic is dominated by the V. cholerae O1 El Tor biotype as the causative agent, a transition occurring between 1923 and 1961. Today, the disease continues to remain a scourge in developing countries, confounded by the fact that V. cholerae is native to estuaries and river systems throughout the world (8).
Over the past 20 years, several new epidemic lineages of V. cholerae O1 El Tor have emerged (or reemerged). For example, in 1992, a new serogroup, namely, O139 of V. cholerae, was identified as the cause of epidemic cholera in India and Bangladesh (25). The initial concern was that a new pandemic was beginning; however, the geographic range of V. cholerae O139 is currently restricted to Asia. Additionally, V. cholerae O1 hybrids and altered El Tor variants have been isolated repeatedly in Bangladesh (Matlab) (23, 24) and Mozambique (1). Altered V. cholerae O1 El Tor isolates produce cholera toxin of the classical biotype but can be biotyped as El Tor by conventional phenotypic assays, whereas V. cholerae O1 hybrid variants cannot be biotyped based on phenotypic tests and can produce cholera toxin of either biotype. These new variants have subsequently replaced the prototype seventh-pandemic V. cholerae O1 El Tor strains in Asia and Africa, with respect to frequency of isolation from clinical cases of cholera (27).
Here, we report the genome sequence of three V. cholerae O1 variants, MJ-1236, a Matlab type I hybrid variant from Bangladesh that cannot be biotyped by conventional methods, CIRS101, an altered O1 El Tor isolate from Bangladesh which harbors ctxB of classical origin, and B33, an altered O1 El Tor isolate from Mozambique which harbors classical CTXΦ, and we compare their genomes with prototype El Tor and classical genomes. From an epidemiological viewpoint, among the three variants characterized in this study, V. cholerae CIRS101 is currently the most “successful” in that strains belonging to this type have virtually replaced the prototype El Tor in Asia and many parts of Africa, notably East Africa. This study, therefore, gives us a unique opportunity to understand why V. cholerae CIRS101 is currently the most successful El Tor variant.
MATERIALS AND METHODS
Genome sequencing.
Draft sequences were obtained from a blend of Sanger and 454 sequences and involved paired-end Sanger sequencing on 8-kb plasmid libraries to 5-fold coverage, 20-fold coverage of 454 pyrosequencing (Roche Diagnostics Corporation, Branford, CT) data. All libraries together provided 6.5-fold coverage. All general aspects of library construction and sequencing performed at the Joint Genome Institute can be found at http://www.jgi.doe.gov/. To finish the genomes, a collection of custom software and targeted reaction types were used. The Phred/Phrap/Consed software package (www.phrap.com) was used for sequence assembly and quality assessment (10-12). After the shotgun stage, reads were assembled with parallel Phrap (High Performance Software, LLC). Possible misassemblies were corrected with Dupfinisher (14) or transposon bombing of bridging clones (Epicentre Biotechnologies, Madison, WI). Gaps between contigs were closed by editing in Consed, custom primer walk, or PCR amplification (Roche Applied Science, Indianapolis, IN). Gene finding and annotation were achieved using the RAST server (2).
Comparative genomics.
Genome-to-genome comparison was performed using three approaches, since completeness and quality of nucleotide sequences varied from strain to strain in the set examined in this study. First, nucleotide sequences as whole contigs were directly aligned using the MUMmer program (20). Second, open reading frames (ORFs) of a given pair of genomes were reciprocally compared with each other using the BLASTN, BLASTP, and TBLASTX programs (ORF-dependent comparison). Third, a bioinformatic pipeline was developed to identify homologous regions of a given query ORF. Initially, a segment on the target contig homologous to a query ORF was identified using the BLASTN program. This potentially homologous region was expanded in both directions by 2,000 bp. Then, nucleotide sequences of the query ORF and selected target homologous region were aligned using a pairwise global alignment algorithm (22), and the resultant matched region in the subject contig was extracted and saved as a homologue (ORF-independent comparison). Orthologs and paralogs were differentiated by reciprocal comparison. In most cases, both ORF-dependent and -independent comparisons yielded the same orthologs, though the ORF-independent method performed better for draft sequences of low quality, in which sequencing errors, albeit rare, hampered identification of correct ORFs.
RESULTS
The combination of traditional Sanger and pyrosequencing resulted in high-quality assemblies of three V. cholerae genomes, B33 (GenBank accession number ACHZ00000000), CIRS101 (accession number ACVW00000000), and MJ-1236 (accession numbers CP001485 and CP001486), which represent altered or hybrid O1 El Tor biotypes (Table 1). One genome, V. cholerae O1 MJ-1236, was closed. However, at approximately nucleotides (nt) 852117 to 852144 of the smaller chromosome, there is a CTXΦ tandem repeat collapse, which could not be resolved by the assembly process. The small chromosome of V. cholerae O1 El Tor CIRS101 was closed and contained on a single contig, 89, and that of V. cholerae O1 El Tor B33 is contained on 2 contigs, 122 and 124, which could not be closed because of the presence of a CTXΦ tandem repeat, as in V. cholerae O1 MJ-1236.
TABLE 1.
General features of V. cholerae CIRS101, MJ-1236, and B33
| Strain | Size (bp) | Size (bp) of chromosome: |
No. of contigs | Statusb | No. of subsystems | No. of coding sequences | No. of RNAs | Suspected no. of missing genes | |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | ||||||||
| CIRS 101 | 4,059,686a | 3,014,033 | 1,045,653 | 18 | HQA | 293 | 3590 | 105 | 11 |
| MJ-1236 | 4,236,368 | 3,149,584 | 1,086,784 | 2 | Closed | 264 | 3810 | 120 | 9 |
| B33 | 4,154,698a | 3,081,617 | 1,073,081 | 16 | HQA | 262 | 3765 | 115 | 11 |
Size is based on combined base pairs of contigs covering each chromosome. The true total is assumed to be larger due to gaps in sequence coverage.
HQA, high-quality assembly. Closed, as deposited in GenBank, there is a tandem CTX prophage repeat that was collapsed into a single copy.
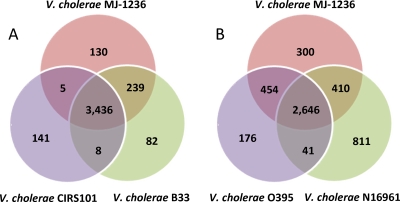
Among the altered and hybrid O1 biotype strains, 3,436 homologous genes were identified as present in all three strains (Fig. 1A), which ranged from 90.2 to 95.7% of the total genome size. Genomic comparison revealed that V. cholerae O1 MJ-1236 is more similar to V. cholerae O1 El Tor B33 than to V. cholerae O1 El Tor CIRS101 (Fig. 1A and 2), a result in agreement with previous findings (7).
FIG. 1.
Venn diagram showing shared and unique ORFs among V. cholerae O1 MJ-1236, O1 El Tor CIRS101, and O1 El Tor B33 (A) and V. cholerae O1 MJ-1236, O1 classical O395, and O1 El Tor N16961 (B).
FIG. 2.
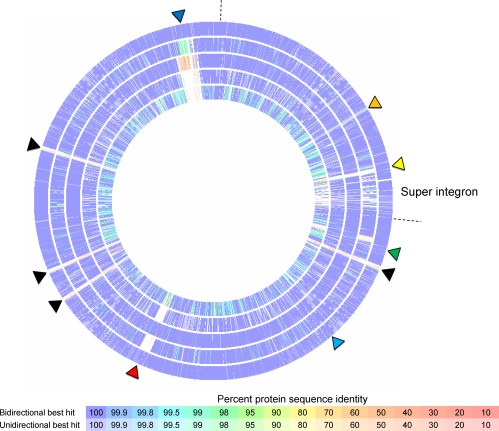
Circular heat map based on bi- and unidirectional best-BLAST-hit similarity using V. cholerae O1 MJ-1236 as the reference genome and five comparison genomes of V. cholerae (from the outermost ring): O1 El Tor B33, O1 El Tor CIRS101, O1 El Tor RC9, O1 El Tor N16961, and O1 classical O395. Regions or islands of interest are indicated by triangles: GI-12 (black), kappa prophage (red), ICE element (dark blue), GI-14 (yellow), GI-15 (light blue), VSP-II (green), and VSP-I on the second chromosome (orange). Chromosomal margins are indicated by dashed lines.
Seventh-pandemic group homology.
Even though the three altered or hybrid strains analyzed in this study display properties that distinguish them from prototype 7th-pandemic V. cholerae O1 El Tor, the genomes are still remarkably similar, with V. cholerae O1 MJ-1236 sharing 3,056 ORFs or genes in common with V. cholerae O1 El Tor N16961 (GenBank accession numbers AE003852 and AE003853), most (2,944) at 100% similarity (Fig. 1B and 2). Surprisingly, V. cholerae O1 MJ-1236 shares more genes in common, 3,100, with V. cholerae O1 classical O395 (accession numbers CP000626 and CP000627) than with V. cholerae O1 El Tor N16961 (Fig. 1B and 2); however, only 2,008 genes are at the 100% similarity level (Fig. 2). The unexpectedly high number of homologous genes is explained by the presence of multiple genomic islands in common with the O1 classical strain (7). Below, we describe in detail the genomic islands that are responsible for the majority of the strain-to-strain variations among the three altered or hybrid strains.
Genomic island 14 of V. cholerae O1 MJ-1236 and O1 El Tor B33.
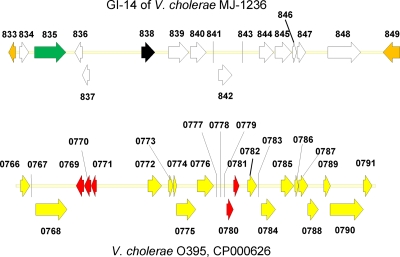
V. cholerae MJ-1236 and B33 contain identical genomic islands of 19,094 bp. Genomic island 14 (GI-14) is inserted between a putative acetyltransferase gene and a GDXG family lipase gene, VCD_000833 and VCD_000849 in V. cholerae O1 MJ-1236 and VCE_000947 and VCD_000932 in V. cholerae O1 El Tor B33, which correspond to VC tags VCA0491 and VCA0493 of V. cholerae O1 El Tor N16961, located in the superintegron region (Fig. 2). It contains coding regions for 13 hypothetical proteins, a homologue of a putative site-specific serine recombinase, PinR (VCD_000835, VCE_000945), and a putative membrane-associated transcriptional regulator (VCD_000838, VCE_000942) and intergenic and flanking regions (Fig. 3). Interestingly, a variant of the island is also found in V. cholerae O1 classical strain O395 (GenBank accession number CP000626), located on the smaller chromosome. In this strain, the island is smaller, 19,028 bp, and includes genes VC0395_0766 to VC0395_0791 and flanking DNA. GI-14 of V. cholerae O1 MJ-1236 and O1 El Tor B33 is approximately 94% identical to that of V. cholerae O395, with over 15,273 bp of homologous nucleotides. The two variants of this island diverge in two regions (Fig. 3). The first divergent region covers genes VC0395_0769 to VC0395_0771, VCD_000836, and VCD_000837 and some flanking DNA. In this region, besides sequence divergence, the V. cholerae O395 island contains additional DNA relative to that of V. cholerae O1 MJ-1236 and O1 El Tor B33. The second divergent region covers genes VC0395_0780, VC0395_0781, and VCD_000842 and flanking DNA. In this region, besides sequence divergence, the V. cholerae O1 MJ-1236 (and B33) island contains additional DNA relative to the V. cholerae O395 island.
FIG. 3.
Genomic island 14 (GI-14) of V. cholerae O1 MJ-1236, which includes genes VCD_000834 to VCD_000848 and flanking genes (orange), and a similar island of V. cholerae O1 classical O395. For GI-14 of V. cholerae O1 MJ-1236, hypothetical protein coding genes are indicated in white, a gene encoding a putative transcriptional regulator in black, and a gene encoding a putative site-specific recombinase in green. For V. cholerae O1 classical and V. cholerae O1 MJ-1236 (and O1 El Tor B33), homologous genes are indicated in yellow, and dissimilar regions are indicated in red.
Kappa phage.
There are two excisable genomic elements on the larger chromosome of V. cholerae MJ-1236 (and B33), identified by elevated coverage of the closed genome and comparative genomics. The first is a 33,109-bp island, located at nt 1260230 to 1293339 and inserted between the flaC and flaD genes (VCD_002149 and VCD_002191, respectively). The island was also present in V. cholerae O1 El Tor B33 and RC9 (GenBank accession number ACHX00000000), but it is missing from the genomes of V. cholerae O1 El Tor CIRS101 and N16961 (Fig. 2; Table 2). It is essentially 100% identical to lysogenic bacteriophages kappa of V. cholerae O1 El Tor strain A107 (accession number AB374228) and K139 of strain O139 MO10 (accession number AF125163 [26]), the latter of which has been well described by Kapfhammer et al. (18). In addition to V. cholerae O1 classical strain O395, as described previously (18), we found that V. cholerae NCTC 8457 (accession number AAWD00000000), AM-19226 (accession number AATY00000000), and 1587 (accession number AAUR00000000) contain genomic islands closely related to lysogenic phage kappa/K139.
TABLE 2.
Unique or altered features of V. cholerae MJ-1236, B33, and CIRS101 with respect to prototypical V. cholerae O1 El Tor strains N16961 and RC9
| Strain | Presence, no., or type of feature |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| TLC | VSP-I | VSP-II | CTXΦ | Kappa phage | GI-14 | GI-15 | GI-12 | ICE-like element | |
| N16961 | + | + | + | CTXETΦ | − | − | − | − | − |
| RC9 | + | + | + | CTXETΦ | + | − | − | − | NDc |
| MJ-1236 | − | 2a | + | CTXHYBΦ | + | + | − | 4 | + |
| B33 | − | + | + | CTXHYBΦ | + | + | + | − | + |
| CIRS101 | + | + | Truncated | CTXHYBΦ | − | − | − | − | + |
| 0395 | + | − | − | CTXclassΦ | + | +b | − | − | − |
ICE element.
The second excisable island in the genome of V. cholerae O1 MJ-1236 is bigger, approximately 100 kb, and located at approximately nt 2960000 to 3060000 of the larger chromosome (Fig. 2 and Table 2). Comparative genomics revealed that this island belonged to the integrative conjugative element (ICE) family of self-transmissible mobile elements. The island was also present in V. cholerae O1 El Tor B33. The two elements were found to be nearly identical and were termed ICEVchB33 (29).
V. cholerae O1 El Tor CIRS101 also contains an ICE-like element (Fig. 2), which is 98 kb in size and inserted into the prfC gene. Based on significant similarity between the integrase gene (99% similar to the int gene of both SXTMO10 and R391, from V. cholerae MO10 and Providencia rettgeri, respectively) and the insertion site, the element was confirmed as belonging to the SXT/R391 family and named ICEVchCIRS101 according to proposed nomenclature (4). The element encompasses genes VCH_000014 to VCH_000067, VCH_000179 to VCH_000188, and VCH_000420 to VCH_000433, which span three contigs of the draft genome, namely, contigs 77, 79, and 82, respectively.
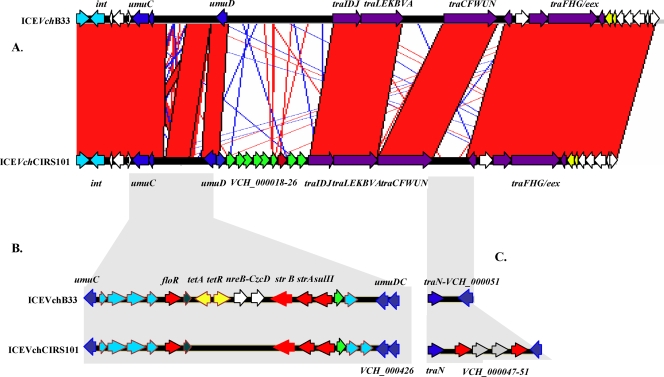
A comparative analysis of ICEVchCIRS101 with ICEVchB33 and other previously described ICEs using the Artemis Comparison Tool (ACT) (www.sanger.ac.uk/Software/ACT) was performed. Overall, the general organization of ICEVchCIRS101 was found to be highly similar to that of ICEVchB33 and other members of the SXT/R391 ICEs. ICEVchCIRS101, like all other related ICEs, showed a highly conserved core of genes responsible for transfer, integration and excision, and control that encompasses ca. 60 kb (Fig. 4A). As noted for other ICEs, specific inserted genes were identified in four hot spots and two variable regions within the core backbone.
FIG. 4.
Schematic representation of ICEVchCIRS101. (A) Alignment of ICEVchCIRS10 with ICEVchB33 generated by the Artemis Comparison Tool (ACT). Red lines/areas indicate homologous regions, and blue lines indicate inversions. Arrows indicate genes or clusters of genes (not scaled) and their orientations. (B) Genetic organization of the resistance clusters of ICEVchCIRS10 and ICEVchB33. (C) Genetic organization of the traN-VCH_000051 insertion of ICEVchCIRS10 and ICEVchB33.
The 5′ region of ICEVchCIRS101, between nt 1 and 7543, shows 99% nucleotide similarity with that of ICEVchB33 (Fig. 4A). The first two ORFs appear to be unique since they are found only in ICEVchB33 and not in either SXTMO10, R391, or ICEPdaSpaI (from Photobacterium damselae) (29).
Like ICEVchB33, SXTMO10, and ICEPdaSpaI, ICEVchCIRS101 contains a cluster of resistance genes inserted at the umuD locus, while in R391 the umuD gene is intact. Insertion of the resistance cluster occurs at the same base pair as in SXTMO10 and ICEVchB33 (Fig. 4A). In ICEVchCIRS101, the cluster is 16,445 bp in length, compared to 19,284 bp in ICEVchB33, and a significant rearrangement was observed (Fig. 4B).
The 5′ end of the resistance cluster is 99% identical to that of ICEVchB33, SXTMO10, and ICEPdaSpaI. Like ICEVchB33, ICEVchInd1, an ICE found in V. cholerae O1 El Tor (15), ICEVchVie1, and ICEVchLaos (16), ICEVchCIRS101 lacks the dfr18 dihydrofolate reductase gene for resistance to trimethoprim, the dCTP deaminase gene, and two other ORFs present in SXTMO10. ICEVchCIRS101 also lacks the efflux protein TetA, the effector of resistance to tetracycline, and its repressor, TetR, found in ICEVchB33 and ICEVchVie1, as well as NreB and the cobalt-zinc-cadmium resistance protein CzcD. The 3′ end of the resistance cluster showed the same genetic arrangement as ICEVchB33 and SXTMO10, namely, strB, strA, sulII, conferring resistance to streptomycin and sulfamethoxazole, respectively, and a transposase gene (Fig. 4B). In addition, ICEVchCIRS101 carries a second transposase gene (VCH_000426) not found in any other ICE belonging to the mutator family of transposases that has 100% nucleotide sequence similarity with the same gene in Shewanella oneidensis MR-1 (gb|AE014299.1|).
Like ICEVchB33, SXTMO10, and ICEPdaSpaI, ICEVchCIRS101 lacks the kanamycin resistance cluster that is present in R391. Interestingly, at this locus, a unique 15,000-bp sequence is inserted that is not found in any other ICE. The left flanking gene is a common gene found among ICEs that separates several members into two groups. The one found in ICEVchCIRS101 has 89% nucleotide similarity with the one found in R391 and ICESpuPO1, while the same gene in ICEVchB33 is 100% similar to that in SXTMO10 and ICEPdaSpaI. Interestingly, while ICEVchB33, SXT, and ICEPdaSpaI do not carry any insertion at this locus, R391 and ICESpuPO1 (from Shewanella putrefaciens) have a kanamycin resistance cluster and a restriction-modification cassette inserted, respectively. Thus, it appears that the two forms of this gene are associated with different insertions. The cluster found in the ICEVchCIRS101 is composed of eight ORFs, (VCH_000019 to VCH_000026), seven of which are annotated as encoding hypothetical proteins and one as encoding the ATP-dependent protease La (VCH_000024); these ORFs are 97% similar, on the nucleotide level, to those encoding similar proteases found in Pelodictyon phaeoclathratiforme BU-1 (GenBank accession number CP001110.1) and Syntrophomonas wolfei subsp. wolfei strain Goettingen (accession number CP000448.1). Proteins encoded by ORFs VCH_000021 to VCH_000026 appear to be shared with S. wolfei subsp. wolfei strain Goettingen and P. phaeoclathratiforme in a similar arrangement. Whatever the function of this cluster, it appears to be a new cluster for ICE and for V. cholerae.
Inserted at the traA-dsbC locus, ICEVchCIRS101 has the same cluster of genes as found in SXTMO10, while ICEVchB33 carries a larger insertion (29). At the 3′ end of the traN gene (nt 64641), ICEVchCIRS101 has a unique 6,140-bp insertion comprised of four genes (VCH_000047 to VCH_000050) (Fig. 4C). Two of the ORFs are annotated as encoding structural maintenance of chromosome (SMC)-like proteins and two as encoding transposases. The SMC proteins are a family of ATPases involved in higher-order chromosome organization and dynamics in prokaryotes and eukaryotes. An ABC motif that is usually found in transporters and permeases and involved in drug resistance mechanisms is present in this protein. The two transposases are found between the SMC-like proteins, suggesting that the two ORFs might have encoded one protein that was interrupted by the insertion of the transposase.
The 3′ end of ICEVchCIRS101 showed 99% nucleotide sequence similarity with that of ICEVchB33 (Fig. 4A), indicating that this element has its integron inserted at the traF locus, as do ICEVchB33 and ICEVchInd1, the two ICEs found in V. cholerae O1 El Tor.
Arrangement of cholera toxin prophage and associated elements.
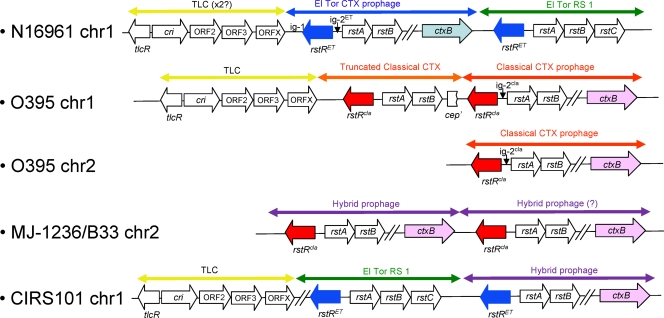
Until characterization of altered and hybrid O1 variants, such as the three in this study, the prototype arrangement of V. cholerae O1 El Tor strains of the 7th pandemic was that of RS1Φ element-CTXETΦ toxin-linked cryptic (TLC) element. In contrast, in toxigenic V. cholerae O1 classical strains, such as O395, the cholera toxin prophage is found on both chromosomes, with a single copy on the smaller chromosome and a TLC element-truncated CTXclassΦ-CTXclassΦ arrangement on the larger chromosome (Fig 5).
FIG. 5.
CTX prophages and associated element arrangements for V. cholerae O1 El Tor N16961, O1 classical O395, O1 MJ-1236, O1 El Tor B33, and O1 El Tor CIRS101. CTX prophage types were determined by nucleotide comparison of core regions and ctxB alleles.
The CTX prophage array in the Mozambique isolate, V. cholerae O1 El Tor B33, proposed by Lee et al. (21) could not be directly confirmed by whole-genome sequencing. This is not surprising since a tandem CTXclassΦ arrangement was proposed and tandem repeats are difficult to resolve unless large insert libraries, such as fosmids, are used. However, PCR using primers ctxB-F (5′ AGATATTTTCGTATACAGAATCTCTAG 3′) and cep-R (5′ AAACAGCAAGAAAACCCCGAGT 3′) confirmed the proposed tandem arrangement by production of a 3.1-kb amplicon (data not shown). In agreement with previous findings (21), there was no El Tor-specific RS1 element adjacent to the prophage(s). Additionally, no homologous ORFs of the TLC element were present in the genome.
Insertion of the prophage(s) was found to be at the same insertion site as that of V. cholerae O1 classical CTXclassΦ, as well as several pre-CTX phages (7), on the smaller chromosome, corresponding to the V. cholerae O1 El Tor N16961 locus tag VCA0569 to -0570 intergenic region. The former gene corresponds to VCE_001020 in V. cholerae O1 El Tor B33.
V. cholerae O1 MJ-1236 appears to have the same tandem CTXΦ arrangement as V. cholerae O1 El Tor B33, as shown by the genome sequence and ctxB-cep PCR (data not shown). As stated earlier, because of the difficulty in resolving large repeats (relative to the insert size library), a single copy of CTXclassΦ appears in the deposited genome, even though there is clonal evidence of a tandem CTXΦ repeat collapse. As in the case of V. cholerae O1 El Tor B33, no RS1Φ or TLC elements are present, and the chromosomal location is the same as that of V. cholerae O1 El Tor B33, which corresponds to genes VCD_000748 to VCD_000758.
For V. cholerae O1 El Tor CIRS101, the El Tor-specific RS1Φ element is present, and the single cholera toxin prophage present is of the El Tor type. However, in this strain, the genomic arrangement of these two elements is opposite that of V. cholerae O1 El Tor N16961 (Fig. 5). Consistent with previous results, the ctxB gene is homologous to that of the classical lineage instead of El Tor. In this draft assembly, the TLC element is present, but it is at the end of one contig, 10, while the RS1Φ element is at the end of another contig, 12. Given that the arrangement of these elements is variable, for example, in V. cholerae O1 El Tor RC9 (7), the exact arrangement could not be deduced from the draft genome sequence.
Within the genomic core regions of the CTX prophages of the three variant strains analyzed in this study, it was surprising to find that all genes besides ctxB are of El Tor lineage, as opposed to classical lineage. The exception is ctxA, which is identical in El Tor and classical strains. In other words, the zot, ace, and cep, etc., genes of V. cholerae MJ-1236, B33, and CIRS101 are identical to those of prototypical El Tor and not those of the classical biotype. Thus, it appears that the CTXΦ core region of these three strains is hybrid in nature, containing the “pre-CTX” core region of the El Tor type, with ctxAB of the classical type. We propose the use of the term CTXHYBΦ to describe these CTX prophages.
For the RS2 region of the cholera toxin prophage, the picture is less clear; i.e., there is more variability. As previously reported, the rstR genes of V. cholerae MJ-1236 and B33 are of the classical biotype. The rstB gene of V. cholerae MJ-1236 is identical to that of prototypical 7th-pandemic El Tor, while the same gene of strain B33 is not identical to that of either the prototypical 7th-pandemic El Tor or the classical biotype. The rstA genes of V. cholerae MJ-1236 and B33 are identical but different from that of prototypical 7th-pandemic El Tor and the classical biotype. The RS2 region of the CTXΦ of V. cholerae CIRS101 is identical to that of prototypical 7th-pandemic El Tor. Taken together, it appears that the RS2 region is too variable to ascribe a single allele for each gene to each biotype. This variability in the RS2 region, as well as in the core region, also exists in environmental isolates of toxigenic V. cholerae, such as strains BX330286 (GenBank accession number ACIA00000000) and V52 (accession number AAKJ02000000). Given these data, we propose that the RS2 region not be used to identify whether CTX prophages are of a certain biotype or type.
Other pathogenicity-related loci.
All three strains contained the prototypical VPI-1 and VPI-2 islands of 7th-pandemic V. cholerae O1 El Tor, with a few single nucleotide polymorphisms (SNPs) observed. V. cholerae O1 El Tor CIRS101 contains an SNP in the tcpA gene at nt 266 (A→G) of VPI-1 and an SNP in the putative membrane protein (gene VC1764, using V. cholerae O1 El Tor N16961 locus tags) of VPI-2 at nt 78 (G→T). Additionally, the putative helicase (VC1760, using V. cholerae O1 El Tor N16961 locus tags) of VPI-2 is disrupted by the insertion of a copy of a strain-specific genomic island in V. cholerae O1 MJ-1236.
As with the VPI islands, all three strains contained the prototypical VSP-I and -II islands of 7th-pandemic V. cholerae O1 El Tor (Table 2), with a few exceptions. V. cholerae O1 MJ-1236 contains a second copy of VSP-I, inserted in the smaller chromosome between genes VCD_000620 and VCD_000630 (corresponding to V. cholerae O1 El Tor N16961 locus tags VCA0095 and VCA0096) (7, 13). Additionally, V. cholerae O1 El Tor CIRS101 has a large deletion in VSP-II (7, 30).
MJ-1236-specific genomic island, GI-12.
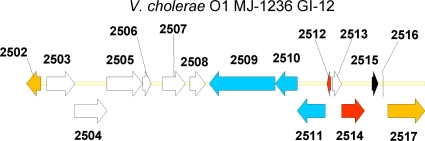
V. cholerae O1 Matlab variant MJ-1236 contains 4 copies of a unique 20-kb repeat on chromosome 1, located at nt 165105 to 184826 (VCD_001183 to VCD_001197, inserted into VPI 2), 1627680 to 1647405 (VCD_002503 to VCD_002516), 1747288 to 1767008 (VCD_002610 to VCD_002623), and 2264456 to 2284175 (VCD_003038 to VCD_003051) (Fig. 2). Interestingly, 3 of the 4 copies are oriented in one direction, and the other copy is oriented on the opposite strand. The genomic islands are almost all identical and are comprised of 14 ORFs (Fig. 6). These include a three-gene type I restriction-modification system, genes coding for 2 predicted transcriptional regulators and 8 hypothetical proteins, and a gene annotated as hipA.
FIG. 6.
Genomic island 12 (GI-12) of V. cholerae O1 MJ-1236 (one of four copies), which includes genes VCD_002503 to VCD_002516 and flanking genes (orange). Hypothetical protein coding genes are indicated in white, a putative transcriptional regulator in black, a three-gene type I restriction-modification system in blue, and hipBA in red.
Type I restriction-modification systems have been described only in bacteria and are important to the process of acquisition and stabilization of foreign DNA. Restriction enzymes are the first defense of a cell against the introduction of foreign DNA. Upon uptake, foreign DNA is rapidly recognized, cleaved by restriction enzymes, and then degraded by cellular exonucleases. However, there are molecular modifications that can protect DNA from being degraded, such as replication-independent methylation. Since the recognition sites are the same, in the recipient cell, methylases are thought to compete with restriction enzymes and foreign DNA is more likely preserved and integrated into the host genome. For the copy of GI-12 shown in Fig. 6, the type I restriction-modification system found in V. cholerae O1 MJ-1236 comprises three proteins: a site-specific DNase encoded by the hsdR gene, VCD_002509 (also called subunit R); a methyltransferase, HsdM (VCD_002511, subunit M); and the signal recognition or domain specificity peptide, HsdS (VCD_002510, subunit S).
The gene product of VCD_002512 was found to belong to the COG1396 superfamily of predicted transcriptional regulators, and that of VCD_002515 was found to belong to the COG4190 superfamily of predicted transcriptional regulators. VCD_002503 to VCD_002505 were annotated as hypothetical; however, the gene products were all found to have partial amino acid identity (approximately 25 to 30%) to phage integrases, with gene VCD_002503 containing the 6 conserved active sites of the XerD site-specific recombinase.
Interestingly, at the 3′ end, the element harbors a toxin/antitoxin module, hipBA, found to be involved in dormancy (9). In Escherichia coli, overexpression of hipA leads to the inhibition of cell growth and, thus, multidrug tolerance or persistence. The HipA protein of V. cholerae O1 MJ-1236 is 88% identical to HipA of V. cholerae RC385 (gi|150419660|gb|EDN11980.1), 40% to HipA of Vibrio fischeri ES114, and 37% to HipA of E. coli F11. In V. cholerae O1 MJ-1236, the HipB protein is annotated as a COG1396 transcriptional regulator and the gene (VCD_002512) is found upstream of hipA and oriented in the opposite direction, while in E. coli the two proteins are cooriented and overlap by 1 bp. Both proteins possess the helix-turn-helix domain found in XRE family-like proteins, which are prokaryotic DNA binding proteins belonging to the xenobiotic response element family of transcriptional regulators. In V. cholerae O1 MJ-1236, there is a 634-bp ORF encoding a hypothetical protein located between hipA and the transcriptional regulator, hipB (Fig. 6).
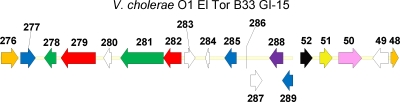
B33-specific genomic island, GI-15.
Among the three altered or hybrid strains, V. cholerae O1 El Tor B33 contains a strain-specific genomic island, GI-15 (7). It is inserted immediately downstream of gene VCE_000276, annotated as a trmE homologue. The size of this island can only be estimated to be at least 21,852 bp due to its being located at the end of a contig (VCE contig 116). The island encompasses genes VCE_000277 to VCE_000289 (Fig. 7) and includes 3 integrase genes (VCE_000277, -285, and -289), homologues of incW (VCE_000278), traN (VCE_000279), incF (VCE_000281), traH (VCE_000282), a restriction endonuclease gene (VCE_000288), and 5 hypothetical protein-coding genes (VCE_000280, -283, -284, -286, and -287). Among genome sequences of other V. cholerae strains, O1 El Tor strain RC9 possesses a homologue of gene VCE_000279, which is 80% identical over 2,312 nt of the total 2,760 nt of the gene. In addition, the first integrase gene, VCE_000277, was found to be approximately 70% identical to integrase genes from V. cholerae strains 623-39 (GenBank accession number NZ_AAWG01000099.1), RC385 (accession number AAKH02000022.1), and TM11079-80 (accession number NZ_ACHW01000015.1). Even though only a few single homologous genes could be found among sequenced V. cholerae genomes, the island was highly similar to a genomic island associated with multidrug resistance (Salmonella genomic island 1 [SGI1]) described for Salmonella enterica subsp. enterica serovar Typhimurium DT104 (accession number AF261825.2) (3). It was 95% identical to SGI1 over 18,460 nt of the 21,852 nt of GI-15. Interestingly, it appears that the island is also present in the genome of Shewanella sp. W3-18-1 (accession number CP000503.1).
FIG. 7.
Genomic island 15 (GI-15) of V. cholerae O1 El Tor B33, which includes genes VCE_000277 to VCE_000289 and flanking genes (orange). Genes encoding the following proteins are represented: hypothetical proteins are indicated in white, putative phage integrases in blue, a putative restriction endonuclease in purple, Tra homologues in red, Inc homologues in green, a transposase in pink, dihydropteroate synthase in yellow, and aminoglycoside (streptomycin/spectinomycin) adenylyltransferase in black.
Because of its location at the end of contig 116, the entire gene content of this island could not be determined. However, in the V. cholerae O1 El Tor N16961 genome, as well as in other V. cholerae genomes, the gene immediately downstream of trmE is the 435-bp gene encoding flavoprotein MioC (VC0002) (Fig. 7). In O1 El Tor B33, the flavoprotein MioC gene is located near the end of another contig, 112, with 4 ORFs located upstream of the gene before the end of the contig, VCE_000049 to VCE_000052, which are annotated as hypothetical protein, transposase, dihydropteroate synthase, and aminoglycoside (streptomycin/spectinomycin) adenylyltransferase (aadA1) genes of a class I integron. It is reasonable to assume that these four genes are also components of this island.
Unique strain-specific genes.
Within the three hybrid or altered O1 strains, there were few genes, outside the genomic islands summarized in Table 2, which distinguished each genome from the other (Fig. 1A). V. cholerae O1 MJ-1236 possesses 130 unique genes relative to V. cholerae O1 El Tor B33 and CIRS101. Fifty-seven of these are accounted for by GI-12, which is a 14-ORF island present in 4 copies (1 copy has a single gene annotated as 2 genes), a second copy of VSP-I on the small chromosome accounts for 9 ORFs, and there are 4 unique ORFs and 22 paralogous ORFs contained in the superintegron. Of the remaining 38 unique ORFs, 19 are unique or paralogous hypothetical ORFs less than 100 nt in length, 11 are unique or paralogous ORFs greater than 100 nt in length, including 6 paralogous transposase ORFs and a second copy of hcp, 4 represent single genes which were annotated as tandem paralogs, 1 (a putative helicase of VPI 2) was interrupted by insertion of GI-12, 2 are ORFs of the ICE element which were later identified as present in O1 El Tor B33 by PCR (29), and the remaining gene is rstR of CTXΦ, which appeared unique since the same gene of O1 El Tor B33 is truncated by its position at the end of a contig.
V. cholerae O1 El Tor B33 possesses 82 unique genes compared to the other two strains included in this study. GI-15 accounts for 16 of those ORFs, and 25 unique or paralogous ORFs reside in the superintegron region. Of the remaining 41 unique genes, 28 are less than 100 nt in length and encode hypothetical proteins, two encode ribosomal proteins (S1p and L5p), one is a truncated copy of ctxA, and 10 are paralogous ORFs larger than 100 nt in length, including two transposase genes and two transposase subunit genes, alpha- and gamma-chain ORFs encoding oxaloacetate decarboxylase, kefC, hcp, and translation elongation factor Tu and integrase genes.
V. cholerae O1 El Tor CIRS101 possesses 141 unique genes compared to O1 MJ-1236 and O1 El Tor B33. Of these, 10 are contained in ICEVchCIRS101, 38 are in the superintegron region, three comprise the RS1Φ element (the rstR gene is missing since the element is at the end of a contig, 88), six comprise the TLC element, and 47 are hypothetical ORFs less than 100 nt in length. Of the 37 remaining unique genes, 25 are accounted for as sequencing, assembly, and gene-finding artifacts; i.e., 22 are single genes that were divided into two ORFs with the same annotation, and three were truncated because of their position at the end of a contig or premature stop codon. The remaining 12 genes included genes encoding five transposases or transposase subunits, a GGDEF family protein, interrupted by a mutator transposase to give two paralogous ORFs, a lipid A biosynthesis lauroyl acyltransferase, and an adenylyl-sulfate kinase, as well as hcp, a gene encoding an ATPase component of a predicted ABC-type transport system, and luxO (located adjacent to luxU).
DISCUSSION
The three genomes that are reported here were sequenced and compared in order to determine factors, in addition to unique arrangements of the cholera toxin prophage and variants of known-virulence-associated islands already identified, which contribute to the epidemicity and environmental fitness of the hybrid and altered O1 El Tor strains and presumably account for their ability to replace prototype El Tor V. cholerae O1, typified by strain N16961.
As previously reported (7), O1 MJ-1236, O1 El Tor B33, and O1 El Tor CIRS101 form a very tight monophyletic group, the 7th-pandemic clade, with V. cholerae O1 El Tor N16961, RC9, and O139 MO10, as determined when more than 2.6 million bp of orthologous protein-coding regions were used. A complete, sequence-based comparison of the genomes of V. cholerae O1 MJ-1236 and O1 El Tor N16961 revealed that 80% and 78% (Fig. 1B) of all genes are shared and that 77% and 75% of all genes are 100% similar, respectively. In contrast, 53% of the genome of V. cholerae O1 MJ-1236 is 100% similar to V. cholerae O1 classical O395, even though these two strains shared more genes in common than MJ-1236 and N16961, such as GI-14 and kappa phage. Despite the fact that each of the three strains sequenced here was originally described using different terminology, i.e., Matlab variant, hybrid, or altered O1 El Tor, based on the presence of classical attributes, such as the presence of CTXclassΦ, all three are clearly 7th-pandemic clade members possessing a genomic backbone of the O1 El Tor/O139 group, recently termed the phylocore genome 1 (PG-1) clade (7). We were unsuccessful in identifying genome-based results to explain the biotyping inconsistencies among these three strains. This should not be surprising given the nature of the biotyping characteristics, such as resistance or sensitivity to polymyxin B and biotype-specific phages. For the latter, the presence of novel lysogenic phage elements in the genomes of these strains may explain the inconclusive biotype-specific phage results, for example, GI-12 in V. cholerae MJ-1236. As for nomenclature, we find the current terms to be appropriate, but it should be understood that the hybrid or altered nature of these strains is due to the presence of a few hybrid or classical-like elements, such as CTXHYBΦ, GI-14, and kappa phage, in the genomic backbone of a 7th-pandemic El Tor, or PG-1, strain.
V. cholerae O1 MJ-1236 was isolated in Matlab, Bangladesh, in 1994, while O1 El Tor CIRS101 was isolated in Dhaka, Bangladesh, in 2002 and O1 El Tor B33 from Beira, Mozambique, in 2004. Based on geography and date of isolation, it is surprising that O1 MJ-1236 is more similar to O1 El Tor B33 than to O1 El Tor CIRS101 (Fig. 1A). In fact, the genomes of B33 and MJ-1236 are 96 and 98% identical, respectively, with differences explained by the presence of a second copy of VSP-I and four copies of GI-12 in O1 MJ-1236 and the presence of GI-15 in O1 El Tor B33 (Table 2). Given these results, we can consider these two strains as belonging to the same clonal complex. Conversely, O1 El Tor CIRS101 lacked several of the genomic islands found in O1 MJ-1236 and O1 El Tor B33, including kappa phage and GI-14, and possessed a truncated variant of VSP-II (Table 2). Additionally, the superintegron region of O1 El Tor CIRS101 was markedly different than that of O1 MJ-1236 and O1 El Tor B33. Based on these results, it seems plausible that the V. cholerae O1 Matlab hybrid emerged and became the established, i.e., prevalent, type of the 7th-pandemic clade in Bangladesh and, later, in Africa, like V. cholerae B33 in Mozambique. In Bangladesh, altered O1 El Tor, typified by CIRS101, emerged after MJ-1236 and became the prevalent type present in Asia and, later, Africa, displacing hybrid variants such as B33. It would appear that strains like MJ-1236 and B33 are transient clones in the genesis of dominant clones such as CIRS101, almost like an experiment in nature in the process of evolution of the fittest clone.
Based on the results presented here, several conclusions can be drawn concerning features presented that are important for endemicity and epidemicity of V. cholerae and how these strains evolve and replace existing clones. As for the displacement of the prototypical 7th-pandemic El Tor by the three strains sequenced, the type of CTXΦ and the presence of an ICE element in the genome appear to be the most likely explanations for these events. It is generally accepted that classical isolates of V. cholerae O1 produce more severe disease, whereas O1 El Tor isolates are better adapted for survival (28). All three strains were isolated from patients with acute cases of watery diarrhea, which were more severe than that typically observed from infection with prototypical V. cholerae O1 El Tor. V. cholerae O1 MJ-1236 and O1 El Tor B33 contain identical hybrid CTX prophage arrangements (Fig. 5), as does O1 El Tor CIRS101, which also contains the RS1Φ and TLC elements. Interestingly, with the exception that it is typed as a CT1 epitype and genotype 1 (the same as classical) element based on ctxB immunology and sequence, respectively, the CTX prophage array of CIRS101 is identical in sequence to that of V. cholerae O1 El Tor N16961. Based on these data, we hypothesize that the ctxB sequence may be a significant factor in determining toxigenic virulence.
The presence of an ICE element in the genomes of all three strains examined in this study also has profound implications with regard to epidemiology. Specifically, these elements harbor antibiotic resistance clusters, which can be gained and lost, via hot spots, in response to contemporary antibiotic therapy to extend the infection rate and course of disease caused by these strains beyond those caused by toxigenic clones not containing the ICE elements. Taken together, these results imply that those elements that impact the epidemiology of the disease contribute greatly to the success of the strain or clone. This should not be surprising, given that this infection spreads effectively through a fecal-oral route of transmission and the high number of cells contained in a typical stool of an infected person.
The more difficult question to answer, on a genomic level, is why clones of V. cholerae CIRS101 have displaced other toxigenic clones, either prototypical or hybrid variants (27), since those aspects of the environment that play a role in the ecology of V. cholerae are typically ignored. The one feature of this genome that stands out, compared to the genomes of V. cholerae MJ-1236 and B33, is the absence of putative prophages or prophage-like elements beyond those elements associated with pathogenic clones of V. cholerae, i.e., CTXΦ, VSP-I and -II, and VPI 1 and 2 (Table 2). The presence or absence of genomic islands such as kappa prophage, GI-12, GI-14, and GI-15 may have ecological significance for V. cholerae MJ-1236 and B33, perhaps in the form of reduced fitness. Another feature that merits comment is the presence of a truncated VSP-II island in the genome of V. cholerae CIRS101. Because many of these genes are hypothetical in function, little that is definitive can be offered as to the effect the loss of this part of the island by insertion of a transposase would have; however, the variant appears to be stable (30).
In conclusion, the unique combination of characteristics of the genome of V. cholerae CIRS101 provides the bacterium with a competitive ecological edge and greater infectivity over that of other pathogenic clones of V. cholerae, such as MJ1236 and B33. Historically, El Tor strains of V. cholerae are considered to have an improved environmental fitness based on the observation that they have displaced classical strains. In turn, classical strains of V. cholerae are believed to produce a more severe form of the disease, cholera (28). The three genomes presented here provide evidence of an amalgamation of environmental fitness of El Tor strains and greater infectivity of classical strains, i.e., a “mixing and matching,” through recombination and lateral gene transfer, resulting in the genesis of new variants of V. cholerae with expanded ecological persistence, infectivity, and dispersion. The success of a clone is a combination of its ability to adapt to changing environmental conditions as a stable inhabitant, evidenced by conservation of the El Tor, or PG-1, genomic backbone, and its ability to transmit progeny through human populations. Causative environmental and/or host factors determining emergence and temporal domination of a particular variant of V. cholerae and displacement of an existing variant(s) through natural selection pose the next challenge.
Acknowledgments
This study was supported by the National Institutes of Health (grant no. 1RO1A139129-01 to R.R.C.) and the National Oceanic and Atmospheric Administration Oceans and Human Health Initiative (grant no. S0660009 to R.R.C.). Funding for genome sequencing and support for C.J.G. was provided by the Office of the Chief Scientist (United States).
Footnotes
Published ahead of print on 26 March 2010.
REFERENCES
- 1.Ansaruzzaman, M., N. A. Bhuiyan, G. B. Nair, D. A. Sack, M. Lucas, J. L. Deen, J. Ampuero, and C. L. Chaignat. 2004. Cholera in Mozambique, variant of Vibrio cholerae. Emerg. Infect. Dis. 10:2057-2059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aziz, R. K., D. Bartels, A. A. Best, M. DeJongh, T. Disz, R. A. Edwards, K. Formsma, S. Gerdes, E. M. Glass, M. Kubal, F. Meyer, G. J. Olsen, R. Olson, A. L. Osterman, R. A. Overbeek, L. K. McNeil, D. Paarmann, T. Paczian, B. Parrello, G. D. Pusch, C. Reich, R. Stevens, O. Vassieva, V. Vonstein, A. Wilke, and O. Zagnitko. 2008. The RAST server: rapid annotations using subsystems technology. BMC Genomics 9:75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boyd, D., G. A. Peters, A. Cloeckaert, K. S. Boumedine, E. Chaslus-Dancla, H. Imberechts, and M. R. Mulvey. 2001. Complete nucleotide sequence of a 43-kilobase genomic island associated with the multidrug resistance region of Salmonella enterica serovar Typhimurium DT104 and its identification in phage type DT120 and serovar Agona. J. Bacteriol. 183:5725-5732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burrus, V., J. Marrero, and M. K. Waldor. 2006. The current ICE age: biology and evolution of SXT-related integrating conjugative elements. Plasmid 55:173-183. [DOI] [PubMed] [Google Scholar]
- 5.Byun, R., L. D. H. Elbourne, R. Lan, and P. R. Reeves. 1999. Evolutionary relationships of pathogenic clones of Vibrio cholerae by sequence analysis of four housekeeping genes. Infect. Immun. 67:1116-1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chatterjee, S. N., and K. Chaudhuri. 2003. Lipopolysaccharides of Vibrio cholerae. I. Physical and chemical characterization. Biochim. Biophys. Acta 1639:65-79. [DOI] [PubMed] [Google Scholar]
- 7.Chun, J., C. J. Grim, N. A. Hasan, J. H. Lee, S. Y. Choi, B. J. Haley, E. Taviani, Y.-S. Jeon, D. W. Kim, J.-H. Lee, T. S. Brettin, D. C. Bruce, J. F. Challacombe, J. C. Detter, C. S. Han, A. C. Munk, O. Chertkov, L. Meincke, E. Saunders, R. A. Walters, A. Huq, G. B. Nair, and R. R. Colwell. 2009. Comparative genomics reveals mechanism for short-term and long-term clonal transitions in pandemic Vibrio cholerae. Proc. Natl. Acad. Sci. U. S. A. 106:15442-15447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Colwell, R. R. 1996. Global climate and infectious disease: the cholera paradigm. Science 274:2025-2031. [DOI] [PubMed] [Google Scholar]
- 9.Correia, F. F., A. D'Onofrio, T. Rejtar, L. Li, B. L. Karger, K. Makarova, E. V. Koonin, and K. Lewis. 2006. Kinase activity of overexpressed HipA is required for growth arrest and multidrug tolerance in Escherichia coli. J. Bacteriol. 188:8360-8367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ewing, B., and P. Greene. 1998. Base-calling of automated sequencer traces using Phred. II. Error probabilities. Genome Res. 8:186-194. [PubMed] [Google Scholar]
- 11.Ewing, B., L. Hillier, M. Wendl, and P. Greene. 1998. Base-calling of automated sequencer traces using Phred. I. Accuracy assessment. Genome Res. 8:175-185. [DOI] [PubMed] [Google Scholar]
- 12.Gordon, D., C. Abajian, and P. Green. 1998. Consed: a graphical tool for sequence finishing. Genome Res. 8:195-202. [DOI] [PubMed] [Google Scholar]
- 13.Grim, C., J. Choi, J. Chun, Y. S. Jeon, E. Taviani, N. A. Hasan, B. Haley, A. Huq, and R. R. Colwell. 2010. Occurrence of the Vibrio cholerae seventh pandemic VSP-I island and a new variant. OMICS 14:1-7. [DOI] [PubMed] [Google Scholar]
- 14.Han, C. S., and P. Chain. 2006. Finishing repetitive regions automatically with Dupfinisher, p. 142-147. In H. R. Arabnia and H. Valafar (ed.), Proceedings of the 2006 International Conference on Bioinformatics and Computational Biology. CSREA Press, Las Vegas, NV.
- 15.Hochhut, B., Y. Lotfi, D. Mazel, S. M. Faruque, R. Woodgate, and M. K. Waldor. 2001. Molecular analysis of antibiotic resistance gene clusters in Vibrio cholerae O139 and O1 SXT constins. Antimicrob. Agents Chemother. 45:2991-3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Iwanaga, M., C. Toma, T. Miyazato, S. Insisiengmay, N. Nakasone, and M. Ehara. 2004. Antibiotic resistance conferred by a class I integron and SXT constin in Vibrio cholerae O1 strains isolated in Laos. Antimicrob. Agents Chemother. 48:2634-2639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kaper, J. B., J. G. Morris, Jr., and M. M. Levine. 1995. Cholera. Clin. Microbiol. Rev. 8:48-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kapfhammer, D., J. Blass, S. Evers, and J. Reidl. 2002. Vibrio cholerae phage K139: complete genome sequence and comparative genomics of related phages. J. Bacteriol. 184:6592-6601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kimsey, H. H., and M. K. Waldor. 1998. CTXphi immunity: application in the development of cholera vaccines. Proc. Natl. Acad. Sci. U. S. A. 95:7035-7039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kurtz, S., A. Phillippy, A. L. Delcher, M. Smoot, M. Shumway, C. Antonescu, and S. L. Salzberg. 2004. Versatile and open software for comparing large genomes. Genome Biol. 5:R12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee, J. H., K. H. Han, S. Y. Choi, M. E. Lucas, C. Mondlane, M. Ansaruzzaman, G. B. Nair, D. A. Sack, L. von Seidlein, J. D. Clemens, M. Song, J. Chun, and D. W. Kim. 2006. Multilocus sequence typing (MLST) analysis of Vibrio cholerae O1 El Tor isolates from Mozambique that harbour the classical CTX prophage. J. Med. Microbiol. 55:165-170. [DOI] [PubMed] [Google Scholar]
- 22.Myers, E. W., and W. Miller. 1988. Optimal alignments in linear space. Comput. Appl. Biosci. 4:11-17. [DOI] [PubMed] [Google Scholar]
- 23.Nair, G. B., S. M. Faruque, N. A. Bhuiyan, M. Kamruzzaman, A. K. Siddique, and D. A. Sack. 2002. New variants of Vibrio cholerae O1 biotype El Tor with attributes of the classical biotype from hospitalized patients with acute diarrhea in Bangladesh. J. Clin. Microbiol. 40:3296-3299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nair, G. B., F. Qadri, J. Holmgren, A. M. Svennerholm, A. Safa, N. A. Bhuiyan, Q. S. Ahmad, S. M. Faruque, A. S. Faruque, Y. Takeda, and D. A. Sack. 2006. Cholera due to altered El Tor strains of Vibrio cholerae O1 in Bangladesh. J. Clin. Microbiol. 44:4211-4213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ramamurthy, T., S. Garg, R. Sharma, S. K. Bhattacharya, G. B. Nair, T. Shimada, T. Takeda, T. Karasawa, H. Kurazano, A. Pal, et al. 1993. Emergence of novel strain of Vibrio cholerae with epidemic potential in southern and eastern India. Lancet 341:703-704. [DOI] [PubMed] [Google Scholar]
- 26.Reidl, J., and J. J. Mekalanos. 1995. Characterization of Vibrio cholerae bacteriophage K139 and use of a novel mini-transposon to identify a phage-encoded virulence factor. Mol. Microbiol. 18:685-701. [DOI] [PubMed] [Google Scholar]
- 27.Safa, A., J. Sultana, P. Dac Cam, J. C. Mwansa, and R. Y. Kong. 2008. Vibrio cholerae O1 hybrid El Tor strains, Asia and Africa. Emerg. Infect. Dis. 14:987-988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Siddique, A. K., G. B. Nair, M. Alam, D. A. Sack, A. Huq, A. Nizam, I. M. Longini, F. Qadri, S. M. Faruque, R. R. Colwell, S. Ahmed, A. Iqbal, N. A. Bhuiyan, and R. B. Sack. 2010. El Tor cholera with severe disease: a new threat to Asia and beyond. Epidemiol. Infect. 138:347-352. [DOI] [PubMed] [Google Scholar]
- 29.Taviani, E., C. Grim, J. Chun, A. Huq, and R. R. Colwell. 2009. Genomic analysis of a novel integrative conjugative element in Vibrio cholerae. FEBS Lett. 583:3630-3636. [DOI] [PubMed] [Google Scholar]
- 30.Taviani, E., C. J. Grim, J. Choi, J. Chun, B. Haley, N. A. Hasan, A. Huq, and R. R. Colwell. 6 May 2010. Discovery of novel Vibrio cholerae VSP II genomic islands using comparative genomic analysis. FEMS Microbiol. Lett. doi: 10.1111/j.1574-6968.2010.02008.x. [DOI] [PMC free article] [PubMed]