Abstract
The β clamp is an essential replication sliding clamp required for processive DNA synthesis. The β clamp is also critical for several additional aspects of DNA metabolism, including DNA mismatch repair (MMR). The dnaN5 allele of Bacillus subtilis encodes a mutant form of β clamp containing the G73R substitution. Cells with the dnaN5 allele are temperature sensitive for growth due to a defect in DNA replication at 49°C, and they show an increase in mutation frequency caused by a partial defect in MMR at permissive temperatures. We selected for intragenic suppressors of dnaN5 that rescued viability at 49°C to determine if the DNA replication defect could be separated from the MMR defect. We isolated three intragenic suppressors of dnaN5 that restored growth at the nonpermissive temperature while maintaining an increase in mutation frequency. All three dnaN alleles encoded the G73R substitution along with one of three novel missense mutations. The missense mutations isolated were S22P, S181G, and E346K. Of these, S181G and E346K are located near the hydrophobic cleft of the β clamp, a common site occupied by proteins that bind the β clamp. Using several methods, we show that the increase in mutation frequency resulting from each dnaN allele is linked to a defect in MMR. Moreover, we found that S181G and E346K allowed growth at elevated temperatures and did not have an appreciable effect on mutation frequency when separated from G73R. Thus, we found that specific residue changes in the B. subtilis β clamp separate the role of the β clamp in DNA replication from its role in MMR.
Replication sliding clamps are essential cellular proteins imparting a spectacular degree of processivity to DNA polymerases during genome replication (24, 39-41). Encoded by the dnaN gene, the β clamp is a highly conserved bacterial sliding clamp found in virtually all eubacterial species (reviewed in reference 7). The β clamp is a head-to-tail, ring-shaped homodimer that encircles double-stranded DNA (1, 39). In eukaryotes and archaea, the analog of the β clamp is proliferating cell nuclear antigen (PCNA) (15, 28, 40, 41). Eukaryotic PCNA is a ring-shaped homotrimer that also acts to encircle DNA, increasing the processivity of the replicative DNA polymerases (40, 41). Although the primary structures of the β clamp and PCNA are not conserved, the tertiary structures of these proteins are very similar, demonstrating structural conservation among bacterial, archaeal, and eukaryotic replication sliding clamps (28, 39-41; reviewed in reference 6).
The function of the β clamp is not limited to its well-defined role in genome replication. The Escherichia coli β clamp binds Hda, which also binds the replication initiation protein DnaA, regulating the active form of DnaA complexed with ATP (19, 37, 43). This allows the β clamp to regulate replication initiation through the amount of available DnaA-ATP. In Bacillus subtilis, the β clamp binds YabA, a negative regulator of DNA replication initiation (12, 29, 52). It has also been suggested that the B. subtilis β clamp sequesters DnaA from the replication origin during the cell cycle through the binding of DnaA to YabA and the binding of YabA to the β clamp (70). Thus, it is hypothesized that in E. coli and B. subtilis, the β clamp influences the frequency of replication initiation through interactions with Hda and YabA, respectively.
The E. coli and B. subtilis β clamp has an important role in translesion DNA synthesis during the replicative bypass of noncoding bases by specialized DNA polymerases belonging to the Y family (20, 33). The roles of the E. coli β clamp in translesion synthesis are well established (5, 8, 30, 31). Binding sites on the E. coli β clamp that accommodate translesion polymerases pol IV (DinB) and pol V (UmuD2′C) have been identified, and the consequence of disrupting their association with the β clamp has illustrated the critical importance of the β clamp to the activity of both of these polymerases (4, 5, 8, 26, 30, 31, 48, 49).
In addition to the involvement of the β clamp in replication initiation, DNA replication, and translesion synthesis, the E. coli and B. subtilis β clamp also functions in DNA mismatch repair (MMR) (45, 46, 64). The MMR pathway recognizes and repairs DNA polymerase errors, contributing to the overall fidelity of the DNA replication pathway (reviewed in references 42 and 60). In both E. coli and B. subtilis, deletion of the genes mutS and mutL increases the spontaneous mutation frequency several hundredfold (13, 25, 63). In E. coli, MutS recognizes and binds mismatches, while MutL functions as a “matchmaker,” coordinating the actions of other proteins in the MMR pathway, allowing the removal of the mismatch and resynthesis of the resulting gap (reviewed in references 42 and 60). MutS and MutL of E. coli and B. subtilis physically interact with the β clamp (45, 46, 51, 64). Interaction between the B. subtilis β clamp and MutS is important for efficient MMR and organization of MutS-green fluorescent protein (GFP) into foci in response to replication errors, while the function of MutL binding to the β clamp is unknown (64).
These studies show that the β clamp is critical for several aspects of DNA metabolism in E. coli and B. subtilis. In E. coli, many dnaN alleles have been examined and used to define the mechanistic roles of the β clamp in vivo (5, 18, 24, 30, 31, 48, 49, 73). A limitation in studying the mechanistic roles of the B. subtilis β clamp is that only two dnaN alleles (β clamp) are available, dnaN5 and dnaN34 (36) (www.bgsc.org/), and both of these alleles do not support growth at temperatures above 49°C, suggesting that they may cause similar defects (36) (www.bgsc.org/). Of these two dnaN alleles, only dnaN5 has been investigated in any detail (36, 53, 64). The mutant β clamp encoded by dnaN5 contains a G73R substitution [dnaN5(G73R)] in a surface-exposed residue located on the outside rim of the β clamp (53, 64). Our previous studies with this allele showed that dnaN5(G73R) confers an increase in mutation frequency at 30°C and 37°C (64). Further characterization of dnaN5(G73R) showed that the increased mutation frequency is caused by a partial defect in MMR (64). Additionally, dnaN5(G73R)-containing cells have a reduced ability to support MutS-GFP focus formation in response to mismatches (64). These results support the hypothesis that G73R in the β clamp causes a defect in DNA replication at 49°C (36) and impaired MMR manifested by a defect in establishing the assembly of MutS-GFP foci in response to replication errors (64).
To understand the roles of the B. subtilis β clamp in MMR and DNA replication, we examined the dnaN5 and dnaN34 alleles. We found that the nucleotide sequences of dnaN5 and dnaN34 and the phenotypes they produce were identical, both producing the G73R missense mutation. We analyzed in vivo β clampG73R protein levels and found that the β clampG73R protein accumulated to wild-type levels at elevated temperatures. To identify amino acid residues that would restore DNA replication at elevated temperatures, we isolated three intragenic suppressors of dnaN5(G73R) that conferred growth of B. subtilis cells at 49°C. Epistasis analysis and determination of the mutation spectrum showed that each dnaN allele isolated in this study caused an MMR-dependent increase in mutation frequency. Additionally, we found that the β clamp binding protein YabA can reduce the efficiency of MMR in vivo when yabA expression is induced. Thus, we have identified residues in the β clamp that are critical for DNA replication and MMR in B. subtilis. We also found that a β clamp binding protein, YabA, can reduce the efficiency of MMR in vivo.
MATERIALS AND METHODS
Bacteriological methods.
The B. subtilis strains used in this study are described in Table 1. Strains were grown in LB medium with the following final antibiotic concentrations as previously described (64-67, 69): erythromycin, 0.5 μg/ml; lincomycin, 12.5 μg/ml; spectinomycin, 100 μg/ml; rifampin, 100 μg/ml; chloramphenicol, 5 μg/ml.
TABLE 1.
Strains used in this study
| Strain | Relevant genotype | Reference or sourcea |
|---|---|---|
| PY79 | SPβ° prototroph | 76 |
| LAS4 | yqjW::kan | 72 |
| LAS5 | yqjH::tet | 72 |
| LAS160 | amyE::PspacmutL (cat) dnaN5(G73R) spoIIIJ::kan mutS-gfp (spc) | 64 |
| LAS169 | dnaN5(G73R); Tn917::mls | 64 |
| LAS360 | amyE::Pspank (hy) yabA (spc) | 27 |
| LAS361 | yabA::cat | 27 |
| LAS393 | ΔmutL::spc | 69 |
| IA24 | dnaN34 ilvA1 metB5 | BGSC |
| NMD18 | spoIIIJ::kan | This work |
| NMD11 | dnaN5(G73R) spoIIIJ::kan | This work |
| NMD22 | dnaN(S22P, G73R) spoIIIJ::kan | This work |
| NMD14 | dnaN(G73R, S181G) spoIIIJ::kan | This work |
| NMD17 | dnaN(G73R, E346K) spoIIIJ::kan | This work |
| NMD32 | dnaN(G73R, S181G) spoIIIJ::kan mutSL::spc | This work |
| NMD33 | dnaN(G73R, E346K) spoIIIJ::kan mutSL::spc | This work |
| NMD34 | dnaN(S22P, G73R) spoIIIJ::kan mutSL::spc | This work |
| NMD35 | yqjW::kan dnaN5(G73R); Tn917::mls | This work |
| NMD36 | yqjH::tet dnaN5(G73R) Tn917::mls | This work |
| NMD38 | dnaN5 mutSL::spc | This work |
| NMD39 | amyE::PspacmutL mutSL::spc | This work |
| NMD45 | dnaN34(G73R) spoIIIJ::kan | This work |
| NMD46 | amyE::PspacmutL (cat) dnaN(G73R, S181G) spoIIIJ::kan mutS-gfp (spc) | This work |
| NMD47 | amyE::PspacmutL (cat) dnaN(G73R, E346K) spoIIIJ::kan mutS-gfp (spc) | This work |
| NMD49 | amyE::Pspank(hy)yabA (spc) mutSL::kan | This work |
| BWW51 | dnaN::PspacdnaN(G73R) (cat) | This work |
| BWW52 | dnaN::PspacdnaN(S181G) (cat) | This work |
| BWW53 | dnaN::PspacdnaN(E346K) (cat) | This work |
| BWW54 | dnaN::PspacdnaN+ (cat) | This work |
All of the strains used in this study are derivatives of PY79, except for IA24, which was obtained from the Bacillus Genetic Stock Center (BGSC).
Spontaneous mutation frequency.
Relative mutation frequencies were determined as described previously (64, 69). The mutation frequency of each strain was normalized to the mutation frequency of the wild-type control (PY79), except for Table 4, where the observed values are reported. The observed mutation frequency for PY79 averaged 3.4 × 10−9 ± 2.9 × 10−9 in all experiments.
TABLE 4.
Missense mutations S181G and E346K show wild-type phenotype for growth at 49°C and mutation frequency
| Allele | dnaN-encoded missense mutation | Frequency of Rifr mutants, 10−9 (fold increase)a | Plating efficiencyb |
|---|---|---|---|
| dnaN::PspacdnaN+ | None | 3.0 ± 0.5 (1) | 0.9 ± 0.075 |
| dnaN::PspacdnaN(G73R) | G73R | 183.7 ± 62 (61) | 0.005 ± 0.0005 |
| dnaN::PspacdnaN(S181G) | S181G | 8.4 ± 6.3 (2.8) | 1.0 ± 0.05 |
| dnaN::PspacdnaN(E346K) | E346K | 9.7 ± 6.2 (3.3) | 0.9 ± 0.07 |
Relative mutation frequency was determined as described in Materials and Methods, except that LB agar plates and plates containing rifampin were supplemented with 25 μM IPTG. The means and standard errors of the means from at least six independent experiments are shown.
Plating efficiency was determined by scoring the number of CFU following serial dilution and incubation at the indicated temperatures. Plates were supplemented with 5 μg/ml chloramphenicol and 25 μM IPTG. The means and standard errors of the means from at least three independent experiments are shown.
Isolation of dnaN5 suppressors.
Over 30 independent cultures with the dnaN5(G73R) allele were grown at 37°C in LB, followed by dilution and plating on LB agar plates and incubation at 49°C overnight. Temperature-resistant colonies that formed were colony purified and reexamined for growth at 49°C. Each isolate that retained the ability to grow at 49°C following colony purification was then grown at 37°C and diluted with a portion plated on LB agar to determine the viable count, while the remaining cells were plated on 100 μg/ml rifampin to determine the spontaneous mutation frequency. Isolates that showed a mutation frequency of ≥10-fold relative to the wild-type control were pursued. Chromosomal DNA was prepared from these isolates, and the dnaN gene from each isolate was PCR amplified, followed by sequence analysis using an Illumina Genome Analyzer (University of Michigan DNA sequencing core, http://seqcore.brcf.med.umich.edu). Primers for PCR amplification of dnaN are as follows: forward, 5′-GGAATTCCATATGAAATTCACGATTCAAAAAGATCGTCTTGTT; reverse, 5′-CGCGGATCCTTAATAGGTTCTGACAGGAAGGATAAG. For sequence analysis of the dnaN gene, we used primers 5′-TTCAATCTGCGGCAAGTGCGGATA, 5′-TGGATAATTCCCGTCCAGAAGCCGTGA, 5′-GTCGTGATTCCGGGAAAAAGTTTA, 5′-ATGAAATTCACGATTCAAAAA, 5′-ATTTTGGACTTCAATTTCTAC, 5′-ATGGCAACTGTAGAAATTGAA, 5′-ATCTTGACAGGTGTAAACTGG, 5′-CTTGTAGATATCGTCATCACA, 5′-AAACTGTCCGCAAAACCGGCT, and 5′-AGTCCAAAATATATGCTGGATGCA.
To determine if the mutations identified in the dnaN gene were responsible for growth at 49°C and an elevated mutation frequency, we integrated a selectable marker (Kanr) by double crossover into the spoIIIJ gene located at 359.9° on the chromosomal linkage map (2, 3). The dnaN gene is located nearby at 1.9° (54) (http://genolist.pasteur.fr/SubtiList/), allowing the cotransformation of each novel dnaN allele with the spoIIIJ::kan allele. We found that the spoIIIJ::kan allele is cotransformed with the dnaN5(G73R) allele at a frequency of ∼71% (by examining 102 colonies; data not shown). We transformed the wild type (PY79) with genomic DNA from each isolate, selecting for the spoIIIJ::kan allele, followed by screening for temperature-resistant growth at 49°C and an increase in mutation frequency, as determined by rifampin resistance, of at least 10-fold relative to the wild-type control. We found that an increase in mutation frequency cotransformed with growth at 49°C in 100% of the colonies examined for each intragenic suppressor isolated (data not shown). The dnaN allele from each newly constructed strain was then PCR amplified and sequenced to verify that each dnaN allele was indeed present in the strain cotransformed with the spoIIIJ::kan marker. After confirmation of the presence of each novel dnaN allele, the strain generated from each cross was used in all subsequent experiments. All of the strains used in this study are described in Table 1, and the missense mutations of intragenic suppressors of dnaN5(G73R) are described in Table 2.
TABLE 2.
Nucleotide sequence changes in dnaN alleles
| Allele and nucleotide change(s)a | Amino acid substitution(s)b | No. of occurrences |
|---|---|---|
| dnaN5 | ||
| 217GGA→217AGA | G73R | NAc |
| 741GGG→741GGA (silent) | ||
| dnaN34 | ||
| 217GGA→217AGA | G73R | NA |
| 741GGG→741GGA (silent) | ||
| dnaN(S22P, G73R) | ||
| 64TCA→64CCA | S22P, G73R | 1 |
| 217GGA→217AGA | ||
| 741GGG→741GGA (silent) | ||
| dnaN(G73R, S181G) | ||
| 217GGA→217AGA | G73R, S181G | 1 |
| 541AGC→541GGC | ||
| 741GGG→741GGA (silent) | ||
| dnaN(G73R, E346K) | ||
| 217GGA→217AGA | G73R, E346K | 1 |
| 1036GAA→1036AAA | ||
| 741GGG→741GGA (silent) |
The 5′ region of the dnaN5 allele was sequenced previously, showing the same missense mutation that we observed, 217GGA→217AGA (G73R) (53). In addition, we found a 741GGG→741GGA silent mutation in the dnaN5 allele. This region of the gene was not sequenced in the previous study (53). The silent mutation is not present in the wild-type dnaN gene that we sequenced from PY79. In addition to sequencing dnaN5, we also sequenced the dnaN34 allele from our lab stock and the dnaN34 allele from strain 1A24 obtained from the Bacillus Genetic Stock Center (www.bgsc.org/).
The amino acid substitutions listed were deduced from the nucleotide sequences.
NA, not available.
Plasmids.
pBW57 (PspacdnaN′) was constructed by placing the 5′ 501 bp of dnaN into plasmid pDH88 by ligation following digestion with HindIII and SphI. The primers used to PCR amplify the 5′ region of dnaN are as follows: forward, 5′-CGCCCCAAGCTTAAGGAGGTATACATATGAAATTCACGATTCAA AAAGATCGTCTT; reverse, 5′-ACATGCATGCCCAGTTTACACCTGTCAAGATAGGGCGTG. Integration of pBW57 into strain NMD11 bearing the dnaN5 allele replaced the G73R missense mutation with the wild-type coding sequence in 95% of the colonies (40/42) examined.
Homology modeling.
Homology modeling was done as previously described (64), except that the structure of the β clamp from Streptococcus pneumoniae was used as a template (Protein Data Bank [PDB] accession number 2awa; http://www.rcsb.org). Each of the amino acid residues examined in this work was highlighted on the homology model of the B. subtilis β clamp by using PyMol (http://pymol.sourceforge.net/).
Mutation spectrum in rpoB.
Independent cultures of each strain indicated were grown in LB medium with appropriate antibiotics until late exponential phase (optical density at 600 nm of 1.0) and then plated on LB agar plates containing 100 μg/ml rifampin. A single colony was removed from each plate and colony purified on LB agar plates containing rifampin; this was followed by DNA purification using either standard phenol-chloroform extraction or the DNeasy DNA isolation kit (Qiagen). A 359-bp PCR product representing cluster 1 of the rpoB gene was amplified, using forward primer 5′-CGTATCGGTTTAAGCCGTATG and reverse primer 5′-AAAACGGTTTACTTTTGCATA, from bp 1261 to bp 1620. The PCR products were purified (Qiagen) and then sequenced (University of Michigan DNA sequencing core; http://seqcore.brcf.med.umich.edu) using forward primer 5′-TTCTTTGGAAGCTCA and reverse primer 5′-TCCGGCACGCTCACG. Sequence data were analyzed using software (Sequencher) to determine the nucleotide substitutions.
Immunoblotting.
Immunodetection of the β clamp was performed as described previously (64, 67, 68). Briefly, 50-ml cultures of the strains indicated were grown in LB to the early exponential phase, an optical density at 600 nm of 0.2 at 30°C. Ten-milliliter volumes of cells were removed prior to the temperature shift as the zero time point. Cells were then shifted to 49°C, followed by the removal of 10 ml of culture at each time point indicated in Fig. 1. Cells were resuspended in lysis buffer as described previously (56), separated by 13% sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and transferred to nitrocellulose as described previously (68). For the immunoblot shown in Fig. 2, each strain was grown to the late exponential phase, an optical density at 600 nm of 0.7 to 1.0, in LB medium for each of the different mutant β clamp proteins shown. For all of the immunoblots, a 1:5,000 dilution of the primary antibody against the β clamp (kindly provided by Alan Grossman, Massachusetts Institute of Technology) and a 1:1,000 dilution of the horseradish peroxidase-conjugated goat anti-rabbit secondary antibody (Pierce) were used.
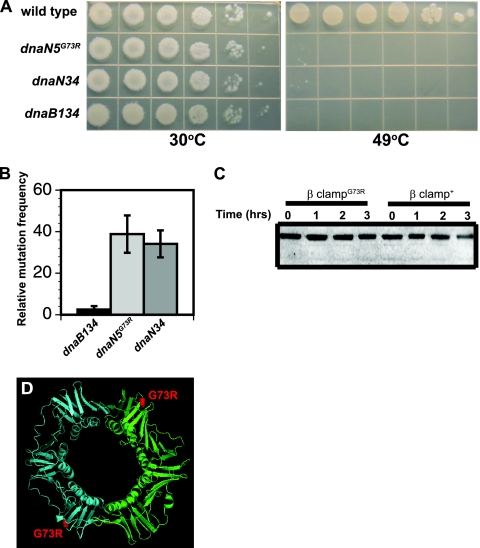
FIG. 1.
The β clampG73R protein accumulates in vivo. (A) Growth of wild-type PY79 and isogenic strains with the dnaN5, dnaN34, and dnaB134 alleles. Tenfold serial dilutions of each strain were plated and grown at the indicated temperatures. Pictures were captured following 14 h of growth. Representative plates from several experiments are shown. (B) Relative mutation frequencies (n-fold increases above the wild-type level) of strains carrying the dnaB134, dnaN5(G73R), and dnaN34 alleles determined at 37°C. The histogram represents the mean ± the standard error of the mean from at least four independent experiments. (C) Representative immunoblot assay of the β clampG73R mutant and wild-type β clamp proteins at 30°C and following a temperature shift to 49°C for the times indicated. The total loaded sample was normalized to cell number based on the optical density of each culture. (D) Ribbon model of the B. subtilis β clamp. One protomer is cyan, and the second protomer is green. The predicated location of the G73R mutation is shown as red spheres.
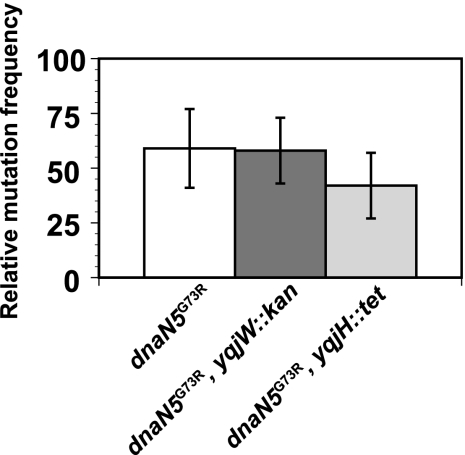
FIG. 2.
Disruption of Y family DNA polymerases does not alter the mutagenesis caused by dnaN5(G73R). The mean relative mutation frequencies of strains with the indicated alleles are shown in the histogram. Error bars show the standard error of the mean for 8 to 17 independent experiments. The mutation frequency of each strain was normalized to the mutation frequency of the wild-type control strain PY79.
Live-cell microscopy.
Cells were prepared for live imaging essentially as described previously (38, 65, 66). Briefly, 300-μl aliquots of cultures grown at 30°C in S750 medium were incubated with the vital membrane stain FM4-64 (1 μg/ml in phosphate-buffered saline; Molecular Probes) (65, 66). Following incubation, cells were allowed to settle on microscope slides containing 1% agarose pads prepared with 1× Spizizen salts (65, 66). Cells were imaged using an Olympus BX61 microscope equipped with a Hamamatsu ORCAR2 cooled charge-coupled device camera and a Lumen 200 arc metal light source (Prior). An Olympus 100× oil immersion 1.45 numerical aperture TIRFM objective lens was used. For detection of GFP and FM4-64, the filter sets used were for fluorescein isothiocyanate excitation at 460 to 500 nm and emission at 510 to 560 nm and tetramethyl rhodamine isocyanate excitation at 510 to 560 nm and emission at 572 to 648 nm. Images were captured and processed using SlideBook 4.2 (Advanced Imaging Software). Cells were scored using Photoshop (Adobe) or the publically available software ImageJ (http://rsbweb.nih.gov/ij/). Images were assembled into figures using Illustrator (Adobe).
RESULTS
dnaN5(G73R) and dnaN34 are identical.
Two dnaN alleles have been isolated in B. subtilis: dnaN5(G73R) and dnaN34 (36) (www.bgsc.org/). We moved the dnaN5(G73R) and dnaN34 alleles into a clean genetic background (PY79) to examine the phenotypes they produce. We found that both dnaN5(G73R) and dnaN34 produced the same level of survival when plated at elevated temperatures (Fig. 1A). The level of survival observed for dnaN5(G73R)- and dnaN34-containing strains was nearly identical to what we observed for the dnaB134(Ts) temperature-sensitive growth allele-carrying strain defective in replication initiation at temperatures of >45°C (9).
We have previously shown that dnaN5(G73R) conferred an increase in mutation frequency (64). We examined the spontaneous mutation frequencies of strains bearing dnaN5(G73R) and dnaN34 with dnaB134 as a negative control. We found that dnaN5(G73R) and dnaN34 conferred nearly identical mutation frequencies at 37°C (Fig. 1B). In contrast, the mutation frequency conferred by dnaB134 was very low (Fig. 1B). Because the phenotypes caused by both dnaN5(G73R) and dnaN34 were so similar, we sequenced both alleles from our lab stocks (Table 2). We found that the dnaN5 and dnaN34 alleles encode a G73R missense mutation at nucleotide 217 (Table 2) (53). The previous study that identified the G73R missense mutation encoded by dnaN5 did not sequence the entire dnaN gene (53). To our knowledge, the dnaN34 allele has not previously been sequenced. Our sequencing of the entire coding region revealed that dnaN5 and dnaN34 also contained a silent mutation of 741GGG to 741GGA (Table 2), and this silent mutation was present in all of the dnaN5 and dnaN34 mutant isolates that we possess. Sequencing of the dnaN34 allele from a strain obtained from the Bacillus Genetic Stock Center (www.bgsc.org) also showed the G73R missense mutation at nucleotide 217, as well as the 741GGG-to-741GGA silent mutation (Table 2). Based on the phenotypes and sequencing of all of the dnaN5 and dnaN34 isolates we possess, we conclude that these alleles are identical (see Discussion).
The β clampG73R protein accumulates in vivo.
As mentioned above, B. subtilis cells harboring the dnaN5(G73R) allele are temperature sensitive for growth at 49°C (Fig. 1A and Table 3). One explanation for the dnaN5(G73R) phenotype is that G73R in the β clamp causes instability, resulting in protein unfolding and proteolytic degradation at elevated temperatures. To address this, we performed immunoblotting of the wild-type β clamp and β clampG73R proteins in whole-cell extracts at 30°C and following a temperature shift to 49°C in 1-h increments over a 3-h time course (see Materials and Methods). We found that the wild-type β clamp and β clampG73R proteins accumulated to similar levels at 30°C. When cells were shifted to 49°C, the β clamp+ and β clampG73R levels showed similar levels of abundance in vivo (Fig. 1C). Thus, the β clampG73R protein accumulates at 49°C. We interpret these results to mean that the β clampG73R protein is stable in vivo.
TABLE 3.
Suppression of the temperature-sensitive growth phenotype caused by dnaN5(G73R)a
| Strain (relevant allele) | Amino acid substitution in β clamp | Plating efficiency |
|---|---|---|
| PY79 (dnaN+) | None (wild type) | 1.2 ± 0.03 |
| LAS393 (ΔmutL) | None (wild type) | 1.1 ± 0.1 |
| NMD11 (dnaN5) | G73R | 9.3 × 10−5 ± 4.2 × 10−5 |
| NMD34 [dnaN(S22P, G73R)] | S22P, G73R | 0.9 ± 0.1 |
| NMD32 [dnaN(G73R, S181G)] | G73R, S181G | 0.8 ± 0.1 |
| NMD33 [dnaN(G73R, E346K)] | G73R, E346K | 0.9 ± 0.1 |
Cells were grown to mid-exponential phase (optical density at 600 nm of 0.5), diluted, and plated on LB agar plates at the indicated temperatures. The plating efficiency reflects the number of observed colonies at 49°C divided by the number of observed colonies at 30°C. The means and standard deviations from at least four independent experiments are shown.
The predicted location of G73R is shown on a homology model of the B. subtilis β clamp (Fig. 1D). The position of this amino acid is in a surface-exposed loop. This position is more consistent with a role in protein-protein interaction than with a role in protein stability. It should be noted that the model of the B. subtilis β clamp is elliptical and is not circular like the crystal structures of the E. coli β clamp (24, 31, 39). This is because our B. subtilis β clamp model is based upon the elliptical Gram-positive S. pneumoniae β clamp structure (www.rcdb.org; PDB accession no. 2awa). In addition, the crystal structure of the β clamp from a second Gram-positive bacterium, S. pyogenes, is also elliptical (1). Therefore, although many of the structural characteristics of the β clamp are conserved, differences in domain placement and domain linkers contribute to the elliptical shape of the β clamp from Gram-positive bacteria (1) (www.rcdb.org; PDB accession no. 2awa).
Translesion DNA polymerases do not contribute to the mutagenesis caused by dnaN5(G73R).
In E. coli, mutation of the β clamp at residue G66 effects utilization of the translesion DNA polymerase pol IV (48, 73). Since B. subtilis dnaN5(G73R) is altered at residue G73, which corresponds to E. coli β clamp residue G66, we investigated the contribution of translesion DNA polymerases to the mutagenesis observed with dnaN5(G73R) (Fig. 2). The B. subtilis homolog of pol IV is polY1 (yqjH), and the homolog of the UmuC component of pol V in B. subtilis is polY2 (yqjW) (17, 72). B. subtilis is not known to have a homolog or analog of E. coli UmuD (17, 72). We combined null alleles of yqjH::tet (polY1) and yqjW::kan (polY2) with dnaN5(G73R) to determine if disruption of either Y family DNA polymerase lowers the mutation frequency of dnaN5(G73R). If so, it would suggest that one or both of the Y family DNA polymerases contributes to the increase in mutagenesis associated with dnaN5(G73R). To the contrary, we found that the mutation frequency of dnaN5(G73R) was unaffected when either polY1 (yqjH) or polY2 (yqjW) was disrupted (Fig. 2). These results show that B. subtilis polY1 and polY2 do not contribute to the mutagenesis conferred by the dnaN5(G73R) allele.
dnaN5(G73R) has a slight effect on replication initiation.
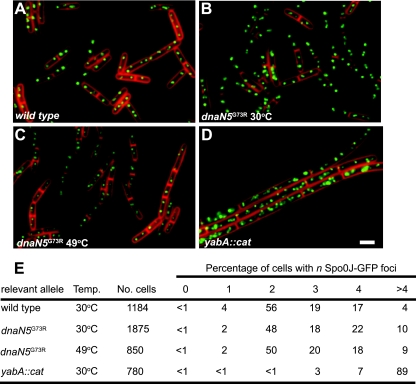
Given that the B. subtilis β clamp binds the YabA protein, which negatively regulates DNA replication initiation (12, 27, 29, 52), we tested whether dnaN5(G73R) alters replication initiation. We examined the number of origins per cell in the dnaN5(G73R) genetic background to determine if the G73R missense mutation has any pleiotropic effects on DNA replication initiation. To determine the number of replication origins per cell, we used Spo0J-GFP as a marker for oriC, because it binds to sequences that flank oriC and Spo0J-GFP foci are a well-established marker for determining origin number per cell (44, 67). We found that the percentage of cells with one origin marked by a Spo0J-GFP focus was lower in a dnaN5(G73R) strain and the percentage of cells with four or more Spo0J-GFP foci was elevated in a dnaN5(G73R)-bearing strain compared with that in the wild-type control (Fig. 3A, B, and C). This result suggests that dnaN5(G73R) causes a mild overinitiation phenotype that is revealed when origins are scored in live cells at 30°C. As a positive control for overinitiation, we scored Spo0J-GFP foci in a yabA::cat mutant strain and found that the percentage of cells with more than four origins per cell was increased over 20-fold and the percentage of cells with one, two, three, and four origins was decreased substantially (Fig. 3D and E) at 30°C. Thus, we find that dnaN5(G73R) causes a slight increase in the percentage of cells with four or more origins, but this allele does not cause an overinitiation phenotype of a magnitude anywhere near that observed in yabA-deficient cells.
FIG. 3.
dnaN5(G73R) causes slight overinitiation of DNA replication. For all of the images, Spo0J-GFP is green and FM4-64 vital membrane staining is red. Cells were imaged during exponential growth (optical density at 600 nm of ∼0.5) according to Materials and Methods. Panels: A, wild-type dnaN+; B, dnaN5(G73R) at 30°C; C, dnaN5(G73R) at 49°C; D, yabA::cat; E, scoring of the percentage of cells with n Spo0J-GFP foci in the indicated genetic backgrounds and temperatures as shown. The number of cells scored is also indicated. Bar, 3 μm.
We considered the possibility that the lethality caused by dnaN5(G73R) was due to unregulated replication initiation at 49°C. We performed a time course experiment at 49°C and found no difference in origin copy number as judged by Spo0J-GFP foci after a shift to 49°C for 30 min (data not shown) and 1 h (Fig. 3C and E). Based on these data, we conclude that dnaN5(G73R) does not cause an increase in replication initiation at 49°C above that observed at 30°C (Fig. 3).
Isolation of intragenic suppressors of dnaN5.
The dnaN5(G73R) allele was isolated and shown to have defects in DNA replication at the nonpermissive temperature of 49°C (36). We have previously shown that the dnaN5(G73R) allele confers an increase in spontaneous mutation frequency that is MMR dependent at the permissive temperatures of 30°C and 37°C (64). As part of our effort to understand the role of the β clamp in MMR, we sought to isolate intragenic suppressors of dnaN5(G73R) that would restore DNA replication at 49°C yet retain an elevated mutation frequency. In doing so, mutant β clamp proteins of this type would cause an MMR deficiency, but unlike dnaN5(G73R), such dnaN alleles would be proficient in DNA replication.
To this end, we selected for, mapped, and sequenced (see Materials and Methods) 30 independent isolates of a dnaN5(G73R)-bearing strain that grew at 49°C. Most of the temperature-resistant isolates acquired mutations that did not map near the dnaN gene, indicating that they were extragenic (data not shown). Of those that did map near the dnaN gene, we identified five isolates that harbored nucleotide changes in dnaN5(G73R). Two were wild-type revertants which changed the arginine of dnaN5(G73R) back to a glycine (data not shown). Three other isolates had a single missense mutation in addition to the G73R missense mutation that is part of dnaN5 (Table 2). We isolated dnaN(S22P, G73R), dnaN(G73R, S181G), and dnaN(G73R, E346K), which suppressed the growth defect of the dnaN5(G73R) mutant at 49°C (Table 2 and 3).
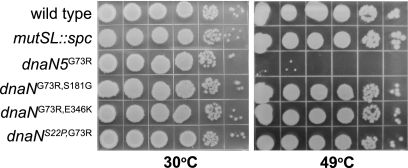
Quantitative plating efficiencies of strains containing each dnaN allele showed that each allele supported the growth of B. subtilis cells to nearly wild-type levels (Table 3). Each dnaN allele was able to support growth at 80% to 90% of the level of the wild type, conferring a plating efficiency of 0.8 to 0.9 when CFU counts were determined for growth at 49°C compared to the CFU counts determined for growth at 30°C (Table 3). Thus, the intragenic suppressors rescued the G73R growth defect by >8,000-fold. Upon serial dilution and plating of strains which contain each dnaN allele, we found that even though the colony size was slightly smaller than that of the wild type, the overall growth and colony morphology returned to nearly wild-type levels, indicating very good suppression of the temperature-sensitive growth phenotype (Fig. 4).
FIG. 4.
Intragenic suppressors restore growth of B. subtilis at 49°C. Shown is the growth of the wild-type and isogenic strains with the indicated alleles [dnaN(G73R) and suppressors] at 30°C and 49°C following 10-fold serial dilution and plating on LB agar plates. Pictures were taken following 14 h of growth. Representative plates from several experiments are shown.
The G73R mutation is located on the outside rim of the β clamp ring (Fig. 1D) in a region of the β clamp to which many proteins are hypothesized to bind, including the clamp loader proteins δ and δ′ (34, 35). The β clamp has an N-terminal face and a C-terminal face, and most current models predict that proteins that bind the β clamp do so on the C-terminal face. The C-terminal face contains the hydrophobic cleft, which serves as a critical binding site for many proteins (5, 8, 14, 18, 30, 31, 48, 49, 62, 73).
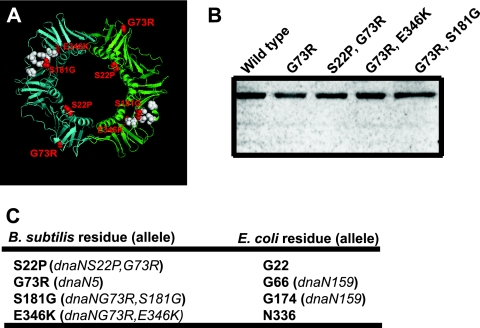
All three missense mutations isolated as intragenic suppressors of the temperature sensitivity caused by dnaN5(G73R) are predicted to change amino acid residues located on the surface of the β clamp, and two suppressors are located near the hydrophobic cleft (Fig. 5A). E346K is predicted to be located at the beginning of a β strand on the N-terminal face of the β clamp directly behind the hydrophobic cleft. S181G is located adjacent to the hydrophobic cleft on the C-terminal face of the β clamp (Fig. 5A). S22P is in a loop located in the inner ring near sites important for DNA interaction at a site distant from the hydrophobic cleft (Fig. 5A). With these data, we speculate that E346K and S181G might be important for interaction between the β clamp and other proteins, while S22P may effect interaction of the β clamp with DNA. We examined the levels of these proteins in vivo and found that each DnaN (β clamp) protein accumulated to wild-type levels (Fig. 5B). Upon aligning the primary structures of the B. subtilis and E. coli β clamp, we found a distinct correlation between the β clamp residues examined in this study and those altered in the E. coli dnaN159 allele (48) (Fig. 5C; see Discussion).
FIG. 5.
Locations of intragenic suppressors of dnaN5(G73R). (A) Ribbon diagram of the B. subtilis β clamp homology model. One protomer is cyan, and the other is green. The predicted positions of S22P, G73R, S181G, and E346K are shown in red, and the amino acids comprising the hydrophobic cleft are white. (B) Representative immunoblot assay of each β clamp protein indicated from cultures grown at 30°C. The loaded sample was normalized to cell number. (C) Comparison of the amino acid substitutions examined in this study and the corresponding E. coli β clamp residue based on alignment of the primary structures. The mutation-bearing allele, if it is known, is in parentheses.
S22P, S181G, and E346K alter the level of spontaneous mutagenesis compared to the G73R missense mutation.
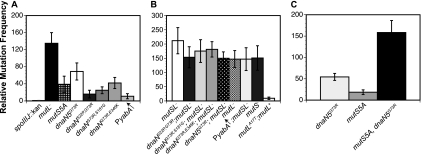
Since the G73R change encoded by dnaN5 affected both DNA replication and MMR, the selection was designed to find suppressors of dnaN(G73R) that would restore growth and, hypothetically, rescue DNA replication. We also asked if the intragenic suppressors of dnaN5(G73R) alter the MMR pathway. When bacteria are grown at 37°C, the relative mutation frequency conferred by dnaN5(G73R) is enhanced by ∼60-fold relative to that of our wild-type control, strain PY79 (Fig. 6A). All three of the dnaN alleles isolated retained an increase in the spontaneous mutation frequency of 20- to 40-fold above that of the wild-type control (Fig. 6A). The dnaN(S22P, G73R) allele was the lowest, showing a 20-fold increase, and dnaN(G73R, E346K) was the highest at ∼40-fold. We found that dnaN(G73R, S181G) conferred an increase in mutation frequency between those obtained with dnaN(S22P, G73R) and dnaN(G73R, E346K). For comparison, we examined the mutation frequency caused by a five-alanine substitution of the β clamp binding motif in MutS (mutS5A) and found that this mutS allele conferred a mutation frequency of ∼40-fold, which is within the error of the dnaN alleles isolated here (Fig. 6A). We speculate that, like the mutS5A allele, each dnaN allele containing the G73R mutation is reduced but not completely defective for interaction with MutS (64).
FIG. 6.
Intragenic suppressors of dnaN5(G73R) maintain an MMR-dependent increase in mutation frequency. (A) The mean relative mutation frequencies of strains with the indicated alleles are presented. The mutation frequency of each strain was normalized to the mutation frequency of wild-type control strain PY79. (B) Relative mutation frequency of each strain indicated after the introduction of an MMR-defective allele (mutSL::spc) in combination with each dnaN allele. For comparison, we show these relative to the mild dominant negative effect of mutL with the A17T missense mutation expressed from the amyE locus under Pspac control. (C) Relative mutation frequencies of the dnaN5(G73R) mutant, the mutS5A mutant, and the double mutant. The bars in the histograms represent the mean ± the standard error of the mean for 6 to 20 independent experiments.
S181G and E346K are similar to dnaN+ for growth at 49°C and mutation frequency.
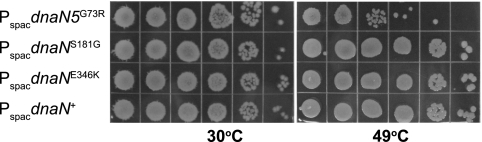
Because S181G and E346K are located near the hydrophobic cleft in the β clamp, these suppressor mutations are the most likely to have roles in protein-protein interactions that have suppressed the replication defect caused by G73R. To examine the roles of S181G and E346K in DNA replication and MMR, we separated these missense mutations from G73R. In order to separate the suppressor mutations, we replaced the 5′ region of the DNA of the alleles dnaN(G73R, S181G) and dnaN(G73R, E346K) with the wild-type dnaN coding sequence, removing the G73R missense mutation and placing the resultant alleles [dnaN(S181G) and dnaN(E346K)] under the control of an isopropyl β-d-thiogalactopyranoside (IPTG)-inducible promoter, Pspac, at their normal chromosomal locations (see Materials and Methods). The proper nucleotide sequence of each allele was then confirmed by PCR amplification and DNA sequence analysis (see Materials and Methods). We also placed dnaN+ and dnaN5(G73R) under Pspac control for comparison (Table 4). We found that the dnaN5(G73R) mutant was temperature sensitive for growth at 49°C and showed an increase in its mutation frequency (Table 4). The temperature-sensitive growth phenotype produced by dnaN5(G73R) is retained but less severe under Pspac control than when dnaN5(G73R) is under native transcriptional control (Fig. 7 and Table 4). We tested dnaN(S181G) and dnaN(E346K) and found that both alleles conferred growth at elevated temperatures (Fig. 7 and Table 4) and produce mutation frequencies very similar to that produced by wild-type dnaN+ (Table 4). We conclude that missense mutations S181G and E346K restore DNA replication at 49°C and have very little effect on MMR when separated from G73R.
FIG. 7.
Missense mutations S181G and E346K restore growth of B. subtilis at 49°C. Shown is the growth of each strain with the dnaN allele indicated under Pspac control at the native locus following 10-fold serial dilutions and at the temperatures indicated. Representative plates from several experiments are shown.
The mutation spectrum resulting from dnaN(G73R, S181G) and dnaN(G73R, E346K) is consistent with an MMR defect.
The antibiotic rifampin impairs transcription by directly binding the β subunit of RNA polymerase encoded by the rpoB gene (21, 61). Mutations in the rpoB gene confer rifampin resistance (Rifr) (22), and in B. subtilis, ∼96% of the rpoB mutations resulting in Rifr are located in a 50-bp region of cluster I. The majority of these mutations alter codons for two amino acids, Q469 and H482 (47, 50). The mutation spectrum of Rifr mutations in rpoB has been widely used as a diagnostic tool to understand the pathway that has caused the Rifr mutagenesis based on the nature of the base pair substitution (10, 22). For example, most of the mutations resulting from defects in MMR are transition mutations. In contrast, SOS mutagenesis generates a higher occurrence of transversion mutations (10, 22).
We selected for and colony purified at least 20 independent Rifr colonies of strains bearing the dnaN5(G73R) allele, an MMR-defective strain (mutSL::spc), and strains with each dnaN allele isolated in this study. We found that strains with either the dnaN5(G73R) or the mutSL::spc allele had a mutation spectrum of primarily AT→GC transition mutations (Table 5), which is indicative of an MMR defect (22, 59). Likewise, we determined the mutation spectrum of each dnaN allele isolated here, and in a pattern similar to that of dnaN5(G73R) and the mutSL-deficient strains, the novel dnaN alleles mostly showed transition mutations with a bias for AT→GC transitions (Table 5). As a control, we determined the mutation spectrum for Rifr colonies from wild-type PY79 and found that ∼20% were transversion mutations (Table 5), suggesting an MMR-independent mechanism. Furthermore, we challenged PY79 with the alkylating agent mitomycin C and also isolated a much higher percentage of transversion mutations relative to the MMR-defective strain or the mutation spectrum conferred by each dnaN allele examined in this study (Table 5). It should be noted that dnaN(S22P, G73R) and dnaN(G73R, S181G) each yielded one transversion mutation out of 20 colonies examined (Table 5), but this frequency of transversion mutations (∼5%) is much lower than the frequency of transversion mutations isolated from untreated wild-type PY79 or PY79 treated with mitomycin C. We conclude that the mutation spectrum in the rpoB gene from each dnaN allele suggests a deficiency in MMR.
TABLE 5.
Mutation spectrum in cluster I of the rpoB genea
| Strain, relevant allele, and rpoB nucleotide change | Deduced amino acid change | No. of occurrences |
|---|---|---|
| NMD11, dnaN5(G73R) | ||
| 1406 AT→GC | Q469R | 2 |
| 1444 CG→TA | H482Y | 1 |
| 1445 AT→GC | H482R | 17 |
| NMD22, dnaN(S22P, G73R) | ||
| 1406 AT→GC | Q469R | 13 |
| 1444 AT→GC | H482R | 2 |
| 1444 CG→GC | H482Db | 1 |
| 1445 AT→GC | H482R | 5 |
| NMD14, dnaN(G73R, S181G) | ||
| 1405 CG→AT | Q469Kb | 1 |
| 1406 AT→GC | Q469R | 2 |
| 1444 CG→TA | H482Y | 2 |
| 1445 AT→GC | H482R | 15 |
| NMD17, dnaN(G73R, E346K) | ||
| 1444 CG→TA | H482Y | 2 |
| 1445 AT→GC | H482R | 18 |
| LAS392, mutSL::spc | ||
| 1406 AT→GC | Q469R | 11 |
| 1444 CG→TA | H482Y | 2 |
| 1445 AT→GC | H482R | 7 |
| PY79 (untreated), wild type | ||
| 1405 CG→AT | Q469Kb | 3 |
| 1406 AT→GC | Q469R | 6 |
| 1444 CG→TA | H482Y | 1 |
| 1445 AT→GC | H482R | 7 |
| 1444 CG→GC | H482Db | 1 |
| PY79 (MMC), wild type | ||
| 1406 AT→GC | Q469R | 12 |
| 1445 AT→GC | H482R | 3 |
| 1444 CG→TA | H482Y | 2 |
| 1405 CG→AT | Q469Kb | 3 |
For mitomycin C treatment, PY79 cells were grown to the mid-exponential phase (optical density at 600 nm of 0.5) and then challenged with 0.2 μg/ml mitomycin C for 1 h, whereupon cells were plated on 100 μg/ml rifampin to obtain Rifr colonies. Colonies were then analyzed as described in Materials and Methods.
Transversion mutation.
dnaN alleles maintain partial defects in MMR.
The rpoB mutation spectrum resulting from all four dnaN alleles [dnaN5(G73R), dnaN(S22P, G73R), dnaN(G73R, S181G), and dnaN(G73R, E346K)] suggests a deficiency in MMR. To determine more directly if the increase in mutation frequency caused by each dnaN allele is MMR dependent, we performed an epistasis analysis. If the increase in mutation frequency observed for each dnaN allele is through the MMR pathway, then the mutation frequency caused by an allele conferring a complete loss of MMR (mutSL::spc) would be epistatic to each dnaN allele. In contrast, if the mutation frequency resulting from each dnaN allele is additive or synergistic with the mutSL::spc allele, then the mutagenesis caused by each dnaN allele would be through a different pathway.
To distinguish between these possibilities, we combined the mutSL::spc allele with each of the dnaN alleles we isolated and determined the mutation frequency relative to that of a strain harboring the mutSL::spc allele alone (Fig. 6B). We found that the mutation frequency conferred by each dnaN allele in combination with an MMR defect did not produce an increase in mutation frequency above the level conferred by the mutSL::spc allele alone (Fig. 6B). Additive or synergistic effects on the frequency of mutagenesis can be detected using this assay (11, 32). Furthermore, we combined the dnaN5(G73R) allele with the mutS5A allele and determined the mutation frequency. Because dnaN5(G73R) and mutS5A each confer partial defects in the MMR pathway, we expected that the combination of these alleles would increase the level of mutagenesis above that conferred by the individual alleles. Indeed, we found that the sum of two partial defects in the MMR pathway was greater than the defects on their own, further confirming that we can detect an increase in mutagenesis with this assay (Fig. 6C). These results show that a complete loss of MMR is epistatic to each dnaN allele, and we conclude that the mutagenesis observed is caused by an MMR deficiency.
In addition to these experiments, we measured the sensitivity of the wild-type control PY79 and stains carrying each of the dnaN alleles to nalidixic acid and mitomycin C. Nalidixic acid inhibits DNA gyrase, causing double-strand breaks (55, 71). Mitomycin C primarily generates bulky N2 adducts on guanine (reviewed in reference 16). We found no difference in sensitivity between strains bearing each dnaN allele and the wild-type control (data not shown). We conclude that dnaN5(G73R) and the dnaN alleles isolated in this study do not cause general defects in DNA repair, as judged by sensitivity to two different DNA-damaging agents.
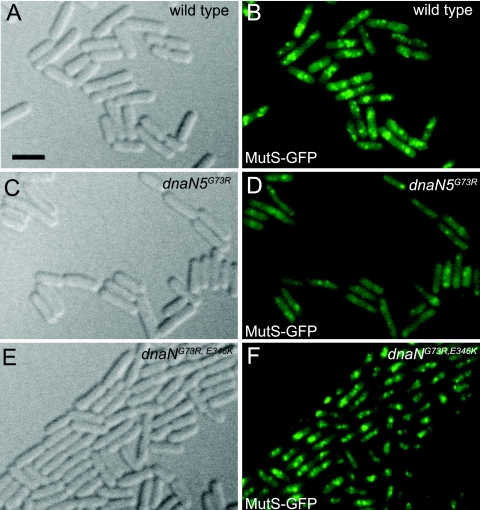
dnaN alleles restore MutS-GFP focus formation.
We have previously shown that MutS-GFP focus formation is reduced ∼3-fold in a dnaN5(G73R)-bearing strain (64). We analyzed MutS-GFP for localization as a focus in response to a 2-aminopurine (2-AP) challenge in the wild-type control dnaN+, dnaN5(G73R), and dnaN(G73R, E346K) backgrounds (Fig. 8). We found that MutS-GFP was reduced for localization as a focus in the dnaN5(G73R) mutant strain, as we previously reported (64) (Fig. 8D). We also found that in the dnaN5(G73R) mutant strain, MutS-GFP fluorescence was more diffuse and cytoplasmic than in the dnaN+ control (compare Fig. 8B to D). Imaging of MutS-GFP foci in the strain bearing dnaN(G73R, E346K) showed qualitatively that repair complex formation was mostly restored (compare Fig. 8D to F). In addition, the dnaN(G73R, S181G) allele, which conferred an increase in mutation frequency that was MMR dependent (Fig. 6), again qualitatively restored MutS-GFP focus formation in a manner similar to focus formation in a strain bearing the dnaN(G73R, E346K) allele (data not shown). Thus, we show that the intragenic suppressor mutations isolated in dnaN5(G73R) support focus formation of MutS-GFP; however, the repair complexes formed in these genetic backgrounds do not function as efficiently in MMR as those formed in the presence of dnaN+. We were not surprised that MutS-GFP focus formation was restored by the presence of the secondary mutations in dnaN5(G73R) because each caused a decrease in mutation frequency compared with the G73R missense mutation alone (Fig. 6A).
FIG. 8.
dnaN alleles are restored for support of MutS-GFP focus formation. Panels A, C, and E are differential interference contrast images showing cell boundaries. Images B, D, and F are MutS-GFP foci following treatment with 2-AP at 600 μg/ml for 1 h. Panels: A and B, MutS-GFP, dnaN+, 2-AP; C and D, MutS-GFP, dnaN5(G73R), 2-AP; E and F, MutS-GFP, dnaN(G73R, E346K), 2-AP. Bar, 3 μm.
Overexpression of YabA causes a mild increase in mutation frequency.
The β clamp is responsible for coordinating many activities at the replication fork. Also, the β clamp interacts with YabA, a negative regulator of replication initiation in B. subtilis (12, 27, 29, 52). Overexpression of the YabA protein causes a decrease in replication initiation, and deletion of yabA increases replication initiation (12, 29, 52) (Fig. 3). It has been shown that mutations in YabA that block interaction with the β clamp cause an increase in replication initiation and mutant GFP-YabA fails to localize with the replisome in live cells (52).
One model predicts that many proteins that bind the β clamp do so at sites that partially overlap. Therefore, we hypothesized that the YabA binding site on the β clamp might overlap with the binding site for MutS or MutL, thus changing MMR efficiency. To address this, we asked if the presence or absence of yabA influences mutation frequency in B. subtilis. We found that overexpression of yabA caused a mild but reproducible increase in mutation frequency of ∼11-fold (Fig. 6A). We found that cells deficient for yabA exhibited a hypomutation frequency as the yabA::cat allele conferred a lower mutation frequency than the wild-type control (data not shown). Moreover, we found that combining yabA overexpression with an MMR defect (mutSL) was not additive or synergistic, suggesting that the elevated mutation frequency caused by yabA was MMR dependent (Fig. 6B). Thus, we speculate that yabA levels can alter the efficiency of MMR, most likely by limiting the access of MutS or MutL to the β clamp in vivo (see Discussion).
DISCUSSION
In this work, we investigated the defects in DNA replication and repair caused by B. subtilis dnaN5 and dnaN34. Strikingly, we found that dnaN5 and dnaN34 were identical as determined by phenotype and nucleotide sequence (Fig. 1A and B and Table 2). It is not clear if these alleles were isolated independently, although our results show that, in addition to encoding the G73R missense mutation, both dnaN5(G73R) and dnaN34(G73R) encode the same silent mutation, suggesting that their isolation may not have been independent. We certainly cannot exclude the possibility that the original alleles isolated were different and at some point since their isolation, the dnaN5 and dnaN34 alleles have been cataloged incorrectly. Since dnaN5(G73R) and dnaN34(G73R) are currently used and widely available, it should be noted that the nucleotide sequences are the same for the strains we have obtained.
The DNA replication defect caused by dnaN5(G73R) at elevated temperatures is unknown. In order to understand the role of amino acid residue G73 in DNA replication and MMR, we selected for suppressors of the temperature-sensitive growth phenotype produced by dnaN5(G73R). We isolated three intragenic suppressors with the missense mutations S22P, S181G, and E346K. S22P is located in a loop in the inner ring of the β clamp near an area of importance for β clamp interaction with DNA (31). Although the corresponding residue in the E. coli β clamp, G22, does not contact DNA in the crystal structure (24, 31), this does not necessarily indicate that G22 does not have a role in DNA interaction in vivo. The S181G missense mutation is located on the edge of the hydrophobic cleft. The hydrophobic cleft is a common interaction site for many proteins that bind the β clamp. The very isolation of S181G as a missense mutation to suppress the temperature-sensitive growth of dnaN5(G73R) strongly suggests that the defect caused by G73R is in protein-protein interaction. It is not known if the G73R missense mutation effects interaction with just a single DNA replication protein or many, but since this location on the β clamp is important for function of the MMR pathway, we suggest that G73 is located in a region of the β clamp where binding sites for several proteins may overlap. In support of this notion, it has been shown that the corresponding residue in the E. coli β clamp, G66, is critical for DNA polymerase switching in vivo and in vitro (48).
The isolation of E346K to suppress G73R is of interest. Most models suggest that the β clamp binding proteins compete for one face of the β clamp. Residue E346 is located on the N-terminal face of the β clamp directly behind the hydrophobic cleft. One explanation is that E346K causes a local alteration in protein structure that changes the C-terminal face of the β clamp in the hydrophobic cleft, altering the way in which proteins bind this region. Another possibility is that the E346K mutation reveals a binding site for proteins on the N-terminal face of the β clamp. Taken together, our findings show that two of the three intragenic suppressor mutations of dnaN5(G73R) reside near the hydrophobic cleft of the β clamp with S181G adjacent to the cleft.
The DNA replication defect caused by the β clampG73R mutation is unknown.
Since the location of the G73R substitution in the B. subtilis protein is near a site identified as critical for the loading of the E. coli β clamp, we sequenced the yqeN (holA) and holB genes coding for δ and δ′, respectively. We sequenced the holA (δ) and holB (δ′) genes from many of the strains we classified as containing extragenic suppressors. We did not identify mutations in either of these genes, suggesting that the defect caused by G73R is not suppressed by mutations in δ or δ′ (data not shown). This does not exclude a defect in clamp loading as contributing to or causing the temperature-sensitive growth of the strain with β clampG73R.
However, since we were unable to find mutations in these genes, we suggest that most of the extragenic suppressors are likely to be informational. Informational suppressors are often obtained as extragenic suppressors of B. subtilis replication initiation alleles, altering either the anticodon of the tRNA (57) or possibly the translation machinery, allowing misreading at a frequency high enough to produce a level of the wild-type protein sufficient to suppress the temperature sensitivity caused by dnaN5(G73R) (23, 58).
The B. subtilis dnaN5(G73R) and E. coli dnaN159 alleles influence different aspects of DNA metabolism.
The E. coli dnaN159 allele encodes two missense mutations, G66E and G174A (73). The dnaN159 allele confers an interesting phenotype characterized by temperature-sensitive growth, chronic SOS induction, and altered DNA polymerase usage (48, 49, 73-75). The temperature sensitivity of the E. coli dnaN159 mutant requires both G66E and G174A, and loss of either suppresses the temperature-sensitive growth phenotype (48). Biochemical analysis of the E. coli β clampG66E and β clampG174A proteins shows that these residues effect replication by pol IV, while G174A affects the replicative DNA polymerase pol III and is required for chronic SOS induction (48). The E. coli G66 and G174 residues that are altered in the dnaN159 mutant correspond to B. subtilis β clamp residues G73 and S181, respectively, and both of these residues are changed in the suppressor allele dnaN(G73R, S181G).
In B. subtilis, mutation of G73 (analogous to E. coli G66) is all that is required for lethality at elevated temperatures, and interestingly, G73R in combination with S181 suppresses the temperature sensitivity caused by G73R. Thus, the dnaN(G73R, S181G) allele isolated in this study is altered at the corresponding residues mutated in E. coli dnaN159, yet the altered β clamp in B. subtilis does not behave in a manner similar to that of the E. coli allele. Furthermore, we examined dnaN5(G73R) for altered usage of Y family polymerases PolY1 (YqjW) and PolY2 (YqjH) and were unable to find an alteration in the mutagenesis caused by dnaN5(G73R) to be linked to either translesion polymerase (Fig. 2). Also, the E. coli dnaN159 allele was shown to be proficient in MMR (10). Thus, critical amino acid residues in the E. coli and B. subtilis β clamp appear to have overlapping functions with respect to causing temperature-sensitive growth; however, these residues impart different deficiencies with respect to MMR and translesion DNA synthesis.
Role of YabA in regulating MMR.
YabA negatively regulates replication initiation through its interactions with the β clamp and DnaA (12, 29, 52). Mutations in YabA have been constructed that alter both interaction with DnaA and the β clamp (12, 29, 52); however, the site on the β clamp that YabA binds is unknown. In this work, we found that overexpression of YabA led to an increase in mutagenesis, which is MMR dependent (Fig. 6B). We speculate that the overexpression of YabA may interfere with MutS or MutL binding to the β clamp, leading to an increase in mutation frequency. Consistent with this hypothesis, we found that the yabA::cat mutant strain had a lower mutation frequency than the wild-type control. Therefore, we speculate that the binding site of YabA on the β clamp may at least partially overlap one of the MMR proteins, either MutS or MutL, as both have been shown to interact with the β clamp in B. subtilis (51, 66).
Acknowledgments
We thank Daniel Zeigler at the Bacillus Genetic Stock Center for strains. We thank Ronald Yasbin and Alexi Goranov for strains and Alan Grossman for strains and antibodies against the β clamp. We thank Julian Adams, Janine Maddock, Jim Bardwell, Robert Bender, Matthew Chapman, and Catherine Collins for the use of equipment and reagents. We thank Bryan Davies and Jeremy Schroeder for critical reading of the manuscript. We also thank two anonymous referees for their insightful suggestions and comments that have improved the quality of this work.
This study was supported by startup funds from the College of Literature, Science, and Arts and from the Department of Molecular, Cellular, and Developmental Biology at the University of Michigan.
Footnotes
Published ahead of print on 7 May 2010.
REFERENCES
- 1.Argiriadi, M. A., E. R. Goedken, I. Bruck, M. O'Donnell, and J. Kuriyan. 2006. Crystal structure of a DNA polymerase sliding clamp from a Gram-positive bacterium. BMC Struct. Biol. 6:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berkmen, M. B., and A. D. Grossman. 2006. Spatial and temporal organization of the Bacillus subtilis replication cycle. Mol. Microbiol. 62:57-71. [DOI] [PubMed] [Google Scholar]
- 3.Berkmen, M. B., and A. D. Grossman. 2007. Subcellular positioning of the origin region of the Bacillus subtilis chromosome is independent of sequences within oriC, the site of replication initiation, and the replication initiator DnaA. Mol. Microbiol. 63:150-165. [DOI] [PubMed] [Google Scholar]
- 4.Beuning, P. J., S. Chan, L. S. Waters, H. Addepalli, J. N. Ollivierre, and G. C. Walker. 2009. Characterization of novel alleles of the Escherichia coli umuDC genes identifies additional interaction sites of UmuC with the beta clamp. J. Bacteriol. 191:5910-5920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beuning, P. J., D. Sawicka, D. Barsky, and G. C. Walker. 2006. Two processivity clamp interactions differentially alter the dual activities of UmuC. Mol. Microbiol. 59:460-474. [DOI] [PubMed] [Google Scholar]
- 6.Bloom, L. B. 2009. Loading clamps for DNA replication and repair. DNA Repair (Amst.) 8:570-578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bruck, I., and M. O'Donnell. 2001. The ring-type polymerase sliding clamp family. Genome Biol. 2:REVIEWS3001. [DOI] [PMC free article] [PubMed]
- 8.Bunting, K. A., S. M. Roe, and L. H. Pearl. 2003. Structural basis for recruitment of translesion DNA polymerase Pol IV/DinB to the beta-clamp. EMBO J. 22:5883-5892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Callister, H., S. Le Mesurier, and R. G. Wake. 1977. Initiation of deoxyribonucleic acid replication in germinating spores of Bacillus subtilis 168 carrying the dnaB (Ts)134 mutation. J. Bacteriol. 130:1030-1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Calmann, M. A., and M. G. Marinus. 2005. Differential effects of cisplatin and MNNG on dna mutants of Escherichia coli. Mutat. Res. 578:406-416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Castellanos-Juárez, F. X., C. Alvarez-Alvarez, R. E. Yasbin, B. Setlow, P. Setlow, and M. Pedraza-Reyes. 2006. YtkD and MutT protect vegetative cells but not spores of Bacillus subtilis from oxidative stress. J. Bacteriol. 188:2285-2289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cho, E., N. Ogasawara, and S. Ishikawa. 2008. The functional analysis of YabA, which interacts with DnaA and regulates initiation of chromosome replication in Bacillus subtilis. Genes Genet. Syst. 83:111-125. [DOI] [PubMed] [Google Scholar]
- 13.Cox, E. C., G. E. Degnen, and M. L. Scheppe. 1972. Mutator gene studies in Escherichia coli: the mutS gene. Genetics 72:551-567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dalrymple, B. P., K. Kongsuwan, G. Wijffels, N. E. Dixon, and P. A. Jennings. 2001. A universal protein-protein interaction motif in the eubacterial DNA replication and repair systems. Proc. Natl. Acad. Sci. U. S. A. 98:11627-11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Doré, A. S., M. L. Kilkenny, S. A. Jones, A. W. Oliver, S. M. Roe, S. D. Bell, and L. H. Pearl. 2006. Structure of an archaeal PCNA1-PCNA2-FEN1 complex: elucidating PCNA subunit and client enzyme specificity. Nucleic Acids Res. 34:4515-4526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dronkert, M. L., and R. Kanaar. 2001. Repair of DNA interstrand cross-links. Mutat. Res. 486:217-247. [DOI] [PubMed] [Google Scholar]
- 17.Duigou, S., S. D. Ehrlich, P. Noirot, and M. F. Noirot-Gros. 2004. Distinctive genetic features exhibited by the Y-family DNA polymerases in Bacillus subtilis. Mol. Microbiol. 54:439-451. [DOI] [PubMed] [Google Scholar]
- 18.Duzen, J. M., G. C. Walker, and M. D. Sutton. 2004. Identification of specific amino acid residues in the E. coli beta processivity clamp involved in interactions with DNA polymerase III, UmuD and UmuD′. DNA Repair (Amst.) 3:301-312. [DOI] [PubMed] [Google Scholar]
- 19.Felczak, M. M., and J. M. Kaguni. 2009. DnaAcos hyperinitiates by circumventing regulatory pathways that control the frequency of initiation in Escherichia coli. Mol. Microbiol. 72:1348-1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Friedberg, E. C., R. Wagner, and M. Radman. 2002. Specialized DNA polymerases, cellular survival, and the genesis of mutations. Science 296:1627-1630. [DOI] [PubMed] [Google Scholar]
- 21.Fukuda, R., and H. Nagasawa-Fujimori. 1983. Mechanism of the rifampicin induction of RNA polymerase beta and beta′ subunit synthesis in Escherichia coli. J. Biol. Chem. 258:2720-2728. [PubMed] [Google Scholar]
- 22.Garibyan, L., T. Huang, M. Kim, E. Wolff, A. Nguyen, T. Nguyen, A. Diep, K. Hu, A. Iverson, H. Yang, and J. H. Miller. 2003. Use of the rpoB gene to determine the specificity of base substitution mutations on the Escherichia coli chromosome. DNA Repair (Amst.) 2:593-608. [DOI] [PubMed] [Google Scholar]
- 23.Garvin, R. T., R. Rosset, and L. Gorini. 1973. Ribosomal assembly influenced by growth in the presence of streptomycin. Proc. Natl. Acad. Sci. U. S. A. 70:2762-2766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Georgescu, R. E., S. S. Kim, O. Yurieva, J. Kuriyan, X. P. Kong, and M. O'Donnell. 2008. Structure of a sliding clamp on DNA. Cell 132:43-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ginetti, F., M. Perego, A. M. Albertini, and A. Galizzi. 1996. Bacillus subtilis mutS mutL operon: identification, nucleotide sequence and mutagenesis. Microbiology 142(Pt. 8):2021-2029. [DOI] [PubMed] [Google Scholar]
- 26.Godoy, V. G., D. F. Jarosz, F. L. Walker, L. A. Simmons, and G. C. Walker. 2006. Y-family DNA polymerases respond to DNA damage-independent inhibition of replication fork progression. EMBO J. 25:868-879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Goranov, A. I., A. M. Breier, H. Merrikh, and A. D. Grossman. 2009. YabA of Bacillus subtilis controls DnaA-mediated replication initiation but not the transcriptional response to replication stress. Mol. Microbiol. 74:454-466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gulbis, J. M., Z. Kelman, J. Hurwitz, M. O'Donnell, and J. Kuriyan. 1996. Structure of the C-terminal region of p21(WAF1/CIP1) complexed with human PCNA. Cell 87:297-306. [DOI] [PubMed] [Google Scholar]
- 29.Hayashi, M., Y. Ogura, E. J. Harry, N. Ogasawara, and S. Moriya. 2005. Bacillus subtilis YabA is involved in determining the timing and synchrony of replication initiation. FEMS Microbiol. Lett. 247:73-79. [DOI] [PubMed] [Google Scholar]
- 30.Heltzel, J. M., R. W. Maul, S. K. Scouten Ponticelli, and M. D. Sutton. 2009. A model for DNA polymerase switching involving a single cleft and the rim of the sliding clamp. Proc. Natl. Acad. Sci. U. S. A. 106:12664-12669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Heltzel, J. M., S. K. Scouten Ponticelli, L. H. Sanders, J. M. Duzen, V. Cody, J. Pace, E. H. Snell, and M. D. Sutton. 2009. Sliding clamp-DNA interactions are required for viability and contribute to DNA polymerase management in Escherichia coli. J. Mol. Biol. 387:74-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ibarra, J. R., A. D. Orozco, J. A. Rojas, K. Lopez, P. Setlow, R. E. Yasbin, and M. Pedraza-Reyes. 2008. Role of the Nfo and ExoA apurinic/apyrimidinic endonucleases in repair of DNA damage during outgrowth of Bacillus subtilis spores. J. Bacteriol. 190:2031-2038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jarosz, D. F., V. G. Godoy, and G. C. Walker. 2007. Proficient and accurate bypass of persistent DNA lesions by DinB DNA polymerases. Cell Cycle 6:817-822. [DOI] [PubMed] [Google Scholar]
- 34.Jeruzalmi, D., M. O'Donnell, and J. Kuriyan. 2001. Crystal structure of the processivity clamp loader gamma (gamma) complex of E. coli DNA polymerase III. Cell 106:429-441. [DOI] [PubMed] [Google Scholar]
- 35.Jeruzalmi, D., O. Yurieva, Y. Zhao, M. Young, J. Stewart, M. Hingorani, M. O'Donnell, and J. Kuriyan. 2001. Mechanism of processivity clamp opening by the delta subunit wrench of the clamp loader complex of E. coli DNA polymerase III. Cell 106:417-428. [PubMed] [Google Scholar]
- 36.Karamata, D., and J. D. Gross. 1970. Isolation and genetic analysis of temperature-sensitive mutants of B. subtilis defective in DNA synthesis. Mol. Gen. Genet. 108:277-287. [DOI] [PubMed] [Google Scholar]
- 37.Kato, J., and T. Katayama. 2001. Hda, a novel DnaA-related protein, regulates the replication cycle in Escherichia coli. EMBO J. 20:4253-4262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Klocko, A. D., K. M. Crafton, B. W. Walsh, J. S. Lenhart, and L. A. Simmons. 2010. Imaging mismatch repair and cellular responses to DNA damage in Bacillus subtilis. J. Vis. Exp. 36(pii):1736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kong, X. P., R. Onrust, M. O'Donnell, and J. Kuriyan. 1992. Three-dimensional structure of the beta subunit of E. coli DNA polymerase III holoenzyme: a sliding DNA clamp. Cell 69:425-437. [DOI] [PubMed] [Google Scholar]
- 40.Krishna, T. S., D. Fenyo, X. P. Kong, S. Gary, B. T. Chait, P. Burgers, and J. Kuriyan. 1994. Crystallization of proliferating cell nuclear antigen (PCNA) from Saccharomyces cerevisiae. J. Mol. Biol. 241:265-268. [DOI] [PubMed] [Google Scholar]
- 41.Krishna, T. S., X. P. Kong, S. Gary, P. M. Burgers, and J. Kuriyan. 1994. Crystal structure of the eukaryotic DNA polymerase processivity factor PCNA. Cell 79:1233-1243. [DOI] [PubMed] [Google Scholar]
- 42.Kunkel, T. A., and D. A. Erie. 2005. DNA mismatch repair. Annu. Rev. Biochem. 74:681-710. [DOI] [PubMed] [Google Scholar]
- 43.Kurz, M., B. Dalrymple, G. Wijffels, and K. Kongsuwan. 2004. Interaction of the sliding clamp beta-subunit and Hda, a DnaA-related protein. J. Bacteriol. 186:3508-3515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lee, P. S., D. C. Lin, S. Moriya, and A. D. Grossman. 2003. Effects of the chromosome partitioning protein Spo0J (ParB) on oriC positioning and replication initiation in Bacillus subtilis. J. Bacteriol. 185:1326-1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.López de Saro, F. J., M. G. Marinus, P. Modrich, and M. O'Donnell. 2006. The beta sliding clamp binds to multiple sites within MutL and MutS. J. Biol. Chem. 281:14340-14349. [DOI] [PubMed] [Google Scholar]
- 46.López de Saro, F. J., and M. O'Donnell. 2001. Interaction of the beta sliding clamp with MutS, ligase, and DNA polymerase I. Proc. Natl. Acad. Sci. U. S. A. 98:8376-8380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Maughan, H., B. Galeano, and W. L. Nicholson. 2004. Novel rpoB mutations conferring rifampin resistance on Bacillus subtilis: global effects on growth, competence, sporulation, and germination. J. Bacteriol. 186:2481-2486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Maul, R. W., S. K. Ponticelli, J. M. Duzen, and M. D. Sutton. 2007. Differential binding of Escherichia coli DNA polymerases to the beta-sliding clamp. Mol. Microbiol. 65:811-827. [DOI] [PubMed] [Google Scholar]
- 49.Maul, R. W., L. H. Sanders, J. B. Lim, R. Benitez, and M. D. Sutton. 2007. Role of Escherichia coli DNA polymerase I in conferring viability upon the dnaN159 mutant strain. J. Bacteriol. 189:4688-4695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nicholson, W. L., and H. Maughan. 2002. The spectrum of spontaneous rifampin resistance mutations in the rpoB gene of Bacillus subtilis 168 spores differs from that of vegetative cells and resembles that of Mycobacterium tuberculosis. J. Bacteriol. 184:4936-4940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Noirot-Gros, M. F., E. Dervyn, L. J. Wu, P. Mervelet, J. Errington, S. D. Ehrlich, and P. Noirot. 2002. An expanded view of bacterial DNA replication. Proc. Natl. Acad. Sci. U. S. A. 99:8342-8347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Noirot-Gros, M. F., M. Velten, M. Yoshimura, S. McGovern, T. Morimoto, S. D. Ehrlich, N. Ogasawara, P. Polard, and P. Noirot. 2006. Functional dissection of YabA, a negative regulator of DNA replication initiation in Bacillus subtilis. Proc. Natl. Acad. Sci. U. S. A. 103:2368-2373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ogasawara, N., S. Moriya, G. Mazza, and H. Yoshikawa. 1986. A Bacillus subtilis dnaG mutant harbours a mutation in a gene homologous to the dnaN gene of Escherichia coli. Gene 45:227-231. [DOI] [PubMed] [Google Scholar]
- 54.Ogasawara, N., S. Moriya, K. von Meyenburg, F. G. Hansen, and H. Yoshikawa. 1985. Conservation of genes and their organization in the chromosomal replication origin region of Bacillus subtilis and Escherichia coli. EMBO J. 4:3345-3350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Peebles, C. L., N. P. Higgins, K. N. Kreuzer, A. Morrison, P. O. Brown, A. Sugino, and N. R. Cozzarelli. 1979. Structure and activities of Escherichia coli DNA gyrase. Cold Spring Harbor Symp. Quant. Biol. 43(Pt. 1):41-52. [DOI] [PubMed] [Google Scholar]
- 56.Rokop, M. E., J. M. Auchtung, and A. D. Grossman. 2004. Control of DNA replication initiation by recruitment of an essential initiation protein to the membrane of Bacillus subtilis. Mol. Microbiol. 52:1757-1767. [DOI] [PubMed] [Google Scholar]
- 57.Rokop, M. E., and A. D. Grossman. 2009. Intragenic and extragenic suppressors of temperature sensitive mutations in the replication initiation genes dnaD and dnaB of Bacillus subtilis. PLoS One 4:e6774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rosset, R., and L. Gorini. 1969. A ribosomal ambiguity mutation. J. Mol. Biol. 39:95-112. [DOI] [PubMed] [Google Scholar]
- 59.Schaaper, R. M., and R. L. Dunn. 1987. Spectra of spontaneous mutations in Escherichia coli strains defective in mismatch correction: the nature of in vivo DNA replication errors. Proc. Natl. Acad. Sci. U. S. A. 84:6220-6224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schofield, M. J., and P. Hsieh. 2003. DNA mismatch repair: molecular mechanisms and biological function. Annu. Rev. Microbiol. 57:579-608. [DOI] [PubMed] [Google Scholar]
- 61.Schulz, W., and W. Zillig. 1981. Rifampicin inhibition of RNA synthesis by destabilisation of DNA-RNA polymerase-oligonucleotide-complexes. Nucleic Acids Res. 9:6889-6906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Scouten Ponticelli, S. K., J. M. Duzen, and M. D. Sutton. 2009. Contributions of the individual hydrophobic clefts of the Escherichia coli beta sliding clamp to clamp loading, DNA replication and clamp recycling. Nucleic Acids Res. 37:2796-2809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Siegel, E. C., and F. Kamel. 1974. Reversion of frameshift mutations by mutator genes in Escherichia coli. J. Bacteriol. 117:994-1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Simmons, L. A., B. W. Davies, A. D. Grossman, and G. C. Walker. 2008. Beta clamp directs localization of mismatch repair in Bacillus subtilis. Mol. Cell 29:291-301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Simmons, L. A., A. I. Goranov, H. Kobayashi, B. W. Davies, D. S. Yuan, A. D. Grossman, and G. C. Walker. 2009. Comparison of responses to double-strand breaks between Escherichia coli and Bacillus subtilis reveals different requirements for SOS induction. J. Bacteriol. 191:1152-1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Simmons, L. A., A. D. Grossman, and G. C. Walker. 2008. Clp and Lon proteases occupy distinct subcellular positions in Bacillus subtilis. J. Bacteriol. 190:6758-6768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Simmons, L. A., A. D. Grossman, and G. C. Walker. 2007. Replication is required for the RecA localization response to DNA damage in Bacillus subtilis. Proc. Natl. Acad. Sci. U. S. A. 104:1360-1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Simmons, L. A., and J. M. Kaguni. 2003. The DnaAcos allele of Escherichia coli: hyperactive initiation is caused by substitution of A184V and Y271H, resulting in defective ATP binding and aberrant DNA replication control. Mol. Microbiol. 47:755-765. [DOI] [PubMed] [Google Scholar]
- 69.Smith, B. T., A. D. Grossman, and G. C. Walker. 2001. Visualization of mismatch repair in bacterial cells. Mol. Cell 8:1197-1206. [DOI] [PubMed] [Google Scholar]
- 70.Soufo, C. D., H. J. Soufo, M. F. Noirot-Gros, A. Steindorf, P. Noirot, and P. L. Graumann. 2008. Cell-cycle-dependent spatial sequestration of the DnaA replication initiator protein in Bacillus subtilis. Dev. Cell 15:935-941. [DOI] [PubMed] [Google Scholar]
- 71.Sugino, A., C. L. Peebles, K. N. Kreuzer, and N. R. Cozzarelli. 1977. Mechanism of action of nalidixic acid: purification of Escherichia coli nalA gene product and its relationship to DNA gyrase and a novel nicking-closing enzyme. Proc. Natl. Acad. Sci. U. S. A. 74:4767-4771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sung, H. M., G. Yeamans, C. A. Ross, and R. E. Yasbin. 2003. Roles of YqjH and YqjW, homologs of the Escherichia coli UmuC/DinB or Y superfamily of DNA polymerases, in stationary-phase mutagenesis and UV-induced mutagenesis of Bacillus subtilis. J. Bacteriol. 185:2153-2160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sutton, M. D. 2004. The Escherichia coli dnaN159 mutant displays altered DNA polymerase usage and chronic SOS induction. J. Bacteriol. 186:6738-6748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sutton, M. D., and J. M. Duzen. 2006. Specific amino acid residues in the beta sliding clamp establish a DNA polymerase usage hierarchy in Escherichia coli. DNA Repair (Amst.) 5:312-323. [DOI] [PubMed] [Google Scholar]
- 75.Sutton, M. D., J. M. Duzen, and R. W. Maul. 2005. Mutant forms of the Escherichia coli beta sliding clamp that distinguish between its roles in replication and DNA polymerase V-dependent translesion DNA synthesis. Mol. Microbiol. 55:1751-1766. [DOI] [PubMed] [Google Scholar]
- 76.Youngman, P., J. B. Perkins, and R. Losick. 1984. Construction of a cloning site near one end of Tn917 into which foreign DNA may be inserted without affecting transposition in Bacillus subtilis or expression of the transposon-borne erm gene. Plasmid 12:1-9. [DOI] [PubMed] [Google Scholar]