Swarming is a particular type of motility that is promoted by flagella and allows bacteria to move rapidly over and between surfaces and through viscous environments. Swarming must confer considerable survival benefit, as a heterogeneous group of bacteria exhibit this form of motility, including strains of Aeromonas, Azospirillum, Bacillus, Burkholderia, Chromobacterium, Clostridium, Escherichia coli, Photobacterium, Proteus, Pseudomonas, Rhizobium, Rhodospirillum, Salmonella, Serratia, Vibrio, and Yersinia (reviewed in references 7 and 19). Although swarming has been described for a long time (8; reviewed in reference 9) and the number of bacterial species known to swarm is increasing, surprisingly little is known about the dynamics or mechanics of swarming cells. This is beginning to change. Swarming collective behavior has significant consequences with respect to bacterial colonization strategies in the environment and in hosts (reviewed in reference 19); moreover, the population behavior of the swarm and the performance of individuals in the swarm provide attractive models for physicists and mathematicians studying group dynamics and self-organizing systems (3, 11, 14, 17). In this issue of Journal of Bacteriology and in other recent work from Howard Berg's laboratory at the Rowland Institute, sophisticated visualization of swarming Escherichia coli cells provides some new and fascinating insights into the phenomenon of swarming (4, 18, 21).
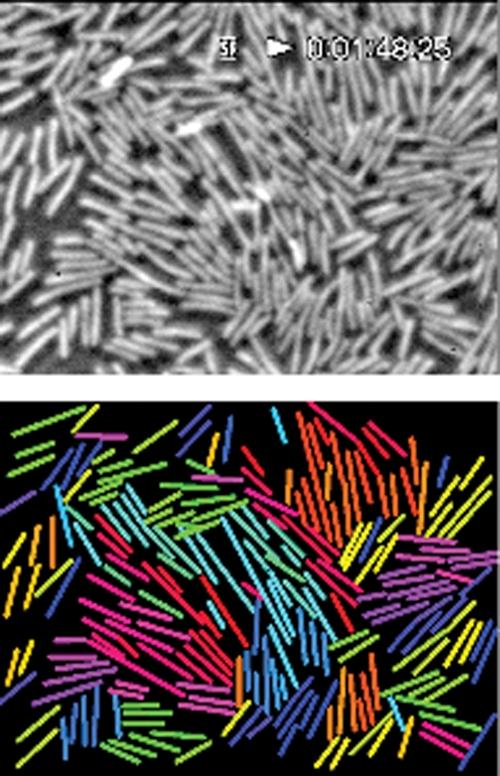
Although diverse types of bacteria exhibit a range of swarming proficiencies, swarm cells are usually (but not always) hyperflagellated and long in comparison to the cell type adapted for swimming in liquid environments (reviewed in reference 6). For example, Proteus mirabilis spectacularly increases its flagellar number, from fewer than 10 to up to 5,000, and its length, from 1 to 2 μm to 20 to 80 μm, upon differentiation from the swimmer to the swarm cell type (reviewed in reference 20). E. coli swarm cell differentiation is more modest: flagellar number and cell length increase 2- to 3-fold (7a). The zone at the edge of an expanding swarm colony is a thin layer of highly motile cells, many of which seem to move together in packs and whirls (14-16). Looking at these vigorously swirling cells prompts numerous questions, many of which have been posed for a long time (e.g., see references 10 and 15). Why are there so many flagella? What do they do? Can a sole bacterium swarm or does it need the pack? How do swarming cells accommodate their neighbor's copious numbers of filaments? Do the swarm cells use each other's filaments to coordinate movement and/or generate thrust? How does a swarm colony advance? What patterns of movement and kinds of acrobatics are observed during swarming? Phase-contrast and fluorescence video microscopy studies are providing some answers to these questions. Individual bacteria can be tracked, and fluorescently labeled flagella allow direct observation of the flagellar movement during swarming. Turner and colleagues used conjugated Alexa Fluor dyes to visualize flagella (18), while Copeland et al. also recently labeled the flagella of swarming cells in another way, by using biarsenical dyes (2). Using these techniques, some old questions are put to rest—as highlighted below—and some new ones can now be addressed. Figure 1 shows a phase-contrast image of cells in a swarm from a tracking video, accompanied by a computer display in which the heading of each cell is indicated by the color of a line running from its head to tail (4, 18).
FIG. 1.
Swarm cell tracking. The top image is a phase-contrast video image of an E. coli swarm. The lower image is a computer display in which the heading of each cell is indicated by the color of a line running from its head to tail. For example, cells moving toward the right are magenta, toward the top are blue, toward the left are green, and toward the bottom are yellow. The coloring changes as a cell's trajectory changes, but each cell's identity remains fixed.
Movement between layers.
Movement on surfaces requires a fluid environment (17). As in swimming, flagellar bundles propel the swarm cell forward; in fact, the speed of translocation of a swarm cell is similar to the swimmer's speed (∼40 μm per s) (4, 18). However, Zhang et al. (21) noticed that the straight trajectory of a swarming cell is markedly different from the curving path observed for bacteria swimming close to a glass surface (12). It seems that the swarm cell's path is straight because the torque generated at the subsurface (agar-fluid interface) is counteracted by the torque generated at the air-fluid interface, i.e., the swarm cell is moving in a fluid environment between 2 surfaces. This was shown for E. coli by demonstrating that small smoke particles floating on the surface of a swarm diffuse only locally—they remain nearly stationary as the cells swarm underneath (21). Thus, swarming occurs between 2 stationary layers. Presumably, some sort of immobile surfactant layer that functions to preserve or promote wetness covers the fluid layer where swarming motility occurs. The nature of the E. coli surfactant is unknown.
Maneuvers in the swarm.
Turner et al. (18) observed four kinds of maneuvers during swarming: forward, essentially straight movement; stalling, which occurs mostly at the advancing edge; lateral movement, which is often caused by collisions with neighboring cells; and reversals. The reversing maneuver is quite amazing: the cell body backs up through its flagellar bundle, and the tail becomes the head. This feat is accomplished when the flagellar motors reverse and the filaments transition from their normal helical shape to the curly polymorphism and then relax again to the normal shape. Such an acrobatic maneuver is unique and probably highly suited to movement in constrained circumstances.
Movement in the pack.
Videos of swarming cells can be viewed online at http://www.rowland.harvard.edu/labs/bacteria/movies_swarmecoli.html. Movies of swarming E. coli and other bacteria have also been captured by the Weibel lab at the University of Wisconsin-Madison and can be seen at http://www.biochem.wisc.edu/faculty/weibel/lab/gallery/default.aspx. The movement is dizzying. Packs of cells form and move together, often looking like streaming swirls. By tracking individual cell paths and measuring velocity, Darnton et al. (4) found that these groups of cells, which are closely packed and aligned, do not move faster but rather move forward for longer periods than do single cells due to a lower rate of deflection by neighboring cells, i.e., there is less jostling in pack movement.
Collisions and cooperativity.
There are many collisions in the densely populated and actively moving swarm. These collisions often result in cell reorientation and realignment (18). Flagellar interactions between cells can occur (2, 18); however, cells do not coordinate movement by forming common bundles. In fact, only rarely are hybrid bundles observed (by using filaments tagged with differently colored labels), and these form transiently, usually occurring between curly flagella upon collision (18). Thus, cell-to-cell flagellar interactions are not crucial for movement: the abundant flagella do not bundle frequently to work together, nor do they tangle.
Swarm expansion and chemotaxis.
Swimming bacteria move in alternating periods of smooth runs and tumbles, which are induced by flagellar reversals or Brownian motion. The tumbles serve to randomly reorient the bacteria. Chemotaxis alters the ratio of the smooth translational period to the tumbling period, e.g., the duration of smooth swimming is prolonged as tumbling is repressed in the presence of attractant and results in the cell's progress up a gradient of attractant (reviewed in reference 1). Chemotaxis is not a requirement for the swarming of E. coli or Salmonella (13). The videos provide some mechanistic explanation for this observation (4). On surfaces, reorientation in a swarm seems to be caused primarily by collisions with neighbors or by jamming at the edge of a swarm. Only rarely does flagellar reversal have an effect on direction (other than reversal). Thus, for E. coli (and Salmonella), Darnton el al. (4) suggest that the chemotaxis system is not important for swarm colony expansion—cell body interactions and flagellar propulsion may do the job.
Encounters at the edge: virgin agar.
Cells at the outermost edge of a swarm become jammed; however, the Berg and Weibel laboratories found that the flagella keep working and can be seen extending over the uncolonized agar (2, 18). The stalled cells eventually manage to reverse back into the swarm, where they are actively motile. Thus, jamming occurs when cells reach unconditioned agar that is prohibitive to movement. Turner et al. (18) suggest that rotation of the flagella of cells that are momentarily trapped at the edge may pump fluid outward from the colony to promote wetness, which eventually allows continued colony expansion.
More questions.
These studies provide fascinating insights into swarming. They also raise many more questions. A variety of bacteria swarm, and they do so with greatly differing abilities. How is this so? What determines the robustness of swarming? Why are so many flagella produced? Could the presence of many flagella be beneficial for moving fluid at the swarm edge? And why are the cells long? Perhaps this too is advantageous for fluid pumping when the cell becomes beached sideways at the swarm edge. The effectiveness of swarming must be dependent to some extent on the ability of the bacteria to create and preserve a fluid environment on the agar, and swarming bacteria have different mechanisms for generating and maintaining wetness. Some swarming bacteria are known to secrete a variety of agents—including surfactin, serawettin, and acidic polysaccharide—that aid movement (reviewed in references 5 and 6). How do these agents affect behavior? Could cell-to-cell flagellar bundling be more significant for bacteria such as P. mirabilis or Vibrio parahaemolyticus, which produce thousands of flagella per cell? Is chemotaxis irrelevant for all swarming bacteria, or might flagellar reversals become important for bacteria that swarm with greater speed (and perhaps are deflected less easily by collision)? With such powerful new visualization techniques, these questions and more shall be very exciting to examine.
Acknowledgments
Work on swarming in my laboratory is supported by NSF grant 0817593.
Figure 1 was kindly provided by H. C. Berg.
The views expressed in this Commentary do not necessarily reflect the views of the journal or of ASM.
Footnotes
Published ahead of print on 30 April 2010.
REFERENCES
- 1.Baker, M. D., P. M. Wolanin, and J. B. Stock. 2006. Systems biology of bacterial chemotaxis. Curr. Opin. Microbiol. 9:187-192. [DOI] [PubMed] [Google Scholar]
- 2.Copeland, M. F., S. T. Flickinger, H. H. Tuson, and D. B. Weibel. 2010. Studying the dynamics of flagella in multicellular communities of Escherichia coli by using biarsenical dyes. Appl. Environ. Microbiol. 76:1241-1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Copeland, M. F., and D. B. Weibel. 2009. Bacterial swarming: a model system for studying dynamic self-assembly. Soft Matter 5:1174-1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Darnton, N. C., L. Turner, S. Rojevsky, and H. C. Berg. 2010. Dynamics of bacterial swarming. Biophys. J. 98:2082-2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Eberl, L., S. Molin, and M. Givskov. 1999. Surface motility of Serratia liquefaciens MG1. J. Bacteriol. 181:1703-1712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fraser, G. M., and C. Hughes. 1999. Swarming motility. Curr. Opin. Microbiol. 2:630-635. [DOI] [PubMed] [Google Scholar]
- 7.Harshey, R. M. 2003. Bacterial motility on a surface: many ways to a common goal. Annu. Rev. Microbiol. 57:249-273. [DOI] [PubMed] [Google Scholar]
- 7a.Harshey, R. M. 1994. Bees aren't the only ones: swarming in Gram-negative bacteria. Mol. Microbiol. 13:389-394. [DOI] [PubMed] [Google Scholar]
- 8.Hauser, G. 1885. Uber Faulnissbacterien und deren Beziehungen zur Septicamie. F. G. W. Vogel, Leipzig, Germany.
- 9.Henrichsen, J. 1972. Bacterial surface translocation: a survey and a classification. Bacteriol. Rev. 36:478-503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hoeniger, J. F. 1964. Cellular changes accompanying swarming of Proteus mirabilis. I. Observations of living cultures. Can. J. Microbiol. 10:1-9. [DOI] [PubMed] [Google Scholar]
- 11.Klausen, M., M. Gjermansen, J. U. Kreft, and T. Tolker-Nielsen. 2006. Dynamics of development and dispersal in sessile microbial communities: examples from Pseudomonas aeruginosa and Pseudomonas putida model biofilms. FEMS Microbiol. Lett. 261:1-11. [DOI] [PubMed] [Google Scholar]
- 12.Lauga, E., W. R. DiLuzio, G. M. Whitesides, and H. A. Stone. 2006. Swimming in circles: motion of bacteria near solid boundaries. Biophys. J. 90:400-412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mariconda, S., Q. Wang, and R. M. Harshey. 2006. A mechanical role for the chemotaxis system in swarming motility. Mol. Microbiol. 60:1590-1602. [DOI] [PubMed] [Google Scholar]
- 14.Mendelson, N. H., A. Bourque, K. Wilkening, K. R. Anderson, and J. C. Watkins. 1999. Organized cell swimming motions in Bacillus subtilis colonies: patterns of short-lived whirls and jets. J. Bacteriol. 181:600-609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Morrison, R. B., and A. Scott. 1966. Swarming of Proteus--a solution to an old problem. Nature 211:255-257. [DOI] [PubMed] [Google Scholar]
- 16.Murray, R. G. E., and R. H. Elder. 1949. The predominance of counterclockwise rotation during swarming of Bacillus species. J. Bacteriol. 58:351-359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rauprich, O., M. Matsushita, C. J. Weijer, F. Siegert, S. E. Esipov, and J. A. Shapiro. 1996. Periodic phenomena in Proteus mirabilis swarm colony development. J. Bacteriol. 178:6525-6538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Turner, L., R. Zhang, N. C. Darnton, and H. C. Berg. 2010. Visualization of flagella during bacterial swarming. J. Bacteriol. 192:3259-3267.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Verstraeten, N., K. Braeken, B. Debkumari, M. Fauvart, J. Fransaer, J. Vermant, and J. Michiels. 2008. Living on a surface: swarming and biofilm formation. Trends Microbiol. 16:496-506. [DOI] [PubMed] [Google Scholar]
- 20.Williams, F. D., and R. H. Schwarzhoff. 1978. Nature of the swarming phenomenon in Proteus. Annu. Rev. Microbiol. 32:101-122. [DOI] [PubMed] [Google Scholar]
- 21.Zhang, R., L. Turner, and H. C. Berg. 2010. The upper surface of an Escherichia coli swarm is stationary. Proc. Natl. Acad. Sci U. S. A. 107:288-290. [DOI] [PMC free article] [PubMed] [Google Scholar]