Abstract
When cells of Escherichia coli are grown in broth and suspended at low density in a motility medium, they swim independently, exploring a homogeneous, isotropic environment. Cell trajectories and the way in which these trajectories are determined by flagellar dynamics are well understood. When cells are grown in a rich medium on agar instead, they elongate, produce more flagella, and swarm. They move in coordinated packs within a thin film of fluid, in intimate contact with one another and with two fixed surfaces, a surfactant monolayer above and an agar matrix below: they move in an inhomogeneous, anisotropic environment. Here we examine swarm-cell trajectories and ways in which these trajectories are determined by flagellar motion, visualizing the cell bodies by phase-contrast microscopy and the flagellar filaments by fluorescence microscopy. We distinguish four kinds of tracks, defining stalls, reversals, lateral movement, and forward movement. When cells are stalled at the edge of a colony, they extend their flagellar filaments outwards, moving fluid over the virgin agar; when cells reverse, changes in filament chirality play a crucial role; when cells move laterally, they are pushed sideways by adjacent cells; and when cells move forward, they are pushed by flagellar bundles in the same way as when they are swimming in bulk aqueous media. These maneuvers are described in this report.
Swarming is a common yet specialized form of surface translocation exhibited by flagellated bacteria and is distinct from swimming (23). When cells are grown on a moist nutrient-rich surface, they differentiate from a vegetative to a swarm state: they elongate, make more flagella, secrete wetting agents, and move across the surface in coordinated packs. Here we focus on the mechanics of bacterial swarming, as exhibited by the model organism Escherichia coli. Others have worked on swarm-cell differentiation in a variety of organisms, including Proteus, Salmonella, Pseudomonas, Serratia, Bacillus, and Vibrio. For example, screens for genes required for swarming in E. coli or Salmonella have been made by Inoue et al. (25) and Wang et al. (40, 41). Vibrio is a special case, because a single polar flagellum enables cells to swim while multiple lateral flagella promote swarming (32). For general reviews, see the work of Allison and Hughes (1), Shapiro (37), Fraser and Hughes (17), and Fraser et al. (16). Also see the work of Eberl et al. (15), Sharma and Anand (38), Harshey (18), Daniels et al. (11), Kaiser (26), O'Toole (33), and Copeland and Weibel (10).
Swarming was first observed with Proteus mirabilis by Hauser (22), who named the genus for a sea god able to change his own form. Proteus is distinctive because cells switch periodically from the vegetative to the swarming state, building terraced colonies (36, 42). This is not observed with E. coli under the conditions used here, where swarms expand at a constant rate propelled by cells swimming vigorously in a monolayer behind a smooth outer boundary.
Swarming in E. coli was discovered by Harshey, who found that K-12 strains, which lack the lipopolysaccharide O antigen, swarmed on Eiken agar (from Japan) but not on Difco agar (from the United States), presumably because the former is more wettable (19, 20). Chemotaxis is not required: cells lacking the chemotaxis response regulator CheY swarm perfectly well, provided that mutations in the motor protein FliM enable transitions between clockwise (CW) and counterclockwise (CCW) rotational states (31). It was suggested that these reversals promote wetness by causing cells to shed lipopolysaccharide.
How do cells in E. coli swarms move across an agar surface? What are their flagella doing? We sought to answer such questions by performing a global analysis of videotaped data (of phase-contrast images) collected from 5 regions of 2 swarms, plotting body lengths, speeds, propulsion angles, local track curvatures, and temporal and spatial correlations, and we found that cells reorient on a time scale of a few tenths of a second, primarily by colliding with one another (13). Our previous report did not describe analyses of individual tracks or visualization of flagella. This aspect of the work is presented here.
Most of the time, cells are driven forwards by a flagellar bundle in the usual way. Flagellar filaments from different cells can intertwine and form common bundles, but this is rare. However, cells in swarms do something not ordinarily seen with swimming cells: they back up. They do this without changing the orientation of the cell body by moving back through the middle of the flagellar bundle. This involves changes in filament shape (in polymorphic form), from normal to curly and back to normal. Polymorphic forms were classified by Calladine (7), on the basis of earlier work by Asakura (3), in terms of the relative lengths of 11 protofilaments, longitudinal arrays of protein subunits that comprise the filament. All polymorphic forms are helical, with some being left-handed (e.g., the normal form) and some being right-handed (e.g., the semicoiled and curly forms, which have half the pitch of the normal filament and half the pitch and half the amplitude, respectively). Transformations from one shape to another can be caused in various ways, e.g., by changes in pH, salinity, or temperature (21, 27, 28) or by application of torque (24). The changes observed with swarm cells are driven by the latter mechanism, when motors switch from CCW to CW rotation. When swimming cells tumble, polymorphic transformations also occur, in the order normal to semicoiled to curly and back to normal (14, 39). But we rarely see the semicoiled form with cells in swarms, and when it appears, it is quite transient. We wonder whether polymorphic transformations evolved to enable cells to escape confined environments when the only way out is to back up, keeping the filaments close to the sides of the cell body.
MATERIALS AND METHODS
Bacteria.
E. coli strain AW405 swims vigorously and is wild type for chemotaxis (2). Strain HCB1668 is a Tn5 fliC null derivative of AW405 in which FliC S353C is expressed from plasmid pBAD33 under the control of the arabinose promoter. This construct was maintained by adding the antibiotics kanamycin (50 μg/ml) and chloramphenicol (34 μg/ml) to the culture medium. A single-colony isolate was grown in T broth (1% Bacto tryptone, 0.5% NaCl) overnight to saturation at 30°C (with gyrorotation at 150 rpm), and dilutions of this culture were used to inoculate swarm plates.
PDMS.
Polydimethylsiloxane (PDMS) (Sylgard 184; Dow Corning) was prepared according to the manufacturer's specifications, spread as a thin sheet (0.17 to 0.20 mm thick) on a polystyrene petri plate coated with a film of Tween 80 (Sigma), and cured for 2 days at room temperature. The resulting product was solid, flexible, transparent, and permeable to oxygen.
Swarm agar.
Swarm agar (0.45% Eiken agar in 1% Bacto peptone, 0.3% beef extract, and 0.5% NaCl), stored in sterile aliquots of 100 ml, was melted completely in a microwave oven and cooled to ∼60°C. Antibiotics were added at the concentrations used in liquid cultures, and arabinose was added to a final concentration of 0.5%. Polystyrene petri plates (150 × 15 mm) were filled with 25 ml of this agar, swirled gently to ensure complete wetting, and then cooled for 15 min (without a lid) inside a large Plexiglas box. Inoculation was done with a 2-μl drop of the saturated culture (described above) at a specified dilution, placed ∼3 cm from the edge of the plate. Plates were air dried for another 15 min (in the Plexiglas box) and then were covered and incubated overnight at 30°C and 100% relative humidity.
Fluorescence labeling.
Swarm cells were collected by gently rinsing the leading edge of a swarm (a region extending ∼1 cm into the colony) with 1 ml of motility medium (0.01 M potassium phosphate, pH 7.0, 0.067 M NaCl, 10−4 M EDTA, and 0.002% Tween 20 [Sigma]) three times. The collected cell suspension was diluted to 10 ml, and the cells were washed three times (by centrifugation at 2,000 × g for 10 min, gentle resuspension of the pellet, and addition of 10 ml of motility medium). The final pellet was adjusted to a volume of ∼250 μl. For HCB1668, a 20-μl solution of a thiol-reactive dye was added (Alexa Fluor 488 or 532 C5 maleimide; Invitrogen-Molecular Probes) (5 mg/ml in dimethyl sulfoxide [DMSO]), and labeling was allowed to proceed for 60 min at room temperature, with gyrorotation at 100 rpm. For AW405, the final wash was performed with motility medium adjusted to pH 7.5, and the cells were labeled with a succinimidyl ester of Cy3 (Amersham Pharmacia) as previously described (39). After labeling of cells with either dye, unused dye was removed by washing cells with motility medium three times, and cells were suspended at a final volume of ∼250 μl for addition to a swarm (see “Fluorescence video microscopy”) or ∼2.0 ml for addition to a tunnel slide (see “Fluorescence digital photomicroscopy”).
Fluorescence digital photomicroscopy.
Tunnels were constructed by placing square coverslips treated with 0.01% poly-l-lysine (Sigma) on two strips of double-sided Scotch tape spaced ∼1 cm apart on microscope slides, and the tunnels were then filled with suspensions of bacteria, inverted, and allowed to stand for ∼10 min in a 100% humidity chamber. Next, the tunnels were rinsed with motility medium and then with motility medium containing 0.25% glutaraldehyde. This stopped flagellar rotation. Slides were viewed with a Nikon Diaphot 200 inverted microscope with a 40× 1.30-numerical-aperture (NA) oil-immersion objective and a 10× relay lens. Illumination was obtained by use of a mercury arc lamp via an R- and B-phycoerythrin fluorescence cube (Chroma 31003). Images were captured with a Nikon D-70 digital camera, using a 10-s exposure time, and were downloaded to a personal computer running Nikon Capture Control 4.0 and Picture Project software. The lengths of the cell bodies and the lengths and numbers of flagella were measured using ImageJ (http://rsb.info.nih.gov/ij/).
Fluorescence video microscopy.
HCB1668 swarm plates were prepared with inoculants diluted 10−3-, 10−5-, and 10−6-fold to stagger the time of swarm development. Cells from the first plate were labeled (see “Fluorescence labeling”) and added (as an ∼3-μl drop) to the advancing edge of the swarm in the second or third plate. In this way, labeled cells were returned to a swarm at approximately the same stage from which they were harvested. In experiments designed to image interactions between flagella of different cells, different Alexa Fluor dyes (usually Alexa Fluor 488 and 532 [Invitrogen-Molecular Probes]) were used on separate cell aliquots, which were mixed and added to the swarm as one ∼3-μl drop. The swarm plates were either returned to the 30°C incubator or left at room temperature just prior to use. Any excess fluid from the drop of labeled cells was absorbed by the agar, and the labeled cells dispersed into the swarm.
Fluorescent cells were imaged in a sandwich between a glass coverslip and a thin sheet of PDMS. PDMS is an ideal substrate for picking up (blotting) swarm cells: it is optically flat, clear, durable, and oxygen permeable. Unlike cells of Serratia marcescens, which adhere to PDMS (12), cells of E. coli are nonadherent and continue to swarm in a thin film of fluid over the PDMS surface. Sheets of cured PDMS (described above) were cut into small rectangles (1 × 1.5 cm), rinsed with motility medium, shaken dry, and then placed in contact with the swarm edge. The PDMS was lifted, removing the bacteria by blotting as described previously (12), and placed cell side down on a rectangular coverslip (previously exposed to a 1% solution of Tween 20, rinsed with water, and shaken dry). This coverslip was mounted on the bottom of an aluminum holder with the PDMS side up, and the holder was covered with a second coverslip. This left a small air gap (∼0.65 mm) above the PDMS, reducing evaporation by leaving only the edges of the preparation exposed to room air. The cells, located between the bottom coverslip and the PDMS, were viewed at room temperature with a Nikon Diaphot 200 inverted microscope equipped with a strobed argon-ion laser (39) tuned to 496 nm, a fluorescence isothiocyanate (FITC)-Texas Red fluorescence cube (Chroma 51006) with a 495/20 excitation filter, a 60× 1.4-NA dark-phase oil-immersion objective, and a 5× relay lens. The excitation light passed through the bottom coverslip to the layer of swarming cells. Phase-contrast illumination from above allowed visualization of unlabeled cells. Such a preparation lasted ∼15 to 40 min before drying reduced cell speed, at which time the preparation was replaced. A low-light black-and-white surveillance camera (Marshall Electronics model V1070) (30 frames/s; 2:1 interlace) or a color surveillance camera (COP model 15-CA35ED) (30 frames/s; 2:1 interlace) was used to acquire images, which were recorded on a digital recorder (Sony model GV-D1000), transferred to a Macintosh G4 or G5 computer using iMovie, and exported to ImageJ for analysis. The cell bodies appeared dark by phase-contrast microscopy, and the flagellar filaments appeared bright by fluorescence microscopy.
Data analysis.
Tracking data consisted of two points (positions of the head and tail) for every cell in a video frame for 30 consecutive frames (1 s) for 5 regions of 2 swarms, as reported previously (13). We distinguished a set of 2,366 individual swarm-cell tracks. Tracks were plotted with blue, red, and green lines showing successive positions of each cell's head, center, and tail positions, respectively. Magenta lines connecting heads and tails displayed cell body axes (long axes). Speed was calculated by dividing the displacement of the cell center in successive frames by the time difference (1/30 s). Propulsion angle was defined as the acute angle between the body axis and the cell's velocity vector (or track) and was considered positive if the track was to the right of the body axis (viewed from above the agar) and negative if it was to the left. Reversals were identified by eye as events during which a cell's head became its tail: cells slowed to a stop and then set off again, moving in the opposite direction. The beginning of a reversal was defined as the time at which the cell's speed fell below 15 μm/s, while the end of the reversal was defined as the time at which it rose above 15 μm/s, with reversal duration defined as the difference between these two times. Reversal angle was defined as the difference in angle between the beginning and ending velocity vectors. The average time between reversals was computed as the total cumulative tracking time (in cell-seconds) divided by the total number of reversals observed. The reversal probability is the inverse of this number.
Tracks were next analyzed for stalls (stops) not associated with reversals. Cells were considered to have stalled if their speed fell below 3 μm/s. Data frames associated with reversals or stalls were excluded from further analysis. The frames that remained spanned cell movements that were forward, sideways, and everything in between: the propulsion angles varied from 0° (forward) to ±90° (sideways). If the propulsion angle was ≤35°, the cells were judged to be moving forward. If the angle was >35°, the cells were judged to be moving laterally.
RESULTS
Flagellar expression.
We wanted to know whether flagellation of the strain constructed for use with thiol-reactive dyes, i.e., HCB1668 (with cysteine-labeled flagella), was significantly different from that of the wild-type strain, AW405. With thiol-reactive dyes, the labeling was more specific: the flagellar filaments were brighter and the cell bodies were dimmer than those observed with amino-reactive dyes. We thus compared 100 cells of each type collected near the edges of swarms: we labeled their flagella (with maleimide and succinimidyl ester dyes, respectively), stopped flagellar rotation with glutaraldehyde, and measured their flagellar filaments (see Materials and Methods). When the strains were grown under identical conditions (on swarm agar containing arabinose), the filaments on the mutant strain were slightly shorter (mean ± standard deviation [SD], 4.5 ± 2.0 μm) but more abundant (7.6 ± 3.0 per cell) than filaments on the wild-type strain (5.1 ± 2.5 μm and 6.6 ± 3.7 per cell, respectively). The total expression levels (length times number) were nearly the same. Cells harvested and labeled this way were about 20% shorter than cells observed in swarms (4.2 rather than 5.2 μm long), probably because the labeling was done in bulk in motility medium at room temperature rather than on an agar surface in a rich medium at 30°C, where cells elongate: divisions that occur during the lengthy labeling procedure would produce offspring that are relatively short. Swarms of strains HCB1668 and AW405 were similar, with colony fronts advancing at rates between 3 and 7 μm/s. Cell densities were ∼0.1 per μm2, about half of what they would be were the cells closely packed (13).
Swarming under PDMS.
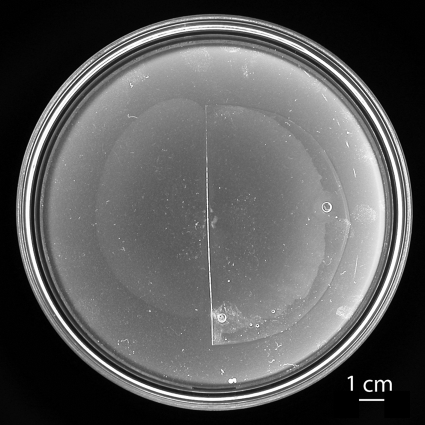
To visualize fluorescently labeled flagella by use of an oil-immersion objective, we needed to view swarming cells through an optically flat, clear preparation. We thus followed the motion of cells in a thin film of fluid (transferred with the cells from a swarm agar blot) between glass and a thin sheet of PDMS (see Materials and Methods). Phase-contrast video images of swarming cells looked the same whether they were made from agar plates or in these glass-PDMS preparations. A more stringent test of whether swarming might be normal under PDMS is illustrated in Fig. 1, which shows a swarm spreading in air (left) and under a semicircular sheet of PDMS (right). Spreading rates were the same in either case (5.6 μm/s). Apparently, PDMS mimics the swarm-air interface. The reason for this turns out to be that the swarm-air interface is stationary; thus, it does not matter whether PDMS (which is oxygen permeable) is added or not. We proved that the swarm-air interface is stationary by recording the motion of small smoke particles deposited on surfaces of swarms (44). The particles diffused freely (but with relatively small diffusion coefficients) in what appeared to be a surfactant monolayer, without being deflected by cells that swarmed underneath. Thus, on agar, swarming occurs between a fixed surfactant monolayer above and a fixed agar surface below. In our glass-PDMS preparations, swarming occurred between a fixed PDMS surface above and a fixed glass surface below. The experiment for Fig. 1 was repeated with sheets of PDMS that had not been exposed to Tween 20 or Tween 80, and the results were the same.
FIG. 1.
Swarming in air or under PDMS. Cells of E. coli strain HCB1668, inoculated at the center of a 150- by 15-mm swarm agar plate (inside diameter, 14 cm), were grown overnight, covered in part by a half-round sheet of PDMS (0.17 mm thick) (right), returned to the incubator for another 1 h, and then photographed. During the final hour, the radius of the swarm increased from ∼2.5 cm to ∼4.5 cm, in essentially the same way whether the swarm was exposed to air or covered by PDMS. The point of inoculation appears as a cluster of small white spots. The larger circular spots (at 3 and 6 o'clock) are air bubbles. The inoculum was a 2-μl droplet of a saturated culture diluted 10−6-fold.
Types of swarm-cell tracks.
In a parallel study (13), we performed a global analysis of data collected from 5 regions of 2 swarms, plotting body lengths, speeds, propulsion angles, local track curvatures, and time and spatial correlations. This analysis was done without particular regard to individual tracks. We returned to this data set and followed the motion of 2,366 cells, distinguishing 4 different track types (see the criteria noted in Materials and Methods)—stalls, reversals, lateral movement, and forward movement. A summary of this analysis is given in Table 1. Since not all cells were in the field of view for the full 30-frame interval, the data set included only 36,505 of a possible 70,980 frames, spanning ∼1,217 s. Since a given cell moving along its track exhibited different kinds of behavior (see below), and relatively few cells exhibited only one kind of behavior, the sum of the number of cells exhibiting different kinds of behaviors was greater than the total number of cells. For example, while 98.2% of the cells (2,323) spent some of their time swimming forward, only 27.7% (655) spent all of their time swimming forward. Note that the percentage of all frames is the same as the percentage of total track time.
TABLE 1.
Track-type distribution
| Track type | No. of frames | % of all frames | No. of cells | % of all cells |
|---|---|---|---|---|
| Stall | 906 | 2.5 | 399 | 16.9 |
| Reverse | 3,110 | 8.5 | 559a | 23.6 |
| Lateral | 6,900 | 18.9 | 1,638 | 69.2 |
| Forward | 25,589 | 70.1 | 2,323b | 98.2 |
There were 796 events; some of the cells reversed more than once.
A total of 655 of these cells moved exclusively forward.
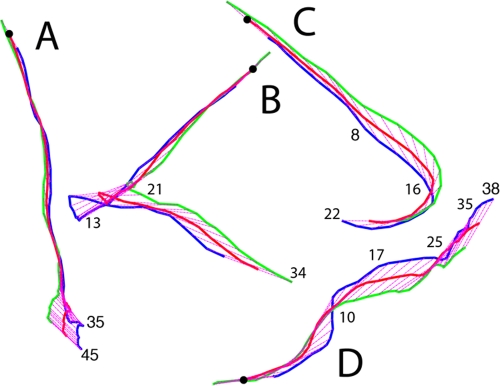
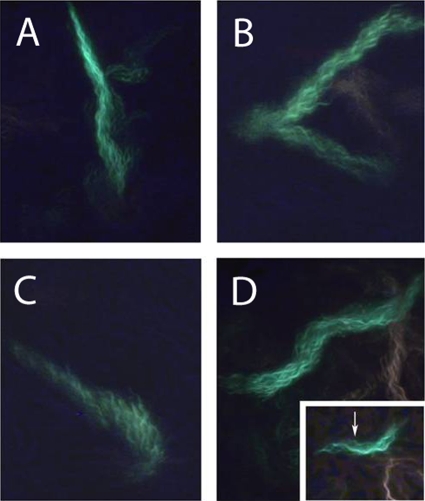
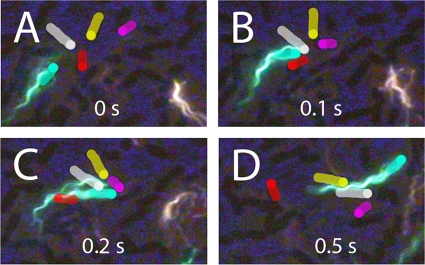
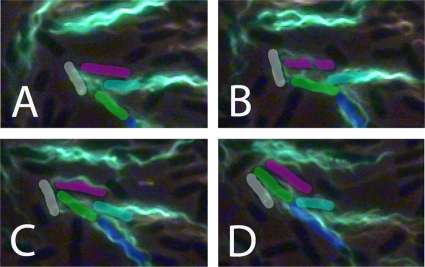
A cell can move forward with the flagellar bundle, the long axis of the cell body, and the direction of motion all aligned, as shown in Fig. 2A and 3A. A cell can reverse (back up) when the cell body changes its direction of motion ∼180°, with the head (as defined at the beginning of the track) now following rather than leading, as shown in Fig. 2B and 3B (note that at the end of this track, the blue line lags behind the green one [Fig. 2B]). A cell can move laterally, with a propulsion angle of >35°. Usually, this is caused by collisions with other cells; sometimes it is caused by a motor reversal. When lateral motion results from collisions, the flagellar bundle and body axis are no longer aligned with the trajectory, as shown in Fig. 2C and 3C. Depending upon the points of impact, the cell body translates sideways (Fig. 2A and B, at the end of the track) or pivots (Fig. 2C, near the middle of the track). When lateral motion results from a motor reversal (Fig. 3D, inset), the flagellar bundle and body axis remain aligned with the trajectory, as shown in Fig. 2D and 3D. Under the crowded conditions of a swarm, it is impossible to distinguish between collisions and a motor reversal without imaging the bundle, unless the motor reversal causes the cell to back up (compare Fig. 2C and D to Fig. 2B). Collisions are common and involve several cells in complex ways, as shown in Fig. 4. All of the cells in this figure were flagellated and motile, but only the flagella on the green cell were labeled.
FIG. 2.
Representative tracks shown within a 42-μm by 57-μm frame. The beginning of each track is indicated by a black dot. The traces of the head, center, and tail (defined at the beginning of the track) are plotted in blue, red, and green, respectively, and the axis of the cell body is shown in magenta (dotted). Frame numbers are indicated at the leading end of the cell. (A) A cell that moved forward for the first 35 frames and then was pushed sideways (exhibited lateral translational movement) for the last 10 frames. (B) A cell that moved forward for the first 13 frames, reversed during the next 8 frames, and finally was pushed sideways (exhibited lateral translational movement) for the last 13 frames. Note that at the end of this track, the blue line lags behind the green line. (C) A cell that moved forward for the first 8 frames, was pushed sideways (exhibited lateral rotational movement) for the next 8 frames, and then continued forward. (D) A cell with two deflections due to motor reversals. The cell moved forward during the first 17 frames (increasing its propulsion angle at frame 10), reoriented its cell body during the next 7 frames in response to the first motor reversal, moved forward for another 10 frames, and then moved laterally during the last 3 frames in response to a second motor reversal. At this point, the cell left the field of view. The frames are 1/30 s apart.
FIG. 3.
Flagellar bundles of cells whose tracks are shown in Fig. 2, labeled with the same letters. The images show the maximum brightness at each pixel over all of the video frames as the cell moved through the field of view. The cells were labeled with Alexa Fluor 488, and the cell bodies are not visible. The inset in panel D is a single video frame (frame 16) showing the curly polymorphic form at the distal end of a normal filament, indicating that a motor has resumed CCW rotation after CW rotation (a motor reversal). The frames are 1/30 s apart. Inset magnification, ×1.3.
FIG. 4.
Collisions. A bacterium labeled with Alexa Fluor 488 (green) collides with several other bacteria. (A) Orientations of cells before the collision. The unlabeled cells are highlighted in white, red, yellow, and pink, with each leading end indicated by a dot. The labeled bacterium is highlighted in green. (B) The labeled bacterium collides with the white and red cells, and this reorients the red cell. (B and C) The pink cell collides first with the white one and then with the labeled cell and reorients by ∼90°. (C) The labeled bacterium's flagellum shows evidence of a motor reversal; one filament has separated from the bundle and lies along the upper surface of the white bacterium. (D) The red cell has reoriented by ∼90° after having moved along the flagellar bundle of the labeled bacterium. The yellow and white cells have aligned and are now moving along the flagellar bundle of the labeled bacterium. The images of other cells in the preparation appear dark.
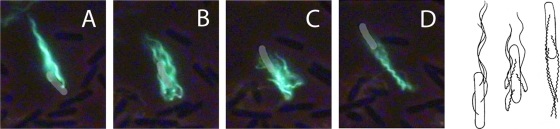
Reversal (Fig. 2B and 3B) is unique to constrained environments. It occurs in swarms and when cells swim in gel-like media (e.g., in high concentrations of methylcellulose) or in narrow constrictions (e.g., see reference 30). However, the flagella have been visualized only in swarms. Reversals of this kind are not seen when cells swim in bulk aqueous media (e.g., see references 4, 14, and 39). When a cell backs up, all of its motors reverse, the flagellar filaments change chirality, and the cell body moves back through the center of the bundle, as shown in Fig. 5. The cell body is pushed through the center of the bundle by the curly section of the filament, either via the propagation of the polymorphic transformation (with the normal ends of the filaments unable to translate) or because the curly section, turning CW, generates thrust. This mode continues until the cell body completes its transit through the bundle, whereupon the curly filaments form a bundle that pushes the cell forward, in the opposite direction from which it started. When the motors again spin CCW, the filaments transform back to the normal form, and the normal bundle continues to push the cell in the new direction. In this phase of the process, one does not see acute angles between normal and curly sections of the filaments; the curly filaments simply transform back to normal and re-form a normal bundle. The filament structure has directionality: curly proximal-normal distal junctions and normal proximal-curly distal junctions are not identical; they appear with acute and obtuse angles, respectively.
FIG. 5.
Typical swarm-cell reversal. A cell labeled with Alexa Fluor 488 is shown to change its direction by backing up. (A) The cell moves downwards and starts the reversal process. (B) The flagellar motors have reversed, and the bright green dots are the filament transformation points between normal (left-handed) and curly (right-handed) forms. The cell body has changed its direction of motion. (C) The loosened bundle appears folded, and the cell body has moved through the center of the loosened bundle to extend past the filaments’ distal tips. Thus, the cell has backed up without changing its orientation. (D) The bundle has reformed with curly filaments, and the cell now swims upwards. This maneuver is depicted schematically in the right panel. Eventually, the flagellar motors switch back to CCW operation, and the filaments transform to normal (not shown). The cell body is highlighted in gray to aid the eye, and the phase-contrast images of other cells in the background appear dark. The frames are 1/10 s apart.
For any given cell, reversals occurred, on average, every 1.5 s and lasted ∼0.09 s, times comparable with those obtained for run and tumble intervals of free-swimming cells (4). The distribution of reversal durations (not shown) was approximately exponential, as found earlier for tumbles. The distribution of reversal angles is shown in Fig. 6. In each event, the head became the tail (at least once). For an angle change of 0°, the cell exhibited successive reversals; this was rare. For an angle change of 180°, the cell backed up without changing the orientation of its long axis (the head became the tail), retracing its earlier path; this was common. The mean change in angle was 128°.
FIG. 6.
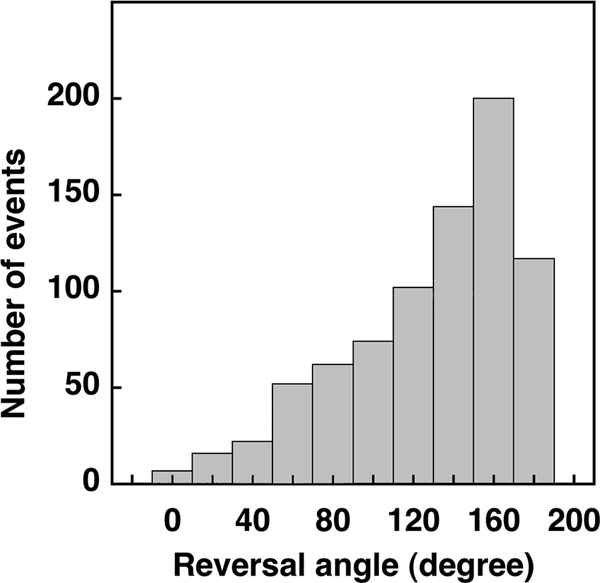
Distribution of reversal angles. In each event, the head became the tail (at least once). For an angle change of 0°, the cell continued in its original direction; this was rare. For an angle change of 180°, the cell backed up without changing the orientation of its long axis, retracing its earlier path; this was common.
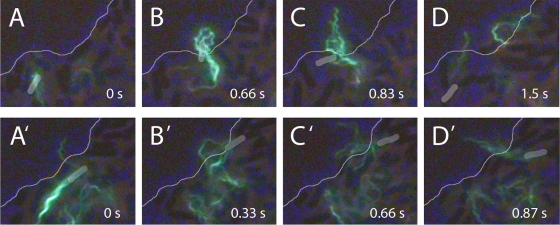
Stalls occurred most frequently at the swarm edge. As described earlier (13), a cell slows as it nears the edge and then stalls, and after a brief pause, it moves away from the edge, either by completely reversing, as shown in Fig. 7A, or by deflecting at a shallow angle, sometimes after traveling along the edge for some distance, as shown in Fig. 7B. Since the majority of cells at the swarm edge reversed their head-tail orientation (13), it is likely that flagellar motion aids in swarm expansion: at the boundary, as cells prepare to swim back into the swarm, their flagella extend out onto the virgin agar (Fig. 7A and B); the rotation of these flagella must pump fluid outward from the colony, aiding in swarm expansion.
FIG. 7.
Cells at swarm edge. (Top) Reversal. (A) A cell labeled with Alexa Fluor 488 (green) moves to the swarm edge (thin line). Between panels A and B, the motors reverse and the bundle comes apart. (B and C) The flagella transform to the curly form and project beyond the swarm edge. (D) The flagella return to the normal form as the motors resume their normal CCW rotation, and the cell swims away from the swarm edge. (Bottom) Deflection. (A′) A cell approaches the swarm edge obliquely. (B′ and C′) The cell pauses at the edge, and its flagella splay outwards. (D′) The cell is deflected inwards and the bundle reforms. The bodies of the labeled cells are highlighted in gray to aid the eye, and the phase-contrast images of other cells in the background appear dark.
In combined fluorescence and phase-contrast video images, we looked for cells that used their flagella to actively reorient themselves. From the phase-contrast images, we measured the cell speed before and after reorientation and the angular change in direction that occurred. From the fluorescence images, we noted the total number of filaments on the cell and the number of filaments that came out of the bundle during the reorientation. There was no change in cell speed. A plot of the change in direction versus the fraction of filaments that came out of the bundle is shown in Fig. 8. If one-half or fewer of the filaments came out of the bundle, then the mean change in direction was 47° ± 28°, similar to the 38° ± 26° found for swimming cells (calculated for Fig. 13 in reference 39). However, when reorientation involved more than half of the filaments, the mean change in direction was 127° ± 37° for swarming cells and 69° ± 42° for swimming cells (calculated for Fig. 13 in reference 39). Thus, swarming cells prefer to back up. The number of events recorded in Fig. 8 is substantially smaller than the number of tracks described in Table 1, because only a small fraction of cells were fluorescently labeled and it was not always possible to visualize all of the flagella on a labeled cell.
FIG. 8.
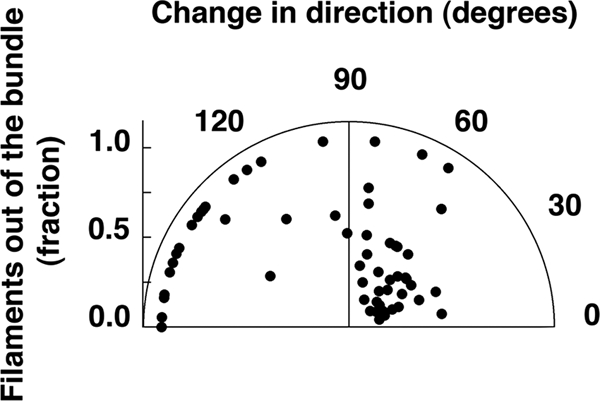
Reorientation driven by flagellar reversals. The polar plot shows the angular change in the trajectories of 60 cells as a function of the fraction of filaments that left the bundle.
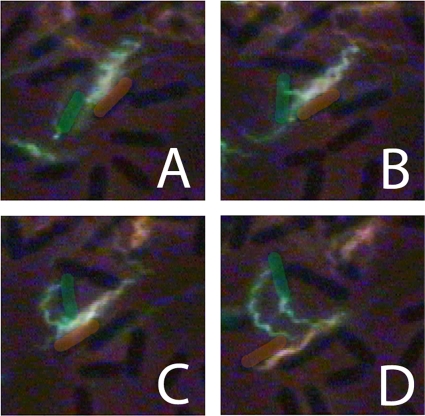
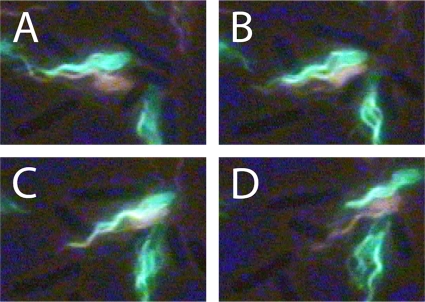
When we began this work, we thought that cell-cell coordination might result from flagellar interactions, with filaments of one cell interacting with the body or filaments of another cell. Hoping to visualize such interactions, we labeled swarm-cell aliquots with dyes of different colors. Although many cells traversed the field of view in tandem, we did not see two bundles of the same polymorphic form entwining. Examples of entwined filaments between cells always involved the curly form in a hybrid bundle (a bundle composed of two different polymorphic forms). An example of such a bundle resulting from motor reversals that occurred in earlier frames is shown in Fig. 9. The two cells traveled together with their filaments entwined and then moved apart. As the cells resumed separate trajectories, the bundle of one unraveled. Normal filament bundles frequently aligned, as shown in Fig. 10, but without forming a common bundle. Sometimes, the bundle of one cell could entrap another cell trailing along behind, aligning two or more cells along parallel trajectories, as shown in Fig. 11.
FIG. 9.
Hybrid flagella. Two bacteria, one with a normal bundle labeled with Alexa Fluor 532 (orange) and the other with a curly bundle labeled with Alexa Fluor 488 (green), traveled together and then moved apart. In panel A, the green filaments are above the orange filaments, and in panel C, they are below the orange filaments. The green filaments appear to be twisted about the orange ones, pinned at their distal ends. (C and D) As the cells move apart, the bundles unravel and the green filaments bend. The cell bodies are highlighted with the color of their filaments to aid the eye. The images of other cells in the background appear dark. The frames are 1/15 s apart.
FIG. 10.
Bundles in phase. (A to C) Two bacteria with normal bundles, one labeled with Alexa Fluor 532 (orange) and the other labeled with Alexa Fluor 488 (green), traveled together and then moved apart. The bundles were in phase but were not wrapped around each other. (D) The bundles moved apart without unraveling. The images of cells in the background appear dark. The frames are 1/15 s apart.
FIG. 11.
Cell alignment. (A) Orientation of cell bodies prior to parallel alignment. An unlabeled cell is highlighted in pink, and labeled cells are highlighted in white, green, aqua, and blue. (B) The filaments of the white cell move over the pink cell. (C) The pink cell nestles between the white cell's filament and its body, pushing the filament away from the cell body at an angle of about 60°. (D) The green cell pushes between the pink and the white cells. The white cell's filament is moved further from the cell body, now at an angle of ∼90°. This filament then bends about the pink cell as the 3 cells align. In subsequent images, the green cell moves ahead and the aqua cell moves into the vacated space. The cell paths then diverge. The images of other cell bodies in the preparation appear dark. The frames are 1/15 s apart.
DISCUSSION
Swarm cells are jostled by their neighbors, which randomizes their directions of motion within a few tenths of a second; run and tumble behavior is not observed (13). In light of this, we wondered whether flagellar filaments play any role other than driving cells forward. Motor reversals are believed to enhance wetness; however, a functional chemotaxis system is not required (6, 31). What do swarm trajectories look like? How might they be affected by motor reversals? We identified four kinds of tracks and found a novel role for motor reversals.
Stalls.
Stalls occurred mostly at the swarm edge. Cells paused, but their flagella continued to spin and thus pump fluid over the agar in front of the swarm. A similar observation was made by Copeland et al. (9). By measuring diffusion coefficients of small particles suspended in the surfactant monolayer on top of the swarm, we found that the fluid in front of the swarm became more shallow as one moved away from the swarm edge, over distances ranging from 10 to 20 μm (44). Pumping is also evident in bulk when large numbers of flagellated cells are adsorbed to glass or PDMS (12, 29). Stalls also occurred briefly when cells backed up (see below).
Reversals.
Cell reversals were easy to spot by eye and were evident in the phase-contrast tracking data. These are dramatic events, but as noted earlier, they do not have a large impact on the average cell behavior, which is dominated by collisions between adjacent cells (13). Reversals occurred, on average, every 1.5 s and required about 0.1 s for completion. They were triggered when motors switched from CCW to CW rotation. Viewed by fluorescence microscopy, the maneuver is exquisite: the bundle that normally pushes the cell forward is loosened by filament transformation to the curly polymorphic form, and the cell body moves backwards along the central axis of the bundle, emerging with a curly bundle behind, as shown in the cartoon that accompanies Fig. 5. This bundle soon relaxes to normal, and the cell continues to swim backwards. Thus, the cell body begins with a normal bundle behind and ends with a normal bundle behind, but swimming in the opposite direction. In the process, the flagellar filaments remain close to the sides of the cell; more often than not, the cell body retains the orientation of its long axis (Fig. 6). This is an ingenious way to escape from confined environments, e.g., from packs of nearby cells aligned in parallel, or from narrow constrictions (30). Given the switching rates measured for motors spinning at speeds observed with swimming cells (14, 43), we do not understand how most or all of the flagellar motors on a given cell manage to switch from CCW to CW rotation at the same time. Reversals of the cell body have also been seen when E. coli tries to swim in 1% methylcellulose (observations made by H.C.B. in 1970, in collaboration with Scott Ramsey and Julius Adler) and with other peritrichously flagellated bacteria, e.g., Bacillus subtilis, encountering obstacles (8). The hallmark of this maneuver, which is evident without flagellar visualization, is that the cell suddenly swims backwards at the same speed at which it was swimming forwards, without changing the orientation of its cell body. It is possible that this maneuver was not seen by Copeland et al. (9) because their cells were swimming in a glass-agar-glass sandwich and might have been oxygen deprived, while our cells were swimming between glass and a thin film of PDMS, which is oxygen permeable.
Lateral motion.
Tracks with propulsion angles of >35° arose primarily from cell collisions but also from motor reversals. Because all of the filaments in a given field could not be imaged at the same time, it was difficult to determine the extent to which each contributed to randomizing cell directions. In a swarm, cells are confined within a thin fluid layer, between a fixed surfactant monolayer above and an agar surface below (44), which limits the ways in which cells can respond when motors reverse. For example, end-over-end motion that can scramble a swimming cell's head-tail orientation (5) does not occur. The flagellar bundle tends to maintain its orientation relative to the cell body during swarming but it appears to bend at times, as shown in Fig. 4B and D (also see reference 9). An important exception is the reversal, as described above. However, when lateral deflections are caused by collisions, the flagellar bundle tends to be deflected as well, retaining its orientation relative to the long axis of the cell, as evident by comparing the orientations of the filaments in Fig. 3C with those of the cell body in Fig. 2C. When lateral deflections are caused by motor reversals, the bundle also tends to remain in line with the cell body, but now these orientations are along the new direction of motion. The larger the fraction of filaments reversing, the larger is the angular deflection (Fig. 8). When the number of motors reversing is relatively small, the angular deflections are similar to those observed with swimming cells (39), but when the number of filaments reversing is large, swarming cells tend to back up rather than choose a new direction at random.
Forward motion.
When swarming cells move forward, they are propelled in the same way as swimming cells. Mean swarming and swimming speeds are about the same, but variations in speed are much larger when cells swarm (13). This appears to be due to cell-cell collisions. As noted above, the cells swim in a thin layer of fluid between a fixed surfactant monolayer above and an agar surface below. Presumably, the fluid layer is at least as thick as the diameter of a cell, which is almost twice as large as the diameter of the flagellar bundle, so there is ample room for filaments to rotate. A study of the hydrodynamics of the motion of a sphere driven by a single helical flagellum between closely opposed fixed plates was made by Ramia et al. (35), who found that swimming speeds were similar to those in bulk: the increase in drag on the cell body was offset by the increase in propulsive force due to an increase in the ratio of the normal to tangential slender-body resistance coefficients.
Comparison of swimming and swarming.
When cells swim, the cell body rolls about the axis of the flagellar bundle. If the axis of the bundle is not parallel to that of the cell body, the cell body appears to wobble. This motion is suppressed within the thin film of fluid in which cells swarm. However, as evident in Fig. 2, cell bodies can slide sideways, often in response to collisions with other cells. They are not free to choose new directions at random, as are swimming cells; the motion is confined to two dimensions. When swimming cells tumble, filaments driven by motors that change their direction of rotation from CCW to CW go through a sequence of polymorphic transformations, from normal to semicoiled to curly and back to normal (14, 39). By the time the transformation to the semicoiled form is complete (about 0.1 s), the cell has chosen a new direction for its next run. The semicoiled filament projects out from the side of the cell. This transformation is rarely seen in swarming cells and appears over only a fraction of the length of a filament, presumably because there is no room for lateral projection. Instead, the transformation from the normal to curly form allows unbundled filaments to remain within the narrow spaces between adjacent cells. When a cell is swimming, the semicoiled filament relaxes to the curly form, and when flagellar motors switch back to CCW rotation, the curly filament relaxes back to normal. This curly-form-to- normal-form transformation occurs with both swimming and swarming cells. The feature of swarming that is unique is cell reversal, as discussed above (Fig. 5).
Interactions between cells.
When neighboring cells travel side by side, one cell can be entrained by the other (Fig. 11). This results from one cell butting against the other and sometimes involves temporary entrapment of one cell by the other's filaments. Hybrid bundles made from filaments of different polymorphic forms wrapped around one another appeared infrequently and tended to involve curly filaments (Fig. 9). Cobundling, with filaments of the same polymorphic form from two cells entwining, was not common and was never seen with normal filaments. One reason that cobundling might be rare is that while a single cell body can counterrotate about its axis, wrapping up the bundle (34), two cell bodies confined to the same plane are not free to rotate about a common axis. The coordinated movements of cells within swarm monolayers do not appear to be stabilized by cobundling.
Conclusions.
Analysis of swarm-cell tracks shows that cells try to swim forward, changing direction in response to cell-cell collisions, as concluded earlier from global averages of behavior computed for the same data set (13). Cells do not coordinate their movements by forming common flagellar bundles; they simply run into one another. Less often, cells change course by switching the direction of flagellar rotation. When all of the flagellar motors switch at about the same time, cells swim back through the flagellar bundle without changing the orientation of the cell body. This maneuver is promoted by polymorphic transformations in filament shape, from normal to curly and eventually back to normal, enabling cells to escape from confined environments.
Acknowledgments
We are grateful to Svetlana Rojevsky for expert technical assistance.
This work was funded by National Institutes of Health grant AI066540 (to H.C.B.).
Footnotes
Published ahead of print on 2 April 2010.
REFERENCES
- 1.Allison, C., and C. Hughes. 1991. Bacterial swarming: an example of prokaryotic differentiation and multicellular behavior. Sci. Prog. 75:403-422. [PubMed] [Google Scholar]
- 2.Armstrong, J. B., and J. Adler. 1967. Genetics of motility in Escherichia coli: complementation of paralyzed mutants. Genetics 56:363-373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Asakura, S. 1970. Polymerization of flagellin and polymorphism of flagella. Adv. Biophys. 1:99-155. [PubMed] [Google Scholar]
- 4.Berg, H. C., and D. A. Brown. 1972. Chemotaxis in Escherichia coli analysed by three-dimensional tracking. Nature 239:500-504. [DOI] [PubMed] [Google Scholar]
- 5.Berg, H. C., and L. Turner. 1995. Cells of Escherichia coli swim either end forward. Proc. Natl. Acad. Sci. U. S. A. 92:477-479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burkart, M., A. Toguchi, and R. M. Harshey. 1998. The chemotaxis system, but not chemotaxis, is essential to swarming motility in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 95:2568-2573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Calladine, C. R. 1978. Change in waveform in bacterial flagella: the role of mechanics at the molecular level. J. Mol. Biol. 118:457-479. [Google Scholar]
- 8.Cisneros, L., C. Dombrowski, R. E. Goldstein, and J. O. Kessler. 2006. Reversal of bacterial locomotion at an obstacle. Phys. Rev. E 73:030901. [DOI] [PubMed] [Google Scholar]
- 9.Copeland, M. F., S. T. Flickinger, H. H. Tuson, and D. B. Weibel. 2010. Studying the dynamics of flagella in multicellular communities of Escherichia coli using biarsenical dyes. Appl. Environ. Microbiol. 76:1241-1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Copeland, M. F., and D. B. Weibel. 2009. Bacterial swarming: a model system for studying dynamic self-assembly. Soft Matter 5:1174-1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Daniels, R., J. Vanderleyden, and J. Michiels. 2004. Quorum sensing and swarming migration in bacteria. FEMS Microbiol. Rev. 28:261-289. [DOI] [PubMed] [Google Scholar]
- 12.Darnton, N., L. Turner, K. Breuer, and H. C. Berg. 2004. Moving fluid with bacterial carpets. Biophys. J. 86:1863-1870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Darnton, N. C., L. Turner, S. Rojevsky, and H. C. Berg. 2010. Dynamics of bacterial swarming. Biophys. J. 98:2082-2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Darnton, N. C., L. Turner, S. Rojevsky, and H. C. Berg. 2007. On torque and tumbling in swimming Escherichia coli. J. Bacteriol. 189:1756-1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eberl, L., S. Molin, and M. Givskov. 1999. Surface motility of Serratia liquefaciens MG1. J. Bacteriol. 181:1703-1712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fraser, G. M., R. B. Furness, and C. Hughes. 2000. Swarming migration by Proteus and related bacteria, p. 381-401. In Y. V. Brun and L. J. Shimkets (ed.), Prokaryotic development. American Society for Microbiology, Washington, DC.
- 17.Fraser, G. M., and C. Hughes. 1999. Swarming motility. Curr. Opin. Microbiol. 2:630-635. [DOI] [PubMed] [Google Scholar]
- 18.Harshey, R. M. 2003. Bacterial motility on a surface: many ways to a common goal. Annu. Rev. Microbiol. 57:249-273. [DOI] [PubMed] [Google Scholar]
- 19.Harshey, R. M. 1994. Bees aren't the only ones: swarming in Gram-negative bacteria. Mol. Microbiol. 13:389-394. [DOI] [PubMed] [Google Scholar]
- 20.Harshey, R. M., and T. Matsuyama. 1994. Dimorphic transition in Escherichia coli and Salmonella typhimurium: surface-induced differentiation into hyperflagellate swarmer cells. Proc. Natl. Acad. Sci. U. S. A. 91:8631-8635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hasegawa, E., R. Kamiya, and S. Asakura. 1982. Thermal transition in helical forms of Salmonella flagella. J. Mol. Biol. 160:609-621. [DOI] [PubMed] [Google Scholar]
- 22.Hauser, G. 1885. Über Fäulnisbakterien und deren Beziehung zur Septicämie. F. G. W. Vogel, Leipzig, Germany.
- 23.Henrichsen, J. 1972. Bacterial surface translocation: a survey and a classification. Bacteriol. Rev. 36:478-503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hotani, H. 1982. Micro-video study of moving bacterial flagellar filaments. III. Cyclic transformation induced by mechanical force. J. Mol. Biol. 156:791-806. [DOI] [PubMed] [Google Scholar]
- 25.Inoue, T., R. Shingaki, S. Hirose, K. Waki, H. Mori, and K. Fukui. 2007. Genome-wide screening of genes required for swarming motility in Escherichia coli K-12. J. Bacteriol. 189:950-957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kaiser, D. 2007. Bacterial swarming: a re-examination of cell-movement patterns. Curr. Biol. 17:R561-R570. [DOI] [PubMed] [Google Scholar]
- 27.Kamiya, R., and S. Asakura. 1977. Flagella transformations at alkaline pH. J. Mol. Biol. 108:513-518. [DOI] [PubMed] [Google Scholar]
- 28.Kamiya, R., and S. Asakura. 1976. Helical transformations of Salmonella flagella in vitro. J. Mol. Biol. 106:167-186. [DOI] [PubMed] [Google Scholar]
- 29.Kim, M. J., and K. S. Breuer. 2007. Use of bacterial carpets to enhance mixing in microfluidic systems. J. Fluids Eng. 129:319-324. [Google Scholar]
- 30.Männik, J., R. Driessen, P. Galadja, J. E. Keymer, and C. Dekker. 2009. Bacterial growth and motility in sub-micron constrictions. Proc. Natl. Acad. Sci. U. S. A. 106:14861-14866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mariconda, S., Q. Wang, and R. M. Harshey. 2006. A mechanical role for the chemotaxis system in swarming motility. Mol. Microbiol. 60:1590-1602. [DOI] [PubMed] [Google Scholar]
- 32.McCarter, L. L. 2004. Dual flagellar systems enable motility under different circumstances. J. Mol. Microbiol. Biotechnol. 7:18-29. [DOI] [PubMed] [Google Scholar]
- 33.O'Toole, G. A. 2008. How Pseudomonas aeruginosa regulates surface behaviors. Microbe 3:65-71. [Google Scholar]
- 34.Powers, T. R. 2002. Role of body rotation in bacterial flagellar bundling. Phys. Rev. E 65:040903. [DOI] [PubMed] [Google Scholar]
- 35.Ramia, M., D. L. Tullock, and N. Phan-Thien. 1993. The role of hydrodynamic interaction in the locomotion of microorganisms. Biophys. J. 65:755-778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rauprich, O., M. Matsushita, C. J. Weijer, F. Siegert, S. E. Esipov, and J. A. Shapiro. 1996. Periodic phenomena in Proteus mirabilis swarm colony development. J. Bacteriol. 178:6525-6538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shapiro, J. A. 1998. Thinking about bacterial populations as multicellular organisms. Annu. Rev. Microbiol. 52:81-104. [DOI] [PubMed] [Google Scholar]
- 38.Sharma, M., and S. K. Anand. 2002. Swarming: a coordinated bacterial activity. Curr. Sci. 83:707-715. [Google Scholar]
- 39.Turner, L., W. S. Ryu, and H. C. Berg. 2000. Real-time imaging of fluorescent flagellar filaments. J. Bacteriol. 182:2793-2801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang, Q., J. G. Frye, M. McClelland, and R. M. Harshey. 2004. Gene expression patterns during swarming in Salmonella typhimurium: genes specific to surface growth and putative new motility and pathogenicity genes. Mol. Microbiol. 52:169-187. [DOI] [PubMed] [Google Scholar]
- 41.Wang, Q., S. Mariconda, A. Suzuki, M. McClelland, and R. M. Harshey. 2006. Uncovering a large set of genes that affect surface motility in Salmonella enterica serovar Typhimurium. J. Bacteriol. 188:7981-7984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Williams, F. D., and R. H. Schwarzhoff. 1978. Nature of the swarming phenomenon in Proteus. Annu. Rev. Microbiol. 32:101-122. [DOI] [PubMed] [Google Scholar]
- 43.Yuan, J., K. A. Fahrner, and H. C. Berg. 2009. Switching of the bacterial flagellar motor near zero load. J. Mol. Biol. 390:394-400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang, R., L. Turner, and H. C. Berg. 2010. The upper surface of an Escherichia coli swarm is stationary. Proc. Natl. Acad. Sci. U. S. A. 107:288-290. [DOI] [PMC free article] [PubMed] [Google Scholar]