Abstract
DcuS is the membrane-integral sensor histidine kinase of the DcuSR two-component system in Escherichia coli that responds to extracellular C4-dicarboxylates. The oligomeric state of full-length DcuS was investigated in vitro and in living cells by chemical cross-linking and by fluorescence resonance energy transfer (FRET) spectroscopy. The FRET results were quantified by an improved method using background-free spectra of living cells for determining FRET efficiency (E) and donor fraction {fD = (donor)/[(donor) + (acceptor)]}. Functional fusions of cyan fluorescent protein (CFP) and yellow fluorescent protein (YFP) variants of green fluorescent protein to DcuS were used for in vivo FRET measurements. Based on noninteracting membrane proteins and perfectly interacting proteins (a CFP-YFP fusion), the results of FRET of cells coexpressing DcuS-CFP and DcuS-YFP were quantitatively evaluated. In living cells and after reconstitution of purified recombinant DcuS in proteoliposomes, DcuS was found as a dimer or higher oligomer, independent of the presence of an effector. Chemical cross-linking with disuccinimidyl suberate showed tetrameric, in addition to dimeric, DcuS in proteoliposomes and in membranes of bacteria, whereas purified DcuS in nondenaturing detergent was mainly monomeric. The presence and amount of tetrameric DcuS in vivo and in proteoliposomes was not dependent on the concentration of DcuS. Only membrane-embedded DcuS (present in the oligomeric state) is active in (auto)phosphorylation. Overall, the FRET and cross-linking data demonstrate the presence in living cells, in bacterial membranes, and in proteoliposomes of full-length DcuS protein in an oligomeric state, including a tetramer.
The DcuSR (dicarboxylate uptake sensor and regulator) system of Escherichia coli is a typical two-component system consisting of a membranous sensor kinase (DcuS) and a cytoplasmic response regulator (DcuR) (11, 26, 48). DcuS responds to C4-dicarboxylates like fumarate, malate, or succinate (19). In the presence of the C4-dicarboxlates, the expression of the genes of anaerobic fumarate respiration (dcuB, fumB, and frdABCD) and of aerobic C4-dicarboxylate uptake (dctA) is activated. DcuS is a histidine protein kinase composed of two transmembrane helices with an intermittent sensory PAS domain in the periplasm (PASP) that was also termed the PDC domain (for PhoQ/DcuS/DctB/CitA domain or fold) (7, 20, 32, 48). The second transmembrane helix is followed by a cytoplasmic PAS domain (PASC) and the C-terminal transmitter domain. PASC functions in signal transfer from transmembrane helix 2 (TM2) to the kinase domain (9). The C-terminal part of the transmitter domain consists of a catalytic or HATPase (histidine kinase/ATPase) subdomain for autophosphorylation of DcuS (16). The N-terminal part of the transmitter contains two conserved α-helical regions, including a conserved His residue which is the site for autophosphorylation. The α-helices serve in dimerization and form a four-helix bundle in the kinase dimer (dimerization and histidine phosphotransfer [DHp] domain) (25, 35, 42, 44).
The dimeric sensor kinases have been supposed to phosphorylate mutually, by the catalytic domain of one monomer, the His residue of the partner monomer (10). The oligomeric state of the membrane-bound sensor kinases EnvZ and VirA was also deduced from in vivo complementation studies (31, 46). In addition, signal transduction across the membrane and along cytoplasmic PAS domains appears to be a mechanical process requiring oligomeric proteins (9, 40). Therefore, His kinases are supposed to be dimeric in the functional state, but a higher oligomeric state has not been tested and is conceivable. Only a limited number of membrane-bound sensor kinases have been studied for their oligomerization in their membrane-bound state. Thus, the oligomeric state of the KdpD and TorS sensor kinases of E. coli have been shown to prevail in the detergent-solubilized state as oligomers, presumably dimers (14, 29). There was indirect information that functional DcuS is a dimer as well. Purified DcuS shows kinase activity only after reconstitution into liposomes, and phosphorylation is stimulated by C4-dicarboxylates (16, 19). Detergent-solubilized DcuS, on the other hand, shows no kinase activity, and it was assumed that reconstituted DcuS prevails as a dimer, whereas the inactivation of the detergent-solubilized form is due to monomerization. Recently, it was suggested that autophosphorylation in a sensor kinase of Thermotoga maritima proceeds by a cis mechanism on DHp and catalytic kinase domains within the same monomer (6). The sensor kinase is supposed to prevail as a dimer for reasons of signal transfer to the sensor domain, but the presence of cis phosphorylation principally brings into question the need for dimers for sensor kinase function.
Overall, it appears that sensor kinases are oligomers for functional reasons. There is, however, no clear evidence for an oligomeric state of full-length sensor kinases in their membrane-embedded state. Moreover, the studies do not address the question of whether the sensor kinases are dimers or higher oligomers. Therefore, several aspects of the oligomeric state of sensor kinases in vivo in bacterial membranes, that is, before solubilization by detergent, are not clear. In this study, the oligomerization of full-length DcuS was examined in vivo in growing bacteria and in bacterial membranes and in vitro after isolation and reconstitution in liposomes by chemical cross-linking and fluorescence resonance energy transfer (FRET) spectroscopy. FRET techniques have been used widely to study intermolecular interactions of biological molecules (1, 4, 18, 21, 23, 34). The sensitivity of fluorescence allows experiments at low concentrations of native proteins, and genetically generated fusions of DcuS with fluorescent proteins ensure site-specific labeling of DcuS for noninvasive and nondestructive measurements in living cells. In particular, it was investigated whether dimers or higher oligomeric states can be detected for DcuS and whether the oligomerization state depends on function-related parameters.
MATERIALS AND METHODS
Bacteria and molecular genetic methods.
The Escherichia coli K-12 strains and plasmids used in this study are listed in Table 1 . Molecular genetic methods were performed according to standard procedures (36). Plasmids were isolated using a QIAprep spin miniprep kit, and PCR products were purified with a QIAquick PCR purification kit (Qiagen, Hilden, Germany). E. coli strains were transformed by electroporation (8). Single and double cysteine mutants of His6-DcuS (pMW324, pMW325, and pMW336) were created in pMW151 using a QuikChange site-directed mutagenesis kit (Stratagene) with primer C199S_cn (5′-CCTTAACCAGAATGCTGGTGCCAATCAG-3′) and the complementary primer C199S_nc or primer C471S_cn (5′-CATCATTAACTTCACTGTGCAGCCAGCC-3′) and the complementary primer C471S_nc. The mutation of dcuS results in the exchange of cysteine to serine residues. For modulation of expression, dcuS was amplified from pMW336 by PCR with oligonucleotide primers dcuS-EcoRI-for (5′-GGATAAGAATTCCCCTCAAG-3′) and dcuS-XbaI-rev (5′-GCCGCAATCTAGATCATC-3′) and cloned via the flanking restriction sites EcoRI and XbaI into the pBAD30-derivative pMW643, resulting in pMW967.
TABLE 1.
Strains of Escherichia coli and plasmids used in this study
| Strain or plasmid | Genotype | Reference or source |
|---|---|---|
| E. coli K-12 strains | ||
| C43(DE3) | Strain for overexpression of membrane proteins carrying a chromosomal T7 polymerase | 28 |
| JM109 | recA1 supE44 endA1 hsdR17 gyrA96 relA1 thi e14− F′ traD36 proAB+lacIq Δ(lacZ)M15 Δ(lac-proAB) | 47 |
| MC4100 | F−araD139 Δ(argF-lac)U169 rpsL150 relA1 flbB530 deoC1 ptsF25 rbsR ΔlacZ | 39 |
| IMW260 | MC4100, but λ[Φ(dcuB′-′lacZ)hyb bla+] dcuS::Camr | 48 |
| Plasmids | ||
| pBAD18-Kan | Expression vector; pBR322 ori, pBAD promoter (Kanr) | 13 |
| pBAD30 | Expression vector; pACYC ori, pBAD promoter (Apr) | 13 |
| pDK108 | Tar1-331-YFP expression plasmid; pBR ori, pTrc promoter, pTrc99a derivative (Apr) | 17 |
| pET28a | Expression vector; pBR322 ori, T7 promoter, His tag (Kanr) | Novagen |
| pECFP | Vector containing enhanced GFP variant CFP (Apr) | Clontech |
| pEYFP | Vector containing enhanced GFP variant YFP (Apr) | Clontech |
| pMW151 | His6-DcuS expression plasmid, pET28a derivative (Kanr) | 16 |
| pMW324 | His6-DcuS (C199S) expression plasmid, pMW151 derivative (Kanr) | This study |
| pMW325 | His6-DcuS (C471S) expression plasmid, pMW151 derivative (Kanr) | This study |
| pMW336 | His6-DcuS (C199S C471S) expression plasmid, pMW151 derivative (Kanr) | This study |
| pMW384 | DcuS-YFP expression plasmid, pMW391 derivative (Kanr) | 37 |
| pMW386 | DcuS-CFP expression plasmid, pMW393 derivative (Kanr) | This study |
| pMW391 | C-terminal YFP protein fusion plasmid, pET28a derivative (Kanr) | 37 |
| pMW393 | C-terminal CFP protein fusion plasmid, pET28a derivative (Kanr) | This study |
| pMW407 | DcuS-YFP expression plasmid, pBAD30 derivative (Apr) | 37 |
| pMW408 | DcuS-CFP expression plasmid, pBAD18-Kan derivative (Kanr) | This study |
| pMW643 | pBAD30 with additional Tetr | This study |
| pMW762 | CFP expression plasmid, pBAD18-Kan derivative (Kanr) | This study |
| pMW765 | YFP expression plasmid, pBAD30 derivative (Apr) | This study |
| pMW766 | CFP-YFP expression plasmid, pBAD18-Kan derivative (Kanr) | This study |
| pMW967 | His6-DcuS (C199S C471S) expression plasmid, pMW643 derivative (Apr Tetr) | This study |
Construction of dcuS-YFP, dcuS-CFP, and CFP-YFP fusions.
The dcuS-YFP fusion (pMW407) was constructed and cloned into pBAD30 as described previously (37). The dcuS-CFP fusion was constructed in the same way, whereas the ECFP gene was amplified from plasmid pECFP (Clontech) and dcuS-CFP was finally cloned from pMW386 via XbaI into pBAD18-Kan. The resulting construct (pMW408) encoded DcuS(1-539)-(Lys)-ECFP(4-240). The fusion protein also carried an N-terminal His6 tag and a thrombin cleavage site. For coexpression of cyan fluorescent protein (CFP) and yellow fluorescent protein (YFP) in one cell, the enhanced fluorescent protein genes were amplified from pECFP or pEYFP (Clontech), respectively, by PCR with oligonucleotide primers for-gfp-NcoI (5′-GGTCGCCACCATGGTGAGC-3′) and HindIII-gfp-rev (5′-CACCAGACAAGAAGCTTGTAATGG-3′). Either PCR fragment was subcloned via the flanking restriction sites NcoI and HindIII into pET28a (Novagen). The ECFP and EYFP genes were subsequently cloned into pBAD18-Kan or pBAD30, respectively, using restriction endonucleases XbaI and HindIII, resulting in pMW762 and pMW765. The CFP-YFP fusion gene was constructed as follows. The ECFP gene was amplified from pECFP with primers for-gfp-NcoI and rev-BamHI-gfp (5′-GGAATTCTAGAGTCGGATCCGCTATACTTG-3′) and the EYFP gene was amplified from pEYFP with primers BamHI-gfp-for (5′-CTCTAGAGGGATCCCGGGTAC-3′) and HindIII-gfp-rev. The ECFP gene fragment was subcloned via NcoI and BamHI into pET28a, and the EYFP gene fragment was then cloned behind the ECFP gene via BamHI and HindIII. The ECFP-EYFP fusion gene was subsequently cloned into pBAD18-Kan using restriction endonucleases XbaI and HindIII. The resulting construct (pMW766) encoded ECFP(1-239)-Linker(9 aa)-EYFP(1-240) (here referred to as CFP-YFP). The sequences of the resulting constructs were verified by DNA sequencing.
E. coli expressing fluorescent proteins.
Expression plasmids of fluorescent protein fusions were transformed into E. coli strain JM109. Bacteria were grown aerobically in LB medium at 30°C for 3 to 4.5 h, corresponding to the mid-exponential phase of growth (36), and induced from the beginning of incubation with 133 to 333 μM l-arabinose as indicated below. Ampicillin or kanamycin were added at a concentration of 100 μg/ml or 50 μg/ml, respectively. When two plasmids were coexpressed in one cell, ampicillin and kanamycin were added to a concentration of 50 μg/ml and 25 μg/ml, respectively. After harvest, E. coli cells were washed twice by spin centrifugation with phosphate-buffered saline (PBS) buffer and then resuspended in PBS buffer, pH 7.5. Absorption spectra were recorded before fluorescence measurements. Cells were diluted to an absorbance of 0.1 at 400 nm to avoid the inner filter effect and signal saturation. Emission spectra were recorded during excitation at 433 nm or 488 nm, respectively. As a protein that does not interact with DcuS-CFP in the membrane, Tar1-331-YFP was expressed under the control of pTrc99a and induced by 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) (pDK108 from V. Sourjik, ZMBH, Heidelberg, Germany). DcuS-CFP and Tar1-331-YFP were coexpressed in E. coli cells in the presence of both l-arabinose and IPTG. The YFP fluorescence of all fusion proteins was comparable to that of YFP.
Absorption and fluorescence spectroscopy.
Absorption and fluorescence spectra were measured in 1-ml quartz cuvettes (semimicro cuvettes) at room temperature with a dual-beam UV-visible spectrophotometer (OMEGA 20; Bruins Instruments, Germany) and a FluoroMax-2 spectrofluorometer (Jobin Yvon-Spex, NJ). Fluorescence spectra were corrected for the wavelength dependence of the fluorometer.
Purification of cytosolic CFP or YFP for reference spectra.
CFP or YFP protein was expressed from a 20-ml culture of E. coli JM109(pECFP) or JM109(pEYFP) grown aerobically in Luria-Bertani medium (LB) for 4 h after 1% inoculation of the overnight culture (36). Cells were harvested by centrifugation (6,300 × g for 10 min at 4°C), washed, and resuspended in 20 ml buffer (50 mM Tris-HCl, pH 7.7, 10 mM MgCl2). Cell supernatants were prepared by 3 cycles of treatment in a French press at 138 × 106 Pa and cleared by centrifugation (8,600 × g for 10 min at 4°C) to remove the cell debris and, subsequently, by ultracentrifugation (200,000 × g for 45 min at 4°C) to separate the cytosolic (soluble protein) fraction from the membrane fraction. The supernatant containing the cytosolic fraction was subjected several times to filter centrifugation (molecular mass cutoff, 10 kDa; Vivascience) to separate fluorescent proteins from small fluorescent molecules. The measured spectra of purified CFP and YFP were normalized to unity by setting the highest fluorescence intensity to 1.0 for use as reference spectra of donor and acceptor, respectively. Emission spectra were recorded at an excitation wavelength of 433 nm or 488 nm, respectively.
Overexpression and purification of His6-DcuS.
Overexpression and isolation of His6-DcuS was performed similarly to the method of Janausch et al. (16). Variants of DcuS encoded by plasmids pMW151, pMW324, pMW325, and pMW336 were expressed in E. coli C43DE3. The membrane fraction was solubilized with 2% Empigen BB. After centrifugation of the solubilized membrane fraction at 300,000 × g for 50 min, the supernatant was applied to a Ni2+-nitrilotriacetic acid (NTA)-agarose column (3 ml; Qiagen) equilibrated in buffer W (50 mM Tris-HCl, pH 7.7, 10% glycerol, 0.5 M NaCl, 0.04% lauryl dimethylamine N-oxide [LDAO, Fluka], 20 mM imidazole). The column was washed with 40 ml of buffer W, and bound His6-DcuS was eluted with 5 ml of buffer E (buffer W with 500 mM imidazole). Samples of 1 ml were collected and analyzed by SDS-PAGE. Elution fractions of >90% purity were pooled and dialyzed against buffer D (buffer W without imidazole). Protein concentrations were determined with RotiQuant (Roth) according to the method of Bradford (5). Samples were frozen in liquid N2 and stored at −80°C.
Reconstitution of His6-DcuS in liposomes.
Liposomes were prepared from E. coli phospholipids (Avanti Polar Lipids, Inc.) as described previously (16). The liposomes were destabilized by the addition of Triton X-100 at an effective detergent/lipid ratio of 2.5. Purified His6-DcuS was added, if not stated otherwise below, at a protein/phospholipid ratio of 1:20 (mg/mg) and stirred gently for 10 to 15 min at 20°C. For every mg of Triton X-100, 5 mg degassed Bio-Beads SM-2 (Bio-Rad), pretreated as described previously (15), was added two times, stepwise, to remove the detergent. The suspension was incubated overnight at 4°C. Subsequently, a further 5 mg of fresh Bio-Beads per mg of Triton X-100 was added to the suspension and incubated for 1 h at 20°C. The supernatant was collected from the Bio-Beads and the volume brought to 1 ml with buffer R (50 mM Tris-HCl, pH 7.7, 10% glycerol). The proteoliposomes were centrifuged at 300,000 × g for 45 min, washed twice, resuspended in buffer R to a volume of 1 ml, frozen for three cycles in liquid N2, and thawed slowly at 20°C. After the final freezing, the proteoliposomes were stored at −80°C until further use. When required, 20 mM Na2-fumarate was added to all buffers.
Fluorescence labeling of His6-DcuS.
Fluorescent dyes (Alexa 488-maleimide and Alexa 594-maleimide; Invitrogen) were dissolved in dimethyl sulfoxide (DMSO) to yield 10 mM stock solutions and freshly diluted by DcuS-labeling buffer (50 mM Tris-HCl, pH 7.2, 500 mM NaCl, 5% glycerol, 0.04% LDAO, and 5 mM imidazole). The reaction mixture, consisting of 1 mg/ml of purified His6-DcuS and 50 μM or 160 μM thiol-reactive dyes in DcuS-labeling buffer, was incubated in the dark overnight at 4°C. Unbound dyes were removed by PD-10 column (Sephadex TMG-25; Amersham Biosciences). Labeled protein was purified and concentrated in a Vivaspin concentrator (molecular mass cutoff, 30 kDa; Vivascience) at 4°C to a volume of 1 ml. The extent of labeling was estimated spectrophotometrically according to the absorption spectrum of labeled His6-DcuS by measuring dye absorbance and protein concentration. Labeled His6-DcuS was kept on ice or reconstituted into liposomes. Emission spectra were recorded at an excitation wavelength of 480 nm (Alexa 488) or 580 nm (Alexa 594).
Chemical cross-linking.
Prior to cross-linking, detergent-solubilized His6-DcuS was dialyzed against 25 mM potassium phosphate, pH 7.4, 300 mM NaCl, 10% glycerol, 0.04% LDAO; reconstituted His6-DcuS was resuspended in 25 mM potassium phosphate, pH 7.4, 10% glycerol; and isolated membranes containing overexpressed His6-DcuS were homogenized in 20 mM morpholinepropanesulfonic acid (MOPS), pH 7.2, 200 mM NaCl. For cross-linking reactions, 3 μg of solubilized His6-DcuS, 6 μg of reconstituted His6-DcuS, or 30 μg of isolated membranes was mixed with cross-linking buffer (20 mM HEPES, pH 7.4, 20 mM KCl, 250 mM sucrose, 1 mM EDTA) to a reaction mixture volume of 20 μl. For in vivo cross-linking, E. coli JM109(pMW967) was grown aerobically in LB medium at 30°C and induced from the beginning of incubation with 0 to 333 μM l-arabinose as indicated below. Tetracycline was added at a concentration of 15 μg/ml. Mid-exponential-phase cells were harvested, washed with PBS buffer, pH 7.5, and resuspended in PBS buffer prior to cross-linking. Disuccinimidyl suberate (DSS; Sigma-Aldrich) dissolved (25 mM) in DMSO was added to a final concentration of 30 μM; in the control reaction mixtures, DMSO was added instead of DSS. The reaction mixtures were incubated with agitation at 20°C for 15 min. The cross-linking reaction was stopped by the addition of 1 M Tris-HCl, pH 7.7, to a final concentration of 100 mM. Samples were dissolved and boiled in 2× SDS sample buffer (22) containing 1 mM dithiothreitol (DTT) for 5 min, subjected to SDS-10% polyacrylamide gel electrophoresis (200 V for 70 min; Bio-Rad Mini-PROTEAN tetra cell system), and transferred to a nitrocellulose membrane (43). The proteins were treated with rabbit polyclonal antiserum (Eurogentec) raised against the periplasmic domain of DcuS and further detected with secondary IgG antibodies coupled to peroxidase (Sigma-Aldrich).
RESULTS
Oligomerization of full-length DcuS was investigated by two different methods for cross-linking of proteins and by FRET spectroscopy of fluorescently labeled samples, both in vitro and in vivo. Cross-linking was achieved by disulfide formation from native Cys residues of DcuS and by the chemical linker disuccinimidyl suberate (DSS) interacting with amino groups. By the combination of the methods, oligomerization was studied in a comparative manner for DcuS present in four different types of preparations, that is, detergent-solubilized His6-DcuS, His6-DcuS reconstituted in liposomes, DcuS in membrane preparations of the bacteria, and DcuS in vivo in growing bacteria.
Dimerization of DcuS by intermolecular Cys471-disulfide formation.
DcuS contains two cysteine residues, one of which (C199) is located in transmembrane helix TM2 and the second (C471) in the cytosolic kinase domain. The oligomeric state of DcuS was analyzed with purified His6-DcuS, single mutants DcuSC199S and DcuSC471S, and the double Cys mutant (DcuSCys−) that lacks both Cys residues. The functional state of the DcuS cysteine mutants was tested in vivo by measuring the induction of the DcuS-dependent dcuB gene using a dcuB′-′lacZ reporter gene fusion (Table 2). The plasmid-encoded cysteine mutants of DcuS were able to complement dcuB-lacZ expression in a dcuS deletion strain. After anaerobic growth in the presence of fumarate, the mutants retained at least 68% of the wild-type activity, demonstrating that the Cys residues are not essential for inducing DcuS-dependent dcuB expression.
TABLE 2.
Functional test for DcuS-CFP, DcuS-YFP, and cysteine mutants of DcuS in vivo by reporter gene measurement of dcuB-lacZ expressiona
| Strain (relevant genotype) | Type of DcuS present in bacteria |
dcuB-lacZ expression (mean ± SD [Miller units]) |
|
|---|---|---|---|
| With fumarate | Without fumarate | ||
| IMW260 (lacking dcuS) | 5 ± 1 | 8 ± 2 | |
| IMW260 pMW151 (dcuS+) | DcuSWT | 279 ± 32 | 2 ± 1 |
| IMW260 pMW324 (dcuS C199S) | DcuSC199S | 261 ± 5 | 3 ± 1 |
| IMW260 pMW325 (dcuS C471S) | DcuSC471S | 191 ± 9 | 3 ± 1 |
| IMW260 pMW336 (dcuS C199S C471S) | DcuSCys− | 189 ± 8 | 3 ± 1 |
| IMW260 pMW384 (dcuS-YFP) | DcuS-YFP | 332 ± 33 | 9 ± 2 |
| IMW260 pMW386 (dcuS-CFP) | DcuS-CFP | 301 ± 30 | 6 ± 3 |
E. coli IMW260 containing the plasmids shown in the table was grown anaerobically in eM9 medium (27) containing glycerol (50 mM) and dimethyl sulfoxide (20 mM) as growth substrates with and without fumarate (20 mM) as effector.
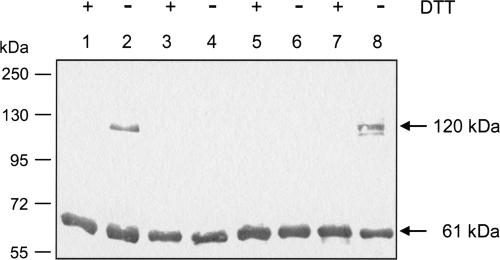
To investigate the oligomeric state in vitro, recombinant DcuS protein was solubilized from the bacterial membranes with nondenaturing detergent Empigen BB and purified in the detergent LDAO (16). The wild-type and Cys mutant forms of DcuS were applied to SDS-PAGE in the presence or absence of the reducing agent DTT. Based on the immunostained SDS-PAGE (Fig. 1) gel, all DcuS variants revealed a band of approximately 61 kDa, corresponding to the apparent molar mass of the monomer. Wild-type DcuS (DcuSWT) and the mutant form DcuSC199S produced an additional band of about 120 kDa responding to anti-DcuS serum under oxidizing conditions (Fig. 1, lanes 2 and 8). The 120-kDa band was completely missing when the same samples were treated with DTT before SDS-PAGE (Fig. 1, lanes 1 and 7). Therefore, the 120-kDa band represents a DcuS dimer that is formed by intermolecular disulfide bonds derived from Cys471. For DcuSCys−, no dimerization was detected because of the lack of oxidizable Cys residues (Fig. 1, lane 4). DcuSC471S that contains only the Cys199 residue showed only a monomeric band, indicating that Cys199 is not accessible to disulfide bond formation (Fig. 1, lane 6). When DcuS was isolated and reconstituted in liposomes before SDS-PAGE, the same response as for the detergent-solubilized DcuS forms was obtained (not shown), and DcuSWT and DcuSC199S formed the Mr 120,000 band under oxidizing conditions. The dimerization is only observed under oxidizing conditions and has presumably no direct role for DcuS function in vivo, since in E. coli, the cytosolic Cys residue is in the reduced state even under aerobic conditions. Moreover, the high activity of the Cys double mutant demonstrates that the Cys residues are not essential for function and dimerization.
FIG. 1.
Influence of oxidizing conditions on the oligomerization of purified His6-DcuS. Detergent-solubilized and purified variants of His6-DcuS (1 μg) were subjected to SDS-PAGE in the presence (+) or absence (−) of DTT. DcuS was detected by Western blotting with antiserum against the periplasmic domain of DcuS. The anti-DcuS-positive bands of 61 and 120 kDa correspond to monomeric and dimeric DcuS. Lanes: 1 and 2, DcuSWT; 3 and 4, DcuSCys−; 5 and 6, DcuSC471S; and 7 and 8, DcuSC199S. For calibration, PageRuler Plus prestained protein ladder (Fermentas) was used.
Determination of the oligomeric state of DcuS by chemical cross-linking.
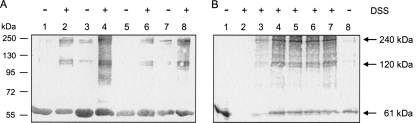
For chemical cross-linking, disuccinimidyl suberate (DSS) was used at low concentrations (30 μM) in order to avoid unspecific cross-linking owing to abundant cross-linker. DcuS samples were prepared either in the detergent-solubilized and purified form, after reconstitution in liposomes, or in bacterial membranes. The presence of DTT disturbed the cross-linking reaction by DSS (not shown), and therefore, cross-linking was performed in the absence of DTT. When purified, detergent-solubilized DcuSWT was used for cross-linking, the immunoblot showed unmodified DcuS (61 kDa) and an additional band of 120 kDa (Fig. 2, lanes 1 and 2). In the same experiment performed with detergent-solubilized DcuSCys−, the 120-kDa DcuS band was not found (lanes 3 and 4). This suggests that DcuSWT forms dimers by intermolecular disulfides prior to the DSS cross-linking procedure, similar to the results of the experiments shown in Fig. 1. In order to avoid interference from cross-linking by disulfide formation between DcuS monomers, all further experiments (Fig. 2, lanes 5 to 9, and Fig. 3) were performed with DcuSCys−. After reconstitution in liposomes and treatment with DSS, DcuSCys− produced a weak and diffuse band of 120 kDa and a clear band of approximately 240 kDa in addition to the 61-kDa band (Fig. 2, lane 6). Under the same conditions, a further band of approximately 90 kDa was formed when DcuS from bacterial membranes or from proteoliposomes was tested. The 90-kDa band, which is of unknown composition, was generally weak but showed various intensities. Overall, the pattern suggests that by incubation with DSS, DcuS from bacterial membranes or proteoliposomes can be detected in a tetrameric state. The 120-kDa band corresponds to a dimer. A very similar pattern comprising the 61-kDa, 120-kDa, and 240-kDa anti-DcuS-responsive bands is found for the cross-linking of DcuSCys− from bacterial membranes (lane 8). A band of 240 kDa is also observed for detergent-solubilized DcuSCys− without cross-linking when higher quantities of the protein are applied to the SDS-PAGE gel (lane 9). A weak band of 240 kDa was also detected in all other forms of detergent-solubilized and reconstituted DcuS when applied in high quantities to the SDS-PAGE gel (not shown). Therefore, at least some DcuS is present in the tetrameric state, and this state is retained in part even after solubilization and after incubation in dilute solutions of SDS.
FIG. 2.
Determination of the oligomeric state of DcuS by chemical cross-linking. Detergent-solubilized His6-DcuS, His6-DcuS reconstituted in liposomes, or preparations of His6-DcuS embedded in bacterial membranes of the bacteria was cross-linked with disuccinimidyl suberate (DSS). Samples were subjected to SDS-PAGE in the presence of DTT, blotted to nitrocellulose membranes (Protran), and immunostained with anti-DcuS. The anti-DcuS-positive bands of 61, 120 and 240 kDa correspond to monomeric, dimeric, and tetrameric DcuS, respectively. Lanes: 1 and 2, solubilized DcuSWT (3 μg); 3 and 4, solubilized DcuSCys− (3 μg); 5 and 6, reconstituted DcuSCys− (3 μg); 7 and 8, bacterial membranes (20 μg) containing DcuSCys−; and 9, solubilized DcuSCys− (7 μg). For calibration, PageRuler Plus prestained protein ladder (Fermentas) was used.
FIG. 3.
Effect of DcuSCys− concentration on dimer and tetramer formation in proteoliposomes (A) and in bacterial membranes of living cells (B). (A) Purified recombinant DcuSCys− (8 μg) was reconstituted in liposomes in the following protein/phospholipid ratios: 1:10 (lanes 1 and 2), 1:20 (lanes 3 and 4), 1:50 (lanes 5 and 6), and 1:100 (lanes 7 and 8). Samples were subjected to SDS-PAGE in the presence of DTT before and after cross-linking with disuccinimidyl suberate (DSS), blotted to nitrocellulose membranes (Protran), and immunostained with anti-DcuS. For calibration, PageRuler Plus prestained protein ladder (Fermentas) was used. (B) E. coli JM109(pMW967) (expressing DcuSCys−) was grown aerobically in LB broth in the presence of 0 μM (lane 2), 10 μM (lane 3), 50 μM (lane 4), 90 μM (lane 5), 133 μM (lanes 6 and 8), or 333 μM (lane 7) arabinose and cross-linked with DSS. Sixty micrograms (lanes 4 to 8), 180 μg (lane 3), or 230 μg (lane 2) of cell lysates were subjected to SDS-PAGE. DcuS was detected by Western blotting with antiserum against the periplasmic domain of DcuS. Lane 1 contains 1 μg of purified DcuSCys−. The anti-DcuS-positive bands of 61, 120, and 240 kDa correspond to monomeric, dimeric, and tetrameric DcuS, respectively.
The oligomeric state of DcuS is independent of DcuS concentration.
To examine the effect of DcuS concentration on the portion of DcuS that can be detected in the oligomeric state, proteoliposomes were produced with increasing contents of DcuSCys− relative to the lipid content. The ratio was varied by a factor of 10 by increasing the protein/lipid ratio from 1:100 to 1:10 (wt/wt) (Fig. 3A). The protein was cross-linked by DSS and analyzed by SDS-PAGE and immunoblotting. All DSS-treated samples contained DcuS of the 120-kDa and 240-kDa forms in addition to the 61-kDa form, even some samples that were not treated with DSS, as is also shown in Fig. 2. The contents of the high-Mr forms of DcuS showed some variation, but there was no systematic increase in the samples with high DcuS contents. In a similar approach, the effect of the DcuS concentration on the amount of oligomers was studied in vivo (Fig. 3B). E. coli containing the low-copy-number DcuSCys− expression plasmid pMW967 was induced for increasing levels of DcuSCys− by increasing the levels of the inducer (0 to 333 μM arabinose), as described earlier (37). The proteins from the cell homogenate were subjected to SDS-PAGE and analyzed for DcuS by immunoblotting. The samples containing 0 to 90 μM arabinose (Fig. 3B, lanes 2 to 5) showed an approximately 8- to 10-fold increase in the content of DcuS (Fig. 3B) (37). But again, all samples with detectable DcuS contained DcuS of the 61-kDa, the 120-kDa, and the major 240-kDa form, and there was no overproportional increase for the high-Mr forms of DcuS with increasing induction of DcuS. Again, the cell homogenate (Fig. 3B, lane 8) displayed weak additional oligomeric bands even when not incubated with DSS.
Quantitative FRET analysis in vivo and in vitro.
The FRET studies were performed with DcuS fused to the CFP or YFP derivative of the GFP protein or with DcuS labeled chemically with fluorescent dyes. DcuS forms stable fusion proteins with CFP or YFP, as tested earlier by immunoblotting (37). For quantitative and reproducible analysis of FRET efficiency, a method described by Gordon et al. (12) was extended. The approach described here is based on intermolecular FRET measurements and does not require any absorbance data. The analysis not only yields transfer efficiencies but also measures for the donor/acceptor concentrations, which allows determination of the donor fraction. This method presumes background-free fluorescence signals of the donor, the acceptor, and mixtures of both. Fluorescence spectra from cellular systems are often strongly contaminated with background signals of different origins. Background subtraction was performed by a multiparameter fitting procedure. The accuracy and reliability of the method was carefully validated by Monte-Carlo simulations and experimental studies of donor/acceptor model systems (see below and the supplemental material). This allows quantitative FRET analysis in living cells.
Alexa-labeled DcuS monomers in detergent and oligomers in liposomes.
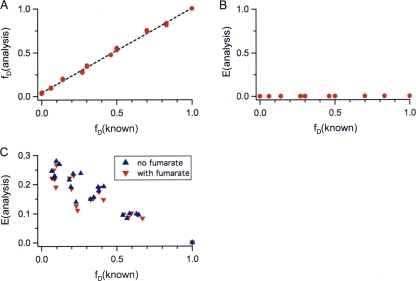
Detergent-solubilized His6-DcuS was tested for FRET in vitro by a mixture series of donor- and acceptor-labeled DcuS monomers in detergent (LDAO) solution. Two aliquots of DcuS protein were labeled with either Alexa 488 (donor) or Alexa 594 (acceptor). Subsequently, donor- and acceptor-labeled DcuS were mixed at different known donor fractions [fD(known)] from 0 to 1. The spectra of these samples revealed background (e.g., scattering due to micelles), which was removed as described in the supplemental material (equation 1, without E. coli background, i.e., parameter d was set to zero and fixed). The values for the donor fraction [fD(analysis)] and transfer efficiency [E(analysis)] were calculated as described in equations 2 to 7 in the supplemental material. In Fig. 4, the values based on these analyses are plotted versus the known experimental values of the donor fraction [fD(known)]. The donor fractions (Fig. 4A) are distributed along a diagonal with a slope of 1, which revealed a very good agreement between known values and those retrieved from the analysis. No FRET was observed in the measured transfer efficiencies (Fig. 4B, E = 0), indicating the absence of oligomerization of detergent-solubilized His6-DcuS.
FIG. 4.
In vitro FRET measurements: mixture series of Alexa-labeled DcuS subunits (A, B) and FRET of reconstituted DcuS (C). (A, B) Detergent-solubilized His6-DcuS was labeled with either Alexa 488 (donor) or Alexa 594 (acceptor) and mixed at different ratios with known donor fractions [fD(known)] of 0 to 1. The final protein concentration was 0.5 μM in DcuS-labeling buffer. The recorded spectra, excited at 480 nm and 580 nm, were analyzed (see the supplemental material) to determine the donor fraction [fD(analysis)] and the transfer efficiency [E(analysis)] of each sample. The values are plotted against the known donor fractions. (C) FRET of reconstituted Alexa-labeled His6-DcuS in liposomes. Detergent-solubilized His6-DcuS was labeled with either Alexa 488 (donor) or Alexa 594 (acceptor) and mixed at different ratios. Subsequently, the mixture of Alexa-labeled His6-DcuS was reconstituted into liposomes. The recorded spectra, excited at 480 nm and 580 nm, were analyzed. Results of FRET of labeled His6-DcuS in liposomes with (red triangles) or without (blue triangles) Na2-fumarate (20 mM) are shown. Spectra were measured in Tris buffer (50 mM, pH 7.7) without fumarate. Results are from five independent test series each (in total, n = 25 data points with or without fumarate).
For studies of the oligomeric state in membranes, recombinant DcuSC199S that retained Cys471 for labeling was used. Detergent-solubilized DcuSC199S was purified, and two aliquots were labeled with either the donor or acceptor (Alexa 488 or Alexa 594). Subsequently, donor- and acceptor-labeled DcuS were mixed at different donor fractions, and each mixture was reconstituted into liposomes separately. A clear FRET signal was detected for reconstituted DcuS (Fig. 4C), whereas the same mixtures of labeled DcuS before reconstitution in detergent showed no energy transfer (Fig. 4B, E = 0). The FRET signal of the reconstituted samples did not change in the absence or presence of fumarate (Fig. 4C). The data indicate oligomerization of DcuS upon reconstitution in liposomes and lack of oligomerization or lack of mixing of the two differently labeled forms in the detergent-solubilized samples.
To study the effect of the DcuS concentration on oligomerization, FRET studies were performed with a mixture of His6-DcuS labeled with either Alexa 488 or Alexa 594 after reconstitution of the mixture in various amounts of phospholipid. The mixture of the Alexa-labeled proteins was reconstituted at protein/phospholipid ratios of 1:10 to 1:160 (mg/mg). FRET was detected at all protein/phospholipid ratios, indicating that DcuS in the sample with low protein content still forms oligomers in highly diluted samples to an extent similar to that in the more concentrated DcuS proteoliposomes. The FRET signal, however, cannot be differentiated for the presence of dimeric and tetrameric DcuS.
DcuS-YFP and DcuS-CFP fusion proteins for in vivo FRET are functional.
For studies on DcuS in growing bacteria, fusions of DcuS to the enhanced cyan and yellow fluorescent proteins (CFP and YFP) were used as a donor-acceptor pair. DcuS was genetically fused with CFP for use as the FRET donor or with YFP as the FRET acceptor. The fusions were placed on different plasmids within E. coli cells. In the DcuS-YFP fusion (encoded by pMW407), the DcuS protein is still functional in sensing and signal transduction in vivo (Table 2), and the YFP protein shows normal fluorescence (37). The DcuS-CFP fusion protein (encoded by pMW408) was tested in a similar way for the functionality of DcuS. Using dcuB-lacZ as a DcuS-DcuR-dependent reporter system, the DcuS-CFP fusion remains fully functional in vivo, comparable to wild-type DcuS (Table 2). The fluorescence of the CFP protein in the fusion was confirmed by fluorescence spectroscopy. Taken together, DcuS, CFP, and YFP were all functional in the fusions used.
Validation of FRET in living cells.
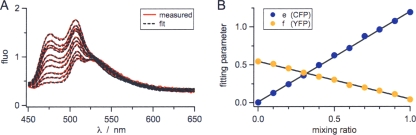
Background subtraction for in vivo FRET by the CFP and YFP proteins (equation 1 in the supplemental material) was evaluated experimentally by using defined mixtures of E. coli cells which expressed cytosolic CFP, YFP, or neither, producing only background FRET due to autofluorescence and Raman and Rayleigh scattering (see Fig. S1 in the supplemental material). Mixtures with various fluorophore concentrations were tested for their effect on the performance of the fitting procedure (an example, a mixture series of CFP-expressing cells and YFP-expressing cells in the presence of a fixed amount of E. coli cells, is shown in Fig. 5). The fitting results showed a linear increase of the CFP amount starting from the origin and a linear decrease for YFP ending at the coordinates 1.0 (Fig. 5B). The linearity of both curves and the absence of any offsets demonstrated that the fitting procedure can be applied to samples with various ratios of CFP and YFP.
FIG. 5.
Validation of FRET in living cells: mixture series of CFP-expressing cells and YFP-expressing cells in the presence of a fixed amount of E. coli cells. (A) In the presence of a constant amount of non-FP-expressing E. coli cells (volume of E. coli [VEcoli], 0.5 ml), CFP-expressing cells and YFP-expressing cells were mixed at different ratios to a total volume (VCFP + VYFP + VEcoli) of 1 ml. Emission spectra were measured in PBS buffer, pH 7.5, with excitation at 433 nm. Measured spectra (red) were analyzed by multiparameter fitting with equation 1 in the supplemental material (black dashed line). The spectra with increasing fluorescence correspond to mixtures with increasing contents of CFP-expressing cells. (B) The fitting results for parameter e (contribution of CFP) and parameter f (contribution of YFP) are plotted versus the mixing ratio [VCFP/(VCFP + VYFP)].
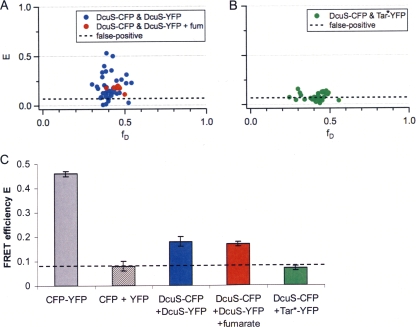
The applicability of the method was also tested in vivo by a tandem fusion of CFP-YFP. The tandem fusion protein is located in the cytoplasm and serves as a positive control for maximal FRET efficiency. Here, the ratio between the donor and acceptor is fixed at 1:1, and therefore, the donor fraction, fD(known), is 0.5. Although the FRET efficiency of the fusion protein is unknown, the observed FRET efficiencies can be estimated based on the distance between the donor and acceptor (r) and the Förster radius (R0) between CFP and YFP (R0 = 4.92 nm) (33). The chromophore is buried in the center of the GFP barrel (30, 45), and the length of an individual CFP or YFP protein is around 4 nm. If CFP and YFP moieties are covalently linked without any spacer in the tandem fusion, the distance, r, between the chromophores is roughly 4 nm, which is smaller than the Förster radius and can result in a FRET efficiency of 0.78 at most, obtained by the equation E = 1/[1 + (r/R0)6] = 0.78. However, a link of 9 amino acid residues between CFP and YFP might increase the distance (r) and in turn lower the FRET efficiency. The experimental results from bacteria expressing the CFP-YFP fusion protein [JM109(pMW766)] confirmed the expectations: a donor fraction, fD, of 0.49 ± 0.01 (mean ± standard deviation of the mean) and a transfer efficiency, E, of 0.46 ± 0.01 were determined (n = 26 data points) (Fig. 6 C).
FIG. 6.
FRET of CFP- and YFP-labeled DcuS in vivo. (A) Coexpression of DcuS-CFP and DcuS-YFP in E. coli cells was induced with arabinose (133 μM) for 1.5 h to 6 h in the absence (blue) or presence (red) of the effector fumarate (fum; 20 mM). Results are from 7 independent test series (in total, n = 52 data points). All samples were measured in PBS buffer, pH 7.5. (B) DcuS-CFP and Tar1-331-YFP (Tar*-YFP) (17) were coexpressed in E. coli cells for 3 to 6 h. Results are from 6 independent test series (in total, n = 39 data points). The emission spectra of all samples were recorded by excitation at 433 nm and 488 nm, respectively, and subsequently analyzed with equations 1 to 7 in the supplemental material. The average of the DcuS/Tar transfer efficiencies was marked as a background for false-positive results due to CFP/YFP interaction. (C) FRET efficiencies (mean ± standard deviation of the mean) of the FRET pairs CFP and YFP directly coupled in the fusion protein encoded by pMW766, the separate CFP and YFP proteins encoded by pMW762 and pMW765, DcuS-CFP and Tar1-331-YFP (Tar*-YFP) (17) encoded by pMW408 and pDK108, and DcuS-CFP and DcuS-YFP encoded by pMW408 and pMW407. The proteins were expressed or coexpressed in E. coli JM109. The average of the DcuS/Tar and CFP/YFP transfer efficiencies was marked as a background (dashed horizontal line) for false-positive results due to CFP/YFP interaction.
DcuS-DcuS interaction.
E. coli JM109(pMW407 pMW408) coexpressing DcuS-YFP and DcuS-CFP was grown in the absence of fumarate and harvested after various times of induction with 133 μM arabinose. The recorded spectra of the samples were evaluated as described in the supplemental material. The expression levels of DcuS-CFP and DcuS-YFP gradually increased with increasing induction time (not shown) (37), but the donor fractions were in a limited range with an average fD of 0.41 ± 0.01 (mean ± standard deviation of the mean). Substantial FRET efficiencies, with an E value of 0.18 ± 0.02, were observed in six independent induction series (n = 44 data points, mean ± standard deviation of the mean) (Fig. 6A and C). For comparison, the FRET signal in bacteria [JM109(pMW762 pMW765)] coexpressing CFP and YFP in the same cells but as independent cytosolic proteins without fusion was distinctly lower (E = 0.08 ± 0.02, fD = 0.47 ± 0.02, n = 16 data points, mean ± standard deviation of the mean) (Fig. 6C). The signal for the cytosolic CFP-YFP fusion protein representing the ideal FRET pair (E = 0.46), on the other hand, was higher than the signal for the DcuS-CFP/DcuS-YFP pair (Fig. 6C).
The GFP protein and its derivatives are known to form dimers (30, 45). Therefore, it was tested whether the oligomerization of DcuS-CFP/DcuS-YFP is independent from a potential interaction of this type. For this purpose, a Tar1-331-YFP fusion protein (17) was analyzed by FRET for interaction with DcuS-CFP. Tar is a membrane protein and functions as a chemotaxis receptor in E. coli. DcuS and Tar are functionally independent. Here, the YFP fusion of a C-terminally truncated Tar1-331, which is homogeneously distributed in the cell membrane, was used (17), in contrast to the polar-located DcuS (37). Donor fractions and transfer efficiencies between DcuS-CFP and Tar1-331-YFP were determined in E. coli JM109(pMW408 pDK108) for 39 samples (Fig. 6 B). The donor fraction, fD, was 0.41 ± 0.01 (mean ± standard deviation of the mean). A minor FRET signal with an E value of 0.07 ± 0.01 was detected (mean ± standard deviation of the mean) (Fig. 6C), which is the same range as for the cytosolic CFP and YFP proteins when present as independent entities (Fig. 6C). FRET at this level may exist without specific interaction between CFP- and YFP-tagged fusions that are colocalized either in the membrane or in the cytosol. Specific FRET signals should be clearly above this background, with an E value of 0.08, which represents a threshold value in our study. Therefore, FRET between DcuS-CFP and DcuS-YFP is well above the background or unspecific level and suggests an oligomeric state of DcuS in living cells.
The FRET efficiencies of the DcuS-CFP/DcuS-YFP pair were already registered in an early phase of induction, indicating that the self-association of DcuS homo-oligomers occurs early or during biosynthesis. When the bacteria were grown in the presence of fumarate (Fig. 6A and C), only minor differences from the FRET under fumarate-deficient conditions were observed. The donor fraction, fD, was 0.45 ± 0.01 and the FRET efficiency, E, was 0.17 ± 0.01 (n = 8 data points, mean ± standard deviation of the mean). No changes in the transfer efficiencies upon the addition of fumarate suggests a preformed or permanent oligomeric state. Thus, signal transduction upon fumarate sensing may be achieved by other mechanisms, like substrate-induced conformational changes, as suggested by structural studies (7, 20, 38), but not by altering the oligomerization state.
DISCUSSION
An improved method for in vivo FRET analysis using flexible background subtraction.
FRET and chemical cross-linking were used to study oligomerization of full-length DcuS in vivo and in vitro, i.e., in bacterial membranes and in proteoliposomes, whereas membrane-bound sensor kinases have mainly been studied before with truncated variants in the detergent-solubilized state. For accurate determination of FRET efficiency, factors like fluorophore concentrations, cross talk, background, or signal-to-noise ratio must be taken into account. Gordon's method (12) was reported as the most reliable approach for FRET quantification in comparison with other methods (2). However, the method requires background-free spectra, whereas living cells usually produce strong background, such as scattering and autofluorescence. FRET studies often neglect the variations among samples, and background correction is conducted by subtracting a constant background, normally a reference containing neither donor nor acceptor, which may not produce background-free data of sufficient quality for further FRET analysis. In this study, the background was corrected for each sample individually by using a multiparameter fitting. This procedure is able to determine the contributions of spectral components and to separate donor and acceptor signals more accurately from the background, even at low signal-to-noise ratios or in complex biological samples. The accuracy and reliability of quantitative FRET analysis was validated with different donor-acceptor pairs for simple model systems but also under complex experimental conditions as in living cells.
Dimeric and tetrameric DcuS in bacteria and proteoliposomes.
In a combined approach, chemical cross-linking and quantitative FRET spectroscopy of full-length DcuS showed consistently that DcuS is an oligomer in vitro after reconstitution in liposomes, in living cells, and in isolated bacterial membranes. The oligomerization is permanent and not affected by the functional state of DcuS, such as the presence of the stimulus fumarate. Cross-linking by DSS identified tetrameric DcuS in significant amounts. Cross-linking generally is not a quantitative method, and it is not possible to estimate the actual content of dimeric and tetrameric DcuS from the cross-linking experiments. It can be assumed, however, that the actual number of oligomeric complexes is higher than represented by the results of the cross-linking experiments, and in particular, the content of the tetrameric DcuS might be an underestimate. The significant amount of tetrameric DcuS in vivo and in vitro and their presence under all test conditions suggest that this form of DcuS is of physiological relevance. The experiments also demonstrated that formation of the dimer and tetramer is not dependent on or stimulated by high concentrations of DcuS. The oligomer of DcuS obviously is not very stable, and most of the detergent-solubilized form of the protein was monomeric, but a small portion remained in the tetrameric state even during SDS-gel electrophoresis.
Cross-linking by disulfide bonding via Cys471 showed a dimeric state for DcuS. The lack of tetramers by this type of connection is explained by the presence of only one cross-linking site per monomer (Cys471), which allows dimer formation but no higher degree of cross-linking. Dimerization by intermolecular disulfide formation of Cys471 residues was observed for the oligomeric DcuS in membranes but also for detergent-solubilized DcuS. The latter showed no FRET and was functionally inactive in autophosphorylation (16), suggesting that this form of DcuS is monomeric. Cross-linking by disulfide formation presumably occurs when monomers interact by chance, which is stimulated by the lack of other reaction partners for Cys471 for disulfide formation.
Significance of the oligomeric state for DcuS function.
In eukaryotic membrane-bound receptor tyrosine kinases, the oligomerization (and activation) is often triggered by the signal molecules, whereas bacterial membrane-bound histidine kinases are supposed to show an oligomeric state independent of the presence of effector (10). This assumption is proven here directly, in vivo and in vitro, for DcuS, which is a permanent oligomer without changes in its oligomerization in the presence of signal molecules. Signal transduction within the DcuS oligomer and in other membrane-bound sensor kinases is mediated instead by conformational changes that are triggered by ligand binding (9, 20, 29, 38, 40). It is supposed that the oligomeric state of DcuS and other sensor kinases is a prerequisite for signal transduction across the membrane. Therefore, the oligomeric state of DcuS appears to be an important parameter for DcuS function, but it has to be tested by further experiments whether the dimer or the tetramer is the functional form, and even-higher oligomeric forms of DcuS cannot be excluded.
There is also genetic and functional evidence for dimerization of periplasmic sensor kinases (10), including DcuS. For dimerization, the DHp domain is of particular significance. The dimeric domain constitutes a four-helix bundle of α-helices, two of which are derived from each monomer in sensor kinase EnvZ and PhoQ of E. coli, a sensor kinase of Thermotoga maritima, and KinB of Geobacillus stearothermophilus (3, 24, 25, 41, 42). The crystal structures of the periplasmic sensor domains of this class of His kinases, including that of DcuS and of the closely related CitA, showed significant contact sites and back-to-back dimerization in the domain (7, 38). The interactions in the sensor domains appear to be weak, and the separate domain of DcuS (DcuS42-181) was monomeric in solution at low concentration with and without ligand. Only high concentrations close to those in the crystal (44 mM) caused dimerization (7).
Supplementary Material
Acknowledgments
We thank the Deutsche Forschungsgemeinschaft (DFG) for financial support.
We thank V. Sourjik (Heidelberg) for plasmid pDK108.
Footnotes
Published ahead of print on 7 May 2010.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Balannik, V., R. A. Lamb, and L. H. Pinto. 2008. The oligomeric state of the active BM2 ion channel protein of influenza B virus. J. Biol. Chem. 283:4895-4904. [DOI] [PubMed] [Google Scholar]
- 2.Berney, C., and G. Danuser. 2003. FRET or no FRET: a quantitative comparison. Biophys. J. 84:3992-4010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bick, M. J., V. Lamour, K. R. Rajashankar, Y. Gordiyenko, C. V. Robinson, and S. A. Darst. 2009. How to switch off a histidine kinase: crystal structure of Geobacillus stearothermophilus KinB with the inhibitor Sda. J. Mol. Biol. 386:163-177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Biener, E., M. Charlier, V. K. Ramanujan, N. Daniel, A. Eisenberg, C. Bjorbaek, B. Herman, A. Gertler, and J. Djiane. 2005. Quantitative FRET imaging of leptin receptor oligomerization kinetics in single cells. Biol. Cell 97:905-919. [DOI] [PubMed] [Google Scholar]
- 5.Bradford, M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principles of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 6.Casino, P., V. Rubio, and A. Marina. 2009. Structural insight into partner specificity and phosphoryl transfer in two-component signal transduction. Cell 139:325-336. [DOI] [PubMed] [Google Scholar]
- 7.Cheung, J. H., and W. A. Hendrickson. 2008. Crystal structures of C4-dicarboxylate ligand complexes with sensor domains of histidine kinases DcuS and DctB. J. Biol. Chem. 283:30256-30265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dower, W. J., J. F. Miller, and C. W. Ragsdale. 1988. High efficiency transformation of Escherichia coli by high voltage electroporation. Nucleic Acids Res. 16:6127-6145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Etzkorn, M. K., P. Dünnwald, V. Vijayan, J. Krämer, C. Griesinger, S. Becker, G. Unden, and M. Baldus. 2008. Plasticity of the PAS domain and a potential role for signal transduction in the histidine kinase DcuS. Nat. Struct. Mol. Biol. 15:1031-1039. [DOI] [PubMed] [Google Scholar]
- 10.Gao, R., and A. M. Stock. 2009. Biological insights from structures of two-component proteins. Annu. Rev. Microbiol. 63:133-154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Golby, P., S. Davies, D. J. Kelly, J. R. Guest, and S. C. Andrews. 1999. Identification and characterization of a two-component sensor-kinase and response regulator system (DcuS-DcuR) controlling gene expression in response to C4-dicarboxylates in Escherichia coli. J. Bacteriol. 181:1238-1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gordon, G. W., G. Berry, X. H. Liang, B. Levine, and B. Herman. 1998. Quantitative fluorescence resonance energy transfer measurements using fluorescence microscopy. Biophys. J. 74:2702-2713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guzman, L. M., D. Belin, M. J. Carson, and J. Beckwith. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 177:4121-4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Heermann, R., K. Altendorf, and K. Jung. 1998. The turgo sensor KdpD of Escherichia coli is a homodimer. Biochim. Biophys. Acta 1415:114-124. [DOI] [PubMed] [Google Scholar]
- 15.Holloway, P. W. 1973. A simple procedure for removal of Triton X-100 from protein samples. Anal. Biochem. 53:304-308. [DOI] [PubMed] [Google Scholar]
- 16.Janausch, I. G., I. Garcia-Moreno, and G. Unden. 2002. Function of DcuS from Escherichia coli as a fumarate-stimulated histidine protein kinase in vitro. J. Biol. Chem. 277:39809-39814. [DOI] [PubMed] [Google Scholar]
- 17.Kentner, D., S. Thiem, M. Hildenbeutel, and V. Sourjik. 2006. Determinants of chemoreceptor cluster formation in Escherichia coli. Mol. Microbiol. 61:407-417. [DOI] [PubMed] [Google Scholar]
- 18.Kentner, D., and V. Sourjik. 2009. Dynamic map of protein interactions in the Escherichia coli chemotaxis pathway. Mol. Syst. Biol. 5:238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kneuper, H., I. G. Janausch, V. Vijayan, M. Zweckstetter, V. Bock, C. Griesinger, and G. Unden. 2005. The nature of the stimulus and of the fumarate binding site of the fumarate sensor DcuS of Escherichia coli. J. Biol. Chem. 280:20596-20603. [DOI] [PubMed] [Google Scholar]
- 20.Kneuper, H., P. D. Scheu, M. Etzkorn, M. Sevvana, P. Dünnwald, S. Becker, M. Baldus, C. Griesinger, and G. Unden. Sensing ligands by periplasmic sensing histidine kinases with sensory PAS domains. In S. Spiro and R. Dixon, (ed.), Sensory mechanisms in bacteria, in press. Caister Academic Press, Norfolk, United Kingdom.
- 21.Kobe, F., U. Renner, A. Woehler, J. Wlodarczyk, E. Papusheva, G. Bao, A. Zeug, D. W. Richter, E. Neher, and E. Ponimaskin. 2008. Stimulation- and palmitoylation-dependent changes in oligomeric conformation of serotonin 5-HT1A receptors. Biochim. Biophys. Acta. 1783:1503-1516. [DOI] [PubMed] [Google Scholar]
- 22.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 23.Li, M., L. G. Reddy, R. Bennett, N. D. Silva, L. R. Jones, and D. D. Thomas. 1999. A fluorescence energy transfer method for analyzing protein oligomeric structure: application to phospholamban. Biophys. J. 76:2587-2599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marina, A., C. Mott, A. Auyzenberg, W. A. Hendrickson, and C. D. Waldburger. 2001. Structural and mutational analysis of the PhoQ histidine kinase catalytic domain. Insight into the reaction mechanism. J. Biol. Chem. 276:41182-41190. [DOI] [PubMed] [Google Scholar]
- 25.Marina, A., C. D. Waldburger, and W. A. Hendrickson. 2005. Structure of the entire cytoplasmic portion of a sensor histidine-kinase protein. EMBO J. 24:4247-4259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mascher, T., J. D. Helman, and G. Unden. 2006. Stimulus perception in bacterial signal-transducing histidine kinases. Microbiol. Mol. Biol. Rev. 70:910-938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miller, J. H. 1992. A short course in bacterial genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 28.Miroux, B., and J. E. Walker. 1996. Over-production of proteins in Escherichia coli: mutant hosts that allow synthesis of some membrane proteins and globular proteins at high levels. J. Mol. Biol. 260:289-298. [DOI] [PubMed] [Google Scholar]
- 29.Moore, J. O., and W. A. Hendrickson. 2009. Structural analysis of sensor domains from the TMAO-responsive histidine kinase receptor TorS. Structure 17:1195-1204. [DOI] [PubMed] [Google Scholar]
- 30.Ormo, M., A. B. Cubitt, K. Kallio, L. A. Gross, R. Y. Tsien, and S. J. Remington. 1996. Crystal structure of the Aequorea victoria green fluorescent protein. Science 273:1392-1395. [DOI] [PubMed] [Google Scholar]
- 31.Pan, S. Q., T. Charles, S. Jin, Z. L. Wut, and E. W. Nester. 1993. Preformed dimeric state of the sensor protein VirA is involved in plant-Agrobacterium signal transduction. Proc. Natl. Acad. Sci. U. S. A. 90:9939-9943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pappalardo, L., I. G. Janausch, V. Vijayan, E. Zientz, J. Junker, W. Peti, M. Zweckstetter, G. Unden, and C. Griesinger. 2003. The NMR structure of the sensory domain of the membraneous two-component fumarate sensor (histidine protein kinase) DcuS of Escherichia coli. J. Biol. Chem. 278:39185-39188. [DOI] [PubMed] [Google Scholar]
- 33.Patterson, G. H., D. W. Piston, and B. G. Barisas. 2000. Forster distances between green fluorescent protein pairs. Anal. Biochem. 284:438-440. [DOI] [PubMed] [Google Scholar]
- 34.Pfleger, K. D. G., and K. A. Eidne. 2005. Monitoring the formation of dynamic G-protein-coupled receptor-protein complexes in living cells. Biochem. J. 385:625-637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Qin, L., R. Dutta, H. Kurokawa, M. Ikura, and M. Inouye. 2000. A monomeric histidine kinase derived from EnvZ, an Escherichia coli osmosensor. Mol. Microbiol. 36:24-32. [DOI] [PubMed] [Google Scholar]
- 36.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 37.Scheu, P., S. Sdorra, Y. F. Liao, M. Wegner, T. Basché, G. Unden, and W. Erker. 2008. Polar accumulation of the metabolic sensory histidine kinases DcuS and CitA in Escherichia coli. Microbiology 154:2463-2472. [DOI] [PubMed] [Google Scholar]
- 38.Sevvana, M., V. Vijayan, M. Zweckstetter, S. Reinelt, D. R. Madden, R. Herbst-Irmer, G. M. Sheldrick, M. Bott, C. Griesinger, and S. Becker. 2008. A ligand-induced switch in the periplasmic domain of sensor histidine kinase CitA. J. Mol. Biol. 377:512-523. [DOI] [PubMed] [Google Scholar]
- 39.Silhavy, T. J., M. L. Berman, and L. W. Enquist. 1984. Experiments with gene fusions. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 40.Szurmant, H., R. A. White, and J. A. Hoch. 2007. Sensor complexes regulating two-component signal transduction. Curr. Opin. Struct. Biol. 17:706-715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tanaka, T., S. K. Saha, C. Tomomori, R. Ishima, D. Liu, K. I. Tong, H. Park, R. Dutta, L. Qin, M. B. Swindells, T. Yamazaki, A. M. Ono, M. Kainosho, M. Inouye, and M. Ikura. 1998. NMR structure of the histidine kinase domain of the Escherichia coli osmosensor EnvZ. Nature 396:88-92. [DOI] [PubMed] [Google Scholar]
- 42.Tomomori, C. T., R. Dutta, H. Park, S. K. Saha, Y. Zhu, R. Ishima, D. Liu, K. I. Tong, H. Kurokawa, H. Qian, M. Inouye, and M. Ikura. 1999. Solution structure of the homodimeric core domain of Escherichia coli histidine kinase EnvZ. Nat. Struct. Biol. 6:729-734. [DOI] [PubMed] [Google Scholar]
- 43.Towbin, H., T. Staehelin, and J. Gordon. 1979. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc. Natl. Acad. Sci. U. S. A. 76:4350-4354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.West, A. H., and A. M. Stock. 2001. Histidine kinases and response regulator proteins in two-component signaling systems. Trends Biochem. Sci. 26:369-376. [DOI] [PubMed] [Google Scholar]
- 45.Yang, F., L. G. Moss, and G. N. Phillips. 1996. The molecular structure of green fluorescent protein. Nat. Biotechnol. 14:1246-1251. [DOI] [PubMed] [Google Scholar]
- 46.Yang, Y., and M. Inouye. 1991. Intermolecular complementation between two defective mutant signal-transducing receptors of Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 88:11057-11061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yanisch-Perron, C., J. Vieira, and J. Messing. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33:103-119. [DOI] [PubMed] [Google Scholar]
- 48.Zientz, E., J. Bongaerts, and G. Unden. 1998. Fumarate regulation of gene expression in Escherichia coli by the DcuSR (dcuSR genes) two-component regulatory system. J. Bacteriol. 180:5421-5425. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.