Abstract
Little is known about the regulation of nitrogenase genes in cyanobacteria. Transcription of the nifH1 and vnfH genes, encoding dinitrogenase reductases for the heterocyst-specific Mo-nitrogenase and the alternative V-nitrogenase, respectively, was studied by using a lacZ reporter. Despite evidence for a transcription start site just upstream of nifH1 and vnfH, promoter fragments that included these start sites did not drive the transcription of lacZ and, for nifH1, did not drive the expression of nifHDK1. Further analysis using larger regions upstream of nifH1 indicated that a promoter within nifU1 and a promoter upstream of nifB1 both contributed to expression of nifHDK1, with the nifB1 promoter contributing to most of the expression. Similarly, while the region upstream of vnfH, containing the putative transcription start site, did not drive expression of lacZ, the region that included the promoter for the upstream gene, ava4055, did. Characterization of the previously reported nifH1 and vnfH transcriptional start sites by 5′RACE (5′ rapid amplification of cDNA ends) revealed that these 5′ ends resulted from processing of larger transcripts rather than by de novo transcription initiation. The 5′ positions of both the vnfH and nifH1 transcripts lie at the base of a stem-loop structure that may serve to stabilize the nifHDK1 and vnfH specific transcripts compared to the transcripts for other genes in the operons providing the proper stoichiometry for the Nif proteins for nitrogenase synthesis.
Anabaena variabilis ATCC 29413 is a filamentous cyanobacterium that fixes atmospheric nitrogen under oxic growth conditions. After removal of fixed nitrogen from the growth medium, ca. 5 to 10% of the vegetative cells differentiate into specialized cells called heterocysts, in which nitrogen fixation occurs (60, 62). Heterocysts protect the oxygen-labile nitrogenase from external oxygen by synthesizing a glycolipid layer that limits oxygen diffusion into the cell (30, 57, 58). Internal oxygen is low in heterocysts because they lack oxygen-evolving photosystem II activity and they have increased respiration (29, 53). A. variabilis has three nitrogenases, but each functions under different environmental conditions (reviewed in reference 46). The primary nitrogenase is the heterocyst-specific Mo-nitrogenase encoded by the nif1 genes (44, 45). A. variabilis also has an alternative heterocyst-specific V-nitrogenase, encoded by the vnf genes, that is only expressed when Mo is limiting (32, 44). A second Mo-nitrogenase, encoded by the nif2 genes (47-49), functions only under anoxic conditions in vegetative cells and heterocysts. Synthesis of all three nitrogenases is repressed in cells grown with a source of fixed nitrogen.
Synthesis of a functional nitrogenase requires the products of many genes, several of which are involved in production and insertion of the FeMo-cofactor that is found in most nitrogenases (reviewed in reference 37). These genes include nifB, nifS, nifU, nifH, nifD, nifK, nifE, nifN, nifX, and nifW. The eight genes of the nifBSUHDKEN locus are expressed on at least three transcripts: nifB-fdxN-nifU-nifS (27), nifHDK (16, 19), and nifEN (reviewed in reference 20). In vegetative cells, the nifHDK operon is interrupted by a 11-kb insertion element that is removed from the chromosome of heterocysts late in the differentiation process to allow transcription of nifHDK (5, 17, 25). Anabaena sp. strain PCC7120 is nearly identical to A. variabilis over the entire length of the nifBSUHDKEN locus except that the locus in Anabaena sp. strain PCC7120 has an additional 55-kb insertion element in fdxN that is not present in A. variabilis (5, 15).
The nifD and nifK genes encode the α-subunit and β-subunit of dinitrogenase, respectively, which together make the heterotetrameric enzyme with two FeMo-cofactors [7Fe-9S-Mo-X-homocitrate] (reviewed in reference 37). NifH, with a [Fe4-S4] cofactor, is the dinitrogenase reductase, which is responsible for transferring electrons to the dinitrogenase (22). NifS catalyzes the removal and transfer of sulfur from cysteine to NifU (63), which acts as a scaffolding protein for the simple [Fe-S] cluster assembly (61). The [Fe-S] clusters are then transferred to NifB, where they are used to generate NifB-co, a [Fe6-S9] cluster that serves as an early precursor to FeMo-co (9). NifE and NifN which form a heterotetramer similar to NifDK, function as a scaffold on which final assembly of the FeMo-co occurs before it is transferred to the apo-nitrogenase (reviewed in reference 37).
Although much is known concerning the function of most of the nif gene products, very little is known about the transcriptional regulation of any of these genes in cyanobacteria. Transcription of the nif genes was first reported over 25 years ago (19, 21); however, almost no progress has been made in identifying any aspects of transcriptional regulation. A putative transcription start site for nifH has been identified at position −123 relative to the start codon in Anabaena sp. strain PCC7120 (19, 21), while the 5′ end of the nifB transcript is −283 relative to the start codon (27). We have recently examined the region immediately upstream of nifH1 in A. variabilis and have found that the intergenic region between nifU1 and nifH1 did not drive expression of nifH1 or a lacZ reporter; thus, we extended the regions for analysis to identify the regions required for regulated expression of nifHDK1 and its paralog, vnfH.
MATERIALS AND METHODS
Strains and growth conditions.
A. variabilis strain FD, a derivative of A. variabilis ATCC 29413 that can grow at 40°C, was maintained on agar-solidified Allen and Arnon (AA) medium (2) supplemented, when appropriate, with 5 mM NH4Cl, 10 mM N-tris (hydroxymethyl)methyl-2-aminoethanesulfonic acid (TES; pH 7.2), 25 to 40 μg of neomycin sulfate ml−1, or 3 μg each of spectinomycin and streptomycin ml−1. Strains were grown photoautotrophically in liquid cultures in an 8-fold dilution of AA medium (AA/8) or in AA/8 supplemented with 5 mM NH4Cl and 10 mM TES (pH 7.2) at 30°C, with illumination at 100 to 120 μE m−2 s−1. Antibiotics, when used, included neomycin (5 μg ml−1) and spectinomycin (0.3 μg ml−1). Strains containing fragments upstream of vnfH were starved of molybdate for at least 10 generations as described previously (44) by growing them in Mo-free conditioned medium (conditioned by growth with strain FD, followed by filtration to remove the cyanobacteria). In some experiments, Mo-starved cells were supplemented with 10−6 M sodium orthovanadate.
Construction of strains.
A 302-bp nifU1-nifH1 intergenic region was amplified from FD DNA, obtained from ammonium grown cultures, using nifH302L/nifH1-R2 primers (Table 1) and cloned into the BglII/SmaI sites of pBP288 (52) to produce pJU362. A 3.8-kb HindIII/SphI fragment of pBR322 was ligated to the HindIII/SphI sites in pJU362 to yield pJU409. A neomycin resistance (Nmr) cassette was amplified from pBP285 (52) with primers nm5′termL/nm5′termR and cloned into the KpnI site of pJU409 in an orientation opposite to lacZ to yield pJU410. The nm5′termL/nm5′termR primers incorporate a terminator at the 5′ end of the Nmr cassette, oriented opposite to the Nmr cassette such that they terminate transcription at the 5′ end of the Nmr cassette to prevent transcriptional readthrough into lacZ. A 1.8-kb PCR fragment made from FD DNA using the primers frtB-L/frtB-R was then cloned into the ScaI/HindIII sites of pJU410 to yield pJU411. Other lacZ fusions were made by cloning various PCR fragments amplified from FD DNA into the BglII/SmaI sites in pJU411 as indicated in Table 2. Promoter fragments were sequenced to verify that they contained no mutations. Recombination of these plasmids with promoter fragments fused to lacZ into the frt region of A. variabilis by single crossover after conjugation (50) resulted in the strains with the same name as the plasmids (Fig. 1 A and B). Strains resulting from a single crossover in the frt region were identified by screening for colonies that were unable to grow in the dark with fructose. Strains resulting from a single crossover in the nif1 region were screened for a Nif− phenotype by their inability to grow on AA agar plates lacking a source of fixed nitrogen.
TABLE 1.
Primers used in this study
| Oligonucleotide | Sequence (5′→3′) |
|---|---|
| DNA | |
| frtB-R | AATAAGCTTCCTTGTCCTAACATCCCGG |
| frtB-L | AATAGTACTTGGCACATTAGCGATCG |
| nifB1RTL | GGCAGCTAGTCCACCGACAT |
| nifB1RTR | ATCCGCAACCACCTGATTTT |
| nifH1RTL | ACAGGCGTGAGATCCAAACA |
| nifH1RTR | CATCAAACGGGTGGAGTCAG |
| nifK1RTL | CTACCTTGAGGAGGAGTGAA |
| nifK1RTR | CTCGGTGTATTCTGGCTGTT |
| nifU3′RTL | CAAAGCCGCAACAAACC |
| nifU3′RTR | GCGTAATCTGGATTCAATCG |
| nifSRTL | CATTCGTGCCGCATTGTTAGCC |
| nifSRTR | ACAGTCCCGGTTTCGTTGTTCG |
| nifH302L | GAAGATCTCAGCCTAGTAGTAGAAGCAGTT |
| nifH1-R2 | ATACCCGGGTCTAATGTTTTCGTCAGTCA |
| nifSUHL1 | TGAAGATCTAAGGTAGATCCAGAGGTTGTAGAGG |
| nifSUHL2 | TGAAGATCTAGCAATGGAATAAGGGCTAATGAG |
| nifBSUHL | TGAAGATCTAGCAACCGCGTCTGATAGTGT |
| nifUH-L | AATAGATCTAGCCCAAGAACAAACATTG |
| nifUH-L2 | AATAGATCTGGGAGTCATTGAAGATAACG |
| nifUH-L3 | AATAGATCTCTGACTTTAGATGAAGCCCTG |
| nifUH-L4 | AATAGATCTGAAAATAAGGTACGTCGCATAG |
| nifUH-L5 | AATAGATCTGGCAAAAACGACCCCTC |
| vnfHBgal-L451 | GAAGATCTTGCATCAATCAAGATATGATTTAGTGATT |
| vnfHBgal-L1380 | GAAGATCTCAGAACGCGCTTAGGGATGAG |
| vnfHBgal-L196 | GAAGATCTAAGACGTTTTCATTGTTTGG |
| vnfHBgal-R1 | ATACCCGGGATCGCGGAAGCCTTTGAGTACTACTT |
| nifBRTR2 | GCAATACGTTCTTGGAGCTTTTC |
| nifURTR | GGTCTTACTTCTTCGTCTAATACTTTTTG |
| nifBPCR1 | GGTAGAATGTGTTTACAGCCAAG |
| nifHout | CCATAGCTGCAAGGGTGTTT |
| Oligop1 | GCGCGAATTCCTGTAGA |
| vnfHPE1R | GCGGAAGCCTTTGAGTAC |
| vnfHPE2R | CCGACAATCAGAATACGTTGT |
| moe2RPE3 | AGCTATGCGTAGTGCGATCGCCACT |
| Moelike2-RPE | AGGAGGCCACTATCCTGCTT |
| nifB1L | AATGTCGACAACAAGATGATTCGGGAACAAGGTGCATTC |
| nifB1R | AATACTAGTCGGTTTCGTTGTTCGCATACATAATTGTCA |
| nifH170L | TGTACATCCGACTAACGAACCCATCATGAACA |
| nifH170R | GCTACATCTGTGATGAGTGCTGAGTCCATA |
| RNA | |
| RNAoligo09 | AUAUGCGCGAAUUCCUGUAGAACGAACACUAGAAGAAA |
TABLE 2.
Strains and plasmids used in this study
| Strain or plasmid | Relevant characteristicsa | Source or reference |
|---|---|---|
| Strains | ||
| FD | Anabaena variabilis ATCC 29413 wild-type parent strain | 10 |
| BP457 | pBP457 integrated into the frtBC region of the chromosome via single crossover | This study |
| BP461 | pBP461 integrated into the frtBC region of the chromosome via single crossover | This study |
| BP462 | pBP462 integrated into the ava4055-vnfH region of the chromosome via single crossover | This study |
| BP469 | pBP462 integrated into the frtBC region of the chromosome via single crossover | This study |
| JU333 | pJU333 integrated into the nifH1 gene via double recombination | This study |
| JU408 | pJU408, with a nifU1-nifH1 deletion (Nmr), integrated into the chromosome of FD via double recombination (Nmr, Nif− after segregation) | This study |
| JU417 | pJU457 integrated into the intergenic nifU1-nifH1 region of the chromosome via single crossover | This study |
| JU425 | pJU402 integrated into the chromosome of strain JU408 via double recombination; resulted in replacement of the nifU1-nifH1 deletion of JU408 with the mutated version of the nifU1-nifH1 intergenic region from pJU402, restoring a Nif+ phenotype | This study |
| JU436 | pJU436 integrated into the nif1 region via double recombination, creating nifB1 deletion mutant | This study |
| JU454 | pJU454 integrated into the frtBC region of the chromosome via single crossover | This study |
| JU466 | pJU466 integrated into the nif1 region via double recombination, creating nifB1 deletion mutant (Nif−) | This study |
| JU467 | pJU467 (containing the promoterless lacZ), integrated into the frtBC region of the chromosome via single crossover | This study |
| JU468 | pJU468 integrated into the nif1 region of the chromosome via single crossover | This study |
| JU469 | pJU469 integrated into the nif1 region of the chromosome via single crossover | This study |
| JU472 | pJU472 integrated into the frtBC region of the chromosome via single crossover | This study |
| JU473 | pJU473 integrated into the frtBC region of the chromosome via single crossover | This study |
| JU476 | pJU476 integrated into the frtBC region of the chromosome via single crossover | This study |
| JU477 | pJU477 integrated into the frtBC region of the chromosome via single crossover | This study |
| JU484 | pJU476 integrated into the nif1 region of the chromosome via single crossover | This study |
| JU485 | pJU477 integrated into the nif1 region of the chromosome via single crossover | This Work |
| Plasmids | ||
| pAAWY3162 | 9-kb library clone of A. variabilis containing ava3910-nifH1 region | JGI |
| pBP285 | Kmr Nmr cassette in a polylinker C.K.3 with a transcriptional terminator at the 3′ end | 52 |
| pBP288 | Cloning vector for integration of transcriptional fusions into the chromosome; Tcr Kmr Nmr Spr Smr Apr | 52 |
| pBP457 | 708-bp promoter fragment containing the ava4055-vnfH intergenic region (primers vnfHBgal-L451 and vnfHBgal-R1) inserted into the BglII/SmaI sites of pJU411 | This study |
| pBP461 | 1.6-kb ava4055-vnfH promoter fragment (primers vnfHBgal-L1380 and vnfHBgal-R1) inserted into the BglII/SmaI sites of pJU411 | This study |
| pBP462 | 2.2-kb ava4055-vnfH PCR fragment (primers vnfHBgal-L1961 and vnfHBgal-R1) inserted into the BglII/SmaI sites of pJU411 | This study |
| pBR322 | Mobilizable plasmid; Apr Tcr | 4 |
| pJU332 | BamHI fragment from pMV2 containing the nifS1-nifD1 region was cloned into the BamHI site of pBR322 | This study |
| pJU333 | KpnI fragment from pPE20 containing lacZ was cloned into the KpnI site of nifH1 in pJU332 | This study |
| pJU362 | 302-bp nifH1 promoter fragment (primers nifH1-R2 and nifH1-302L) inserted into the BglII/SmaI sites of pBP288 | This study |
| pJU375 | pEL1 with the Nmr cassette from pBP285 cloned into the EcoRV site and orientated toward the 5′ end of nifU | This study |
| pJU376 | 4.5-kb EcoRV fragment from pRL2948a cloned into the SmaI site on pJU375 | This study |
| pJU408 | pJU376 with the 400-bp AgeI fragment deleted | This study |
| pJU409 | 5.2-kb HindIII-SphI fragment from pJU362, containing the 302-bp nifH1 promoter driving lacZ, cloned into the HindIII-SphI sites of pBR322 | This study |
| pJU410 | Nmr cassette from pBP285 with a 5′ terminator (primers Nm5′TermL and Nm5′TermR), inserted into KpnI site of pJU409 to inhibit plasmid readthrough | This study |
| pJU411 | 1.8-kb frtBC PCR fragment (primers frtB-L and frtB-R), used as region of homology for recombination, inserted into the HindIII site of pJU410 | This study |
| pJU402 | pEL1 (33) with a mutation in the nifH1 upstream region that abolishes the putative NtcA binding site. | This study |
| pJU429 | 1.6-kb PCR fragment of nifB using the primers nifB1L and nifB1R cloned into the ScaI/SpeI sites on pEL1 | This study |
| pJU436 | 5.4-kb NruI/ScaI fragment containing sacB and Emr from pRL2948a cloned into the ZraI site in pJU429 | This study |
| pJU454 | 1.3-kb nifU1-nifH1 promoter fragment (primers nifUH-L and nifH1-R2) inserted into the BglII/SmaI sites of pJU411 | This study |
| pJU457 | 302-bp intergenic nifU1-nifH1 promoter fragment, inserted into BglII-SmaI sites of pJU411 | This study |
| pJU463 | 3.1-kb BsrGI-MscI fragment of pJU455 self-ligated, creates a nifB1 deletion | This study |
| pJU466 | 5.4-kb BglII fragment containing sacB and Emr from pRL2948a inserted into the BamHI site of pJU463 | This study |
| pJU468 | 1.6-kb nifB1 promoter fragment (primers pnifB-L and pnifB-R) inserted into the BglII/SmaI sites of pJU410 | This study |
| pJU469 | 1.3-kb nifH1-nifH1 promoter fragment (primers nifUH-L and nifH1-R2) inserted into the BglII/SmaI sites of pJU410 | This study |
| pJU472 | 743-bp nifH1-nifH1 promoter fragment (primers nifH1-R2 and nifUH-L4) inserted into the BglII/SmaI sites of pJU411 | This study |
| pJU473 | 544-bp nifH1-nifH1 promoter fragment (primers nifH1-R2 and nifUH-L5) inserted into the BglII/SmaI sites of pJU411 | This study |
| pJU476 | 3.0-kb fdxN-nifH1 promoter fragment (nifSUHL2 and nifH1-R2) inserted into the BglII/SmaI sites of pJU411 | This study |
| pJU477 | 6.0-kb nifB1-nifH1 promoter fragment (primers nifBSUHL and nifH1-R2) inserted into the BglII/SmaI sites of pJU411 | This study |
| pMV2 | nifH1 region with Smr Spr cassette inserted at the AgeI site in nifH1 | 32 |
| pPE20 | Source of lacZ for transcriptional fusions | 48 |
| pRL2948a | Source of mobilization site, oriT, and sacB gene, which confers sucrose sensitivity; Cmr Emr | C. P. Wolk |
| pBP285 | Kmr Nmr cassette in a polylinker C.K.3 with a transcriptional terminator at the 3′ end | 52 |
Cmr, chloramphenicol resistance; Tcr, tetracycline resistance; Apr, ampicillin resistance; Smr, streptomycin resistance; Kmr, kanamycin resistance; Spr, spectinomycin resistance; Emr, erythromycin resistance; Nmr, neomycin resistance.
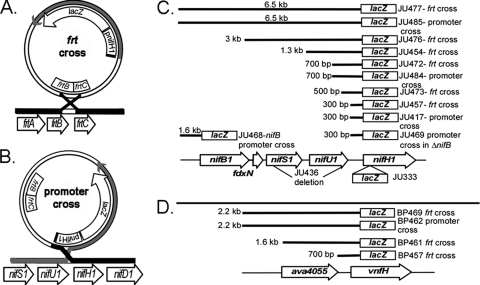
FIG. 1.
Map of the genes analyzed in these studies. Two possible single-crossover events between plasmids bearing the promoter-lacZ fusions are shown. (A) Recombination between the frtBC genes on the vector and in the chromosome resulted in a strain in which only the promoter fragment provided in the plasmid drove expression of lacZ. The chromosomal nifHDK1 structural gene region was unchanged. (B) Recombination between the nifH1 promoter fragment in the plasmid and the chromosomal promoter placed lacZ under the control of the full, normal nifH1 upstream region, including nifBSU1. The chromosomal nifHDK1 structural genes are under the control of only the plasmid-borne promoter fragment. (C) Diagram of the nifH1 region with the strain names and sizes of the tested promoter fragments (not drawn to scale). Strain JU436 has a deletion of the nifSU coding region as indicated. (D) Diagram of the vnfH region with the strain names and sizes of the tested promoter fragments.
Isolation of double recombinant mutants, JU425 (NtcA-binding site mutant), JU333 (lacZ inserted in the nifH1 gene), JU436 (nifS1-nifU1 deletion), and JU466 (nifB1 deletion) from single-recombinant exconjugant colonies was accomplished by using sacB selection (6) on AA plates supplemented with 5 mM NH4Cl, 10 mM TES, and 10% sucrose. These mutants were constructed as follows. Plasmid pJU445 was constructed by cloning a 9.5-kb fragment containing the nif1 region (ava3910-nifH1) from pAAWY3162 (a plasmid made by JGI for sequencing the A. variabilis genome) into pBR322 using SalI/BamHI. pJU445 was then digested with BsrGI/MscI (nonmethylated MscI site), blunted, and religated to create a 3.1-kb deletion of nifB1 and upstream sequences, producing strain pJU363. A 5.4-kb BglII fragment containing sacB and erythromycin resistance (Emr) from pRL2948a (52) was cloned into the BamHI site in pJU463 to produce pJU466. JU466 resulted from a double crossover of pJU466 into FD. After growth and segregation JU466 was Nif−. A 6-kb nifS1-nifD1 fragment of pMV2 (52) was cloned into pBR322 using BamHI to produce pJU332. A 5-kb fragment of pPE20 (48) containing a promoterless lacZ was cloned into nifH1 of pJU332 at the internal KpnI site to yield pJU333. Integration of pJU333 into the chromosome of FD by double crossover yielded JU333. Segregation of JU333 was determined by its Nif− phenotype. Mutant JU425 was created by replacing the deleted nifH1-nifUH1 intergenic region (with a Nmr cassette in the deleted region) in JU408 (making it Nif−) with the mutated version of the NtcA-binding site present in pJU402 by double-crossover events upstream of nifU1 and downstream of nifH1. This resulted in loss of the Nmr cassette in JU408 after segregation; thus, JU425 was Nms and Nif+ when fully segregated. pJU429 was made by cloning a 1.6-kb PCR fragment of nifB1 into the ScaI/SpeI sites of pEL1 (32). A 5.4-kb NruI/ScaI fragment containing sacB and Emr from pRL2948a was cloned into the ZraI site in pJU429 to produce pJU436. JU436 was made by using strain JU408 as the parent strain in the same manner as JU425, but using pJU436 to create the nifS1-nifU1 deletion, and was Nms when fully segregated.
RNA isolation, RT-PCR, and 5′ RACE (rapid amplification of cDNA ends).
RNA was isolated from 50-ml cultures grown in AA/8 or Mo-free AA/8 containing 10−6 M sodium orthovanadate. Cells were harvested, the media were removed, and the cells were resuspended in 400 μl of Tri-Reagent (Sigma) with 200 mg of 150-μm glass beads. Cells were lysed by 2 min of amalgamation using a Wig L Bug dental amalgamator, followed by a 5-min incubation at 55°C. After centrifugation, the Tri-Reagent layer was removed to a new tube, and the lysis step without the 55°C incubation was repeated. The two organic phases were combined and extracted twice with chloroform. The RNA was then isopropanol precipitated and resuspended in 34 μl of water plus 1 μl of RNasin (Promega). Then, 10 μg of total nucleic acid was subjected to DNase digestion by using a Turbo DNA-free kit (Ambion, Austin, TX). Reverse transcription-PCR (RT-PCR) was performed as previously described (33) and as modified by Ungerer et al. (52) with primers specific for each gene: nifB, nifBRTL/nifBRTR; nifS, nifSRTL/nifSRTR; nifU, nifU3′RTL/nifU3′RTR; nifH, nifHRTL/nifHRTR; and nifK, nifKRTL/nifKRTR.
5′ RACE was performed as described previously (3) with the following modifications. A total of 20 μg of RNA, treated with DNase, was extracted with phenol-chloroform-isoamyl alcohol and then with chloroform, followed by ethanol precipitation. The RNA was resuspended in 50 μl, and half was treated with 20 U of tobacco acid pyrophosphatase (TAP; Epicentre, Madison, WI) for 60 min at 37°C. The remaining half of the RNA was not treated with TAP, but all subsequent treatments were performed on both samples. The RNA was extracted with phenol-chloroform-isoamyl alcohol, followed by extraction with chloroform. Next, 200 pmol of the RNA adapter, RNAoligo09, was added to each tube before they were ethanol precipitated. The pellet was resuspended in 14 μl of water, heated to 90°C for 5 min, and ligated to the adapter overnight at 17°C using T4 single-stranded RNA ligase (NEB). The ligated RNA was extracted with organic solvents and ethanol precipitated as described above, resuspended in 20 μl of water, and reverse transcribed using Superscript III (Invitrogen) according to their protocol using the following primers: nifH, nifHRTR; nifU, nifU3′RTR; nifB, nifBRTR2; ava4055, moe2-RPE2; and vnfH, vnfHPE1R. PCR was performed using the left primer oligoP1 and the following right primers: nifH, nifHout; nifU, nifURTR; nifB, nifBPCR1; ava4055, Moelike2-RPE; and vnfH, vnfHPE2R.
β-Galactosidase and acetylene reduction assays.
For nitrogen stepdown experiments, cells grown in AA/8 (with or without Mo or V) supplemented with 5 mM NH4Cl and 10 mM TES at an optical density at 720 nm (OD720) of 0.08 to 0.10 were washed three times with 25 ml of AA/8 (with or without Mo or V) and resuspended at an OD720 of 0.025, without antibiotics. The cultures were split, and 5 mM NH4Cl-10 mM TES was added to half. After 24 h, the cells were harvested for the assay. β-Galactosidase assays were performed as previously described (26). Acetylene reduction assay was performed as previously described (32, 41).
RESULTS
Sequences essential for nifH1 or vnfH expression are far upstream from these genes.
To identify regions upstream of nifH1 that are essential for transcription, we constructed transcriptional fusions to lacZ using DNA fragments of various sizes upstream from nifH1. These constructs were made in a plasmid that contained an internal fragment of the fructose transport operon (frtBC) (52) for integration of the plasmid in the chromosome by single crossover (Fig. 1). Depending on whether the crossover event between the plasmid and chromosome occurred in the nifH1 promoter region or in the frtBC region, lacZ expression would be driven either by the entire normal chromosomal region upstream of nifH1 (Fig. 1B) or only the shorter nifH1 upstream fragment (Fig. 1A). Conversely, expression of the chromosomal copy of nifHDK1 would be driven either by the normal upstream region (Fig. 1A) or the truncated plasmid-borne upstream region (Fig. 1B). Measuring β-galactosidase or nitrogenase activity in strains in which expression of lacZ or nifHDK1 was driven by promoter fragments of different sizes (Fig. 1C) allowed us to determine the approximate location of the essential promoter elements.
We first examined strains containing the 300-bp fragment that comprised the entire nifU1-nifH1 intergenic region fused to lacZ (Fig. 2 A). Strain JU457, in which the plasmid recombined into the frtBC region of homology (resulting in two defective frtABC operons) was identified by its Frt− phenotype (inability to grow heterotrophically in the dark with fructose). In JU457, the 300-bp nifU1-nifH1 intergenic region fused to lacZ did not produce β-galactosidase (Fig. 2A). The strain resulting from the alternative single crossover within the 300-bp nifH1 upstream region, JU417 (Frt+ and thus able to grow heterotrophically in the dark with fructose), provided all of the nifH1 upstream region driving expression of lacZ. JU417 gave β-galactosidase levels comparable to the positive control, JU333, in which a promoterless lacZ gene was inserted into nifH1 in the chromosome by double crossover (Fig. 2A). These data suggested that essential transcriptional elements were farther upstream than the 300-bp nifU1-nifH1 intergenic region.
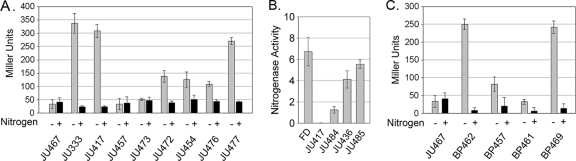
FIG. 2.
Expression from promoter regions. Expression of lacZ, by β-galactosidase activity of various-sized promoter fragments (A and C), or nitrogenase activity, by acetylene reduction (B) from various promoter fragments or a nifSU deletion as shown in Fig. 1C. Nitrogenase is expressed as nmol of ethylene mg OD720−1 h−1. Strain JU467 is a promoterless lacZ fusion used to measure background β-galactosidase.
A. variabilis has an alternative V-nitrogenase, encoded by the vnf genes, that, like the nif genes, is repressed by fixed nitrogen and may share a similar mode of regulation with the principal Mo-nitrogenase (32, 44). We examined the intergenic region between vnfH and the upstream gene, ava4055, for its ability to drive expression of lacZ as described above for nifH1. In the strain in which the crossover occurred in the frtBC region, BP457, the ava4055-vnfH intergenic region provided a modest increase in lacZ expression compared to the negative control, JU467, but expression in BP457 was less than one-third of the level in the positive control strain, BP469. In contrast, the strain with the alternative crossover within the ava4055-vnfH intergenic region, BP462, gave high levels of lacZ expression (Fig. 2C). These results suggested that essential promoter elements for vnfH lie upstream of the ava4055-vnfH intergenic region.
To identify the regions required for nitrogenase expression, plasmids with larger fragments extending farther upstream from nifH1 or vnfH were constructed (Fig. 1C and D). Strains in which these plasmids recombined using the frtBC region of homology so that the truncated nifH1 or vnfH upstream regions drove lacZ expression were identified by their Frt− phenotype. JU473, containing a 500-bp nifH1 upstream fragment that extended into nifU1, did not drive the expression of lacZ; however, JU472, with a 700-bp nifH1 upstream fragment extending ∼400 bp into the nifU1 coding region, provided ∼25% of the β-galactosidase activity measured in strains in which lacZ expression was driven by the complete normal nifH1 upstream region (JU333 and JU417) (Fig. 2A). Fragments of larger sizes (JU454, 1.3 kb; JU476, 3 kb) did not further increase β-galactosidase activity (Fig. 2A). These results suggested that there was an essential transcriptional element in the 700-bp fragment (JU472) that was missing in the 500-bp region (JU473), which placed it in the nifU1 coding region. Strain JU477, containing a 6.5-kb region, which extended from nifH1 to 1.6 kb upstream of nifB1, provided levels of β-galactosidase activity similar to, but somewhat lower than, the strains in which lacZ expression was driven by the complete, normal nifH1 upstream region (JU333 and JU417) (Fig. 2A).
Strains in which recombination occurred in the promoter region of the plasmid were identified by their Frt+ phenotype and verified by PCR (Fig. 1B). Expression of the chromosomal copy of nifHDK1 from the truncated promoter region was measured by nitrogenase activity. A 300-bp fragment (the nifU1-nifH1 intergenic region) driving expression of nifHDK1 (JU417) gave no nitrogenase activity (Fig. 2B), a finding consistent with the results for lacZ expression from that same 300-bp fragment (Fig. 2A). A 3-kb fragment (extending from nifH1 through nifU1) driving the expression of nifHDK1 (JU484) gave ∼25% of the nitrogenase activity of the control strain (JU485) in which the large 6.5-kb fragment (extending from nifH1 to 1.6 kb upstream of nifB1) drove expression of nifHDK1 at a level comparable to, but somewhat lower than, the wild-type strain (FD) (Fig. 2B). The level of nitrogenase activity observed for the 3-kb fragment (JU484) was similar to the level of expression of lacZ driven by the 700-bp region that extended into nifU1; thus, together, these data suggested that there was a weak promoter in nifU1. The strain with a deletion of nifS1-nifU1 (JU436), but with an otherwise complete wild-type upstream region, had ∼75% of the nitrogenase activity of JU485 (Fig. 2B). These data suggested that the expression of nifH1 required two regions: a weak promoter in the nifU1 coding region and a strong promoter upstream of fdxN, possibly in the nifB1 promoter region.
Essential sequences for the expression of vnfH were identified in the region upstream of ava4055. Two vnfH promoter fragments were constructed with the entire ava4055 coding region. The first, BP461, extended only to the start of the ava4055 coding region, while the second, BP469, also included the ava4055 promoter region (Fig. 1D). Only BP469, which included the putative ava4055 promoter region, had β-galactosidase activity that was comparable to the control strain, BP462, that had crossed over in the vnfH region (Fig. 2C). Thus, the data indicated that the ava4055 promoter drives expression of both itself and vnfH.
Previously published data have indicated that nifH1 and vnfH have their own promoters. Two pieces of supporting evidence have been published. (i) Transcription start sites have been determined for nifH in other closely related cyanobacteria (19) and for vnfH (31), and (ii) Northern blots show a strong ∼1.1-kb transcript corresponding to the size of the nifH1 and vnfH genes alone (31). In the case of nifHDK, a stable transcript corresponding to the entire operon has also been reported (36). However, the data shown in Fig. 2 indicated that the intergenic regions between nifU1 and nifH1 and between ava4055 and vnfH were unable to initiate transcription.
Together, these findings lead to two possible hypotheses for nifH1. (i) Full transcriptional activation of nifH1 requires upstream activation sequences. One activation element is in the nifU1 coding region, and the other element is shared with the nifB1 promoter. In this model, transcription of nifHDK1 originates from the putative transcriptional start site in the nifU1-nifH1 intergenic region (19). However, the upstream activator elements work with this nifH1 promoter to activate transcription at this start site and each upstream activator contributes to the expression. (ii) Transcription does not originate at the previously identified nifH1 transcription start site but rather from two separate, upstream promoters and continues into the nifHDK1 operon. One promoter is in the nifU1 coding region and the other is likely the nifB1 promoter. The transcripts are then further processed in the intergenic region to produce discrete nifBSU1 and nifHDK1 transcripts.
Identification of dual nifH1 promoters.
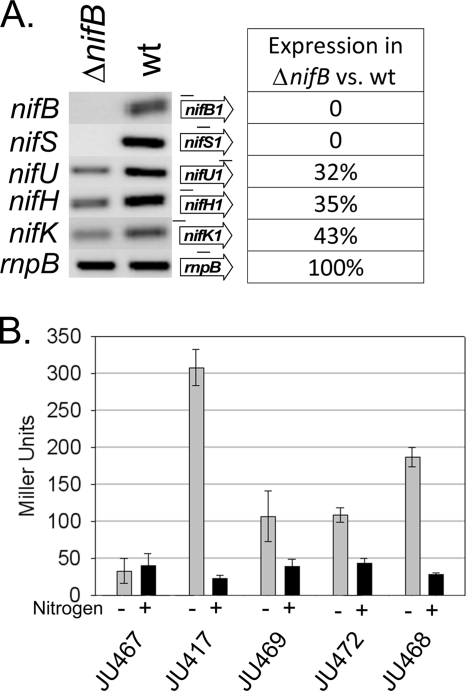
We have shown that a region upstream of nifB1 is essential for high-level expression of nifH1. If the nifB1 promoter were required for expression of nifH1, then we would expect a decrease in nifHDK1 expression when nifB1 and its promoter are deleted. Using semiquantitative RT-PCR, we measured the expression of the nif1 genes in a strain in which nifB1 and its promoter were deleted. The nifHDK1 genes were expressed in the nifB1 mutant, although at a much lower level than in the wild-type strain (Fig. 3 A). However, nifU1, which is believed to be under the control of the nifB1 promoter and was shown by Northern blot analysis to be on the nifBSU1 transcript (27), was also expressed in the nifB1 deletion mutant (Fig. 3A). This indicated that there was an additional nifU1 transcript originating from within nifBSU1 and supports the hypothesis that the essential element in nifU1 is a true promoter and is not an activator for a promoter in the nifU1-nifH1 intergenic region. These results indicated that the nifB1 promoter is required for high-level expression of nifHDK1. Thus, it appears that expression of nifHDK1 depends on dual promoters that initiate transcription upstream of nifB1 and within nifU1.
FIG. 3.
Expression of nif1 genes in a nifB1 deletion mutant. (A) RT-PCR of genes of the nif1 cluster in the wild-type (wt) strain and in a nifB1 promoter deletion strain. The rnpB gene, which is constitutively expressed, was used as a control to show equal amounts of RNA (56). The percent expression was calculated from the intensity of the band in the nifB1 deletion strain compared to the wild-type strain, after normalizing each band first to rnpB. The two rnpB bands differed by <5%. The region of the gene that was amplified is denoted as a line over the corresponding gene next to the gel. (B) β-Galactosidase activity in a nifB1 deletion strain.
The roles of the nifB1 promoter and the internal nifU1 promoter in expression of nifH1 were examined separately. A plasmid with the 300-bp nifU1-nifH1 intergenic region fused to lacZ was integrated into the chromosome at the nifU1-nifH1 intergenic region in a nifB1 deletion strain. This strain, JU469, had the nifH1 upstream region, from the start of nifH1 up to the end of nifB1, driving the expression of lacZ. Thus, it had the internal nifU1 promoter but not the nifB1 promoter. Expression of lacZ in JU469 was about one-third that of the control strain, JU417 (Fig. 3B). This level of expression was similar to expression from the 700-bp nifH1 promoter fragment that contained only the promoter in nifU1 (JU472). A plasmid with a 1.6-kb region upstream of nifB1 fused to lacZ was integrated into the chromosome at the nifB1 region to determine lacZ expression from the nifB1 promoter alone. Expression in this strain, JU468, was about two-thirds of the control strain, JU417 (Fig. 3B). Moreover, the sum of the expression from the internal nifU1 promoter and the nifB1 promoter was very similar to the level of expression of the control strain, JU417, which had both promoters. Thus, two promoters, the nifB1 promoter and a promoter within nifU1, are necessary and sufficient for expression of nifHDK1, but the primary promoter for nifHDK1 is the nifB1 promoter.
Processing of the nifBSUHDK1 transcript.
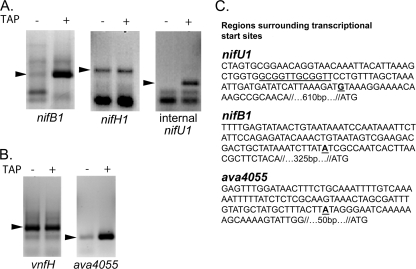
The nifB1 promoter contributes substantially to the expression of nifHDK1; however, Northern blot analysis indicates that in other cyanobacteria the nifBSU and nifHDK transcripts are separate (21, 27, 36). This suggested that the larger nifBSUHDK1 transcript may be efficiently cleaved posttranscriptionally. We verified the apparent transcription start site of nifH1 using 5′ RACE, a method that can distinguish between processed and primary transcripts (3). This technique requires the ligation of an RNA adapter to the 5′ end of the transcript. The ligation is impaired by the 5′ triphosphate present on primary transcripts; thus, ligation requires treatment of the sample with tobacco acid phosphatase (TAP), which hydrolyzes the triphosphate to a monophosphate. Processed transcripts already have a 5′ monophosphate and, thus, TAP is not required for ligation of a processed transcript to the adapter. If the ligation reaction works equally well with or without TAP, then the transcript is processed. We performed RNA ligase-mediated RT-PCR and then recovered and sequenced the cDNA bands to determine the nifH1, nifB1, and internal nifU1 transcription start sites (Fig. 4 C). The transcription start site of nifH1, as determined by 5′ RACE, was within a few nucleotides of the previously determined nifH start sites for Anabaena sp. strain PCC 7120 (19) and Anabaena azollae (21); however, the same product was made in RNA samples treated with or without TAP (Fig. 4A). This indicated that the putative transcription start site in the nifU1-nifH1 intergenic region is actually a site at which the larger transcript is processed rather than a transcription start site. The transcription start sites identified in nifU1 and upstream of nifB1 were primary transcript start sites, since the reactions gave a strong product only after treatment of the RNA with TAP (Fig. 4A).
FIG. 4.
5′ RACE was performed to determine transcripts beginning upstream of nifB1, nifH1, and an internal region of nifU1 (A) and vnfH and ava4055 transcripts (B). Arrows indicate the products that were sequenced. The region amplified is shown as a black line over the gene next to the corresponding RT lanes. (C) The transcription start sites in nifU1 and upstream of nifB1 and ava4055. The distance from the transcription start sites to the start codons of nifH1, nifB1, and ava4055 are shown for each strain. Putative −10 regions (underlined) were selected based only on their location for nifU1 and ava4055 because a consensus −10 site is not present upstream of these transcription start sites. The putative −10 region for nifB was selected due to its similarity to a consensus −10. A direct repeat located approximately −35 to the internal nifU transcriptional start site is also underlined. Transcriptional start sites are shown in boldface and underlined.
Based on sequencing of the nifU1 5′ RACE reaction product, the transcriptional start site in nifU1 is 320 nucleotides upstream from the 3′ end of nifU1, within the 700-bp promoter fragment, which was the smallest promoter fragment that drove expression of lacZ. We identified a pair of direct repeats, GCGGTT, −35 to this transcriptional start site that might serve as a binding site for a regulator or heterocyst-specific sigma factor (Fig. 4C). The sequence of the 5′ RACE reaction product for nifB1 placed the transcription start site within a few nucleotides of the published transcription start site for nifB in A. azollae, a strain whose sequence in the entire nif region is identical to A. variabilis (27). Alignment of the nifB1 and nifU1 promoters yielded no significant similarities between the two nif1 promoters, suggesting that the two promoters do not share a similar mode of regulation.
Processing of the ava4055-vnfH transcript.
The expression of vnfH was shown to require only the ava4055 promoter; however, vnfH and ava4055 are found on separate transcripts (32), suggesting that the apparent vnfH transcript may also result from processing. The bands obtained by 5′ RACE, using RNA treated with TAP or without, for the vnfH transcript were of equal intensity, indicating that vnfH is a processed transcript (Fig. 4B). The sequence of the RNA ligase-mediated RT-PCR product revealed a start site within a few nucleotides of the transcription start site that was identified by primer extension (Fig. 4C) (32). Amplification of the ava4055 transcript by 5′ RACE, using RNA either treated with or without TAP, yielded a strong product only when the RNA was treated with TAP, indicating that it is a primary transcript (Fig. 4B and C).
Regulation of nifH1 by NtcA.
The global regulator of nitrogen status, NtcA, has been proposed to bind a noncanonical NtcA-binding site −40 to the originally reported nifH transcriptional start site (processing site) to activate transcription of nifHDK in Anabaena sp. strain PCC 7120 (7, 34, 54) (Fig. 5 A); however, the reported interaction between NtcA and the putative promoter region upstream of nifH was weak (7, 34, 54). We were able to produce a mobility shift using purified NtcA with the nifU1-H1 intergenic region from A. variabilis, but a nonspecific competitor efficiently competed for binding, indicating that the binding in this strain was not specific (data not shown). However, given the differences in the putative binding-site sequences between Anabaena sp. strain PCC 7120 and A. variabilis (Fig. 5A), it is hardly surprising that binding of NtcA to this region in the two strains would be different. For A. variabilis the data provided here suggest that because there is no promoter or transcription start site in the nifU1-nifH1 intergenic region, NtcA could not activate transcription at this location. To determine whether NtcA regulates nifH1 transcription by binding to this region, we mutated the putative NtcA-binding site in the chromosome. Abolishing the putative NtcA-binding site, JU425, did not affect nitrogenase activity (Fig. 5B) or diazatrophic growth (data not shown). Thus, NtcA does not activate expression of nifHDK1 from this region.
FIG. 5.
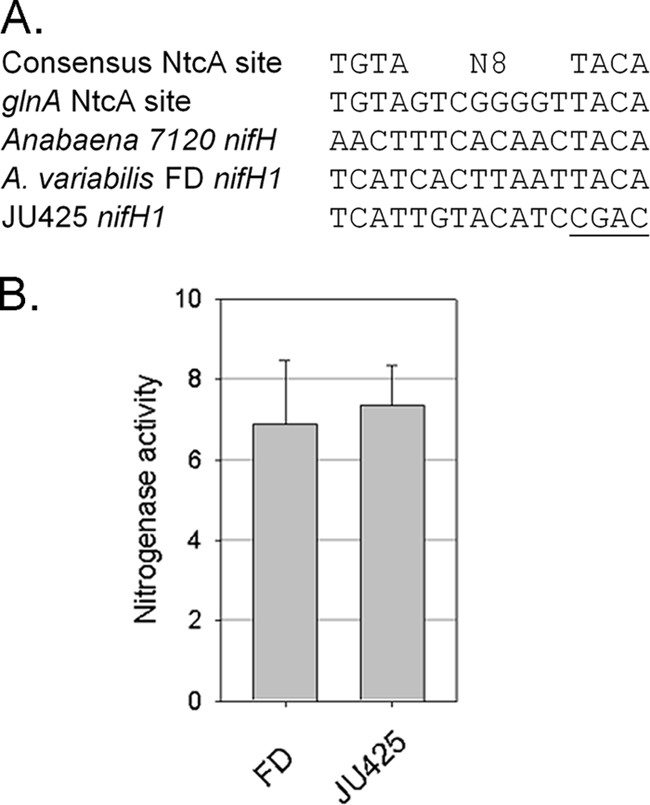
NtcA regulation of nifH1. (A) Comparison of the putative NtcA-binding sites from glnA (55) and various nifH1 genes to the altered putative NtcA-binding site that is −40 to the nifH1 processing site in JU425. (B) Comparison of nitrogenase activity between the wild type and the putative NtcA-binding site mutant (JU425). Nitrogenase is expressed as nmol of ethylene mg OD720−1 h−1.
DISCUSSION
Since the initial identification of the putative nifH transcription start site almost 3 decades ago (19, 21), little progress has been made in identifying the sites or factors that lead to heterocyst-specific, nitrogen-regulated expression of nifHDK in cyanobacteria. Our attempts to identify any fragment in the nifU1-nifH1 intergenic region of A. variabilis that could drive wild-type levels of expression of nifH1 or a lacZ reporter were unsuccessful. We demonstrate here that the reasons are 2-fold. First and foremost, the nifHDK1 transcript is actually a cleavage product of a larger transcript, thus it does not have its own promoter. Second, normal expression of nifHDK1 is a result of at least two promoters that function together to provide high-level expression of the nitrogenase structural genes. The transcript is then cleaved in the nifU1-nifH1 intergenic region to produce two distinct mRNAs. Cleavage of the nifBSUHDK1 transcript is likely to be very efficient, perhaps cotranscriptionally, since it is difficult to detect the full-length transcript by Northern analysis (16, 36). The existence of not only nifHDK, but also nifH and nifHD transcripts, in Anabaena sp. strain PCC 7120 (21) suggests that there may be additional processing events from the nifBSUHDK transcript to produce the final transcripts. The processing of polycistronic mRNA is a common method of posttranscriptional regulation that is used by bacteria to allow coordinated expressions of several genes from a single promoter while providing nonstoichiometric expression of individual genes of the operon (18, 31).
The differential stability of transcript segments is a result of stem-loop structures at the extreme 5′ or 3′ ends of the RNA (18). These structures have been reported to stabilize specific regions of the transcript relative to the whole (38). In order to afford stability to an mRNA, a stem-loop must be positioned no more than two nucleotides from the 5′ or 3′ end (13). The initial cleavage of a polycistronic mRNA often occurs in an intercistronic region and is often mediated by RNase E (1). RNase P then trims the 5′ leader to the base of a stem-loop, a position at which the stem-loop can protect the mRNA from degradation by acting to block the initiation of degradation (1, 23). In E. coli, an otherwise unstable transcript can be stabilized by fusing a stem-loop structure to the extreme 5′ end of the transcript (1). Stem-loop structures have also been observed to act as degradation barriers when present at the 3′ end of the transcript. In Rhodobacter capsulatus the photosynthetic genes pufBALMX are arranged on a single operon; however, the half-life of the pufLMX segment is 3 min, while the half-life of the pufBA segment is 20 min, resulting in large differences in protein expression from the two transcript segments (8). In this case, an intercistronic stem-loop at the 3′ end of pufBA acts as a decay terminator that prevents degradation of the pufLMX segment from extending into the pufBA genes.
The putative transcription start site originally mapped to the intergenic region upstream of nifH (19, 21) does not result from the initiation of transcription. This finding explains the failure to identify the key regulatory regions controlling nifHDK1 expression. The data presented here show that the nifBSUHDK1 genes are coregulated under the control of the nifB1 promoter and the internal nifU1 promoter; however, the contribution to nifHDK1 expression from the two promoters was not equal. The nifB1 promoter was responsible for 70 to 75% of the nifHDK1 transcript, while the internal nifU1 promoter produced ca. 25 to 30%, as evidenced by reporter expression from strains containing either nifB1 (JU468) or internal nifU1 (JU469) promoters. Furthermore, when the nifHDK1 genes were expressed from only the internal nifU1 promoter (JU484), nitrogenase activity was 25% of the level observed when both promoters drove expression of these genes. When the nifHDK1 genes were expressed from only the nifB1 promoter (JU436) in a nifU1-nifS1 deletion strain, nitrogenase activity was 75% of the level observed when both promoters drove expression of these genes (JU485). We showed previously that neither NifS1 nor NifU1 is required for nitrogenase activity, presumably because other proteins, perhaps those that make Fe-S clusters for photosynthesis, function in their place (24). Together, these findings suggest that the nifB1 promoter is the primary promoter driving expression of nifHDK1.
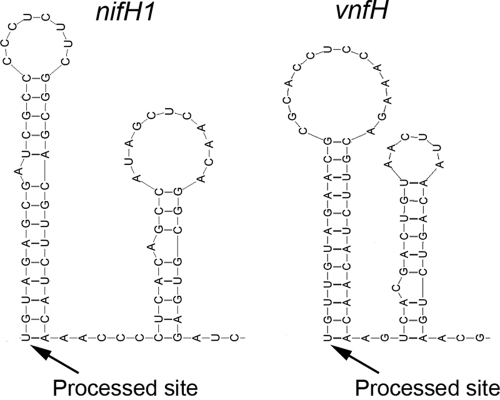
Microarray data indicate that the nifHDK genes of Anabaena sp. strain PCC7120 are expressed more strongly than the nifBSU genes (12). This is due, at least in part, to the second promoter in nifU. However, the low activity from the internal nifU1 promoter in A. variabilis would be insufficient to account for the large difference in expression between the nifBSU and nifHDK operons observed in Anabaena sp. strain PCC7120 (12). Therefore, we hypothesize that RNA processing could also contribute to increased expression of nifHDK relative to nifBSU. If a processing event places a stem-loop structure at the 5′ end of the transcript, then that transcript will have increased stability (1). Processing of a nifHDK transcript in Rhodobacter (59) and of a nifH transcript in Heliobacterium chlorum (14) at the base of a stem-loop structure has been reported, and there appears to be processing of nifHDK transcripts in Trichodesmium as well (11). We investigated the 5′ untranslated regions of the nifH1 and vnfH genes for potential secondary structure close to the 5′ end of the transcript. The 5′ untranslated regions of both nifH1 and vnfH can potentially fold into very similar secondary structures and they share some sequence identity, particularly in the folded region (Fig. 6). The base of the first stem is conserved, except that the fourth and sixth nucleotides of the stem are different between nifH1 and vnfH; however, evolution has created compensating mutations in the stem to retain base pairing, which further supports a function for the structure. Moreover, the position of the 5′ end of the transcript at the base of the first stem is conserved for both genes, and this is the specific position that is required for a 5′ hairpin structure to afford stability to the transcript (1) (Fig. 6). This suggests that processing of the transcript at the stem could provide additional stability to the nifHDK1 and vnfH segments of the transcript. The additional stability of the nifHDK1 segment relative to the nifBSU1 segment may provide the proper ratio of nitrogenase proteins that is required for nitrogen fixation. These findings suggest that processing of nif transcripts may be a common mode of gene regulation in cyanobacteria.
FIG. 6.
Predicted secondary structures (64) near the processed 5′ start sites of the nifH1 and vnfH transcripts.
Suzuki et al. (43) found evidence that nifBSU and nifHDK are coregulated in Anabaena sp. strain PCC7120. Mutants that lost the transcriptional activator AnCrpA, which was shown to bind to the nifB promoter, but not nifH, showed decreased expression of both the nifBSU and nifHDK operons (43). Thus, the decreased expression of nifHDK may result from decreased nifB promoter activity in this ancrpA mutant. In addition to the nifB promoter, we also identified a promoter in the nifU coding region. Although this promoter cannot drive high-level expression of nifHDK, it likely contributes to the increased level of expression of nifHDK relative to nifBSU. Several genes exhibiting dual promoters have been identified recently in Anabaena spp. such as devB, hetR, hetC, and coxBAC (28, 40). In fact, the coxBAC operon of A. variabilis utilizes dual promoters and processing of the mRNA to achieve proper regulation (40). The zwf operon of Nostoc punctiforme is another interesting example of multiple promoters. This operon has four genes on a transcript; however, there are additional promoters internal to the operon that are differentially regulated depending on the carbon and nitrogen sources (42). In the case of nifH1, dual promoters are used to produce higher levels of expression than can be achieved using a single promoter. This is similar to the coxBAC operon in that both promoters must be functioning simultaneously to provide the maximum level of expression.
Development requires complex changes in gene expression and precise, coordinated timing of gene expression. Multicellular organisms utilize multiple promoters to allow for different levels of expression of the same gene in different cell types, during various stages of development, or under different environmental conditions (reviewed in reference 39). The leader sequences of multiple transcripts can significantly affect the stability leading to differences in gene expression (13). Anabaena is a model organism for the study of development and the origins of multicellularity. Recently, several key developmental regulators of heterocyst differentiation have been found to have multiple promoters including ntcA, hetR, and hetC (28, 35). Also, genes that are differentially expressed between vegetative cells and heterocysts, such as glnA, petH, and ntcA, have been shown to accomplish this through the use of multiple promoters (35, 51, 54, 55). The data presented here indicate that the nifHDK1 operon is also controlled by multiple promoters. This suggests that the use of multiple promoters to coordinate changes in gene expression during development may be common to organisms that undergo cellular development.
Acknowledgments
Support for this research was provided by National Science Foundation grants MCB-0416663 and CHE-610177.
We thank Lucy Kastner for technical help with the β-galactosidase assays.
Footnotes
Published ahead of print on 30 April 2010.
REFERENCES
- 1.Alifano, P., F. Rivellini, C. Piscitelli, C. M. Arraiano, C. B. Bruni, and M. S. Carlomagno. 1994. Ribonuclease E provides substrates for ribonuclease P-dependent processing of a polycistronic mRNA. Genes Dev. 8:3021-3031. [DOI] [PubMed] [Google Scholar]
- 2.Allen, M. B., and D. I. Arnon. 1955. Studies on nitrogen-fixing blue-green algae. I. Growth and nitrogen fixation by Anabaena cylindrica Lemm. Plant Physiol. 30:366-372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bensing, B. A., B. J. Meyer, and G. M. Dunny. 1996. Sensitive detection of bacterial transcription initiation sites and differentiation from RNA processing sites in the pheromone-induced plasmid transfer system of Enterococcus faecalis. Proc. Natl. Acad. Sci. U. S. A. 93:7794-7799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bolivar, F., R. L. Rodriguez, P. J. Greene, M. C. Betlach, H. L. Heynker, and H. W. Boyer. 1977. Construction and characterization of new cloning vehicles. II. A multipurpose cloning system. Gene 2:95-113. [PubMed] [Google Scholar]
- 5.Brusca, J. S., M. A. Hale, C. D. Carrasco, and J. W. Golden. 1989. Excision of an 11-kilobase-pair DNA element from within the nifD gene in Anabaena variabilis heterocysts. J. Bacteriol. 171:4138-4145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cai, Y., and C. P. Wolk. 1990. Use of a conditionally lethal gene in Anabaena sp. strain PCC 7120 to select for double recombinants and to entrap insertion sequences. J. Bacteriol. 172:3138-3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chastain, C. J., J. S. Brusca, T. S. Ramasubramanian, T. F. Wei, and J. W. Golden. 1990. A sequence-specific DNA-binding factor (VF1) from Anabaena sp. strain PCC 7120 vegetative cells binds to three adjacent sites in the xisA upstream region. J. Bacteriol. 172:5044-5051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen, C.-Y. A., J. T. Beatty, S. N. Cohen, and J. G. Belasco. 1988. An intercistronic stem-loop structure functions as an mRNA decay terminator necessary but insufficient for puf mRNA stability. Cell 52:609-619. [DOI] [PubMed] [Google Scholar]
- 9.Curatti, L., J. A. Hernandez, R. Y. Igarashi, B. Soboh, D. Zhao, and L. M. Rubio. 2007. In vitro synthesis of the iron molybdenum cofactor of nitrogenase from iron, sulfur, molybdenum, and homocitrate using purified proteins. Proc. Natl. Acad. Sci. U. S. A. 104:17626-17631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Currier, T. C., and C. P. Wolk. 1979. Characteristics of Anabaena variabilis influencing plaque formation by cyanophage N-1. J. Bacteriol. 139:88-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dominic, B., Y.-B. Chen, and J. P. Zehr. 1998. Cloning and transcriptional analysis of the nifUHDK genes of Trichodesmium sp. IMS101 reveals stable nifD, nifDK, and nifK transcripts. Microbiol. 144:3359-3368. [DOI] [PubMed] [Google Scholar]
- 12.Ehira, S., M. Ohmori, and N. Sato. 2003. Genome-wide expression analysis of the responses to nitrogen deprivation in the heterocyst-forming cyanobacterium Anabaena sp. strain PCC 7120. DNA Res. 10:97-113. [DOI] [PubMed] [Google Scholar]
- 13.Emory, S. A., P. Bouvet, and J. G. Belasco. 1992. A 5′-terminal stem-loop structure can stabilize mRNA in Escherichia coli. Genes Dev. 6:135-148. [DOI] [PubMed] [Google Scholar]
- 14.Enkh-Amgalan, J., H. Kawasaki, H. Oh-oka, and T. Seki. 2006. Cloning and characterization of a novel gene involved in nitrogen fixation in Heliobacterium chlorum: a possible regulatory gene. Arch. Microbiol. 186:327-337. [DOI] [PubMed] [Google Scholar]
- 15.Golden, J. W., C. D. Carrasco, M. E. Mulligan, G. J. Schneider, and R. Haselkorn. 1988. Deletion of a 55-kilobase-pair DNA element from the chromosome during heterocyst differentiation of Anabaena sp. strain PCC 7120. J. Bacteriol. 170:5034-5041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Golden, J. W., L. L. Whorff, and D. R. Wiest. 1991. Independent regulation of nifHDK operon transcription and DNA rearrangement during heterocyst differentiation in the cyanobacterium Anabaena sp. strain PCC 7120. J. Bacteriol. 173:7098-7105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Golden, J. W., and D. R. Wiest. 1988. Genome rearrangement and nitrogen fixation in Anabaena blocked by inactivation of xisA gene. Science 242:1421-1423. [DOI] [PubMed] [Google Scholar]
- 18.Grunberg-Manago, M. 1999. Messenger RNA stability and its role in control of gene expression in bacteria. Annu. Rev. Genet. 33:193-227. [DOI] [PubMed] [Google Scholar]
- 19.Haselkorn, R., D. Rice, S. E. Curtis, and S. J. Robinson. 1983. Organization and transcription of genes important in Anabaena heterocyst differentiation. Ann. Microbiol. 134B:181-193. [DOI] [PubMed] [Google Scholar]
- 20.Herrero, A., A. M. Muro-Pastor, and E. Flores. 2001. Nitrogen control in cyanobacteria. J. Bacteriol. 183:411-425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jackman, D. M., and M. E. Mulligan. 1995. Characterization of a nitrogen-fixation (nif) gene cluster from Anabaena azollae 1a shows that closely related cyanobacteria have highly variable but structured intergenic regions. Microbiology 141:2235-2244. [DOI] [PubMed] [Google Scholar]
- 22.Lawson, D. M., and B. E. Smith. 2002. Molybdenum nitrogenases: a crystallographic and mechanistic view. Met. Ions Biol. Syst. 39:75-119. [PubMed] [Google Scholar]
- 23.Li, Y., and S. Altman. 2003. A specific endoribonuclease, RNase P, affects gene expression of polycistronic operon mRNAs. Proc. Natl. Acad. Sci. U. S. A. 100:13213-13218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lyons, E. M., and T. Thiel. 1995. Characterization of nifB, nifS, and nifU genes in the cyanobacterium Anabaena variabilis: NifB is required for the vanadium-dependent nitrogenase. J. Bacteriol. 177:1570-1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Meeks, J. C., E. L. Campbell, and P. S. Bisen. 1994. Elements interrupting nitrogen fixation genes in cyanobacteria: presence and absence of a nifD element in clones of Nostoc sp. strain Mac. Microbiol. 140:3225-3232. [Google Scholar]
- 26.Miller, J. 1992. A short course in bacterial genetics, p. 72-74. Cold Spring Harbor Laboratory Press, Plainview, NY.
- 27.Mulligan, M. E., and R. Haselkorn. 1989. Nitrogen fixation (nif) genes of the cyanobacterium Anabaena species strain PCC 7120: the nifB-fdxN-nifS-nifU operon. J. Biol. Chem. 264:19200-19207. [PubMed] [Google Scholar]
- 28.Muro-Pastor, A. M., E. Flores, and A. Herrero. 2009. NtcA-regulated heterocyst differentiation genes hetC and devB from Anabaena sp. strain PCC 7120 exhibit a similar tandem promoter arrangement. J. Bacteriol. 191:5765-5774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Murry, M. A., A. J. Horne, and J. R. Benemann. 1984. Physiological studies of oxygen protection mechanisms in the heterocysts of Anabaena cylindrica. Appl. Environ. Microbiol. 47:449-454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Murry, M. A., and C. P. Wolk. 1989. Evidence that the barrier to the penetration of oxygen into heterocysts depends upon two layers of the cell envelope. Arch. Microbiol. 151:469-474. [Google Scholar]
- 31.Newbury, S. F., N. H. Smith, and C. F. Higgins. 1987. Differential mRNA stability controls relative gene expression within a polycistronic operon. Cell 51:1131-1143. [DOI] [PubMed] [Google Scholar]
- 32.Pratte, B. S., K. Eplin, and T. Thiel. 2006. Cross-functionality of nitrogenase components NifH1 and VnfH in Anabaena variabilis. J. Bacteriol. 188:5806-5811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pratte, B. S., and T. Thiel. 2006. High-affinity vanadate transport system in the cyanobacterium Anabaena variabilis ATCC 29413. J. Bacteriol. 188:464-468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ramasubramanian, T. S., T. F. Wei, and J. W. Golden. 1994. Two Anabaena sp. strain PCC 7120 DNA-binding factors interact with vegetative cell- and heterocyst-specific genes. J. Bacteriol. 176:1214-1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ramasubramanian, T. S., T. F. Wei, A. K. Oldham, and J. W. Golden. 1996. Transcription of the Anabaena sp. strain PCC 7120 ntcA gene: multiple transcripts and NtcA binding. J. Bacteriol. 178:922-926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ramirez, M. E., P. B. Hebbar, R. Zhou, C. P. Wolk, and S. E. Curtis. 2005. Anabaena sp. strain PCC 7120 gene devH is required for synthesis of the heterocyst glycolipid layer. J. Bacteriol. 187:2326-2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rubio, L. M., and P. W. Ludden. 2008. Biosynthesis of the iron-molybdenum cofactor of nitrogenase. Annu. Rev. Microbiol. 62:93-111. [DOI] [PubMed] [Google Scholar]
- 38.Sawers, R. G. 2006. Differential turnover of the multiple processed transcripts of the Escherichia coli focA-pflB operon. Microbiol. 152:2197-2205. [DOI] [PubMed] [Google Scholar]
- 39.Schibler, U., and F. Sierra. 1987. Alternative promoters in developmental gene expression. Annu. Rev. Genet. 21:237-257. [DOI] [PubMed] [Google Scholar]
- 40.Schmetterer, G., A. Valladares, D. Pils, S. Steinbach, M. Pacher, A. M. Muro-Pastor, E. Flores, and A. Herrero. 2001. The coxBAC operon encodes a cytochrome c oxidase required for heterotrophic growth in the cyanobacterium Anabaena variabilis strain ATCC 29413. J. Bacteriol. 183:6429-6434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shah, V. K., L. C. Davis, and W. J. Brill. 1975. Nitrogenase. VI. Acetylene reduction assay: dependence of nitrogen fixation estimates on component ratio and acetylene concentration. Biochim. Biophys. Acta 384:353-359. [DOI] [PubMed] [Google Scholar]
- 42.Summers, M. L., and J. C. Meeks. 1996. Transcriptional regulation of zwf, encoding glucose-6-phosphate dehydrogenase, from the cyanobacterium Nostoc punctiforme strain ATCC 29133. Mol. Microbiol. 22:473-480. [DOI] [PubMed] [Google Scholar]
- 43.Suzuki, T., H. Yoshimura, S. Ehira, M. Ikeuchi, and M. Ohmori. 2007. AnCrpA, a cAMP receptor protein, regulates nif-related gene expression in the cyanobacterium Anabaena sp. strain PCC 7120 grown with nitrate. FEBS Lett. 581:21-28. [DOI] [PubMed] [Google Scholar]
- 44.Thiel, T. 1993. Characterization of genes for an alternative nitrogenase in the cyanobacterium Anabaena variabilis. J. Bacteriol. 175:6276-6286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Thiel, T. 1996. Isolation and characterization of the vnfEN genes of the cyanobacterium Anabaena variabilis. J. Bacteriol. 178:4493-4499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Thiel, T. 2004. Nitrogen fixation in heterocyst-forming cyanobacteria, p. 73-110. In W. Klipp, B. Masepohl, J. R. Gallon, and W. E. Newton (ed.), Genetics and regulation of nitrogen fixing bacteria. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 47.Thiel, T., E. M. Lyons, and J. C. Erker. 1997. Characterization of genes for a second Mo-dependent nitrogenase in the cyanobacterium Anabaena variabilis. J. Bacteriol. 179:5222-5225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Thiel, T., E. M. Lyons, J. C. Erker, and A. Ernst. 1995. A second nitrogenase in vegetative cells of a heterocyst-forming cyanobacterium. Proc. Natl. Acad. Sci. U. S. A. 92:9358-9362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Thiel, T., and B. Pratte. 2001. Effect on heterocyst differentiation of nitrogen fixation in vegetative cells of the cyanobacterium Anabaena variabilis ATCC 29413. J. Bacteriol. 183:280-286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Thiel, T., and C. P. Wolk. 1987. Conjugal transfer of plasmids to cyanobacteria. Methods Enzymol. 153:232-243. [DOI] [PubMed] [Google Scholar]
- 51.Tumer, N. E., S. J. Robinson, and R. Haselkorn. 1983. Different promoters for the Anabaena glutamine synthetase gene during growth using molecular or fixed nitrogen. Nature 306:337-342. [Google Scholar]
- 52.Ungerer, J. L., B. S. Pratte, and T. Thiel. 2008. Regulation of fructose transport and its effect on fructose toxicity in Anabaena spp. J. Bacteriol. 190:8115-8125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Valladares, A., A. Herrero, D. Pils, G. Schmetterer, and E. Flores. 2003. Cytochrome c oxidase genes required for nitrogenase activity and diazotrophic growth in Anabaena sp. PCC 7120. Mol. Microbiol. 47:1239-1249. [DOI] [PubMed] [Google Scholar]
- 54.Valladares, A., A. M. Muro-Pastor, M. F. Fillat, A. Herrero, and E. Flores. 1999. Constitutive and nitrogen-regulated promoters of the petH gene encoding ferredoxin:NADP+ reductase in the heterocyst-forming cyanobacterium Anabaena sp. FEBS Lett. 449:159-164. [DOI] [PubMed] [Google Scholar]
- 55.Valladares, A., A. M. Muro-Pastor, A. Herrero, and E. Flores. 2004. The NtcA-dependent P1 promoter is utilized for glnA expression in N2-fixing heterocysts of Anabaena sp. strain PCC 7120. J. Bacteriol. 186:7337-7343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vioque, A. 1992. Analysis of the gene encoding the RNA subunit of ribonuclease P from cyanobacteria. Nucleic Acids Res. 20:6331-6337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Walsby, A. E. 2007. Cyanobacterial heterocysts: terminal pores proposed as sites of gas exchange. Trends Microbiol. 15:340-349. [DOI] [PubMed] [Google Scholar]
- 58.Walsby, A. E. 1985. The permeability of heterocysts to the gases nitrogen and oxygen. Proc. R. Soc. Lond. B 226:345-366. [Google Scholar]
- 59.Willison, J. C., J. Pierrard, and P. Hübner. 1993. Sequence and transcript analysis of the nitrogenase structural gene operon (nifHDK) of Rhodobacter capsulatus: evidence for intramolecular processing of nifHDK mRNA. Gene 133:39-46. [DOI] [PubMed] [Google Scholar]
- 60.Wolk, C. P., J. Zhu, and R. Kong. 1999. Genetic analysis of heterocyst formation, p. 509-515. In G. A. Peschek, W. Loeffelhardt, and G. Schmetterer (ed.), The phototrophic prokaryotes. Kluwer Academic/Plenum Publishers, New York, NY.
- 61.Yuvaniyama, P., J. N. Agar, V. L. Cash, M. K. Johnson, and D. R. Dean. 2000. NifS-directed assembly of a transient [2Fe-2S] cluster within the NifU protein. Proc. Natl. Acad. Sci. U. S. A. 97:599-604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhang, C. C., S. Laurent, S. Sakr, L. Peng, and S. Bedu. 2006. Heterocyst differentiation and pattern formation in cyanobacteria: a chorus of signals. Mol. Microbiol. 59:367-375. [DOI] [PubMed] [Google Scholar]
- 63.Zheng, L., R. H. White, V. L. Cash, R. F. Jack, and D. R. Dean. 1993. Cysteine desulfurase activity indicates a role for NifS in metallocluster biosynthesis. Proc. Natl. Acad. Sci. U. S. A. 90:2754-2758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zuker, M. 2003. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 31:3406-3415. [DOI] [PMC free article] [PubMed] [Google Scholar]