Abstract
Replication and segregation of the Rhizobium etli symbiotic plasmid (pRetCFN42d) depend on the presence of a repABC operon, which carries all the plasmid-encoded elements required for these functions. All repABC operons share three protein-encoding genes (repA, repB, and repC), an antisense RNA (ctRNA) coding gene, and at least one centromere-like region (parS). The products of repA and repB, in conjunction with the parS region, make up the segregation system, and they negatively regulate operon transcription. The last gene of the operon, repC, encodes the initiator protein. The ctRNA is a negative posttranscriptional regulator of repC. In this work, we analyzed the secondary structures of the ctRNA and its target and mapped the motifs involved in the complex formed between them. Essential residues for the effective interaction localize at the unpaired 5′ end of the antisense molecule and the loop of the target mRNA. In light of our results, we propose a model explaining the mechanism of action of this ctRNA in the regulation of plasmid replication in R. etli.
Small noncoding RNAs are widespread in nature and play essential roles in regulating gene expression, mostly at a posttranscriptional level. Prokaryotic regulatory small RNAs fall into two classes: cis-encoded RNAs and trans-encoded RNAs. The first, also known as countertranscript RNAs (ctRNAs) (17), are frequently found in accessory genetic elements, including plasmids, phages, and transposons (5). They are encoded by the complementary strands of their target genes, and for this reason, the ctRNAs and the target mRNAs can form extended-pairing hybrids (3). In contrast, trans-encoded RNAs and their targets are encoded at separate chromosomal loci, with their complementary regions being limited to short stretches. Moreover, trans-encoded RNAs frequently require the action of the RNA chaperone protein Hfq (6) to improve their stability or to facilitate RNA-RNA complex formation.
The participation and mechanisms of action of ctRNAs in the control of plasmid replication are well established: ctRNAs prevent plasmid replication by inhibiting the production of plasmid-encoded molecules (RNAs or proteins) involved in the initiation of plasmid replication. The plasmid replication rate is inversely proportional to the intracellular concentrations of the corresponding ctRNAs. ctRNAs are constitutively expressed and diffusible, usually have short half-lives, and interact rapidly with their targets, thereby effectively correcting fluctuations in plasmid copy number (4). However, the sequences of ctRNAs, the structural basis of the interaction between ctRNAs and their targets, and the relationship between ctRNAs and plasmid replication control have been analyzed for only a small number of plasmid systems (5). Here we describe the molecular details of the mechanism of action of a ctRNA in the replication control of a plasmid belonging to the repABC family.
repABC plasmids, commonly found in alphaproteobacteria, are characterized by the presence of a repABC operon, which carries all the elements required for plasmid replication and segregation (8, 23). In general, repABC operons consist of three protein-encoding genes, an antisense RNA (ctRNA) gene, and at least one centromere-like region (parS). The products of the first two genes, repA and repB, in conjunction with the parS region, compose the segregation system, and repC, the last gene of the operon, encodes the initiator protein (2, 25). The ctRNA gene is located in the large repB-repC intergenic region and is encoded on the strand complementary to that encoding the operon mRNA (Fig. 1 a) (9, 18, 30). The symbiotic plasmid, formally pRetCFN42d (p42d), of Rhizobium etli CFN42 carries a repABC operon whose expression depends on a single promoter (11, 26). The transcription of the repABC operon is negatively autoregulated by RepA and RepB (26, 22), and in all of them the ctRNA (55 to 59 nucleotides [nt]) is a negative posttranscriptional regulator of repC. These molecules also act as strong incompatibility factors, since it was shown that genes encoding ctRNAs introduced in trans displace their cognate plasmids (30, 18, 9).
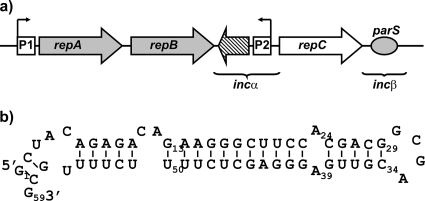
FIG. 1.
(a) Schematic representation of the Rhizobium etli p42d repABC operon. Gray arrows represent genes encoding proteins involved in plasmid segregation and in the negative regulation of the operon. The white arrow shows the position of the gene encoding RepC, the initiator protein. The dashed arrow indicates the position of the gene encoding the small antisense RNA (ctRNA). Squares mark the positions of the promoters: P1, the operon promoter, and P2, the ctRNA gene promoter. The oval shows the position of parS. Brackets enclose two regions involved in plasmid incompatibility. (b) ctRNA folding prediction obtained with the Mfold software (33).
The tumor-inducing plasmid (pTiR10) from Agrobacterium tumefaciens belongs to the repABC family. Chai and Winans (9) described a transcriptional/translational attenuation theoretical model explaining how the ctRNA (repE) of pTiR10 acts; this model is easily applicable to other repABC operons. In the model, the repABC mRNA exhibits two alternative secondary structures, depending on whether or not the ctRNA is paired with the repABC mRNA. In the absence of the ctRNA, the section corresponding to the repB-repC intergenic region of the repABC mRNA folds into a large stem-loop structure, but the predicted repC Shine-Dalgarno (SD) sequence and the repC initiation codon remain single stranded, allowing repC translation. When the ctRNA interacts with repABC mRNA, the repC leader sequence folds into a stem-loop structure that resembles an intrinsic terminator; however, this structure also occludes the SD sequence and the repC initiation codon, thereby blocking repC translation. These observations suggest that repC levels could be modulated by a transcriptional/translational attenuation mechanism.
In this work, we analyzed the in vitro secondary structures of the following: (i) the ctRNA encoded in the repB-repC intergenic region of the R. etli CFN42 symbiotic plasmid p42d, (ii) the repABC mRNA (the target) in the repB-repC intergenic region (mIncα) of the same plasmid; and (iii) the complex formed between the ctRNA and mIncα, using a combination of RNA probing assays and an in silico RNA secondary structure prediction algorithm (Mfold) (33). Also, we calculated the kinetic parameters of hybrid formation between mIncα, the ctRNA, and various ctRNA mutants and the half-life of the ctRNA. Further, we conducted an in vivo incompatibility analysis of mutant ctRNAs. Our findings indicate that the interaction between the ctRNA and repABC mRNA is very fast and requires the nonpaired 5′ end of the ctRNA and a loop in the target mRNA. This system operates by a novel mechanism in the plasmid replication system, contrasting with the loop-loop kissing previously reported (31). Nevertheless, our observations are generally consistent with the model proposed by Chai and Winans (9).
MATERIALS AND METHODS
DNA templates and RNA synthesis.
Oligodeoxyribonucleotides were synthesized in a 3400 DNA synthesizer (Applied Biosystems, Massachusetts); the DNA sequences of the oligonucleotides used in this work are listed in Table 1. The wild-type ctRNA (ctRNAwt) was generated by annealing and extension of the oligonucleotides ctRNA-T7-L and ctRNA-T7-U, essentially as described previously (1). Similarly, the ctRNA variant templates ctRNA-CGCU, ctRNA-mut2-5, ctRNA-mut2-5-CGCU, ctRNA-Δ(2-11), and ctRNA-Δ(1-8) were created by annealing and extension of the primers ctRNAasaCGCU, ctRNAmut2-5, ctRNAmut2yloop, ctRNAdel11, and ctRNAdel8, respectively, with ctRNA-T7-U. Other ctRNA templates—ctRNA-Δ(2-11)(52-59) and ctRNA-Δ(52-59)—were synthesized by annealing and extension of the primers ctRNAdel10-5′,8-3′ and ctRNAdel8-3′ with the primer Complement 3′. The template for transcribing the repBC intergenic region was obtained by PCR using the primers RNAT7-Bam-Alfa-U and MluAlfa-L. In competition binding assays, an oligonucleotide complementary to the 5′ end of the ctRNA (ODN5′) and another oligonucleotide complementary to the 3′ end of the same molecule (ODN3′) were used.
TABLE 1.
Oligonucleotides used in this work
| Oligonucleotide | Sequencea |
|---|---|
| ctRNA-T7-L | 5′-TAA TAC GAC TCA CAT AGC TAC AGA GAC AGAAGG CTT CCA CGCAGG CGA CGT TGA GGG AGCTCTTTT CTT TTGCG |
| ctRNA-T7-U | 5′-CGC AAA AGA AAA GAG CTC CCT |
| ctRNAasaCGCU | 5′-TAA TAC GAC TCA CTA TAG CTA CAG AGA CAGAAG GGC TTC CAC GAC GCG CTC GTT GAG GGA GCTCTT TTC TTT TGC G |
| ctRNAmut2-5 | 5′-TAA TAC GAC TCA CTA TAG AAT GAG AGA CAG AAG GGC TTC CAC GAC GGC GAC GTT GAG GGA GCT CTT TTC TTT TGC G |
| ctRNAmut2yloop | 5′-TAA TAC GAC TCA CTA TAG AAT GAG AGA CAG AAG GGC TTC CAC GAC GCGCTC GTT GAG GGA GCT CTT TTC TTT TGC G |
| ctRNAdel11 | 5′-TAA TAC GAC TCA CTA TAG AGA AGG GCT TCC ACG ACG GCG ACG TTG AGG GAG CTC TTT TCT TTT GCG |
| ctRNAdel8 | 5′-TAA TAC GAC TCA CTA TAG ACA GAA GGG CTT CCA CGA CGG CGA CGT TGA GGG AGC TCT TTT CTT TTG CG |
| ctRNAdel10-5′,8-3′ | 5′-TAA TAC GAC TCA CTA TAG AGA AGG GCT TCC ACG ACG GCG ACG TTG AGG GAG CTC TTT TC |
| ctRNAdel8-3′ | 5′-TAA TAC GAC TCA CTA TAG CTA CAG AGA CAG AAG GGC TTC CAC GAC GGC GAC GTT GAG GGA GCT CTT TTC |
| Complement 3′ | 5′-GAA AAG AGC TCC CTC AAC GT |
| RNAT7Bam-Alfa-U | 5′-GGA TCC TAA TAC GAC TCA CTA TAG CCC GCA AAA GAA AAG AGC TCC CTC |
| MluAlfa-L | 5′-ACG CGT TGT CTC TCA CCT TTC AGC |
| ODN5′ | 5′-GTC TCT GTA GC |
| ODN3′ | 5′-CGC AAA AG |
| tRNA-Met | 5′-GCC TTC AGG TTA TGA GCC TGA CG |
| S-SymA-U | 5′-CCG CGG CCC GCA AAA GAA AAA GGC |
| K-SymA-L | 5′-GGT ACC TCA CGA CAC CCC CCG CCC |
| AlfaDHind-U | 5′-AAG CTT CCC GCA AAA GAA AAG AGC TCC |
| AlfaD-L | 5′-TGT CTC TCA CCT TTC AGC AGG CAA |
| ctRNADqS-U1 | 5′-GGA AGC CTT CCT CTC TGT CTC TGT AGC AAG AAC AGA ATC GCA TT |
| ctRNADqS-U2 | 5′-GGA TCC GCC CGC AAA AGA AAA AGG CCC CCA ACG AAC AAG CCG TGG AAG CCT TCC TCT CTG |
| ctRNA5n-U1 | 5′-GAA GCC CTT CTC TCT GTC TGG CAA GAA CAG AAT CGC ATT TCC |
| ctRNA5n-U2 | 5′-GGA TCC CCG GCA AAA GAA AAG AGC TCC CTC AAC GTC GCC GTC GTG GAA GCC CTT CTC TCT GTC TGG |
| ctRNAart-U1 | 5′-ACC AGG ATT CTG TCT CTG TAG CAA GAA CAG AAT CGC ATT TCC |
| ctRNAart-U2 | 5′-GGA TCC CCC GCA AAA GAA AAT CTT GGT TCA CCT AGT ATT AGG TCA ACC AAG ATT CTG TCT CTG |
| RepC-HL | 5′-TTC GAG GGG AAG ACA ATC AAT TGT |
| RepB-HL | 5′-CGC CTA CGG CCG CCA CTC CCT GTT TCC GTT G |
Underlined and bold sequences correspond to the T7 promoter.
RNAs were synthesized by in vitro transcription and purified as previously described (1). The transcription reactions were carried out in volumes of 100 μl, with 3 μg of template DNA in a reaction buffer containing 40 mM Tris-HCl (pH 7.9), 6 mM MgCl2, 2 mM spermidine, 10 mM NaCl, 20 mM dithiothreitol (DTT), 0.01% Triton X-100, 1 mM nucleoside triphosphates (NTPs), 0.5 U of RNase inhibitor (Ambion Inc.), and 0.5 U of T7 RNA polymerase purified as previously described (20). Reaction mixtures were incubated at 37°C for 2 h, and the template DNA was then eliminated with 3 U RQ1 DNase (Promega). Samples were cleaned in Sephadex G-25 (GE) columns and then purified from 10% denaturing polyacrylamide gels.
RNA-RNA interaction assays. (i) Affinity assays.
Binding reactions were carried out with 5 nM 32P-labeled ctRNAwt or its variants with various amounts of unlabeled mRNA-incα (mIncα) (0.625, 1.25, 2.5, 3.74, 5.0, 12.5, 25.0, 50.0, or 125.0 nM) in TMN binding buffer (1×) [20 mM M Tris acetate (OAc) (pH 7.5), 10 mM Mg(OAc)2, 100 mM NaCl]. The RNAs were incubated separately at 65°C for 10 min, followed by incubation at 37°C for 10 min to allow the attainment of the native conformation. RNAs were mixed in the proportions described above and then incubated at 37°C for 10 min. The reactions were stopped by diluting the samples with an equal volume of denaturing application buffer: 94% formamide, 17 mM Na2EDTA, 0.025% Xylene cyanol, and 0.025% bromophenol blue (24). Hybrids were analyzed in 10% polyacrylamide-7 M urea gels run at 12 W for 90 min at room temperature. Gels were quantified using the Image Quant 5.2 software program (GE Healthcare).
(ii) Time courses.
Binding reactions were carried out in TMN buffer at 37°C with 5 nM 32P-labeled ctRNA and 50 nM unlabeled mIncα. Samples were withdrawn at the times indicated, and the reactions were stopped by the addition of one volume of application buffer. Samples were analyzed as described above.
(iii) Competition assays.
Labeled ctRNAwt, mIncα, and the primers ODN5′and ODN3′ were first denatured and renatured in TMN 1× buffer as described above. Subsequently, 5 nM labeled ctRNAwt was mixed with 760 pM of ODN5′ or ODN3′ or both in TMN (1×) buffer, and incubated at 37°C for 10 min. Finally, 50 nM mIncα was added, and the binding reaction mixes were incubated for 10 min at 37°C. Reactions were stopped by addition of one volume of application buffer. Samples were analyzed in 10% polyacrylamide-7 M urea gels, and images of dried gels were obtained and quantified as described above.
Secondary structure probing assays.
Fifty femtomoles of labeled ctRNAwt, its variants, or their target (mIncα) were used in the RNA probing assays. Partial digestion reactions were performed with 0.1 U of RNase T1 (Industrial Research, New Zealand) in TMN (1×) buffer containing 2 μg tRNA (Ambion Inc.) and incubated at 37°C for 2 min. Chemical partial hydrolysis was performed at 37°C for 10 min, using 30 mM lead acetate (Merck) in TMN (1×) buffer supplemented with 1 μg of tRNA. Partial digestion reactions were carried out with 0.1 U of RNase V1 (Industrial Research) in TMN (1×) buffer containing 2 μg tRNA at 4°C for 30 min. The secondary structures of the ctRNAwt/mIncα complex were analyzed by mixing 50 fmol of labeled RNA with 5 pmol of its nonlabeled counterpart in TMN (1×) buffer at 37°C for 10 min. Enzymatic and chemical hydrolysis were performed as described above. Chemical hydrolysis was stopped by adding EDTA to a final concentration of 10 mM. Samples were extracted with phenol-chloroform and precipitated with ethanol. Products were separated on high-resolution denaturing polyacrylamide gels (12%). Images were obtained as described above.
Bacterial strains and growth conditions.
Escherichia coli DH5α (12) and S17-1 (27) were grown at 37°C in Luria-Bertani medium, and Sinorhizobium meliloti 1021 and CFNX101, a Rhizobium etli recA::ΩSpcr derivative containing p42d (19), were grown at 30°C in PY medium supplemented with 10 mM CaCl2 (21). Nalidixic acid (20 μg/ml) and kanamycin (30 μg/ml) were added as appropriate.
Site-directed mutagenesis and plasmid incompatibility.
To determine the incompatibility phenotype of the ctRNA variants studied in vitro, we introduced the same changes into a recombinant plasmid containing the whole repB-repC intergenic region (incα) (Table 2) by site-directed mutagenesis, using the QuikChange II kit according to the manufacturer's instructions (Stratagene). Additionally, we made three constructs: one of them expressing a chimeric ctRNA gene consisting of the left arm from p42d ctRNA and the stem-loop and right arm from the ctRNA from Sinorhizobium meliloti 1021 pSymA (ctRNADqS). The second construct expresses a ctRNA possessing a left arm with a nonrelated sequence of 15 nucleotides (5′-GCCAGACAGAGAGGG), but the rest of the molecule is identical to p42d ctRNA (ctRNA5N). Finally, we also constructed a mutant ctRNA with its left and right arms identical in sequence to the arms present in the p42d ctRNA but with a stem-loop structure that is completely different in sequence although identical in size and structure (ctRNAart) (see Fig. S1 in the supplemental material).
TABLE 2.
Plasmids used in this work
| Plasmid | Relevant characteristics | Reference |
|---|---|---|
| pBBR1MCS2 | Kmr cloning vector replicable in Rhizobium | Kovach et al., 1995 (16) |
| pCR-ctRNAwt 42d | pBBR1MCS2 carrying a ctRNA wild type from p42d R. etli | This work |
| pCR-ctRNAwt SymA | pBBR1MCS2 carrying a ctRNA wild type from pSymA S. meliloti 1021 | This work |
| pCR-ctRNA-CGCU | pBBR1MCS2 carrying a ctRNA with modified loop (from 5′-GCGA to 5′-CGCU) | This work |
| pCR-ctRNAmut2-5 | pBBR1MCS2 carrying a ctRNA with nucleotides 2-5 changed in the left arm from CUAC to AAUG | This work |
| pCR-ctRNAmut2-5 CGCU | pBBR1MCS2 carrying a ctRNA with both mutations: modified loop (from 5′-GCGA to 5′-CGCU) and nucleotides 2-5 changed in the left arm from CUAC to AAUG | This work |
| pCR-ctRNA-Δ(1-8) | pBBR1MCS2 carrying a ctRNA with lack eight of the first nucleotide of the left arm | This work |
| pCR-ctRNADqS | pBBR1MCS2 carrying a ctRNA with stem-loop and 3′ end from incα region of S. meliloti pSymA and 5′ end (13 nt) from incα region of R. etli p42d | This work |
| pCR-ctRNA5N | pBBR1MCS2 carrying a ctRNA with stem-loop and 3′ end from incα region of R. etli p42d and 5′ end with nonrelated nucleotides | This work |
| pCR-ctRNAart | pBBR1MCS2 carrying a ctRNA with stem-loop with nonrelated nucleotides, 3′ end and 5′ end from ctRNA of p42d | This work |
All of these mutants were constructed using PCR technology. The mutant ctRNADqS was obtained in two successive PCRs: in the first amplification reaction, a plasmid carrying the ctRNA from pSymA was used as a template and the oligonucleotides ctRNADqS-U1 and K-SymA-L were used as primers. In the second PCR, the product of the first PCR was used as a template and ctRNADqS-U2 and K-SymA-L as primers. Mutant ctRNA5N was also obtained by two successive PCRs: primers ctRNA5n-U1 and AlfaD-L were used to raise the first amplification product, using a plasmid carrying the ctRNA gene from p42d as a template. In the second PCR, the oligonucleotides ctRNA5n-U2 and AlfaD-L were used as primers and the amplification product of the first PCR was used as a template. The mutant pCR-ctRNAart was obtained in a similar way: In the first PCR, the oligonucleotides ctRNAart-U1 and AlfaD-L were used as primers, and a plasmid with the ctRNA gene from p42d was used as a template. In the second PCR, the product of the first PCR was used as a template, and the oligonucleotides ctRNAart-U2 and AlfaD-L were used as primers. As controls, p42d ctRNA was obtained by PCR using the primers AlfaDHind-U and AlfaD-L, and a plasmid carrying the p42d ctRNA was used as a template. The ctRNA from pSymA was obtained by PCR using the primers S-SymA-U and K-SymA-L, and a plasmid carrying the pSymA ctRNA was used as a template. Amplification products were inserted into pBBR1MCS2 (Kmr), a vector that replicates in Rhizobium (16). All constructs were confirmed by DNA sequencing, performed at the DNA sequence facility of the Biotechnology Institute (IBT-UNAM).
The pBBR1MCS2 derivatives were introduced into R. etli CFNX101 using E. coli S17-1 as a donor strain. Strains were grown in the appropriate antibiotic-free liquid medium to stationary phase, mixed in a donor-recipient ratio of 1:1 on PY plates, and incubated overnight at 30°C. Colonies were resuspended in PY medium, and serial dilutions were plated on PY agar containing nalidixic acid and kanamycin. The plasmid profiles of at least four transconjugants per conjugation were determined by the in-gel lysis procedure described by Wheatcroft et al. (32) and analyzed applying the considerations described by Ramírez-Romero et al. (25).
Northern blot analysis.
An overnight culture of Rhizobium etli CFNX101 was diluted in PY to an optical density at 620 nm (OD620) of 0.2. This culture was further grown to an OD620 of 0.7 on a shaker, supplemented with nalidixic acid at a final concentration of 20 μg/ml, and sampled 2 ml. Sodium azide was added a final concentration of 100 mM and immediately transferred to new tubes containing a chilled solution of 0.2 M EDTA in glycerol (2:1). Total RNA was isolated from the samples with Trizol as recommended by the manufacturer (Invitrogen) and resolved on a 1% agarose gel with formaldehyde at a final concentration of 1% by electrophoresis. Gels were blotted onto Nylon membranes (Hybond-N+; GE) using a semidry transfer cell (Bio-Rad). Membranes were probed with a 32P-labeled oligonucleotide complementary to the repB coding sequence (RepB-HL) and an oligonucleotide complementary to the repC coding sequence (RepC-HL); the autoradiographic images were obtained with a phosphorimager (Storm 820; GE Healthcare) and quantified using Image Quant 5.2 software (GE Healthcare). Samples were normalized with tRNAMet (primer tRNA-Met).
Determination of ctRNA half-life.
An overnight culture of Rhizobium etli CFNX101 was diluted in PY (21) to an OD620 of 0.2. This culture was further grown to an OD620 of 0.7 on a shaker, supplemented with rifampin (dissolved in dimethyl sulfoxide [DMSO]) at a final concentration of 50 μg/ml, and sampled (25 ml) at various intervals (0, 1, 5, 10, 15, 30, and 60 min). Sodium azide was added to each sample to a final concentration of 100 mM, and the samples were immediately transferred to new tubes containing a chilled solution of 0.2 M EDTA in glycerol (2:1). Total RNA was isolated from the samples with Trizol as recommended by the manufacturers (Invitrogen) and resolved on 10% polyacrylamide-7 M urea gels by electrophoresis. Gels were blotted onto Nylon membranes (Hybond-N+; GE) using a semidry transfer cell (Bio-Rad). Membranes were probed with a 32P-labeled oligonucleotide complementary to the ctRNA (Complement 3′); the autoradiographic images were obtained with a phosphorimager (Storm 820; GE Healthcare) and quantified using Image Quant 5.2 software (GE Healthcare). Samples were normalized with tRNAMet (primer tRNA-Met). Images were obtained and quantified as described above. R. etli cells subjected to a treatment with DMSO (1:1,000) alone served as a control for ctRNA analysis.
RESULTS
Binding assays of ctRNA with repABC incα mRNA (mIncα).
The 155-bp repB-repC intergenic region, or incα region, of R. etli p42d contains an antisense RNA (ctRNA) gene. The promoter of this gene, including its −10 and −35 hexameric elements, was mapped experimentally (30). The size, 59 nt, of the ctRNA was calculated by Venkova-Canova et al., using in vitro and in vivo techniques (30). An in silico analysis of its secondary structure predicted a single stem-loop-structured domain (Fig. 1b). Genetic analysis suggested that this structure is essential for the termination of its own transcription (30).
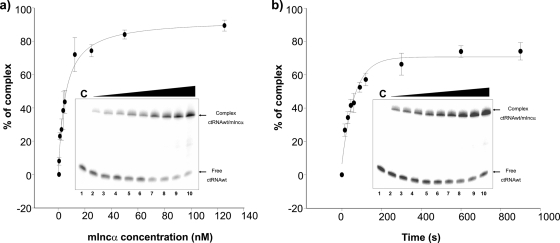
The ctRNA/mIncα duplex affinity was determined by gel shift analysis. Various concentrations of unlabeled mIncα were tested with a fixed concentration of labeled ctRNA to determine the dissociation constant (Kd) of the ctRNA/mIncα complex. This analysis gave a Kd value of 5.17 nM (Fig. 2 a and Table 3). To determine the binding rate constant (ka) of the ctRNA/mIncα complex, a constant amount of mIncα was incubated with labeled ctRNA, and the time course of the binding between these molecules was followed (Fig. 2b and Table 3). The rate constant was calculated to be ≅3.16 × 105 M−1 s−1 at 37°C.
FIG. 2.
ctRNA binding kinetics. Binding assays were performed as described in Material and Methods. (a) A graphic representation of the relative complex formation between the ctRNA and mIncα as s function of the mIncα concentration. (b) A graphic representation of the relative complex formation between the ctRNA and mIncα as a function of the time of incubation.
TABLE 3.
Binding affinity constants and Kd pairing rate constants (ka) of the ctRNA and its derivativesa
| ctRNA or derivative | Binding affinity parameterb |
Pairing rate constant (ka)c |
||
|---|---|---|---|---|
| Bmax ± SD (%) | Kd ± SD (nM) | a ± SD | ka (M−1 s−1) | |
| ctRNAwt | 93.09 ± 2.62 | 5.17 ± 0.51 | 64.162 ± 4.770 | 3.160 × 105 |
| ctRNA-CGCU | 103.96 ± 3.28 | 10.29 ± 1.03 | 72.709 ± 3.642 | 2.200 × 105 |
| ctRNA-Δ(52-59) | 96.09 ± 1.17 | 2.60 ± 1.29 | 84.065 ± 4.184 | 18.300 × 105 |
| ctRNA-mut2-5 | 59.92 ± 15.33 | 161.41 ± 64.11 | 18.157 ± 3.916 | 0.260 × 105 |
| ctRNA-mut2-5-CGCU | 41.02 ± 9.49 | 163.22 ± 58.41 | 5.109 ± 0.447 | 0.780 × 105 |
| ctRNA-D(2-11) | 13.40 ± 0.88 | 244.25 ± 22.45 | ND | ND |
| ctRNA-D(1-8) | 46.39 ± 7.53 | 118.90 ± 32.73 | 11.395 ± 0.928 | 0.680 × 105 |
| ctRNA-D(2-11)(52-59) | NDd | ND | ND | ND |
Values are the means of results from three independent assays ± SD.
Binding affinity parameters were calculated according to the equation y = (Bmax·x)/(Kd + x), where y is the percentage of complexed RNA, Bmax is the amplitude of the reaction, x is the concentration of the target RNA, and Kd is the dissociation constant.
Binding rate constants were determined from time courses assays. Data were fitted to the equation y = y0 + a·(1 − e−bt), where y is the percentage of complexed RNA, a represents the amplitude of the binding reaction, b is the reaction rate constant, and bt is the time.
ND, not determined.
In vitro secondary structure probing of the ctRNA, mIncα region, and ctRNA/mIncα complex.
The ctRNA is encoded in the complementary strand to the repB-repC intergenic region. This fact allows the assumption of an interaction involving both domains. RNase and lead probing assays were undertaken to further study this hypothesis.
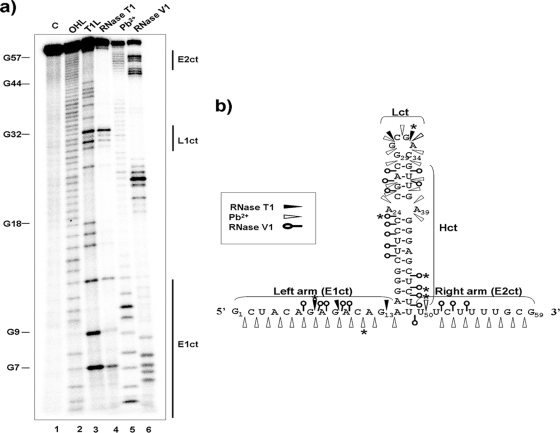
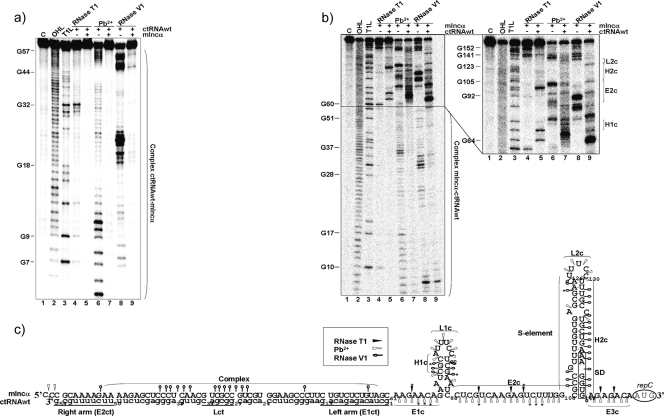
For studying the ctRNA structure and the residues involved in the association with its target, it was 32P 5′ end labeled and partially digested with RNase T1, RNase V1, or lead acetate. RNase T1 preferentially cuts at 5′ of guanosines in single-stranded regions; lead cleaves 3′ to any nucleotide in nonpaired RNA regions, and the RNase V1 preferentially digests double-stranded or stacked RNA regions. The resulting degradation pattern was used for secondary structure prediction with MFold software (Fig. 1b). The analysis showed that the ctRNA is composed of a 14-nt-long helix (Hct) and capped by an apical loop containing a GNRA motif (Lct) (14). Two single-stranded regions flank this stem-loop: the left arm, of 13 nt (E1ct), and a U-rich right arm of 9 nt (E2ct) (Fig. 3). In summary, it contains the essential features of an intrinsic transcriptional terminator. Partial digestions of the 5′-labeled ctRNA in the presence of the mIncα RNA showed a clear resistance to cleavage by both RNase T1 and lead, suggesting the involvement of the whole antisense molecule in the interaction with its target.
FIG. 3.
ctRNA secondary structure. (a) Autoradiogram of a polyacrylamide gel used to resolve 5′-labeled ctRNA after treatment with RNase T1, lead acetate (Pb2+), and RNase V1. Lane 1 (C), undigested probe; lane 2 (OHL), alkaline ladder; lane 3 (T1L), RNase T1 partial digestion of denatured ctRNA used as a ladder; lane 4, RNase T1 partial digestion of ctRNA; lane 5, Pb2+ partial degradation of ctRNA; lane 6, RNase V1 partial digestion of ctRNA. Vertical bars indicate loop and arms of the ctRNA. (b) Secondary structure model for ctRNA. Relevant loops and linear and helix regions are marked with the letters L, E, and H, followed by a number and the letters “ct.” Major cuts are indicated by asterisks.
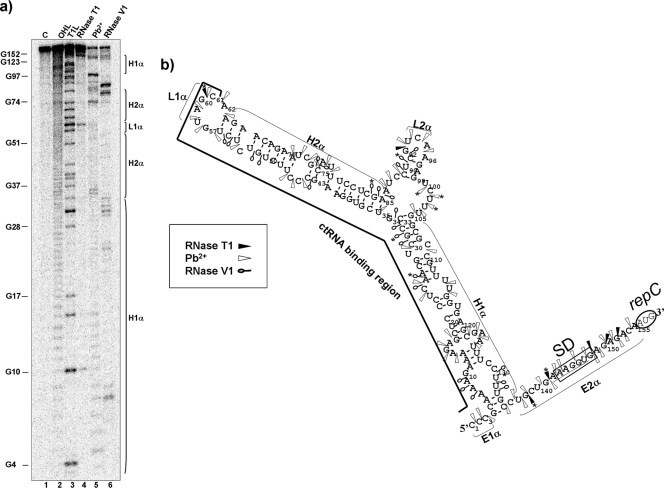
The secondary structure of the mIncα RNA was subsequently analyzed to identify the nucleotides involved in the interaction with the ctRNA. Target RNA was 5′ end labeled and subjected to treatment with nucleases and lead, as described above. The resulting degradation pattern was employed for secondary structure prediction with MFold software (Fig. 4). These studies indicated that mIncα folds into a complex structure that contains two major paired regions, H1α and H2α: H1α is a 20-bp-long helical region interrupted by four internal loops; H2α presents two helixes, connected by an internal loop and closed by a six-nucleotide apical loop (L1α). H1α and H2α are joined by a 20-nt-long linker that can be folded in a short stem-loop domain (L2α). The loop sequence (5′-GUAGCA) of this structure is complementary to the first 5 nt of the ctRNA left arm. The 3′ end of mIncα is a single-stranded region of 20 nt containing the SD sequence, the repC initiation codon remains unpaired, and the region complementary to the ctRNA is completely paired (Fig. 4b). Partial digestions of the 5′-end-labeled mIncα in the presence of the ctRNA revealed a patent conformational change (Fig. 5). This new folding includes two stem-loop domains separated by a 18-nt-long single-stranded region (E2c) (Fig. 5b and c). The first stem-loop motif contains 5-paired nucleotide (H1c), a bulge of two unpaired nucleotides (A71 and A86), and a loop (L1c) of 8 nt. The second stem-loop motif or S-element contains 17 paired nucleotides (H2c), a 5-nt loop (L2c), one bulged nucleotide (G111), and a bubble of two unpaired nucleotides (A120 and C136); the putative repC SD sequence is occluded within the S element.
FIG. 4.
Secondary structure of the target mIncα. (a) Autoradiogram of a polyacrylamide gel used to resolve 5′-labeled mIncα after treatment with RNase T1, lead acetate (Pb2+), and RNase V1. Lane 1 (C), undigested probe; lane 2 (OHL), alkaline ladder; lane 3 (T1L), RNase T1 partial digestion of denatured mIncα used as a ladder; lane 4, RNase T1 partial digestion of mIncα; lane 5, Pb2+ partial degradation of mIncα; lane 6, RNase V1 partial digestion of mIncα. Vertical bars indicate loop and helixes of the mIncα. (b) mIncα secondary structure consistent with cleavage patterns. Black arrowheads indicate RNase T1 sites, white arrowheads indicate Pb2+ sites, and open circles indicate RNase V1 cleavages. Major cuts are indicated by asterisks (*). Relevant loop and helix regions are marked with the letters L and H, followed by a number and the Greek letter, “α.” The mIncα region complementary to the ctRNA is marked with a black line. “SD” indicates the position of repC Shine-Dalgarno sequence. The repC initiation codon (AUG) is encircled.
FIG. 5.
ctRNA/mIncα complex secondary structure. (a) Autoradiogram of a polyacrylamide gel of 5′-labeled ctRNA in the presence or absence of the target RNA (mIncα) treated with RNase T1, lead acetate (Pb2+), and RNase V1. Lane 1 (C), undigested probe; lane 2 (OHL), alkaline ladder; lane 3, (T1L), RNase T1 partial digestion of denatured ctRNA used as a ladder; lane 4, RNase T1 partial digestion of ctRNA; lane 5, RNase T1 partial digestion of ctRNA in the presence of a 10× excess of target mIncα; lane 6, Pb2+ partial degradation of ctRNA; lane 7, Pb2+ partial degradation of ctRNA in the presence of a 10× excess of target mIncα; lane 8, RNase V1 partial digestion of ctRNA; lane 9, RNase V1 partial digestion of ctRNA in the presence of a 10× excess of target mIncα. (b) Autoradiograms of polyacrylamide gels of 5′-labeled mIncα in the presence or absence of the target ctRNA treated with RNase T1, Pb2+, and RNase V1. Lane 1 (C), undigested probe; lane 2 (OHL), alkaline ladder; lane 3, (T1L), RNase T1 partial digestion of denatured mIncα used as a ladder; lane 4, RNase T1 partial digestion of mIncα; lane 5, RNase T1 partial digestion of mIncα in the presence of a 10× excess of target ctRNA; lane 6, Pb2+ partial degradation of mIncα; lane 7, Pb2+ partial degradation of mIncα in the presence of a 10× excess of target ctRNA; lane 8, RNase V1 partial digestion of mIncα; lane 9, RNase V1 partial digestion of mIncα in the presence of a 10× excess of target ctRNA. (c) ctRNA/mIncα complex secondary structure model consistent with cleavage patterns. Uppercase letters indicate sequence corresponding to mIncα, and lowercase letters indicate the ctRNA sequence. Black arrowheads indicate RNase T1 sites, white arrowheads indicate the Pb2+ sites, and open circles indicate RNase V1 cleavage. Relevant loops, helixes, and linear regions are marked with letters L, H, and E, followed by a number and a second letter, c. “SD” indicates the position of repC Shine-Dalgarno sequence. Major cuts are indicated by asterisks (*).
Relative abundances of repB and repC transcripts.
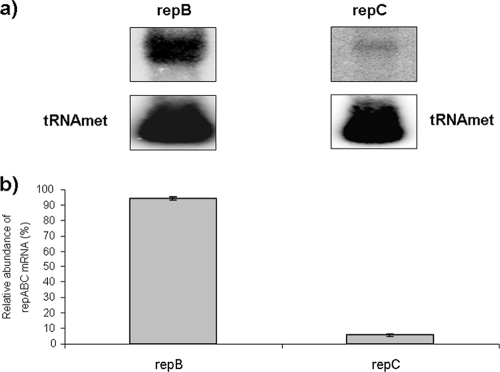
ctRNA binding to its target RNA induces in the latter the formation of a stem-loop structure (the S element), which can be interpreted as an intrinsic terminator or, taking into account that the S element occludes the repC Shine-Dalgarno sequence, as a repC translational attenuator. To discriminate between these two hypotheses, the relative abundances of the repB and repC transcripts—up- and downstream from the S-element—were calculated through a Northern blot analysis, using as probes two oligonucleotides, one of them (RepB-HL) complementary to the repB 3′ end and the other (REpC-HL) complementary to the repC 5′ end. If the relative abundances of repB and repC are the same, it would indicate that the S element works as a translational attenuator. On the other hand, the repB transcript being more abundant than that of repC would indicate that the S element works as an intrinsic transcriptional terminator. As shown in Fig. 6, repB is approximately 17-fold more abundant that repC, indicating that the S element works as an intrinsic terminator.
FIG. 6.
Relative abundances of repB and repC transcripts. (a) Autoradiogram of a Northern blot of Rhizobium etli CFNX101 total RNA hybridized against two oligonucleotides: one complementary to the repB 3′ end and the other complementary to the repC 5′ end. The RNA load was normalized, hybridizing the same samples against an oligonucleotide complementary to tRNAMet. (b) Graphic representation of the normalized hybridization signals.
Binding assays of ctRNA mutant derivatives with mIncα.
Most cis-encoded antisense RNAs have a complex secondary structure containing one or several stem-loop elements. Antisense RNAs initiate the interaction with their cognate RNA through complementary apical loops. However, in some cases, the initial interaction between an antisense RNA and its target involves a single-stranded region and a stem-loop motif: examples include the toxin-antitoxin stabilization system (hok/sok) of plasmid R1 and RNA-IN/RNA-OUT of insertion sequence IS10 (28, 15).
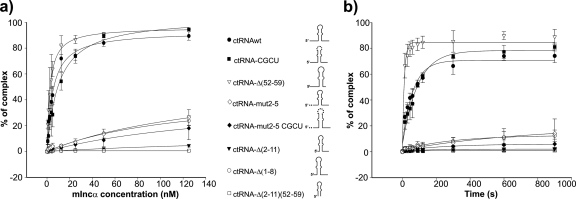
The ctRNA secondary structure described here possesses three candidate regions for initiation of the interaction with its mIncα target site: the left arm, the right arm, and the apical loop. To determine which regions are required to initiate binding to mIncα, we synthesized a collection of truncated versions of the ctRNA, as well as versions carrying point mutations. The collection of constructs tested included the following: (i) a deletion mutant lacking nucleotides 2 to 11 in the left arm [ctRNA-Δ(2-11)], (ii) a deletion mutant lacking nucleotides 1 to 8 in the left arm [ctRNA-Δ(1-8)], (iii) a ctRNA derivative in which nucleotides 2 to 5 in the left arm were changed from CUAC to AAUG [ctRNAmut2-5], (iv) a ctRNA in which the loop was changed from 5′-GGCGAC to 5′-GCGCUC [ctRNA-CGCU]; (v) a deletion mutant without nucleotides 52 to 59 in the right arm [ctRNA-Δ(52-59)]; (vi) a deletion mutant lacking nucleotides 2 to 11 and 52 to 59 [ctRNA-Δ(1-11)(52-59)]; and (vii) a mutant in which nucleotides 2 to 5 and those located in the loop were changed as described above (ctRNA-mut2-5CGCU). Structure probing assays indicated that all RNA variants conserved the central stem-loop motif of the wild-type ctRNA (data not shown). We then studied the interaction kinetics of these derivatives for the target mRNA. The thermodynamic and kinetic parameters of the interaction between these derivatives and their targets are shown in Fig. 7 and Table 3. These results indicate that the terminal nucleotides of the ctRNA left arm are crucial for the interaction with the target mRNA. Only those ctRNA variants carrying nucleotide sequence changes or truncations affecting the left arm severely affect the complex formation. Little effect is observed when the apical loop sequence is altered, indicating that it is not required for the interaction with the target. An analysis of the sequence and secondary structure of the target RNA shows that the complementary region corresponding to the first few nucleotides of the ctRNA left arm lies within the loop L1α, suggesting that this loop is probably involved in the first steps of the ctRNA-target interaction (Fig. 4b).
FIG. 7.
Binding kinetics of the ctRNA and its truncated or mutant derivatives for the target RNA (mIncα). The binding assays were performed as described in Materials and Methods. (a) A graphic representation of the relative complex formation between the ctRNA or its derivatives and mIncα, as a function of the mIncα concentration. (b) A graphic representation of the relative complex formation between the ctRNA or its derivatives and mIncα as a function of the incubation time.
An oligonucleotide complementary to the left arm of the ctRNA strongly inhibits duplex formation between ctRNA and its target site.
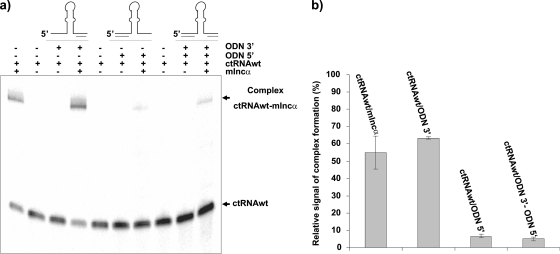
To analyze the role of the left arm of the ctRNA in duplex formation, two deoxyribooligonucleotides were synthesized, one of them complementary to the left arm of the ctRNA (ODN5′) and the other to the right arm (ODN3′), and were treated individually or together as competitors for ctRNA/mIncα hybrid formation (Fig. 8). Under the conditions used in this experiment, around 50% of the labeled ctRNA was able to bind its target. A similar percentage of ctRNA/mIncα hybrids was observed when the reaction was done in the presence of ODN3′. In contrast, the yield of ctRNA/mIncα duplex was substantially reduced in the presence of ODN5′. Binding assays in the presence of both ODN5′ and ODN3′ resulted in percentages of ctRNA/mIncα complex very similar to that in the presence of ODN5′ alone (Fig. 8). Therefore, these results suggest that complex formation strongly requires the ctRNA left arm and that the right arm does not participate significantly during the first steps of the ctRNA/mIncα interaction.
FIG. 8.
Inhibition of ctRNA/mIncα complex formation by specific oligonucleotides. (a) Electrophoretic mobility shift assay (EMSA) of 32P-labeled ctRNA and its target (mIncα) in the presence of oligonucleotides complementary to the ctRNA left arm (ODN5′), complementary to the ctRNA right arm (ODN3′), or both, as described in Materials and Methods. (b) Graphic representation of the relative complex formation obtained from three independent EMSA assays.
Mutations in the left arm of the ctRNA but not those located in the loop change its incompatibility properties.
We used a plasmid incompatibility assay to confirm our in vitro observations that the left arm of ctRNA is essential for the interaction with mIncα. If this also applies in vivo, mutations or deletions in the left arm of the ctRNA gene might abolish incompatibility with p42d, the parental plasmid. In contrast, the incompatibility properties of ctRNA might be unaffected by mutations in regions not involved in the first steps of the RNA-RNA interaction.
To test this hypothesis, we constructed plasmids with the repBC intergenic sequence carrying mutations in the ctRNA gene to produce ctRNA variants and employed them in our studies (Table 2; see also Fig. S1 in the supplemental material). These constructs were introduced into CFNX101, an R. etli recA derivative containing p42d, and the plasmid profiles of the transconjugants were evaluated. The mutant with the modified loop, pCR-ctRNA-CGCU, was as incompatible as the wild-type ctRNA gene. In contrast, the constructs pCR-ctRNAmut2-5, pCR-ctRNAmut2-5CGCU, pCR-ctRNA-Δ(1-8), and pCR-ctRNA5N, carrying ctRNA genes with modified left arms, were compatible. In addition, plasmid pCR-ctRNAart, expressing the ctRNA gene with the left and right arms identical in sequence to the arms present in the wild-type ctRNA but containing a stem-loop of the same size with a different sequence, was found to be compatible. However, pCR-ctRNADqS, a plasmid with a chimeric gene containing the p42d ctRNA left arm and the stem-loop and right arm from pSymA ctRNA, was incompatible. These results show that in vivo, the left arm is essential but not sufficient for the ctRNA-target interactions and the involvement of the stem-loop domain is also needed to exert incompatibility.
Half-life of the ctRNA.
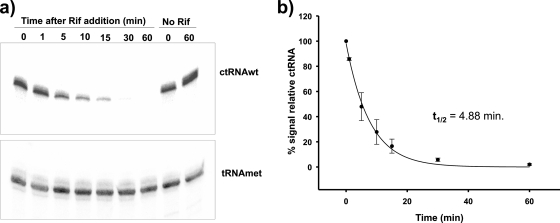
cis-encoded small antisense RNAs involved in the regulation of plasmid replication are generally short-lived, consistent with efficient regulation of plasmid copy number. To determine the half-life of the ctRNA, R. etli CFNX101 (with p42d) was cultivated in rich medium to an OD620 of 0.7. Cell culture was stopped by the addition of rifampin. R. etli CFNX101, treated similarly but without the addition of rifampin, served as a control. Samples were then taken at different time points, and the total RNA was isolated from the samples. The abundance of the ctRNA was quantified by Northern blot analysis using a ctRNA probe and a tRNAMet probe as a loading control. The half-life of the ctRNA was calculated to be around 5 min (Fig. 9).
FIG. 9.
ctRNA half-life determination. (a) Northern blots of total RNA from samples taken at the indicated times (min) after rifampin addition and controls without rifampin, hybridized with an oligonucleotide complementary to the ctRNA and with an oligonucleotide complementary to tRNA-Met, as described in Materials and Methods. (b) Graphic representation of ctRNA decay observed in three independent experiments.
DISCUSSION
A common feature of prokaryotic antisense RNAs is the presence of well-defined secondary structures that act as key structure elements, which play essential roles in the rapid and efficient interaction with their targets. The secondary structure of the ctRNA encoded in the repABC operon of plasmid p42d consists of a single stem-loop flanked by two unpaired regions; the loop in this molecule lacks the U-turn (5′-YUNR) motif frequently found in other cis-encoded antisense RNAs (10, 13) but contains a GNRA motif usually used as a stabilizing motif in RNA-RNA interactions. We have previously shown that the stem-loop structure and the 3′ U-rich tail of the ctRNA are essential for transcriptional termination (30). On the other hand, the target RNA (mIncα) forms a large stem-loop structure interrupted by several bulges and internal loops, but the region containing the repC Shine-Dalgarno sequence remains unpaired. The interaction of the ctRNA with the target RNA (mIncα) induces a refolding of the sequence placed downstream of the anchoring site, resulting in the formation of two new stem-loop structures in the unpaired region of the target RNA; one of them, the S element, behaves as an intrinsic transcriptional terminator.
The ability of ctRNAs to correct fluctuations in plasmid copy number appropriately depends on the efficiency of binding to their targets and on their short half-lives. The ctRNA present in the p42d repABC operon fulfills these requirements: its half-life was calculated to be around 5 min, which is in the same range (1 to 5 min) as those of the ctRNAs from other plasmids (4, 7), and its pairing rate constant was 3.1 × 105 M−1 s−1, again in the same range as those of other sense-antisense interactions (i.e., RNA-OUT/RNA-IN and RNAI/RNAII) (15, 29).
Several lines of evidence indicate that the left arm of the ctRNA is responsible for efficient interaction with the target. First, ctRNA derivatives carrying changes or deletions in the first few nucleotides of the left arm show substantially impaired binding abilities. In contrast, mutations in other regions of the ctRNA do not affect binding properties. Second, oligonucleotides blocking the left arm of the ctRNA severely affect its capacity to bind the target RNA. In contrast, oligonucleotides blocking the right arm of the ctRNA do not affect the ability of the ctRNA to bind its target. Third, plasmid incompatibility tests also indicated that the left-arm ctRNA is essential to exert this phenotype: the constructs expressing ctRNAwt or a ctRNA derivative with an extensive change in the loop were equally effective at displacing p42d, whereas the constructs expressing a ctRNA with mutations in the left arm or carrying deletions in this region were compatible with p42d. However, a construct expressing a ctRNA with both arms identical to the arms of p42d but harboring a stem-loop structure similar in size but not in sequence to that present in p42d ctRNA was compatible. These results suggest that some nucleotides of the stem-loop are also required to exert incompatibility against p42d.
As mentioned earlier, the pSymA ctRNA is compatible with p42d and shares an overall 70% of sequence identity with p42d ctRNA. However, sequence identity between ctRNA stem-loops is only 63%. A plasmid expressing a pSymA ctRNA but with a left-arm sequence identical to the same region of p42d ctRNA is capable of displacing p42d, supporting the notion that besides the interaction between the ctRNA left arm and mIncα, some additional pairing between these two molecules is required to exert incompatibility.
A reexamination of the target RNA structure revealed that the loop (L1α) is complementary to the first nucleotides of the ctRNA. It is therefore possible that under in vitro conditions, the interaction begins at this location and propagates, with a zipper-like mechanism, until the formation of the duplex is complete. However, our in vivo experiments suggest that extended pairing is not a requirement to induce the mIncα structural change needed to occlude the repC Shine-Dalgarno sequence. Similar observations indicating that full complex formation between a ctRNA and its target is not required to exert their regulatory functions have also been made for other plasmid systems (4).
As far as we know, this is the first time this type of RNA-RNA mechanism (apical-loop/single-stranded region) in a replication control system has been reported. However, this mechanism has been found previously in the transposition regulation system of IS10 (15) and in the postsegregational killing system hok/sok of the plasmid R1 (28).
Our findings are consistent with following model of regulation: the target RNA, the mIncα encoded between repB and repC, adopts either of two radically different conformations, one in the absence of the ctRNA and the other in its presence. In the absence of the ctRNA, the target RNA folds in such way that the Shine-Dalgarno sequence of repC is free and ready for translation; when the critical concentration of RepC is reached in the cell, plasmid replication begins. In contrast, when the ctRNA pairs with the target, it induces the formation of two stem-loop structures; one of them operates as an intrinsic terminator, aborting repC transcription and, in consequence, plasmid replication.
An in silico analysis of the repB-repC intergenic region of other repABC plasmids showed that the ctRNA gene is in the same relative position in most of them (8). Moreover, in all cases, the ctRNA has a similar stem-loop structure (30, 18, 9). These observations suggest that all ctRNAs in repABC plasmids have a similar mechanism of action.
Supplementary Material
Acknowledgments
This work was supported by the Consejo Nacional de Ciencia y Tecnología (México) (grant number 46738-Q to M.A.C.), the Ministerio de Educación y Ciencia (Spain) (grant number BFU2006-02508 to A.B.-H.), and the Programa de Apoyo a Proyectos de Investigación e Inovación Tecnológica (México) (grant number IN20580 to M.A.C.). R.C.-R. was supported during the Ph.D. program (Programa de Doctorado en Ciencias Biomédicas-Universidad Nacional Autónoma de México) by scholarships from the Consejo Nacional de Ciencia y Tecnología and Dirección General de Estudios de Posgrado (México).
We are greatly indebted to Ángeles Pérez-Oseguera, América Rivera-Urbalejo, Francisco Pedraza-López, and Vicente Augustin-Vacas for their technical support.
Footnotes
Published ahead of print on 30 April 2010.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Barroso-delJesus, A., M. Tabler, and A. Berzal-Herranz. 1999. Comparative kinetic analysis of structural variants of the hairpin ribozyme reveals further potential to optimize its catalytic performance. Antisense Nucleic Acid Drug Dev. 9:433-440. [DOI] [PubMed] [Google Scholar]
- 2.Bartosik, D., J. Baj, and M. Wlodarczyk. 1998. Molecular and functional analysis of pTAV320, a repABC-type replicon of the Paracoccus versutus composite plasmid pTAV1. Microbiology 144:3149-3157. [DOI] [PubMed] [Google Scholar]
- 3.Brantl, S. 2002. Antisense RNAs in plasmids: control of replication and maintenance. Plasmid 48:165-173. doi: 10.1016/S0147-619X(02)00108-7. [DOI] [PubMed] [Google Scholar]
- 4.Brantl, S. 2004. Plasmid replication control, p. 47-59. In E. Funnell and G. J. Phillips (ed.), Plasmid biology. ASM Press, Washington, DC.
- 5.Brantl, S. 2007. Regulatory mechanisms employed by cis-encoded antisense RNAs. Curr. Opin. Microbiol. 10:102-109. doi: 10.1016/j.mib.2007.03.012. [DOI] [PubMed] [Google Scholar]
- 6.Brennan, R. G., and T. M. Link. 2007. Hfq structure, function and ligand binding. Curr. Opin. Microbiol. 10:125-133. doi: 10.1016/j.mib.2007.03.015. [DOI] [PubMed] [Google Scholar]
- 7.Brenner, M., and J. Tomizawa. 1991. Quantitation of ColE1-encoded replication elements. Proc. Natl. Acad. Sci. U. S. A. 88:405-409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cevallos, M. A., R. Cervantes-Rivera, and R. M. Gutiérrez-Rios. 2008. The repABC plasmid family. Plasmid 60:19-37. doi: 10.1016/j.plasmid.2008.03.001. [DOI] [PubMed] [Google Scholar]
- 9.Chai, Y., and S. C. Winans. 2005. A small antisense RNA downregulates expression of an essential replicase protein of an Agrobacterium tumefaciens Ti plasmid. Mol. Microbiol. 56:1574-1585. doi: 10.1111/j.1365-2958.2005.04636.x. [DOI] [PubMed] [Google Scholar]
- 10.Franch, T., and K. Gerdes. 2000. U-turns and regulatory RNAs. Curr. Opin. Microbiol. 3:159-164. doi: 10.1016/S1369-5274(00)00069-2. [DOI] [PubMed] [Google Scholar]
- 11.González, V., P. Bustos, M. A. Ramírez-Romero, A. Medrano, H. Salgado, I. Hernández-González, C. Hernández-Celis, V. Quintero, G. Moreno-Hagelsieb, L. Girard, O. Rodríguez, M. Flores, M. A. Cevallos, J. Collado-Vides, D. Romero, and G. Dávila. 2003. The mosaic structure of the symbiotic plasmid of Rhizobium etli and its relations with other symbiotic genomic compartments. Genome Biol. 4:R36. doi: 10.1186/gb-2003-4-6-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hanahan, D. 1983. Studies of transformation of E. coli with plasmids. J. Mol. Biol. 166:557-560. [DOI] [PubMed] [Google Scholar]
- 13.Heidrich, N., and S. Brantl. 2003. Antisense-RNA mediated transcriptional attenuation: importance of a U-turn loop structure in the target RNA of plasmid pIP501 for efficient inhibition by the antisense RNA. J. Mol. Biol. 333:917-929. doi: 10.1099/mic.0.2006/002329-0. [DOI] [PubMed] [Google Scholar]
- 14.Jucker, F. M., and A. Pardi. 1995. GNRA tetraloops make a U-turn. RNA 1:219-222. [PMC free article] [PubMed] [Google Scholar]
- 15.Kittle, J. D., R. W. Simons, J. Lee, and N. Kleckner. 1989. Insertion sequence IS10 anti-sense pairing initiates by an interaction between the 5′ end of the target RNA and a loop in the anti-sense RNA. J. Mol. Biol. 210:561-572. [DOI] [PubMed] [Google Scholar]
- 16.Kovach, M. E., P. H. Elzer, D. S. Hill, G. T. Robertson, M. A. Farris, R. M. Roop, and K. M. Peterson. 1995. Four new derivatives of the broad-host range cloning vector pBBR1MCS, carrying different antibiotic resistance cassettes. Gene 166:175-176. doi: 10.1016/0378-1119(95)00584-1. [DOI] [PubMed] [Google Scholar]
- 17.Kumar, C. C., and R. P. Novick. 1985. Plasmid pT181 replication is regulated by two countertranscripts. Proc. Natl. Acad. Sci. U. S. A. 82:638-642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.MacLellan, S. R., L. A. Smallbone, C. D. Sibley, and T. M. Finan. 2005. The expression of a novel antisense gene mediates incompatibility within the large repABC family of alpha-proteobacterial plasmids. Mol. Microbiol. 55:611-623. doi: 10.1111/j.1365-2958.2004.04412.x. [DOI] [PubMed] [Google Scholar]
- 19.Martínez-Salazar, J., D. Romero, M. L. Girard, and G. Dávila. 1991. Molecular cloning and characterization of the recA gene of Rhizobium phaseoli and construction of recA mutants. J. Bacteriol. 173:3035-3040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nacheva, G. A., and A. Berzal-Herranz. 2003. Preventing nondesired RNA-primed RNA extensión catalyzed by T7 RNA polymerase. Eur. J. Biochem. 270:1458-1465. doi: 10.1046/j.1432-1033.2003.03510.x. [DOI] [PubMed] [Google Scholar]
- 21.Noel, K. D., A. Sánchez, L. Fernández, J. Leemans, and M. A. Cevallos. 1984. Rhizobium phaseoli symbiotic mutants with transposon Tn5 insertions. J. Bacteriol. 158:48-155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pappas, K. M., and S. C. Winans. 2003. The RepA and RepB autorepressors and TraR play opposing roles in the regulation of a Ti plasmid repABC operon. Mol. Microbiol. 49:441-455. doi: 10.1046/j.1365-2958.2003.03560.x. [DOI] [PubMed] [Google Scholar]
- 23.Pappas, K. M. 2008. Cell-cell signaling and the Agrobacterium tumefaciens Ti plasmid copy number fluctuations. Plasmid 60:89-107. doi: 10.1016/j.plasmid.2008.05.003. [DOI] [PubMed] [Google Scholar]
- 24.Persson, C., E. G. Wagner, and K. Nordström. 1988. Control of replication of plasmid R1: kinetics of in vitro interaction between the antisense RNA, CopA, and its target, CopT. EMBO J. 7:3279-3288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ramirez-Romero, M. A., N. Soberon, A. Perez-Oseguera, J. Tellez-Sosa, and M. A. Cevallos. 2000. Structural elements required for replication and incompatibility of the Rhizobium etli symbiotic plasmid. J. Bacteriol. 182:3117-3124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ramirez-Romero, M. A., J. Tellez-Sosa, H. Barrios, A. Perez-Oseguera, V. Rosas, and M. A. Cevallos. 2001. RepA negatively autoregulates the transcription of the repABC operon of the Rhizobium etli symbiotic plasmid basic replicon. Mol. Microbiol. 42:195-204. doi: 10.1111/j.1365-2958.2001.02621.x. [DOI] [PubMed] [Google Scholar]
- 27.Simon, R., U. Priefer, and A. Pühler. 1983. A broad host-range mobilization system for in vivo genetic engineering transposon mutagenesis in Gram negative bacteria. Biol. Technol. 1:784-791. [Google Scholar]
- 28.Thisted, T., N. S. Sørensen, E. G. Wagner, and K. Gerdes. 1994. Mechanism of postsegregational killing: Sok antisense RNA interacts with Hok mRNA via its 5′-end single-stranded leader and competes with the 3′-end of Hok mRNA for binding to the mok translational initiation region. EMBO J. 13:1960-1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tomizawa, J., and T. Itoh. 1981. Plasmid ColE1 incompatibility determined by interaction of RNA I with primer transcript. Proc. Natl. Acad. Sci. U. S. A. 78:6096-6100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Venkova-Canova, T., N. E. Soberón, M. A. Ramírez-Romero, and M. A. Cevallos. 2004. Two discrete elements are required for the replication of a repABC plasmid: an antisense RNA and a stem-loop structure. Mol. Microbiol. 54:1431-1444. 10.1111/j.1365-2958.2004.04366.x. [DOI] [PubMed] [Google Scholar]
- 31.Wagner, E. G., and S. Brantl. 1998. Kissing and RNA stability in antisense control of plasmid replication. Trends Biochem. Sci. 23:451-454. doi: 10.1016/S0968-0004(98)01322-X. [DOI] [PubMed] [Google Scholar]
- 32.Wheatcroft, R., G. D. McRae, and R. W. Miller. 1990. Changes in the Rhizobium meliloti genome and the ability to detect supercoiled plasmids during bacteroid development. Mol. Plant Microbe Interact. 3:9-17. [Google Scholar]
- 33.Zuker, M., and P. Stiegler. 1981. Optimal computer folding of large RNA sequences using thermodynamics and auxiliary information. Nucleic Acids Res. 9:133-148. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.