Abstract
The isolated ligand binding domain of the chemotaxis aspartate receptor is the focus of the present study, which both (a) identifies structural regions involved in the attractant-induced conformational change and (b) investigates the kinetic parameters of attractant binding. To analyze the attractant-induced conformational change within the homodimeric domain, 19F NMR is used to monitor six para-fluorophenylalanine (4-F-Phe) positions within each identical subunit of the homodimer. The binding of one molecule of aspartate to the homodimer perturbs three of the 4-F-Phe resonances significantly: 4-F-Phe150 in the attractant binding site, 4-F-Phe107 located 26 Å from the site, and 4-F-Phe180 at a distance of 40 Å from the site. Comparison of the frequency shifts triggered by aspartate and glutamate reveals that these attractants generate different conformations in the vicinity of the attractant site but trigger indistinguishable long-range conformational effects at distant positions. This long-range conformational change is specific for attractant binding, since formation of the Cys36–Cys36′ disulfide bond or the nonphysiological binding of 1,10-phenanthroline to an aromatic pocket distal to the attractant site each yield conformational changes which are significantly more localized. The attractant-triggered perturbations detected at 4-F-Phe107 and 4-F-Phe180 indicate that the structural change includes an intrasubunit component communicated through the domain to its C-terminal region, which, in the full-length receptor, continues through the membrane as the second membrane-spanning helix. It would thus appear that the transmembrane signal is transmitted through this helix. The 19F NMR results also reveal the association rate constant for aspartate binding to the isolated periplasmic domain (kon ~ 109 M−1 s−1), enabling deduction of the dissociation rate constant (k off ~ 103 s−1). Aspartate binding thus approaches the diffusion-controlled limit. The observed binding equilibrium and resulting conformational changes are rapid on the time scale of the chemotactic response.
The ability to alter internal functions in response to external factors such as environmental conditions and hormonal signals is essential to all cells. The primary step in a signal transduction pathway mediating the cellular response to an external cue is generally a transmembrane signal generated by a cell-surface receptor. The aspartate receptor of Escherichia coli and Salmonella typhimurium provides an unusually accessible model system in which to probe the nature of transmembrane signaling. This receptor belongs to a widely distributed group of prokaryotic transmembrane receptors, each of which enables a physiological response to a chemical or physical stimulus (Adler, 1969; Russo & Koshland, 1983; Boyd et al., 1983; Nowlin et al., 1985; Dahl et al., 1989; Utsumi et al., 1989; Hazelbauer et al., 1990; Shaw, 1991; Collins et al., 1992; McBride et al., 1992). Like the other prokaryotic receptors, the aspartate receptor is characterized by a topology possessing an external (periplasmic) ligand binding domain, a cytoplasmic signaling domain, and a pair of transmembrane α-helices connecting the two domains. Such topology is similar to that predicted for a large class of eukaryotic receptors including the tyrosine-kinase-linked growth hormone receptors [e.g., epidermal growth factor, insulin, nerve growth factor (Ullrich et al., 1984, 1985; Johnson et al., 1986)], suggesting that these prokaryotic and eukaryotic receptors may share important structural (Russo & Koshland, 1983) and mechanistic (Moe et al., 1989) features.
In its physiological role, the aspartate receptor is regulated by the binding of such attractants as aspartate, aspartate analogues (Clark & Koshland, 1979), and phenol (Imae et al., 1987) to the periplasmic ligand binding domain. The resulting transmembrane signal to the cytoplasmic domain in turn regulates a cytoplasmic phosphorylation pathway which controls the swimming behavior of the cell [for reviews, see Bourret et al. (1991), Stock et al. (1992), Armitage (1992), Parkinson and Kofoid (1992), and Hazelbauer et al. (1993)]. Signaling proteins homologous to components of the phosphorylation pathway appear to be ubiquitous in prokaryotic cells and have recently been detected in eukaryotic cells as well (Chang et al., 1993; Ota & Varshavsky, 1993).
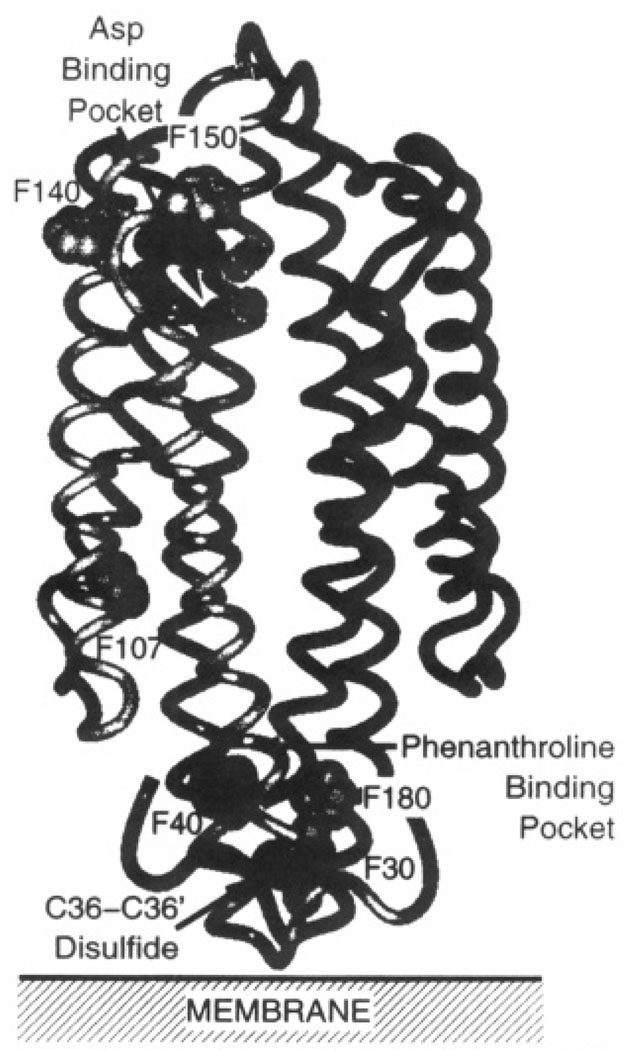
The structure of the aspartate receptor has been extensively characterized, particularly that of the periplasmic and transmembrane domains. The 120-kDa receptor is a homodimer of identical subunits in both the presence and absence of ligand (Falke & Koshland, 1987; Milligan & Koshland, 1988). The three-dimensional structure of the soluble periplasmic domain has been determined to 2.0-Å resolution by X-ray crystallography (Milburn et al., 1991; Yeh et al., 1993; Scott et al., 1993). The architecture (Figure 1) consists of a dimer of four-helix bundles, with two symmetric attractant binding sites at the dimer interface. At the opposite end of the molecule, near the predicted location of the bilayer in the native receptor, an engineered Cys36–Cys36′ disulfide bond covalently links the two subunits. In this same region, 3 Å from the disulfide, a single molecule of the nonphysiological ligand 1,10-phenanthroline lies bound in an aromatic pocket. In the intact receptor, the structure of the transmembrane domain has been characterized by targeted disulfide mapping, yielding a model for the packing of the membrane-spanning α-helices through which the transmembrane signal is communicated (Falke & Koshland, 1987; Falke et al., 1988; Lynch & Koshland, 1991; Pakula & Simon, 1992).
FIGURE 1.
Structure of the apo periplasmic ligand binding domain of the S. typhimurium aspartate receptor (Milburn et al., 1991). Shown is a backbone ribbon diagram, where black and gray ribbons indicate the two different subunits. The gray CPK atoms are the Phe residues of subunit 1. Also shown are the atoms which coordinate aspartate in one of the two symmetry-related attractant binding sites (black from subunit 1, black with stipples from subunit 1′). Finally, the 1,10-phenanthroline binding pocket and the Cys36–Cys36′ disulfide, both located near the end of the domain distal from the attractant binding site, are indicated.
The targeted disulfide approach has revealed the presence of a transmembrane conformational change triggered by attractant binding (Falke & Koshland, 1987), but the size of the aspartate receptor and the fact that it is an integral membrane protein have hindered further structural and kinetic studies of the transmembrane signal. Even the water-soluble periplasmic domain, which is 36 kDa as a dimer, is too large for solution structure determination by existing NMR methodology. One method useful in such an application is 19F NMR of the protein labeled with fluorine at specific aromatic residues [reviewed in (Luck and Falke 1991a–c) and Drake et al. (1993)]. The utility of this technique stems from the inherent qualities of the 19F nucleus, including high sensitivity (0.833 that of 1H) and natural abundance (100%), lack of background resonances, ease of biosynthetic incorporation, and the nonperturbing nature of the fluorine substitution at aromatic hydrogen positions (Gammon et al., 1972; Sykes et al., 1974; Pratt & Ho, 1975; Lu et al., 1976; Post et al., 1984; Wilson & Dahlquist, 1985; Rule et al., 1987; Gerig, 1989, 1994; Peersen et al., 1990; Luck & Falke, 1991 a,b; Gregory & Gerig, 1991; Drake et al., 1993). The shielding of the 19F nucleus depends strongly on the symmetry of the lone pair electrons in the atom; this symmetry is easily perturbed by packing forces within the local van der Waals environment and local electrostatic forces. Thus, the 19F NMR chemical shift is among the most sensitive detectors of structural changes at specific labeling positions in a macromolecule (Gerig, 1989, 1994; Augspurger et al., 1992; deDios et al., 1993; Chambers et al., 1994). Moreover, NMR methods provide unique kinetic information regarding the rate at which structural changes occur (Wagner & Wütrich, 1986). Finally, the power of NMR is extended through the use of protein engineering, which allows the assignment of resonances and the resolution of ambiguities in interpretation (Rule et al., 1987; Drake et al., 1993; Bourret et al., 1993).
In a previous study, 5-fluorotryptophan (5-F-Trp) was successfully incorporated into the intact, membrane-bound receptor, and the 19F NMR resonance of a lone mobile 5-F-Trp residue was detected (Falke et al., 1992). A transmembrane conformational change was observed in that study, but resonances from multiple probe sites are needed to map out the regions of the protein participating in transmembrane signaling. In the current work, para-fluorophenylalanine (4-F-Phe)1 has been incorporated into the phenylalanine positions of the soluble ligand binding domain fragment (residues 25–188), and 19F NMR spectra have been obtained. Each subunit possesses six phenylalanines which are fortuitously located, as illustrated in Figure 1 and Table 1, thereby providing probes in several important regions of the domain structure. Phe150 is in the immediate vicinity of the attractant binding site. Phe140 is a solvent-exposed residue near the attractant binding site. Phe107 lies in the core of a subunit 26 Å from the attractant binding site, where it provides sensitive detection of intrasubunit conformational changes. The remaining three Phe residues, Phe30, Phe40, and Phe180, are members of an aromatic cluster which surround the phenanthroline binding pocket at the dimer interface, proximal to the Cys36–Cys36′ disulfide bond. To complement the NMR data, attractant binding studies using intrinsic tryptophan fluorescence have also been carried out. Together, the results both (a) place strong constraints on the cooperativity and kinetics of aspartate binding and (b) reveal regions of the ligand binding domain involved in the resulting conformational change. Finally, the study addresses the effect of the intersubunit Cys36–Cys36′ disulfide bond on the conformation and dynamics of the ligand binding domain.
Table 1.
Positional Parameters of Phenylalanine Residues in the Ligand Binding Domaina
| position | distance (Å) from bound aspartateb |
distance (Å) from position 33c |
distance (Å) from Cys36 disulfided |
|---|---|---|---|
| 30 | 45 | 9 | 4 |
| 40 | 40 | 13 | 5 |
| 107 | 2o | 29 | 25 |
| 140 | 10 | 55 | 52 |
| 150 | 7 | 57 | 53 |
| 180 | 40 | 16 | 7 |
Calculated from the crystallographic coordinates of Milburn et al. (1991).
Distance from the α-carbon of bound aspartate to the para-carbon position of the indicated phenylalanine ring in subunit1.
Distance from the α-carbon of Leu33, near the predicted membrane interface, to the para-carbon position of the indicated phenylalanine in the same subunit 1.
Distance from the thiol-sulfur of Cys36 to the para-carbon position of the nearest indicated phenylalanine in the dimer.
MATERIALS AND METHODS
Materials
The expression strain used was E. coli RP3808 [Δ(che A-che Z) DE2209 tsr-1 leuB6 kis-4 eda-50 rps L136 [thi-1 Δ (gal-attL) DE99 ara-14 lacY1 mt1-1 xyl-5 tonA31 tsx-78] /mks/], kindly provided by Sandy Parkinson (University of Utah). 4-Fluorophenylalanine (the para-fluoro isomer, 4-F-Phe) was purchased as a racemic mixture from Sigma. Glyphosate was purchased as solid tablets of Roundup Herbacide (60% glyphosate, Monsanto Laboratories). l-Aspartate and l-glutamate of >99.9% purity were purchased from Sigma.
Cloning and Mutagenesis
The gene encoding the S. typhimurium aspartate receptor under control of the trc promotor, with the first transmembrane segment replaced by the cleavable signal sequence of E. coli alkaline phosphatase and a stop codon inserted after residue 188, was cloned into the vector pBLUESCRIPT KS+, giving the plasmid pMK155 as previously described (Milligan & Koshland, 1993). A cysteine codon at position 36 was incorporated into the plasmid by oligonucleotide directed mutagenesis (Kunkel, 1987) to give the gene for the N36C codon in plasmid pMK155.N36C. Plasmids which express the N36C mutant bearing an additional Phe to Tyr substitution were constructed in a similar fashion.
Isolation of the Fluorine-Labeled Ligand Binding Domain
Wild-type and mutant periplasmic-domain proteins were expressed in the E. coli strain RP3808 bearing the appropriate version of pMK155. No induction was necessary as the trc promoter is constitutively expressed in this strain. The labeling medium contained 2.5 g/L bactotryptone, 1.25 g/L yeast extract, 5 g/L NaCl, and 0.75 g/L 4-fluoro-d,l-Phe. Immediately prior to innoculation, filter-sterilized stock solutions were used to yield the following final concentrations of additional components: 1 g/L glyphosate, 0.04% glucose, and 0.1 g/L ampicillin. The glyphosate served to inhibit aromatic amino acid biosynthysis (Kim et al., 1990). Cultures (500 mL per 2-L flask) were grown with vigorous aeration at 37 °C for 10 h. A 10-L prep typically yielded 9 g of cells.
The ligand binding domain was purified using a variation of a gentle osmotic shock procedure previously described (Milligan & Koshland, 1993). Cells were harvested from media by centrifugation [Beckman JA-10 rotor, 6000 rpm (6400g), 5 min] and washed twice in a buffer containing 10 mM Tris, pH 7.4 with HCl, 30 mM NaCl, and 0.5 mM EDTA with pelleting by centrifugation after each wash. The cells were then resuspended in ice-cold spheroplast buffer containing 100 mM Tris, pH 8.0 with HCl, 500 mM sucrose, 0.5 mM EDTA, and 0.2 mM PMSF followed by incubation at room temperature for 10 min. Finally, the cells were harvested by centrifugation [Beckman JA-14 rotor, 10 000 rpm (15000g), 10 min] and gently resuspended in ice-cold 1 mM MgCl2 to lyse the outer membrane, where the divalent cation serves to maintain integrity of the cytoplasmic membrane. The suspension was incubated on ice for 15 min. The spheroplasted cells were removed by centrifugation [Beckman JA-14 rotor, 12 000 rpm (22000g), 10 min]. To the supernatant were added the following final concentrations of constituents: 50 mM Tris, pH 7.4 with HCl, 2 mM EDTA, 2 mM 1,10-phenanthroline, and 0.2 mM PMSF. The protein was recovered by ammonium sulfate precipitation. In this step, 0.26 g of solid ammonium sulfate was added per mL of starting solution. After stirring at 4 °C for 1 h, the precipitated protein was collected by centrifugation [Beckman JA-14 rotor, 12 000 rpm (22000g), 30 min] and resuspended in 10 mL of 10 mM Tris, pH 8.0 with HCl. The protein was dialyzed overnight against 1 L of 10 mM Tris, pH 8.0 with HCl, including at least two exchanges with fresh buffer.
The dialyzed sample was loaded onto a 75-mL Q-Sepharose column (2.5-cm diameter) equilibrated with 10 mM Tris, pH 8.0 with HCl. A gradient was applied of 0–300 mM NaCl across a total of 600 mL of 10 mM Tris, pH 8.0 with HCl: the ligand binding domain eluted between 100 and 150 mM NaCl. Fractions collected during the gradient were analyzed by monitoring the absorbance at 280 nm. The peak fractions were pooled and concentrated by ultrafiltration (Amicon, YM10 membrane) to a volume of ~10 mL. The pool was then dialyzed for 2–3 days against 1 L of 10 mM Tris, pH 8.0 with HCl, 50 mM NaCl, 50 mM KCl, and 1 mM MgCl2, including at least three changes of buffer. A typical 10-L prep yielded between 10 and 30 mg of protein of ≥85% purity. This procedure yielded the oxidized homodimer cross-linked by the Cys36–Cys36′ disulfide bond; for reduced N36C domain the sample was incubated overnight under a nitrogen atmosphere, with 50 mM DTT and 0.5 mM EDTA at 4 °C.
Measurement of the Concentration of the Purified Ligand-Binding Domain
The total protein concentration was determined using a bicinchonic acid (BCA) colorimetric assay (Stoscheck, 1990). To improve accuracy and precision, sodium dodecyl sulfate (SDS) was added to the reaction mixture to a final concentration of 0.1% (w/v). The resulting concentration was corrected for the purity of the ligand binding domain, as determined by SDS–PAGE (15% acrylamide gel).
In some cases the concentration of the ligand binding domain was determined by measuring the absorbance of the sample at 280 nm. An extinction coefficient for the ligand binding domain was determined by the method of Gill and von Hippel (1989), yielding ε280 = 29 200 M−1 cm−1 for the wild-type dimer, ε280 = 29 400 M−1 cm−1 for the disulfide-linked dimer, and ε280 = 32 000 M−1 cm−1 for the tyrosine replacement mutants of the disulfide-linked dimer. This method gave a calculated ligand binding domain concentration which agreed, to within an error of 10%, with the value determined using the BCA assay.
Measurement of Ligand Binding
The binding of aspartate or glutamate to the ligand binding domain was monitored at 25 °C by the resulting increase in intrinsic tryptophan steady-state fluorescence using an SLM 48000S spectrofluorimeter (λex = 283 nm, 4-nm bandwidth; λem = 350 nm, 8-nm bandwidth). Samples contained 2.5 µM dimeric ligand binding domain for determination of the glutamate dissociation constant (K D) and 0.1 µM dimeric ligand binding domain for determination of the aspartate K D. The low-affinity ligand glutamate was routinely used to measure the binding constants of the replacement mutants because its larger K D eliminated the need to correct the total ligand concentration for bound ligand, as was required for aspartate binding. To eliminate the possibility of effects due to a shifting monomer–dimer equilibrium, the disulfide-cross-linked N36C domain was used in all cases. Though the emission maximum is at 336 nm, the greatest change upon the binding of aspartate or glutamate was observed at an emission wavelength of 350 nm. Nonlinear least-squares analysis was used to determine K D from the untransformed binding titration:
| (1) |
where F is the observed fluorescence intensity, F 0 is the fluorescence intensity before addition of ligand, ΔF max is the ligand, and [L] is the free ligand concentration.
19F NMR Measurements
19F NMR spectra were obtained at 470 MHz on a Varian VXR 500 spectrometer equipped with a 5-mm 1H/19F probe. Samples contained 0.3–1.5 mM dimeric ligand binding domain in 10 mM Tris, pH 8.0 with HCl, 50 mM NaCl, 50 mM KCl, and 1 mM MgCl2. Samples were prepared by concentrating, using ultrafiltration (Amicon, YM10 membrane), the final dialysate from the purification to a volume of 600 µL and then adding D2O and 5-fluorotryptophan, the latter as an internal frequency standard (referenced to −49.5 ppm, its known chemical shift relative to TFA at 0 ppm), thereby enabling direct comparison of chemical shifts in different spectra. Standard undecoupled spectral parameters were as follows: 12 000-Hz spectral width, 16K data points, 80° pulse width, 0.68-s aquisition time, 1.2-s relaxation delay, 20-Hz line broadening, and temperature control at 25 °C. For quantitative integrations, spectra were obtained with the pulse width decreased to 60° and the relaxation delay increased to 5.0 s. T 1 relaxation measurements, utilizing a 180°–τ−90° pulse sequence and a 4.0-s relaxation delay, yielded T 1 values ranging from 0.7 to 1.4 s, within the typical range observed for 4-F-Phe resonances (Drake et al., 1993).
Determination of the Extent of Fluorine Incorporation
The 19F NMR spectrum of the apo-domain was integrated, yielding four peaks of equal intensity and one of double intensity (the result of two overlapping resonances). The average intensity of the individual 4-F-Phe resonances was then compared to that of the internal standard of known concentration and this information, together with the known protein concentration, provided the extent of fluorine incorporation.
Molecular Graphics
Crystallographic coordinates of the apo- and aspartate-occupied conformations of the ligand binding domain were graciously supplied by Kim and coworkers (Milburn et al., 1991) and were visualized using Biosym Technologies Insight II graphics software running on a Silicon Graphics Personal Iris workstation. Surface accessibility calculations were carried out using the program Accessibility developed by Handschumacher and Richards (Richards, 1977; version 1983).
Error Estimates
Integrations of individual 19F NMR resonances exhibited a relative error of ± 10%, determined by comparing the relative integrals from four independent spectra. The standard deviation inherent in measurements of 19F frequency shifts is ±0.1 ppm, ascertained by comparing the chemical shifts of identical resonances in four independent spectra. Errors for parameters obtained in binding curve fits were determined by nonlinear least-squares error analysis.
RESULTS
Affinity of the Ligand Binding Domain for Aspartate
In order to investigate the structural integrity of the ligand binding domain, the affinity of the oxidized N36C domain for the attractant aspartate was compared to that of the intact receptor. The affinity of the soluble domain for aspartate was measured via titration of intrinsic tryptophan fluorescence, yielding KD = 1.4 ± 0.5 µM, a value within the range previously determined for the intact wild-type receptor in the membrane (KD = 0.1–6 µM; Clark & Koshland, 1979; Biemann & Koshland, 1994) as well as for the isolated domain (KD = 1–2 µM; Milligan & Koshland, 1993). This result indicates that aspartate binds to the isolated domain and triggers a conformational change which is essentially native in its thermodynamics, suggesting that the engineered Cys36–Cys36′ intersubunit disulfide bond is relatively nonperturbing. Additional evidence that the engineered disulfide is nonperturbing is provided by its minimal effect on transmembrane signaling in the intact receptor as assayed by (a) methylation rates (Falke et al., 1988; Milligan & Koshland, 1991) and (b) transmembrane regulation of kinase activity (Chervitz and Falke, unpublished data). In contrast, many point mutations generate large perturbations in transmembrane signaling assays (Ames et al., 1988; Yaghmai & Hazelbauer, 1992; Chervitz and Falke, unpublished).
Incorporation of 4-F-Phe
The ligand binding domain was fluorine-labeled for 19F NMR studies by overexpression of the cloned gene in the presence of glyphosate, which supresses the synthesis of aromatic amino acids (Kim et al., 1990). A 4:1 ratio of para-fluoro-l-phenyalanine(4-F-Phe) to unlabeled l-phenylalanine was added to the growth medium to optimize production of fluorine-labeled protein. Since E. coli possesses the ability to discriminate against substitution at the para position of the Phe ring, the resulting efficiency of 4-F-Phe labeling was 7%. Thus, at any given Phe position within the fluorine-labeled ligand binding domain, there was a 7% probability of finding a 4-F-Phe residue.
Due to the low efficiency of fluorine labeling it was not possible to test directly the effect of the fluorine incorporation on receptor activity; however, the effect of a more extreme perturbation was accessible to quantitation. In protein structures, hydroxyl substitution for aromatic hydrogen is expected to be more perturbing than fluorine substitution: this stems from the ~50% greater volume of the hydroxyl group relative to fluorine and the propensity of hydroxyl to form hydrogen bonds, while aromatic fluorine is a poor hydrogen bond acceptor and is incapable of donating a hydrogen bond (Pauling, 1960; Gerig, 1989). To implement the test, a set of six mutant proteins was generated, each possessing a single tyrosine substitution at a different phenylalanine position. The resulting mutants, which provide 100% substituted hydroxyl at the para position of a single phenylalanine residue, enabled estimation of the maximum perturbation due to fluorine incorporation at that position. The attractant affinities of the resulting mutants were compared using a fluorescence assay to monitor glutamate binding (see Materials and Methods).
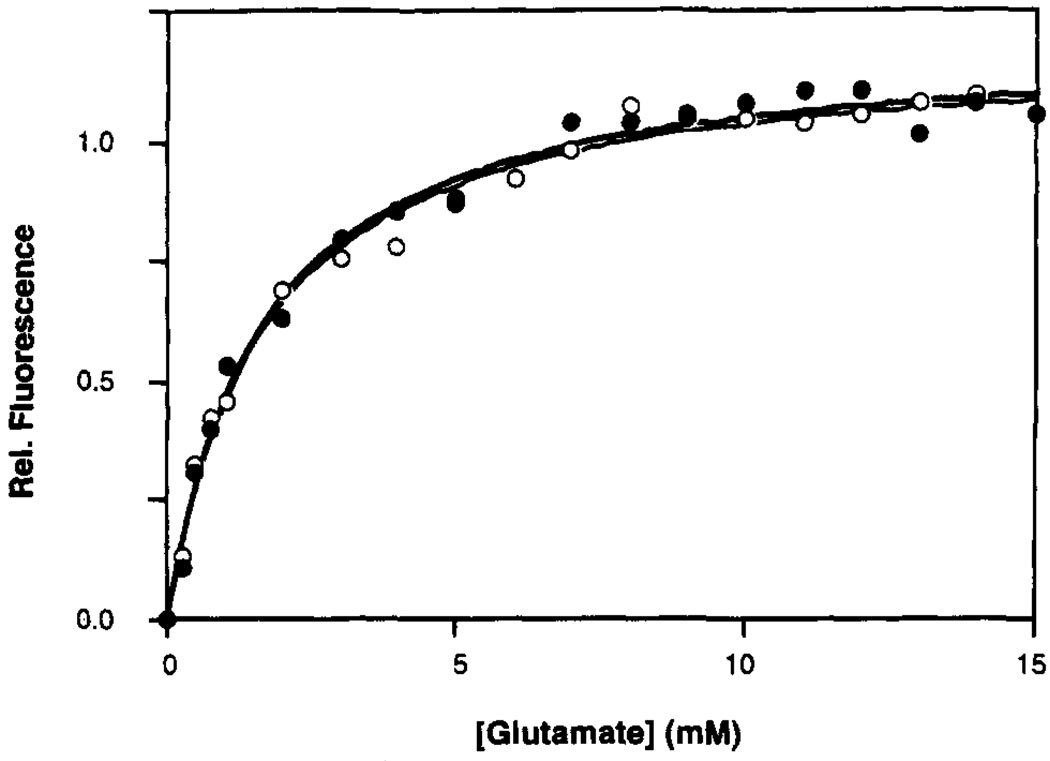
Figure 2 compares the glutamate binding curves, monitored by intrinsic tryptophan fluorescence, of the disulfide-linked ligand binding domain (N36C) and a representative Phe to Tyr replacement mutant (N36C/F107Y). The curves are indistinguishable. Table 2 shows the calculated K Ds of each replacement mutant for glutamate. The affinities of the mutant proteins for glutamate range from 4-fold tighter to 1.6-fold weaker binding. These effects are quite small on a free energy scale (≤0.8 kcal mol−1), suggesting that hydroxyl substitutions at the para position of each phenylalanine residue cause at most minor perturbation of the domain. Therefore the even smaller perturbations due to 4-F-Phe incorporation are likely to be negligible.
FIGURE 2.
Glutamate binding curves at 25 °C for the oxidized N36C (closed circles) and N36C/F107Y (open circles) ligand binding domains, obtained by monitoring intrinsic tryptophan fluorescence. A nonlinear least squares best-fit curve generated for a homogeneous population of sites is shown for each domain (N36C = bold line; N36C/F107Y = narrow line; best fit KD values summarized in Table 2). The buffer was 10 mM Tris, pH 8.0 with HCl, 50 mM NaCl, 50 mM KCl, and 1 mM MgCl2 The concentration of the dimeric domain was 2.5 µM (or 5.0 µM monomer).
Table 2.
Tyrosine Substitutions at Phenylalanine Positions: Effect on Ligand Binding
| Mutant | glutamate (mM) KDa |
|---|---|
| N36C | 2.0 ± 0.2 |
| N36C/F30Y | 3.2 ± 0.3 |
| N36C/F40Y | 2.2 ± 0.4 |
| N36C/F107Y | 1.4 ± 0.1 |
| N36C/F140Y | 2.2 ± 0.6 |
| N36C/F150Y | 0.5 ± 0.3 |
| N36C/F180Y | 1.3 ± 0.8 |
Measured by monitoring intrinsic tryptophan fluorescence during glutamate titration at 25 °C in 10 mM Tris, pH 8.0 with HCl, 50 mM NaCl, 50 mM KCl, and 1 mM MgCl2. The concentration of the dimeric domain was 2.5 µM.
Assignment of the 19F NMR Resonances to Specific 4-F-Phe Residues
Figure 3A presents the 19F NMR spectrum of 4-F-Phe-labeled N36C ligand binding domain in the apo conformation. Six resonances are discernible, two of which overlap to a large degree. The observation of only six resonances from the homodimer indicates that the structures of the two subunits are the same on average, such that the six 4-F-Phe residues in subunit 1 are equivalent to the corresponding residues in subunit 1′ Thus the two subunits rapidly sample the same range of conformations, yielding fully overlapping spectra (see below). Integration reveals that the 4-F-Phe resonances each possess the same intensity, verifying that the fluorine labeling efficiency is the same at all six phenylalanine positions of each subunit.
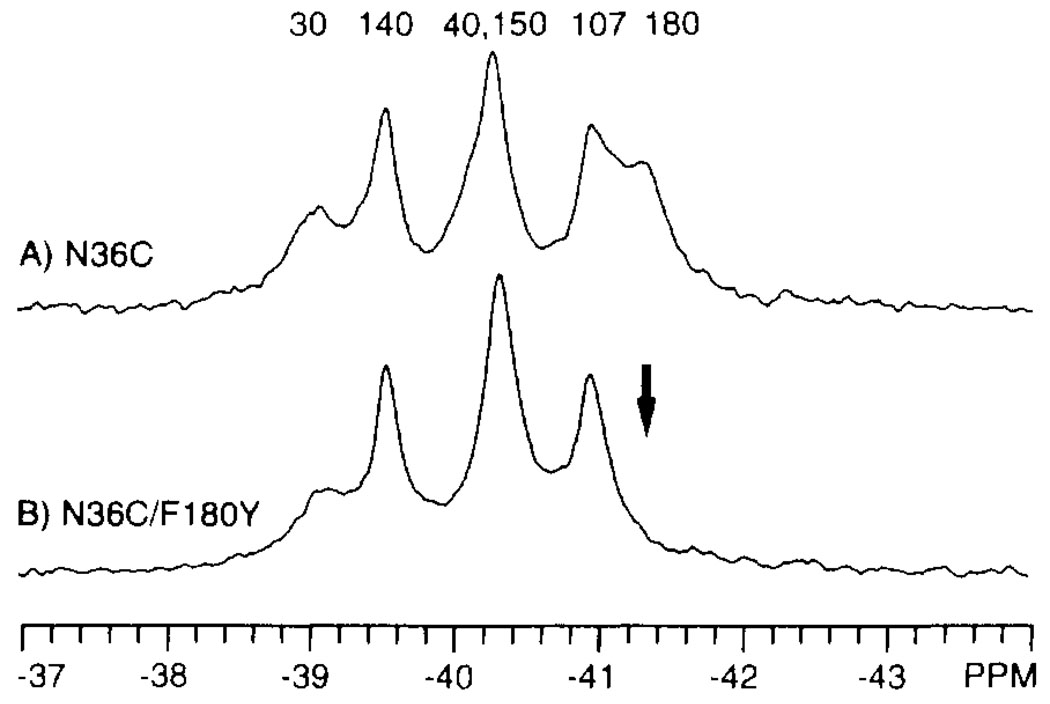
FIGURE 3.
Assignment of 4-F-Phe 19F NMR resonances by site-directed mutagenesis. Shown are the 19F NMR spectra of two 4-F-Phe-labeled domains: N36C (A) and N36C/F180Y (B). The arrow indicates the resonance deleted by the mutation. The final assignments provided by replacement and nudge mutational analysis are indicated (see text). Spectra were obtained at 470 MHz and 25°C in the same buffer as in Figure 2, with the addition of 10% D2O and 50 µM 5-F-Trp. The concentration of dimeric domain was 0.6 mM.
The six resonances were assigned to specific 4-F-Phe residues by two methods. Where possible, direct replacement of individual Phe residues was used. In this method, a mutant bearing a Tyr substitution at a single Phe position is labeled with 4-F-Phe at the remaining Phe positions and used to generate a 19F NMR spectrum, in which the 19F NMR resonance from the targeted position disappears. As an example, Figure 3B illustrates the 19F NMR spectrum of a mutant bearing a Tyr substitution at the Phe180 position (N36C/F180Y); here five resonances are unchanged while the sixth is obliterated. The missing resonance is directly assigned to the 4-F-Phe180 residue. This procedure was systematically repeated for the other phenylalanine positions to complete the majority of the assignments in the spectra of different ligation and oxidation states.
In several cases, an additional method was needed to achieve unambiguous assignment. This method utilized a nudge mutation, wherein a residue in van der Waals contact with a single Phe residue is identified in the crystal structure and then substituted with a residue of different size (Drake et al., 1993). When the chemical shift of a specific resonance was altered, this resonance was directly assigned to the nudged 4-F-Phe residue.
Together these two assignment methods allowed the unambiguous assignment of 33 l9F NMR resonances associated with different ligation and oxidation states of the 4-F-Phe labeled N36C domains. Of these resonance assignments, 28 were obtained by direct replacement, while five required nudge mutational analysis. The assignment of one resonance, 4-F-Phe140, was independently confirmed by paramagnetic broadening studies. The 4-F-Phe140 residue is the only one of the six phenylalanine positions in the crystal structure which is accessible to a solvent probe the size of Gd(III)-EDTA (4.8-Å radius; Luck & Falke, 1991c). As expected, the resonance assigned to 4-F-Phe140 yielded the largest resonance broadening upon addition of the aqueous probe Gd(III)-EDTA to either the apo- or aspartate-occupied conformations (data not shown).
Conformational Effects of Attractant Binding
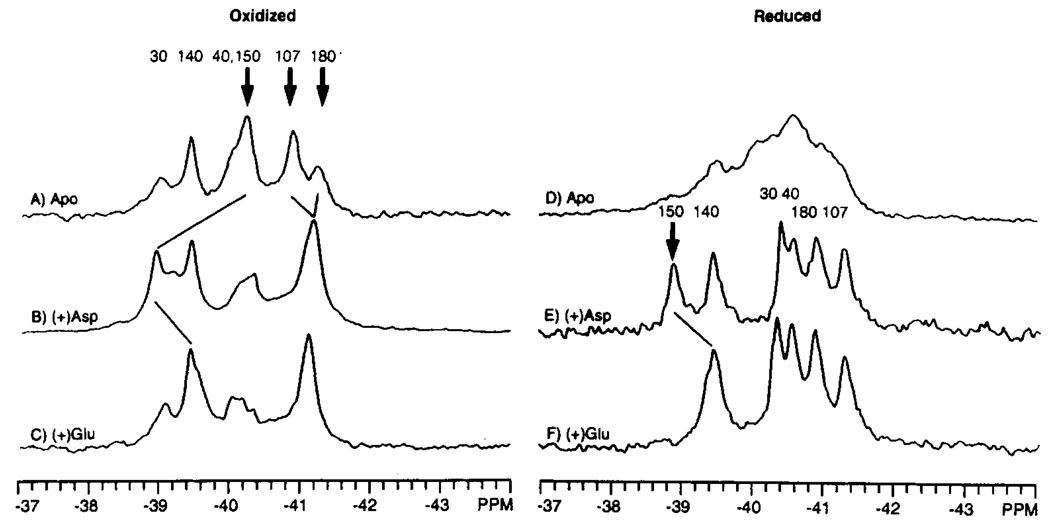
The addition of aspartate or glutamate, both of which act as attractants through the aspartate transducer (Clarke & Koshland, 1979), induces a structural change in the oxidized N36C domain which was detected by 19F NMR, as shown in Figure 4A–C. Significant attractant-induced chemical shift changes are observed in these spectra for the 4-F-Phe150, 4-F-Phe107, and 4-F-Phe180 resonances. The 4-F-Phe150 resonance exhibits different chemical shifts in the apo, aspartate-bound, and glutamate-bound states (−40.3, −39.0, and −39.5 ppm, respectively) consistent with this residue being adjacent to the attractant binding site. Most importantly, aspartate and glutamate each cause the same chemical shift change at the 4-F-Phe107 position (−0.3 ppm), as well as a small but significant change at the 4-F-Phe180 position (+0.1 to +0.2 ppm), suggesting that these attractants generate a similar long-range conformational change. Similarly, when attractant is added to the reduced N36C domain, the 4-F-Phe150 resonances of the aspartate- and glutamate-bound states differ by 0.5 ppm, but the 4-F-Phe107 and 4-F-Phe180 resonances are shifted by either attractant the same degree from the apo-state (Figure 4D–F). Together, these results suggest that aspartate and glutamate generate different local conformations at the attractant binding site but yield the same long-range conformational change detected at the 4-F-Phe107 and 4-F-Phe180 positions. Despite its long-range nature, this attractant-induced conformational change remains localized within specific regions of the molecule, since conformational changes are not detected at the 4-F-Phe30, 4-F-Phe40, and 4-F-Phe140 positions (Figure 4A–C).
FIGURE 4.
Effect of attractant ligands on the 19F NMR spectrum of the 4-F-Phe labeled ligand binding domain. Shown are the spectra of the apo (A, D), aspartate bound (B, E), and glutamate-bound (C, F) states of the oxidized and reduced N36C ligand binding domain. The bold arrows indicate which of the assigned resonances are shifted by attractant binding; the new positions of these resonances are indicated by the light diagonal lines. Sample conditions were as in Figure 3; where indicated, 5 mM aspartate or 25 mM glutamate was also present. The concentration of dimeric domain was 0.7–2 mM.
Additional evidence for a long-range attractant-induced conformational change was provided by paramagnetic broadening studies. The 4-F-Phe180 residue, which lies 45 Å away from the attractant binding site, yielded significant resonance broadening by aqueous Gd(III)-EDTA only in the attractant-occupied conformation of the oxidized N36C domain (data not shown). Thus attractant binding significantly increases the solvent exposure of this residue.
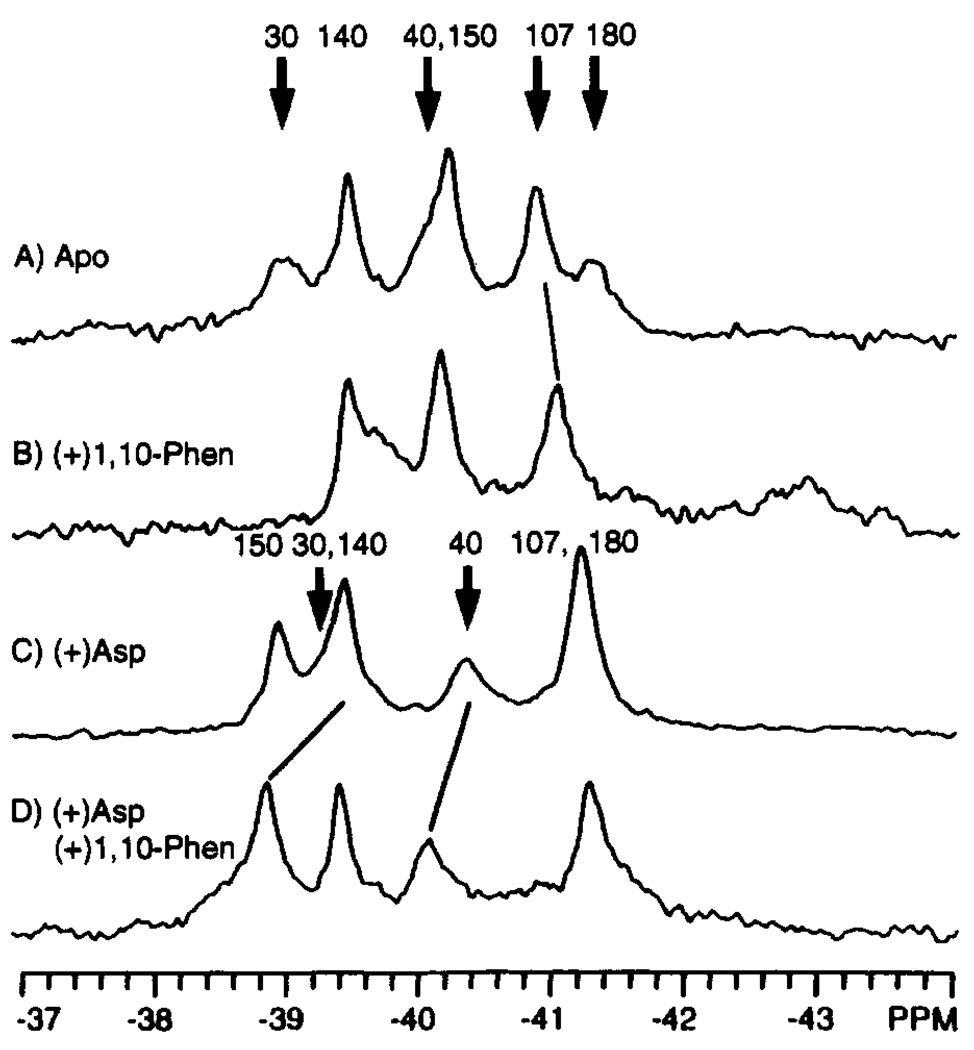
The binding of 1,10-phenanthroline to the oxidized N36C domain was also studied as a control, since this ligand is thought to be neither an attractant nor a repellent in chemotaxis. In contrast to the long-range effects of aspartate or glutamate binding, 1,10-phenanthroline binding was observed to cause a more localized structural change. The effect of 1,10-phenanthroline on the 19F NMR spectrum of the apo domain is summarized in Figure 5A,B. As anticipated, the 4-F-Phe30, 4-F-Phe40, and 4-F-Phe180 resonances, which originate from the 1,10-phenanthroline binding site observed in the crystal structure (Milburn et al., 1991), are significantly shifted by 1,10-phenanthroline binding. A minimal shift is also observed for the next closest label, namely, 4-F-Phe107 (−0.1 ppm, equal to the error level of the measurement). In contrast, the more distant 4-F-Phe140 and 4-F-Phe150 resonances show no detectable change. Similarly, when 1,10-phenanthroline is added to the aspartate-bound domain, the only resonances affected are 4-F-Phe30 and 4-F-Phe40 in the 1,10-phenanthroline binding site (Figure 5C,D). Thus the observed effects of 1,10-phenanthroline binding are confined to the vicinity of its aromatic binding pocket near the subunit interface.
FIGURE 5.
Effect of 1,10-phenanthroline on the 19F NMR spectrum of the 4-F-Phe-labeled ligand binding domain. Shown are the spectra of the apo (A), phenanthroline-bound (B), aspartate-bound (C), and aspartate and phenanthroline-bound (D) states of the oxidized N36C ligand binding domain. The bold arrows indicate the assigned resonances which are shifted by ligand binding; where known, the new positions of these resonances are indicated by the light diagonal lines. Sample parameters were as in Figure 3; also present were 3 mM 1,10-phenanthroline and 5 mM aspartate where indicated. The concentration of dimeric domain was 0.4 mM.
Stoichiometry of the Attractant-Induced Conformational Change
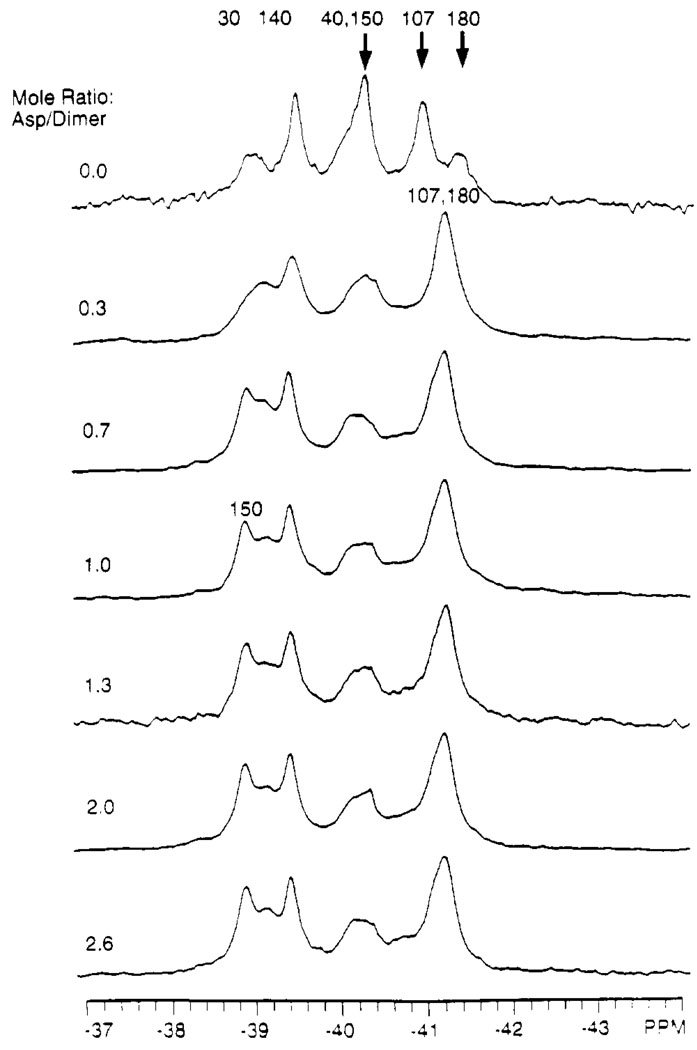
The two empty attractant-binding sites of the apo domain are virtually identical due to the C 2 symmetry axis of the homodimeric structure, but crystallographic evidence suggests that only one site is occupied by aspartate at saturation (Milburn et al., 1991; Yeh et al., 1993). Titration of the 19F NMR spectrum of 4-F-Phe labeled domain (oxidized N36C) with aspartate yields information concerning the stoichiometry of aspartate required to trigger the maximal conformational change (Figure 6). No further change in the 19F NMR spectrum is observed after the first aspartate binding site in the dimer becomes occupied. In particular, the 4-F-Phe150 resonance has shifted to a new frequency and attains its full magnitude when the mole ratio of [aspartate] / [receptor dimer] reaches 1.0 (Figure 6). It should be noted that although these results are consistent with half-of-sites binding, they do not rule out the subsequent binding of an aspartate at the second site without further conformational change.
FIGURE 6.
Aspartate titration of the 19F NMR spectrum of the 4-F-Phe-labeled ligand binding domain. The mole ratio [total aspartate]/[total oxidized N36C dimer] is indicated for each spectrum. Bold arrows highlight the resonances shifted by aspartate binding; the final position of these resonances in the aspartate-saturated state are indicated by the revised assignments. Sample parameters were as in Figure 3. The free aspartate concentration in the final sample (mole ratio = 2.6) was > 1 mM, since the total aspartate concentration (4 mM) significantly exceeded the concentration of attractant binding sites (3 mM, assuming two binding sites per dimeric domain).
Kinetics of Aspartate Binding
The kinetics of aspartate binding can be probed using the 19F NMR titration data of Figure 6 and eqs 2a–c, which describe the general relationship between the rate of interconversion of two conformations and the character of the NMR spectrum (Wagner & Wütrich, 1986):
| (2a) |
| (2b) |
| (2c) |
If two conformations which exhibit resonance frequencies νa and νb are allowed to interconvert at the rate νic, the resulting line shapes will reveal which of the indicated limits applies. In the slow interconversion limit, νic is slow relative to the frequency difference between the two conformations, thus distinct resonances are observed at νa and νb. In the intermediate interconversion limit, νic approximates the frequency difference and the resonances are significantly broadened such that they disappear. In the rapid interconversion limit, νic is rapid relative to the frequency difference, and a single average resonance is observed at a frequency that is the population-weighted mean of νa and νb.
The rate constant for aspartate binding was determined by titrating the domain with this ligand, while noting the ligand concentration that yielded the intermediate interconversion limit. At that point the rate of conformational interconversion caused by aspartate binding approaches the frequency shift generated by the aspartate-induced conformational change. The analysis focused on the 4-F-Phe1 50 resonance of the oxidized N36C domain, which exhibits a frequency of −40.2 ppm in the absence of aspartate, as illustrated in Figure 6. When aspartate is added, the structure begins to interconvert between the apo and aspartate-occupied conformations. As the aspartate concentration increases, the rate of aspartate binding increases accordingly, and the interconversion rate υic passes from the slow limit into the intermediate limit. Figure 6 reveals that this intermediate limit is reached when the [aspartate]/[receptor dimer] ratio nears 0.3, where the 4-F-Phe150 resonance is observed to disappear. Further addition of aspartate continues to increase the interconversion rate νic until it enters the rapid limit, where the 4-F-Phe150 resonance reappears at its new chemical shift of −38.9 ppm. The overall frequency shift of the 4-F-Phe150 resonance between the apo and aspartate-saturated conformations is 1.3 ppm ~ 610 Hz, corresponding to an NMR time scale of ~ 2 ms.
Using this aspartate-induced frequency shift and the parameters of the intermediate interconversion limit (eq 2b), the rate constants for aspartate binding and dissociation are estimated as follows. In the intermediate limit the interconversion rate, or the aspartate on-rate, equals the net frequency shift of the 4-F-Phe150 resonance (or, υic = k on [aspartate] ~ 610 Hz). This on-rate, together with the estimated concentration of free aspartate ([aspartate] ~ 0.4 µM) in the intermediate exchange limit, yield the pseudo-first-order rate constant for aspartate binding k on ~109 M−1 s−1. Subsequently, this on-rate constant, together with the known dissociation constant (K D = k off / k on ~ 1 µM) can be used to deduce the off-rate for aspartate dissociation, yielding k off ~ 103 s−1. (Note that no corrections are needed for population differences between the apo and aspartate-occupied conformations, since their populations are approximately equal in the intermediate limit, where the aspartate association and dissociation rates, as well as the NMR frequency difference, are all ~103 s−1.)
Kinetics of Subunit Resonance-Averaging
The 19F NMR results indicate that the structures of the two subunits rapidly sample the same range of conformations, (eq 2c), so that their time-averaged spectra are similar or identical. Thus, only six resonances are observed for the 12 4-F-Phe residues of the oxidized N36C apo-dimer (Figure 4A), and again for the aspartate-saturated dimer only six resonances are detected (Figure 4B). It follows that in both of these ligation states the two subunits are equivalent on average. In contrast, if the two subunits had different conformations which interchanged slowly or were different on average, then up to 12 4-F-Phe resonances would be observed for a given ligation state. For the apo-dimer, the C 2 symmetry axis proposed from the crystal structure may account for the observed equivalence of the two subunits (Milburn et al., 1991). However, for the dimer containing one molecule of bound aspartate, the observed equivalence implies that the aspartate occupancy of the two binding sites must alternate rapidly. Since the frequency differences between the instantaneous resonances of the two subunits in the dimer cannot be measured, the time scale of the observed resonance-averaging cannot be calculated directly. Instead these dynamics can be inferred from the deduced aspartate dissociation rate k off ~ 103 s−1, which guarantees that the aspartate occupancy of the two sites will randomly alternate on the millisecond time scale.
Effects of the Cys36–Cys36′ Intersubunit Disulfide Bond
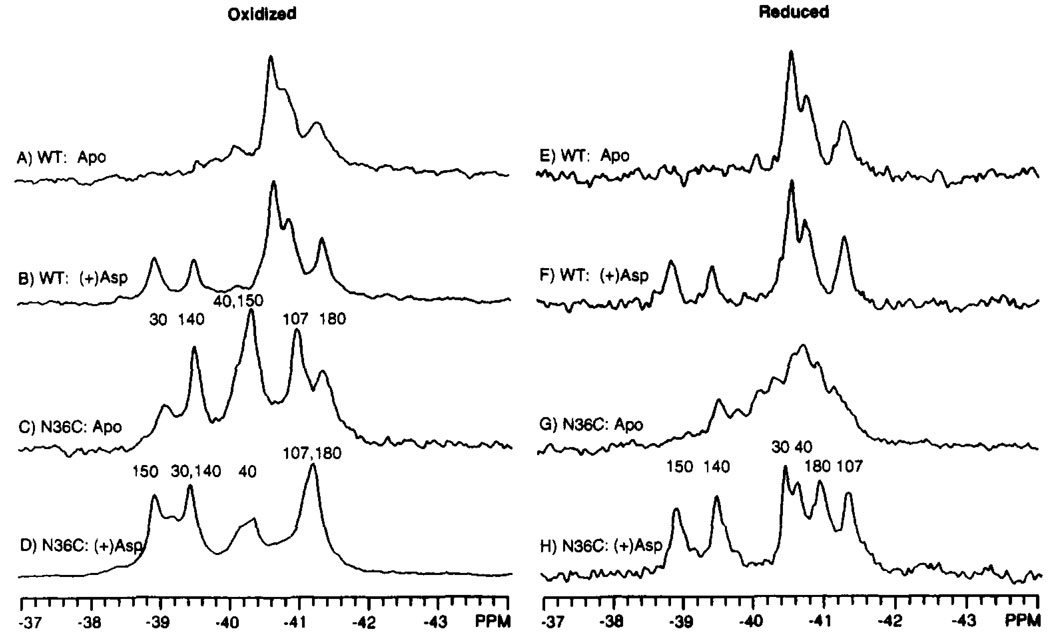
Shown in Figure 7 are the spectra of the wild-type domain lacking the Cys36–Cys36′ disulfide, as well as the reduced and oxidized N36C domains, each in the presence and absence of aspartate. The Cys36–Cys36′ disulfide was observed to be formed upon isolation and was reduced by overnight incubation in the presence of 50 mM DTT. As indicated in Figure 7, DTT had no effect on the structure of the wild-type ligand binding domain, indicating that spectral changes caused by DTT in the N36C domain stemmed specifically from reduction of the disulfide.
FIGURE 7.
Effect of the Cys36–Cys36′ disulfide on the 19F NMR spectrum of the 4-F-Phe-labeled ligand binding domain. Shown are the spectra of domains under oxidizing (A–D) or reducing (E–H, 50 mM DTT) conditions for both the wild-type and N36C mutant domains. The known assignments are indicated. Sample parameters were as in Figure 3; also present was 5 mM aspartate, where indicated. The concentration of dimeric domain was 0.4–1.0 mM.
When the Cys36–Cys36′ disulfide is absent, either in the wild-type domain or the reduced N36C domain, the 19F NMR spectrum exhibited poorly resolved or missing resonances (Figure 7 A,E,G), indicating a heterogeneous or dynamic structure. Addition of aspartate or formation of the Cys36–Cys36′ disulfide serves to stabilize the structure, as demonstrated by the dramatic increase in the resolution of the six resonances (Figure 7B–D,F,H).
In order to further probe the structural effect of the disulfide, the resonances of the aspartate-occupied N36C domain were assigned in the reduced and oxidized conformations, as illustrated in Figure 7D,H. Three resonances, 4-F-Phe30, 4-F-Phe40, and 4-F-Phe180, are significantly shifted (−1.3, −0.4 and +0.3 ppm, respectively) by formation of the Cys36–Cys36′ disulfide. In the crystal structure, each of these three residues lie within 7 Å of the Cys36 position (Figure 1 and Table 1). A small shift (−0.1 to −0.2 ppm) is also observed for the 4-F-Phe107 resonance at a distance of 25 Å from Cys 36. The remaining 4-F-Phe140 and 4-F-Phe150 resonances, originating from residues >50 Å from Cys 36, are unaffected by disulfide formation. Thus the structural changes associated with Cys36 disulfide formation do not extend to the aspartate binding region of the molecule.
DISCUSSION
The current 19F NMR titration data reveal that the binding of one aspartate molecule per homodimer triggers the maximum conformational change in the isolated ligand binding domain. This is consistent with the existence of negative cooperativity between the two attractant binding sites (Milligan & Koshland, 1993; Biemann & Koshland, 1994). Positive cooperativity and noncooperative binding are ruled out, since each would require a higher aspartate binding stoichiometry to generate the maximal conformational change. The observed negative cooperativity requires that the binding of aspartate destroys the C 2 symmetry of the ligand binding domain. Such an observation leads to a model in which the binding of an attractant molecule to the first site generates an allosteric conformational change which repositions residues in the second site such that the ligand affinity of that site is reduced or abolished. It should be noted that aspartate binds to either site in the apo-domain, since the 19F NMR resonances of the two subunits in the dimer are identical, indicating that aspartate binding alternates rapidly between the two sites on the NMR time scale to yield resonance averaging. In contrast, if one of the two sites in the apo-domain was permanently incapable of binding aspartate, the 19F NMR resonances from different subunits would be twinned at saturating aspartate, since the two subunits would be permanently inequivalent.
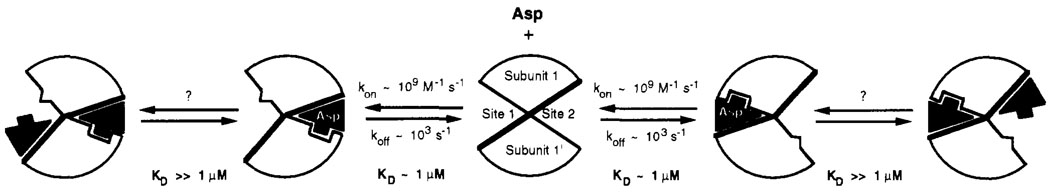
The 19F NMR results demonstrate that the conformational activation of the ligand binding domain is extremely rapid on the time scale of the chemotaxis response, which for attractant is ~0.2 s (Segall et al., 1982; Khan et al., 1992). For the isolated ligand binding domain, the estimated on-rate constant for aspartate is k on ~ 109 s−1 M−1. This on-rate constant is 1–2 orders of magnitude slower than diffusion-controlled binding (Fersht, 1985); thus binding is somewhat slowed by steric factors or the accompanying conformational change. Yet by the standards of protein binding sites the attractant site is highly accessible, since the observed on-rate constant is comparable to that observed for enzymes such as malate dehydrogenase, lactate dehydrogenase, and chymotrypsin (Czerlinski & Schreck, 1964a,b; Quast et al., 1974). Using the on-rate constant for aspartate binding and the known aspartate dissociation constant, the off-rate constant is estimated to be k off ~ 103s−1. In principle, the conformational changes associated with attractant binding and release could be slower in the intact transmembrane receptor where motions in the transmembrane and cytoplasmic domains are coupled to attractant binding. However, the aspartate K D of the ligand binding domain does not dramatically change when it is severed from the rest of the protein, indicating that k on and k off are unaltered in the related domain or that they change by the same factor. It seems likely that even in the intact receptor, attractant binding, dissociation, and the coupled conformational changes will also be rapid and will not contribute significantly to the chemotactic response time. Further evidence supporting the proposal that the intact receptor is a highly dynamic molecule is provided by its extensive flexibility, as observed in disulfide-trapping studies of thermal backbone motions (Falke & Koshland, 1987). A kinetic model for aspartate binding is presented in Figure 8.
FIGURE 8.
Schematic model of the structural and kinetic aspects of aspartate binding. In the absence of bound aspartate, the ligand binding domain has, on average, a symmetrical structure possessing a C2 axis lying between the two identical subunits (center). The binding of the first aspartate molecule can occur at either of the two equivalent attractant binding sites, with the indicated association and dissociation rate constants. This binding event causes a conformational change within at least one subunit and destroys the symmetry of the dimer. The conformational change is communicated to the empty site, where the structure is altered such that the affinity for aspartate is significantly reduced (negative cooperativity). The binding of a second aspartate to this empty site was neither detected nor excluded by the present study; however, if it occurs it generates no detectable conformational change within the protein at the positions monitored by 19F NMR.
Complementing this kinetic scheme, the 19F NMR data reveal important spatial features of attractant-induced activation. Attractant binding generates significant changes in the chemical shifts of the resonances 4-F-Phe150, 4-F-Phe107, and 4-F-Phe180; in addition, the solvent exposure of the latter resonance changes. These long-range conformational effects are observed at distances 7, 26, and 40 Å from the bound attractant, respectively, and are specific for attractants; for example, the binding of the control molecule 1,10-phenan-throline or the formation of the Cys36–Cys36′ disulfide yield more localized conformational changes. Moreover, since 4-F-Phe107 lies buried within the core of a single subunit at least 10 Å from the subunit interface, there must be an intrasubunit component of the attractant-triggered conformational change. This intrasubunit component is observed both in the presence and absence of the Cys36–Cys36′ disulfide bond: Figure 7 panels C,D and G,H show that the 4-F-Phe107 resonance is shifted by aspartate both in the oxidized and reduced forms of the N36C domain. Independent evidence for an intrasubunit conformational change has been provided by studies of the membrane-bound receptor: when the transmembrane and cytoplasmic domains of one subunit in the dimer are removed, the remaining intact subunit can still generate a transmembrane signal, albeit reduced in magnitude (Milligan & Koshland, 1991). Crystallographic evidence has produced a model in which the subunits are rigid and simply rotate relative to one another upon attractant binding (Milburn et al., 1991). However, such results do not rule out a conformational change within the individual subunits, since the intrasubunit rearrangements could be too subtle to detect at the currently available crystallographic resolution or could be suppressed by crystal packing forces.
The attractant-induced conformational change detected by 19F NMR appears to targetted toward the C-terminus, rather than the N-terminus, of the ligand binding domain. The environmental changes detected by the 4-F-Phe180 probe originate eight residues from the C-terminus. In contrast, no detectable environmental perturbations are revealed by the 4-F-Phe30 and 4-F-Phe40 probes residing five and 15 residues from the N-terminus, respectively. In the intact receptor, the C-terminal region of the ligand binding domain extends into the bilayer as the second transmembrane helix (TM2, residues 189–212) and ultimately joins the signaling domain in the cytoplasm. Thus the NMR results are consistent with a transmembrane signaling model in which attractant binding triggers a change in the position or orientation of TM2, thereby communicating directly with the signaling domain on the distal side of the bilayer (Lynch & Koshland, 1991).
Finally, the available evidence suggests that the Cys36–Cys36′ disulfide bond stabilizes a structural interaction which anchors together helices α1 and α1′ in the native receptor structure. In this picture α1 and α1′, which continue through the bilayer as the transmembrane helices TM1 and TM1′, remain fixed during the transmembrane signal while TM2 and TM2′ move relative to them. Such a model explains the ability of the receptor containing a disulfide bond between α1 and α1′ to generate a transmembrane signal (Falke et al., 1988; Lynch & Koshland, 1991; Chervitz and Falke, unpublished results). When the isolated ligand binding domain is generated by removal of the transmembrane helices, the model proposes that unless the disulfide is present, the interactions stabilizing the dimer will be weakened due to the shortening of a 1 and α1′. Consistent with this picture are the observations that (a) the isolated domain possesses an increased propensity to dissociate into subunits (Milligan & Koshland, 1993) and (b) incorporation of the Cys36–Cys36′ disulfide into the isolated domain improves the contact between helices α1 and αl′ in the crystal structure (Yeh et al., 1993). It follows that the enhanced structural stabilization observed in 19F NMR spectra when the disulfide is formed may well stem from restoration of native interactions between helices α1 and αl′.
ACKNOWLEDGMENT
We thank Drs. Sung-Hou Kim and Sandy Parkinson for helpful discussions.
Footnotes
Support was provided by NIH Grants GM40731 (to J.J.F.) and DK09765 (to D.E.K.).
Abbreviations: 4-F-Phe, para-fluorophenylalanine; EDTA, (ethylenedinitri1o)tetraacetic acid; Tris, tris(hydroxymethy1)aminomethane; DTT, dithiothreitol; PMSF, phenylmethanesulfonylfluoride; BCA, bicinchonic acid; SDS, sodium dodecyl sulfate.
REFERENCES
- Adler J. Science. 1969;166:1588–1597. doi: 10.1126/science.166.3913.1588. [DOI] [PubMed] [Google Scholar]
- Ames P, Chen J, Wolff C, Parkinson JS. Cold Spring Harbor Symp. Quant. Biol. 1988;53:59–65. doi: 10.1101/sqb.1988.053.01.010. [DOI] [PubMed] [Google Scholar]
- Armitage JP. Annu. Rev. Physiol. 1992;54:683–714. doi: 10.1146/annurev.ph.54.030192.003343. [DOI] [PubMed] [Google Scholar]
- Augspurger J, Pearson J, Oldfield E, Dykstra CE, Park KD, Schwarz D. J. Magn. Reson. 1992;100:342–357. [Google Scholar]
- Biemann H-P, Koshland DE., Jr Biochemistry. 1994;33:629–634. doi: 10.1021/bi00169a002. [DOI] [PubMed] [Google Scholar]
- Bourret RB, Borkovich KA, Simon MI. Annu. Rev. Biochem. 1991;60:401–441. doi: 10.1146/annurev.bi.60.070191.002153. [DOI] [PubMed] [Google Scholar]
- Bourret RB, Drake SK, Chervitz SA, Simon MI, Falke JJ. J. Biol. Chem. 1993;268:13089–13096. [PMC free article] [PubMed] [Google Scholar]
- Boyd A, Kendall K, Simon MI. Nature. 1983;301:623–626. doi: 10.1038/301623a0. [DOI] [PubMed] [Google Scholar]
- Chambers SE, Lau EY, Gerig JT. J. Am. Chem. Soc. 1994 (in press) [Google Scholar]
- Chang C, Kwok SF, Bleecker AB, Meyerowitz EM. Science. 1993;262:539–544. doi: 10.1126/science.8211181. [DOI] [PubMed] [Google Scholar]
- Clarke S, Koshland DE., Jr J. Bacteriol. 1979;254:9695–9702. [PubMed] [Google Scholar]
- Collins LA, Egan SM, Stewart V. J. Bacteriol. 1992;174:3667–3675. doi: 10.1128/jb.174.11.3667-3675.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czerlinski GH, Schreck G. Biochemistry. 1964a;3:89–100. doi: 10.1021/bi00889a015. [DOI] [PubMed] [Google Scholar]
- Czerlinski GH, Schreck G. J. Biol. Chem. 1964b;239:913. [PubMed] [Google Scholar]
- Dahl MK, Simon MI. J. Bacteriol. 1989;171:2361–2371. doi: 10.1128/jb.171.5.2361-2371.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- deDios AC, Pearson JG, Oldfield E. Science. 1993;260:1491–1496. doi: 10.1126/science.8502992. [DOI] [PubMed] [Google Scholar]
- Drake SK, Bourret RB, Luck LA, Simon MI, Falke JJ. J. Biol. Chem. 1993;268:13081–13088. [PMC free article] [PubMed] [Google Scholar]
- Falke JJ, Koshland DE., Jr Science. 1987;273:1596–1600. doi: 10.1126/science.2820061. [DOI] [PubMed] [Google Scholar]
- Falke JJ, Dernburg AF, Sternberg DA, Zalkin N, Milligan DL, Koshland DE., Jr J. Biol. Chem. 1988;263:14850–14858. [PubMed] [Google Scholar]
- Falke JJ, Luck LA, Sherrer J. Biophys. J. 1992;62:3495–3499. doi: 10.1016/S0006-3495(92)81787-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fersht A. Enzyme Structure and Mechanism. New York: W. H. Freeman & Co.; 1985. [Google Scholar]
- Gammon KL, Smallcombe SH, Richards JH. J. Am. Chem. Soc. 1972;94:4573–4580. doi: 10.1021/ja00768a027. [DOI] [PubMed] [Google Scholar]
- Gerig JT. Methods Enzymol. 1989;177:3–23. doi: 10.1016/0076-6879(89)77003-8. [DOI] [PubMed] [Google Scholar]
- Gerig JT. Prog. Nucl. Magn. Reson. 1994 (in press) [Google Scholar]
- Gill SC, von Hippel PH. Anal. Biochem. 1989;182:319–326. doi: 10.1016/0003-2697(89)90602-7. [DOI] [PubMed] [Google Scholar]
- Gregory DH, Gerig JT. J. Comput. Chem. 1991;12:180–185. [Google Scholar]
- Hazelbauer GL, Yaghmai R, Burrows GG, Baumgartner JW, Dutton DP, Morgan DG. In: Biology of the Chemotactic Response. Armitage JB, Lackie JM, editors. Cambridge University Press; 1990. pp. 107–134. [Google Scholar]
- Hazelbauer GL, Berg HC, Matsumura P. Cell. 1993;73:15–22. doi: 10.1016/0092-8674(93)90156-k. [DOI] [PubMed] [Google Scholar]
- Imae Y, Oosawa K, Mizund T, Kihara M, Macnab RM. J. Bacteriol. 1987;169:371–379. doi: 10.1128/jb.169.1.371-379.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson D, Lanahan A, Buck CR, Sehgal A, Morgan C, Mercer E, Bothwell M, Chao M. Cell. 1986;47:545–554. doi: 10.1016/0092-8674(86)90619-7. [DOI] [PubMed] [Google Scholar]
- Khan S, Amoyaw K, Spudich JL, Reid GP, Trantham DR. Biophys. J. 1992;62:67–68. doi: 10.1016/S0006-3495(92)81781-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HW, Perez JA, Ferguson SJ, Campbell ID. FEBS. 1990;272:34–36. doi: 10.1016/0014-5793(90)80442-l. [DOI] [PubMed] [Google Scholar]
- Kunkel TA. Methods Enzymol. 1987;154:367. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- Long DG, Weis RM. Biochemistry. 1992;31:9904–9911. doi: 10.1021/bi00156a007. [DOI] [PubMed] [Google Scholar]
- Lu P, Jarema MC, Mosar K, Daniel WE. Proc. Natl. Acad. Sei. U.S.A. 1976;73:3471–3475. doi: 10.1073/pnas.73.10.3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luck LA, Falke JJ. Biochemistry. 1991a;30:4248–4256. doi: 10.1021/bi00231a021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luck LA, Falke JJ. Biochemistry. 1991b;30:4257–4261. doi: 10.1021/bi00231a022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luck LA, Falke JJ. Biochemistry. 1991c;30:6484–6490. doi: 10.1021/bi00240a019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch BA, Koshland DE., Jr Proc. Natl. Acad. Sci. U.S.A. 1991;88:10402–10406. doi: 10.1073/pnas.88.23.10402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBride MJ, Köhler T, Zusman DR. J. Bacteriol. 1992;174:4246–4257. doi: 10.1128/jb.174.13.4246-4257.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milburn MV, Prive GG, Milligan DL, Scott WG, Yeh J, Jancarik J, Koshland DE, Jr, Kim S-H. Science. 1991;254:1342–1347. doi: 10.1126/science.1660187. [DOI] [PubMed] [Google Scholar]
- Milligan DL, Koshland DE., Jr J. Biol. Chem. 1988;263:6268–6275. [PubMed] [Google Scholar]
- Milligan DL, Koshland DE., Jr Science. 1991;254:1651–1654. doi: 10.1126/science.1661030. [DOI] [PubMed] [Google Scholar]
- Milligan DL, Koshland DE., Jr J. Biol. Chem. 1993;268:19991–19997. [PubMed] [Google Scholar]
- Moe GR, Bollag E, Koshland DE., Jr Proc. Natl. Acad. Sci. U.S.A. 1989;86:5683–5687. doi: 10.1073/pnas.86.15.5683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowlin DM, Nettleton DO, Ordal GW, Hazelbauer GL. J. Bacteriol. 1985;163:262–266. doi: 10.1128/jb.163.1.262-266.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ota IM, Varshavsky A. Science. 1993;262:566–569. doi: 10.1126/science.8211183. [DOI] [PubMed] [Google Scholar]
- Pauling L. The Nature of the Chemical Bond and the Structure of Molecules and Crystals. Ithaca, NY: Cornell University Press; 1960. [Google Scholar]
- Pakula AA, Simon MI. Proc. Natl. Acad. Sci. U.S.A. 1992;89:4144–4148. doi: 10.1073/pnas.89.9.4144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkinson JS, Kofoid EC. Annu. Rev. Genet. 1992;26:71–112. doi: 10.1146/annurev.ge.26.120192.000443. [DOI] [PubMed] [Google Scholar]
- Peersen OB, Pratt EA, Truong HTN, Ho C, Rule GS. Biochemistry. 1990;29:3256–3262. doi: 10.1021/bi00465a017. [DOI] [PubMed] [Google Scholar]
- Post JFM, Cottam PF, Simplaceau V, Ho C. J. Mol. Biol. 1984;179:729–743. doi: 10.1016/0022-2836(84)90164-5. [DOI] [PubMed] [Google Scholar]
- Pratt EA, Ho C. Biochemistry. 1975;14:3035–3040. doi: 10.1021/bi00684a037. [DOI] [PubMed] [Google Scholar]
- Quast U, Engel J, Heumann H, Krause G, Steffen E. Biochemistry. 1974;13:2512. doi: 10.1021/bi00709a600. [DOI] [PubMed] [Google Scholar]
- Richards FM. Annu. Rev. Biophys. Bioeng. 1977;6:151–176. doi: 10.1146/annurev.bb.06.060177.001055. [DOI] [PubMed] [Google Scholar]
- Rule GS, Pratt EA, Simplaceau V, Ho C. Biochemistry. 1987;26:549–556. doi: 10.1021/bi00376a029. [DOI] [PubMed] [Google Scholar]
- Russo AF, Koshland DE., Jr Science. 1983;220:1016–1020. doi: 10.1126/science.6302843. [DOI] [PubMed] [Google Scholar]
- Scott WG, Milligan DL, Milburn MV, Prive GG, Yeh J, Koshland DE, Jr, Kim S-H. J. Mol. Biol. 1993;232:555–573. doi: 10.1006/jmbi.1993.1411. [DOI] [PubMed] [Google Scholar]
- Segall JE, Manson MD, Berg HC. Nature. 1982;296:855–857. doi: 10.1038/296855a0. [DOI] [PubMed] [Google Scholar]
- Shaw CH. BioEssays. 1991;13:25–29. doi: 10.1002/bies.950130105. [DOI] [PubMed] [Google Scholar]
- Stock JB, Surette MG, McCleary WR, Stock AM. J. Biol. Chem. 1992;267:19753–17956. [PubMed] [Google Scholar]
- Stoscheck CM. Methods Enzymol. 1990;182:50–69. doi: 10.1016/0076-6879(90)82008-p. [DOI] [PubMed] [Google Scholar]
- Sykes BD, Wingarten HJ, Schlesinger M. Proc. Natl. Acad. Sci. U.S.A. 1974;71:469–473. doi: 10.1073/pnas.71.2.469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullrich AL, Coussens L, Hayflick JS, Dull TJ, Gray A, Tam AW, Yarden Y, Liberman TA, Schlessinger J, Downward J, Mayes ELV, Whittle N, Waterfield MD, Seeburg PH. Nature. 1984;309:418–425. doi: 10.1038/309418a0. [DOI] [PubMed] [Google Scholar]
- Ullrich A, Bell JR, Chen EY, Herrera R, Petruzzelli LM, Dull TJ, Gray A, Coussens L, Liao YC, Tsubokawa M, Mason A, Seeburg PH, Grunfeld C, Rosen OM, Ramachandran JR. Nature. 1985;313:418–425. doi: 10.1038/313756a0. [DOI] [PubMed] [Google Scholar]
- Utsumi R, Brissette RE, Rampersaud A, Forst SA, Oosawa K, Inouye M. Science. 1989;246:1246–1249. doi: 10.1126/science.2476847. [DOI] [PubMed] [Google Scholar]
- Wagner G, Wütrich K. Methods Enzymol. 1986;131:307–326. doi: 10.1016/0076-6879(86)31047-4. [DOI] [PubMed] [Google Scholar]
- Wilson ML, Dahlquist FW. Biochemistry. 1985;24:1920–1928. doi: 10.1021/bi00329a018. [DOI] [PubMed] [Google Scholar]
- Yaghmai R, Hazelbauer GL. Proc. Natl. Acad. Sci. U.S.A. 1992;89:7890–7894. doi: 10.1073/pnas.89.17.7890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh JI, Biemann HP, Pandit J, Koshland DE, Jr, Kim S-H. J. Biol. Chem. 1993;268:9787–9792. [PubMed] [Google Scholar]