Abstract
Tyrosine phosphorylation is important in signaling pathways underlying tumorigenesis. A mutational analysis of the Protein Tyrosine Kinase (PTK) gene family in cutaneous metastatic melanoma identified 30 somatic mutations in the kinase domain of 19 PTKs. The whole of the coding region of these 19 PTKs was further evaluated for somatic mutations in a total of 79 melanoma samples. This analysis revealed novel ERBB4 mutations in 19% of melanoma patients and that an additional two kinases (FLT1 and PTK2B) are mutated in 10% of melanomas. Seven missense mutations in the most commonly altered PTK (ERBB4) were examined and found to increase kinase activity and transformation ability. Melanoma cells expressing mutant ERBB4 had reduced cell growth after shRNA–mediated knockdown of ERBB4 or treatment with the ERBB inhibitor lapatinib. These studies might lead to personalized therapeutics specifically targeting the kinases that are mutationally altered in individual melanomas.
Malignant melanoma is the most fatal skin cancer 1,2. To develop personalized treatments for advanced disease, it is important to identify genetic alterations leading to melanoma. Protein tyrosine kinases (PTKs) are frequently mutated in cancer (http://www.sanger.ac.uk/genetics/CGP/Census/), and since they are amenable to pharmacologic inhibition 3,4, further analysis of the PTK gene family may identify new therapeutic strategies. In this study, we used high-throughput gene sequencing to analyze the entire PTK gene family in melanoma, and have identified many novel somatic alterations.
We initially sequenced the coding exons comprising the kinase domains of all 86 members of this gene superfamily in 29 melanomas (Supplementary Table 1). A total of 593 exons were extracted from genomic databases and amplified by polymerase chain reaction (PCR) from cancer genomic DNA samples using specific primers (Supplementary Table 2) and directly sequenced with dye-terminator chemistry. We determined whether a mutation was somatic (i.e., tumor-specific) by examining the sequence of the gene in genomic DNA from normal tissue of the relevant patient. From the ~12 Mb of sequence information obtained, we identified 19 genes containing a total of 30 somatic mutations within their kinase domains. All coding exons of these 19 genes were then analyzed for mutations in a total of 79 melanoma samples using specific primers (Supplementary Table 3).
We identified 99 non-synonymous, somatic mutations in 19 PTK genes (Table 1 and examples in Supplementary Figure 1 and 3). Only three genes (EPHA6, PDGFRA and PTK2) out of the 19 have previously been reported to be mutated in melanoma (http://www.sanger.ac.uk/genetics/CGP/Census/). The clinical information associated with the melanoma tumors containing somatic PTK mutations is provided in Supplementary Table 4.
Table 1.
Somatic mutations Identified in PTK-s
| Gene | CCDS accession* | Ref Seq accession* | No. of mutations# | % of cases affected# | Exon | Nucleotide† | Amino Acid† | Functional Domain | Tumor | NRAS/BRAF mutation** |
|---|---|---|---|---|---|---|---|---|---|---|
| DDR1 | CCDS4690.1 | NM_001954.3 | 2 | 2.5 | 8 | C1115T | S372F | None | 6T | BRAF |
| 11 | G1709A | R570Q | Protein Tyrosine Kinase | 43T | BRAF | |||||
| FER | CCDS4098.1 | NM_005246.1 | 2 | 2.5 | 11 | T1594C | Y532H | SH2 Motif | 58T | BRAF |
| 13 | G1739A | G580D | Protein Tyrosine Kinase | 30T | BRAF | |||||
| FLT1 | CCDS9330.1 | NM_002019.3 | 8 | 10.1 | 7 | G842A | R281Q | IG | 37T | BRAF |
| 7 | C860T | S287F | IG | 7T | NRAS | |||||
| 12 | −9 Intronic C>A | Splice Site | N/A | 20T | BRAF | |||||
| 13 | G1767A | W589X | IGc2 | 13T | None | |||||
| 17 | C2440T | P814S | None | 39T | None | |||||
| 21 | G2827A | E943K | Protein Tyrosine Kinase | 44T | NRAS | |||||
| 24 | G3241A | D1081N | Protein Tyrosine Kinase | 78T | BRAF | |||||
| 28 | G3667A | E1223K/LOH | None | 85T | BRAF | |||||
| EPHA6 | NM_001080448.2 | 5 | 6.3 | 1 | C1202G | T307S | None | 30T | BRAF | |
| 4 | G1763T | R494M | Protein Tyrosine Kinase | 36T | BRAF | |||||
| 4 | G1891A | E537K | Protein Tyrosine Kinase | 32T | BRAF | |||||
| 8 | A2246T | K655I | Protein Tyrosine Kinase | 29T | BRAF | |||||
| 8 | G2320A | E680K | None | 21T | BRAF | |||||
| EPHA10 | CCDS41305.1 | NM_001099439.1 | 7 | 6.3 | 3 | G235A | V79M | Ephrin Receptor | 52T | BRAF |
| 3 | T236C | V79A | Ephrin Receptor | 52T | BRAF | |||||
| 3 | G370A | E124K | Ephrin Receptor | 55T | None | |||||
| 3 | G649A | G217S | None | 71T | BRAF | |||||
| 3 | G650A | G217D | None | 71T | BRAF | |||||
| 13 | G2369A | G790E | Protein Tyrosine Kinase | 63T | NRAS | |||||
| 14 | G2528C | G843A | Protein Tyrosine Kinase | 37T | BRAF | |||||
| EPHB1 | NM_004441 | 4 | 5.1 | 3 | C235T | R79W | Ephrin Receptor | 39T | None | |
| 12 | G2311A | D771N | Protein Tyrosine Kinase | 60T | NRAS | |||||
| 13 | G2432A | G811E | Protein Tyrosine Kinase | 44T | NRAS | |||||
| 15 | G2757A | W919X | Sterile Alpha Motif | 63T | NRAS | |||||
| EPHB2 | CCDS229.2 | NM_017449.1 | 7 | 8.9 | 3 | G325A | E109K | Ephrin Receptor | 4T | BRAF |
| 3 | C614T | A205V | None | 72T | None | |||||
| 4 | G952A | D318N | Fibronectin Type 3 Domain | 71T | BRAF | |||||
| 7 | C1535T | T512I | Fibronectin Type 3 Domain | 83T | Both | |||||
| 10 | G1846A | E615K | Protein Tyrosine Kinase | 29T | BRAF | |||||
| 10 | G1846A | E615K | Protein Tyrosine Kinase | 68T | BRAF | |||||
| 14 | C2663T | P887L | None | 77T | None | |||||
| EPHB6 | CCDS5873.1 | NM_004445.1 | 7 | 8.9 | 3 | C392T | S131F | Ephrin Receptor | 60T | NRAS |
| 3 | C455T | S152F | Ephrin Receptor | 55T | None | |||||
| 5 | G1210A | G404S | Fibronectin Type 3 Domain | 50T | BRAF | |||||
| 11 | G2036A | R679Q | Protein Tyrosine Kinase | 5T | BRAF | |||||
| 11 | C2063G | A688G | Protein Tyrosine Kinase | 54T | BRAF | |||||
| 11 | C2110T | R704W | Protein Tyrosine Kinase | 26T | BRAF | |||||
| 13 | −5 Intronic C>T | Splice Site | N/A | 18T | None | |||||
| ERBB4 | CCDS2394.1 | NM_005235.2 | 24 | 19.0 | 2 | C113T | L39F | Receptor L Domain | 71T | BRAF |
| 3 | T331C | Y111H | Receptor L Domain | 13T | None | |||||
| 8 | G939A | M313I | Growth Factor Receptor | 63T | NRAS | |||||
| 8 | G949A | E317K | Growth Factor Receptor | 17T | NRAS | |||||
| 9 | C1022T | S341L | Receptor L Domain | 96T | None | |||||
| 10 | C1177T | R393W | Receptor L Domain | 49T | BRAF | |||||
| 11 | C1226T | P409L | Receptor L Domain | 76T | None | |||||
| 12 | G1354A | E452K | Receptor L Domain | 7T | NRAS | |||||
| 12 | G1354A | E452K/LOH | Receptor L Domain | 55T | None | |||||
| 12 | G1472A | R491K/LOH | Growth Factor Receptor | 34T | BRAF | |||||
| 14 | G1624A | E542K | Growth Factor Receptor | 63T | NRAS | |||||
| 14 | C1630T | R544W | Growth Factor Receptor | 56T | BRAF | |||||
| 14 | G1687A | E563K | Growth Factor Receptor | 12T | NRAS | |||||
| 15 | −10 Intronic C>T | Splice Site/LOH | N/A | 68T | BRAF | |||||
| 15 | G1825A | D609N | Growth Factor Receptor | 76T | None | |||||
| 18 | C2098T | P700S | None | 24T | NRAS | |||||
| 21 | G2506A | E836K | Protein Tyrosine Kinase | 86T | BRAF | |||||
| Protein Tyrosine | ||||||||||
| 21 | G2614A | E872K | Kinase/Activation Loop | 63T | NRAS | |||||
| 23 | G2806A | G936R | Protein Tyrosine Kinase | 24T | NRAS | |||||
| 24 | −4 Intronic C>T | Splice Site | N/A | 13T | None | |||||
| 25 | C3097T | P1033S | None | 76T | None | |||||
| 26 | −1 Intronic G>A | Splice Site | N/A | 76T | None | |||||
| 28 | G3521A | R1174Q | None | 63T | NRAS | |||||
| 28 | G3737A | S1246N | His-Me Finger Endonucleases | 71T | BRAF | |||||
| MATK | CCDS12113.1 | NM_002378.2 | 1 | 1.3 | 12 | G1248A | W416X | Protein Tyrosine Kinase | 13T | None |
| MET | CCDS43636.1 | NM_000245 | 3 | 3.8 | 5 | G1829A | C610Y/LOH | IPT | 1T | BRAF |
| 14 | A3176G | N1059S | None | 13T | None | |||||
| 16 | G3509A | R1170Q | Protein Tyrosine Kinase | 29T | BRAF | |||||
| NTRK1 | CCDS1161.1 | NM_002529.2 | 2 | 2.5 | 8 | G1137A | M349I | None | 18T | None |
| 14 | C1747G | R547G | Protein Tyrosine Kinase | 13T | None | |||||
| PDGFRA | CCDS3495.1 | NM_006206.2 | 5 | 5.1 | 3 | G571A | A191T | IG | 64T | BRAF |
| 9 | G1375A | E459K/LOH | None | 32T | BRAF | |||||
| 18 | C2669T | S890F | Protein Tyrosine Kinase | 41T | BRAF | |||||
| 20 | C2810T | P937L/LOH | Protein Tyrosine Kinase | 32T | BRAF | |||||
| 21 | G3070A | D1024N | None | 63T | NRAS | |||||
| PTK2 | CCDS6381.1 | NM_153831.2 | 1 | 1.3 | 15 | C1481T | A494V | Protein Tyrosine Kinase | 13T | None |
| PTK2B | CCDS6057.1 | NM_173176.1 | 8 | 10.1 | 5 | −4 Intronic C>T | Splice Site | N/A | 79T | BRAF |
| 8 | G818A | W273X | FERM | 76T | None | |||||
| 13 | G1241A | G414E | None | 95T | NRAS | |||||
| 14 | C1285T | R429C | Protein Tyrosine Kinase | 17T | NRAS | |||||
| 16 | G1480A | E494K | Protein Tyrosine Kinase | 26T | BRAF | |||||
| 24 | G2374A | E792K | None | 36T | BRAF | |||||
| 29 | G2753A | R918Q | Focal AT | 85T | BRAF | |||||
| 29 | G2812A | E938K | Focal AT | 83T | Both | |||||
| PTK6 | CCDS13524.1 | NM_005975.2 | 2 | 2.5 | 4 | G629A | W210X | Protein Tyrosine Kinase | 12T | NRAS |
| 5 | −7 Intronic C>T | Splice Site | N/A | 51T | BRAF | |||||
| PTK7 | CCDS4884.1 | NM_002821.3 | 1 | 1.3 | 7 | C1054T | P352S | IGc2 | 84T | BRAF |
| Frizzled Cysteine-Rich | ||||||||||
| ROR2 | CCDS6691.1 | NM_004560.2 | 4 | 5.1 | 5 | T574C | Y192H | Domain | 71T | BRAF |
| 7 | T1172C | V391A | Kringle | 72T | None | |||||
| 9 | C1670T | S557L | Protein Tyrosine Kinase | 5T | BRAF | |||||
| 9 | G2377T | A793S | None | 81T | BRAF | |||||
| TIE1 | CCDS482.1 | NM_005424.2 | 6 | 7.6 | 2 | G139A | E47K | None | 13T | None |
| 2 | C161T | S54L | None | 16T | BRAF | |||||
| 2 | C266T | T89M | None | 52T | BRAF | |||||
| 2 | G292A | D98N | None | 43T | BRAF | |||||
| 11 | G1598A | G533E | Fibronectin Type 3 Domain | 39T | None | |||||
| 22 | C3281T | P1094L/LOH | Protein Tyrosine Kinase | 12T | NRAS |
Accession numbers for mutated PTK-s in Santa Cruz and GenBank.
Number of non-synonymous and splice site mutations observed and percent of tumors affected for each of the 19 genes in the panel of 79 melanoma cancers.
Nucleotide and amino acid change resulting from mutation.
“X” refers to stop codon. “LOH” refers to cases wherein the wild-type allele was lost and only the mutant allele remained. ”Splice site” refers to a case wherein the alteration affected ten bases spanning the exon.
Mutations previously observed in NRAS, or BRAF.
“None” refers to no mutation observed. SH2 Motif, Src homology 2 domain; IG, Immunoglobin; IGc2, Immunoglobin C-2 Type; IPT, IG-like, plexins, transcription factors; Focal AT, Focal Adhesion Targeting Region; FERM, Protein 4.1, Ezrin, Radixin, Moesin Domain. Domains were found using Ensembl and InterPro.
The observed somatic mutations could either be “driver” mutations that play a functional role in promoting the neoplastic process or nonfunctional “passenger” changes. In the 19 genes found to be mutated, 99 non-synonymous and 17 synonymous mutations were identified, yielding a N:S (non-synonymous:synonymous) ratio of 5.8:1, significantly higher than the N:S ratio of 2.5:1 predicted for nonselected passenger mutations (p<1×10−5)5, suggesting that many of these are likely to be “driver” mutations. The number of C>T mutations was significantly greater than other nucleotide substitutions resulting in a high prevalence of C:G>T:A transitions (p<0.0001) (Supplementary Figure 2A), confirming previously reported melanoma signatures 6.
To evaluate the effect of some of these mutations on kinase function, we focused on ERBB4, the most highly mutated gene in the screen, which harbored 24 somatic mutations (19%). Interestingly, five of the 15 samples with ERBB4 mutations contained more than one somatic mutation in ERBB4, which may act synergistically as previously seen for EGFR 7. The large number of mutations identified in ERBB4 strongly suggests that these mutations may be functionally important in melanoma.
Interestingly, 7 out of the 24 non-synonymous somatic mutations discovered in ERBB4 occurred at Glu (E) residues (p<0.00005, binomial test), all of which resulted in changes to Lys (K), causing a charge reversal. The underlying reason for this might be due to the high frequency of C:G>T:A transitions (Supplementary Figure 2B). Clustering of somatic mutations is seen in various functional domains of ERBB4 (Figure 1 and Supplementary Figure 3), with mutations in the kinase domain co-localizing with previously described mutations (found in various cancer types at frequencies ranging from 1.1–4.7% 8,9) and occurring at highly conserved residues. These genetic data suggest that mutant ERBB4 is likely to function as an oncogene in melanoma.
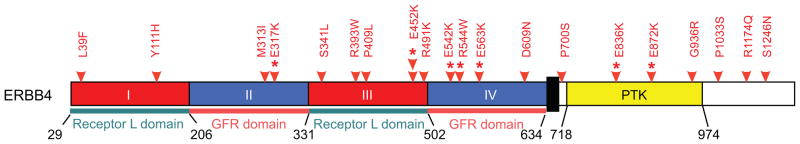
Figure 1. Distribution of mutations in ERBB4.
Red arrows indicate the location of ERBB4 somatic mutations found in this screen. Red stars indicate ERBB4 mutants used in subsequent analysis. Boxes represent functional domains (I, Extracellular Domain Subregion I; II, Extracellular Domain Subregion II; III, Extracellular Domain Subregion III; IV, Extracellular Domain Subregion IV; Kinase, Tyrosine Kinase Domain).
To prioritize ERBB4 missense mutations for further characterization, we assessed the positions of the mutations in its crystal structure10,11 and found that some of our observed alterations had similar positioning to mutations reported in the ERBB family members EGFR and ERBB2 in lung cancer, glioblastoma and gastric cancer 12 (Supplementary Figure 4). Based on this analysis, we chose to evaluate the E317K mutation in the extracellular domain, which is near the EGFR R324L mutation; the E542K, R544W, and E563K mutations which co-localize; the E452K mutation, which was found in two patients; and two mutations in the kinase domain: E836K, which is found near the ERBB2 N857S mutation; and the E872K alteration.
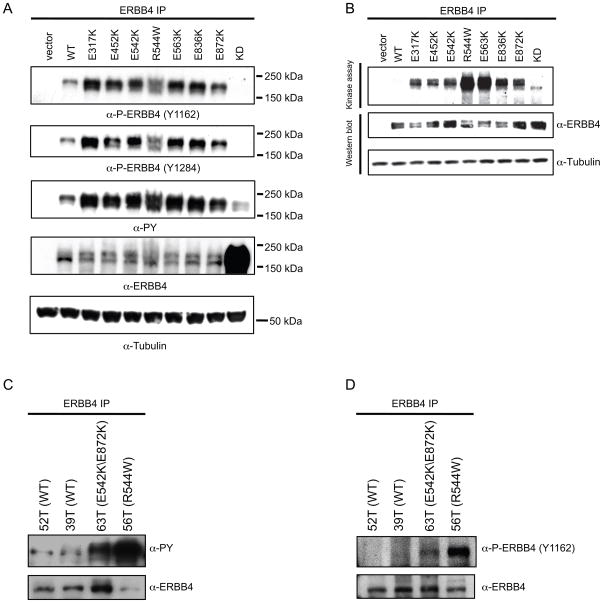
To determine whether the ERBB4 mutations had enhanced kinase activity, we transiently expressed wild type (WT) ERBB4 or the seven mutants (E317K, E452K, E542K, R544W, E563K, E836K, E872K) as well as a kinase dead (KD) version of ERBB4 (K751M) in HEK 293T cells and assessed catalytic activity using ERBB4 autophosphorylation (both total and residues Y1162 or Y1284) as a measure of receptor activation. Compared to WT ERBB4, all the missense mutants showed increased phosphorylation of the receptor (Figure 2A). No site-specific phosphorylation was seen in cells exogenously expressing the KD ERBB4. Similar levels of total ERBB4 protein were observed except for KD ERBB4, which was higher (Figure 2A). To determine if the increased tyrosine phosphorylation of the ERBB4 mutants correlates with increased kinase activity, a kinase assay using the same set of ERBB4 mutants was performed. The ERBB4 mutants showed a marked increase in kinase activity compared to WT ERBB4 and expression levels of total ERBB4 protein were comparable (Figure 2B). As in transfected cells, ERBB4 autophosphorylation was markedly elevated in the melanoma lines harboring ERBB4 mutations compared to melanoma lines harboring endogenous WT ERBB4 (Fig 2C–D).
Figure 2. ERBB4 mutants exhibit increased basal activation.
A. ERBB4 mutants have increased tyrosine phosphorylation. HEK 293T cells were transiently transfected with the indicated constructs. 24 hrs after transfection, cells were serum starved, lysed and immunoprecipitated with α-ERBB4. After immunoprecipitation of ERBB4, immunoblots were performed using specific antibodies, as indicated. Lysates were immunoblotted with an α-tubulin antibody. B. ERBB4 mutants exhibit increased in vitro kinase activity. HEK 293T cells were transiently transfected as in A. Equivalent amounts of protein from cell lysates were immunoprecipitated and used in a kinase assay to measure receptor autophosphorylation. The same samples that were used in the kinase assay were immunoblotted with ERBB4 antibody and lysates were blotted with α-tubulin. KD: kinase dead. C. Increased basal activation of endogenous mutant ERBB4. Melanoma lines that harbor either WT or mutant ERBB4 were serum starved and then lysed, immunoprecipitated for ERBB4, then immunoblotted withα-PY20 or α-ERBB4. D. Mutant ERBB4 has increased basal activity. Melanoma lines harboring either WT or mutant ERBB4 were serum deprived, lysed, immunoprecipitated for ERBB4, and analyzed by immunoblotting with α-P-ERBB4 (P-Y1162) or α-ERBB4.
ERBB4 is known to activate several downstream signaling pathways including the ERK and AKT pathways 13. To evaluate which of these signaling pathways is activated by the ERBB4 mutations, we performed immunoblot analysis of melanoma cell lines harboring endogenous ERBB4 mutations. Phosphorylation of AKT was elevated in cells expressing any of the three evaluated mutant ERBB4s, whereas ERK showed similar activation in cells expressing WT or mutant ERBB4 (Supplementary Figure 5).
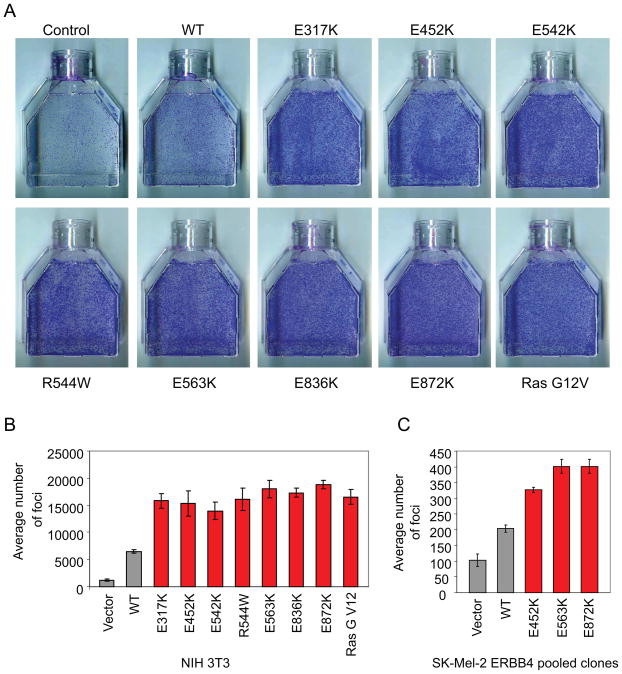
To determine whether the ERBB4 variants are transforming, NIH 3T3 cells were transiently transfected with vector, WT ERBB4, one of the seven constitutively active ERBB4 mutants, or oncogenic K-RasG12V. Ten days after transfection, all ERBB4 mutations transformed NIH 3T3 cells more efficiently than WT ERBB4. Strikingly, the transformation ability of the ERBB4 mutations was similar to oncogenic K-RasG12V (Figure 3A–B). Similarly, expression of mutant ERBB4 significantly increased anchorage-independent growth as assessed by colony formation in soft agar (Supplementary Figure 6Ap<0.05, t-test). Similar results were seen for several mutants expressed in the human melanoma cell line SK-Mel-2, which expresses WT ERBB4. Levels of ERBB4 were comparable in all clones (Supplementary Figure 6B).
Figure 3. Mutant ERBB4 induces cell transformation and anchorage-independent growth in NIH 3T3 and SK-Mel-2 cells.
A. NIH 3T3 cells were transfected with the indicated constructs. The photographs show foci stained with Hema3 after 10 days. Ras G12V was included as a positive control for cell transformation. B. The graph indicates the average number of foci visualized in A. C. Transformation ability of melanoma SK-Mel-2 cells stably expressing either vector, WT ERBB4 or various ERBB4 missense mutants was assessed by foci formation in tissue culture flasks. The graph indicates the number of foci formed after 14 days.
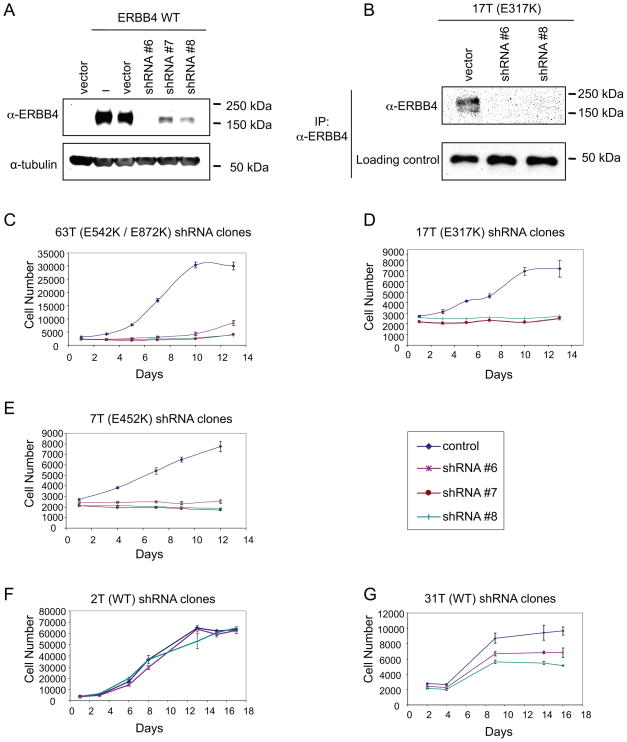
In order to assess if melanoma cells harboring endogenous ERBB4 mutations are dependent on ERBB4 signaling for proliferation, we used short hairpin RNA (shRNA) to stably knockdown ERBB4 protein levels in melanoma lines harboring either WT (2T and 31T) or mutant ERBB4 (17T, E317K; 63T, E542K/E872K; or 7T, E452K). Specific targeting of ERBB4 by shRNAs was confirmed both in transfected HEK 293 cells and in one of the melanoma cell lines by immunoblotting (Figure 4A–B). Three unique shRNA constructs targeting ERBB4 had minimal effect on the proliferation of cells expressing WT receptor but significantly reduced the growth of melanoma lines containing mutant ERBB4 (Figure 4C–G). Thus, mutant ERBB4 is essential for growth of melanomas harboring these mutations. Evaluation of the effects of ERBB4 knockdown on downstream signaling pathways revealed that down-regulation of ERBB4 in cells harboring mutant versions of the gene reduces levels of endogenous, phosphorylated AKT, but not of phosphorylated ERK. In contrast, inhibition of ERBB4 expression in cells harboring WT versions of the gene showed similar levels of AKT and ERK activation (Supplementary Figure 7).
Figure 4. Expression of mutant ERBB4 provides an essential cell survival signal in melanoma.
A. HEK 293 cells were transiently co-transfected with either vector or WT ERBB4 together with either control vector or shRNAs that target ERBB4. Cell lysates were analyzed by immunoblotting using α-ERBB4. For normalization, lysates were analyzed in parallel by α-tubulin immunoblotting. B. Cells transduced with shRNA targeting ERBB4 were lysed and immunopreciptitated using α-ERBB4 beads. Immunoprecipitates were blotted with specific antibodies, as indicated. C–G. shRNA-mediated ERBB4 knockdown in melanoma lines containing ERBB4 mutations results in reduced cell growth. Cells were seeded in 96-well plates and incubated for 13–17 days. Plates were analyzed every other day for cell proliferation where the average cell number at each time point was measured by determining DNA content using SYBR Green I. Melanoma cells harboring ERBB4 mutations stably transduced with shRNA constructs targeting ERBB4, but not those stably transduced with the control vector only, showed decreased growth relative to control. This did not occur in melanoma cells harboring WT ERBB4.
Because shRNA-mediated cell death could result from specific or nonspecific effects, we examined the ability of an exogenous, non-targetable WT ERBB4 construct (NT ERBB4), engineered to be resistant to knockdown by the introduction of three silent mutations in the region of ERBB4 targeted by shRNA #6, to rescue the effects of knockdown of endogenous ERBB4. Melanoma cells harboring the E317K mutation stably expressing either control or ERBB4 shRNA #6 construct were transduced with the lentiviral NT ERBB4 construct or empty vector as control. Similar phosphotyrosine content is seen in both WT and NT ERBB4 constructs, demonstrating that the silent mutations in the NT construct do not affect the ability of the receptor to be phosphorylated to wild-type levels (Supplementary Figure 8A). Importantly, pooled clones of NT reconstituted cells were markedly more resistant to growth inhibition induced by ERBB4 knockdown (#6/NT) than shRNA control-infected cells (Vect/Vect).
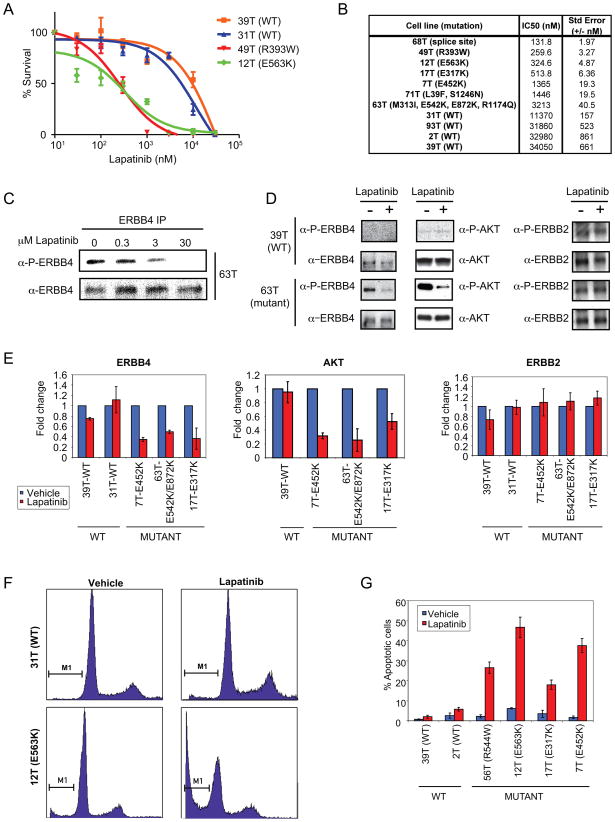
To evaluate mutant ERBB4 as a potential target for specific inhibition of melanoma cell survival, we targeted the ERBB4 pathway with the FDA-approved pan-ERBB pharmacologic inhibitor, lapatinib (GW2016) 14. Exposure of melanoma cells to lapatinib resulted in reduced cell proliferation to a greater extent in cells containing endogenous ERBB4 mutations than in cells containing endogenous WT ERBB4 (Figure 5A). An IC50 calculation revealed that melanoma cells harboring ERBB4 mutations were 10–250 fold more sensitive to lapatinib than cells with WT receptor (Figure 5B) and treatment with lapatinib inhibited receptor autophosphorylation in a dose-dependent manner (Figure 5C). This increased sensitivity to lapatinib was accompanied by specific inhibition of ERBB4 and AKT activation in cells harboring mutant ERBB4 (Figure 5D–E). Activation of other downstream elements, such as ERK, was also slightly inhibited by lapatinib (Supplementary Figure 9 A–B). Thus, although signaling by mutant ERBB4 demonstrates selective activation of AKT, lapatinib treatment of cells harboring mutant ERRB4 results in uniform inhibition of downstream signaling pathways. Only mutant ERBB4 was inhibited by lapatinib in our melanoma cell lines. No inhibition of its family member ERBB2 was seen (Figure 5D–E) and no phosphorylation of EGFR was observed in any of these cells (data not shown). The observed reduced proliferation occurred in cells harboring BRAF, NRAS, ARAF or CRAF mutations in addition to the ERBB4 mutations (Supplementary Tables 4 and 5).
Figure 5. Melanoma lines expressing ERBB4 mutants exhibit increased sensitivity to ERBB inhibition by lapatinib.
A. Representative dose response curves showing lapatinib efficacy against ERBB4 mutant lines compared to WT ERBB4 lines. Cells were treated for 72 hours in the presence of increasing concentrations (0.01–30 μM) of lapatinib, and relative cell number was estimated by methylene blue protein staining and plotted as percent survival when compared to vehicle-treated control versus Log (lapatinib) nM (where 1 is 10 nM lapatinib). Fitted lines were generated using 4-parameter nonlinear regression via GraphPad Prism. B. ERBB4 mutant lines have increased sensitivity to lapatinib compared to WT ERBB4 lines. The IC50 values for inhibition of cell growth by 72 hr treatment with lapatinib of a larger panel of lines harboring WT and mutant ERBB4 were analyzed using GraphPad Prism v.5 (n=3). C. Immunoprecipitation and western blot analysis of ERBB4 autophosphorylation in cells treated with lapatinib. Cells were treated for 1 hr with lapatinib or vehicle alone as control. Lysates were immunoprecipitated with α-ERBB4 followed by western blot analysis with α-ERBB4 and α-P-ERBB4 (Y1162). D. Melanoma lines expressing mutant ERBB4 exhibit increased lapatinib sensitivity with respect to ERBB4 as well as AKT phosphorylation. The activity of ERBB4, AKT as well as ERBB2 was determined by immunoblotting with phospho-specific antibodies. Cells were treated for 1 hr with 5 μM lapatinib or vehicle alone. Lysates were immunoprecipitated using α-ERBB2 or α-ERBB4. Lysates and immunoprecipitates were analyzed by western blotting using the indicated antibodies. Shown are representative blots. E. Quantitative assessment of data from 2 lines harboring WT ERBB4 and 3 lines harboring mutant ERBB4 that were performed similarly to D. D. The ratio of band intensities of (P-Y1162)-ERBB4/ERBB4, (P)-S473-AKT/AKT and (P-Y1248)-ERBB2/ERBB2 for each cell line are shown. F. Mutant ERBB4 cells have increased sub-G1 population in the presence of lapatinib compared to WT ERBB4 cells. FACS analysis of 31T (WT) and 12T (E563K) showing cell cycle distribution (PI staining, x-axis) vs cell counts (y-axis). Shown are representative plots. G. Quantitation of FACS-sorted lapatinib-treated cells. The percent apoptotic cells were determined based on the sub G1 population for vehicle treated cells or lapatinib treated cells.
To elucidate the mechanism of decreased growth of cells expressing mutant ERBB4 following lapatinib treatment, we examined cells for cell cycle perturbations or apoptosis by flow cytometry. Lapatinib markedly increased apoptosis of melanoma cells harboring mutant ERBB4 compared to lines harboring WT ERBB4 (Figure 5F–G). Thus, expression of mutant ERBB4 appears essential for suppression of pro-apoptotic signals in melanoma cells harboring these mutations, which is consistent with the selective activation of AKT in ERBB4 mutant cells (Supplementary Figure 5) and previous results demonstrating an anti-apoptotic role for AKT 15. These results suggest that lapatinib preferentially inhibits mutant ERBB4 signaling and that cells with ERBB4 mutations are subject to “oncogene addiction” 16. Moreover, the enhanced AKT signaling in cells with mutant ERBB4 might provide an additional therapeutic target in these tumors.
Previous studies have shown that lapatinib is a much more potent inhibitor of EGFR and ERBB2 (>10X more potent) than ERBB411,17–19. Although lapatinib is clearly leading to a loss of ERBB4 phosphorylation, it is not clear that this is through direct inhibition of ERBB4 kinase activity. It is possible that the inhibitory effects seen by lapatinib are due to ERBB4 transphosphorylation by EGFR and/or ERBB2, and that lapatinib blocks ERBB4 phosphorylation by directly inhibiting EGFR or ERBB2. Alternatively, it is possible that mutant ERBB4 proteins have higher affinity for binding of lapatinib than WT ERBB4. Future work to investigate the mechanism by which lapatinib exerts increased specificity of mutant ERBB4 is warranted.
Here we describe the identification of 99 novel somatic mutations in 19 PTKs in melanoma, few of which had previously been linked to melanoma. The high frequency of mutations identified in ERBB4, their co-localization to particular functional domains, as well as the functional studies described above, suggests that these mutations are oncogenic. In contrast to oncogenes with mutational hotspots, such as PIK3CA, BRAF and NRAS, ERBB4 mutations occur throughout the gene. Our data and previously reported heterogeneous mutational activation of another oncogene, FLT3, definitively demonstrate that not all mutations in oncogenes must be clustered to be functionally important20. Changes that effect enzyme activity can result from single or multiple mutations within a gene that increase activity or abrogate negative regulatory domains. Interestingly, sample 63T harbored two somatic mutations (E542K and E872K) for which the biochemical effects were assessed separately. Both mutations showed increased receptor autophosphorylation and increased kinase activity. These data demonstrate that both mutations exhibit independent, gain-of-function effects, suggesting that the mutations might be synergistic as has been described previously for EGFR 7,21.
Our findings indicate that if future experiments verify that mutational activation of ERBB4 is essential for tumor growth in vivo, targeting of ERBB4 with small-molecule inhibitors should be considered for the large number of patients with these mutations. Broad spectrum ERBB inhibitors, such as lapatinib and canertinib 14,22,23 have already been developed. Our results suggest that further development of such inhibitors is warranted and the clinical utility of this class of compounds be explored in the treatment of melanoma.
Methods
Tumor Tissues
Tissue and melanoma cell lines used in this study were described previously24.
PCR, sequencing and mutational analysis
PCR and sequencing was done as previously described24. The kinase domain mutation screen was analyzed using Consed 25. Variants were called using Polyphred 6.1126 and DIP Detector (Hansen N., unpublished), an in del detector for improved sensitivity in finding insertions and deletions.
Sequence traces of the secondary screen were analyzed using the Mutation Surveyor software package (SoftGenetics, State College, PA).
Construction of wild-type and mutant ERBB4 expression vector
Human ERBB4 (NM_005235) was cloned by PCR as previously described24 using a clone (#8327667) purchased from Open Biosystems with primers in Supplemental Table 5. The PCR product was cloned into the mammalian expression vector pCDF-MCS2-EF1-Puro™ (Systems Biosciences, Inc., Mountain View, CA) via the XbaI and NotI restriction sites. The E317K, E452K, E542K, R544W, E563K, K751M, E836K, E872K and non-targetable ERBB4 point mutants were made using Phusion PCR for site-directed mutagenesis.
Cell culture and transient expression
Metastatic melanoma tumor lines were maintained as previously described 27. HEK 293T cells as well as NIH 3T3 were purchased from ATCC (Manassas, VA) and maintained in complete Dulbecco’s Modified Eagles Medium (DMEM) supplemented with 10% Fetal Bovine Serum (FBS), 1X non-essential amino acids, 2mM L-glutamine, and 0.75% sodium bicarbonate. HEK 293T cells were transfected with Lipfectamine 2000 reagent (Invitrogen, Carlsbad, CA) at a 6:1 ratio with DNA (μl:μg) using 3–5 μg of plasmid DNA.
Immunoprecipitation and western blotting
Transfected cells were gently washed 3X in PBS and then lysed using 0.5–1.0 ml 1% NP-40 lysis buffer (1% NP-40, 50mM Tris-HCl pH 7.5, 150mM NaCl, Complete Protease Inhibitor tablet, EDTA-free (Roche, Indianapolis, IN), 1μM sodium orthovanadate, 1 mM sodium fluoride, and 0.1% β-mercaptoethanol) per T-75 flask for 20 minutes on ice. Lysed cells were scraped and transferred into a 1.5 mL microcentrifuge tube. Extracts were centrifuged for 10 minutes at 14,000 rpm at 4°C. 500 μl of supernatant was immunoprecipitated overnight using 20 μl of anti-ERBB4 agarose-conjugated beads (Santa Cruz Biotechnology, Inc., Santa Cruz, CA). The immunoprecipitates were washed and subjected to SDS-PAGE and western blotting as previously described 28. Primary antibodies used in our analysis were anti-ERBB4 (Santa Cruz Biotechnology), anti-P-ERBB4 (Y1162) (Abgent), anti-P-ERBB4 (Y1284) (Cell Signaling, Danvers, MA), anti-PY20 (Zymed-Invitrogen), anti-P-ERK1/2 (T202/Y204), anti-ERK1/2, anti-P-AKT (S473), anti-AKT (Cell Signaling, Danvers, MA), anti-P-STAT5A/B (Y694/Y699) (Upstate Biotech-Millipore), anti-STAT5 (Cell Signaling), and anti-α-tubulin (Calbiochem-EMD Biosciences, Gibbstown, NJ).
Pooled stable expression
To make lentivirus, ERBB4 constructs were co-transfected into HEK 293T cells seeded at 1.5×106 per T75 flask with pVSV-G and pFIV-34N (kind gifts from Todd Waldman, Georgetown University) helper plasmids using Lipofectamine 2000 as described by the manufacturer. Virus-containing media was harvested 48–60hr after transfection, filtered, aliquoted and stored at −80°C.
SK-Mel-2 cells (National Cancer Institute, Division of Cancer Treatment, Developmental Therapeutics Program, Frederick, MD) were grown in RPMI-1640 (Lonza, Walkersville, MD) and supplemented with 10% fetal bovine serum (HyClone, Logan, UT) SK-Mel-2 and NIH 3T3 cells were seeded at 1.5 × 106 cells per T75 flask 24 hr prior to infection. Lentivirus for ERBB4 (WT, E317K, E452K, E542K, R544W, E563K, K751M, E836K, and E872K point mutants) and empty vector control were used to infect SK-Mel-2 cells or NIH 3T3 cells as previously described29. Stable expression of ERBB4 proteins (WT and mutants) was determined by SDS-PAGE analysis followed by immunoblotting with anti-ERBB4 and anti-tubulin to show equivalent expression among pools.
Lentiviral shRNA
Constructs for stable depletion of ERBB4 were obtained from Open Biosystems (Huntsville, AL) and three were confirmed to efficiently knockdown ERBB4 at the protein level. Lentiviral stocks were prepared as previously described24. Melanoma cell lines (2T, 7T, 17T, 31T, and 63T) were infected with shRNA lentiviruses for each condition (vector and scrambled controls and three independent ERBB4-specific shRNAs). Selection and growth were done as described above. Stably infected pooled clones were tested in functional assays.
To rescue shRNA-mediated knock-down of ERBB4 in melanoma cell lines the non-targetable ERBB4 lentivirus was made as described above and used to infect the melanoma cell line 17T. After infection, cells were given 48 to 72 hours to recover from infection prior to testing in functional assays.
Proliferation and growth inhibition assays
To examine growth potential, melanoma cell lines (2T, 7T, 17T, 31T, and 63T) stably infected with either vector or scrambled controls or ERBB4-specific shRNAs were seeded into 96 well plates at 2,500 cells per well and incubated for 13–17 days. Samples were analyzed every 48 hr by lysing cells in 50 μl 0.2% SDS/well and incubating for 2 hour at 37°C prior to addition of 150 μl/well of SYBR Green I solution (1:750 SYBR Green I (Invitrogen-Molecular Probes) diluted in dH20).
The effects of tyrosine kinase inhibitors (TKIs) on the proliferation of melanoma cell lines were tested by seeding 96-well plates at 5,000 cells/well in the presence or absence of serum containing media and incubated for 24 hr prior to addition of TKIs. Increasing concentrations of lapatinib (Tykerb-GlaxoSmithKline) were added to each well in four replicates with DMSO as negative control. Plates were analyzed 72 hr post-addition of TKIs using the SYBR Green I proliferation assay described above.
To further test TKIs on melanoma cell lines we seeded 96-well plates at 5,000 cells per well and incubated 24 hr prior to addition of TKIs (e.g. lapatinib) at concentrations from 10 nM to 30 μM. Once inhibitors were added, cells were incubated for 72 hr at 37°C. Cells were then analyzed as previously described18. Plates were read at 650nm on a Molecular Devices (Spectra Max) Plate Reader and analyzed using SoftMax v5 and GraphPad Prism v5.
Soft agar assay
SK-Mel-2 pooled ERBB4 clones were plated in duplicate at 1000 cells/well and NIH 3T3 pooled ERBB4 clones were plated in duplicate at 5000 cells/well in top plugs consisting of sterile 0.33% Bacto-Agar (BD, Sparks, MD) and 10% fetal bovine serum (HyClone, Logan, UT) in a 24-well plate. The lower plug contained sterile 0.5% Bacto-Agar and 10% fetal bovine serum. After two weeks, the colonies were photographed and counted.
NIH 3T3 transformation assay
150 ng of each plasmid was transfected by the calcium phosphate precipitation method into NIH 3T3 cells cultured in 12 well plates. 24hr after transfection, 5% of transfected cells were seeded into T-25 flasks and cultured in normal growth medium for 10 days. The cells were stained with Hema3 (Sigma St. Louis, MO) and analyzed for the presence of foci.
Analysis of ERBB4 kinase activity
HEK 293T cells were transiently transfected with ERBB4 (WT, E317K, E452K, E542K, R544W, E563K, E836K, E872K and kinase-dead (K751M)) or empty vector and incubated for 18–24 hr at 37°C in reduced (0.5%) serum containing medium prior to immunoprecipitation. Cells were harvested and ~3 mg of lysate were used in each immunoprecipitation reaction. Immunoprecipitates were performed as described above. Immune complexes were washed three times in lysis buffer followed by two washes in kinase buffer (20 mM HEPES pH 7.4, 50 mM NaCl, 3 mM MnCl2, 20 mM MgCl2, 1 mM sodium orthovanadate, 1 mM sodium fluoride, and 1X complete protease inhibitor tablet). Immune complexes were then resuspended in 50μl kinase buffer and 10μl incubated in the presence of [γ-32P]ATP (3μCi per reaction) for 15 min at 37°C. Kinase reactions were stopped by the addition of 2X SDS sample buffer and phosphorylated samples were resolved on 8% Tris-Glycine gels. Gels were stained and destained prior to autoradiography.
Immunoblot quantitation analysis
Scanned films from western blot analysis of SDS-PAGE were analyzed using ImageJ (NIH software). Individual bands were quantitated and plots were generated to determine the intensities in each band. The data was then exported to Microsoft Excel and analyzed further for phospho:total ratios of protein.
Flow cytometry analysis
Melanoma cells were seeded into T-25 flasks at densities of 3×105 cells per flask in normal complete T2 medium and incubated at 37°C for 24 hr prior to addition of lapatinib. Lapatinib or vehicle was added 72 hr at a concentration of 5 μM. Cells were then harvested for FACS analysis by first removing the medium into a new conical tube followed by trypsinizing of attached cells in T-25 flasks. Trypsinized cells and those from the medium were combined and washed in ice-cold PBS. Cells were collected by centrifugation at 1,000 rpm at 4°C. Ice-cold 70% ethanol was added to cell pellets and allowed to fix overnight at 4°C followed by washing in ice-cold PBS. DNase-free RNase (Roche) was to cells resuspended in 0.5–1ml PBS and incubated at 37°C for 30 min before adding 50–100 μl of Propidium Iodide (PI-0.5 mg/ml) (Roche). Cellular DNA content was analyzed on Becton Dickinson FACSCalibur using CellQuest software.
X-ray crystal structure assembly
The X-ray crystal structures of the ERBB4 extracellular and kinase domains were used as templates in the program SWISS-MODEL. Location of EGFR and ERBB2 mutations in the crystal were found by aligning the protein sequences for EGFR, ERBB2, ERBB3, and ERBB4 using ClustalW 30. Previously known mutations in EGFR and ERBB2 were matched to the sequence of ERBB4 using the ClustalW alignment.
Statistical analysis
To determine whether the ratio of nonsynonymous to synonymous mutations observed was statistically significant, the exact binomial test was used, with an expected ratio of 2.5:1. All the statistical calculations were performed in the R statistical environment (http://www.r-project.org) 5. Further statistical analyses were performed using Microsoft Excel to generate p-values to determine significance (two-tailed t-test). Inhibition curves (IC50) were analyzed and plotted using GraphPad Prism v5.
Supplementary Material
Acknowledgments
We thank Drs. Bert Vogelstein, Todd Waldman, Daphne Bell, Paul Meltzer, Larry Brody, Glenn Merlino Silvio Gutkind, and Isabel Cardenas-Navia for their helpful comments on the manuscript. We also thank members of the NISC Comparative Sequencing Program for generating the sequence data analyzed here. We also would like to thank Stacie Anderson and Martha Kirby for assistance with FACS analysis. This work supported by the Intramural Research Programs of the National Human Genome Research Institute and National Cancer Institute, National Institutes of Health
Footnotes
Author contributions
T.DP. and Y.S. designed the study; J.R.W. and S.A.R. collected and analyzed the melanoma samples, N.S.A., J.C.C., K.E.Y., J.C.L., NISC., P.C and Y.S. analyzed the genetic data; T.D.P., X.W. and K.E.Y., performed and analyzed the functional data. All authors contributed to the final version of the paper.
References
- 1.Jermal A. Cancer Statistics, 2006. CA: A Cancer J Clin. 2006;56:106–30. doi: 10.3322/canjclin.56.2.106. [DOI] [PubMed] [Google Scholar]
- 2.Tsao H, Atkins MB, Sober AJ. Management of cutaneous melanoma. N Engl J Med. 2004;351:998–1012. doi: 10.1056/NEJMra041245. [DOI] [PubMed] [Google Scholar]
- 3.Futreal PA, et al. A census of human cancer genes. Nat Rev Cancer. 2004;4:177–83. doi: 10.1038/nrc1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sawyers C. Targeted cancer therapy. Nature. 2004;432:294–7. doi: 10.1038/nature03095. [DOI] [PubMed] [Google Scholar]
- 5.Sjoblom T, et al. The consensus coding sequences of human breast and colorectal cancers. Science. 2006;314:268–74. doi: 10.1126/science.1133427. [DOI] [PubMed] [Google Scholar]
- 6.Greenman C, et al. Patterns of somatic mutation in human cancer genomes. Nature. 2007;446:153–8. doi: 10.1038/nature05610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Godin-Heymann N, et al. Oncogenic activity of epidermal growth factor receptor kinase mutant alleles is enhanced by the T790M drug resistance mutation. Cancer Res. 2007;67:7319–26. doi: 10.1158/0008-5472.CAN-06-4625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Soung YH, et al. Somatic mutations of the ERBB4 kinase domain in human cancers. Int J Cancer. 2006;118:1426–9. doi: 10.1002/ijc.21507. [DOI] [PubMed] [Google Scholar]
- 9.Ding L, et al. Somatic mutations affect key pathways in lung adenocarcinoma. Nature. 2008;455:1069–75. doi: 10.1038/nature07423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bouyain S, Longo PA, Li S, Ferguson KM, Leahy DJ. The extracellular region of ErbB4 adopts a tethered conformation in the absence of ligand. Proc Natl Acad Sci U S A. 2005;102:15024–9. doi: 10.1073/pnas.0507591102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Qiu C, et al. Mechanism of activation and inhibition of the HER4/ErbB4 kinase. Structure. 2008;16:460–7. doi: 10.1016/j.str.2007.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Riese DJ, 2nd, Gallo RM, Settleman J. Mutational activation of ErbB family receptor tyrosine kinases: insights into mechanisms of signal transduction and tumorigenesis. Bioessays. 2007;29:558–65. doi: 10.1002/bies.20582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Frey MR, Edelblum KL, Mullane MT, Liang D, Polk DB. The ErbB4 growth factor receptor is required for colon epithelial cell survival in the presence of TNF. Gastroenterology. 2009;136:217–26. doi: 10.1053/j.gastro.2008.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Heymach JV, Nilsson M, Blumenschein G, Papadimitrakopoulou V, Herbst R. Epidermal growth factor receptor inhibitors in development for the treatment of non-small cell lung cancer. Clin Cancer Res. 2006;12:4441s–4445s. doi: 10.1158/1078-0432.CCR-06-0286. [DOI] [PubMed] [Google Scholar]
- 15.Grant S, Qiao L, Dent P. Roles of ERBB family receptor tyrosine kinases, and downstream signaling pathways, in the control of cell growth and survival. Front Biosci. 2002;7:d376–89. doi: 10.2741/grant. [DOI] [PubMed] [Google Scholar]
- 16.Weinstein IB. Cancer. Addiction to oncogenes--the Achilles heal of cancer. Science. 2002;297:63–4. doi: 10.1126/science.1073096. [DOI] [PubMed] [Google Scholar]
- 17.McHugh LA, et al. Lapatinib, a dual inhibitor of ErbB-1/-2 receptors, enhances effects of combination chemotherapy in bladder cancer cells. Int J Oncol. 2009;34:1155–63. doi: 10.3892/ijo_00000244. [DOI] [PubMed] [Google Scholar]
- 18.Rusnak DW, et al. The effects of the novel, reversible epidermal growth factor receptor/ErbB-2 tyrosine kinase inhibitor, GW2016, on the growth of human normal and tumor-derived cell lines in vitro and in vivo. Mol Cancer Ther. 2001;1:85–94. [PubMed] [Google Scholar]
- 19.Xia W, et al. Combining lapatinib ( GW572016), a small molecule inhibitor of ErbB1 and ErbB2 tyrosine kinases, with therapeutic anti-ErbB2 antibodies enhances apoptosis of ErbB2-overexpressing breast cancer cells. Oncogene. 2005;24:6213–21. doi: 10.1038/sj.onc.1208774. [DOI] [PubMed] [Google Scholar]
- 20.Shih LY, et al. Heterogeneous patterns of FLT3 Asp(835) mutations in relapsed de novo acute myeloid leukemia: a comparative analysis of 120 paired diagnostic and relapse bone marrow samples. Clin Cancer Res. 2004;10:1326–32. doi: 10.1158/1078-0432.ccr-0835-03. [DOI] [PubMed] [Google Scholar]
- 21.Pao W, et al. Acquired resistance of lung adenocarcinomas to gefitinib or erlotinib is associated with a second mutation in the EGFR kinase domain. PLoS Med. 2005;2:e73. doi: 10.1371/journal.pmed.0020073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Allen LF, Eiseman IA, Fry DW, Lenehan PF. CI-1033, an irreversible pan-erbB receptor inhibitor and its potential application for the treatment of breast cancer. Semin Oncol. 2003;30:65–78. doi: 10.1053/j.seminoncol.2003.08.009. [DOI] [PubMed] [Google Scholar]
- 23.Sharma SV, Bell DW, Settleman J, Haber DA. Epidermal growth factor receptor mutations in lung cancer. Nat Rev Cancer. 2007;7:169–81. doi: 10.1038/nrc2088. [DOI] [PubMed] [Google Scholar]
- 24.Palavalli LH, et al. Analysis of the matrix metalloproteinase family reveals that MMP8 is often mutated in melanoma. Nat Genet. 2009;41:518–20. doi: 10.1038/ng.340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gordon D, Abajian C, Green P. Consed: a graphical tool for sequence finishing. Genome Res. 1998;8:195–202. doi: 10.1101/gr.8.3.195. [DOI] [PubMed] [Google Scholar]
- 26.Bhangale TR, Stephens M, Nickerson DA. Automating resequencing-based detection of insertion-deletion polymorphisms. Nat Genet. 2006;38:1457–62. doi: 10.1038/ng1925. [DOI] [PubMed] [Google Scholar]
- 27.Chappell DB, Zaks TZ, Rosenberg SA, Restifo NP. Human melanoma cells do not express Fas (Apo-1/CD95) ligand. Cancer Res. 1999;59:59–62. [PMC free article] [PubMed] [Google Scholar]
- 28.Samuels Y, et al. High frequency of mutations of the PIK3CA gene in human cancers. Science. 2004;304:554. doi: 10.1126/science.1096502. [DOI] [PubMed] [Google Scholar]
- 29.Solomon DA, et al. Mutational inactivation of PTPRD in glioblastoma multiforme and malignant melanoma. Cancer Res. 2008;68:10300–6. doi: 10.1158/0008-5472.CAN-08-3272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guex N, Peitsch MC. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis. 1997;18:2714–23. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.